Abstract
Even in high-quality X-ray crystal structures of oligonucleotides determined at a resolution of 1 Å or higher, the orientations of first-shell water molecules remain unclear. We used cryo neutron crystallography to gain insight into the H-bonding patterns of water molecules around the left-handed Z-DNA duplex [d(CGCGCG)]2. The neutron density visualized at 1.5 Å resolution for the first time allows us to pinpoint the orientations of most of the water molecules directly contacting the DNA and of many second-shell waters. In particular, H-bond acceptor and donor patterns for water participating in prominent hydration motifs inside the minor groove, on the convex surface or bridging nucleobase and phosphate oxygen atoms are finally revealed. Several water molecules display entirely unexpected orientations. For example, a water molecule located at H-bonding distance from O6 keto oxygen atoms of two adjacent guanines directs both its deuterium atoms away from the keto groups. Exocyclic amino groups of guanine (N2) and cytosine (N4) unexpectedly stabilize waters H-bonded to O2 keto oxygens from adjacent cytosines and O6 keto oxygens from adjacent guanines, respectively. Our structure offers the most detailed view to date of DNA solvation in the solid-state undistorted by metal ions or polyamines.
INTRODUCTION
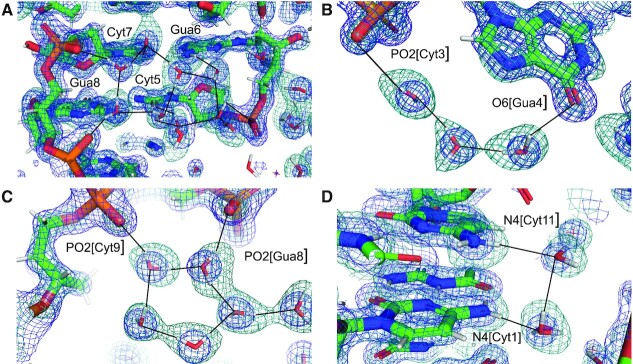
X-ray crystal structures of double helical nucleic acid fragments have revealed intricate hydration patterns inside the grooves and around the sugar-phosphate backbones. Among them are the water spine or ribbon in the minor groove of the DNA duplex [d(CGCGAATTCGCG)]2 (1,2), water tandems that link 2′-OH groups across the minor groove of double-stranded RNA (3), and distinct water bridges between intra-strand phosphate groups in A-form and B-form duplexes as a result of different sugar puckers and consequently phosphate distances (4). The water content of even tightly packed crystals of duplexes and other DNA and RNA folding motifs that diffract to high resolution is typically 25% or higher. X-ray crystallography can then pinpoint the locations of water molecules in the first and often also the second hydration shell. Examples of the quality of the electron density computed at ca. 1 Å around first-shell waters forming H-bonds to base and backbone atoms of a left-handed Z-DNA duplex are depicted in Figure 1. However, even in this high-quality X-ray structure and a few others determined to resolutions beyond 1 Å, donor-acceptor patterns of water H-bonds remain hidden. One might speculate that water molecules bridging oxygen atoms, as seen in Figure 1B and C, act as donors in both H-bonds, but there is no experimental evidence in support of this. Further, it is readily apparent that trying to reliably assign the roles of donors and acceptors for H-bonds such as those shown in Figure 1D and E is futile, especially if one considers bifurcated cases in addition to more common unidirectional H-bonding.
Figure 1.
Prominent first-shell hydration patterns around [d(CGCGCG)]2 (PDB ID 1ICK (47)). (A) Standard C:G base pair. Longitudinal patterns bridging base atoms from opposite strands include (B) a water spine inside the minor groove that links cytosine O2 atoms, (C) waters between guanine O6 atoms on the convex surface and (D) water tandems that link cytosine N4 atoms on the convex surface. (E) Water molecules at the periphery of the minor groove linking guanine N2 atoms to 5′- and 3′-adjacent phosphate groups in a transverse fashion. Fourier 2Fo-Fc sum electron density drawn at the 1σ level surrounds water molecules and individual nucleotides. Water molecules are cyan spheres, atoms engaged in H-bonds are highlighted as spheres and H-bonds are drawn with thin solid lines.
The first single crystal structure reported for an oligo-2′-deoxynucleotide, the hexamer d(CGCGCG), revealed a left-handed duplex (5). Z-DNA is stabilized by negative supercoiling during transcription when polymerases and helicases in their wake generate underwound DNA (6–11). The transition from B- to Z-DNA (B−Z junction) occurs within a single A−T pair, whereby both bases are extruded from the duplex when the phosphate backbone reverses direction (12). The discovery of the Zα protein domain that specifically binds to Z-DNA left little doubt as to a biological significance of left-handed DNA (13). The domain recognizes left-handed DNA without specific contacts to bases by following the zig-zag conformation of the backbone (14). The Zα domain binds both Z-DNA and Z-RNA and is notably associated with double-stranded RNA-specific adenosine deaminase (ADAR (15)), zipcode binding protein 1 (ZBP1 (16)) and viral orthologs that regulate innate immunity. In recent years, the case for an involvement of Z-DNA and Z-RNA in human disease has been strengthened considerably (17,18).
In Z-DNA, bases form standard Watson-Crick pairs, but cytosine and guanosine adopt different sugar puckers (C2′-endo and C3′-endo, respectively). Phosphates exhibit a zig-zag like arrangement along the two backbones such that adjacent base pairs display alternating, low (−10°, CpG steps) and high twists (−50°, GpC steps). Infinite stacks of duplexes exhibit close lateral spacings in Z-DNA crystals, with a record low volume per base pair of ca. 1000 Å3. The orthorhombic crystals belonging to space group P212121 grow under various conditions and include the Mg2+, spermine, and mixed spermine/Mg2+ forms (19). The ordered water contents (w/w) for these three range from 21% (spermine form) to 29% (Mg2+ form), with calculated crystal densities of between 1.37 (Mg2+ form) and 1.43 g/cm3 (spermine form). The density of mixed spermine/Mg2+ form crystals was experimentally determined to be 1.49 g/cm3. The calculated density, including 74 refined water positions per asymmetric unit, amounts to 1.41 g/cm3. The difference between the experimental and calculated densities of this Z-DNA crystal form thus accounts for another 16 water molecules, consistent with 90 water molecules surrounding an individual duplex in the asymmetric unit. The exceptional quality of the diffraction data and the level of hydration, with up to 85 water molecules visualized in the Mg2+ form (19), render Z-DNA a perfect target for an in-depth analysis of the water structure around DNA using neutron diffraction.
Neutron crystallography provides insight into the positions of hydrogen (deuterium) atoms in the structures of nucleic acids and proteins (20–24). A key difference between oligonucleotide and protein crystals in regard to neutron diffraction experiments is that proteins can be recombinantly expressed in their perdeuterated form. The synthesis of perdeuterated DNA oligos using solid phase phosphoramidite approaches would be prohibitively expensive by comparison. Synthetic strategies for the preparation of nucleosides and nucleotides deuterated at single or multiple sites have been reported (25). However, facile access to phosphoramidite building blocks for solid-phase oligonucleotide synthesis remains currently out of reach. The preparation of perdeuterated DNA polynucleotides for neutron fiber diffraction experiments has also been described (26), but the approach cannot be applied to produce milligram amounts of an oligonucleotide for single crystal neutron diffraction studies. As with all other previously reported neutron structures of DNA oligonucleotides, a limitation of the present analysis of the Z-DNA hexamer is that the majority of backbone and base hydrogen atoms is not exchanged by deuterium in D2O-based crystallization solutions. Thus, the levels of background scattering are expected to be higher in neutron diffraction experiments with DNA compared to protein crystals.
Previously reported neutron structures of DNA fragments revealed the orientations of a number of water molecules, but did not provide a complete picture of the first hydration shell around DNA duplexes (20,21,27,28). Compared to establishing protonation states of nucleobases and protein sidechains or the orientation of water molecules in channels or cavities by neutron crystallography, reliable determination of H-bond donor-acceptor patterns for water molecules on the surface of DNA duplexes is more challenging. This is because the mobility of such water molecules is typically increased relative to those situated in sheltered regions such as protein active sites or inside the deep (major) grooves of A- and B-form DNA duplexes.
Neutron diffraction experiments are mostly conducted at room temperature (RT) with crystals sealed in capillaries as neutrons, unlike X-rays, do not induce radiation damage. However, given the expected increased mobility of first-shell and certainly higher shell water compared to DNA atoms, RT neutron data are unlikely to provide detailed insight into the water structure around a DNA duplex. Indeed, the neutron density for the Z-DNA duplex computed using RT data at ca. 1.6 Å showed deuterium atoms for bases, but did not unambiguously resolve the orientation of many water molecules even in the first hydration shell (our unpublished data). Also supporting the notion that RT data may be insufficient for a detailed analysis of the solvent structure in DNA crystals are the RT and low temperature (LT) X-ray crystal structures of the Z-DNA spermine form. Thus, in the RT structure, separate electron density peaks between adjacent cytosines in the minor groove were assigned to a string of water molecules (29). However, the LT structure clearly showed a second spermine molecule inside the minor groove (30). A comparison of the mobilities of water and spermine molecules in the RT and LT structures demonstrates the benefits of data collected at cryogenic temperature in that the B-factors of water molecules are distinctly lower in the latter (31). More evidence that cryo-cooling of crystals can reduce the mobility of water molecules is provided by the LT neutron structure (15K) of concanavalin A that led to the identification of twice as many water molecules (32) compared to the RT structure (33). In addition, the B-factors of water molecules in the LT structure were around fourfold lower than those of waters in the RT structure. The benefits of cryo-neutron crystallography were recently also demonstrated in studies investigating transient protein ligand complexes in β-lactamases (34) and aimed at clarifying the protonation states of cytochrome c peroxidase (35).
In order to analyze the water structure that surrounds Z-DNA, we crystallized the duplex without divalent metal ions and polyamines from an (ND4)2SO4/D2O solution (36). This prevents the disruption of the first and the second hydration shell around the DNA by metal ions and organic polycations. The crystal used had an approximate volume of 1 mm3 (31), and cryo neutron diffraction data were collected at 100K on the Macromolecular Neutron Diffractometer (MaNDi (37)) at the Spallation Neutron Source (SNS), Oak Ridge National Laboratory (ORNL, Oak Ridge, TN) to a resolution of 1.5 Å. Here, we report the most detailed analysis yet of the orientation of water molecules that surround a DNA double helix.
MATERIALS AND METHODS
Synthesis and purification of d(CGCGCG)
The Z-DNA hexamer d(CGCGCG) was synthesized and HPLC purified by Integrated DNA Technologies (Coralville, IA, USA; www.idtdna.com). Following lyophilization, a D2O stock solution of the DNA hexamer with a concentration of 60 mg/ml was prepared.
Crystallization
We used previously reported conditions without spermine to grow crystals of the Z-DNA hexamer (31,36). Briefly, crystals were obtained by mixing equal volumes of DNA with a D2O crystallization buffer containing 2.5 M ammonium sulfate (ND4)2SO4, 10 mM magnesium acetate, and 50 mM perdeuterated 2-(N-morpholino)ethanesulfonic acid (MES), pH 6.1, and equilibrating droplets by vapor diffusion against a reservoir of the crystallization buffer. Crystallization experiments were assembled using Nextal EasyXtal Tools with sitting-drops bridges inserted. The greaseless closures of the EasyXtal Tool allow for efficient modification of the reservoir conditions. Crystallization drop volumes were 30 μl. The reservoir was filled with 500 μl of D2O crystallization buffer diluted to 50% with D2O and allowed to equilibrate for 3–4 days. The strength of the crystallization buffer in the reservoir was then gradually increased over a period of weeks by adjusting the strength of the buffer by 5% and allowing for equilibration for 3–4 days before again increasing the buffer strength. This procedure was continued until a crystal appeared.
Crystal mounting and cryo-protection
Cryoprotection was achieved by transferring the crystal into a large drop of Paratone-N oil. Mother-liquor adhering to the surface of the crystal was removed using a nylon loop to swiftly shift the crystal in the drop allowing the viscosity of the oil to pull the mother liquor away. The crystal was then transferred to a fresh drop of Paratone-N oil from which the crystal was mounted on a MiTeGen dual-thickness micromount of appropriate size. Flash-cooling was done by plunging into liquid nitrogen. Crystals were stored in liquid nitrogen before transporting them by hand to the SNS. No modifications to the cryoprotection protocol were required even given the large crystal volume.
Neutron data collection and processing
Neutron diffraction data were collected using the macromolecular neutron diffractometer (MaNDi) instrument (37,38) at the SNS. A total of 15 diffraction images were collected with an exposure time of 12 h each. The crystal was held static during each diffraction image and was rotated 10° between images. Data collection was completed in 7.5 days, yielding a dataset of good completeness to a resolution of 1.50 Å (Table 1). These data were reduced with the Mantid program (39) using newly developed 3D profile fitting for time-of-flight diffraction data (40,41). Integrated intensities were then scaled using Lauenorm from the Lauegen suite (42,43). The reduced and corrected data statistics are shown in Table 1.
Table 1.
Selected crystal data, data collection and processing, and refinement statistics
| Diffraction method | Cryo X-ray data | Cryo neutron data |
|---|---|---|
| PDB ID code | 7JY2 | |
| Space group | P212121 | |
| a, b, c [Å] | 17.99, 31.16, 44.12 | |
| α, β, γ [°] | 90, 90, 90 | |
| Resolution range [Å] | 16.66–1.00 (1.08–1.00) | 14.69–1.50 (1.55–1.50) |
| Total no. of reflections | 140 000 (18 713) | 21 134 (1 182) |
| No. of unique reflections | 13 361 (2 407) | 3587 (307) |
| Completeness [%] | 95.9 (88.6) | 84.2 (78.52) |
| Multiplicity | 10.5 (7.6) | 5.89 (3.85) |
| 〈I/σ(I)〉 | 36.9 (12.6) | 13.8 (4.2) |
| R-merge [%] | 4.1 (14.8) | 27.7 (38.1) |
| R-meas [%] | 4.3 (15.9) | 30.3 (43.2) |
| R-pim [%] | 1.3 (5.5) | 11.60 (19.7) |
| CC1/2 [%] | 99.9 (98.5) | 91.80 (48.2) |
| Wilson B factor [Å2] | 3.05 | n/a |
| R-work | 0.182 (0.424) | 0.277 (0.390) |
| R-free | 0.204 (0.456) | 0.303 (0.389) |
| R-free test set size (%) | 10.26 | 4.49 |
| R-free test set count | 2 507 | 161 |
| No. of non-hydrogen atoms | 321 | 321 |
| No. of DNA atoms | 244 | 244 |
| No. of solvent molecules | 77 | 77 |
| R.m.s.d. bonds (Å) | 0.016 | 0.012 |
| R.m.s.d. angles (°) | 1.65 | 1.44 |
| Avg. B-factor, DNA (Å2) | 25.4 | 25.4 |
| Avg. B-factor, water (Å2) | 33.4 | 33.4 |
X-ray data collection and processing
The crystal used for neutron diffraction was returned to the laboratory but did not survive the journey. A very similar crystal from the same batch and with nearly identical unit cell parameters was used. X-ray diffraction data were collected using the D8 Venture (Bruker AXS, Madison, WI, USA) system in the Biomolecular Crystallography Facility in the Vanderbilt University Center for Structural Biology. The system includes an Excillum D2+ MetalJet X-ray source with Helios MX optics providing Ga Kα radiation at 1.3418 Å wavelength. The crystal was mounted on a kappa axis goniometer and maintained at 100 K using an Oxford Cryosystems Cryostream 800 cryostat. The detector was a PHOTON III charge-integrating pixel array detector. Data collection was performed in shutterless mode. Data were reduced using Proteum3 software (Bruker AXS, Madison, WI, USA). Selected crystal data and data collection statistics are summarized in Table 1.
Joint XN structure refinement
Initial phases were obtained by the molecular replacement technique, using the high-resolution model of the Z-DNA duplex based on the X-ray structure of crystals grown under identical conditions (PDB ID 3QBA). All crystallographic refinements were carried out with the program PHENIX (44,45), and model building was performed using the COOT molecular graphics program (46). Nonexchangeable H atoms were treated with a group occupancy in the joint refinement (X-ray term), typically resulting in values that are below 1. In the case of partially exchanged hydrogens, occupancies were handled differently: they add up to 1 for H and D.
Separate X-ray and neutron refinements
To investigate potential variations in the water positions and orientations as a result of using different crystals and slightly different cryoprotection protocols, we carried out separate X-ray and neutron refinements of the Z-DNA structure. For details see the Supplementary Data (Table S1).
RESULTS
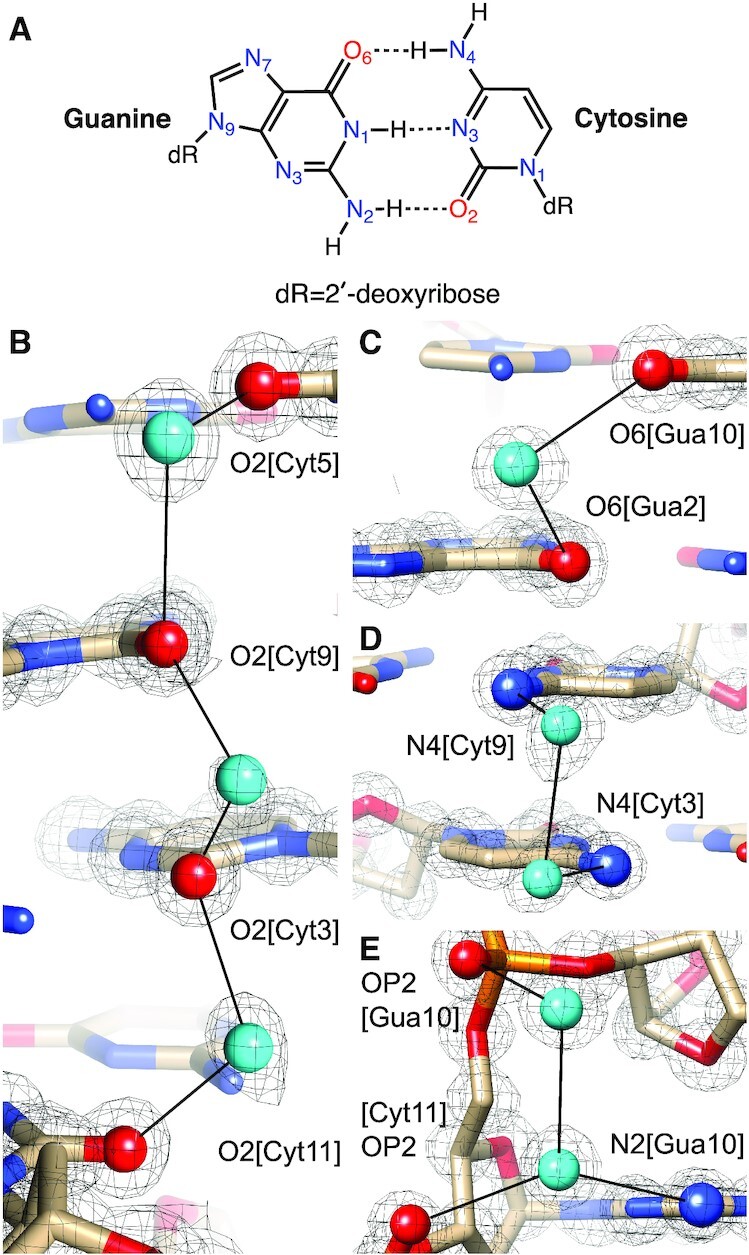
Crystals of the Z-DNA duplex [d(CGCGCG)]2 were grown by sitting drop vapor diffusion in deuterium oxide at pH 6.1 (50 mM MES buffer) using as precipitant (ND4)2SO4 at a concentration of 2.5 M. Neutron diffraction data were collected at 100 K on the MaNDi/SNS instrument for one week and yielded a data set with a resolution of 1.5 Å and good completeness. A crystal from the same drop and of a similar size was used for X-ray diffraction data collection on an in-house Bruker MetalJet instrument to a resolution of 1.0 Å. Coordinates based on the X-ray structure of Z-DNA crystals grown under identical conditions (21) served as the starting model for a joint cryo X-ray/neutron refinement of the left-handed duplex. Crystal data, neutron, and X-ray data collection statistics and refinement parameters are summarized in Table 1. The asymmetric unit of the final joint X-ray/neutron Z-DNA structure comprises a single DNA duplex, 64 D2O molecules, and an additional 14 water oxygen atoms, the latter surrounded by electron density but no or insufficient neutron density to establish water orientation. Examples of the quality of the superimposed electron and neutron densities around water molecules are depicted in Figure 2. For the following description of the water structure, nucleotides in the first strand are numbered C1 to G6, and nucleotides in the second strand are numbered C7 to G12. Refinement using the neutron data indicates near complete H to D exchange for nucleobase N2(G) and N4(C) amino groups (Table 2) and 5′- and 3′-terminal hydroxyls. The N2 amino group of G4 constitutes an outlier in that D21 and D22 exhibit occupancies of 0.23 and 0.53, respectively. Similarly, most D1(G) occupancies are quite high (between 0.5 and 0.94), but G4 again constitutes an exception in that the occupancy of its D1 is only 0.25. Guanine H8 hydrogen atoms are also partially replaced by deuterium, whereby the exchange is highest for 3′-terminal guanosines (G6 0.33, G12 0.76; Table 2). Superimposed electron and neutron density maps provide no evidence of ammonium or sulfate ions binding to the Z-DNA duplex.
Figure 2.
Quality of the final joint cryo X-ray/neutron crystal structure of Z-DNA. Examples of the superimposed 2Fo-Fc electron (1.0 Å, blue) and neutron (1.5 Å, green) densities drawn at the 1.5σ threshold. (A) Water pentagons spanning the minor groove at one end of the duplex. (B) Three water molecules linking guanine to the phosphate of the adjacent residue on the convex surface. (C) Water pentagon nestled against the phosphate backbone in the minor groove. (D) Water tandem bridging exocyclic amino groups of adjacent cytosines on the convex surface. Selected residues and/or atoms are labeled and H-bonds are drawn with thin solid lines.
Table 2.
Deuterium occupancies for exchangeable hydrogen atoms of G and C nucleobases
| Base pair | D1(G) | D21(G) | D22(G) | D8(G) | D41(C) | D42(C) |
|---|---|---|---|---|---|---|
| C1 : G12 | 0.50 | 0.46 | 1.00 | 0.76 | 1.00 | 0.39 |
| G2 : C11 | 0.93 | 1.00 | 0.95 | 0.04 | 1.00 | 1.00 |
| C3 : G10 | 0.94 | 0.87 | 0.56 | 0.27 | 0.70 | 0.62 |
| G4 : C9 | 0.25 | 0.23 | 0.53 | 0.00 | 0.99 | 1.00 |
| C5 : G8 | 0.58 | 0.89 | 0.84 | 0.05 | 0.95 | 0.71 |
| G6 : C7 | 0.61 | 1.00 | 1.00 | 0.33 | 1.00 | 1.00 |
Water structure in the minor groove
In the minor groove, the most prominent H-bond donor and acceptor atoms presented by nucleobases are N2(G) and O2(C), respectively, and they form H-bonds to water molecules in all cases. By comparison, N3(G) is not contacted by water as it lies in close proximity of the sugar as a result of the syn conformation of guanosine in Z-DNA. The C2′, C3′ and C5′ atoms are positioned between 3.2 and 3.5 Å from N3 and hydrogens of these sugar atoms are directed toward the base nitrogen. Thus, the N3(G) acceptor is tucked away at the border of the minor groove and inaccessible to water. Instead, its lone electron pair is surrounded by three sugar H atoms with average N3…H2′/H3′/H5″ distances of 2.9 Å (G4) and 2.8 Å (G10) for central guanosines. The syn orientation of guanosine brings N2 in relatively close proximity of the 3′ and 5′ phosphates of the nucleotide, such that two water molecules can link the exocyclic amino group to both. A single water accepts a hydrogen from N2 and in turn donates one of its hydrogens to the 3′-phosphate (OP2). Its other hydrogen is used for a H-bond to a second water that itself donates a hydrogen to the 5′ phosphate (Figure 3). This water structure is observed quite systematically in the Z-DNA duplex, but is disrupted for G6 and G12 at the ends that do not carry a 3′ phosphate. A ZII conformation occasionally adopted by a phosphate and observed (partial occupancy) between G10 and C11 also disrupts the pattern as the distance between OP2 and N2 becomes too long to be bridged by a single water (Figure 3B).
Figure 3.
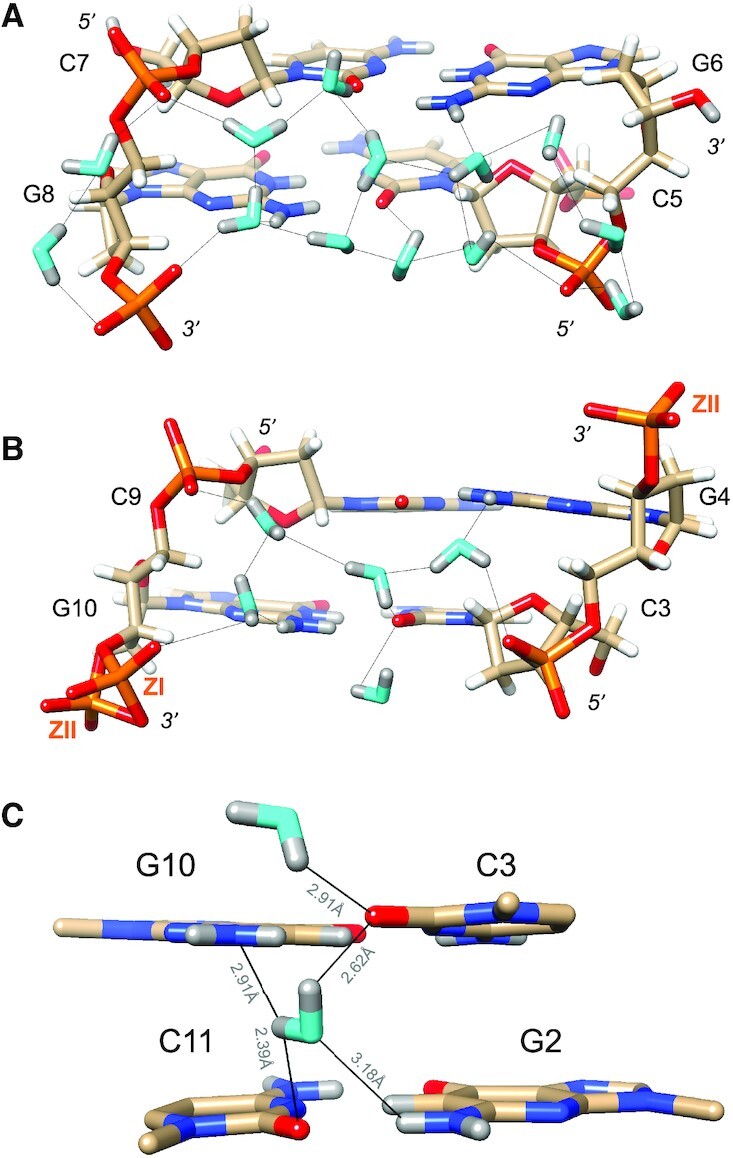
Water structure in the minor groove. Transverse hydration patterns that link cytosine O2 keto oxygens and guanine N2 amino groups to backbone phosphates. (A) Base pairs C5:G8 and G6:C7 and (B) base pairs C3:G10 and C4:C9. Water oxygen and deuterium atoms are colored in cyan and gray, respectively, and DNA deuterium and hydrogen atoms are colored in gray and white, respectively. H-bonds (D…O) are drawn as thin solid lines, and strand polarities are indicated. ZI and ZII refer to two different phosphate orientations that are characterized by distinct ϵ and ζ backbone torsion angles. (C) The sole example of a water molecule bridging O2 keto oxygens of adjacent cytosines in the minor groove. Electrostatically favorable interactions by this water are not limited to O2 (cytosine) but also involve N2 (guanine), i.e. the lone electron pair of N2(G10) and one of the deuterium atoms of N2(G2). D…N and D…O distances are indicated by thin solid lines.
Owing to the syn conformation of guanosine in Z-DNA, N2 atoms lie closer to 5′- and 3′-phosphate groups on average than O2 atoms of cytidine. Thus, the average distance of N2 to 5′- and 3′-phosphates (OP2 atoms) is 6.1 and 5.1 Å, respectively. The average distance of O2 to 5′- and 3′-phosphates (OP2 atoms) is 6.8 and 6.6 Å, respectively. The shorter distances between N2 and OP2 atoms of 3′-phosphates enable a single water bridge between them as discussed above. However, distances closer to 6 Å and beyond preclude the formation of a one-water bridge between guanine base and phosphate. Hence the two-water bridge linking N2(G) to OP2 from the 5′-phosphate and O2(C) to OP2 atoms from both the 5′-phosphate and the 3′-phosphate (Figure 3). With water bridges involving O2 and OP2, both waters have to act as H-bond donors to DNA atoms. These hydration patterns on the borders of the minor groove are then extended across the entire groove. At one end of the duplex, an intricate ribbon involving two hexagons and two pentagons links backbones and nucleobase edges (Figure 3A). The hexagon on one side of the minor groove is composed of two phosphate groups (G8 and C9) and four waters. On the other side, the hexagon involves five waters and the phosphate of C5. The two hexagons are linked by central water pentagons, with the entire hydration network being composed of first and second shell water molecules.
There is only a single example of a water molecule linking O2 atoms of cytosines from adjacent base pairs in the minor groove. The water sits halfway between O2 atoms of C3 and C11, with OW…O2 distances of 2.80 and 2.79 Å, respectively (Figure 3C). Despite the fact that the two cytosine keto oxygens are the water's closest neighbors, its orientation suggests a more complicated involvement with the G2:C11 and C3:G10 base pairs than simply as a bridge between O2(C3) and O2(C11). The deuterium atoms of the water do not lie in the plane defined by OW, O2(C3) and O2(C11), and are thus not pointing straight at the keto oxygens. Instead, the water molecule is oriented more sideways vis-à-vis C:G base pair edges, with OW—DW—O(C) angles of ca. 100° (Figure 3C). The N2(D)2 amino nitrogen of G10 is positioned 3.60 Å from the water oxygen. The DW atom close to O2(C11) (distance of 2.39 Å) is positioned 2.91 Å from N2(G10), leading us to believe that the particular orientation of the water molecule enables an electrostatically favorable interaction between water deuterium and nitrogen lone electron pair. The N2(D)2 amino nitrogen of G2 is positioned 3.70 Å from the water oxygen with a lone pair of the latter directed toward the amino group. The distance between OW and the amino deuterium participating in G2:C11 pairing is 3.18 Å and thus somewhat too long to contribute favorably to the water-base pair interaction. However, given the orientation of the water and the observed distances to acceptor and donor atoms of the two C:G pairs, the notion that the water molecule interacts with O2(C3), O2(C11) and N2(G10) is reasonable. Thus, water in the central minor groove appears to optimize H-bonds with the edges of base pairs rather than solely interacting with cytosine O2 keto groups (Figure 3C).
Water structure on the convex surface
Z-DNA lacks a major groove, and instead, the O6 and N7 (guanine) acceptors and the N4 amino group (cytosine) donor are presented on the so-called convex surface. The particular orientation of backbones relative to base pairs in Z-DNA renders water-mediated bridges between base edges and phosphate groups much less common than in the narrow minor groove. Exceptions can occur when a phosphate flips from the ZI to the ZII conformation, thereby rotating non-bridging OP1 and OP2 oxygen atoms away from the minor groove (Figures 3B and 4A). The alternating positive and negative twists between adjacent base pairs in Z-DNA shift pairs of N4 amino groups roughly above one another on the convex surface. The distances between N4 atoms in the three pairs range from 3.6 to 3.9 Å. Amino pairs are linked by water tandems such that water oxygen atoms are positioned more or less in the planes defined by cytosine bases (Figure 4A, B). Both waters accept in H-bonds with amino groups, and one donates, and the other accepts in the inter-water H-bond. However, water orientations vis-à-vis cytosines differ somewhat between pairs of N4 amino groups as the examples in Figure 4A, and B demonstrate. These differences can likely be traced to the hydration networks that individual waters participate in, as orientations of waters are not simply dictated by their interactions with DNA atoms, but are also influenced by other water molecules that are part of the first and second hydration shells.
Figure 4.
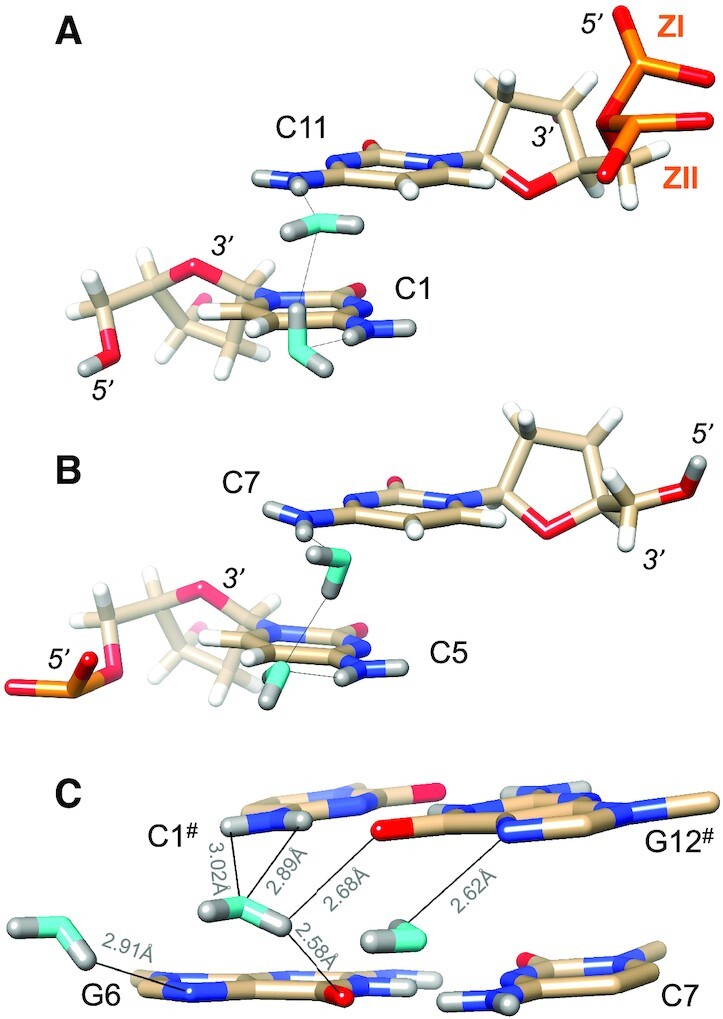
Water molecules bridging either pairs of cytosine N4 amino groups or pairs of guanine O6 keto groups on the convex surface. The Z-DNA neutron structure reveals H-bond donor-acceptor patterns for water bridges between cytosines (A) C1 and C11, and (B) C5 and C7. Water oxygen and deuterium atoms are colored in cyan and gray, respectively, and DNA deuterium and hydrogen atoms are colored in gray and white, respectively. H-bonds (D…O) are drawn as thin solid lines. (C) Water molecule bridging O6 keto oxygens of adjacent guanines on the convex surface (center), flanked by water molecules forming H-bonds to N7 of G6 and G12# (# marks a symmetry-related residue). Electrostatically favorable interactions by the central water are not limited to O6 (guanine) but also involve N4(C1#), i.e. a lone electron pair of water and the deuterium atoms of the amino group. D…N and D…O distances are indicated by thin solid lines.
The particular orientation of a water molecule linking O6 keto oxygens of adjacent guanines on the convex surface is somewhat reminiscent of the situation in the minor groove. As in the case of the aforementioned water linking O2 atoms of cytosines from adjacent base pairs there, the water situated between guanine keto groups does not project its deuterium atoms in the direction of O6 atoms. Rather, it turns away slightly so as to share one of its deuteriums with both guanine keto oxygens. The distances between the water oxygen and O6(G6) and O6(C12#) are 3.02 and 3.00 Å, respectively, and the corresponding DW…O6 distances are 2.58 and 2.68 Å, respectively (Figure 4C). This orientation of the water results in one of its lone electron pairs being directed toward the deuterium atoms of the N4 amino group of cytosine C1#. The OW…N4 distance is 3.25 Å, and the OW…D(N4) distances are 2.89 Å (D41) and 3.02 Å. These distances are not significantly different from those between water OW/DW and O6 keto oxygens (Figure 4C). Just like with the water bridging O2 keto oxygens of cytosine in the minor groove, a water on the convex surface can interact with two O6 keto oxygens of guanosines and an amino group of one of the cytidines. Therefore, water in the central portion of the convex surface does not just engage with O6 keto groups of adjacent guanines but interacts with the DNA by H-bonding to acceptors and donors of G:C pairs.
The water molecule interacting with O6 keto oxygens of G6 and G12#, as well as the N4 amino group of C1#, is flanked by water molecules that form H-bonds to N7 of adjacent guanines (Figure 4C). The waters contacting N7(G6) and N7(G12#) are too far removed from the central water for effective interactions (3.5 Å and 4.0 Å, respectively), but are embedded in water networks spanning the convex surface. Except for one guanine, all N7 acceptors form H-bonds to a water molecule, but in some cases, the neutron density did not permit us to settle the orientations of these water molecules. In cases where the orientations were established, for example, the water molecule associated with N7 of G10, it is H-bonded to a water from a pair of solvent molecules that bridges N4 amino groups (C1/C11). The water molecule H-bonded to N7 of G12 is linked to the water tandem associated with the N4 amino groups of C7# and C5# via an intermediate water.
Water networks on the convex surface include longitudinal and transverse patterns. An example of the former is depicted in Figure 5, with a string of water molecules spanning four layers of bases along the edge of the convex surface. As in the minor groove, pentagons constitute the predominant ring-shaped arrangement of waters that are part of the first and second hydration shells. The participation of either a DNA base or backbone atom in the pentagons is common. An example that includes O6(G8) is shown at the top of Figure 5, although one side of that pentagon is not constituted by an actual H-bond. Instead, the water molecule, unlike in the case of that between O6 atoms of G6 and G12# (Figure 4C), turns its back—i.e. lone pairs—toward the O6 keto groups of G4 and G8. The distances between water and the pair of O6 atoms (3.07 and 2.95 Å, Figure 5) are not incompatible with H-bond formation, but the relative orientations of lone pairs among the three oxygen atoms are. Perhaps this is an indication that waters participating in networks around DNA occasionally prefer interactions with neighboring solvent molecules instead of an H-bond to a DNA atom. In any case, the water molecule located below the base plane of G4, as depicted in Figure 5, does form an H-bond to the O6 keto oxygen of that guanine and then bridges the oxygen to another water pentagon. Here, the pentagon is composed of four water molecules and OP1 of the phosphate group linking G2 and C3. An example of a transverse hydration network on the convex surface is shown in Figure 6. Nine water molecules span the entire width of the duplex, starting with a water that forms an H-bond to the 5′-hydroxyl group of C7 and ending with waters that either H-bond to N7 of G6 or the phosphate group from a neighboring duplex. The neutron density did not allow a definitive assignment of the orientations of all waters in the network (cyan spheres in Figure 6). However, it is encouraging that neutron crystallography allows visualization of donor-acceptor patterns in water networks around a DNA duplex, even on the more exposed convex surface compared to the narrow minor groove. Higher resolution data might reveal the orientations of the remaining waters that lack neutron density for deuterium atoms in the present structure.
Figure 5.
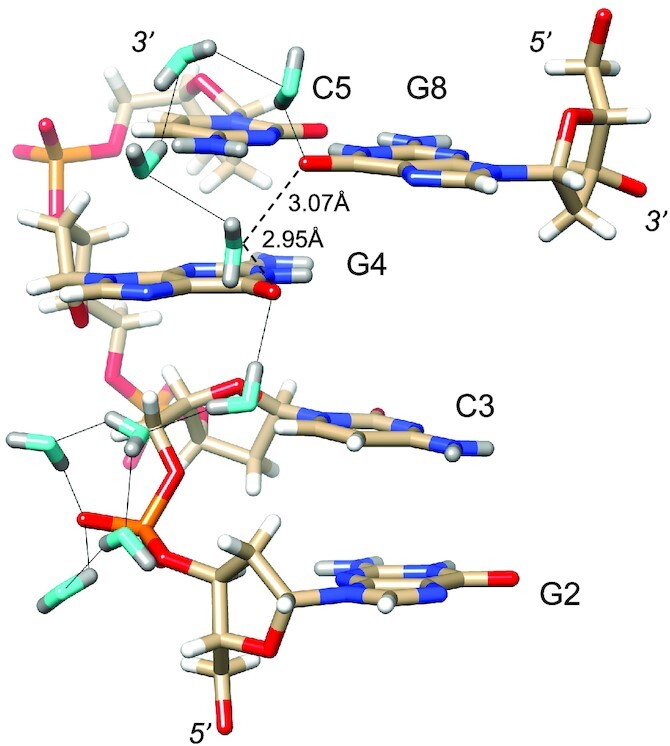
Longitudinal hydration pattern linking O6 keto oxygen of G and backbone phosphates on one side of the convex surface. Water oxygen and deuterium atoms are colored in cyan and gray, respectively, and DNA deuterium and hydrogen atoms are colored in gray and white, respectively. H-bonds (D…O) are drawn as thin solid lines. Note the formation of pentagons involving four water molecules and either a phosphate oxygen or an O6 keto oxygen. A water molecule positioned between G4 and G8 lies within H-bonding distance from O6 keto oxygens (dashed lines). However, its particular orientation, i.e. both deuterium atoms are directed away from O6 atoms, renders H-bond formation unlikely.
Figure 6.
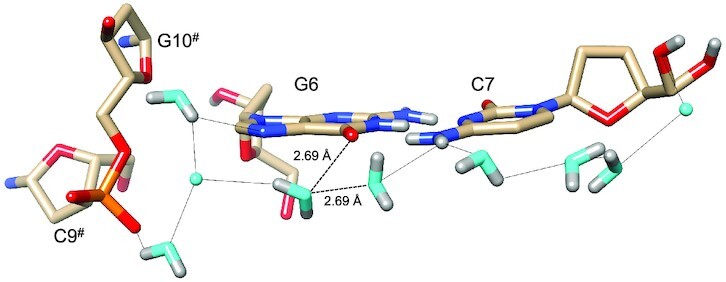
Transverse hydration pattern linking base edges and backbones across the convex surface. Water oxygen and deuterium atoms are colored in cyan and gray, respectively, and H-bonds (D…O/N) are drawn as thin solid lines. Dashed lines link a water molecule to an adjacent one as well as to O6(G6), whereby the distances are compatible with H-bond formation. However, the relative orientations of the two water molecules and O6 render H-bonding unlikely. Symmetry-related residues are marked by #.
Phosphate hydration
The shortest distances between non-bridging oxygen atoms of adjacent intra-strand phosphate groups are typically around 4.5 Å with phosphate pairs that both assume the more common ZI orientation. Such distances become much longer (6–7 Å) in cases where the local backbone conformation flips to ZII, thereby rotating the phosphate group outwards and away from the minor groove (Figure 3). Despite the relatively short spacing between adjacent phosphates with a regular ZI backbone conformation, they are most often linked by two water molecules (Figures 2A, C and 3A, B). Both waters act as donors in H-bonds to phosphate oxygens and either the water closer to the 5′-end or that closer to the 3′-end accepts/donates in the inter-water H-bond (Figures 3A,B and 7B). Such waters are then embedded in hydration motifs that extend across the minor groove (Figure 3A) or along the convex surface (Figure 5) and commonly entail pentagon motifs. Patterns seen in the water structure around phosphates, among others, include 5-membered (Figure 5), 7-membered (Figure 3A) and even 10- (Figure 7B) and 12-membered rings (Figure 7A). In regions with ZII backbone conformations and thus longer distances between adjacent phosphates, two water molecules are not sufficient to bridge the former. Thus, the phosphates of nucleotides C5 and C11 exhibit ZII conformations (mixed ZI/ZII in the latter case; Figures 3B, 4A, 7). As a consequence, the phosphates of G4 and G6 are close enough to be linked by two water molecules (Figure 7A). Conversely, the ZI phosphate of G4 and the ZII phosphate of C5 are linked by four water molecules. A further four waters then connect the C5 phosphate to the O4′ atom of G6. The sugar oxygen occasionally accepts in H-bonds to water but is poorly hydrated compared to base edges and phosphate groups. The shortest distance between adjacent intra-strand phosphate groups in the Z-DNA duplex studied here is found between OP2(C11) and OP1(G12): 4.25 Å (Figure 7B). This pair is joined by a single water molecule, the only example of a one-water bridge between phosphates observed in the DNA structure.
Figure 7.
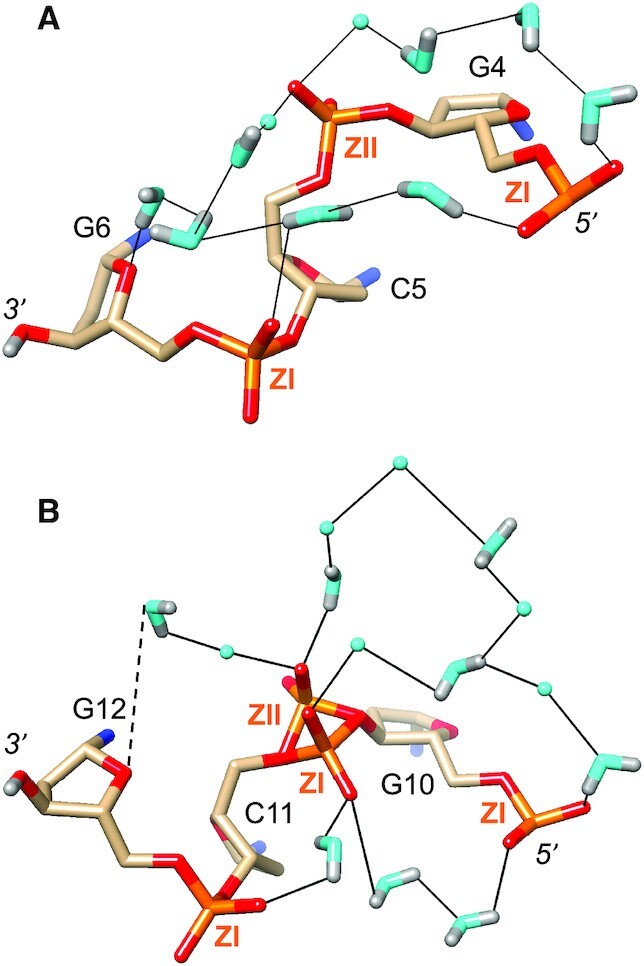
Phosphate hydration. Water structure around the backbones of nucleotides (A) G4 to G6, and (B) G10 to G12. Water molecules without significant neutron density for deuterium atoms are drawn as cyan spheres, and H-bonds are thin solid lines. ZI and ZII designate particular conformations of phosphate groups in Z-DNA.
DISCUSSION
The joint X-ray and neutron refinement of the Z-DNA duplex has thus far revealed the orientations of 64 D2O molecules that are part of the first and second hydration shells. As the examples depicted in Figure 2 demonstrate, the overlaid neutron and electron densities around DNA atoms and water molecules are of good quality and the maps complement each other well. The neutron diffraction data collected at cryo temperature on MaNDi/SNS at ORNL are of high completeness to a maximum resolution of 1.5 Å. The dataset was collected in a week-long experiment using a crystal that was quite large with a volume of ca. 1 mm3. We believe that further optimization (e.g. wavelength) and a somewhat longer exposure can yield complete data with a resolution around 1.2 Å. It is likely that the orientations of many of the water molecules that currently lack neutron density for deuterium atoms can then also be established. The neutron data were initially processed to 1.4 Å resolution and the <I/σ(I)> in the 1.45–1.40 Å resolution bin was 3.6 with a completeness of 62.6%. We decided to cut the data at 1.5 Å (<I/σ(I)> of 4.2 and completeness of 78.5% in the 1.55–1.50 Å resolution bin; Table 1) in order to work with a more complete data set. Still, it should be possible to collect neutron data to higher resolution with a Z-DNA crystal of this size as the mean intensity did not drop to a level of, say, <I/σ(I)> = 2 at 1.4 Å. The MaNDi setup is such that the detector cannot be moved; instead it was necessary to use a more optimal wavelength and collect longer frames.
As expected, hydrogen atoms in the Z-DNA duplex that are exchanged with deuterium are limited to exocyclic amino groups in cytosine and guanine and 5′- and 3′-terminal hydroxyls. We also found an indication for partial H-D exchange by H8 hydrogen atoms in some guanines, as reported by Niimura et al. (28). It is possible that different crystallization conditions can affect exchange to some degree. However, the level of background scattering in neutron experiments with crystals of synthetic oligonucleotides is bound to be higher than with crystals of perdeuterated proteins because none of the H atoms in the DNA backbone are exchanged. The Z-DNA crystal form described here was grown using ammonium sulfate as the precipitant to avoid binding of divalent metal ions or polyamines to the DNA, thereby displacing first and second shell water molecules and distorting the water structure around the duplex. It is noteworthy that the density maps did not reveal any coordinated sodium, ammonium, or sulfate ions. The ammonium sulfate crystal form displays unit cell constants that are very similar to those grown from magnesium, spermine, or mixed magnesium/spermine conditions (19) and all these Z-DNA crystals belong to space group P212121.
X-ray crystal structures of Z-DNA at atomic resolution revealed regular longitudinal and transverse hydration patterns (19), and the former included a chain of waters in the minor groove that linked cytosine O2 atoms, and water molecules bridging pairs of guanine O6 keto oxygens and cytosine N4 amino groups (Figure 1). By comparison, transversal hydration patterns both in the minor groove and on the convex surface appear to be more common in the cryo-neutron structure of Z-DNA analyzed here. In particular, repetitive patterns involving water molecules linking the edges of guanine and cytosine bases to phosphate groups are commonly seen in the minor groove (A, B). Longitudinal water motifs include water tandems bridging adjacent pairs of cytosine N4 amino groups (Figure 4A,B) and adjacent phosphate groups (Figures 3A, B and 7). Hydration motifs such as single waters linking O2 keto oxygens and O6 keto oxygens of neighboring cytidines in the minor groove (Figure 3C) and guanosines on the convex surface (Figure 4C), respectively, are not observed repeatedly.
The cryo-neutron structure at high resolution has revealed an interesting hydration pattern that involves amino groups of either cytosine or guanine and pairs of keto oxygens in both the minor groove and on the convex surface. In this motif, a water donates H-bonds to two keto oxygens and also establishes electrostatically favorable interactions with adjacent amino groups. In the minor groove, the water interacts with the N2 amino group of guanine in addition to forming H-bonds to O2 keto oxygens of stacked cytosines (Figure 3C). On the convex surface, the water interacts with the N4 amino group of cytosine in addition to forming H-bonds to O6 keto oxygens of stacked guanines (Figure 4C). The neutron density does not indicate a change in the sp2 hybridization of the nitrogen atom with a concomitant loss of conjugation with the nucleobase π system as a result of this hydration motif. The orientation of water molecules vis-à-vis C:G pairs observed here likely serves the optimization of H-bonding interactions with the edges of DNA bases and perhaps other water molecules. Thus, this hydration motif may be energetically beneficial compared to simply bridging adjacent pairs of O2 and O6 keto oxygens in the minor groove and on the convex surface, respectively, as implied by electron density alone (Figure 1). More work using high-resolution cryo neutron crystallography is necessary to establish whether such features are commonplace in the water structure that surrounds DNA duplexes.
CONCLUSIONS
We used cryo neutron crystallography to analyze the water structure around the DNA duplex [d(CGCGCG)]2 at near-atomic resolution. The resulting neutron density reveals the orientation of the majority of first-shell water molecules. Cryo neutron data allow insight into acceptor and donor functions for water molecules that are part of intricate hydration patterns in the minor groove and on the Z-DNA convex surface, thus complementing information previously obtained from room temperature neutron structures or X-ray crystal structures determined to ultra-high resolution. The discovery based on the determination of deuterium positions that water is shared among guanine and cytosine keto and amino groups in the minor groove and on the convex surface rather than just clamping keto oxygens of adjacent cytidines (minor groove) or guanosines (convex surface) is particularly noteworthy.
Several challenges remain to be overcome in order to visualize at truly atomic resolution—1 Å or higher—the water structure around oligonucleotides based on joint neutron and X-ray diffraction data collected at cryogenic temperature. Even with the higher flux of the neutron beam at MaNDi/SNS, a crystal with a volume of, say, 0.1 mm3 will not deliver atomic resolution neutron data. Obviously, the resolution of the X-ray data that can be achieved even with small Z-DNA crystals or other well diffracting DNA and RNA oligo crystals is not an issue. As well, crystals of Z-DNA with volumes of 1 mm3 and larger can be grown within weeks as we have shown. Cryoprotection and freezing of very large crystals requires special caution and transport using a dryshipper via courier is not encouraged. The crystal has to remain cooled and stable for two weeks and more in the neutron beam and then recovered for X-ray data collection. Hence, collecting the latter data on a home source nearby like the MetalJet in our case—without the need to ship the crystal to an X-ray synchrotron—is beneficial. The opportunities to collect neutron diffraction data at cryogenic temperature worldwide are very scarce. Once data collection time is granted, conducting an optimal experiment in order to push the resolution limit closer to 1 Å is expected to take at least two weeks and may require changing the wavelength to avoid limiting data resolution. Finally, joint refinement of cryogenic X-ray and neutron data for oligonucleotides is not a routine affair and more experience needs to be gained to understand differences seen in the temperature factors of ‘X-ray’ and ‘neutron’ water molecules.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structure have been deposited with the Protein Data bank under accession number 7JY2.
Supplementary Material
ACKNOWLEDGEMENTS
The research at O.R.N.L.’s Spallation Neutron Source was sponsored by the Scientific User Facilities Division, Office of Basic Energy Sciences, US Department of Energy.
Contributor Information
Joel M Harp, Department of Biochemistry and Center for Structural Biology, Vanderbilt University, School of Medicine, Nashville, TN 37232, USA.
Leighton Coates, Neutron Scattering Division, Oak Ridge National Laboratory, 1 Bethel Valley Road, Oak Ridge, TN 37831, USA.
Brendan Sullivan, Neutron Scattering Division, Oak Ridge National Laboratory, 1 Bethel Valley Road, Oak Ridge, TN 37831, USA.
Martin Egli, Department of Biochemistry and Center for Structural Biology, Vanderbilt University, School of Medicine, Nashville, TN 37232, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
L.C. and B.S. acknowledge support from the National Institutes of Health under grant number [R01 GM071939]. The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal
Conflict of interest statement. None declared.
REFERENCES
- 1. Drew H., Dickerson R.E.. Structure of a B-DNA dodecamer. III. Geometry of hydration. J. Mol. Biol. 1981; 151:535–556. [DOI] [PubMed] [Google Scholar]
- 2. Tereshko V., Minasov G., Egli M.. The Dickerson-Drew B-DNA dodecamer revisited - at atomic resolution. J. Am. Chem. Soc. 1999; 121:470–471. [Google Scholar]
- 3. Egli M., Portmann S., Usman N.. RNA hydration: a detailed look. Biochemistry. 1996; 35:8489–8494. [DOI] [PubMed] [Google Scholar]
- 4. Egli M., Tereshko V., Teplova M., Minasov G., Joachimiak A., Sanishvili R., Weeks C.M., Miller R., Maier M.A., An H.et al.. X-ray crystallographic analysis of the hydration of A- and B-form DNA at atomic resolution. Biopolymers (Nucleic Acid Sci.). 2000; 48:234–252. [DOI] [PubMed] [Google Scholar]
- 5. Wang A.H.-J., Quigley G.J., Kolpak F.J., Crawford J.L., van Boom J.H., van der Marel G.A., Rich A.. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979; 282:680–686. [DOI] [PubMed] [Google Scholar]
- 6. Peck L.J., Wang J.C.. Energetics of B-to-Z transition in DNA. Proc. Natl. Acad. Sci. U.S.A. 1983; 80:6206–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rich A., Nordheim A., Wang A.H.-J.. The chemistry and biology of left-handed Z-DNA. Ann. Rev. Biochem. 1984; 53:791–846. [DOI] [PubMed] [Google Scholar]
- 8. Lancillotti F., Lopez M.C., Arias P., Alonso C.. Z-DNA in transcriptionally active chromosomes. Proc. Natl. Acad. Sci. USA. 1987; 84:1560–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wittig B., Dorbic T., Rich A.. Transcription is associated with Z-DNA formation in metabolically active permeabilized mammalian cell nuclei. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:2259–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wittig B., Wölfl S., Dorbic T., Vahrson W., Rich A.. Transcription of human C-MYC in permeabilized nuclei is associated with formation of Z-DNA in three discrete regions of the gene. EMBO J. 1992; 11:4653–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rich A., Zhang S.. Z-DNA: the long road to biological function. Nat. Rev. Genet. 2003; 4:566–572. [DOI] [PubMed] [Google Scholar]
- 12. Ha S.C., Lowenhaupt K., Rich A., Kim Y.G., Kim K.K.. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005; 437:1183–1186. [DOI] [PubMed] [Google Scholar]
- 13. Herbert A., Alfken J., Kim Y.G., Mian I.S., Nishikura K., Rich A.. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:8421–8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwartz T., Rould M.A., Lowenhaupt K., Herbert A., Rich A.. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999; 284:1841–1845. [DOI] [PubMed] [Google Scholar]
- 15. Bass B.L. RNA editing by adenosine deaminases that act on RNA. Ann. Rev. Biochem. 2002; 71:817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kesavardhana S., Kuriakose T., Guy C.S., Samir P., Malireddi R.K., Mishra A., Kanneganti T.D.. ZBP1/DAI ubiquitination and sensing of influenza vRNPs activate programmed cell death. J. Exp. Med. 2017; 214:2217–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ravichandran S., Subramani V.K., Kim K.K.. Z-DNA in the genome: from structure to disease. Biophys. Rev. 2019; 11:383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Herbert A. Z-DNA and Z-RNA in human disease. Commun. Biol. 2019; 2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gessner R.V., Quigley G.J., Egli M.. Comparative studies of high-resolution Z-DNA crystal structures. Part 1: common hydration patterns of alternating dC-dG. J. Mol. Biol. 1994; 236:1154–1168. [DOI] [PubMed] [Google Scholar]
- 20. Leal R.M.F., Callow S., Callow P., Blakeley M.P., Cardin C.J., Denny W.A., Teixeira S.C.M., Mitchell E.P., Forsyth V.T.. Combined neutron and X-ray diffraction studies of DNA in crystals and solution. Acta Cryst. D. 2010; 66:1244–1248. [DOI] [PubMed] [Google Scholar]
- 21. Fenn T.D., Schnieders M.J., Mustyakimov M., Wu C., Langan P., Pande V.S., Brunger A.T.. Reintroducing electrostatics into macromolecular crystallographic refinement: application to neutron crystallography and DNA hydration. Structure. 2011; 19:523–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen J.C.-H., Hanson L., Fisher S.C., Langan P., Kovalevsky A.Y.. Direct observation of hydrogen atom dynamics and interactions by ultrahigh resolution neutron protein crystallography. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:15301–15306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cuypers M.G., Mason S.A., Blakeley M.P., Mitchell E.P., Haertlein M., Forsyth V.T.. Near-atomic resolution neutron crystallography on perdeuterated Pyrococcus furiosus rubredoxin: implication of hydronium ions and protonation state equilibria in redox changes. Angew. Chem. Int. Ed. 2013; 52:1022–1025. [DOI] [PubMed] [Google Scholar]
- 24. Chen J.C.-H., Unkefer C.J.. Fifteen years of the protein crystallography station: the coming of age of macromolecular neutron crystallography. IUCrJ. 2017; 4:72–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Földesi A., Trifonova A., Kundu M.K., Chattopadhyaya J.. The synthesis of deuterionucleosides. Nucleot. Nucleos. Nucleic Acids. 2000; 19:1615–1656. [DOI] [PubMed] [Google Scholar]
- 26. Fuller W., Forsyth T., Mahendrasingam A.. Water–DNA interactions as studied by X-ray and neutron fibre diffraction. Phil. Trans. R. Soc. Lond. B. 2004; 359:1237–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arai S., Chatake T., Ohhara T., Kurihara K., Tanaka I., Suzuki N., Fujimoto Z., Mizuno H., Niimura N.. Complicated water orientations in the minor groove of the B-DNA decamer d(CCATTAATGG)2 observed by neutron diffraction measurements. Nucleic Acids Res. 2005; 33:3017–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chatake T., Tanaka I., Umino H., Arai S., Niimura N.. The hydration structure of a Z-DNA hexameric duplex determined by a neutron diffraction technique. Acta Cryst. D. 2005; 61:1088–1098. [DOI] [PubMed] [Google Scholar]
- 29. Egli M., Williams L. D., Gao Q., Rich A.. Structure of the pure-spermine form of Z-DNA (magnesium free) at 1Å resolution. Biochemistry. 1991; 30:11388–11402. [DOI] [PubMed] [Google Scholar]
- 30. Bancroft D., Williams L. D., Rich A., Egli M.. The low temperature crystal structure of the pure-spermine form of Z-DNA reveals binding of a spermine molecule in the minor groove. Biochemistry. 1994; 33:1073–1086. [DOI] [PubMed] [Google Scholar]
- 31. Harp J., Coates L., Sullivan B., Egli M.. Cryo-neutron crystallographic data collection and preliminary refinement of left-handed Z-DNA d(CGCGCG). Acta Cryst. F. 2018; 74:603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blakeley M.P., Kalb A.J., Helliwell J.R., Myles D.A.A.. The 15-K neutron structure of saccharide-free concanavalin A. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:16405–16410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Habash J., Raftery J., Nuttall R., Price H.J., Wilkinson C., Kalb A.J., Helliwell J.R.. Direct determination of the positions of the deuterium atoms of the bound water in -concanavalin A by neutron Laue crystallography. Acta Cryst. D. 2000; 56:541–550. [DOI] [PubMed] [Google Scholar]
- 34. Coates L., Tomanicek S., Schrader T.E., Weiss K.L., Ng J.D., Juttner P., Ostermann A.. Cryogenic neutron protein crystallography: routine methods and potential benefits. J Appl. Cryst. 2014; 47:1431–1434. [Google Scholar]
- 35. Casadei C.M., Gumiero A., Metcalfe C.L., Murphy E.J., Basran J., Concilio M.G., Teixeira S.C.M., Schrader T.E., Fielding A.J., Ostermann A.et al.. Heme enzymes. Neutron cryo-crystallography captures the protonation state of ferryl heme in a peroxidase. Science. 2014; 345:193–197. [DOI] [PubMed] [Google Scholar]
- 36. Langan P., Li X., Hanson B.L., Coates L., Mustyakimov M.. Synthesis, capillary crystallization and preliminary joint X-ray and neutron crystallographic study of Z-DNA without polyamine at low pH. Acta Cryst. F. 2006; 62:453–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Coates L., Cuneo M.J., Frost M.J., He J.H., Weiss K.L., Tomanicek S.J., McFeeters H., Vandavasi V.G., Langan P., Iverson E.B.. The macromolecular neutron diffractometer MaNDi at the spallation neutron source. J. Appl. Cryst. 2015; 48:1302–1306. [Google Scholar]
- 38. Coates L., Cao H.B., Chakoumakos B.C., Frontzek M.D., Hoffmann C., Kovalevsky A.Y., Liu Y., Meilleur F., dos Santos A.M., Myles D.A.A.et al.. A suite-level review of the neutron single-crystal diffraction instruments at Oak Ridge National Laboratory. Rev. Sci. Instrum. 2018; 89:092802. [DOI] [PubMed] [Google Scholar]
- 39. Arnold O., Bilheux J.C., Borreguero J.M., Buts A., Campbell S.I., Chapon L., Doucet M., Draper N., Leal R.F., Gigg M.A.et al.. Mantid - data analysis and visualization package for neutron scattering and SR experiments. Nucl. Instrum. Methods A. 2014; 764:156–166. [Google Scholar]
- 40. Sullivan B., Archibald R., Langan P.S., Dobbek H., Bommer M., McFeeters R.L., Coates L., Wang X.P., Gallmeier F., Carpenter J.M.et al.. Improving the accuracy and resolution of neutron crystallographic data by three-dimensional profile fitting of Bragg peaks in reciprocal space. Acta. Cryst. D. 2018; 74:1085–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sullivan B., Archibald R., Vandavasi V., Langan P., Langan P.S., Coates L., Azadmanesh J., Lynch V.E.. BraggNet: integrating Bragg peaks using neural networks. J. Appl. Cryst. 2019; 52:854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Campbell J.W., Hao Q., Harding M.M., Nguti N.D., Wilkinson C.. LAUEGEN version 6.0 and INTLDM. J. Appl. Cryst. 1998; 31:496–502. [Google Scholar]
- 43. Helliwell J.R., Habash J., Cruickshank D.W.J., Harding M.M., Greenhough T.J., Campbell J.W., Clifton I.J., Elder M., Machin P.A., Papiz M Z.et al.. The recording and analysis of synchrotron X-radiation Laue diffraction photographs. J. Appl. Cryst. 1989; 22:483–497. [Google Scholar]
- 44. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Afonine P.V., Mustyakimov M., Grosse-Kunstleve R.W., Moriarty N. W., Langan P., Adams P.D.. Joint X-ray and neutron refinement with phenix.refine. Acta Cryst. D. 2010; 66:1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Cryst. D. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dauter Z., Adamiak D.A.. Anomalous signal of phosphorus used for phasing DNA oligomer: importance of data redundancy. Acta Cryst. D. 2001; 57:990–995. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structure have been deposited with the Protein Data bank under accession number 7JY2.