Abstract
Sirtuin 3 (SIRT3) is an NAD+-dependent deacetylase linked to a broad range of physiological and pathological processes, including aging and aging-related diseases. However, the role of SIRT3 in regulating human stem cell homeostasis remains unclear. Here we found that SIRT3 expression was downregulated in senescent human mesenchymal stem cells (hMSCs). CRISPR/Cas9-mediated depletion of SIRT3 led to compromised nuclear integrity, loss of heterochromatin and accelerated senescence in hMSCs. Further analysis indicated that SIRT3 interacted with nuclear envelope proteins and heterochromatin-associated proteins. SIRT3 deficiency resulted in the detachment of genomic lamina-associated domains (LADs) from the nuclear lamina, increased chromatin accessibility and aberrant repetitive sequence transcription. The re-introduction of SIRT3 rescued the disorganized heterochromatin and the senescence phenotypes. Taken together, our study reveals a novel role for SIRT3 in stabilizing heterochromatin and counteracting hMSC senescence, providing new potential therapeutic targets to ameliorate aging-related diseases.
INTRODUCTION
Mammalian Sirtuins are homologs of Saccharomyces cerevisiae Sir2 (silent information regulator 2), and require NAD+ as a cofactor that drives deacetylase activity (1–4). From unicellular yeast to mammals, the lifespan of organisms is positively regulated by Sir2 or Sirtuins (5–11). Mammalian SIRT3, for example, is found primarily in the mitochondria (12). SIRT3 plays crucial roles in mitochondrial homeostasis, metabolic modulation, gene transcription, stress response and genomic stability (13). Human longevity has been linked to a single nucleotide polymorphism (SNP), G477T, in exon 3 of SIRT3, as well as a variable number of tandem repeats (VNTR) polymorphism in intron 5 (14,15). SIRT3 is also implicated in various age-related pathologies, including osteoporosis, osteoarthritis, cardiovascular diseases, metabolic syndrome and neurodegenerative diseases (2,16). Sirt3-deficient mice have a shorter lifespan compared to wild-type (WT) mice (10). This is partially due to heart failure caused by cardiac mitochondrial dysfunction and reactive oxygen species (ROS) dyshomeostasis in hematopoietic stem cells (HSCs) (10,17). In addition, increased expression of SIRT3 upon caloric restriction in mice has been associated with the reduction of oxidative damage and prevention of age-related disorders (18,19). However, the roles of SIRT3 in regulating human stem cell aging remain largely unexplored.
Mitochondrial-localized SIRT3 has been reported to regulate mitochondrial metabolism via its deacetylation activity. For instance, SIRT3 deacetylates and activates AceCS2 and HMGCS2 in the mitochondria (20–22), which regulate the formation of epigenetic modulators acetyl-CoA and β-hydroxybutyrate. SIRT3 is also found in the nucleus to deacetylate histones (23,24). For example, SIRT3 promotes non-homologous end joining (NHEJ)-dependent DNA damage repair via the deacetylation of H3K56ac (23). Although SIRT3 has been implicated in a variety of biological processes, little is known about the role of SIRT3 in the regulation of higher-order heterochromatin organization.
Stem cell senescence and exhaustion contribute to compromised regenerative capability, resulting in tissue dysfunction and organismal aging (25–27). Stem cell senescence is accompanied by a variety of epigenetic changes including disorganized heterochromatin, aberrant DNA methylation and histone modifications, as well as alterations in nuclear envelope maintenance and mitochondrial homeostasis (25,28,29). The discovery of the molecular mechanisms underlying stem cell senescence has provided novel insights into the process of aging and allowed the development of intervention strategies for aging-associated disorders (30,31). Given our current understanding of SIRT3, it potentially plays an important role in regulating human stem cell senescence and its molecular mechanism has yet to be unraveled.
Here, we revealed a novel role of SIRT3 in preventing human mesenchymal stem cell (hMSC) senescence, possibly in part via heterochromatin stabilization. We observed that SIRT3 interacted with nuclear envelope proteins and heterochromatin-associated proteins. In addition, loss of SIRT3 led to several heterochromatin-associated abnormalities including aberrant nuclear envelope morphology, heterochromatin loss, the detachment of genomic lamina-associated domains (LADs) from the nuclear lamina, increased chromatin accessibility and the activation of repetitive sequence expression. Consequently, accelerated senescence was observed in hMSCs lacking SIRT3. The re-introduction of SIRT3 was sufficient to rescue the loss of heterochromatin and alleviate hMSC senescence phenotypes. Collectively, we found a new geroprotective role of SIRT3 in safeguarding heterochromatin of hMSCs.
MATERIALS AND METHODS
Animal experiments
Animal experiments performed in this study were approved by the Chinese Academy of Sciences Institutional Animal Care and Use Committee. For teratoma formation assay, 5 × 106SIRT3+/+ or SIRT3–/– human embryonic stem cells (hESCs) on Matrigel (BD Biosciences)-coated plates were collected and mixed with Matrigel: mTeSR (STEMCELL Technologies) (1:4) solution. Then, the mixture was injected subcutaneously into the inguinal region of NOD/SCID mice (male, 6 weeks old). At about 12 weeks after the injection, the teratoma was collected for further analysis. The in vivo hMSC transplantation assay was conducted as previously described (32). In brief, 1 × 106SIRT3+/+ or SIRT3–/– hMSCs expressing luciferase (Luc) were injected into the tibialis anterior (TA) muscle of nude mice (male, 6 weeks old). An in vivo imaging system (IVIS) spectrum imaging system (XENOGEN, Caliper) was used for luciferase activity analysis at 1, 3, and 5 days after hMSC injection.
Cell culture
SIRT3+/+ hESCs (H9 hESCs, WiCell Research) and SIRT3–/- hESCs were cultured on mouse embryonic fibroblast (MEF, inactivated with mitomycin C) feeder cells in hESC culture medium (DMEM/F12 (Thermo Fisher Scientific), supplemented with 20% Knockout Serum Replacement (Thermo Fisher Scientific), 1% penicillin/streptomycin (Thermo Fisher Scientific), 0.1 mM non-essential amino acids (NEAA, Thermo Fisher Scientific), 2 mM GlutaMAX (Thermo Fisher Scientific), 55 μM β-mercaptoethanol (Thermo Fisher Scientific), and 10 ng/ml bFGF (Joint Protein Central)), or maintained on Matrigel-coated plates with mTeSR medium. hMSCs differentiated from hESCs and primary hMSCs were cultured in hMSC culture medium (Minimum Essential Medium α (MEMα) basal medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS, Gemcell, Cat# 100-500, Lot# A77E01F), 0.1 mM NEAA, 1% penicillin/streptomycin, and 1 ng/ml bFGF).
Generation of SIRT3–/– hESCs
CRISPR/Cas9-mediated gene knockout was performed following previously reported methods with minor modifications (32,33). In brief, sgRNAs were designed using the website http://crispr.mit.edu. Sequences representing sgRNAs targeting exon 1 or exon 3 of SIRT3 were cloned into pCAG-mCherry-sgRNA vector (Addgene, #87110). After treatment with ROCK inhibitor (Y-27632, TOCRIS) for 24 h, pCAG-mCherry-SIRT3 sgRNAs and pCAG-1BPNLS-Cas9-1BPNLS-2AGFP were simultaneously electroporated into SIRT3+/+ hESCs by 4D-Nucleofector (Lonza). Cells were then seeded on Matrigel-coated six-well plates and cultured with mTeSR medium. After 48 h, EGFP/mCherry-dual-positive cells were sorted by fluorescent-activated cell sorting (FACS) system (BD FACS Aria II) and seeded on MEF feeder cells with hESC culture medium. Emerging clones were manually picked out for further verification. Genomic DNAs from these hESC clones were extracted for PCR and DNA sequencing. sgRNA sequences for gene editing and primers for clone identification and off-target detection are listed in Supplementary Table S1. SIRT3–/– hESCs generated by sgRNA #4-mediated gene editing were used for further analysis in this study.
Generation and characterization of hMSCs
hMSC differentiation from hESCs was performed as previously described (34,35). In brief, hESC-derived embryoid bodies were seeded on Matrigel-coated six-well plates in hMSC differentiation medium containing MEMα (Thermo Fisher Scientific), 10% fetal bovine serum (FBS, Gemcell), 1% penicillin/streptomycin (Thermo Fisher Scientific), 1 ng/ml bFGF (Joint Protein Central) and 5 ng/ml TGFβ (Humanzyme) for 10 days. The fibroblast-like cells were then transferred into hMSC culture medium, and derived-hMSCs were purified by FACS system (BD FACS Aria II) via selecting CD73, CD90 and CD105 triple-positive cells. The triple-lineage differentiation potentials of hESC-derived hMSCs were evaluated by von Kossa staining (osteogenesis), Saffron O staining (chondrogenesis), and Oil red O staining (adipogenesis) (36).
Isolation of primary hMSCs
Primary hMSCs were isolated from an old individual (76 years old) as previously reported (37). In brief, primary hMSCs were isolated under the approval of the ethics committee of 306th Hospital of PLA in Beijing. The gingiva tissues were processed into small pieces and then digested in TrypLE™ Express Enzyme (Thermo Fisher Scientific) plus Dispase IV (Thermo Fisher Scientific) at 37°C for 30 min. After digestion, the mixture was filtered with a 70-μm cell strainer (Falcon) and then centrifuged at 500 g for 10 min. The resulting pellet was resuspended in hMSC culture medium and transferred into a gelatin-coated 6-well plate for culture. After about two weeks, the primary hMSCs were collected using TrypLE™ Express Enzyme (Thermo Fisher Scientific) for subculture.
DNA methylation analysis of the OCT4 promoter region
DNA methylation analysis of the OCT4 promoter region was carried out as previously reported (38,39). In brief, genomic DNAs were extracted using a DNA Extraction Kit (TIANGEN) and processed for bisulfite treatment using an EZ DNA Methylation Kit (Zymo Research). Genomic DNA fragment of the OCT4 promoter region was amplified using LA Taq DNA Polymerase Hot-Start Version (TaKaRa), and subsequently cloned into the pMD20 T vector (TaKaRa). Six clones from each sample were sequenced with the universal primer M13. Primers used for DNA methylation analysis are listed in Supplementary Table S1.
Hematoxylin and eosin (H&E) staining
H&E staining was performed as previously described with modifications (40). In brief, teratomas were fixed in 4% paraformaldehyde (PFA) and embedded in Tissue-Tek O.C.T. Compound (Sakura) for cryosections. The cryosections of teratomas were air-dried for 10 min to remove moisture, washed with PBS and fixed with 4% PFA for 5 min. The nuclei were stained by the hematoxylin solution (Servicebio) for 5 min and rinsed in running tap water. Then, the sections were stained by eosin solution (Servicebio) for 2 min and dipped in 80%, 90% and 100% alcohol. Finally, the slides with teratoma sections were cleaned with 100% xylene and mounted with Cytoseal-60 (Stephens Scientific).
Immunofluorescence microscopy
For immunofluorescence analysis, cells seeded on coverslips (Thermo Fisher Scientific) with a suitable confluence were fixed in 4% PFA for 15 min, washed with PBS at least three times, permeabilized using 0.4% Triton X-100 in PBS for 30 min, and then blocked with 10% donkey serum (Jackson ImmunoResearch) in PBS for 1 h at room temperature. Cells were then incubated with indicated primary antibodies in 10% donkey serum in PBS at 4°C overnight. Cells were washed at least three times with PBS and incubated with secondary antibodies and Hoechst 33342 (Invitrogen, H3570) at room temperature for 1 h, then washed at least three times with PBS. Finally, the coverslips were mounted using mounting medium (Vector Labs), and images were captured with a Leica SP5 confocal system.
Antibodies used for immunofluorescence analysis are as follows: anti-SOX2 (Santa Cruz, sc-17320), anti-OCT4 (Santa Cruz, sc-5279), anti-NANOG (Abcam, ab21624), anti-Ki67 (ZSGB-BIO, ZM-0166), anti-γH2AX (Millipore, 05-636), anti-53BP1 (BETYHL, A300-273A), anti-Lamin B1 (Abcam, ab16048), anti-FLAG (Sigma, F1804), anti-H3K9me3 (Abcam, ab8898), anti-Lamin A/C (Santa Cruz, sc-376248), anti-KAP1 (Abcam, ab10483), anti-HP1α (Cell Signaling Technology, 2616), Alexa 488 donkey anti-mouse IgG (Invitrogen, A21202), Alexa 568 donkey anti-rabbit IgG (Invitrogen, A10042) and Alexa 647 donkey anti-goat IgG (Invitrogen, A21447).
Transmission electron microscopy (TEM)
Transmission electron microscopy was performed as previously reported (41). In brief, 5 × 106 cells of SIRT3+/+ and SIRT3–/– hMSCs were collected enzymatically by TrypLE™ Express Enzyme (Thermo Fisher Scientific), washed once with PBS, and pelleted by centrifugation. Cell pellets were fixed with 2.5% (vol/vol) glutaraldehyde with Phosphate Buffer (PB) (0.1 M, pH 7.4), washed four times with PB, postfixed with 1% (wt/vol) osmium tetroxide in PB for 2 h at 4°C, dehydrated through a graded ethanol series (30%, 50%, 70%, 80%, 90%, 100%, 100%, 7 min of immersion each) into pure acetone (10 min of immersion, twice), infiltrated in a graded mixture (3:1, 1:1, 1:3) of acetone and SPI-PON812 resin (16.2 g SPI-PON812, 10 g DDSA and 8.9 g NMA), embedded in pure resin with 1.5% BDMA and polymerized for 12 h at 45°C and 48 h at 60°C. The 70-nm ultrathin sections were obtained using a Leica EM UC6 ultramicrotome, and then double-stained with uranyl acetate and lead citrate. Images were captured with a Spirit Transmission Electron Microscope (FEI Company) at 100 kV. Mitochondrial number per cell and individual mitochondrial size were analyzed from 30 to 40 electron micrographs using ImageJ-based Fiji software (https://imagej.net/Fiji) (42).
Senescence-associated-β-galactosidase (SA-β-gal) staining assay
SA-β-gal staining of hMSCs was performed as previously reported (43). In brief, cells were washed twice with PBS and fixed in fixation buffer (2% formaldehyde and 0.2% glutaraldehyde) for 5 min at room temperature. Fixed cells were stained with fresh staining buffer (40 mM citric acid/Na phosphate buffer, 5 mM K4[Fe(CN)6], 5 mM K3[Fe(CN)6], 150 mM NaCl, 2 mM MgCl2, 1 mg/ml X-gal (AMRESCO, 0428-25G)) at 37°C overnight. Images were captured with a microscope digital camera (Olympus). The numbers of SA-β-gal-positive cells were determined with ImageJ.
Clonal expansion assay
Briefly, 2 000 cells were seeded in each well of a 12-well plate, and cultured for 12-14 days before cells were completely confluent. Then, cells were fixed with 4% PFA for 30 min and stained with 0.2% crystal violet (Biohao, C0520) for 1 h at room temperature and washed with running tap water. The relative cell density was quantified using ImageJ.
Lentivirus production
HEK293T cells were co-transfected with indicated lentiviral overexpression plasmids and lentiviral packing vectors including psPAX2 (Addgene, #12260) and pMD2.G (Addgene, #12259). The medium containing lentiviral particles were harvested at 48 h and 72 h after transfection, and lentiviral particles were concentrated by ultracentrifugation at 19 400 g at 4°C for 2.5 h.
Plasmid construction
SIRT3-FLAG cDNA was generated from the cDNA of SIRT3+/+ hMSCs via PCR amplification and then cloned into the pLE4 empty vector (a kind gift from Dr. Tomoaki Hishida) by using NovoRec® plus One-step PCR Cloning Kit (Novoprotein, #NR005) according to the manufacturer's instructions and confirmed by DNA sequencing. Cloning primers are listed in Supplementary Table S1.
Co-immunoprecipitation (co-IP)
Co-IP assay was carried out as previously reported (37). In brief, HEK293T cells transfected with plasmids expressing SIRT3-FLAG or Luc-FLAG were lysed in CHAPS lysis buffer (120 mM NaCl, 0.3% CHAPS (Sigma, C900480), 1 mM EDTA, 40 mM HEPES, pH 7.5, and cOmpleteTM protease inhibitor cocktail (Roche)) at 4°C for 2 h with rotation, and then centrifuged at 12 000 g at 4°C for 30 min to collect the supernatants. The supernatants were mixed with anti-FLAG Affinity Gel (Sigma, A2220) and rotated at 4°C overnight. The resulting immunocomplexes were washed with CHAPS lysis buffer three times, incubated with FLAG peptide in CHAPS lysis buffer at 4°C for 2 h with rotation to competitively elute SIRT3-interacting protein complexes by anti-FLAG Affinity Gel, and then centrifuged at 3 000 g at 4°C for 5 min. Finally, the resultant supernatants were boiled in 1× SDS loading buffer for 10 min and processed for western blot analysis.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis
LC-MS/MS was performed as previously reported (37). Briefly, eluted proteins from the co-IP assay were separated using 12% SDS-PAGE gel and then stained with Coomassie blue. Protein bands stained by Coomassie blue were cut from the SDS-PAGE gel, subjected to decoloration and dehydration, and then digested with sequencing-grade Trypsin (Worthington). Peptides were extracted from the resulting gel pieces, dried, and then dissolved in 0.1% formic acid. Subsequently, the dissolved peptides were separated with a C18 reverse phase column and then eluted for mass spectrometry analysis using a nanoLC-Q EXACTIVE Mass Spectrometer (Thermo Fisher Scientific, San Jose, CA, USA). Raw data were processed with the MaxQuant (Version 1.3) and Proteome Discoverer (Version 1.4). False discovery rate (FDR) was analyzed with Percolator and set to 1% as the threshold of the peptide confidence parameter. Proteins with the log2 peak area ratio (44) of SIRT3-FLAG and Luc-FLAG [log2 (SIRT3-FLAG/Luc-FLAG)] > 3 were considered as potential SIRT3-interacting proteins. Gene Ontology Cellular Component (GO-CC) terms analysis was performed using Metascape (45). These potential SIRT3-interacting proteins and enriched GO-CC terms are listed in Supplementary Table S2.
Western blot analysis
Cells were lysed in 1× SDS lysis buffer (62.5 mM Tris–HCl (pH 6.8), 2% (wt/vol) SDS) and boiled for 10 min at 105°C. Whole cell lysates were quantified using a Pierce® Microplate BCA Protein Assay Kit (Thermo Fisher Scientific, #23252), and then mixed with 1× SDS loading buffer. The prepared samples were subjected to SDS-PAGE separation and electrotransferred to PVDF membranes (Millipore). Membranes were then blocked with 5% non-fat milk at room temperature and incubated with the indicated primary antibodies overnight at 4°C. After several washes, the membranes were incubated with appropriate secondary antibodies conjugated with horseradish peroxidase (HRP) for 1 h at room temperature. The visualization and data processing of western blot results were obtained using a ChemiDoc XRS+ system (Bio-Rad) and analyzed with ImageJ.
Antibodies used for western blot analysis are as follows: anti-SIRT3 (Cell Signaling Technology, 2627), anti-SIRT3 (Cell Signaling Technology, 5490), anti-SIRT3 (Abcam, ab189860), anti-SIRT3 (Sigma, S4072), anti-SIRT3 (Abgent, AP6242a), anti-P16 (BD Bioscience, 550834), anti-P21 (Cell Signaling Technology, 2947), anti-β-Tubulin (Immunoway, YM3030), anti-GAPDH (Santa Cruz, sc-365062), anti-β-Actin (Santa Cruz, sc-69879), anti-COX IV (Cell Signaling Technology, 4850), anti-KAP1 (Abcam, ab10483), anti-Lamin B1 (Abcam, ab16048), anti-FLAG (Sigma, F1804), anti-LAP2 (BD Bioscience, 611000), anti-HP1α (Cell Signaling Technology, 2616), anti-HP1γ (Cell Signaling Technology, 2619), anti-LBR (Abcam, ab32535), HRP-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, 115-035-003) and HRP-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, 111-035-003).
RNA and DNA purification and analyses
For RNA analysis, total RNAs were first extracted using TRIzol Reagent (Thermo Fisher Scientific) and genomic DNAs were removed with a DNA-free Kit (Thermo Fisher Scientific). Then, cDNAs were reverse-transcribed using the GoScript Reverse Transcription system (Promega) and quantitative PCR (qPCR) was conducted using the SYBR Green qPCR Mix (TOYOBO) in a CFX384 Real-Time system (Bio-Rad). Primers used for qPCR analysis are listed in Supplementary Table S1.
For DNA analysis, genomic DNAs were first extracted using a DNA Extraction Kit (TIANGEN) and PCR was then performed using PrimeSTAR DNA Polymerase (TaKaRa) following the manufacturer's protocols. Primers used for PCR amplification are listed in Supplementary Table S1.
Quantification of relative mtDNA content
Mitochondrial DNA (mtDNA) content was measured according to the method described by Venegas et al. (46) with minor modifications. The mtDNA content was defined as the relative ratio between mitochondrial gene (tRNALeu(UUR)) copy number and simultaneously measured copy number of a nuclear gene (β-2-microglobulin, β2M) using qPCR analysis. The qPCR analysis was conducted using a Bio-Rad CFX384 Real-Time system with the SYBR Green qPCR Mix (TOYOBO). Briefly, total hMSC DNAs were purified using Genomic DNA Buffer Set/Genomic-tip 20/G Kit (QIAGEN). 0.5 ng genomic DNAs were used to perform qPCR assays, which generated a datasheet with the CT value for each qPCR. Relative mtDNA content was measured by the difference in ΔCT between the tRNALeu(UUR) and β2M genes, which was calculated as (β2M average CT) – (tRNALeu(UUR) average CT) = ΔCT, 2 × 2(ΔCT) = mtDNA content. qPCR primers used to calculate mtDNA content are listed in Supplementary Table S1.
Three-dimensional (3D) imaging analysis
3D image reconstruction was conducted using Imaris software as previously reported (47–49). In brief, SIRT3-deficient hMSCs were transduced with lentiviruses expressing SIRT3-FLAG, and then prepared for immunofluorescence analysis. Primary antibodies used were anti-FLAG (Sigma, F1804) and anti-Lamin B1 (Abcam, ab16048). Imaging was captured with a Leica SP5 confocal system. All images were acquired in an 8-bit TIFF format at 512 × 512 pixels resolution. Z-stacks were taken at 200-nm intervals. The image stacks were delivered into the Imaris software package (Bitplane Scientific Solutions) to generate 3D surface reconstruction images of confocal z-stacks. Using Imaris software, the ‘Surfaces’ tool was used to create a 3D rendering of each channel. Using these rendered surfaces, a surface–surface colocalization function was used to visualize and calculate the amount of overlapping area between the SIRT3-FLAG surface and Lamin B1 surface.
Measurement of mitochondrial mass, reactive oxygen species (ROS) and mitochondrial membrane potential (MMP)
For mitochondrial mass measurement, cells were incubated with 10 μM of nonyl acridine orange (NAO, Invitrogen, #A1372) for 10 min at 37°C and then measured by flow cytometry (BD LSRFortessa).
Total cellular ROS levels were measured using 2.5 μM H2DCFDA (Invitrogen, #C6827) and mitochondrial ROS levels were determined using 2.5 μM MitoSOX Red (Invitrogen, #M36008). Cells were stained with H2DCFDA or MitoSOX Red for about 20 min at 37°C in the dark, and then analyzed by flow cytometry (BD LSRFortessa).
MMPs of hMSCs were measured with the Cell MeterTM JC-10 Mitochondrial Membrane Potential Assay Kit (AAT Bioquest, #22801) following the manufacturer's instructions and a previous study (50). In brief, cells were incubated with 1× JC-10 dye-loading solution at 37°C for 15-30 min, then analyzed by flow cytometry (BD LSRFortessa). The ratio of fluorescence intensities Ex/Em = 490/590 (JC-10 aggregate emission) and 490/530 nm (JC-10 monomer emission) (FL590/FL530) was calculated to define MMP levels.
H2O2 concentration measurement
To determine the total H2O2 levels in the cells, Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Invitrogen, #A22188) was used according to manufacturer's instructions. Cells were lysed in lysis buffer and then centrifugated at 20 000 g at 4°C for 10 min. The resultant supernatants were plated in a black, clear bottom 96-well plate. The samples along with serial dilutions of H2O2 for the standard curve were reacted with the working solution at room temperature for 30 min, protected from light. Fluorescence measurements of reaction product were performed (excitation = 530 nm, emission = 590 nm) using a BioTek spectrophotometer. Each point of the standard curve and the cell samples were measured in triplicate.
Whole genome sequencing
Whole genome sequencing was performed as previously described (37,51). Briefly, the total genomic DNAs per duplicate were extracted from 1 × 106 cells using a DNeasy Blood & Tissue Kit (QIAGEN). DNA quality control, library construction and sequencing on Illumina HiSeq X Ten platforms were performed in Novogene Bioinformatics Technology Co., Ltd following standard protocols.
CNV analysis
Briefly, after quality control and adaptor trimming, reads were aligned to the human hg19 genome. Then the number of reads for each consecutive 500-kb window was counted using readCounter function in hmmcopy_utils (https://github.com/shahcompbio/hmmcopy_utils). The R/Bioconductor package HMMcopy (v1.26.0) (52) was used to correct copy number, GC content and mappability.
RNA-seq
Library preparation and sequencing were performed as previously reported (53). Briefly, total RNAs were extracted using TRIzol Reagent, genomic DNAs were removed using the DNA-free Kit and mRNAs were purified by Poly (A) enrichment. Library construction was conducted using a NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina (NEB) according to the manufacturer's instruction. Library quality control and sequencing on Illumina HiSeq X Ten platforms were performed by Novogene Bioinformatics Technology Co., Ltd.
RNA-seq data processing
RNA-seq data processing was performed as previously described (32). First, low-quality reads and adaptors were removed using TrimGalore (v0.4.4_dev) (54). Clean reads were aligned to the UCSC human hg19 genome using HISAT2 software (v2.1.0) (54). Counts for annotated genes were generated using HTSeq (v0.6.1) (55). The R/Bioconductor package DESeq2 (56) was used to identify differentially expressed genes with a cutoff Benjamini-Hochberg adjust P-value less than 0.05 and |log2(fold change)| > 0.58. Enrichment analysis was performed using Toppgene (57) (https://toppgene.cchmc.org/) with adjusted P-value < 0.05.
For repetitive elements analysis, after quality control and adaptor trimming, reads were aligned to the human hg19 genome (RepEnrich2 provided setup) using Bowtie2 (v2.2.9). Unique and multi-mapped reads were assigned to repetitive elements using RepEnrich2 software. Differentially expressed repetitive elements were analyzed with edgeR (58) on the fractional counts data. Repetitive elements with a Benjamini-Hochberg adjusted P-value of less than 0.05 and |log2(fold change)| > 0.5 were considered as significant (59).
DamID-seq
Library preparation and sequencing for DamID-seq were performed as previously described (37). In brief, SIRT3+/+ or SIRT3–/– hMSCs were transduced with lentiviruses expressing Dam-only or Dam-Emerin (EMD). Cells were collected at 72 h after transduction and genomic DNAs were isolated using a DNeasy Blood & Tissue Kit (QIAGEN). Genomic DNAs were subjected to DpnI digestion, adaptor ligation, DpnII digestion, PCR amplification, purification, sonication, and AlwI digestion. DNA library construction was conducted using a NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs). Library quality control and sequencing using Illumina HiSeq X Ten platforms were performed by Novogene Bioinformatics Technology Co., Ltd.
DamID-seq data processing
DamID-seq data processing was performed as previously described (37,51). First, low-quality reads were removed and Dam and Dam-EMD adaptor sequences were trimmed using TrimGalore (v0.4.4_dev) software. Clean reads were aligned to the UCSC human hg19 genome using bowtie2 (v2.2.9) with default parameters. Reads aligned to mitochondrial DNA (chrM) or with low mapping quality (MAPQ score < 10) were removed using samtools (v1.9). Duplicate reads were also removed using Picard for future analysis. To eliminate bias introduced by sequencing depth, replicates for each sample were merged and the same numbers of high-quality reads were randomly sampled for merged bams. To quantify the DamID signals, we calculated the log2 ratio of Dam-EMD and Dam signals [log2 (Dam-EMD/Dam)] for each 10-base-pair (bp) bin using deepTools (version 3.3.0) software. The R/Bioconductor package HMMt was used to identify LADs (60).
Identification of LAD-localized repetitive elements and protein-coding genes
Analysis of LAD-localized repetitive elements and protein-coding genes was performed as previously described (51) with some modifications. Briefly, the annotations of repetitive elements (from HOMER) or protein-coding genes were intersected with LAD regions using the findOverlapPairs function in R/Bioconductor package GenomicRanges (v1.36.0). Repetitive elements with DamID signals ≥ 0 and protein-coding genes with overlapping ratios ≥ 0.8 were identified as LAD-localized repetitive elements and protein-coding genes, respectively.
Determination of expression levels of LAD-localized repetitive elements
To determine the expression levels of repetitive elements within LAD regions, cleaned reads from RNA-seq were realigned to the human hg19 genome using STAR (v5.21) as previously described (61). The normalized read counts were calculated by RPKM (reads per kilobase per million mapped reads) for each 10 bp using the bamCoverage function and then the expression level of each repetitive element within LAD regions was calculated using the multiBigwigSummary function.
ATAC-seq
Library preparation and sequencing were performed as previously reported (51). Briefly, 50 000 SIRT3+/+ or SIRT3–/– hMSCs were washed twice with 500 μl ice-cold PBS and dissociated in 50 μl lysis buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% (v/v) Nonidet P-40 Substitute). The sample was then centrifuged at 500 g for 10 min at 4°C, followed by incubation at 37°C for 30 min supplemented with 50 μl transposition reaction mix (10 μl 5× TTBL buffer, 4 μl TTE mix and 36 μl nuclease-free H2O) from the TruePrep DNA Library Prep Kit V2 for Illumina (Vazyme Biotech). After tagmentation, fragmented DNAs were purified with AMPure XP beads (Beckman Coulter). TruePrep DNA Library Prep Kit V2 for Illumina and TruePrep Index Kit V2 for Illumina (Vazyme Biotech) were used to amplify the library. Libraries were purified and selected using AMPure XP beads. Library quality control and sequencing using Illumina HiSeq X Ten platforms were performed by Novogene Bioinformatics Technology Co., Ltd.
ATAC-seq data processing
For ATAC-seq data analysis, quality control and adaptor trimming were first performed by TrimGalore (v0.4.4_dev). Clean reads were then aligned to the human hg19 genome using Bowtie2 (v2.2.9) with default parameters. Reads aligned to mitochondrial DNA (chrM) or with low mapping quality (MAPQ score < 10) were removed using samtools (v1.9) (62). Duplicate reads were also removed using Picard software (v1.113). To avoid the bias introduced by sequencing depth, replicates for each sample were merged and the same numbers of high-quality reads were randomly sampled for merged bams. Each read was extended by 250 bp and normalized by RPKM for each 10-bp using bamCoverage function in deepTools (v3.3.0) software (63).
After excluding blacklisted regions, peak calling was performed using MACS2 (v2.1.2) with parameter “–nomodel –shift 0 –extsize 250” (64). Genome annotations for ATAC-seq peaks were performed using the annotatePeaks function in HOMER software (65). The R/Bioconductor package DiffBind (v2.12.0) (66) was used to identify differential peaks occupancy with the DESeq2 method. Consensus peaks with absolute fold difference > 1 and FDR < 0.05 were considered as differentially enriched peaks. Remaining peaks were classified as shared peaks.
Statistical analysis
Statistical data are presented as the means ± SEM. Two-tailed Student's t-test was conducted using the GraphPad Prism 8 software. P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***) are considered statistically significant.
RESULTS
Downregulation of SIRT3 during hMSC senescence
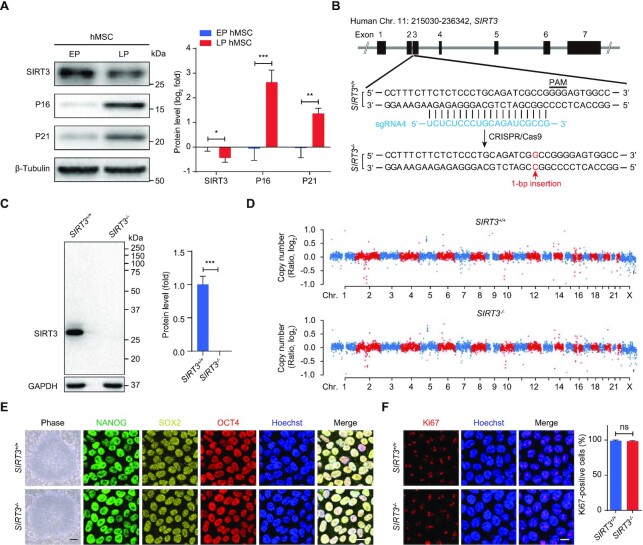
SIRT3 downregulation has been put forward as an indicator of aging in mice (17,67). To determine the expression pattern of SIRT3 during human stem cell senescence, we first evaluated SIRT3 expression in replicative senescent hMSCs that were characterized by the upregulation of classical senescence markers P16 and P21 and the downregulation of nuclear lamina and heterochromatin-associated proteins including Lamin B1, KAP1, LAP2, HP1α and HP1γ (Figure 1A and Supplementary Figure S1A). We observed that the protein levels of SIRT3 were decreased in replicative senescent hMSCs (Figure 1A).
Figure 1.
Downregulation of SIRT3 in senescent hMSCs and generation of SIRT3-deficient hESCs. (A) Western blot analysis of SIRT3, P16 and P21 expression in replicative senescent hMSCs. Early passage (EP), passage 4 (P4); late passage (LP), P14. β-Tubulin was used as loading control. Data are presented as the means ± SEM. n = 3. *P < 0.05; **P < 0. 01; ***P < 0. 001. (B) Schematic diagram of SIRT3 gene editing strategy using CRISPR/Cas9- mediated non-homologous end-joining (NHEJ) in hESCs. The SIRT3 sgRNA is shown in blue. 1-bp insertion (shown in red) was identified by DNA sequencing. (C) Western blot analysis of SIRT3 in SIRT3+/+ and SIRT3–/– hESCs. GAPDH was used as a loading control. Data are presented as the means ± SEM. n = 3. ***P < 0.001. (D) Copy number variation (CNV) analysis of SIRT3+/+ and SIRT3–/– hESCs by whole genome sequencing. (E) Immunofluorescent (IF) images of pluripotency markers NANOG, SOX2 and OCT4 and phase-contrast images for SIRT3+/+ and SIRT3–/– hESCs. Scale bar, 25 μm (IF images) and 250 μm (phase-contrast images). (F) Immunofluorescence analysis of Ki67 in SIRT3+/+ and SIRT3–/– hESCs. Scale bar, 10 μm. The statistical analysis of Ki67-positive cells is shown on the right. Data are presented as the means ± SEM. n = 3. ns, not significant.
SIRT3 deficiency accelerates hMSC senescence
To explore a potential geroprotective role of SIRT3 in hMSCs, we knocked out SIRT3 in hESCs using CRISPR/Cas9-based gene editing (Figure 1B and Supplementary Figure S1B). Gene editing using sgRNAs led to a single base-pair insertion (G/C) in exon 3 of SIRT3, resulting in the depletion of SIRT3 (Figure 1B-C and Supplementary Figure S1B-D). No off-target cleavage was detected at the top ten predicted sites (Supplementary Figure S1E). Genome-wide copy number variation (CNV) and karyotype analysis demonstrated that genomic stability and integrity were maintained in SIRT3–/– hESCs (Figure 1D and Supplementary Figure S1F). As observed in SIRT3+/+ hESCs, SIRT3–/– hESCs expressed pluripotency markers including NANOG, SOX2 and OCT4 (Figure 1E and Supplementary Figure S1G). In addition, SIRT3 deficiency did not affect DNA hypomethylation at the OCT4 promoter in hESCs (Supplementary Figure S1H). To evaluate the differentiation potential of SIRT3–/– hESCs, we conducted teratoma formation analysis and observed comparable differentiation capacities into three germ layers relative to SIRT3+/+ hESCs (Supplementary Figure S1I). Immunofluorescence staining and flow cytometry analysis further showed comparable Ki67-positive cells, cell cycle status, and mitochondrial mass in both SIRT3+/+ and SIRT3–/– hESCs (Figure 1F and Supplementary Figure S1J and K). Altogether, these data indicate that SIRT3 may be dispensable for the maintenance of genomic stability and pluripotency in hESCs.
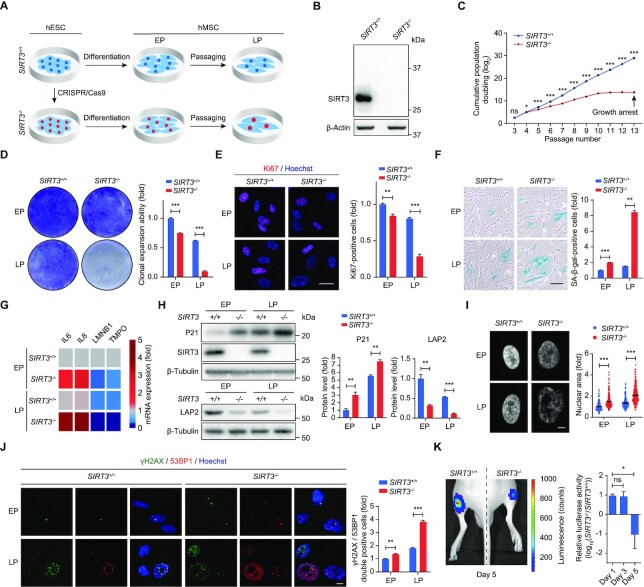
We then differentiated SIRT3+/+ and SIRT3–/– hESCs into hMSCs (Figure 2A). Both SIRT3+/+ and SIRT3–/– hMSCs expressed canonical hMSC markers such as CD73, CD90 and CD105 (Supplementary Figure S2A). SIRT3 ablation in hMSCs was verified by western blot analysis (Figure 2B). The genomic integrity of SIRT3–/– hMSCs was well maintained as shown by CNV analysis (Supplementary Figure S2B). Compared with SIRT3+/+ hMSCs, SIRT3–/– hMSCs were able to differentiate into all three lineages including adipocytes, chondrocytes and osteoblasts, though with a reduced ability to differentiate toward adipocytes or chondrocytes (Supplementary Figure S2C-E). SIRT3–/– hMSCs exhibited retarded cellular growth as indicated by impaired hMSC proliferation, restricted clonal expansion, decreased Ki67-positive cells, reduced S phase and prolonged G2/M phase (Figure 2C-E and Supplementary Figure S2F). Moreover, SIRT3 deficiency accelerated hMSC senescence, as manifested by increased senescence-associated β-galactosidase (SA-β-gal) staining, upregulated expression of IL6, IL8 and P21, downregulated expression of Lamin B1 and LAP2, and larger nuclei and increased DNA damage (Figure 2F-J). We next evaluated the in vivo retention ability of SIRT3–/– hMSCs and found accelerated decay of SIRT3–/– hMSCs at the engrafted sites (Figure 2K). These findings indicate that SIRT3 prevents hMSCs from accelerated senescence.
Figure 2.
SIRT3 deficiency accelerates hMSC senescence and cellular dysfunction. (A) Schematic diagram showing the generation of SIRT3+/+ and SIRT3–/– hMSCs from hESCs. (B) Western blot analysis of SIRT3 in SIRT3+/+ and SIRT3–/– hMSCs at EP (P4). β-Actin was used as a loading control. (C) Growth curve showing cumulative population doubling of SIRT3+/+ and SIRT3–/– hMSCs. Data are presented as the means ± SEM. n = 3. ns, not significant; *P < 0.05; ***P < 0.001. (D) Clonal expansion analysis of SIRT3+/+ and SIRT3–/– hMSCs at EP (P4) and LP (P9). Data are presented as the means ± SEM. n = 3. ***P < 0.001. (E) Immunofluorescence analysis of Ki67 in SIRT3+/+ and SIRT3–/– hMSCs at EP (P4) and LP (P9). Scale bar, 25 μm. The statistical analysis of Ki67-positive cells is shown on the right. Data are presented as the means ± SEM. n = 3. **P < 0.01; ***P < 0.001. (F) SA-β-gal staining of SIRT3+/+ and SIRT3–/– hMSCs at EP (P4) and LP (P9). Scale bar, 50 μm. Data are presented as the means ± SEM. n = 3. **P < 0.01; ***P < 0.001. (G) RT-qPCR analysis for the expression of IL6, IL8, LMNB1 (Lamin B1) and TMPO (LAP2) in SIRT3+/+ and SIRT3–/– hMSCs at EP (P4) and LP (P9). Data are representative of three independent experiments. (H) Western blot analysis of P21, LAP2 and SIRT3 in SIRT3+/+ and SIRT3–/– hMSCs at EP (P4) and LP (P9). β-Tubulin was used as loading control. Data are presented as the means ± SEM. n = 3. **P < 0.01; ***P < 0.001. (I) Nuclear area analysis in SIRT3+/+ and SIRT3–/– hMSCs at EP (P4) and LP (P9). Nuclei were stained with Hoechst 33342 and nuclear area was measured with ImageJ. Scale bar, 5 μm. Data are presented as the means ± SEM. n ≥ 150. ***P < 0.001. (J) Immunofluorescence analysis of γH2AX and 53BP1 in SIRT3+/+ and SIRT3–/– hMSCs at EP (P4) and LP (P9). Scale bar, 10 μm. The statistical analysis of γH2AX and 53BP1 double-positive cells is shown on the right. Data are presented as the means ± SEM. n = 3. **P < 0.01; ***P < 0.001. (K) Photon flux from tibialis anterior (TA) muscles of nude mice transplanted with SIRT3+/+ (left) or SIRT3–/– hMSCs (right) expressing luciferase. Luciferase activity in TA tissue was detected using an in vivo imaging system (IVIS). Data are presented as the means ± SEM. n = 5. ns, not significant; *P < 0.05.
SIRT3 interacts with nuclear lamina proteins and heterochromatin-associated proteins
Given that SIRT3 has been implicated in regulating mitochondrial homeostasis (68), we evaluated mitochondrial function in SIRT3–/– hMSCs using a series of indices. Electron microscopy analysis revealed that SIRT3 knockout in hMSCs led to an increase in mitochondrial number and a decrease in mitochondrial size (Supplementary Figure S2G). Consistently, mitochondrial mass and mitochondrial DNA copy number were also increased in SIRT3–/– hMSCs (Supplementary Figure S2H and I). In addition, flow cytometry analysis showed that the absence of SIRT3 in hMSCs resulted in increased mitochondrial and cellular ROS levels (Supplementary Figure S2J-L), along with decreased mitochondrial membrane potential (Supplementary Figure S2M). These findings suggest that SIRT3 plays an important role in the maintenance of mitochondrial homeostasis in hMSCs.
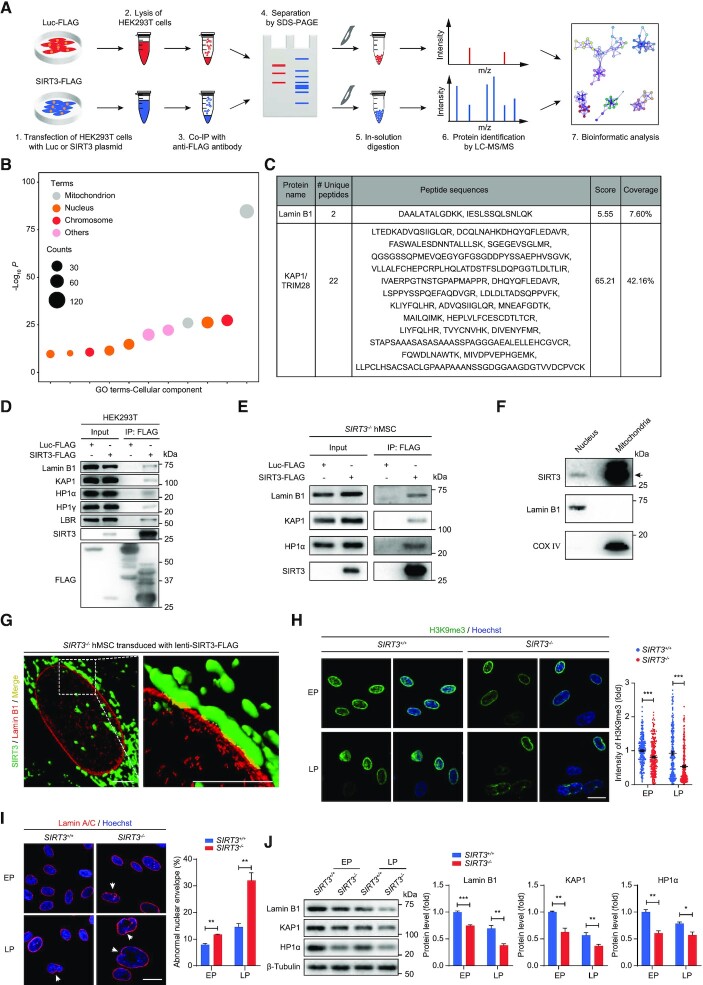
Previous studies have indicated the important function of nuclear-localized SIRT3 in a set of biological processes (24). To investigate the molecular mechanism underlying hMSC senescence phenotypes caused by SIRT3 deficiency, especially its function in the nucleus, we sought to identify novel SIRT3-interacting nuclear proteins. To this end, FLAG-tagged SIRT3 was expressed in HEK293T cells and immunoprecipitation (IP) was performed using an anti-FLAG antibody-conjugated affinity gel followed by LC-MS/MS (IP-MS) analysis (Figure 3A). As expected, many SIRT3-interacting proteins were mitochondrial-localized proteins identified previously (Figure 3B and Supplementary Table S2), such as IDH2 (Supplementary Figure S3A), a known SIRT3 substrate (18). Intriguingly, there were also many SIRT3-interacting proteins located in the nucleus including the nuclear envelope protein, Lamin B1, and the heterochromatin-associated protein KAP1 (Figure 3B and C, Supplementary Figure S3A and Table S2). Co-immunoprecipitation (co-IP) verified the interaction between SIRT3 and Lamin B1 (Figure 3D and E), as well as other nuclear-localized proteins including the heterochromatin proteins KAP1, HP1α and HP1γ, and the nuclear envelope protein LBR (Figure 3D-E and Supplementary Figure S3B). Interactions between SIRT3 and these nuclear proteins were further supported by the presence of SIRT3 protein in the nuclear fraction in subcellular fractionation assay (Figure 3F). To further interrogate the newly identified interactions between SIRT3 and these nuclear proteins in hMSCs, we transduced SIRT3–/– hMSCs with lentiviral vectors expressing FLAG-tagged SIRT3. 3D reconstruction of z-stack images capturing FLAG and Lamin B1 immunofluorescence revealed that SIRT3 and Lamin B1 co-localized at the nuclear periphery in addition to its primary localization in the mitochondria (Figure 3G). Taken together, these findings imply a potentially uncharacterized role of SIRT3 in the nucleus, likely via the interaction with nuclear lamina and heterochromatin-associated proteins.
Figure 3.
SIRT3 interacts with nuclear lamina and heterochromatin-associated proteins. (A) Schematic workflow of co-IP followed by LC-MS/MS analysis for identifying SIRT3-interacting proteins. FLAG-tagged Luc was used as negative control. (B) Gene Ontology Cellular Component (GO-CC) enrichment analysis of SIRT3-interacting proteins identified by mass spectrometry. (C) Lamin B1 and KAP1 were identified as novel interacting proteins of SIRT3 by mass spectrometry. (D) Co-IP analysis for the interactions between indicated proteins and SIRT3-FLAG in HEK293T cells. (E) Co-IP analysis for the interactions of KAP1, Lamin B1, HP1α with SIRT3 in SIRT3–/–hMSCs. (F) Western blot analysis of SIRT3 in the fractions of nucleus and mitochondria isolated from SIRT3+/+hMSCs. The black arrow indicates the expected position of SIRT3. Lamin B1 and COX IV were used as marker proteins for nuclear and mitochondrial fractions, respectively. (G) Three-dimensional reconstruction images showing the colocalization of SIRT3 with Lamin B1 in SIRT3–/– hMSCs expressing FLAG-tagged SIRT3. SIRT3 (FLAG) is shown in green, Lamin B1 is marked in red and the colocalization of SIRT3 and Lamin B1 is shown in yellow. Scale bar, 4 μm. (H) Immunofluorescence analysis of H3K9me3 in SIRT3+/+and SIRT3–/– hMSCs at EP (P4) and LP (P9). Scale bar, 25 μm. Data are presented as the means ± SEM. n > 200. ***P < 0.001. (I) Immunofluorescence analysis of Lamin A/C in SIRT3+/+and SIRT3–/– hMSCs at EP (P4) and LP (P9). Scale bar, 25 μm. The white arrow indicates abnormal nuclear envelope. The statistical analysis of abnormal nuclear envelope is shown on the right. Data are presented as the means ± SEM. n = 3. **P < 0.01. (J) Western blot analysis of Lamin B1, KAP1 and HP1α in SIRT3+/+ and SIRT3–/– hMSCs at EP (P4) and LP (P9). β-Tubulin was used as loading control. Data are presented as the means ± SEM. n = 3. *P < 0.05; **P < 0.01; ***P < 0.001.
SIRT3 deficiency leads to loss of heterochromatin and derepression of repetitive sequences
To specifically examine the effects of SIRT3 depletion on the regulation of nuclear envelope and heterochromatin organization in hMSCs, we further conducted electron microscopy analysis, and examined the heterochromatin mark H3K9me3 (69). Both approaches showed that SIRT3 deficiency accelerated the loss of heterochromatin in hMSCs over passaging (Figure 3H and Supplementary Figure S3C). Meanwhile, the percentage of cells with an irregular nuclear envelope in SIRT3–/– hMSCs was much higher compared with that in SIRT3+/+ hMSCs (Figure 3I). In addition, western blot analysis demonstrated decreased expression of nuclear envelope and heterochromatin-associated proteins including Lamin B1, KAP1 and HP1α in SIRT3–/– hMSCs (Figure 3J).
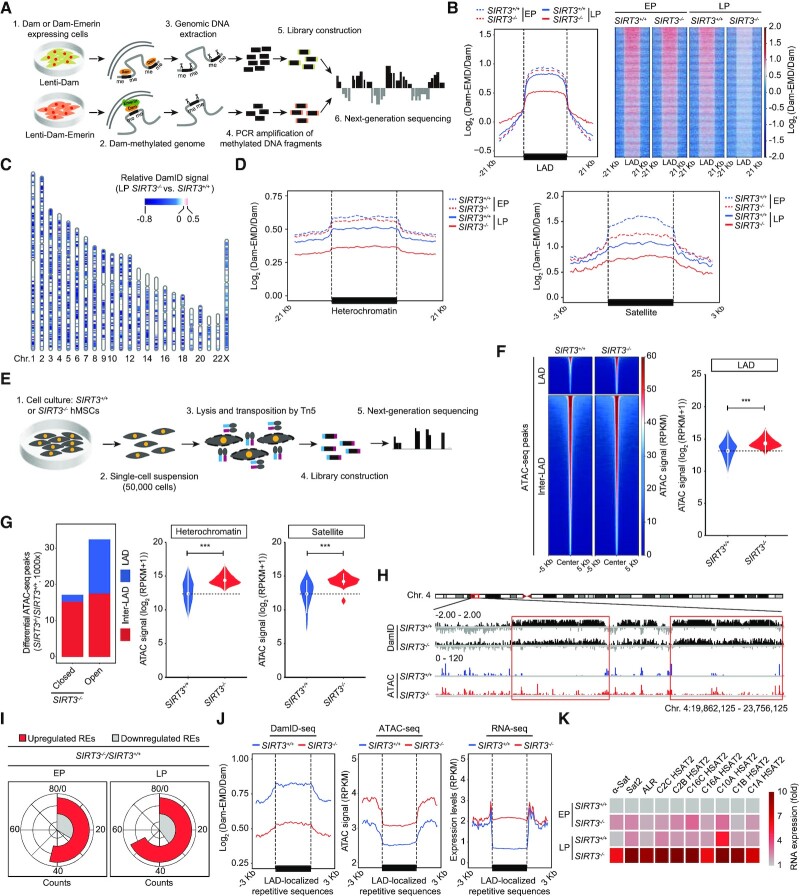
Lamina-associated domains (LADs) comprise the genomic heterochromatin regions associated with the nuclear lamina (70,71). It has been reported that heterochromatin disorganization and loss of LADs are defined drivers of aging (29,31,37,51,69). To further dissect the role of SIRT3 in heterochromatin regulation, we used DNA adenine methyltransferase identification with high-throughput sequencing (DamID-seq) to map LAD regions in SIRT3+/+ and SIRT3–/– hMSCs (Figure 4A and Supplementary Figure S4A and B, Table S3). We found that the DamID signals were decreased in SIRT3–/– hMSCs (Figure 4B-C and Supplementary Figure S4C), especially those at heterochromatin regions and satellite, rRNA and LTR repetitive sequences within LADs (Figure 4D and Supplementary Figure S4D and E). These data indicate detachment of LADs from the nuclear lamina associated with heterochromatin loss, consistent with morphological anomalies in the nuclei of SIRT3–/– hMSCs. In line with the DamID-seq results, data from assay for transposase accessible chromatin using sequencing (ATAC-seq) revealed a more open chromatin state in the absence of SIRT3 (Figure 4E–H, Supplementary Figure S4F–I and Table S3), especially in LADs and LAD-localized repetitive sequences (satellite, rRNA, LTR) (Figure 4F–H and Supplementary Figure S4H and I). Consistent with the changes in chromatin state, further RNA sequencing (RNA-seq) and reverse transcription and quantitative PCR (RT-qPCR) analysis showed that the expression of most repetitive elements including satellite, rRNA and LTR, was increased in SIRT3–/– hMSCs compared with SIRT3+/+ hMSCs (Figure 4I-K, Supplementary Figure S4J-O, and Table S4). These findings demonstrate that SIRT3 deficiency in hMSCs leads to loss of heterochromatin, detachment of LADs from the nuclear lamina, and enhancement of transcription from genomic repetitive sequences. Meanwhile, we observed decreased DamID signals, increased chromatin accessibility and transcription of LAD-localized protein-coding genes in SIRT3–/– hMSCs compared with those in SIRT3+/+ hMSCs (Supplementary Figure S4P-R), further indicating a more open and active chromatin state of LAD regions upon SIRT3 deficiency.
Figure 4.
DamID-seq and ATAC-seq analyses in SIRT3+/+ and SIRT3–/– hMSCs. (A) Schematic diagram showing the workflow for DamID-seq library preparation and sequencing. Dam-Emerin expression in hMSCs drives the methylation of adenines near the nuclear envelope by the DNA adenine methyltransferase (Dam). Genomic DNA sequence containing Dam-methylated sites was specifically cut using Dpn I, PCR amplified, and then used for library construction and high-throughput sequencing. A parallel control ‘Dam only’ was used to account for background Dam signals. (B) Line plot (left) and heatmaps (right) showing reduced DamID signals [log2 (Dam-EMD/Dam)] at LAD regions in SIRT3–/– hMSCs compared to SIRT3+/+ hMSCs at EP (P4) and LP (P9). (C) Ideogram showing the loss of DamID signals in SIRT3–/– hMSCs compared to those in SIRT3+/+ hMSCs at LP (P9) over 23 chromosomes. The color key from blue to red represents relatively low to high DamID signals in SIRT3–/– hMSCs compared to those in SIRT3+/+ hMSCs. (D) Line plots showing the loss of DamID signals at heterochromatin regions and LAD-localized satellite repetitive elements in SIRT3–/– hMSCs compared to those in SIRT3+/+ hMSCs at EP (P4) and LP (P9). (E) Schematic diagram showing the workflow for ATAC-seq library preparation and sequencing. (F) Left, heatmaps showing the ATAC signals within LAD and inter-LAD regions in SIRT3+/+ hMSCs and SIRT3–/– hMSCs at LP (P9). Right, violin plot showing the ATAC signals within LADs in SIRT3+/+ hMSCs and SIRT3–/– hMSCs at LP (P9). ***P < 0.001. (G) Left, the distribution of differential ATAC-seq peaks within LAD or inter-LAD regions of SIRT3–/– hMSCs compared to those of SIRT3+/+ hMSCs at LP (P9). Right, violin plots showing increased accessibility at heterochromatin regions and LAD-localized satellite repetitive elements in SIRT3–/– hMSCs compared to those in SIRT3+/+ hMSCs at LP (P9). ***P < 0.001. (H) Representative tracks of DamID-seq and ATAC-seq signals showing decreased DamID signals along with increased ATAC-seq signals within LAD regions in SIRT3–/– hMSCs compared to those in SIRT3+/+ hMSCs at LP (P9). (I) RNA-seq analysis showing repetitive elements (REs) that were differentially expressed in SIRT3–/– hMSCs compared to those in SIRT3+/+ hMSCs at EP (P3) and LP (P7). (J) Line plots showing decreased DamID signals, increased chromatin accessibility and upregulated expression levels of LAD-localized repetitive sequences in SIRT3–/–hMSCs at LP (P7 for RNA-seq and P9 for DamID-seq and ATAC-seq) compared to SIRT3+/+ hMSCs using a conjoint analysis of DamID-seq, ATAC-seq and RNA-seq. LAD-localized repetitive sequences with higher expression levels in SIRT3–/– hMSCs at LP (P7) were processed for conjoint analysis. (K) Heatmap showing the relative RNA expression levels of repetitive elements in SIRT3–/–hMSCs compared to those in SIRT3+/+ hMSCs at EP (P4) and LP (P9) using RT-qPCR analysis. Expression levels of repetitive elements were normalized to SIRT3+/+ hMSCs at EP (P4). Data are representative of three independent experiments.
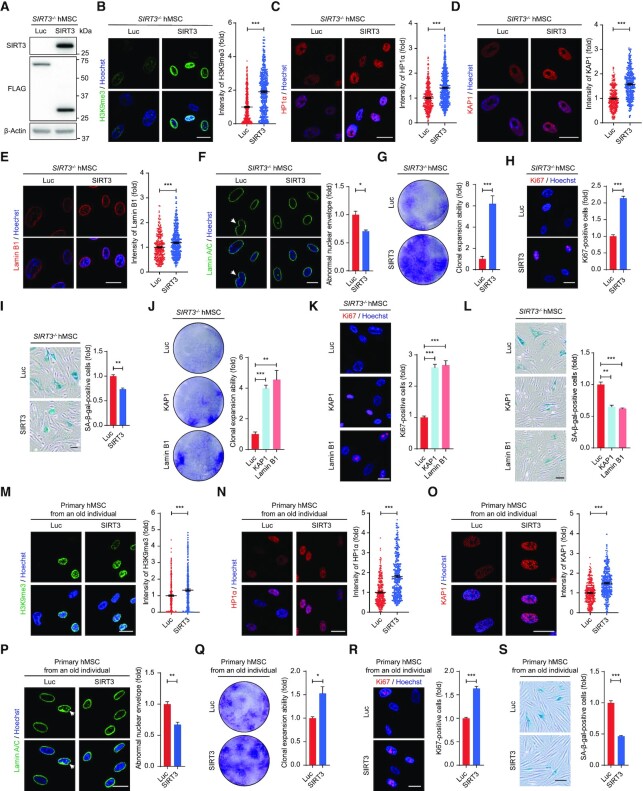
Re-introduction of SIRT3 attenuates heterochromatin loss and hMSC senescence
To further evaluate the role of SIRT3 in regulating heterochromatin homeostasis and hMSC senescence, we re-introduced SIRT3 in SIRT3–/– hMSCs (Figure 5A). The re-introduction of SIRT3 not only promoted the expression of heterochromatin markers H3K9me3, HP1α, KAP1, but also increased the protein level of Lamin B1 and reduced nuclear envelope irregularity (Figure 5B-F). Concomitantly, increased proliferation ability and decreased SA-β-gal-positive cells were observed in SIRT3–/– hMSCs upon the re-introduction of SIRT3 (Figure 5G-I). Likewise, promoting heterochromatin condensation by overexpression of heterochromatin proteins KAP1 or nuclear lamina protein Lamin B1 also attenuated the senescence of SIRT3-deficient hMSCs (Figure 5J-L).
Figure 5.
Overexpression of SIRT3 alleviates hMSC senescence. (A) Western blot analysis showing the levels of SIRT3 and FLAG-tagged proteins in SIRT3–/– hMSCs (P6) expressing Luc-FLAG or SIRT3-FLAG. β-Actin was used as loading control. (B) Immunofluorescence analysis of H3K9me3 in SIRT3–/– hMSCs (P6) expressing Luc or SIRT3. Scale bar, 25 μm. Data are presented as the means ± SEM. n > 300. ***P < 0.001. (C) Immunofluorescence analysis of HP1α in SIRT3–/– hMSCs (P6) expressing Luc or SIRT3. Scale bar, 25 μm. Data are presented as the means ± SEM. n > 300. ***P < 0.001. (D) Immunofluorescence analysis of KAP1 in SIRT3–/– hMSCs (P6) expressing Luc or SIRT3. Scale bar, 25 μm. Data are presented as the means ± SEM. n > 300. ***P < 0.001. (E) Immunofluorescence analysis of Lamin B1 in SIRT3–/– hMSCs (P6) expressing Luc or SIRT3. Scale bar, 25 μm. Data are presented as the means ± SEM. n > 300. ***P < 0.001. (F) Immunofluorescence analysis of Lamin A/C in SIRT3–/– hMSCs (P6) expressing Luc or SIRT3. Scale bar, 25 μm. The white arrow indicates abnormal nuclear envelope. The statistical analysis of abnormal nuclear envelope is shown on the right. Data are presented as the means ± SEM. n = 3. *P < 0.05. (G) Clonal expansion analysis of SIRT3–/– hMSCs (P6) expressing Luc or SIRT3. Data are presented as the means ± SEM. n = 3. ***P < 0.001. (H) Immunofluorescence analysis of Ki67 in SIRT3–/– hMSCs (P6) expressing Luc or SIRT3. Scale bar, 25 μm. The statistical analysis of Ki67-positive cells is shown on the right. Data are presented as the means ± SEM. n = 3. ***P < 0.001. (I) SA-β-gal staining of SIRT3–/– hMSCs (P6) expressing Luc or SIRT3. Scale bar, 50 μm. Data are presented as the means ± SEM. n = 3. **P < 0.01. (J) Clonal expansion analysis of SIRT3–/– hMSCs (P6) expressing Luc, KAP1 or Lamin B1. Data are presented as the means ± SEM. n = 3. **P < 0.01; ***P < 0.001. (K) Immunofluorescence analysis of Ki67 in SIRT3–/– hMSCs (P6) expressing Luc, KAP1 or Lamin B1. Scale bar, 25 μm. The statistical analysis of Ki67-positive cells is shown on the right. Data are presented as the means ± SEM. n = 3. ***P < 0.001. (L) SA-β-gal staining of SIRT3–/– hMSCs (P6) expressing Luc, KAP1 or Lamin B1. Scale bar, 50 μm. Data are presented as the means ± SEM. n = 3. **P < 0.01; ***P < 0.001. (M) Immunofluorescence analysis of H3K9me3 in primary hMSCs (P10) expressing Luc or SIRT3. Scale bar, 25 μm. Data are presented as the means ± SEM. n > 300. ***P < 0.001. (N) Immunofluorescence analysis of HP1α in primary hMSCs (P10) expressing Luc or SIRT3. Scale bar, 25 μm. Data are presented as the means ± SEM. n > 300. ***P < 0.001. (O) Immunofluorescence analysis of KAP1 in primary hMSCs (P10) expressing Luc or SIRT3. Scale bar, 25 μm. Data are presented as the means ± SEM. n > 300. ***P < 0.001. (P) Immunofluorescence analysis of Lamin A/C in primary hMSCs (P10) expressing Luc or SIRT3. Scale bar, 25 μm. The white arrow indicates abnormal nuclear envelope. The statistical analysis of abnormal nuclear envelope is shown on the right. Data are presented as the means ± SEM. n = 3. **P < 0.01. (Q) Clonal expansion analysis of primary hMSCs (P10) expressing Luc or SIRT3. Data are presented as the means ± SEM. n = 3. *P < 0.05. (R) Immunofluorescence analysis of Ki67 in primary hMSCs (P10) expressing Luc or SIRT3. Scale bar, 25 μm. The statistical analysis of Ki67-positive cells is shown on the right. Data are presented as the means ± SEM. n = 3. ***P < 0.001. (S) SA-β-gal staining of primary hMSCs (P10) expressing Luc or SIRT3. Scale bar, 50 μm. Data are presented as the means ± SEM. n = 3. ***P < 0.001.
Next, we overexpressed SIRT3 in primary hMSCs isolated from an old individual, and observed increased expression of heterochromatin-associated markers and reduced abnormal nuclear envelope (Figure 5M-P). Furthermore, overexpression of SIRT3 alleviated senescence in physiologically aged hMSCs (Figure 5Q-S). Altogether, these results indicate that SIRT3 exerts a geroprotective role in hMSCs at least in part by stabilizing heterochromatin.
DISCUSSION
In this study, we have uncovered a novel role for SIRT3 in safeguarding hMSCs from senescence. First, we found reduced SIRT3 expression in hMSCs undergoing senescence. Secondly, SIRT3 depletion accelerated senescence in hMSCs. Thirdly, SIRT3 interacted with nuclear lamina proteins and heterochromatin-associated proteins, thus involved in nuclear envelope and heterochromatin organization. Finally, the re-introduction of SIRT3 restored heterochromatin and rescued hMSC senescence. In summary, these observations suggest a new role of SIRT3 in the stabilization of heterochromatin architecture and the alleviation of hMSC senescence (Supplementary Figure S5), thus providing novel insights into the epigenetic mechanisms regulating human stem cell homeostasis.
We gained these insights through generating SIRT3-deficient hESCs using CRISPR/Cas9-editing and obtained SIRT3-null hMSCs through directed differentiation, which provides a valuable platform for current and future investigations into the role of SIRT3 in regulating human stem cell homeostasis. Based on these models, we found that SIRT3 was dispensable for the maintenance of hESC pluripotency. Besides, SIRT3 deficiency in hESCs did not induce any abnormality in the nuclear envelope, consistent with previous studies describing the tolerance of cellular defects in pluripotent stem cells (72). In contrast, SIRT3-deficient hMSCs (a type of somatic stem cells) exhibited accelerated senescence with a variety of cellular dysfunctions, indicating a cell-type specific role of SIRT3 in regulating stem cell homeostasis. In addition to SIRT3, other Sirtuins, such as SIRT6 and SIRT7, have been implicated in protecting hMSCs from accelerated senescence (37,73), suggesting the conservation of different Sirtuin family members in regulating hMSC homeostasis.
As an important epigenetic layer in aging regulation, heterochromatin is characterized by extensive enrichment of repetitive sequences, which are often repressed under normal conditions (69,74). By contrast, their aberrant activation has been implicated in senescence and in aging-related disorders (51,69,75–79). By showing that SIRT3 interacted with nuclear envelope proteins and heterochromatin-associated proteins and that SIRT3 deficiency resulted in reduced heterochromatin, decreased association between LADs and nuclear lamina, increased heterochromatin accessibility, and aberrant activation of repetitive sequences, our study revealed a novel role of nuclear SIRT3 in stabilizing heterochromatin organization and counteracting hMSC senescence. Likewise, a nucleolar Sirtuin, SIRT7 has recently been reported as a geroprotective heterochromatin stabilizer (37). Furthermore, previous studies have reported that SIRT6 and SIRT7 deficiency both result in the derepression of LINE1 repetitive sequences, thus triggering the innate immune response and accelerating cellular senescence (37,80–82).
Previous studies have investigated the role of SIRT3 in the regulation of cellular senescence, and most of them attribute it to the modulation of mitochondrial functions (17,67,83). Consistently, we observed profound mitochondrial defects including increased mitochondrial ROS levels in SIRT3-deficient hMSCs. Mitochondrial ROS may not only affect DNA and histone modifications via the modulation of epigenetic modifiers (84,85), but also lead to heterochromatin loss (86). Therefore, it is possible that mitochondrial-localized SIRT3 may also contribute to safeguarding hMSCs from senescence.
Taken together, we identified SIRT3 as a novel heterochromatin safeguard to counteract hMSC senescence. Together with the known knowledge of SIRT3 in the mitochondria, our study uncovered a new nuclear activity of SIRT3 in rejuvenating aged human stem cells. These findings not only strengthen our understanding of the epigenetic regulation of heterochromatin during human cellular senescence but also provide potential druggable targets for the development of novel therapeutic strategies to prevent stem cell decay and ameliorate aging-related diseases.
DATA AVAILABILITY
Whole genome sequencing, RNA-seq, ATAC-seq, DamID-seq data generated in this study have been deposited in the Genome Sequence Archive (87) in the National Genomics Data Center (88), Beijing Institute of Genomics (China National Center for Bioinformation) of the Chinese Academy of Sciences, under accession number HRA000466 that are publicly accessible at http://bigd.big.ac.cn/gsa-human. Mass spectrometry data have been deposited in the ProteomeXchange Consortium via the PRIDE (89) partner repository with the dataset identifier PXD023479. RNA-seq data have also been deposited in the Aging Atlas database (90) (https://bigd.big.ac.cn/aging/index), Beijing Institute of Genomics (China National Center for Bioinformation) of the Chinese Academy of Sciences.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Lei Bai, Qun Chu, Ruijun Bai, Shikun Ma, Jing Lu and Ying Yang for their administrative assistance, Wei Li, Jingyi Jia and Junyan Jiao for their help in animal experiments, Xiaoqing Ren, Jiping Yang, Lina Fu, Jinghui Lei, Hongkai Zhao and Shunlei Duan for their technical support, Jifeng Wang for his help in LC–MS/MS assay, Yun Feng for her help in 3D image reconstruction, Junying Jia and Shuang Sun for their help in FACS experiments, Can Peng for her help in TEM analysis, Junfeng Hao for her help in H&E staining analysis. The processing of high-throughput sequencing data was conducted on the ‘Era’ petascale supercomputer of the Computer Network Information Center of CAS.
Contributor Information
Zhiqing Diao, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Qianzhao Ji, University of Chinese Academy of Sciences, Beijing 100049, China; State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China.
Zeming Wu, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; Beijing Institute for Stem Cell and Regenerative Medicine, Beijing 100101, China.
Weiqi Zhang, University of Chinese Academy of Sciences, Beijing 100049, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China.
Yusheng Cai, State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; Beijing Institute for Stem Cell and Regenerative Medicine, Beijing 100101, China.
Zehua Wang, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Jianli Hu, University of Chinese Academy of Sciences, Beijing 100049, China; CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China.
Zunpeng Liu, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Qiaoran Wang, University of Chinese Academy of Sciences, Beijing 100049, China; CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China.
Shijia Bi, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Daoyuan Huang, Advanced Innovation Center for Human Brain Protection, National Clinical Research Center for Geriatric Disorders, Xuanwu Hospital Capital Medical University, Beijing 100053, China.
Zhejun Ji, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; Beijing Institute for Stem Cell and Regenerative Medicine, Beijing 100101, China.
Guang-Hui Liu, University of Chinese Academy of Sciences, Beijing 100049, China; State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; Beijing Institute for Stem Cell and Regenerative Medicine, Beijing 100101, China; Advanced Innovation Center for Human Brain Protection, National Clinical Research Center for Geriatric Disorders, Xuanwu Hospital Capital Medical University, Beijing 100053, China.
Si Wang, State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; Beijing Institute for Stem Cell and Regenerative Medicine, Beijing 100101, China; Advanced Innovation Center for Human Brain Protection, National Clinical Research Center for Geriatric Disorders, Xuanwu Hospital Capital Medical University, Beijing 100053, China.
Moshi Song, University of Chinese Academy of Sciences, Beijing 100049, China; State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; Beijing Institute for Stem Cell and Regenerative Medicine, Beijing 100101, China.
Jing Qu, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; Beijing Institute for Stem Cell and Regenerative Medicine, Beijing 100101, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2018YFC2000100]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDA16010100]; National Key Research and Development Program of China [2017YFA0103304, 2017YFA0102802, 2018YFA0107203, 2020YFA0804000, 2020YFA0112201, 2020YFA0113400]; National Natural Science Foundation of China [81921006, 81625009, 91749202, 81861168034, 91949209, 31671429, 92049304, 81671377, 81701388, 91749123, 81822018, 81870228, 81922027, 82071588, 92049116]; Program of the Beijing Municipal Science and Technology Commission [Z191100001519005]; Beijing Natural Science Foundation [Z190019, JQ20031]; Key Research Program of the Chinese Academy of Sciences [KFZD-SW-221]; K. C. Wong Education Foundation [GJTD-2019-06, GJTD-2019-08]; Science and Technology Service Network Initiative of Chinese Academy of Sciences [KFJ-STS-QYZD-2021-08-001]; Youth Innovation Promotion Association of CAS [2016093, 2021078]; Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences, the State Key Laboratory of Stem Cell and Reproductive Biology and the State Key Laboratory of Membrane Biology. Funding for open access charge: National Natural Science Foundation of China [91949209].
Conflict of interest statement. None declared.
REFERENCES
- 1. Houtkooper R.H., Pirinen E., Auwerx J.. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012; 13:225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McDonnell E., Peterson B.S., Bomze H.M., Hirschey M.D.. SIRT3 regulates progression and development of diseases of aging. Trends Endocrinol. Metab. 2015; 26:486–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang X., Cao R., Niu J., Yang S., Ma H., Zhao S., Li H.. Molecular basis for hierarchical histone de-β-hydroxybutyrylation by SIRT3. Cell Discov. 2019; 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wei Z., Song J., Wang G., Cui X., Zheng J., Tang Y., Chen X., Li J., Cui L., Liu C.Y.et al.. Deacetylation of serine hydroxymethyl-transferase 2 by SIRT3 promotes colorectal carcinogenesis. Nat. Commun. 2018; 9:4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaeberlein M., McVey M., Guarente L.. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999; 13:2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tissenbaum H.A., Guarente L.. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001; 410:227–230. [DOI] [PubMed] [Google Scholar]
- 7. Rogina B., Helfand S.L.. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. U. S. A. 2004; 101:15998–16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Satoh A., Brace C.S., Rensing N., Cliften P., Wozniak D.F., Herzog E.D., Yamada K.A., Imai S.. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013; 18:416–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kanfi Y., Naiman S., Amir G., Peshti V., Zinman G., Nahum L., Bar-Joseph Z., Cohen H.Y.. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012; 483:218–221. [DOI] [PubMed] [Google Scholar]
- 10. Benigni A., Cassis P., Conti S., Perico L., Corna D., Cerullo D., Zentilin L., Zoja C., Perna A., Lionetti V.et al.. Sirt3 deficiency shortens life span and impairs cardiac mitochondrial function rescued by opa1 gene transfer. Antioxid. Redox. Signal. 2019; 31:1255–1271. [DOI] [PubMed] [Google Scholar]
- 11. Tian X., Firsanov D., Zhang Z., Cheng Y., Luo L., Tombline G., Tan R., Simon M., Henderson S., Steffan J.et al.. SIRT6 is responsible for more efficient DNA double-strand break repair in long-lived species. Cell. 2019; 177:622–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giralt A., Villarroya F.. SIRT3, a pivotal actor in mitochondrial functions: metabolism, cell death and aging. Biochem. J. 2012; 444:1–10. [DOI] [PubMed] [Google Scholar]
- 13. Lombard D.B., Zwaans B.M.M.. SIRT3: as simple as it seems?. Gerontology. 2014; 60:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rose G., Dato S., Altomare K., Bellizzi D., Garasto S., Greco V., Passarino G., Feraco E., Mari V., Barbi C.et al.. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp. Gerontol. 2003; 38:1065–1070. [DOI] [PubMed] [Google Scholar]
- 15. Bellizzi D., Rose G., Cavalcante P., Covello G., Dato S., De Rango F., Greco V., Maggiolini M., Feraco E., Mari V.et al.. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics. 2005; 85:258–263. [DOI] [PubMed] [Google Scholar]
- 16. Almeida M., Porter R.M.. Sirtuins and FoxOs in osteoporosis and osteoarthritis. Bone. 2019; 121:284–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown K., Xie S., Qiu X., Mohrin M., Shin J., Liu Y., Zhang D., Scadden D.T., Chen D.. SIRT3 reverses aging-associated degeneration. Cell Rep. 2013; 3:319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Someya S., Yu W., Hallows W.C., Xu J., Vann J.M., Leeuwenburgh C., Tanokura M., Denu J.M., Prolla T.A.. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010; 143:802–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qiu X., Brown K., Hirschey M.D., Verdin E., Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010; 12:662–667. [DOI] [PubMed] [Google Scholar]
- 20. Hirschey M.D., Shimazu T., Capra J.A., Pollard K.S., Verdin E.. SIRT1 and SIRT3 deacetylate homologous substrates: AceCS1,2 and HMGCS1,2. Aging (Albany NY). 2011; 3:635–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hallows W.C., Lee S., Denu J.M.. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. U. S. A. 2006; 103:10230–10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shimazu T., Hirschey M.D., Hua L., Dittenhafer-Reed K.E., Schwer B., Lombard D.B., Li Y., Bunkenborg J., Alt F.W., Denu J.M.et al.. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010; 12:654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sengupta A., Haldar D.. Human sirtuin 3 (SIRT3) deacetylates histone H3 lysine 56 to promote nonhomologous end joining repair. DNA Repair (Amst.). 2018; 61:1–16. [DOI] [PubMed] [Google Scholar]
- 24. Scher M.B., Vaquero A., Reinberg D.. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007; 21:920–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lopez-Otin C., Blasco M.A., Partridge L., Serrano M., Kroemer G.. The hallmarks of aging. Cell. 2013; 153:1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oh J., Lee Y.D., Wagers A.J.. Stem cell aging: mechanisms, regulators and therapeutic opportunities. Nat. Med. 2014; 20:870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goodell M.A., Rando T.A.. Stem cells and healthy aging. Science. 2015; 350:1199–1204. [DOI] [PubMed] [Google Scholar]
- 28. Ren R., Ocampo A., Liu G.H., Izpisua Belmonte J.C.. Regulation of stem cell aging by metabolism and epigenetics. Cell Metab. 2017; 26:460–474. [DOI] [PubMed] [Google Scholar]
- 29. Zhang W., Qu J., Liu G.H., Belmonte J.C.I.. The ageing epigenome and its rejuvenation. Nat. Rev. Mol. Cell Biol. 2020; 21:137–150. [DOI] [PubMed] [Google Scholar]
- 30. Kubben N., Zhang W., Wang L., Voss T.C., Yang J., Qu J., Liu G.H., Misteli T.. Repression of the antioxidant NRF2 pathway in premature aging. Cell. 2016; 165:1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liang C., Liu Z., Song M., Li W., Wu Z., Wang Z., Wang Q., Wang S., Yan K., Sun L.et al.. Stabilization of heterochromatin by CLOCK promotes stem cell rejuvenation and cartilage regeneration. Cell Res. 2020; 31:187–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang S., Min Z., Ji Q., Geng L., Su Y., Liu Z., Hu H., Wang L., Zhang W., Suzuiki K.et al.. Rescue of premature aging defects in Cockayne syndrome stem cells by CRISPR/Cas9-mediated gene correction. Protein Cell. 2020; 11:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H., Wu Z., Liu X., Zhang S., Ji Q., Jiang X., Liu Z., Wang S., Qu J., Zhang W.et al.. ALKBH1 deficiency leads to loss of homeostasis in human diploid somatic cells. Protein Cell. 2020; 11:688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Geng L., Liu Z., Zhang W., Li W., Wu Z., Wang W., Ren R., Su Y., Wang P., Sun L.et al.. Chemical screen identifies a geroprotective role of quercetin in premature aging. Protein Cell. 2019; 10:417–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheng F., Wang S., Song M., Liu Z., Liu P., Wang L., Wang Y., Zhao Q., Yan K., Chan P.et al.. DJ-1 is dispensable for human stem cell homeostasis. Protein Cell. 2019; 10:846–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fu L., Hu Y., Song M., Liu Z., Zhang W., Yu F.X., Wu J., Wang S., Izpisua Belmonte J.C., Chan P.et al.. Up-regulation of FOXD1 by YAP alleviates senescence and osteoarthritis. PLoS Biol. 2019; 17:e3000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bi S., Liu Z., Wu Z., Wang Z., Liu X., Wang S., Ren J., Yao Y., Zhang W., Song M.et al.. SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein Cell. 2020; 11:483–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu Z., Zhang W., Song M., Wang W., Wei G., Li W., Lei J., Huang Y., Sang Y., Chan P.et al.. Differential stem cell aging kinetics in Hutchinson-Gilford progeria syndrome and Werner syndrome. Protein Cell. 2018; 9:333–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ling C., Liu Z., Song M., Zhang W., Wang S., Liu X., Ma S., Sun S., Fu L., Chu Q.et al.. Modeling CADASIL vascular pathologies with patient-derived induced pluripotent stem cells. Protein Cell. 2019; 10:249–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang S., Zheng Y., Li J., Yu Y., Zhang W., Song M., Liu Z., Min Z., Hu H., Jing Y.et al.. Single-Cell transcriptomic atlas of primate ovarian aging. Cell. 2020; 180:585–600. [DOI] [PubMed] [Google Scholar]
- 41. McLelland G.L., Goiran T., Yi W., Dorval G., Chen C.X., Lauinger N.D., Krahn A.I., Valimehr S., Rakovic A., Rouiller I.et al.. Mfn2 ubiquitination by PINK1/parkin gates the p97-dependent release of ER from mitochondria to drive mitophagy. Elife. 2018; 7:e32866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B.et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang X., Liu Z., Liu X., Wang S., Zhang Y., He X., Sun S., Ma S., Shyh-Chang N., Liu F.et al.. Telomere-dependent and telomere-independent roles of RAP1 in regulating human stem cell homeostasis. Protein Cell. 2019; 10:649–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bondarenko P.V., Chelius D., Shaler T.A.. Identification and relative quantitation of protein mixtures by enzymatic digestion followed by capillary reversed-phase liquid chromatography-tandem mass spectrometry. Anal. Chem. 2002; 74:4741–4749. [DOI] [PubMed] [Google Scholar]
- 45. Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K.. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019; 10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Venegas V., Halberg M.C.. Measurement of mitochondrial DNA copy number. Methods Mol. Biol. 2012; 837:327–335. [DOI] [PubMed] [Google Scholar]
- 47. Costes S.V., Daelemans D., Cho E.H., Dobbin Z., Pavlakis G., Lockett S.. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 2004; 86:3993–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Paolicelli R.C., Bolasco G., Pagani F., Maggi L., Scianni M., Panzanelli P., Giustetto M., Ferreira T.A., Guiducci E., Dumas L.et al.. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011; 333:1456–1458. [DOI] [PubMed] [Google Scholar]
- 49. Valm A.M., Cohen S., Legant W.R., Melunis J., Hershberg U., Wait E., Cohen A.R., Davidson M.W., Betzig E., Lippincott-Schwartz J.. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature. 2017; 546:162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gong S.S., Peng Y.Y., Jiang P.P., Wang M., Fan M.J., Wang X.J., Zhou H., Li H.W., Yan Q.F., Huang T.S.et al.. A deafness-associated tRNA(His) mutation alters the mitochondrial function, ROS production and membrane potential. Nucleic Acids Res. 2014; 42:8039–8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hu H., Ji Q., Song M., Ren J., Liu Z., Wang Z., Liu X., Yan K., Hu J., Jing Y.et al.. ZKSCAN3 counteracts cellular senescence by stabilizing heterochromatin. Nucleic. Acids. Res. 2020; 48:6001–6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ha G., Roth A., Lai D., Bashashati A., Ding J.R., Goya R., Giuliany R., Rosner J., Oloumi A., Shumansky K.et al.. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res. 2012; 22:1995–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu Z., Shi Y., Lu M., Song M., Yu Z., Wang J., Wang S., Ren J., Yang Y.G., Liu G.H.et al.. METTL3 counteracts premature aging via m6A-dependent stabilization of MIS12 mRNA. Nucleic Acids Res. 2020; 48:11083–11096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Anders S., Pyl P.T., Huber W.. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen J., Bardes E.E., Aronow B.J., Jegga A.G.. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009; 37:W305–W311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang Y.J., Guo L., Gonzales P.K., Gendron T.F., Wu Y., Jansen-West K., O’Raw A.D., Pickles S.R., Prudencio M., Carlomagno Y.et al.. Heterochromatin anomalies and double-stranded RNA accumulation underlie C9orf72 poly(PR) toxicity. Science. 2019; 363:eaav2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Filion G.J., van Bemmel J.G., Braunschweig U., Talhout W., Kind J., Ward L.D., Brugman W., de Castro I.J., Kerkhoven R.M., Bussemaker H.J.et al.. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010; 143:212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jin Y., Hammell M.. Analysis of RNA-Seq Data Using TEtranscripts. Methods Mol. Biol. 2018; 1751:153–167. [DOI] [PubMed] [Google Scholar]
- 62. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Proc G.P.D.. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ramirez F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R.et al.. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012; 481:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Son M.J., Kwon Y., Son T., Cho Y.S.. Restoration of mitochondrial NAD(+) levels delays stem cell senescence and facilitates reprogramming of aged somatic cells. Stem Cells. 2016; 34:2840–2851. [DOI] [PubMed] [Google Scholar]
- 68. Lombard D.B., Tishkoff D.X., Bao J.. Mitochondrial sirtuins in the regulation of mitochondrial activity and metabolic adaptation. Handb. Exp. Pharmacol. 2011; 206:163–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang W., Li J., Suzuki K., Qu J., Wang P., Zhou J., Liu X., Ren R., Xu X., Ocampo A.et al.. Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science. 2015; 348:1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kind J., Pagie L., Ortabozkoyun H., Boyle S., de Vries S.S., Janssen H., Amendola M., Nolen L.D., Bickmore W.A., van Steensel B.. Single-Cell dynamics of genome-nuclear lamina interactions. Cell. 2013; 153:178–192. [DOI] [PubMed] [Google Scholar]
- 71. van Steensel B., Belmont A.S.. Lamina-Associated Domains: Links with chromosome architecture, heterochromatin, and gene repression. Cell. 2017; 169:780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang W., Qu J., Suzuki K., Liu G.H., Izpisua Belmonte J.C.. Concealing cellular defects in pluripotent stem cells. Trends Cell Biol. 2013; 23:587–592. [DOI] [PubMed] [Google Scholar]
- 73. Pan H., Guan D., Liu X., Li J., Wang L., Wu J., Zhou J., Zhang W., Ren R., Zhang W.et al.. SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res. 2016; 26:190–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Liu J., Ali M., Zhou Q.. Establishment and evolution of heterochromatin. Ann. N. Y. Acad. Sci. 2020; 1476:59–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. De Cecco M., Criscione S.W., Peterson A.L., Neretti N., Sedivy J.M., Kreiling J.A.. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany NY). 2013; 5:867–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gorbunova V., Boeke J.D., Helfand S.L., Sedivy J.M.. Human Genomics. Sleeping dogs of the genome. Science. 2014; 346:1187–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wood J.G., Jones B.C., Jiang N., Chang C., Hosier S., Wickremesinghe P., Garcia M., Hartnett D.A., Burhenn L., Neretti N.et al.. Chromatin-modifying genetic interventions suppress age-associated transposable element activation and extend life span in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2016; 113:11277–11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. LaRocca T.J., Cavalier A.N., Wahl D.. Repetitive elements as a transcriptomic marker of aging: evidence in multiple datasets and models. Aging Cell. 2020; 19:e13167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhang W., Qu J., Liu G.H., Belmonte J.C.I.. The ageing epigenome and its rejuvenation. Nat. Rev. Mol. Cell Biol. 2020; 21:137–150. [DOI] [PubMed] [Google Scholar]
- 80. Simon M., Van Meter M., Ablaeva J., Ke Z., Gonzalez R.S., Taguchi T., De Cecco M., Leonova K.I., Kogan V., Helfand S.L.et al.. LINE1 derepression in aged wild-type and SIRT6-Deficient mice drives inflammation. Cell Metab. 2019; 29:871–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. De Cecco M., Ito T., Petrashen A.P., Elias A.E., Skvir N.J., Criscione S.W., Caligiana A., Brocculi G., Adney E.M., Boeke J.D.et al.. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature. 2019; 566:73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Van Meter M., Kashyap M., Rezazadeh S., Geneva A.J., Morello T.D., Seluanov A., Gorbunova V.. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 2014; 5:5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wiley C.D., Velarde M.C., Lecot P., Liu S., Sarnoski E.A., Freund A., Shirakawa K., Lim H.W., Davis S.S., Ramanathan A.et al.. Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metab. 2016; 23:303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gu X., Sun J., Li S., Wu X., Li L.. Oxidative stress induces DNA demethylation and histone acetylation in SH-SY5Y cells: potential epigenetic mechanisms in gene transcription in Aβ production. Neurobiol. Aging. 2013; 34:1069–1079. [DOI] [PubMed] [Google Scholar]
- 85. Schroeder E.A., Raimundo N., Shadel G.S.. Epigenetic silencing mediates mitochondria stress-induced longevity. Cell Metab. 2013; 17:954–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Frost B., Hemberg M., Lewis J., Feany M.B.. Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci. 2014; 17:357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wang Y., Song F., Zhu J., Zhang S., Yang Y., Chen T., Tang B., Dong L., Ding N., Zhang Q.. GSA: genome sequence archive. Genomics Proteomics Bioinformatics. 2017; 15:14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. National Genomics Data Center, M. and Partners Database resources of the National Genomics Data Center in 2020. Nucleic Acids Res. 2020; 48:D24–D33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M.et al.. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019; 47:D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Aging Atlas Consortium Aging Atlas: a multi-omics database for aging biology. Nucleic Acids Res. 2021; 49:D825–D830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole genome sequencing, RNA-seq, ATAC-seq, DamID-seq data generated in this study have been deposited in the Genome Sequence Archive (87) in the National Genomics Data Center (88), Beijing Institute of Genomics (China National Center for Bioinformation) of the Chinese Academy of Sciences, under accession number HRA000466 that are publicly accessible at http://bigd.big.ac.cn/gsa-human. Mass spectrometry data have been deposited in the ProteomeXchange Consortium via the PRIDE (89) partner repository with the dataset identifier PXD023479. RNA-seq data have also been deposited in the Aging Atlas database (90) (https://bigd.big.ac.cn/aging/index), Beijing Institute of Genomics (China National Center for Bioinformation) of the Chinese Academy of Sciences.