Abstract
Crosslinking and immunoprecipitation (CLIP) methods are powerful techniques to interrogate direct protein-RNA interactions and dissect posttranscriptional gene regulatory networks. One widely used CLIP variant is photoactivatable ribonucleoside enhanced CLIP (PAR-CLIP) that involves in vivo labeling of nascent RNAs with the photoreactive nucleosides 4-thiouridine (4SU) or 6-thioguanosine (6SG), which can efficiently crosslink to interacting proteins using UVA and UVB light. Crosslinking of 4SU or 6SG to interacting amino acids changes their base-pairing properties and results in characteristic mutations in cDNA libraries prepared for high-throughput sequencing, which can be computationally exploited to remove abundant background from non-crosslinked sequences and help pinpoint RNA binding protein binding sites at nucleotide resolution on a transcriptome-wide scale. Here we present a streamlined protocol for fluorescence-based PAR-CLIP (fPAR-CLIP) that eliminates the need to use radioactivity. It is based on direct ligation of a fluorescently labeled adapter to the 3′end of crosslinked RNA on immobilized ribonucleoproteins, followed by isolation of the adapter-ligated RNA and efficient conversion into cDNA without the previously needed size fractionation on denaturing polyacrylamide gels. These improvements cut the experimentation by half to 2 days and increases sensitivity by 10–100-fold.
INTRODUCTION
The fate of all eukaryotic RNAs is controlled by their protein partners which accompany them from transcription to turnover (1). RNA binding proteins (RBPs) participate in all processes involving RNA, including RNA biogenesis, processing, modification, translation and finally turnover. The importance of posttranscriptional gene regulatory networks is highlighted by the fact that >1500 human genes encode for proteins known or predicted to bind RNA (2) and that multiple diseases are associated with RBPs (3). RBPs typically recognize their targets at short, degenerate sequence-motifs (4–6), complicating prediction of RBP–RNA interactions and calling for systems-wide experimental methods to dissect the complex posttranscriptional gene regulatory networks controlled by RBPs. Crosslinking and Immunoprecipitation (CLIP) (7) and its many variants (8) address this need by allowing to identify not only the RNA bound by the RBP in living cells, but also its binding sites. In CLIP approaches, protected footprints of RNA irreversibly crosslinked to interacting RBPs are sequenced to precisely identify RBP binding sites on a target RNA. This in turn reveals critically important details of RBP function, such as the location of its binding site relative to other cis-acting elements or its recognized sequence motifs, which often resemble but may not be identical to high-affinity motifs determined in vitro by e.g. SELEX (9,10), RNA Bind’n'seq (5), RNAcompete (4).
All CLIP protocols share the same basic workflow: (i) RNA and interacting proteins are irreversibly crosslinked in intact cells by UV light and the RNA is digested to short footprints by RNases. (ii) The crosslinked ribonucleoprotein (RNP) of interest is immunopurified and RNA footprints are isolated. (iii) RNA footprints are converted into cDNA suitable for next generation sequencing. (iv) Sequenced reads are aligned to the genome and high-confidence RBP binding sites are computationally separated from noise derived from fragments of abundant cellular RNAs (11,12).
One widely used CLIP variant is photoactivatable ribonucleoside enhanced CLIP (PAR-CLIP) (13). In PAR-CLIP, cells are incubated prior to crosslinking with modified nucleosides, 4-thiouridine (4SU) or 6-thioguanisine (6SG) that get incorporated into nascent RNAs. The exocyclic thione group increases the photoreactivity of the base, allowing lower energy UV light (312 ≤ λ ≤ 365 nm) to be used for crosslinking. In the case of 4SU, the photoaddition of reactive amino acids leaves a lesion at position 4 of the base and changes its base-pairing properties (14). Crosslinked 4SU preferentially pairs with guanosine during reverse transcription, which results in a characteristic T-to-C transition (or G-to-A when using 6SG) in the sequenced cDNA. Computationally scoring these transitions determines the precise location and intensity of the RBP interaction and importantly, removes background from contaminating non-crosslinked RNAs derived from fragments of abundant transcripts (11,12).
A typical challenge for CLIP methods is the low yield of recovered RNA footprints that hovers in the nanogram range and complicates cDNA library construction efforts. For PAR-CLIP, the small RNA cDNA library construction procedure is derived from protocols designed for the characterization of abundant cellular small RNAs (miRNAs, piRNAs) (15,16) and requires successive ligations of 3′ and 5′ adapters to the RNA footprints in order to introduce primer-binding sites for reverse transcription and PCR. To avoid amplification of unwanted sequences, such as adapter-adapter ligation products, multiple denaturing gel purification steps are required, resulting in loss of material. The use of radioactive, P-32 RNA labeling aids tracking of the RNA footprints through the stringent size-selection and purification steps. However, the use of radioisotopes provides unique challenges with regard to laboratory licensing, variable labeling intensity, and long imaging times. Here, we present non-radioactive, fluorescence-based PAR-CLIP (fPAR-CLIP) that relies on ligation of fluorescent 3′ adapters to the crosslinked RNP while immobilized on beads to allow stringent purification and minimize gel electrophoresis-based size selections, thereby halving experimentation time and cutting the amount of necessary input material by ∼10–100-fold.
MATERIALS AND METHODS
A step-by-step protocol for fPAR-CLIP is included in the Supplementary Methods accompanying this manuscript.
Fluorescent 3′ adapter synthesis and purification
DNA precursors of fluorescently-labeled, barcoded, preadenylated 3′-sequencing adapters (single fluorescently-labeled: 5′-PO4-NNTGACTGTGGAATTCTCGGGTGCCAAGG-3′-aminolinker; double fluorescently-labeled: 5′-PO4-NNTGACTGTGGAATTCTCGGGT(NH2)GCCAAGG-3′-aminolinker; underlined sequences represent a unique barcode and can be varied) were synthesized at a 1.0 μmol scale on an ABI 3900 Upgrade DNA synthesizer (Biolytic). The following reagents were used for synthesis: PT-Amino-Modifier C6 CPG solid glass support (Glen Research), DNA phosphoramidites with base-labile deprotection groups (Glen Research), Solid Chemical Phosphorylation Reagent II (Glen Research) enabling 5′-phosphate modification and Amino-Modifier C6 dT phosphoramidite (Glen Research) allowing conjugation of a second fluorophore. The last two nucleotides were synthesized by mixing DNA phosphoramidites in the following ratio: dA:dC:dG:dT = 3:3:2:2 in order to ensure all bases were present in equal ratios. After completion of DNA synthesis, the oligonucleotides were deprotected according to the manufacturer's instructions (Glen Research). The crude DNAs were analyzed, purified by denaturing polyacrylamide gel electrophoresis, desalted, dried and QC-ed as previously described (17). Pure phosphorylated, amino-modified oligonucleotides were conjugated to either ATTO NHS ester (ATTO-TEC) or Alexa NHS ester (ThermoFisher) according to a previously described protocol (17). The crude fluorescently-labeled oligonucleotides were EtOH precipitated and analyzed by DPAGE and HPLC. The crude DNAs were preadenylated and purified as described before (18). Pure fluorescently-labeled, barcoded, preadenylated 3′-sequencing adapters were analyzed by denaturing polyacrylamide gel electrophoresis (DPAGE) and electrospray ionization mass spectrometry (ESI-MS).
Cell culture and immunoprecipitation
Flp-In™ 293 T-REx cells with stably integrated transgenic FLAG-HA tagged RBP of interest were cultured at 80% confluency in DMEM supplemented with 10% FBS and 1x Pen/Strep (ThermoFisher). 8–12 h before crosslinking, 4-thiouridine (4SU) was added to the cell culture medium at a final concentration of 100 μM. For U2AF1 PAR-CLIP, 4SU-labeling was reduced to 2 h at a final concentration of 250 μM. Cells were washed once with 1x PBS and crosslinked with 0.3 J/cm2 of 312 or 365 nm UV light for PAR-CLIP or 0.3 J/cm2 of 254 nm UVC light for regular CLIP in a Spectrolinker XL-1500 (Spectronics Corporation) or comparable instrument equipped with the appropriate UV lamps. Following crosslinking, cells were collected in PBS and centrifuged at 500 × g for 5 min at 4°C. For chromatin associated RNPs (e.g. U2AF1) cells were lysed by gentle resuspension in a modified HLB buffer (10 mM Tris at pH 7.5, 50 mM NaCl, 3 mM MgCl2, 0.1% NP-40, 10% glycerol) for 2 min and spun for 2 min at 200 × g. The nuclei were washed twice with the same buffer and extracted using MWS buffer (10 mM Tris–HCl at pH 7.0, 4 mM EDTA, 0.3 M NaCl, 1 M urea, and 1% [v/v] NP-40). The chromatin pellet was washed twice in MWS buffer and resuspended in RIPA buffer (10 mM Tris–HCl (pH 8.0), 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1%, sodium deoxycholate, 0.1% SDS, 140 mM NaCl supplemented with protease inhibitor cocktail (cOmplete, Sigma Aldrich 4693159001), followed by sonication in a bath sonicator (Bioruptor 300, Diagenode) three times for 30 s at medium amplitude at 4°C. For KHSRP, the cell pellet was lyzed in 3 volumes of RIPA buffer and sonicated. For cytoplasmic RBPs, cells were lysed in 3 volumes of NP40 lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% (v/v) NP40, 0.5 mM DTT) on ice for 15 min with optional addition of 0.1 U/μl RNase (T1, I or A/T1mix) and incubation for 10 min at 22°C. The cell extract was cleared at 15 000 × g for 15 min in a chilled centrifuge. Immunoprecipitation was done by incubating the cleared extracts with M2 anti-FLAG magnetic beads (20 μl of beads per ml of lysate) (Millipore Sigma, M8823). Binding was performed for 2 h at 4°C with rotation and beads were washed three times with 1 ml IP buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% (v/v) NP40).
RNase digestion, dephosphorylation, 3′ adapter ligation, phosphorylation
Beads were treated with RNase (T1, I or A/T1 mix) at a final concentration ranging from 0.1 to 1.5 U/μl in 1× bead volume IP buffer at 22°C for 10 min. Next, beads were washed twice with 1 ml of each of the following: high salt wash buffer (20 mM Tris, pH 7.5, 500 mM NaCl, 2 mM EDTA, 1% (v/v) NP40), RIPA buffer and dephosphorylation buffer (50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2). Dephosphorylation was achieved using Quick CIP (NEB, M0525S) according to manufacturer's instructions adjusted to 1 x bead volume. The reaction was supplemented with 1 U/μl RNase inhibitor (SUPERase•In™, Thermo Scientific, AM2694) and performed at 37°C for 10 min with shaking. Beads were washed twice with dephosphorylation buffer and twice with ligation buffer without DTT. RNA footprints were ligated to the 3′ adapter (5′-rAppNNTGACTGTGGAATTCTCGGGT(fl)GCCAAGG-fl, underlined sequence represents a barcode that can be changed in order to allow multiplexed sequencing, a full list of oligonucleotide sequences used for fPAR-CLIP is available in Supplementary Table S1) using T4 Rnl2(1–249)K227Q (NEB, M0351). 1× bead volume of the ligation mix (1× T4 RNA Ligase Reaction Buffer, 15% PEG8000, 0.5 μM of adapter, 10 U/μl Rnl2(1–249)K227Q ligase, 1 U/μl RNase inhibitor) was added to the beads and the ligation was performed at 4°C for 16 h with slow rotation. Next, beads were washed three times with 1 ml of ligation buffer without DTT and footprints were phosphorylated using T4 PNK (NEB, M0201S) according to manufacturer's instructions adjusted to 1× bead volume. The reaction was supplemented with 1 U/μl RNase inhibitor and performed at 37°C for 30 min with shaking.
SDS PAGE separation, gel excision and proteinase K digestion
Following phosphorylation, beads were washed twice with high salt buffer (20 mM Tris, pH 7.5, 500 mM NaCl, 2 mM EDTA, 1% (v/v) NP40) and twice with IP buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% (v/v) NP40, 0.5 mM DTT (for lysis only, added fresh) Protease inhibitors cocktail (for lysis only, added fresh, 1 tablet per 20 ml)) and RNPs were recovered with addition of 1 bead volume of 2× SDS loading buffer (2× NuPAGE LDS Sample Buffer (Thermo Fisher Scientific, NP0008) and 50 mM DTT) and incubated at 95°C for 5 min. 10% of the eluted RNPs was used for western blot with anti-FLAG M2 antibody. The rest 90% of the eluted RNPs were separated on 4–12% SDS-PAGE alongside 20 μl of 1/10 dilution of Prestained Protein Ladder, 10 to 250 kDa (PageRuler Plus, Thermo Scientific, 26620). Gel was visualized directly using an Azure c600 imaging system at the IR 660 nm detection channel or GE Typhoon 9500 scanner at the AF647 detection channel and bands corresponding to the RNP-fluorescent 3′adapter ligation products were excised and shredded using gel breaker tubes (IST Engineering BioTech, 3388). The shredded gel was incubated with 100 μl (1× shredded gel volume) 2× proteinase K buffer (100 mM Tris–HCl, pH 7.5, 150 mM NaCl, 12.5 mM EDTA, 2% (w/v) SDS) supplemented with 50 mM DTT, at 90°C for 3 min. Proteolysis was achieved with three successive proteinase K treatments each performed by adding 300 μl of 1 mg/ml proteinase K in 2× proteinase K buffer and incubating at 50°C for 30 min each time (final volume is ∼10 times that of the typical gel piece (∼100 μl volume)). For larger gel pieces volume of proteolysis mix may need to be adjusted so that the final volume relative to the gel piece is >3×. Shredded gel was removed using 5 μm filter tubes (IST Engineering BioTech, 5388). Note that the column may need to be loaded multiple times.
RNA was purified by phenol/chloroform/IAA extraction and was precipitated using isopropanol and 15 μg Glycoblue (Thermo Fisher Scientific/Ambion, AM9516) and redissolved 9 μl of water.
5′ Adapter Ligation
2 μl 10× RNA ligase buffer containing ATP (provided with Rnl1 T4 RNA ligase, Thermo Fischer Scientific, EL0021), 6 μl 50% PEG-8000, and 1 μl of 10 μM 5′ adapter (5′(aminolinker)-GTTCAGAGTTCTACAGTCCGACGATCrNrNrNrN, a full list of oligonucleotide sequences used for fPAR-CLIP is available in Supplementary Table S1) were added to the sample RNA was denatured at 90°C for 1 min and immediately cooled on ice for 2 min. 2 μl of Rnl1 T4 RNA ligase (Thermo Fischer Scientific, EL0021) was added and the reaction mixture was incubated at 37°C for 1 h. RNA was purified using Oligo Clean & Concentrator (Zymo Research, D4060) and eluted with 11 μl of water.
Reverse transcription
cDNA was synthesized using RT primer (GCCTTGGCACCCGAGAATTCCA, a full list of oligonucleotide sequences used for fPAR-CLIP is available in Supplementary Table S1) and SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific, 18090010). Reaction mix was prepared according to manufacturer's instructions. Briefly, 1 μl of RT primer and 1 μl of 10 mM dNTP(each) mix was added to 11 μl of adapter ligated sample, heated at 65°C for 5 min and chilled on ice. 4 μl of the 5× SSIV Buffer, 1 μl of 100 mM DTT, 1 μl of RNase inhibitor and 1 μl of SuperScript® IV Reverse Transcriptase (200 U/μl) were added and the reaction mix was incubated at 56°C for 30 min.
First low cycle PCR amplification and pilot PCR
Low cycle PCR was performed using Platinum Taq DNA polymerase (Thermo Fiscer Scientific, 10966034), a short 5′ PCR (CTTCAGAGTTCTACAGTCCGACGA) primer and the RT primer according to manufacturer's instructions (10 μl of cDNA in 400 μl total, 8 PCR tubes). The PCR cycler was programmed as follows. 94°C, 2 min; 6 cycles of 94°C, 30 s; 60°C, 30 s; 72°C, 15 sec; followed by 1 min at 72°C. Amplified DNA was purified and concentrated using DNA Clean & Concentrator kit (Zymo Research, D4013). PCR products were size selected, for a range of 78–98 bp, on a 3% agarose gel (3% Pippin gel cassettes, Sage Science, CSD3010). For the pilot PCR, 25% of the first low cycle PCR was used as template for a 1 × 100 μl PCR reaction with 5′ long PCR primer: AATGATACGGCGACCACCGAGATCTACACGTTCAGAGTTCTACAGTCCGA and 3′ RNA PCR primer index: CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA (underlined index sequence may vary, a full list of oligonucleotide sequences used for fPAR-CLIP is available in Supplementary Table S1). After 8 cycles, 15 μl of product was removed every two cycles (8, 10, 12, …, 18 cycles). Products were visualized on a 2.5% agarose gel. The lowest cycle number with visible amount of product was chosen for a final large-scale PCR (30 μl of size selected in 300 μl total) with the same primers. The PCR products were cleaned and concentrated using DNA Clean & Concentrator kit (Zymo Research, D4013) and size selected (∼140–170 bp) to remove primers and adapter-adapter byproducts using a 3% agarose gel (3% Pippin Prep). Quality of the final library was assessed using TapeStation system (Agilent). Sequencing was performed on an Illumina HiSeq 3000 or NovaSeq 6000 platform.
Data processing
Bcl2fastq (v2.20.0) was used to demultiplex and convert BCL basecall files from the sequencer to fastq files. Cutadapt (cutadapt 1.15 with Python 3.6.4) (19) was used to demultiplex adapter barcoded samples (without removing adapters). For demultiplexing libraries with a 5′ adapter barcode, the 5′ adapter barcode anchored to the 5′ end of the read was removed during demultiplexing (without removing the degenerate unique molecular identifiers (UMIs)). PARpipe (https://github.com/ohlerlab/PARpipe) was used for further pre-processing, implementation of Paralyzer (20) and annotation of groups and clusters. To allow the use of additional UMIs in the new protocol and preserve the true number of reads in the final bam file, during preprocessing fastq sequences were collapsed prior to adapter and UMI removal to remove potential PCR duplicates.
RESULTS
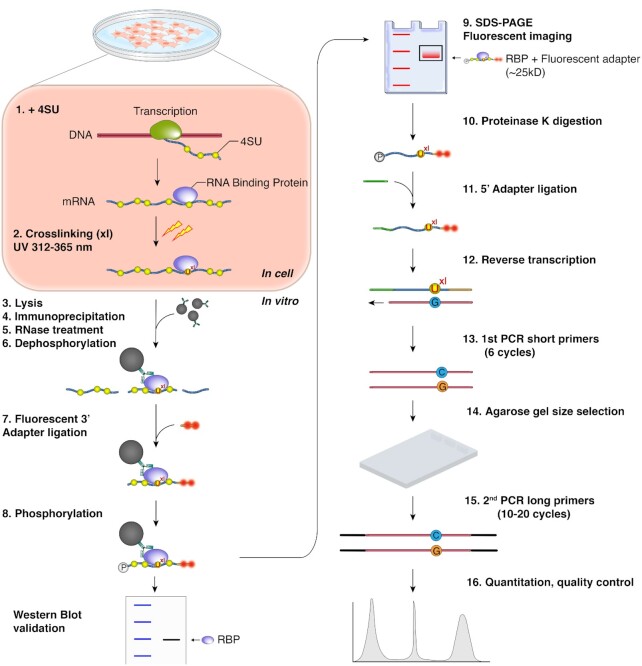
Our goals were to simplify and thus reduce technical challenges to increase reliability of the PAR-CLIP protocol, as well as to eliminate the need to use radioactivity. Below we describe the optimization of (a) the 3′ adapter ligation step, (b) the 5′ adapter ligation step and (c) reverse transcription and cDNA amplification (Figure 1). Labeling of nascent RNA with photoreactive nucleosides, 4-thiouridine (4SU) or 6-thioguanosine (6SG), UVA/B light (312 ≤ λ ≤ 365 nm)-mediated crosslinking, and immunoprecipitation of the ribonucleoprotein (RNP) of interest are unchanged compared to the original protocol (13). We prefer to use transgenically expressed or endogenously FLAG- or V5-tagged RBPs, allowing immunoprecipitation (IP) with validated, high-affinity antibodies that withstand stringent wash steps. The RNase digestion after lysis and IP can be performed with any RNase (e.g. RNase I, T1, or A/T1 mix) and details were discussed in depth previously (11,21). Nevertheless, we provide information regarding the impact of RNase concentrations on the complexity of PAR-CLIP datasets after describing our streamlined cDNA library construction. A detailed protocol can be found in the Materials and Methods section and as a step-by-step instruction in the Supplementary Methods.
Figure 1.
fPAR-CLIP schematic.
3′ Adapter ligation and SDS polyacrylamide-gel fractionation of crosslinked ribonucleoproteins
In order to minimize the number of gel-purification steps of the protocol, we decided to build upon a previous PAR-CLIP protocol that called for direct ligation of a 3′ adapter to the protected RNA footprints on the immunoprecipitated RNP immobilized to magnetic beads (22) (Figure 1). This approach allows for convenient removal of excess unligated adapter by stringent washing and thus minimizes the formation of adapter-adapter ligation products, which contaminate low-input small RNA cDNA libraries.
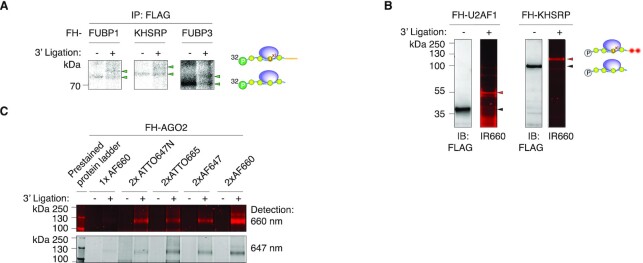
We previously observed that the crosslinked RNA footprint-RBP complex typically migrated at the predicted size of the unmodified RBP (14). Nevertheless, one nt of RNA adds on average 345 Da, or the equivalent of ∼3 amino acids (aa), to the apparent weight of the RBP and thus, the addition of the 26 nucleotides (nt) long 3′ adapter could result in an electrophoretic mobility shift of the crosslinked RNP. Most of the established CLIP protocols that include on-beads adapter ligation steps, including HiTS-CLIP (23), iCLIP (24), irCLIP (25) and eCLIP (26), circumvent this problem by isolating a larger area above the unligated RBP (27), which may include non-specific RNAs or RNAs interacting with additional, co-purified RBPs fractionated in the gel, thereby sacrificing some purification stringency. We wanted to quantify the effect of 3′ adapter addition to the crosslinked RNP on electrophoretic mobility using a set of three stable HEK293 cell lines expressing FLAG-HA (FH) tagged FUBP1, KHSRP or FUBP3. After 4SU-labeling, lysis, FLAG immunoprecipitation and limited RNase T1 digestion, we labeled RNA 5′ ends with P-32. Next, we ligated the standard DNA 3′ adapter and fractionated the RNP by SDS-polyacrylamide gel electrophoresis (PAGE) and compared electrophoretic mobility of unligated and ligated RNPs (Figure 2A). In all three cases, the adapter was ligated at an approx. 50% efficiency and 3′ adapter ligated RNPs migrated ∼25 kDa above the unligated RNPs, indicating that addition of 26 nt of DNA to the RBP-protected footprint resulted in a predictable shift in apparent weight of the crosslinked RNP.
Figure 2.
Ligation of a 3′ adapter to crosslinked RNPs results in a predictable electrophoretic mobility shift. (A) SDS-PAGE of crosslinked, radiolabled, and immunoprecipitated FLAG-HA-tagged (FH) FUBP1, FH-KHSRP, and FH-FUBP3 RNPs before and after ligation to a 3′ adapter. (B) Immunoblots and fluorescent images of SDS-PAGE separating fluorescent adapter-ligated crosslinked FH-U2AF1 (left panels) and FH-KHSRP (right panels) RNPs. Fluorescent images were detected at 700 nm wavelength. (C) Comparison of FH-AGO2 fPAR-CLIP band intensity using different fluorescent adapters labeled with the indicated dyes and imaged at 647 or 660 nm detection wavelength.
Next, we explored the possibility of adapting fluorescent labeling instead of radioactive labeling for tracking RNPs after 3′ adapter ligation, analogous to a previously published iCLIP variant, irCLIP (25). Initially we tested a 3′ adapter coupled to a single Alexa Fluor dye emitting at a wavelength of 690 nm (AF660) and thus detectable on an infrared fluorescence scanner. We tested this adapter with the FH-KHSRP cell line, as well as a FH-U2AF1 cell line (28). Adapter-ligated RNPs were clearly detectable in the IR 660 nm channel and as before, adapter-ligated RNPs migrated ∼20 kDa above the non-crosslinked RBPs (Figure 2B). Fluorescent labeling thus exhibited the sensitivity to allow visualizing crosslinked RBPs of various size. We refer to the adaptation of the PAR-CLIP protocol using fluorescent dyes as fPAR-CLIP.
The fluorescent signal intensity is expected to correlate with the number of fluorophores, as well as the extinction coefficient of the dye. To optimize visualization of fPAR-CLIP, we generated a set of 3′ adapters labeled with either one or two Alexa or ATTO dyes of different emission wavelength in the far red or near infrared spectrum (λ > 647 nm). We tested the sensitivity of fPAR-CLIP using approximately 106 FH-AGO2 expressing cells and visualized crosslinked and 3′ adapter-ligated FH-AGO2-RNPs at 647 and 700 nm detection wavelengths. We detected robust signal from all double labeled fluorescent adapters (Figure 2C). The adapter carrying 2× AF660 dyes at 660 nm detection wavelength showed the highest sensitivity and was chosen for further experiments. Taken together, our data indicate that 3′ adapter ligation on immobilized RNPs and fluorescent labeling may represent a viable strategy to label RNPs and streamline the PAR-CLIP protocol.
We decided to ligate only the 3′ fluorescent adapter and not additionally the 5′adapter on beads to limit fluorescent RNPs visualized by SDS-PAGE to a single species. Additional ligation of the 5′ adapter would create fluorescent bands corresponding to 5′-3′-, as well as 3′-ligated RNP subpopulations. Particularly in the case of poorly crosslinked RBPs that may be difficult to detect in the first place, the interpretation of a gel-image containing patterns of singly- and doubly ligated RNP complexes could prove to be complicated and result in the inadvertent isolation of the wrong complex.
Recovery of 3′ adapter ligated crosslinked footprint
Crosslinked, adapter-ligated RNPs migrate as discrete bands by SDS-PAGE in fPAR-CLIP. We thus reasoned that it is possible to eliminate the transfer of fractionated RNPs for all the RBPs from gel to nitrocellulose that most CLIP protocols include to improve visualization of crosslinked RNPs. For fPAR-CLIP, we image the gels at detection wavelengths of 647 and 660 nm and excise only the discrete band migrating ∼25 kDa above the predicted weight of the RBP of interest (Figure 2B, C). Next, RNA footprints are isolated from the crosslinked RNP by in-gel digestion of the RBP itself by Proteinase K and used as input in a 5′ adapter ligation step.
5′ Adapter ligation
T4 RNA ligases prefer RNA 3′ ends as substrates for ligation (16,29). Therefore, 5′ adapters in commercial and homemade small RNA cDNA library construction approaches are typically RNA molecules. This can lead to substantial contamination of the library with partial or complete 5′ adapter sequences due to their concatenation, especially with low input amounts of small RNA in CLIP protocols. We examined whether using chimeric 5′ adapters of the same sequence composed of DNA with either ten or four RNA nucleotides at their 3′ end would impact adapter ligation efficiency, or subsequent reverse transcription and amplification (sequences are found in the Materials and Methods section). We introduced an aminolinker to the 5′ end of the adapter to suppress any ligation of 5′ adapter to itself (30). The last four nucleotides of the adapter were degenerate (either A, G, C, U at equal probability) to reduce ligation biases. Together with the additional two degenerate nucleotides in the 3′ adapter, these can also serve as unique molecular identifiers (UMI) to allow computational elimination of PCR duplicates in the final sequenced cDNA (31,32). We found that using these adapters consisting mostly of DNA neither impaired reverse transcription nor showed any ligation substrate sequence bias (Supplementary Figure S1) and eliminated any sequence reads derived from fragments of 5′ adapters.
Size selection
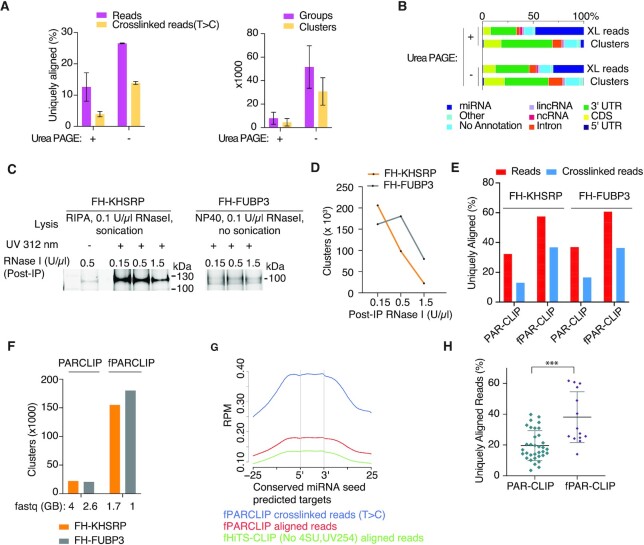
Small RNA cDNA library construction procedures typically contain denaturing polyacrylamide gel-based size selection steps to minimize amplification of unwanted side products. We reasoned that our stringent removal of excess 3′ adapter by washing could obviate some of the size selection steps. To avoid the use of denaturing polyacrylamide gels (DPAGE), reduce time, and increase recovery we tested the utility of an automated agarose gel apparatus (Pippin Prep) for size selection and removal of adapter-adapter ligation products in the cDNA. We compared FH-AGO2 PAR-CLIP libraries where the 5′adapter ligated product was either (i) used directly as input in reverse transcription (RT), followed by low-cycle PCR, size-selection on a Pippin Prep, and another round of PCR, or (ii) as in our previous protocols, size selected on DPAGE, followed by RT, PCR, and another round of size-selection by Pippin Prep to remove adapter-adapter ligation products. Omitting the traditional DPAGE purification before RT increased the complexity of PAR-CLIP cDNA libraries as measured by number and fraction of uniquely mapping crosslinked reads, as well as increased numbers of clusters (regions of overlapping sequence reads with robust evidence of crosslinked reads with T-to-C mutations) (Figure 3A, B).
Figure 3.
Streamlined cDNA library construction in fPAR-CLIP increases cDNA library complexity. (A) Left panel: Percentage of uniquely aligned reads from PAR-CLIP libraries size selected after 5′adapter ligation by denaturing polyacrylamide gel electrophoresis (DPAGE) or used directly as input in RT and low cycle PCR followed by size selection by automated agarose gel electrophoresis. Right panel: comparison of groups and clusters identified in both libraries. (B) Distribution of crosslink reads across different annotation categories for the samples from (A). (C) Fluorescent image of SDS PAGE using different concentration of RNase I after KHSRP and FUBP3 IP. (D) Number of clusters for FH-KHSRP and FH-FUBP3 fPAR-CLIPs from (C) using different concentration of RNase I. (E) Percentage of uniquely aligned sequence reads (red bars) or crosslinked reads (blue bars) from PAR-CLIP and fPAR-CLIP experiments for FH-KHSRP and FUBP3. (F) Number of binding sites identified from PAR-CLIP and fPAR-CLIP experiments for FH-KHSRP (orange) and FH-FUBP3 (grey). The size of the raw sequencing file is indicated below. (G) Enrichment of FH-AGO2 fPARCLIP and HiTS-CLIP footprints across predicted, conserved miRNA target sites. (H) Comparison of uniquely aligned reads from all published PAR-CLIP libraries with at least 20,000 RBP binding sites compared to fPAR-CLIP libraries produced in our laboratory (* P < 0.05, Mann–Whitney U test). See Supplementary Table S2 for the list of analyzed PAR-CLIP and fPAR-CLIP experiments.
Furthermore, we optimized PCR amplification itself in our fPAR-CLIP protocol, opting for a two-step strategy, with a first PCR using the shortest possible primer pairs, resulting in footprint-containing products of 73–100 bp that can be easily separated by agarose gel purification from adapter-adapter ligation products of 54 bp, followed by a second PCR with the longer Illumina-sequencing compatible primers (Figure 1, steps 13–15, Supplementary Figure S1A).
Impact of RNase concentration on PAR-CLIP cDNA library complexity
With our streamlined fPAR-CLIP cDNA library construction protocol in hand, we wanted to revisit the influence of RNase digestion after cell lysis and IP on library complexity and binding site identification and chose the nuclear RBP KHSRP as our test protein. Crosslinked FH-KHSRP HEK293 cells were lysed with RIPA buffer, treated with 0.1 U/μl RNase I and sonicated to break up chromatin. After FLAG-IP, additional ribonuclease treatment was performed with increasing concentrations (0.15, 0.5 and 1.5 U/μl) of RNase I. Fluorescent imaging of 3′adapter ligated and gel-fractionated RNPs showed that with increasing amounts of RNase I, the amount of recovered crosslinked RNA decreased (Figure 3C), also reflected in a concomitant overall reduced number of binding sites from prepared cDNA libraries (Figure 3D). Shearing of RNA by sonication during cell lysis can have a considerable effect on the optimal RNase concentration; while for KHSRP 0.15 U/μl of RNase I during IP resulted in the most complex cDNA libraries, for the highly related FUBP3 where we lysed the cells without sonication using 0.1% NP40 detergent, 0.5 U/μl RNase I during IP were optimal (Figure 3C, D).
The necessity of careful adjustment of RNase concentrations is even more important when small RNPs are purified from the chromatin fraction. Previously we used ∼3 U/ml of an RNase A/T1 mix for a U2AF1 PARCLIP from purified chromatin (28). We now performed fPARCLIP for U2AF1 following the same protocol for chromatin enrichment and treated the IP’ed RNPs with either ∼1.5 or ∼15 U/ml RNase A/T1 mix and observed highest performance at the lowest RNase concentration (Supplementary Figure S1D) (28). Nevertheless, it has to be noted that not all RBPs showed pronounced sensitivity to RNase conditions, e.g. AGO2 did not show significant difference in performance when titrating 0.15–1.5 U/μl RNase I after IP (Supplementary Figure S1E), presumably reflecting their ability to efficiently protect an RNA footprint. Furthermore, aligned reads from our RNase titration experiments described above showed a length distribution unique for each protein that was independent of RNase concentrations. This suggested that overdigested footprints were not accessible to 3′ ligation on beads and that the desired length distribution strictly relied on the structural properties of the RNA–protein interaction and the size of the protein.
In summary, our data underscore the importance of optimizing RNase concentrations and lysis conditions for each RBP to obtain the most complex fPAR-CLIP (or any other CLIP) libraries (11,21). Overdigested, short RNA fragments, may also prevent unambiguous mapping to the genome after sequencing. The sequence specificity of the used RNase should also be considered; e.g. in the case of RNase T1 that cuts after G’s, we observed a pronounced G-depletion in observed binding sites (13). While RNase T1 itself does not preclude the recovery of binding sites for RBPs binding G-rich motifs, e.g. CNBP and DHX36 (33,34), it needs to be carefully titrated in order to avoid biased enrichment of G-poor regions. This may be most evident when studying RBPs binding within the already G-poor 3′UTR, where a potential G-containing binding site will be cut and removed by the RNase, but G-depleted sequences that were close enough to be crosslinked can be recovered and considered as binding sites.
Simplified library preparation protocol in fPAR-CLIP increases library complexity
A direct comparison of results from fPAR-CLIP and the previous PAR-CLIP protocol for our test RBPs, KHSRP and FUBP3, showed a 2–3-fold increase in uniquely mapped sequence reads with a concomitant ∼10-fold increase in overall binding site numbers (Figure 3E and F). In order to demonstrate that our protocol can be adapted to other CLIP procedures, we compared AGO2 fPARCLIP with HiTS-CLIP (23) performed with UV 254 nm crosslinking and without 4SU treatment, but using the same streamlined library preparation method. As an indicator of specificity for our AGO2 CLIPs, we examined the enrichment of footprints across predicted miRNA target sites (35). fPARCLIP showed a 50% higher coverage compared to HiTS-CLIP on predicted miRNA target sites, which increased to 200% when only considering those containing the characteristic T-to-C mutation (Figure 3G), highlighting the importance of restricting CLIP analyses to crosslinked reads in order to separate signal from noise.
Finally, we wanted to document that the increased performance of the fPAR-CLIP protocol is generalizable. We quantified the fraction of uniquely aligned sequences from all available PAR-CLIP experiments (from our group as well as available on GEO, see Supplementary Table S2) that yielded at least 20 000 clusters compared to all fPAR-CLIP experiments performed in our group. On average, fPAR-CLIP significantly outperformed standard PAR-CLIP (Figure 3H).
DISCUSSION
Traditional PAR-CLIP, like other CLIP methods, is time consuming and technically challenging, resulting in high rates of experimental failure, most often due to difficulties in generating complex cDNA libraries from the low amounts of recovered crosslinked RNA. Compared to the previous protocols, our streamlined fPAR-CLIP allows for a 10–100-fold decrease in required input material. To assess sensitivity of fPAR-CLIP, we chose AGO2 as our test RBP, considering that it crosslinks with much poorer efficiency than RBPs typically used to benchmark other CLIP approaches, e.g. HNRNPC and RBFOX2, which may be less representative of the wide range of RNPs. For AGO2 PAR-CLIP experiments, we previously required 107–108 cells to obtain robust crosslinking signal and complex cDNA libraries (with at least 20 000 clusters) (13,36) and with fPAR-CLIP we were able to use <106 cells. Increased sensitivity was also observed using other RBPs, such as U2AF1, FUBP1, KHSRP or FUBP3. This improvement in performance was achieved by optimization of multiple steps.
First, we were able to eliminate the use of radioactivity for PAR-CLIP by using fluorescent adapters analogous to irCLIP (25). Use of ATTO and Alexa dyes with strong fluorescent signal improved the signal to noise ratio of detection compared to radioactive labeling. Detection of blue pre-stained protein markers at the same wavelength with the fluorescent adapter simplifies alignment of the gel to the printed fluorescent image and increases specificity of crosslinked RNA footprint retrieval. As an added benefit, fPAR-CLIP removes the need for access to a phosphorimager and can be performed in laboratories that are not licensed for the use of radioactivity.
Second, we removed the transfer of gel-fractionated, crosslinked RNPs to nitrocellulose that is part of most CLIP protocols. This step is typically included to obtain a clean gel image and to possibly reduce background from non-crosslinked RNA. We previously noticed severe loss of material on nitrocellulose that in some extreme cases, e.g. U2AF1, even reproducibly precluded the generation of high-quality cDNA libraries and required the extraction of RBP protected RNA footprints directly from the shredded SDS-PAGE band (28).
Third, our optimized cDNA library preparation protocol reduces the time necessary to generate an fPAR-CLIP library from 4–5 days to 2 days. Mainly, we removed three out of four gel-based size-selection steps from the PAR-CLIP protocol. Originally, size selections by denaturing urea PAGE were performed after isolation of radiolabeled RNA footprints, 3′ adapter ligation, and 5′ adapter ligation, in addition to an agarose-gel based size selection after reverse transcription and PCR. Each of these size selection steps routinely results in an ∼50% loss of material (37), reducing the efficiency of library construction to <10% of the input molecules. The Illumina TruSeq compatible primers introduce altogether 124 bp on 3′ and 5′ end of the library during PCR, resulting in a small relative size difference of footprint-containing cDNA at 140–150 bp from empty, adapter-adapter ligation products at 124 bp, complicating the agarose-gel based cleanup of the cDNA library. With stringent washes on beads and a two-step PCR strategy with a first amplification using the shortest possible primers, we were able to limit size selection to an agarose gel after the first step of PCR. The sequences necessary for sequencing flow cell attachment and additional barcodes are only introduced in a following PCR step using the longer Illumina Truseq compatible primers.
In summary, fPAR-CLIP retains all of the characteristics of PAR-CLIP, while cutting sample preparation time in half and increasing sensitivity up to 100-fold. Furthermore, our approach can be extended to related CLIP variants, such as HiTS-CLIP (23) and CRAC (38). The streamlined small RNA cDNA library preparation at the core of the protocol can be conveniently adapted for profiling of small RNAs, such as miRNAs, piRNAs or tRNAs, as well as ribosome profiling.
DATA AVAILABILITY
fPAR-CLIP data have been deposited to the Gene Expression Omnibus (GEO) under the accession number GSE163925. Accession numbers for PAR-CLIP and additional fPAR-CLIP experiments analyzed in Figure 3H are listed in Supplementary Table S2.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the members of the Hafner and Haase groups for helpful discussions. We want to thank Faiza Naz, Gustavo Gutierrez-Cruz and Dr Stefania dell’Orso (NIAMS Genomics and Technology Section) for sequencing support.
Author contributions: D.A, P.K., K.M., P.C., A.H. and M.H. designed experiments. P.K performed AGO2 PAR-CLIPs. D.A. and K.M. performed all other experiments. D.A., A.J. and D.C. analyzed data. P.C. designed and synthesized fluorescent adapters. D.A. and M.H. wrote the manuscript. D.A., A.J., P.K., A.H. and M.H. performed final edits.
Contributor Information
Dimitrios G Anastasakis, Laboratory of Muscle Stem Cells and Gene Regulation, National Institute for Arthritis and Musculoskeletal and Skin Disease, National Institutes of Health, Bethesda, 20892 MD, USA.
Alexis Jacob, Laboratory of Muscle Stem Cells and Gene Regulation, National Institute for Arthritis and Musculoskeletal and Skin Disease, National Institutes of Health, Bethesda, 20892 MD, USA.
Parthena Konstantinidou, Laboratory of Cellular and Molecular Biology, National Institutes of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, 20892 MD, USA.
Kazuyuki Meguro, Laboratory of Clinical Immunology & Microbiology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, 20892 MD, USA.
Duncan Claypool, Laboratory of Muscle Stem Cells and Gene Regulation, National Institute for Arthritis and Musculoskeletal and Skin Disease, National Institutes of Health, Bethesda, 20892 MD, USA.
Pavol Cekan, MultiplexDX s.r.o., 841 04 Bratislava, Slovakia.
Astrid D Haase, Laboratory of Cellular and Molecular Biology, National Institutes of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, 20892 MD, USA.
Markus Hafner, Laboratory of Muscle Stem Cells and Gene Regulation, National Institute for Arthritis and Musculoskeletal and Skin Disease, National Institutes of Health, Bethesda, 20892 MD, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institute for Arthritis and Musculoskeletal and Skin Diseases; National Institute of Diabetes and Digestive and Kidney Diseases. Funding for open access charge: Intramural Research Program of NIAMS and NIDDK.
Conflict of interest statement. P.C. is the founder and CEO of MultiplexDX s.r.o.
REFERENCES
- 1. Singh G., Pratt G., Yeo G.W., Moore M.J.. The clothes make the mRNA: past and present trends in mRNP fashion. Annu. Rev. Biochem. 2015; 84:325–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gerstberger S., Hafner M., Tuschl T.. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014; 15:829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gerstberger S., Hafner M., Ascano M., Tuschl T.. Evolutionary conservation and expression of human RNA-binding proteins and their role in human genetic disease. Adv. Exp. Med. Biol. 2014; 825:1–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ray D., Kazan H., Chan E.T., Pena Castillo L., Chaudhry S., Talukder S., Blencowe B.J., Morris Q., Hughes T.R.. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 2009; 27:667–670. [DOI] [PubMed] [Google Scholar]
- 5. Lambert N., Robertson A., Jangi M., McGeary S., Sharp P.A., Burge C.B.. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell. 2014; 54:887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lunde B.M., Moore C., Varani G.. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007; 8:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ule J., Jensen K.B., Ruggiu M., Mele A., Ule A., Darnell R.B.. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003; 302:1212–1215. [DOI] [PubMed] [Google Scholar]
- 8. Lee F.C.Y., Ule J.. Advances in CLIP technologies for studies of protein-RNA interactions. Mol. Cell. 2018; 69:354–369. [DOI] [PubMed] [Google Scholar]
- 9. Ellington A.D., Szostak J.W.. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990; 346:818–822. [DOI] [PubMed] [Google Scholar]
- 10. Tuerk C., Gold L.. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990; 249:505–510. [DOI] [PubMed] [Google Scholar]
- 11. Kishore S., Jaskiewicz L., Burger L., Hausser J., Khorshid M., Zavolan M.. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods. 2011; 8:559–564. [DOI] [PubMed] [Google Scholar]
- 12. Friedersdorf M.B., Keene J.D.. Advancing the functional utility of PAR-CLIP by quantifying background binding to mRNAs and lncRNAs. Genome Biol. 2014; 15:R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr, Jungkamp A.C., Munschauer M.et al.. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ascano M., Hafner M., Cekan P., Gerstberger S., Tuschl T.. Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip Rev RNA. 2012; 3:159–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P.. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001; 294:858–862. [DOI] [PubMed] [Google Scholar]
- 16. Hafner M., Renwick N., Brown M., Mihailovic A., Holoch D., Lin C., Pena J.T., Nusbaum J.D., Morozov P., Ludwig J.et al.. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011; 17:1697–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Renwick N., Cekan P., Bognanni C., Tuschl T.. Multiplexed miRNA fluorescence in situ hybridization for formalin-fixed paraffin-embedded tissues. Methods Mol. Biol. 2014; 1211:171–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hafner M., Landgraf P., Ludwig J., Rice A., Ojo T., Lin C., Holoch D., Lim C., Tuschl T.. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008; 44:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011. 2011; 17:3. [Google Scholar]
- 20. Corcoran D.L., Georgiev S., Mukherjee N., Gottwein E., Skalsky R.L., Keene J.D., Ohler U.. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011; 12:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garzia A., Meyer C., Morozov P., Sajek M., Tuschl T.. Optimization of PAR-CLIP for transcriptome-wide identification of binding sites of RNA-binding proteins. Methods. 2017; 118–119:24–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Benhalevy D., McFarland H.L., Sarshad A.A., Hafner M.. PAR-CLIP and streamlined small RNA cDNA library preparation protocol for the identification of RNA binding protein target sites. Methods. 2017; 118–119:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Licatalosi D.D., Mele A., Fak J.J., Ule J., Kayikci M., Chi S.W., Clark T.A., Schweitzer A.C., Blume J.E., Wang X.et al.. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008; 456:464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Konig J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B., Turner D.J., Luscombe N.M., Ule J.. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010; 17:909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zarnegar B.J., Flynn R.A., Shen Y., Do B.T., Chang H.Y., Khavari P.A.. irCLIP platform for efficient characterization of protein–RNA interactions. Nat. Methods. 2016; 13:489–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Van Nostrand E.L., Pratt G.A., Shishkin A.A., Gelboin-Burkhart C., Fang M.Y., Sundararaman B., Blue S.M., Nguyen T.B., Surka C., Elkins K.et al.. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods. 2016; 13:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ule J., Jensen K., Mele A., Darnell R.B.. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005; 37:376–386. [DOI] [PubMed] [Google Scholar]
- 28. Palangat M., Anastasakis D.G., Fei D.L., Lindblad K.E., Bradley R., Hourigan C.S., Hafner M., Larson D.R.. The splicing factor U2AF1 contributes to cancer progression through a noncanonical role in translation regulation. Genes Dev. 2019; 33:482–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nandakumar J., Shuman S.. How an RNA ligase discriminates RNA versus DNA damage. Mol. Cell. 2004; 16:211–221. [DOI] [PubMed] [Google Scholar]
- 30. Belair C.D., Hu T., Chu B., Freimer J.W., Cooperberg M.R., Blelloch R.H.. High-throughput, efficient, and unbiased capture of small RNAs from Low-input samples for sequencing. Sci. Rep. 2019; 9:2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McCloskey M.L., Stoger R., Hansen R.S., Laird C.D.. Encoding PCR products with batch-stamps and barcodes. Biochem. Genet. 2007; 45:761–767. [DOI] [PubMed] [Google Scholar]
- 32. Miner B.E., Stoger R.J., Burden A.F., Laird C.D., Hansen R.S.. Molecular barcodes detect redundancy and contamination in hairpin-bisulfite PCR. Nucleic Acids Res. 2004; 32:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sauer M., Juranek S.A., Marks J., De Magis A., Kazemier H.G., Hilbig D., Benhalevy D., Wang X., Hafner M., Paeschke K.. DHX36 prevents the accumulation of translationally inactive mRNAs with G4-structures in untranslated regions. Nat. Commun. 2019; 10:2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Benhalevy D., Gupta S.K., Danan C.H., Ghosal S., Sun H.W., Kazemier H.G., Paeschke K., Hafner M., Juranek S.A.. The human CCHC-type Zinc finger nucleic Acid-Binding protein binds G-Rich elements in target mRNA coding sequences and promotes translation. Cell Rep. 2017; 18:2979–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Agarwal V., Bell G.W., Nam J.W., Bartel D.P.. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015; 4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lipchina I., Elkabetz Y., Hafner M., Sheridan R., Mihailovic A., Tuschl T., Sander C., Studer L., Betel D.. Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev. 2011; 25:2173–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kieft J.S., Batey R.T.. A general method for rapid and nondenaturing purification of RNAs. RNA. 2004; 10:988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Granneman S., Kudla G., Petfalski E., Tollervey D.. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:9613–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
fPAR-CLIP data have been deposited to the Gene Expression Omnibus (GEO) under the accession number GSE163925. Accession numbers for PAR-CLIP and additional fPAR-CLIP experiments analyzed in Figure 3H are listed in Supplementary Table S2.