Abstract
Tumours with mutations in the BRCA1/BRCA2 genes have impaired double-stranded DNA break repair, compromised replication fork protection and increased sensitivity to replication blocking agents, a phenotype collectively known as ‘BRCAness’. Tumours with a BRCAness phenotype become dependent on alternative repair pathways that are error-prone and introduce specific patterns of somatic mutations across the genome. The increasing availability of next-generation sequencing data of tumour samples has enabled identification of distinct mutational signatures associated with BRCAness. These signatures reveal that alternative repair pathways, including Polymerase θ-mediated alternative end-joining and RAD52-mediated single strand annealing are active in BRCA1/2-deficient tumours, pointing towards potential therapeutic targets in these tumours. Additionally, insight into the mutations and consequences of unrepaired DNA lesions may also aid in the identification of BRCA-like tumours lacking BRCA1/BRCA2 gene inactivation. This is clinically relevant, as these tumours respond favourably to treatment with DNA-damaging agents, including PARP inhibitors or cisplatin, which have been successfully used to treat patients with BRCA1/2-defective tumours. In this review, we aim to provide insight in the origins of the mutational landscape associated with BRCAness by exploring the molecular biology of alternative DNA repair pathways, which may represent actionable therapeutic targets in in these cells.
INTRODUCTION
Modern DNA sequencing techniques have allowed systematic analyses of the seemingly random distribution of somatic passenger mutations across the cancer genome, revealing distinct patterns known as ‘mutational signatures’ (1). In general, mutational signatures are shaped by the joint effects of DNA damage and erroneous DNA damage repair (2). Based on size and complexity of the mutations, three classes of mutational signatures can be distinguished: single-base substitutions, small insertions and deletions (indels) and genomic rearrangements or structural variations (3). Single-base substitution signatures are based on six subtypes of single nucleotide substitutions (C>A, C>G, C>T, T>A, T>C and T>G). When viewed in the context of their 3′ and 5′ flanking nucleotides, these six substitutions generate 96 unique trinucleotide mutations (1). The ‘Catalogue Of Somatic Mutations In Cancer’ (COSMIC) currently lists more than 30 single-base substitution signatures, identified by the PCAWG consortium, across the spectrum of human cancers. Signatures based on indels and genomic rearrangements have been described as well, but the number of studies to characterize these more complex signatures have been relatively limited (4–6). Although the molecular backgrounds that underlie many mutational signatures are still unknown, some have been associated with specific types of DNA damage, such as tobacco smoke-induced and UV-induced DNA lesions, or with certain DNA maintenance of damage repair defects, including mismatch repair deficiency and homologous recombination (HR) deficiency.
Mono-allelic deleterious germline mutations in the DNA maintenance genes BRCA1 or BRCA2 predispose to hereditary breast and ovarian cancer (7–9). Loss of the second allele of BRCA1 or BRCA2, either through somatic mutations in BRCA1 or BRCA2 or through BRCA1 promoter hypermethylation, is considered a major oncogenic event that drives carcinogenesis in these individuals (7). Deleterious germline variants in genes acting in the same DNA maintenance pathways as BRCA1 and BRCA2, such as PALB2, RAD51B, RAD51C, RAD51D, XRCC2, XRCC3, and to a lesser extend BARD1, have also been associated with an increased breast and ovarian cancer risk (9–11). BRCA1/2-deficient tumours are characterized by high genomic instability and hypersensitivity to DNA damaging agents, in particular cisplatin and poly ADP-ribose polymerase (PARP) inhibitors (12–14). The BRCA1/2 proteins play essential roles in protecting cells against toxic double-stranded DNA breaks (DSBs) through HR, and thereby help preserve genome integrity. Additionally, BRCA1 and BRCA2 ensure faithful DNA replication through the protection of stalled replication forks (15). The replication fork protection functions of BRCA1 and BRCA2 become increasingly important during conditions of replication stress, a state of global replication fork slowing and stalling (16). Together, defects in HR repair, impaired fork protection, and the subsequent hypersensitivity to DNA damaging agents are referred to as ‘BRCAness’.
The BRCAness phenotype is reflected in multiple mutational signatures, including base substitution signatures SBS3 and SBS8, indel signatures ID6 and ID8, and rearrangement signatures RS1, RS3 and RS5 (Table 1) (4,5,17). The two most prominent mutations observed in the BRCAness mutational signatures include 4–50 bp indels flanked by short regions of sequence microhomology (reflected in signatures SBS3 and ID6), and small <10 kb tandem duplications (reflected in RS3, mostly prominent in BRCA1-deficient tumours) (4,5). Interestingly, many of the genomic scars observed in BRCA1/2-deficient tumours are also observed in tumours that do not show apparent mutations in one of the BRCA1/2 genes, indicating that the concept of BRCAness extends beyond BRCA1 and BRCA2 mutations (18). In line with this notion, BRCA-like tumours without known driver mutations may also respond favourably to PARP inhibitor treatment (5,19–21), and the identification of these tumours based on mutational signature analyses could therefore have important implications for the treatment of these patients. In this review, we will discuss various DNA maintenance pathways in the context of cell cycle regulation, thereby providing a better understanding of their contributions to the BRCAness mutational landscape.
Table 1.
Mutational signatures associated with BRCAness
| Signature | Characteristics | Affected genes | Aetiology | Reference |
|---|---|---|---|---|
| SBS3 | Uniform distribution of mutations across all 96 possible base substitution types | BRCA1, BRCA2, PALB2, RAD51C, CHK2 | Possibly associated with deletions introduced by Pol θ-mediated processing of DSBs | (4,79,81,220) |
| SBS8 | CC>AA double nucleotide substitutions | BRCA1, BRCA2 | Unknown | (4,79) |
| ID6 | ≥5 bp deletions, flanked by ≥2 bp microhomology at breakpoint junctions | BRCA2, PALB2, (BRCA1) | Pol θ-mediated processing of DSBs | (4,17,81) |
| ID8 | ≥5 bp deletions, flanked by 0–3 bp microhomology at breakpoint junctions | BRCA1, BRCA2 | NHEJ-mediated repair of DSBs, possibly a contribution of Pol θ-mediated repair | (4,17) |
| RS3 | 1–100 kb tandem duplications | BRCA1 | Pol θ-mediated processing of DSBs and stalled forks | (5,145) |
| RS5 | <100 kb deletions, flanked by >10 bp microhomology at breakpoint junctions | BRCA2, PALB2, (BRCA1) | SSA-mediated processing of DSBs | (5,79,81) |
Associated gene mutations that have been linked to mutational signature in cell lines or patients, and their corresponding genomic scars are indicated.
DOUBLE-STRANDED DNA BREAK REPAIR PATHWAY CHOICE: THE SWITCH BETWEEN HR AND NHEJ
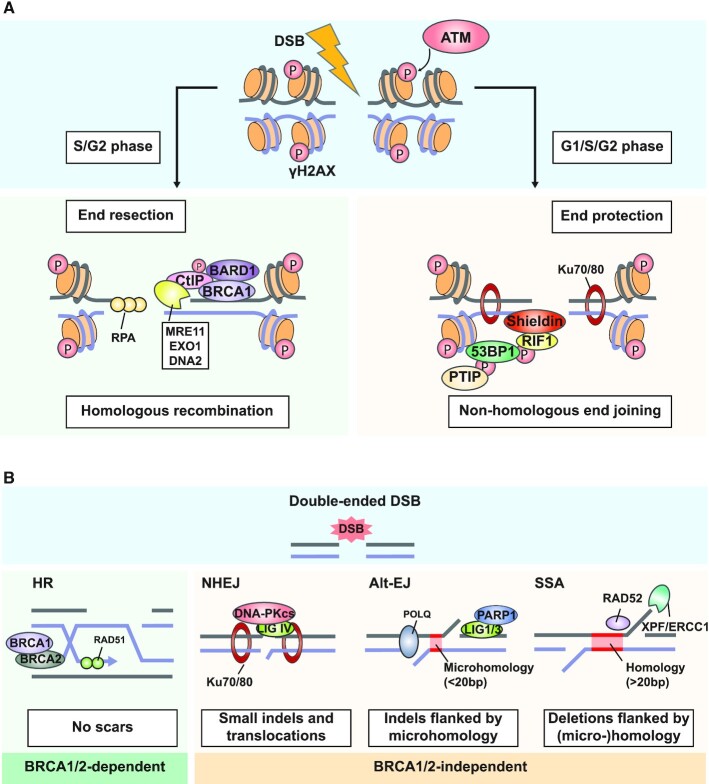
Historically, the best described functions of BRCA1 and BRCA2 are their roles in the repair of DSBs (22). Failure to efficiently process DSBs results in severe genomic instability, characterized by frequent chromosome breakage (23). Cells are equipped with a number of pathways for the repair of DSBs, including HR, non-homologous end-joining (NHEJ), single-strand annealing (SSA) and alternative end joining (Alt-EJ) (Figure 1A, B). NHEJ and HR are generally considered the two main pathways of choice for the repair of DSBs, with SSA and Alt-EJ providing back-up repair capacity (24). HR uses the intact sister chromatid as a template for the repair of DSBs, and relies on processing of the ends of the DSB to generate a 3′ single-stranded DNA overhang, a process known as ‘end resection’ (25). DSB repair by HR leaves no visible genomic scars and therefore fully preserves genome integrity, making it the preferred repair pathway. However, cells cannot use HR throughout the cell cycle, as the presence of sister chromatids is limited to S and G2-phase (26). In the absence of an intact repair template in G1-phase, cells mostly rely on non-homologous end joining (NHEJ) for the repair of DSBs.
Figure 1.
Repair pathways of DNA double stranded breaks and their effects on genome integrity. (A) DNA double stranded breaks (DSBs) are recognized by the ATM serine/threonine kinase which phosphorylates H2AX to γH2AX. Repair pathway choice is influenced by blocking or initiating end-resection. At the molecular level, pathway choice is determined by either 53BP1 or BRCA1 and CtIP. When 53BP1 is phosphorylated, PTIP, RIF1 and the Shieldin complex are recruited to the break-site and protect the two DNA ends from end resection, and conversely promote Ku70/80-mediated non-homologous end joining (NHEJ) repair. Phosphorylated CtIP forms a complex with BRCA1 and BARD1, promoting end resection by CtIP and the MRE11, EXO1 and DNA2 nucleases. The resulting stretches of single-stranded DNA (ssDNA) are subsequently coated by RPA. (B) BRCA1/2-proficient cells can utilize HR to resolve DSBs. Following BRCA1-promoted end resection, BRCA2 facilitates loading of RAD51, which replaces RPA. RAD51 catalyses strand invasion and homology search on the sister chromatid, allowing the error free repair. BRCA1/2-deficient cells can use BRCA-independent pathways: NHEJ, alternative end joining (Alt-EJ) and single-strand annealing (SSA). NHEJ requires Ku70/80 proteins, which facilitate the localization of DNA-PKcs and ligase IV (LIG IV) to non-resected ends, that ligate the DNA ends independently of sequence homology, frequently resulting in small deletions. During Alt-EJ, Pol θ catalyses the annealing of small sequences of microhomology (<20 bp) to promote ligation of resected ends, possibly requiring PARP1, Ligase 1 or Ligase 3. Alt-EJ repair results in deletion of the sequence located between regions with microhomology, and occasional Pol θ-mediated base insertions. SSA requires sequence homology of >20 bp, which are annealed by RAD52. The 3′ flaps are cleaved by the XPF/ERCC1 endonuclease complex resulting in deletions of the sequence in between the homology regions.
The switch between NHEJ to HR is regulated by 53BP1, which prevents DNA end resection (Figure 1A). Phosphorylation of 53BP1 by the ATM serine/threonine kinase promotes complex formation of 53BP1 with PTIP and RIF1 (27–30). RIF1 has been shown to directly interact with the SHLD1-SHLD2-SHLD3-REV7 (Shieldin) complex (31). Together with 53BP1, the shieldin complex protects DNA ends from end resection, by sterically shielding the DNA ends from nuclease activity, thereby promoting NHEJ (Figure 1A) (31). During S-phase, high CDK-levels catalyse the phosphorylation of CtIP, which promotes complex formation of BRCA1 and BARD1, and subsequently leads to the removal of the 53BP1-RIF1 complex from the break site and access of nucleases to DNA ends (27,28,32–35). Interestingly, BRCA1 is generally considered a key regulator of end resection (35–38), but as end resection has been observed in the absence of BRCA1, the exact requirement of BRCA1 is unclear (28,39–42). When the DNA ends at a DSB are not protected, they are processed by CtIP and the MRE11–RAD50–NBS1 (MRN), creating 3′ ssDNA overhangs (Figure 1A). Long-range DNA end resection is further catalysed by either the EXO1 endonuclease or the DNA2–BLM complex (Figure 1A) (25). Resected DSBs are no longer a substrate for NHEJ, and instead HR is now the preferred repair pathway.
The stretches of ssDNA generated by end resection are rapidly coated by RPA, protecting them from degradation. BRCA2 subsequently promotes the loading of the RAD51 recombinase, replacing RPA (43–45). The RAD51 molecules form nucleoprotein filaments, which catalyse invasion and homology search on the sister chromatid (Figure 1B). Once homology is found, DNA is synthesized using the homologous template, either via synthesis-dependent strand annealing (SDSA) or through the formation of a double Holliday junction (46). During SDSA, one end of the DSB is displaced and annealed onto the sister chromatid, allowing for DNA synthesis using the intact template DNA, exclusively resulting in non-crossover events (46,47). Alternatively, the second end of the DSB may be captured and annealed to the displaced template strand, resulting in the formation of a double Holliday junction, and allowing DNA synthesis in two directions across the break site (46,47). When synthesis is completed, the majority of Holliday junctions are ‘dissolved’ by the BLM-TOP3A-RMI complex, resulting in non-crossover events (48). Holliday junctions that are not processed by the BLM–TOP3A–RMI complex before S-phase is completed, are ‘resolved’ by the resolvases SLX1–SLX4, MUS81–EME1 or GEN1, which can result in crossover events, also called sister chromatid exchanges (SCEs) (46,47,49,50). Although HR is generally an error-free pathway, loss-of-heterozygosity may occur when the homologous chromosome is used as a template, followed by a crossover event (49,50).
ERRONEOUS DSB REPAIR IN BRCA1/2-DEFICIENT CELLS INTRODUCES INDELS FLANKED BY MICROHOMOLOGY
DSB repair in BRCA1/2-deficient cells relies entirely on the DSB repair pathways NHEJ, SSA and Alt-EJ, which are more error prone than HR (49). NHEJ is active during G1, S and G2 phase of the cell cycle, and is initiated by the binding of the KU-complex to the ends of the break, followed by activation of the DNA-PKcs kinase. Activation of DNA-PKcs sets the stage for blunt-end ligation of the DNA ends by ligase IV, independent of any sequence homology (51) (Figure 1A, B). In the presence of two compatible ends, NHEJ is able to repair a DSB without introducing errors (52). However, repair of DSBs by NHEJ may result in small deletions, which show no to very limited homology around the break sites (Figure 1B). Mutational signature ID8, characterized by small (<5 bp) deletions flanked by no to minor (>3 bp) microhomology at the break site, may be associated with DSB repair through NHEJ (Table 1) (4,17). ID8 is only mildly enriched in BRCA1/2-deficient tumours, indicating that NHEJ may only have a minor contribution to the repair of DSBs in the absence of HR (4,17,53).
Instead, BRCA1/2-deficient cells frequently show an enrichment for mutational signature SBS3, characterized by uniform base substitutions across all trinucleotides, and ID6, characterized by ≥ 5 bp deletions, commonly flanked by ≥2 bp microhomology at breakpoint junctions (Table 1) (4,17). The SBS3 and ID6 signatures have not only been identified in cells with mutant BRCA1 or BRCA2, but also in patient samples and cell line models with mutations in PALB2 or in the RAD51 paralogs RAD51B, RAD51C, RAD51D, XRCC2 and XRCC3 (10,54,55). SBS3 and ID6 are highly correlated, and may be the direct result of DSB repair by Alt-EJ, sometimes also referred to as theta-mediated end joining or microhomology-mediated end joining (4,17). The key enzyme in this pathway is DNA polymerase θ (POLQ), which contains both helicase and polymerase activity and can effectively catalyse the pairing of small microhomology regions within long stretches of ssDNA (Figure 1B). Like HR, Alt-EJ generally acts on resected ends with 15–100 nucleotide 3′ overhangs, generated by CtIP/MRE11-mediated end resection, which can occur in the absence of BRCA1 (56–58). DNA pol θ only requires 1–20 bp of microhomology to anneal the two ends of the DSB (56,59,60). PARP1 may play a role in the annealing of the ssDNA and the recruitment of other repair factors (61,62). In the final step, DNA ligase I and III may catalyse the ligation of the annealed ends (Figure 1B) (63). The exact roles of PARP1, DNA ligase I and III in Alt-EJ are however still unclear (64). Genomic scars left by Alt-EJ mostly consist of small deletions of 20–200 bp located in between the microhomology regions, which correlate well with the mutations observed in signature ID6 (4,17,65,66). Occasionally, Pol θ incorporates so-called template insertions (‘delins’) of 3–30 bp at the deletion junction, which are present at high frequency in tumours with BRCA1/2 mutations (65,67). As a result, DNA damage repaired by Pol θ leaves both deletions and insertions in the genome flanking the site of the DNA lesion (Figure 1B). Moreover, Pol θ is a low fidelity polymerase, and has a preference for inserting adenine opposite abasic sites, as well as a tendency for incorporating guanine or thymidine opposite a thymidine, thereby generating single base substitutions (68).
The large contribution of signatures SBS3 and ID6 to the BRCAness mutational landscape suggests that BRCA1/2-deficient tumours rely heavily on Alt-EJ for the repair of DSBs. Therefore, the Alt-EJ addiction of BRCA1/2-deficient tumours may provide interesting therapeutic options for cancers displaying BRCAness mutational signatures, for example through pharmacological inhibition of Pol θ. Indeed, BRCA1/2-deficient tumours display increased sensitivity to Pol θ inhibition, including those that show resistance to PARP inhibitor treatment (69–71). Notably, both the polymerase and helicase enzymatic domains of Pol θ could be targeted by small molecule inhibitors, providing multiple opportunities for the development of pharmacological inhibitors of Pol θ (72).
In addition to creating a dependence on Pol θ, loss of BRCA1 or BRCA2 is also synthetic lethal with RAD52, the main protein involved in SSA (73,74). The SSA pathway is similar to Alt-EJ in that it requires CtIP and the MRN complex for the processing of DNA ends. However, SSA involves more extensive end resection, and requires EXO1 or DNA2 nuclease activity (25). Moreover, SSA utilizes larger homology regions (>20 bp), which naturally occur throughout our genome and are sometimes referred to as ‘short interspersed nuclear elements’ (Figure 1B) (75). Annealing of the homology repeats is catalysed by the ssDNA-binding protein RAD52, followed by cleavage of the non-homologous 3′ flaps by the XPF-ERCC1 endonuclease complex (57). SSA competes with HR for the repair of resected breaks, and the BRCA1/PALB2 complex directly inhibits SSA activity (76). Similar to Alt-EJ, the process of SSA results in the formation of deletions between the homology regions, thereby leaving scars in the genome (75). Deletions induced by the SSA machinery are generally larger than those that arise as a result of Alt-EJ (77). Such deletions of up to 10 kb in size have indeed been described in BRCA1/2-deficient cells (78). Moreover, mutational signature RS5, which is characterized by large deletions with long (>10 bp) microhomologies that span short-interspersed nuclear elements and is likely the result of SSA activity (Table 1) (79), has been observed in both BRCA1 and BRCA2 deficient tumours (5,79,80). Remarkably, others described an absence of RS5 in BRCA1-deficient cells (81). As SSA is dependent on end-resection, these observations further point to BRCA1 being dispensable for end resection. Although the link between SSA and BRCAness at the mutation level has not been as well established as for Alt-EJ, the synthetic lethality between RAD52 and BRCA1/2 provides interesting therapeutic options, with RAD52 inhibitors currently being in development (82).
Overall, the activity of SSA and Alt-EJ may account for the mutations observed in the BRCAness signatures SBS3, ID6 and RS5. However, not all the observed mutations in BRCAness tumours can be explained solely by defects in DSB repair. In order to understand the complex genomic scars observed in BRCA1/2-deficient tumours, other processes should be considered, including the role of BRCA1 and BRCA2 during DNA replication.
SOURCES OF REPLICATION STRESS: A NEW CONTEXT FOR THE BRCA1 AND BRCA2 PROTEINS
Besides their functions in DSB repair, BRCA1, BRCA2 and RAD51 are key guardians of genome integrity in S-phase of the cell cycle, during which DNA replication takes place. DNA is replicated by the replisome, which consists of the DNA polymerases α, ϵ and δ, the Cdc45-MCM2–7-Gins (CMG) helicase complex and the PCNA DNA clamp, among others (83). Following DNA unwinding by MCM2–7 helicase, the polymerases incorporate deoxyribonucleotide triphosphate (dNTPs) into a newly synthesized strand of DNA, forming a three-way structure known as the replication fork (Figure 2A). At the leading strand, DNA synthesis occurs continuously by DNA polymerase ϵ in the 3′ to 5′ direction. On the lagging strand, DNA synthesis is performed discontinuously by DNA polymerase δ, generating short fragments known as ‘Okazaki fragments’ that require recurrent repriming of DNA synthesis.
Figure 2.
Sources of replication stress and the replication stress response. (A) Replication forks can be slowed down or halted by many different sources of DNA damage, including difficult to replicate loci, depletion of free nucleotides (dNTPs), DNA gaps, interstrand crosslinks (ICLs), secondary structures such as G4-structures, single-stranded DNA (ssDNA), DNA–RNA hybrids (R-loops or ribonucleotide incorporation), obstruction of the replication machinery by DNA–protein complexes (DPCs) and torsional stress. (B) Stalled replication forks expose ssDNA stretches, which are bound by RPA and activate a cell cycle checkpoint via ATRIP, ATR and CHK1. The stalled fork is reversed by fork remodellers SMARCAL1, HLTF and ZRANB3, and involves RAD51-mediated annealing of nascent strands. In BRCA1/2-proficient cells, RAD51-coated DNA stretches are protected against nucleolytic degradation by BRCA1 and BRCA2 to allow time for excision repair and fork restart. In BRCA1/2-deficient cells, protection of stalled fork is defective, which results in nucleolytic degradation of nascent DNA by MRE11, DNA2 or EXO1. Extended nucleolytic degradation leads to the collapse of the replication fork and formation of a single-ended DSB.
Conditions that result in slowing or stalling of the replication fork are collectively referred to as ‘replication stress’. The continuous unwinding of the DNA by the helicase after the polymerases have stalled, results in the accumulation of long stretches of fragile single-stranded DNA (ssDNA). In general, replication stress is caused by local nucleotide pool depletions or physical obstructions in the DNA that block replication forks (16). In vitro, these phenotypes can be mimicked using small molecules, such as hydroxyurea (HU), which depletes the nucleotide pool, or aphidicolin, which inhibits the DNA polymerase. Replication stress is considered a common driver of genome instability and may provide potential targets for therapeutic intervention (84). The BRCA1/2 proteins protect cells against replication stress, and BRCA1/2-deficient cells frequently display high levels of replication stress and DNA under-replication (85).
DNA replication origin are sequences in the genome at which the replisome assembles. During S-phase, licensed origins are activated (or ‘fired’) by the phosphorylation of the MCM2–7 helicase by Cyclin E1/CDK2 and CDC7/BDF4 (DDK) (86–89). The firing of the replication origins is tightly orchestrated by the ATR kinase, which functions as the conductor by determining when and which origins fire during S-phase (90). Overexpression of oncogenes, such as HPV-16 E6/E7, Cyclin E, MYC and H/KRAS (84) disturbs the timing of DNA replication through premature firing of replication origins (91–94). As a consequence of dysregulated origin firing, an excess of origins is fired simultaneously, resulting in local nucleotide pool depletions (Figure 2A) (92–94). Interestingly, Cyclin E1 overexpression is mutually exclusive with BRCA1/2 mutations, suggesting that the DNA lesions that arise upon oncogene-induced replication stress require BRCA1/2-mediated processing (95).
Some replication origins are located within gene bodies, and their untimely firing results in collisions of the replication and transcription machinery (91). Oncogene overexpression has also been demonstrated to directly enhance transcription (96,97), which further increases the number of collisions between the replication and transcription machinery (98,99). Upon collision, newly transcribed RNA can rehybridize with DNA behind the transcription complex, which results in the formation of RNA–DNA hybrid structures, referred to as R-loops (Figure 2A). R-loops expose a displaced ssDNA strand, which is vulnerable to DNA damage. Persistent R-loops block replication fork progression, providing a source of replication stress and ultimately resulting in genomic instability (100,101). BRCA2-deficient cells show an increase in R-loops, providing a potential source of replication stress in these tumours (102).
Replication stress can also occur independently of oncogene overexpression, for example by distortion of the DNA helix, thereby blocking replication fork progression. Examples of helix-distorting lesions include secondary DNA structures, interstrand crosslinks (ICLs), torsional stress and DNA-protein cross-links (DPCs) (Figure 2A). The formation of secondary structures in the DNA, such as G-quadruplexes (G4), often occurs at regions of the genome that are intrinsically more difficult to replicate, known as common fragile sites (CFSs). CFSs are hotspots for genomic scars and rearrangements, especially in cells with defective DNA damage repair and impaired fork protection (103). ICLs occur throughout the genome and result from endogenous sources, such as reactive aldehydes formed in the process of alcohol catabolism, but also from chemotherapeutic agents, such as cisplatin or mitomycin C (Figure 2A) (104). Torsional stress is another class of replication-blocking lesions, which often occurs as a result of transcription and replication machinery collisions, and requires the activity of topoisomerases to be resolved (105). The use of topoisomerase inhibitors, such as etoposide and doxorubicin, strongly increases the amount of unresolved torsional stress, resulting in increased replication stress (106). Some topoisomerase inhibitors, including those targeting topoisomerase I (TOP I), also trap the topoisomerase enzyme to the DNA, forming a DPC (107). Similarly, PARP inhibitors, such as olaparib, trap the enzyme PARP1 to the DNA, thereby blocking DNA replication (108,109) (Figure 2A). Additionally, PARP inhibitors may interfere with Okazaki fragment processing, hampering the progression of lagging strand synthesis (110,111), which also perturbs replication progression. Sensitivity to replication-blocking agents, including cisplatin and PARP inhibitors, is a defining feature of the BRCAness phenotype, and highlights the importance of the function of BRCA1 and BRCA2 in the protection against replication stress and the repair of the lesions that arise as a consequence of replication stress.
BRCA1 AND BRCA2 PROTECT STALLED REPLICATION FORKS
During DNA replication, BRCA1, BRCA2 and RAD51 play essential roles in the protection of stalled replication forks, independent of their functions in HR (85,112). Prolonged stalling of replication forks upon replication stress results in the accumulation of long stretches of ssDNA, which are vulnerable for breaking or forming secondary structures. To prevent this from occurring, these stretches of ssDNA are quickly covered by RPA (113). RPA then stimulates the binding of the ATR-ATRIP complex to stalled forks (114). Once localized to the stalled fork, ATR phosphorylates a large number of downstream targets, including CHK1, which prevents cell cycle progression. Moreover, in response to replication stress, ATR-CHK1 prevent firing of distant dormant origins, while activating those in the close vicinity of the blocked replication fork (115,116). The ATR-mediated cell cycle arrest provides time for resolving the replication-blocking lesion and restarting the forks, with BRCA1, BRCA2 and RAD51 as key players.
Protection and restart of stalled forks is initiated by RAD51-mediated annealing of the nascent strands to form a ‘chicken foot’-shaped reversed fork (Figure 2B) (117,118). This initial step in fork reversal occurs independently of BRCA2 (119,120), but requires fork remodellers, including SMARCAL1 (121,122), HLTF (123) and ZRANB3 (124). Subsequently, the RAD51-coated reversed DNA strands are protected by BRCA1 and BRCA2 against uncontrolled nucleolytic degradation by MRE11 (15,112), EXO1 (125) or DNA2 (126,127). The main goal of fork reversal may be the repositioning of the replication-blocking lesion into the double strand helix, allowing for excision repair (128). Additionally, reversed forks provide a starting point for HR-mediated fork restart (Figure 2B) (112,125,129). In the absence of BRCA1 or BRCA2, reversed forks are no longer protected, resulting in the nucleolytic degradation of nascent DNA by MRE11, EXO1 and DNA2 (15,112,125–127). Degradation of nascent DNA at forks exposes ssDNA stretches, which are prone to collapse into a one-ended DSB (Figure 2B) (130). Intriguingly, cells defective for fork protection did not show extensive accumulation of DSBs (85), suggesting that degradation of nascent DNA does not always lead to collapsed forks, or that compensatory pathways are involved in the repair of these single-ended DSBs, as described below.
The relative contributions of the HR and fork protection functions of BRCA1 and BRCA2 to the BRCAness phenotype are still debated, and various separation-of-function models have been designed to address this question (Table 2). Restoration of fork protection in cells deficient for HR, for example by inhibition of MRE11 or depletion of SMARCAL1, PARP1, PTIP, CHD4 or RADX, renders cells less sensitive to chemotherapy and PARP inhibition (131–136). In line with these findings, loss of fork protection in cells proficient for HR, for example through deletion of RIF1, increases cisplatin sensitivity and results in genome instability (127). Thus, the observations that cells expressing separation-of-function mutations that selectively impair fork protection displayed increased sensitivity to cisplatin (127), while restoration of fork protection in BRCA1/2-deficient cells resulted in reduced cisplatin sensitivity (131–136), suggest that fork protection—at least in part— determines response to DNA damaging agents. Indeed, restoration of fork protection in BRCA-deficient cells has been suggested as a potential mechanism for tumours to acquire PARP inhibitor resistance (137).
Table 2.
Overview of current separation-of-function models for HR and fork protection deficiency and their phenotypes related to genomic instability

|
‘HR’ = homologous recombination, ‘FP’ = fork protection, ‘+’ indicates proficiency, ‘–' indicates deficiency.
However, restoration of fork protection in HR-deficient cells usually does not completely restore cell viability (131–134), suggesting that loss of fork protection is not the major determining factor for cisplatin and PARP inhibitor response in BRCA1/2 mutant cells. Indeed, two studies with separation-of-function mutants indicated that loss of the HR-function of BRCA2 determines efficacy of DNA damaging drugs. Firstly, hamster cells expressing an HR-proficient but fork protection-deficient BRCA2 mutant (BRCA2S3291A) exhibit high levels of spontaneous chromosomal aberrations, but these cells are far less sensitive to cisplatin and PARP inhibition compared to cells with complete loss of BRCA2 (Table 2) (85,112). Secondly, mice with mutations in the Brca1-interacting protein Bard1 (Bard1S563F and Bard1K607A) fail to recruit Brca1 to stalled forks, which inhibits fork protection without compromising HR-mediated DNA repair (138). These cells did exhibit an accumulation of DNA damage but only showed mild sensitivity to PARP inhibition (138).
Overall, we hypothesize that both loss of HR and loss of fork protection contribute to genome instability, each leaving distinct scars in the genome that are a direct consequence of compensating DNA repair pathways. The capacity of these compensating DNA repair pathways likely determines the viability of BRCA1/2-deficient cells, depending on the context (e.g. untreated versus treatment with DNA damaging agents, including PARP inhibitors or cisplatin). Restoration of either HR or fork protection in BRCA1/2-deficient cells may relieve the dependence on the compensating DNA pathways, partly improving viability and reducing sensitivity to chemotherapeutic agents. Interestingly, recent efforts to extract mutational signature data from cell line models with DNA repair defects demonstrated the feasibility of validating BRCAness mutational signatures in cell lines (139). Such in-depth genomic analysis of the above-described separation-of-function models is warranted to shed light on the relative contribution of fork protection and HR deficiency to the mutational signatures observed in BRCAness tumours.
INTER-CHROMOSOMAL TRANSLOCATIONS RESULT FROM ERRONEOUS PROCESSING OF COLLAPSED REPLICATION FORKS
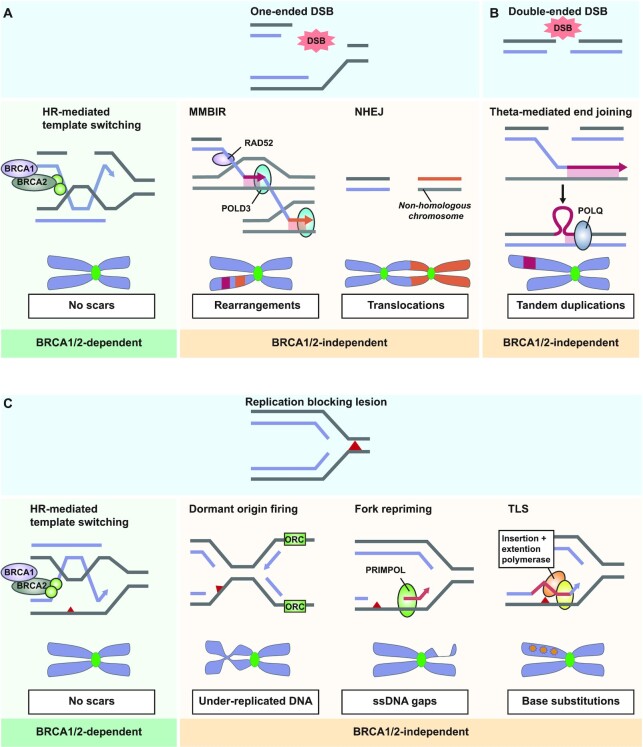
The collapse of replication forks results in the formation of one-ended DSBs that lack a proximal DNA end for ligation (Figure 3A). Due to the absence of the proximal second DNA end at the break site, these one-ended breaks are poor substrates for NHEJ (140–142). In BRCA1/2-proficient cells, one-ended DSBs can be repaired by RAD51-mediated template switching followed by synthesis-dependent repair, leaving no genomic scars (Figure 3A). It is therefore tempting to speculate that erroneous repair of these one-ended DSBs may be the source of the more complex rearrangements observed in BRCA1/2-deficient tumours. Indeed, erroneous recombination events at these collapsed forks in yeast, were shown to result in gross chromosomal rearrangements (143). Moreover, processing of one-ended DSBs by NHEJ in BRCA1/2-deficient cells results in chromosomal rearrangements and chromosome fusions (Figure 3A) (140–142). Moreover, inter-chromosomal translocations are frequently observed in BRCA1-deficient cells (78).
Figure 3.
Repair of DNA lesions resulting from stalled replication forks and their associated genomic scars. (A) Single-ended DSBs are repaired in BRCA1/2-proficient cells through error-free HR-mediated template switching. In BRCA1/2-deficient cells, microhomology-mediated break-induced replication (MMBIR) is used, which employs RAD52-mediated homology search and POLD3-dependent DNA synthesis. Multiple rounds of RAD52/POLD3-mediated template switching events lead to complex inter-chromosomal rearrangements. Repair of single-ended DSBs by NHEJ involves ligation of the DNA end with a nearby non-compatible DNA end, possibly resulting in chromosome fusions. (B) Pol θ-mediated strand invasion at single-ended or double-ended DSBs may result in the formation of tandem duplications. Limited end resection hampers reannealing of the invading strand, resulting in excess DNA synthesis, and consequently duplications flanked by areas with limited microhomology. (C) Replication-blocking lesions are repaired by HR-mediated template switching in BRCA1/2-proficient cells, which repairs the lesion in an error-free way. In absence of BRCA1/2, firing of dormant origins results in bypass of the lesion, leaving gaps of under-replicated DNA. Alternatively, repriming of the replication downstream of the lesion by PRIMPOL-mediated bypass leaves behind ssDNA gaps. Lastly, translesion synthesis (TLS) can be used to bypass the lesion, which incorporates random nucleotides across the site of lesion, resulting in base substitutions.
As an additional underlying mechanism, RAD52-dependent microhomology-mediated break-induced replication (MMBIR) has been implicated in the formation of these inter-chromosomal translocations (144). During MMBIR, the 3′ ssDNA overhang at the one-ended DSB anneals with a region of microhomology at any nearby ssDNA region, either at the same chromosome or at any nearby chromosome. Subsequently, POLD3-mediated DNA synthesis is performed using the intact DNA as a template. Successive rounds of MMBIR, involving multiple template switching events, may result in complex rearrangement patterns often observed BRCAness tumours, a process known as chromoanasynthesis (Figure 3A) (144). No unique mutational signature has been directly linked to chromoanasynthesis to this date.
POL Θ-MEDIATED DSB REPAIR PROMOTES THE FORMATION OF TANDEM DUPLICATIONS
In addition to inter-chromosomal translocations, BRCA1-deficient cells display <100 kb repeated sequences known as tandem duplications (53,145). Tandem duplications in BRCA1-deficient cells are often flanked by short regions of microhomology at the breakpoints, and are represented in mutational signature RS3 (Table 1) (78). Multiple models have been suggested for the formation of tandem duplications. In the simplest model, both sister chromatids are broken and erroneously fused together in a head-to-tail orientation by NHEJ or Pol θ-mediated end joining (145,146). Alternatively, tandem repeats may arise when end resection at two-ended DSBs is limited due to absence of BRCA1 (53). Resected ends with short 3′ ssDNA tails are a poor substrate for HR, but can be efficiently bound by Pol θ (58,147). In this model, Pol θ catalyses strand invasion of the sister chromatid, followed by DNA synthesis (53). Due to limited end resection at both ends of the break, the extended invaded strand fails to reanneal with the proximal DNA end (Figure 3B) (53). After some attempts, Pol θ may reanneal with the sister based on small microhomology, introducing duplications in one of the two sister chromatids (Figure 3B) (53).
A similar mechanism may be activated at stalled forks. Recent evidence demonstrated microhomology-mediated strand annealing may play an important role in repair of one-ended DSBs that arise from collapsed forks (148). A complex ‘restart-bypass’ model has been suggested that describes the formation of tandem repeats by Pol θ at stalled forks (145). In this model, a stalled fork encounters a fork approaching in opposite direction. Attempts by the stalled fork to restart using an upstream sequence, results in a small part of the DNA which is now synthesized by both the stalled fork and the approaching fork (145). When both forks collide, Pol θ may incorrectly fuse the duplicated piece of DNA, generating a tandem duplication (145). Despite the different starting points, the latter two models both require Pol θ-mediated end-joining, which is in agreement with the observed microhomology flanking the tandem repeats (5).
DAMAGE BYPASS INTRODUCES GAPS AND BASE SUBSTITUTIONS
In addition to repairing one-ended DSB repair, cells can also use DNA damage tolerance and bypass mechanisms to complete DNA replication. If nearby origins are present, replication can simply be re-initiated at nearby dormant origins downstream of the obstructive lesion, potentially leaving a small under-replicated region in the DNA (Figure 3C) (116,149–152). Alternatively, various translesion synthesis (TLS) polymerases allow completion of DNA replication by directly traversing a replication-blocking lesion. These polymerases lack proofreading activity and are intrinsically more error-prone than the canonical S-phase polymerase, resulting in a decrease in replication fidelity (153). A multitude of human TLS polymerases have been described, and can be divided into Y-family polymerases (Pol η, Pol ι, Pol κ and Rev1) and B-family polymerase (Pol ζ). Each TLS polymerase has different substrate specificities and nucleotide misincorporation rates (154). BRCA1/2-deficient cells increasingly rely on DNA damage tolerance and DNA bypass mechanisms to ensure completion of DNA synthesis before the onset of mitosis. Although TLS polymerases play an important role in lesion bypass and gap filling in BRCAness tumours (155), the base substitutions that are induced by TLS polymerases are not consistent with COSMIC signature SBS3 (156). It is tempting to speculate that less well-characterized signatures that show specific base substitutions, such as SBS8, may in fact be associated with TLS-like mechanisms.
Additionally, cells can replicate past an obstruction through de novo repriming of DNA synthesis (157). One of the proteins responsible for repriming is PRIMPOL, which contains both DNA primase and DNA polymerase activity (158,159). Repriming by PRIMPOL during replication may leave behind gaps of ssDNA in the DNA template. As a result, the mutation profile of PRIMPOL differs from that of Y- and B-family TLS polymerases, as it generates insertions and deletions rather than base misincorporations (158). Recent data demonstrated the presence of large amounts of ssDNA gaps in BRCA-deficient cells, especially upon treatment with replication stress-inducing agents (119,135). The accumulation of ssDNA gaps appears to be an important characteristic of the BRCAness phenotype, as suppression of gap formation in BRCA1/2-deficient cells, rescued the sensitivity to chemotherapeutic agents (135,136). PARP inhibitors also induce large amounts of ssDNA gaps (160), and PARP inhibitor response correlates well with ability of cells to suppress gap formation (160). Entering mitosis with under-replicated or damaged DNA is detrimental for faithful chromosome segregation, and an accumulation of ssDNA gaps may therefore promote mitotic failure.
MiDAS LIMITS GENOME INSTABILITY IN CELLS WITH HIGH LEVELS OF REPLICATION STRESS
Cells rely on cell cycle checkpoint kinases to prevent cells from prematurely entering into mitosis. In response to damaged or under-replicated DNA, ATR/CHK1 is activated to prevent mitotic entry and to allow time for filling in persistent DNA gaps in regions where replication stress occurred (161–163). However, limited amounts of gaps and breaks may escape detection and fail to sufficiently activate ATR, which prevents repair and causes these lesions to be transmitted into mitosis. As discussed above, BRCA-like tumours have compromised repair of S-phase lesions, and are therefore strongly dependent on a functioning ATR checkpoint to prevent entry into mitosis with under-replicated DNA. Indeed, HR-deficient tumours have elevated ATR activity (164), and inhibition of ATR in BRCA1/2-deficient tumours increases the amounts of DNA damage that gets transferred into mitosis (165).
During mitosis, most canonical DNA damage repair pathways are inactive (166–168). However, in the early stages of mitosis, DNA synthesis at regions of under-replicated DNA can be finalized by mitotic DNA synthesis (MiDAS) (169). Sites of MiDAS can be detected by the incorporation of the thymidine analogue EdU during mitosis (170). The MiDAS pathway uses a microhomology-based form of DNA synthesis, similar to the mechanism of break-induced replication to repair one-ended DSBs (169). POLD3 is the main polymerase responsible for DNA synthesis at collapsed forks in mitosis, highlighting the similarity with the break-induced replication pathway. Furthermore, MiDAS requires the SLX4 scaffold protein, the MUS81-EME1 endonuclease complex, the RAD52 recombinase and the RECQL5 helicase (169,171,172). FANCD2 is not directly required for the process of MiDAS, however FANCD2 localizes to MiDAS foci (172). Inhibition of the key components of the MiDAS pathway results in increased mitotic aberrations (172), underscoring the importance of MiDAS as a last resort mechanism to prevent loss of genetic information (173). Currently, it remains unclear whether MiDAS is error-free, and its contribution to the formation of genomic scars is not yet understood. Efforts to sequence MiDAS repair sites may elucidate the contribution of MiDAS to mutational landscapes in tumours (174).
Most HR proteins, including RAD51 and BRCA2, are not required for MiDAS directly, but prevent the accumulation of under-replicated DNA in S-phase (172). Consequently, BRCA2-deficient cells accumulate unresolved S-phase intermediates and show increased number of EdU foci in prometaphase (85). Moreover, PARP inhibitor treatment in S-phase increases FANCD2 foci in prometaphase in BRCA2-deficient cells (85,165,175). These observations support a role for MiDAS in dealing with unrepaired S-phase damage that results from BRCAness, thereby maintaining genome integrity throughout the remaining phases of mitosis.
S-PHASE LESIONS ARE TRANSFERRED INTO MITOSIS IN BRCA1/2-DEFICIENT CELLS
If MiDAS fails to resolve S-phase lesions during the early phases of mitosis, physical linkages and entanglements between chromosomes are persisting into anaphase (Figure 4) (169,172). If unresolved, these aberrancies may trigger mitotic cell death, sometimes referred to as ‘mitotic catastrophe’ (176). Generally, mitotic cells are resilient towards apoptosis, due to high concentrations of anti-apoptotic proteins (177). Mitotic aberrancies may prolong mitotic duration, allowing time for phosphorylation and degradation of the anti-apoptotic proteins, triggering cell cycle arrest pathways (178,179). BRCA1/2-deficient cells frequently manifest increased mitotic aberrancies, which were originally attributed to loss of a direct role for BRCA2 at regulating mitotic spindle integrity and cytokinesis (180–183). However, when BRCA1/2-deficient cells are treated with agents that induce S-phase-specific DNA damage, including HU or PARP inhibitors, the amount of anaphase bridges and lagging chromosomes strongly increased. These observations suggest that many mitotic aberrancies in BRCA1/2-deficient cells in fact originate from unrepaired S-phase damage (175,184,185). Using genome-wide synthetic lethality screens, BRCA1/2-deficient cells were recently shown to become dependent on CIP2A, which participates in tethering of mitotic DNA lesions (186). These findings underscore that mitotic DNA damage is a characteristic feature as well as a vulnerability of BRCA1/2-deficient cells.
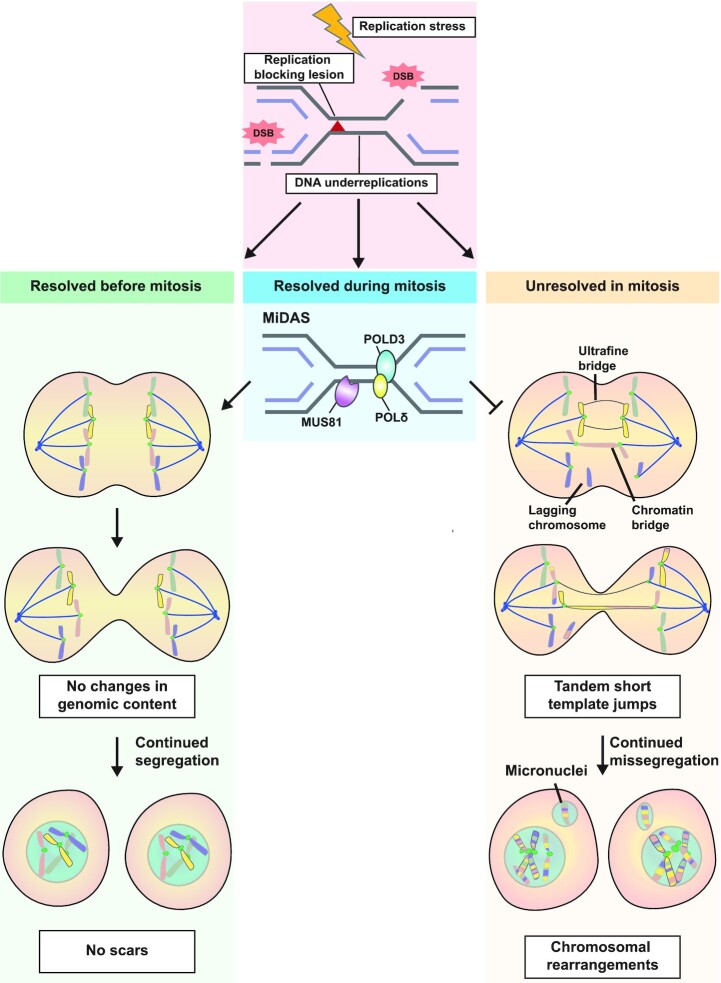
Figure 4.
Consequences of unresolved replication stress in mitosis. Cell are equipped with various pathways to resolve DNA lesions before the onset mitosis to ensure equal segregation of the DNA content over the emerging daughter cells. When cells enter mitosis with under-replicated DNA, POLD3-mediated Mitotic DNA synthesis (MiDAS) can be activated to synthesize new DNA during the early stages of mitosis, preventing further mitotic aberrations. Persistent DNA lesions induce the formation of chromosome bridges, lagging chromosomes or ultrafine DNA bridges. Breakage of these bridges introduces tandem short template (TST) jumps in the genome. Missegregated chromosomes can end up in micronuclei, where DNA synthesis is often compromised and causes induction of chromothripsis and complex intrachromosomal rearrangements.
Unresolved DNA lesions in mitosis appear as bulky chromatin bridges, ultrafine DNA bridges (UFBs) or lagging chromosome fragments (‘laggards’) in anaphase (Figure 4). Bulky DNA bridges are abnormal DNA structures that form physical linkages between the chromosome packs during mitosis, usually resulting from improper attachment of the mitotic spindle. DNA in bulky bridges remains packaged around histones and can be detected using conventional DNA dyes such as DAPI. Bulky DNA bridges can be a product of end-to-end fusions of multiple chromosome fragments by NHEJ or Alt-EJ, resulting in the formation of multi-centric chromosomes, as described above (Figure 3A) (187). Chromosome fusions are especially prevalent in cells with compromised telomere protection, a phenotype often observed in BRCA1/2-deficient cells (188–190). A chromatin bridge is formed when multi-centric chromosomes are linked to opposite spindle poles while remaining connected at the fused ends (187). Originally, bulky chromatin bridges were proposed to contribute to chromosomal instability through breakage-fusion-bridge (BFB) cycles (191,192). In this process, the connected chromosomes break unevenly as a result of the pulling forces in anaphase, resulting in unequal separation of chromosome fragments among the daughter cells. These unprotected chromosome fragments may again fuse in the next cell cycle, generating new multi-centric chromosomes that form new chromatin bridges in the subsequent anaphase (Figure 4) (192,193). The degree to which bulky DNA bridges break in mitosis is debated, with recent evidence suggesting that many DNA bridges remain intact during anaphase (194). The fused chromosomes may lag and become segregated into one of the two daughter cells, resulting in whole-chromosome aneuploidy (195). Classically, BFB cycles have been associated with intrachromosomal fusions, which leave scars known as fold-back inversions (196). Fold-back inversions, characterized by head-to-head inter-chromosomal rearrangements of duplicated segments, have been observed in BRCA1/2-deficient cells, but are not strongly enriched (197). Mutations that arise from BFB cycles are often more complex, comprising of ∼200 bp insertions termed ‘Tandem Short Template (TST) jumps’ at the break sites, combined with massive chromosomal rearrangements caused by chromothripsis (194). Chromothripsis involves the pulverization and subsequent random re-ligation of DNA fragments, and may be promoted by erroneous rounds of DNA replication at broken chromosome ends in the S-phase following bridge breakage (Figure 4) (194). BRCA1/2-deficient cells are characterized by large amounts of bulky chromatin bridges and micronuclei, and consequently, loss of BRCA1 or BRCA2 is associated with an increased frequency of chromothripsis (198). The complex architecture of the rearrangements in genomes subjected to chromothripsis, which differ significantly from the reference genome, make it difficult to define distinct mutational signatures associated with the phenomenon. Moreover, although chromothripsis may play a role in shaping the genome of BRCA1/2-deficient cancers, chromothripsis-associated mutations may not be exclusive to BRCAness tumours only. Chromothripsis has for example been linked with local hypermutation (‘kataegis’) a phenotype observed in signatures SBS2 and SBS13, which are not enriched in BRCA1/2-deficient tumours (199,200).
Like bulky DNA bridges, lagging chromosomes are the result of an improper spindle assembly checkpoint. Whole-chromosome lagging chromosomes often result from spindle assembly checkpoint defects, whereas acentric chromosome fragments often result from DNA breaks and DNA damage defects. BRCA2-deficiciency results in both an increase in DNA breaks and spindle assembly errors, resulting in the presence of both classes of lagging chromosomes (23). Fragments of broken chromosomes that lack centromeres do not attach to the mitotic spindle, but migrate with chromosome packs to daughter cells (Figure 4). As a consequence, lagging chromosomes separate randomly into either of the two daughter cells, resulting to loss of genetic information in one of the daughters and a gain in the other (201). Lagging chromosomes may present themselves as micronuclei in the following G1 phase (Figure 4). The chromatin in micronuclei is not properly organized, and does not adequately support DNA replication and repair. As a consequence, micronuclei containing chromosome fragments are generally very unstable and may result in chromothripsis in the subsequent S-phase (192,201). Like with bulky bridges, these chromothripsis events will eventually result in major genetic alterations (194) (Figure 4).
A final class of mitotic aberrancies are UFBs, which differ from bulky chromatin bridges in their lack of histones. They can only be detected using immunofluorescence analysis of UFB-binding proteins, such as PICH, BLM and RIF1 (202,203). Different types of UFBs have been described which differ in their genomic location, including centromeric UFBs, telomeric UFBs and CFS-associated UFBs (203). Recently, a new class of UFBs has been described that results from accumulation of recombination intermediates that join the sister chromatids (204,205). In HR-deficient cells, UFBs are often associated with under-replicated DNA at CFSs resulting from high levels of replication stress. UFBs occur directly at sites of prolonged fork stalling, and often show signs of persistent ssDNA (184,206,207). It is unclear how UFBs are processed exactly, and some evidence suggests that broken UFBs may end up in so-called ‘53BP1 bodies’ in the subsequent G1 phase (208–210). In addition to the broken DNA, these 53BP1 bodies contain various DNA damage markers (208,209), and are shielded from erroneous repair by NHEJ in G1 phase to promote a new chance of completing DNA synthesis at this region in the subsequent S-phase when HR proteins are present (211). It is currently unknown whether or how breakage of UFBs results in genome instability, and it is therefore not yet possible to find genomic scars of UFBs.
CONCLUDING REMARKS
The discovery that specific DNA damage repair defects translate to distinct patterns of genomic scars, allows characterization of tumours based on whole genome sequencing (WGS) data. This is especially interesting in the context of BRCAness, since it opens up possibilities to identify tumours with the BRCAness phenotype, but that lack mutations in BRCA1, BRCA2 or other HR genes. Indeed, the BRCAness-associated signature SBS3 is not exclusively found in tumours with mutations in BRCA1 or BRCA2, and provides a good predictive marker for PARP inhibitor sensitivity (81). However, some unexplained differences in chemotherapy response may still exist between true BRCA1/2-deficient tumours and tumours that show the BRCAness phenotype without mutations in the BRCA1/2 genes (212). Nowadays, two major algorithms are available to predict BRCAness, known as HRDectect (5) and CHORD (213). Both HRDetect and CHORD algorithms have proven to be useful for the selection of patients eligible for PARP inhibitor treatment (5,213). For example, the HRDetect algorithm recently identified patients with triple-negative breast cancer (TNBC) that would benefit from PARP inhibitor treatment using mutational signature analysis of whole genome sequencing (WGS) data (19,214). A current limitation in the use of genome-wide mutational signature mapping in patient tumour samples is the relatively low number of passenger mutations that can be detected (215) and the costs required for WGS. A new computational tool called Signature Multivariate Analysis (SigMA) shifts the focus back from whole genome sequencing to the analysis of targeted gene panels combined with machine-learning techniques to detect mutational signatures similar to signature SBS3, possibly reducing the costs (215). Most studies on PARP inhibitors now focus on breast, ovarian and prostate cancer, but our understanding of mutational signatures will aid treatment decision and mechanistic insight into the tumorigenesis of other cancer types. For example, some Ewing's sarcomas were demonstrated to phenocopy BRCA1-deficient tumours, despite being proficient for BRCA1 itself (20). These Ewing's sarcomas are highly sensitive to olaparib (20), and we hypothesize that these tumours will show similar mutational profiles as true BRCA-deficient tumours.
As discussed above, mutations in BRCA1/2-deficient tumours are the result of alternative repair of replication stress-associated DNA damage and the consequential problems of unresolved replication intermediates in mitosis, indicating that induction of replication stress may be key in the induction of cell death in BRCA-deficient tumours. Moreover, the genomic scars resulting from the increased dependence of BRCAness tumours on alternative repair pathways indicate that targeting these pathways, for example through RAD52 or Pol θ inhibition, may further sensitize BRCAness tumours to DNA damaging agents (216,217). Moreover, forcing cells with DNA damage into mitosis before DNA lesions are resolved, for example by inhibition of ATR, may provide an alternative therapeutic vulnerability (165,218,219). Our understanding of mutational signatures therefore not only has the potential to expand patient selection possibilities, but may also aid in the development of new therapeutic strategies.
ACKNOWLEDGEMENTS
We are grateful to van Vugt Lab members for constructive comments and the fruitful discussions in het ‘Noorderplantsoen’ during COVID-19 times.
Contributor Information
Colin Stok, Department of Medical Oncology, University Medical Center Groningen, University of Groningen, Hanzeplein 1, 9713GZ, Groningen, The Netherlands.
Yannick P Kok, Department of Medical Oncology, University Medical Center Groningen, University of Groningen, Hanzeplein 1, 9713GZ, Groningen, The Netherlands.
Nathalie van den Tempel, Department of Medical Oncology, University Medical Center Groningen, University of Groningen, Hanzeplein 1, 9713GZ, Groningen, The Netherlands.
Marcel A T M van Vugt, Department of Medical Oncology, University Medical Center Groningen, University of Groningen, Hanzeplein 1, 9713GZ, Groningen, The Netherlands.
FUNDING
Netherlands Organization for Scientific Research [NWO-VIDI 917.13334 to M.A.T.M.v.V.]; European Research Council [ERC-Consolidator grant ‘TENSION’ to M.A.T.M.v.V.]. Funding for open access charge: ERC.
Conflict of interest statement. M.A.T.M.v.V. has acted on the Scientific Advisory Board of Repare Therapeutics, which is unrelated to this work. The other authors declare no conflict of interest.
REFERENCES
- 1. Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A.J.R., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.-L.et al.. Signatures of mutational processes in human cancer. Nature. 2013; 500:415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Volkova N.V., Meier B., González-Huici V., Bertolini S., Gonzalez S., Vöhringer H., Abascal F., Martincorena I., Campbell P.J., Gartner A.et al.. Mutational signatures are jointly shaped by DNA damage and repair. Nat. Commun. 2020; 11:2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Helleday T., Eshtad S., Nik-Zainal S.. Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 2014; 15:585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alexandrov L., Kim J., Haradhvala N., Huang M.N., Ng A.W., Wu Y., Boot A., Covington K., Gordenin D., Bergstrom E.et al.. The repertoire of mutational signatures in human cancer. Nature. 2020; 578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davies H., Glodzik D., Morganella S., Yates L.R., Staaf J., Zou X., Ramakrishna M., Martin S., Boyault S., Sieuwerts A.M.et al.. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat. Med. 2017; 23:517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yi K., Ju Y.S.. Patterns and mechanisms of structural variations in human cancer. Exp. Mol. Med. 2018; 50:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Antoniou A., Pharoah P.D.P., Narod S., Risch H.A., Eyfjord J.E., Hopper J.L., Loman N., Olsson H., Johannsson O., Borg Å.et al.. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am. J. Hum. Genet. 2003; 72:1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rahman N., Seal S., Thompson D., Kelly P., Renwick A., Elliott A., Reid S., Spanova K., Barfoot R., Chagtai T.et al.. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat. Genet. 2007; 39:165–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Antoniou A.C., Casadei S., Heikkinen T., Barrowdale D., Pylkäs K., Roberts J., Lee A., Subramanian D., De Leeneer K., Fostira F.et al.. Breast-cancer risk in families with mutations in PALB2. N. Engl. J. Med. 2014; 371:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Golmard L., Castéra L., Krieger S., Moncoutier V., Abidallah K., Tenreiro H., Laugé A., Tarabeux J., Millot G.A., Nicolas A.et al.. Contribution of germline deleterious variants in the RAD51 paralogs to breast and ovarian cancers. Eur. J. Hum. Genet. 2017; 25:1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Suszynska M., Kluzniak W., Wokolorczyk D., Jakubowska A., Huzarski T., Gronwald J., Debniak T., Szwiec M., Ratajska M., Klonowska K.et al.. Bard1 is a low/moderate breast cancer risk gene: Evidence based on an association study of the central European p.q564x recurrent mutation. Cancers. 2019; 11:740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T.. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005; 434:913–917. [DOI] [PubMed] [Google Scholar]
- 13. Farmer H., McCabe N., Lord C., Tutt A., Johnson D., Richardson T., Santarosa M., Dillon K.J., Hickson I., Knights C.et al.. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005; 434:917–921. [DOI] [PubMed] [Google Scholar]
- 14. Fong P.C., Boss D.S., Yap T.A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O’Connor M.J.et al.. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009; 361:123–134. [DOI] [PubMed] [Google Scholar]
- 15. Schlacher K., Wu H., Jasin M.. A distinct replication fork protection pathway connects fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012; 22:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zeman M.K., Cimprich K.A.. Causes and consequences of replication stress. Nat. Cell Biol. 2014; 16:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hwang T., Reh S., Dunbayev Y., Zhong Y., Takata Y., Shen J., McBride K.M., Murnane J.P., Bhak J., Lee S.et al.. Defining the mutation signatures of DNA polymerase θ in cancer genomes. NAR Cancer. 2020; 2:zcaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Menghi F., Barthel F.P., Yadav V., Tang M., Ji B., Tang Z., Carter G.W., Ruan Y., Scully R., Verhaak R.G.W.et al.. The tandem duplicator phenotype is a prevalent genome-wide cancer configuration driven by distinct gene mutations. Cancer Cell. 2018; 34:197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chopra N., Tovey H., Pearson A., Cutts R., Toms C., Proszek P., Hubank M., Dowsett M., Dodson A., Daley F.et al.. Homologous recombination DNA repair deficiency and PARP inhibition activity in primary triple negative breast cancer. Nat. Commun. 2020; 11:2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gorthi A., Romero J.C., Loranc E., Cao L., Lawrence L.A., Goodale E., Iniguez A.B., Bernard X., Masamsetti V.P., Roston S.et al.. EWS-FLI1 increases transcription to cause R-Loops and block BRCA1 repair in Ewing sarcoma. Nature. 2018; 555:387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lips E.H., Mulder L., Oonk A., Van Der Kolk L.E., Hogervorst F.B.L., Imholz A.L.T., Wesseling J., Rodenhuis S., Nederlof P.M.. Triple-negative breast cancer: BRCAness and concordance of clinical features with BRCA1-mutation carriers. Br. J. Cancer. 2013; 108:2172–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roy R., Chun J., Powell S.N.. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer. 2012; 12:68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tutt A., Gabriel A., Bertwistle D., Connor F., Paterson H., Peacock J., Ross G., Ashworth A.. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr. Biol. 1999; 9:1107–1110. [DOI] [PubMed] [Google Scholar]
- 24. Wang H., Wang H., Rosidi B., Rosidi B., Perrault R., Perrault R., Wang M., Wang M., Zhang L., Zhang L.et al.. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005; 65:4020–4030. [DOI] [PubMed] [Google Scholar]
- 25. Cejka P. DNA end resection: Nucleases team up with the right partners to initiate homologous recombination. J. Biol. Chem. 2015; 290:22931–22938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johnson R.D., Jasin M.. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000; 290:22931–22938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bothmer A., Robbiani D.F., Di Virgilio M., Bunting S.F., Klein I.A., Feldhahn N., Barlow J., Chen H.T., Bosque D., Callen E.et al.. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol. Cell. 2011; 42:319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Escribano-Díaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T.F., Tkáč J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D.et al.. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 2013; 49:872–883. [DOI] [PubMed] [Google Scholar]
- 29. Zimmermann M., Lottersberger F., Buonomo S.B., Sfeir A., De Lange T.. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 2013; 339:700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Di Virgilio M., Callen E., Yamane A., Zhang W., Jankovic M., Gitlin A.D., Feldhahn N., Resch W., Oliveira T.Y., Chait B.T.et al.. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013; 339:711–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Noordermeer S.M., Adam S., Setiaputra D., Barazas M., Pettitt S.J., Ling A.K., Olivieri M., Álvarez-Quilón A., Moatti N., Zimmermann M.et al.. The shieldin complex mediates 53BP1-dependent DNA repair. Nature. 2018; 560:117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Greenberg R.A., Sobhian B., Pathania S., Cantor S.B., Nakatani Y., Livingston D.M.. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 2006; 20:34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P.. Human CtIP promotes DNA end resection. Nature. 2007; 450:509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen L., Nievera C.J., Lee A.Y.L., Wu X. Cell cycle-dependent complex formation of BRCA1·CtIP·MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008; 283:7713–7720. [DOI] [PubMed] [Google Scholar]
- 35. Bunting S.F., Callén E., Wong N., Chen H.T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L.et al.. 53BP1 inhibits homologous recombination in brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010; 141:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schlegel B.P., Jodelka F.M., Nunez R.. BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res. 2006; 66:5181–5189. [DOI] [PubMed] [Google Scholar]
- 37. Densham R.M., Garvin A.J., Stone H.R., Strachan J., Baldock R.A., Daza-Martin M., Fletcher A., Blair-Reid S., Beesley J., Johal B.et al.. Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat. Struct. Mol. Biol. 2016; 23:647–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu G., Ross Chapman J., Brandsma I., Yuan J., Mistrik M., Bouwman P., Bartkova J., Gogola E., Warmerdam D., Barazas M.et al.. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature. 2015; 521:541–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Patel D.S., Misenko S.M., Her J., Bunting S.F.. BLM helicase regulates DNA repair by counteracting RAD51 loading at DNA double-strand break sites. J. Cell Biol. 2017; 216:3521–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Polato F., Bunting S., Wong N., Chen H.-T., Kozak M., Kruhlak M., Reczek C., Lee W., Baer R., Ludwig T.et al.. CtIP-mediated resection is essential for viability and can operate independently of BRCA1. J. Cell Biol. 2014; 205:1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schmidt C.K., Galanty Y., Sczaniecka-Clift M., Coates J., Jhujh S., Demir M., Cornwell M., Beli P., Jackson S.P.. Systematic E2 screening reveals a UBE2D-RNF138-CtIP axis promoting DNA repair. Nat. Cell Biol. 2015; 17:1458–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ochs F., Somyajit K., Altmeyer M., Rask M.B., Lukas J., Lukas C.. 53BP1 fosters fidelity of homology-directed DNA repair. Nat. Struct. Mol. Biol. 2016; 23:714–721. [DOI] [PubMed] [Google Scholar]
- 43. Liu J., Doty T., Gibson B., Heyer W.D.. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010; 17:1260–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. San Filippo J., Sung P., Klein H.. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008; 77:229–257. [DOI] [PubMed] [Google Scholar]
- 45. Thorslund T., McIlwraith M.J., Compton S.A., Lekomtsev S., Petronczki M., Griffith J.D., West S.C.. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 2010; 17:1263–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Scully R., Panday A., Elango R., Willis N.A.. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019; 20:698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nassif N., Penney J., Pal S., Engels W.R., Gloor G.B.. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 1994; 14:1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu L., Hickson I.O.. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003; 426:870–874. [DOI] [PubMed] [Google Scholar]
- 49. Ceccaldi R., Rondinelli B., D’Andrea A.D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016; 26:52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sarbajna S., Davies D., West S.C.. Roles of SLX1-SLX4, MUS81-EME1, and GEN1 in avoiding genome instability and mitotic catastrophe. Genes Dev. 2014; 28:1124–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010; 79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bétermier M., Bertrand P., Lopez B.S.. Is non-homologous end-joining really an inherently error-prone process?. PLos Genet. 2014; 10:e1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kamp J.A., van Schendel R., Dilweg I.W., Tijsterman M.. BRCA1-associated structural variations are a consequence of polymerase theta-mediated end-joining. Nat. Commun. 2020; 11:3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Póti Á., Gyergyák H., Németh E., Rusz O., Tóth S., Kovácsházi C., Chen D., Szikriszt B., Spisák S., Takeda S.et al.. Correlation of homologous recombination deficiency induced mutational signatures with sensitivity to PARP inhibitors and cytotoxic agents. Genome Biol. 2019; 20:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li A., Geyer F.C., Blecua P., Lee J.Y., Selenica P., Brown D.N., Pareja F., Lee S.S.K., Kumar R., Rivera B.et al.. Homologous recombination DNA repair defects in PALB2-associated breast cancers. npj Breast Cancer. 2019; 5:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017; 18:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Verma P., Greenberg R.A.. Noncanonical views of homology-directed DNA repair. Genes Dev. 2016; 30:1138–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yun M.H., Hiom K.. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009; 459:460–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Roerink S.F., Schendel R., Tijsterman M.. Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome Res. 2014; 24:954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kelso A.A., Lopezcolorado F.W., Bhargava R., Stark J.M.. Distinct roles of RAD52 and POLQ in chromosomal break repair and replication stress response. PLos Genet. 2019; 15:e1008319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Audebert M., Salles B., Calsou P.. Effect of double-strand break DNA sequence on the PARP-1 NHEJ pathway. Biochem. Biophys. Res. Commun. 2008; 369:982–988. [DOI] [PubMed] [Google Scholar]
- 62. Chan S.H., Yu A.M., McVey M.. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLos Genet. 2010; 6:e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Paul K., Wang M., Mladenov E., Bencsik-Theilen A., Bednar T., Wu W., Arakawa H., Iliakis G.. DNA ligases I and III cooperate in alternative non-homologous end-joining in vertebrates. PLoS One. 2013; 8:e59505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zelensky A.N., Schimmel J., Kool H., Kanaar R., Tijsterman M.. Inactivation of Pol θ and C-NHEJ eliminates off-target integration of exogenous DNA. Nat. Commun. 2017; 8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. van Schendel R., van Heteren J., Welten R., Tijsterman M.. Genomic scars generated by polymerase theta reveal the versatile mechanism of alternative end-joining. PLoS Genet. 2016; 12:e1006368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. McVey M., Lee S.E.. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008; 24:529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Carvajal-Garcia J., Cho J.E., Carvajal-Garcia P., Feng W., Wood R.D., Sekelsky J., Gupta G.P., Roberts S.A., Ramsden D.A.. Mechanistic basis for microhomology identification and genome scarring by polymerase theta. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:8476–8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Seki M., Masutani C., Yang L.W., Schuffert A., Iwai S., Bahar I., Wood R.D.. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004; 23:4484–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhou J., Gelot C., Pantelidou C., Li A., Yucel H., Davis R.E., Farkkila A., Kochupurakkal B., Syed A., Shapiro G.I.et al.. Polymerase theta inhibition kills homologous recombination deficient tumors. 2020; bioRxiv doi:26 May 2020, preprint: not peer reviewed 10.1101/2020.05.23.111658. [DOI] [PMC free article] [PubMed]
- 70. Dai C.H., Chen P., Li J., Lan T., Chen Y.C., Qian H., Chen K., Li M.Y.. Co-inhibition of pol θ and HR genes efficiently synergize with cisplatin to suppress cisplatin-resistant lung cancer cells survival. Oncotarget. 2016; 7:65157–65170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mateos-Gomez P.A., Gong F., Nair N., Miller K.M., Lazzerini-Denchi E., Sfeir A.. Mammalian polymerase theta promotes alternative-NHEJ and suppresses recombination. Nature. 2015; 518:254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wood R.D., Doublié S.. DNA polymerase θ (POLQ), double-strand break repair, and cancer. DNA Repair (Amst). 2016; 44:22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Toma M., Sullivan-Reed K., Śliwiński T., Skorski T.. RAD52 as a potential target for synthetic lethality-based anticancer therapies. Cancers (Basel). 2019; 11:1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Feng Z., Scott S.P., Bussen W., Sharma G.G., Guo G., Pandita T.K., Powell S.N.. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bhargava R., Onyango D.O., Stark J.M.. Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 2016; 32:566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Anantha R.W., Simhadri S., Foo T.K., Miao S., Liu J., Shen Z., Ganesan S., Xia B.. Functional and mutational landscapes of BRCA1 for homology-directed repair and therapy resistance. Elife. 2017; 6:e21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Reh W.A., Nairn R.S., Lowery M.P., Vasquez K.M.. The homologous recombination protein RAD51D protects the genome from large deletions. Nucleic Acids Res. 2017; 45:1835–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lal A., Ramazzotti D., Weng Z., Liu K., Ford J.M., Sidow A.. Comprehensive genomic characterization of breast tumors with BRCA1 and BRCA2 mutations. BMC Med. Genomics. 2019; 12:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nik-Zainal S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X., Martincorena I., Alexandrov L.B., Martin S., Wedge D.C.et al.. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016; 534:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Vanderstichele A., Busschaert P., Olbrecht S., Lambrechts D., Vergote I.. Genomic signatures as predictive biomarkers of homologous recombination deficiency in ovarian cancer. Eur. J. Cancer. 2017; 86:5–14. [DOI] [PubMed] [Google Scholar]
- 81. Póti Á., Gyergyák H., Németh E., Rusz O., Tóth S., Kovácsházi C., Chen D., Szikriszt B., Spisák S., Takeda S.et al.. Correlation of homologous recombination deficiency induced mutational signatures with sensitivity to PARP inhibitors and cytotoxic agents. Genome Biol. 2019; 20:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sullivan K., Cramer-Morales K., McElroy D.L., Ostrov D.A., Haas K., Childers W., Hromas R., Skorski T.. Identification of a Small Molecule Inhibitor of RAD52 by Structure-Based Selection. PLoS One. 2016; 11:e0147230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Burgers P.M.J., Kunkel T.A.. Eukaryotic DNA replication fork. Annu. Rev. Biochem. 2017; 86:417–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Schoonen P.M., Guerrero Llobet S., van Vugt M.A.T.M.. Replication stress: driver and therapeutic target in genomically instable cancers. Adv. Protein Chem. Struct. Biol. 2019; 115:157–201. [DOI] [PubMed] [Google Scholar]
- 85. Feng W., Jasin M.. BRCA2 suppresses replication stress-induced mitotic and G1 abnormalities through homologous recombination. Nat. Commun. 2017; 8:525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tanaka T., Knapp D., Kim N.. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997; 90:649–660. [DOI] [PubMed] [Google Scholar]
- 87. Heller R.C., Kang S., Lam W.M., Chen S., Chan C.S., Bell S.P.. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011; 146:80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tanaka S., Umemori T., Hirai K., Muramatsu S., Kamimura Y., Araki H.. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007; 445:328–332. [DOI] [PubMed] [Google Scholar]
- 89. Moyer S.E., Lewis P.W., Botchan M.R.. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. U. S. A. 2006; 103:10236–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fragkos M., Ganier O., Coulombe P., Méchali M.. DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol. 2015; 16:360–374. [DOI] [PubMed] [Google Scholar]
- 91. Macheret M., Halazonetis T.D.. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature. 2018; 555:112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bester A.C., Roniger M., Oren Y.S., Im M.M., Sarni D., Chaoat M., Bensimon A., Zamir G., Shewach D.S., Kerem B.. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011; 145:435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Maya-Mendoza A., Ostrakova J., Kosar M., Hall A., Duskova P., Mistrik M., Merchut-Maya J.M., Hodny Z., Bartkova J., Christensen C.et al.. Myc and Ras oncogenes engage different energy metabolism programs and evoke distinct patterns of oxidative and DNA replication stress. Mol. Oncol. 2015; 9:601–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Srinivasan S.V., Dominguez-Sola D., Wang L.C., Hyrien O., Gautier J.. Cdc45 is a critical effector of Myc-dependent DNA replication stress. Cell Rep. 2013; 3:1629–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Etemadmoghadam D., Weir B.A., Au-Yeung G., Alsop K., Mitchell G., George J., Davis S., D’Andrea A.D., Simpson K., Hahn W.C.et al.. Synthetic lethality between CCNE1 amplification and loss of BRCA1. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:19489–19494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gaillard H., García-Muse T., Aguilera A.. Replication stress and cancer. Nat. Rev. Cancer. 2015; 15:276–289. [DOI] [PubMed] [Google Scholar]
- 97. Jones R.M., Mortusewicz O., Afzal I., Lorvellec M., García P., Helleday T., Petermann E.. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 2013; 32:3744–3753. [DOI] [PubMed] [Google Scholar]
- 98. Bermejo R., Lai M.S., Foiani M.. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol. Cell. 2012; 45:710–718. [DOI] [PubMed] [Google Scholar]
- 99. Helmrich A., Ballarino M., Nudler E., Tora L.. Transcription-replication encounters, consequences and genomic instability. Nat. Struct. Mol. Biol. 2013; 20:412–418. [DOI] [PubMed] [Google Scholar]
- 100. Aguilera A., García-Muse T.. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012; 46:115–124. [DOI] [PubMed] [Google Scholar]
- 101. Gan W., Guan Z., Liu J., Gui T., Shen K., Manley J.L., Li X.. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011; 25:2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bhatia V., Barroso S.I., García-Rubio M.L., Tumini E., Herrera-Moyano E., Aguilera A.. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014; 511:362–365. [DOI] [PubMed] [Google Scholar]
- 103. Durkin S.G., Glover T.W.. Chromosome fragile sites. Annu. Rev. Genet. 2007; 41:169–192. [DOI] [PubMed] [Google Scholar]
- 104. Vesela E., Chroma K., Turi Z., Mistrik M.. Common chemical inductors of replication stress: Focus on cell-based studies. Biomolecules. 2017; 7:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Vos S.M., Tretter E.M., Schmidt B.H., Berger J.M.. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011; 12:827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Loegering D., Arlander S.J.H., Hackbarth J., Vroman B.T., Roos-Mattjus P., Hopkins K.M., Lieberman H.B., Karnitz L.M., Kaufmann S.H.. Rad9 protects cells from topoisomerase poison-induced cell death. J. Biol. Chem. 2004; 279:18641–18647. [DOI] [PubMed] [Google Scholar]
- 107. Pommier Y., Laco G.S., Kohlhagen G., Sayer J.M., Kroth H., Jerina D.M.. Position-specific trapping of topoisomerase I-DNA cleavage complexes by intercalated benzo[a]-pyrene diol epoxide adducts at the 6-amino group of adenine. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:10739–10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Pommier Y., O’Connor M.J., De Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci. Transl. Med. 2016; 8:362. [DOI] [PubMed] [Google Scholar]
- 109. Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: Clearing up the misunderstandings. Mol. Oncol. 2011; 5:387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Maya-Mendoza A., Moudry P., Merchut-Maya J.M., Lee M., Strauss R., Bartek J.. High speed of fork progression induces DNA replication stress and genomic instability. Nature. 2018; 559:279–284. [DOI] [PubMed] [Google Scholar]
- 111. Hanzlikova H., Kalasova I., Demin A.A., Pennicott L.E., Cihlarova Z., Caldecott K.W.. The importance of poly(ADP-ribose) polymerase as a sensor of unligated Okazaki fragments during DNA replication. Mol. Cell. 2018; 71:319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M.. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011; 145:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Byun T.S., Pacek M., Yee M.C., Walter J.C., Cimprich K.A.. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005; 19:1040–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zou L., Elledge S.J.. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003; 300:1542–1548. [DOI] [PubMed] [Google Scholar]
- 115. Ge X.Q., Blow J.J.. Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories. J. Cell Biol. 2010; 191:1285–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ge X.Q., Jackson D.A., Blow J.J.. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007; 21:3331–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kolinjivadi A.M., Sannino V., de Antoni A., Técher H., Baldi G., Costanzo V.. Moonlighting at replication forks – a new life for homologous recombination proteins BRCA1, BRCA2 and RAD51. FEBS Lett. 2017; 591:1083–1100. [DOI] [PubMed] [Google Scholar]
- 118. Quinet A., Lemaçon D., Vindigni A.. Replication fork reversal: players and guardians. Mol. Cell. 2017; 68:830–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Mijic S., Zellweger R., Chappidi N., Berti M., Jacobs K., Mutreja K., Ursich S., Ray Chaudhuri A., Nussenzweig A., Janscak P.et al.. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat. Commun. 2017; 8:859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Zellweger R., Dalcher D., Mutreja K., Berti M., Schmid J.A., Herrador R., Vindigni A., Lopes M.. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 2015; 208:563–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Bétous R., Mason A.C., Rambo R.P., Bansbach C.E., Badu-Nkansah A., Sirbu B.M., Eichman B.F.. SMARCAL1 catalyzes fork regression and holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012; 26:151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Bétous R., Couch F.B., Mason A.C., Eichman B.F., Manosas M., Cortez D. Substrate-selective repair and restart of replication forks by DNA translocases. Cell Rep. 2013; 3:1958–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kile A.C., Chavez D.A., Bacal J., Eldirany S., Korzhnev D.M., Bezsonova I., Eichman B.F., Cimprich K.A.. HLTF’s ancient HIRAN domain binds 3′ DNA ends to drive replication fork reversal. Mol. Cell. 2015; 58:1090–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Ciccia A., Nimonkar A.V., Hu Y., Hajdu I., Achar Y.J., Izhar L., Petit S.A., Adamson B., Yoon J.C., Kowalczykowski S.C.et al.. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol. Cell. 2012; 47:396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Lemaçon D., Jackson J., Quinet A., Brickner J.R., Li S., Yazinski S., You Z., Ira G., Zou L., Mosammaparast N.et al.. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 2017; 8:860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Przetocka S., Porro A., Bolck H.A., Walker C., Lezaja A., Trenner A., von Aesch C., Himmels S.F., D’Andrea A.D., Ceccaldi R.et al.. CtIP-mediated fork protection synergizes with BRCA1 to suppress genomic instability upon DNA replication stress. Mol. Cell. 2018; 72:568–582. [DOI] [PubMed] [Google Scholar]
- 127. Mukherjee C., Tripathi V., Manolika E.M., Heijink A.M., Ricci G., Merzouk S., de Boer H.R., Demmers J., van Vugt M.A.T.M., Ray Chaudhuri A.. RIF1 promotes replication fork protection and efficient restart to maintain genome stability. Nat. Commun. 2019; 10:3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Cortez D. Replication-coupled DNA repair. Mol. Cell. 2019; 74:866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Rondinelli B., Gogola E., Yücel H., Duarte A.A., Van De Ven M., Van Der Sluijs R., Konstantinopoulos P.A., Jonkers J., Ceccaldi R., Rottenberg S.et al.. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat. Cell Biol. 2017; 19:1371–1378. [DOI] [PubMed] [Google Scholar]
- 130. Cortez D. Preventing replication fork collapse to maintain genome integrity. DNA Repair (Amst.). 2015; 32:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Taglialatela A., Alvarez S., Leuzzi G., Sannino V., Ranjha L., Huang J.W., Madubata C., Anand R., Levy B., Rabadan R.et al.. Restoration of replication fork stability in BRCA1- and BRCA2-Deficient cells by inactivation of SNF2-family fork remodelers. Mol. Cell. 2017; 68:414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Ding X., Chaudhuri A.R., Callen E., Pang Y., Biswas K., Klarmann K.D., Martin B.K., Burkett S., Cleveland L., Stauffer S.et al.. Synthetic viability by BRCA2 and PARP1/ARTD1 deficiencies. Nat. Commun. 2016; 7:12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Chaudhuri A.R., Callen E., Ding X., Gogola E., Duarte A.A., Lee J.E., Wong N., Lafarga V., Calvo J.A., Panzarino N.J.et al.. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016; 535:382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Dungrawala H., Bhat K.P., Le Meur R., Chazin W.J., Ding X., Sharan S.K., Wessel S.R., Sathe A.A., Zhao R., Cortez D. RADX promotes genome stability and modulates chemosensitivity by regulating RAD51 at replication forks. Mol. Cell. 2017; 67:374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Panzarino N.J., Krais J., Cong K., Peng M., Mosqueda M., Nayak S., Bond S., Calvo J.A., Doshi M.B., Bere M.et al.. Replication gaps underlie BRCA-deficiency and therapy response. Cancer Res. 2021; doi:10.1158/0008-5472.CAN-20-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Guillemette S., Serra R.W., Peng M., Hayes J.A., Konstantinopoulos P.A., Green M.R., Green M.R., Cantor S.B.. Resistance to therapy in BRCA2 mutant cells due to loss of the nucleosome remodeling factor CHD4. Genes Dev. 2015; 29:489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Haynes B., Murai J., Lee J.M.. Restored replication fork stabilization, a mechanism of PARP inhibitor resistance, can be overcome by cell cycle checkpoint inhibition. Cancer Treat. Rev. 2018; 71:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Billing D., Horiguchi M., Wu-Baer F., Taglialatela A., Leuzzi G., Nanez S.A., Jiang W., Zha S., Szabolcs M., Lin C.S.et al.. The BRCT domains of the BRCA1 and BARD1 tumor suppressors differentially regulate homology-directed repair and stalled fork protection. Mol. Cell. 2018; 72:127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Zou X., Owusu M., Harris R., Jackson S.P., Loizou J.I., Nik-Zainal S.. Validating the concept of mutational signatures with isogenic cell models. Nat. Commun. 2018; 9:1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Balmus G., Pilger D., Coates J., Demir M., Sczaniecka-Clift M., Barros A.C., Woods M., Fu B., Yang F., Chen E.et al.. ATM orchestrates the DNA-damage response to counter toxic non-homologous end-joining at broken replication forks. Nat. Commun. 2019; 10:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Roseaulin L., Yamada Y., Tsutsui Y., Russell P., Iwasaki H., Arcangioli B.. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 2008; 27:1378–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zhu M., Zhao H., Limbo O., Russell P.. Mre11 complex links sister chromatids to promote repair of a collapsed replication fork. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:8793–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Lambert S., Watson A., Sheedy D.M., Martin B., Carr A.M.. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005; 121:689–702. [DOI] [PubMed] [Google Scholar]
- 144. Hastings P.J., Ira G., Lupski J.R.. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLos Genet. 2009; 5:e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Willis N.A., Frock R.L., Menghi F., Duffey E.E., Panday A., Camacho V., Hasty E.P., Liu E.T., Alt F.W., Scully R.. Mechanism of tandem duplication formation in BRCA1-mutant cells. Nature. 2017; 551:590–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Schimmel J., Kool H., Schendel R., Tijsterman M.. Mutational signatures of non-homologous and polymerase theta-mediated end-joining in embryonic stem cells. EMBO J. 2017; 36:3634–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Heyer W.D., Ehmsen K.T., Liu J.. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010; 44:113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Wang Z., Song Y., Li S., Kurian S., Xiang R., Chiba T., Wu X.. DNA polymerase (POLQ) is important for repair of DNA double-strand breaks caused by fork collapse. J. Biol. Chem. 2019; 294:3909–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Woodward A.M., Göhler T., Luciani M.G., Oehlmann M., Ge X., Gartner A., Jackson D.A., Blow J.J.. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J. Cell Biol. 2006; 173:673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. McIntosh D., Blow J.J.. Dormant origins, the licensing checkpoint, and the response to replicative stresses. Cold Spring Harb. Perspect. Biol. 2012; 4:a012955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Elvers I., Johansson F., Groth P., Erixon K., Helleday T.. UV stalled replication forks restart by re-priming in human fibroblasts. Nucleic. Acids. Res. 2011; 39:7049–7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Lopes M., Foiani M., Sogo J.M.. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006; 21:15–27. [DOI] [PubMed] [Google Scholar]
- 153. Sale J.E., Lehmann A.R., Woodgate R.. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012; 13:141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Vaisman A., Woodgate R.. Translesion DNA polymerases in eukaryotes: what makes them tick?. Crit. Rev. Biochem. Mol. Biol. 2017; 52:274–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Zámborszky J., Szikriszt B., Gervai J.Z., Pipek O., Póti A., Krzystanek M., Ribli D., Szalai-Gindl J.M., Csabai I., Szallasi Z.et al.. Loss of BRCA1 or BRCA2 markedly increases the rate of base substitution mutagenesis and has distinct effects on genomic deletions. Oncogene. 2017; 36:746–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Ma J., Setton J., Lee N.Y., Riaz N., Powell S.N.. The therapeutic significance of mutational signatures from DNA repair deficiency in cancer. Nat. Commun. 2018; 9:3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Schiavone D., Jozwiakowski S.K., Romanello M., Guilbaud G., Guilliam T.A., Bailey L.J., Sale J.E., Doherty A.J.. PrimPol is required for replicative tolerance of G quadruplexes in vertebrate cells. Mol. Cell. 2016; 61:161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Boldinova E.O., Wanrooij P.H., Shilkin E.S., Wanrooij S., Makarova A.V.. DNA damage tolerance by eukaryotic DNA polymerase and primase primpol. Int. J. Mol. Sci. 2017; 18:1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Piberger A.L., Bowry A., Kelly R., Walker A.K., Gonzalez D., Bailey L.J., Doherty A.J., Mendez J., Morris J.R., Bryant H.E.et al.. PrimPol-dependent single-stranded gap formation mediates homologous recombination at bulky DNA adducts. Nat. Commun. 2019; 11:5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Cong K., Kousholt A.N., Peng M., Panzarino N.J., Lee W.T.C., Nayak S., Krais J., Calvo J., Bere M., Rothenberg E.et al.. PARPi synthetic lethality derives from replication-associated single-stranded DNA gaps. 2019; bioRxiv doi:25 September 2019, preprint: not peer reviewed 10.1101/781989. [DOI]
- 161. Eykelenboom J.K., Harte E.C., Canavan L., Pastor-Peidro A., Calvo-Asensio I., Llorens-Agost M., Lowndes N.F.. ATR activates the S-M checkpoint during unperturbed growth to ensure sufficient replication prior to mitotic onset. Cell Rep. 2013; 5:1095–1107. [DOI] [PubMed] [Google Scholar]
- 162. Saldivar J.C., Hamperl S., Bocek M.J., Chung M., Bass T.E., Cisneros-Soberanis F., Samejima K., Xie L., Paulson J.R., Earnshaw W.C.et al.. An intrinsic S/G2 checkpoint enforced by ATR. Science. 2018; 361:806–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Lemmens B., Lindqvist A.. DNA replication and mitotic entry: a brake model for cell cycle progression. J. Cell Biol. 2019; 218:3892–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Krajewska M., Fehrmann R.S.N., Schoonen P.M., Labib S., De Vries E.G.E., Franke L., Van Vugt M.A.T.M. ATR inhibition preferentially targets homologous recombination-deficient tumor cells. Oncogene. 2015; 34:3474–3481. [DOI] [PubMed] [Google Scholar]
- 165. Schoonen P.M., Kok Y.P., Wierenga E., Bakker B., Foijer F., Spierings D.C.J., van Vugt M.A.T.M. Premature mitotic entry induced by ATR inhibition potentiates olaparib inhibition-mediated genomic instability, inflammatory signaling, and cytotoxicity in BRCA2-deficient cancer cells. Mol. Oncol. 2019; 13:2422–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Heijink A.M., Krajewska M., Van Vugt M.A.T.M.. The DNA damage response during mitosis. Mutat. Res. 2013; 750:45–55. [DOI] [PubMed] [Google Scholar]
- 167. Orthwein A., Fradet-Turcotte A., Noordermeer S.M., Canny M.D., Brun C.M., Strecker J., Escribano-Diaz C., Durocher D.. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science. 2014; 344:189–193. [DOI] [PubMed] [Google Scholar]
- 168. Giunta S., Belotserkovskaya R., Jackson S.P.. DNA damage signaling in response to double-strand breaks during mitosis. J. Cell Biol. 2010; 190:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Minocherhomji S., Ying S., Bjerregaard V.A., Bursomanno S., Aleliunaite A., Wu W., Mankouri H.W., Shen H., Liu Y., Hickson I.D.. Replication stress activates DNA repair synthesis in mitosis. Nature. 2015; 528:286–290. [DOI] [PubMed] [Google Scholar]
- 170. Garribba L., Wu W., Özer Ö., Bhowmick R., Hickson I.D., Liu Y.. Inducing and detecting mitotic DNA synthesis at difficult-to-replicate loci. Methods Enzymol. 2018; 601:45–58. [DOI] [PubMed] [Google Scholar]
- 171. Di Marco S., Hasanova Z., Kanagaraj R., Chappidi N., Altmannova V., Menon S., Sedlackova H., Langhoff J., Surendranath K., Hühn D.et al.. RECQ5 helicase cooperates with MUS81 endonuclease in processing stalled replication forks at common fragile sites during mitosis. Mol. Cell. 2017; 66:658–671. [DOI] [PubMed] [Google Scholar]
- 172. Bhowmick R., Minocherhomji S., Hickson I.D.. RAD52 facilitates mitotic DNA synthesis following replication stress. Mol. Cell. 2016; 64:1117–1126. [DOI] [PubMed] [Google Scholar]
- 173. Özer Ö., Hickson I.D.. Pathways for maintenance of telomeres and common fragile sites during DNA replication stress. Open Biol. 2018; 8:180018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Macheret M., Bhowmick R., Sobkowiak K., Padayachy L., Mailler J., Hickson I.D., Halazonetis T.D.. High-resolution mapping of mitotic DNA synthesis regions and common fragile sites in the human genome through direct sequencing. Cell Res. 2020; 30:997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Schoonen P.M., Talens F., Stok C., Gogola E., Heijink A.M., Bouwman P., Foijer F., Tarsounas M., Blatter S., Jonkers J.et al.. Progression through mitosis promotes PARP inhibitor-induced cytotoxicity in homologous recombination-deficient cancer cells. Nat. Commun. 2017; 8:15981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Vitale I., Galluzzi L., Castedo M., Kroemer G.. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat. Rev. Mol. Cell Biol. 2011; 12:385–392. [DOI] [PubMed] [Google Scholar]
- 177. Ruan W., Lim H.H., Surana U.. Mapping mitotic death: functional integration of mitochondria, spindle assembly checkpoint and apoptosis. Front. Cell Dev. Biol. 2019; 6:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Ganem N.J., Pellman D. Limiting the proliferation of polyploid cells. Cell. 2007; 131:437–440. [DOI] [PubMed] [Google Scholar]
- 179. Hyun S.Y., Jang Y.J.. p53 activates G1 checkpoint following DNA damage by doxorubicin during transient mitotic arrest. Oncotarget. 2015; 6:4804–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Choi E., Park P.G., Lee H.O., Lee Y.K., Kang G.H., Lee J.W., Han W., Lee H.C., Noh D.Y., Lekomtsev S.et al.. BRCA2 Fine-tunes the spindle assembly checkpoint through reinforcement of BubR1 acetylation. Dev. Cell. 2012; 22:295–308. [DOI] [PubMed] [Google Scholar]
- 181. Ehlen A., Martin C., Miron S., Julien M., Theillet F.X., Boucherit V., Ropars V., Duchambon P., Marjou A.El, Justin S.Z.et al.. Proper chromosome alignment depends on BRCA2 phosphorylation by PLK1. Nat. Commun. 2018; 11:1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Futamura M., Arakawa H., Matsuda K., Katagiri T., Saji S., Miki Y., Nakamura Y.. Potential role of BRCA2 in a mitotic checkpoint after phosphorylation by hBUBR1. Cancer Res. 2000; 60:1531–1535. [PubMed] [Google Scholar]
- 183. Daniels M.J., Wang Y., Lee M.Y., Venkitaraman A.R.. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science. 2004; 306:876–879. [DOI] [PubMed] [Google Scholar]
- 184. Ait Saada A., Teixeira-Silva A., Iraqui I., Costes A., Hardy J., Paoletti G., Fréon K., Lambert S.A.E. Unprotected replication forks are converted into mitotic sister chromatid bridges. Mol. Cell. 2017; 66:398–410. [DOI] [PubMed] [Google Scholar]
- 185. Laulier C., Cheng A., Stark J.M.. The relative efficiency of homology-directed repair has distinct effects on proper anaphase chromosome separation. Nucleic. Acids. Res. 2011; 39:5935–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Adam S., Rossi S.E., Moatti N., Zompit M.D.M., Ng T.F., Álvarez-Quilón A., Desjardins J., Bhaskaran V., Martino G., Setiaputra D.et al.. CIP2A is a prime synthetic-lethal target for BRCA-mutated cancers. 2021; bioRxiv doi:08 February 2021, preprint: not peer reviewed 10.1101/2021.02.08.430060. [DOI] [PubMed]
- 187. Ly P., Cleveland D.W.. Rebuilding chromosomes after catastrophe: emerging mechanisms of chromothripsis. Trends Cell Biol. 2017; 27:917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. McPherson J.P., Hande M.P., Poonepalli A., Lemmers B., Zablocki E., Migon E., Shehabeldin A., Porras A., Karaskova J., Vukovic B.et al.. A role for Brca1 in chromosome end maintenance. Hum. Mol. Genet. 2006; 15:831–838. [DOI] [PubMed] [Google Scholar]
- 189. Min J., Choi E.S., Hwang K., Kim J., Sampath S., Venkitaraman A.R., Lee H.. The breast cancer susceptibility gene BRCA2 is required for the maintenance of telomere homeostasis. J. Biol. Chem. 2012; 287:5091–5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Kargaran P.K., Yasaei H., Anjomani-Virmouni S., Mangiapane G., Slijepcevic P.. Analysis of alternative lengthening of telomere markers in BRCA1 defective cells. Genes Chromosom. Cancer. 2016; 55:864–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. McClintock B. The production of homozygous deficient tissues with mutant characteristics by means of the aberrant mitotic behavior of ring-shaped chromosomes. Genetics. 1938; 23:315–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Crasta K., Ganem N.J., Dagher R., Lantermann A.B., Ivanova E.V., Pan Y., Nezi L., Protopopov A., Chowdhury D., Pellman D.. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012; 482:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. McClintock B. The fusion of broken ends of chromosomes following nuclear fusion. Proc. Natl. Acad. Sci. U.S.A. 1942; 28:458–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Umbreit N.T., Zhang C.Z., Lynch L.D., Blaine L.J., Cheng A.M., Tourdot R., Sun L., Almubarak H.F., Judge K., Mitchell T.J.et al.. Mechanisms generating cancer genome complexity from a single cell division error. Science. 2020; 368:eaba0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Pampalona J., Roscioli E., Silkworth W.T., Bowden B., Genescà A., Tusell L., Cimini D.. Chromosome bridges maintain kinetochore-microtubule attachment throughout mitosis and rarely break during anaphase. PLoS One. 2016; 11:e0147420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Zakov S., Kinsella M., Bafna V.. An algorithmic approach for breakage-fusion-bridge detection in tumor genomes. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:5546–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Wang Y.K., Bashashati A., Anglesio M.S., Cochrane D.R., Grewal D.S., Ha G., McPherson A., Horlings H.M., Senz J., Prentice L.M.et al.. Genomic consequences of aberrant DNA repair mechanisms stratify ovarian cancer histotypes. Nat. Genet. 2017; 49:856–864. [DOI] [PubMed] [Google Scholar]
- 198. Ratnaparkhe M., Wong J.K.L., Wei P.C., Hlevnjak M., Kolb T., Simovic M., Haag D., Paul Y., Devens F., Northcott P.et al.. Defective DNA damage repair leads to frequent catastrophic genomic events in murine and human tumors. Nat. Commun. 2018; 9:4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Petljak M., Alexandrov L.B., Brammeld J.S., Price S., Wedge D.C., Grossmann S., Dawson K.J., Ju Y.S., Iorio F., Tubio J.M.C.et al.. Characterizing mutational signatures in human cancer cell lines reveals Episodic APOBEC mutagenesis. Cell. 2019; 176:1282–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Nik-Zainal S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L.A.et al.. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012; 149:979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Zhang C.Z., Spektor A., Cornils H., Francis J.M., Jackson E.K., Liu S., Meyerson M., Pellman D.. Chromothripsis from DNA damage in micronuclei. Nature. 2015; 522:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Hengeveld R.C.C., de Boer H.R., Schoonen P.M., de Vries E.G.E., Lens S.M.A., van Vugt M.A.T.M.. Rif1 Is Required for Resolution of Ultrafine DNA Bridges in Anaphase to Ensure Genomic Stability. Dev. Cell. 2015; 34:466–474. [DOI] [PubMed] [Google Scholar]
- 203. Liu Y., Nielsen C.F., Yao Q., Hickson I.D.. The origins and processing of ultra fine anaphase DNA bridges. Curr. Opin. Genet. Dev. 2014; 26:1–5. [DOI] [PubMed] [Google Scholar]
- 204. Tiwari A., Addis Jones O., Chan K.L.. 53BP1 can limit sister-chromatid rupture and rearrangements driven by a distinct ultrafine DNA bridging-breakage process. Nat. Commun. 2018; 9:677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Chan Y.W., Fugger K., West S.C.. Unresolved recombination intermediates lead to ultra-fine anaphase bridges, chromosome breaks and aberrations. Nat. Cell Biol. 2018; 20:92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Laulier C., Cheng A., Stark J.M.. The relative efficiency of homology-directed repair has distinct effects on proper anaphase chromosome separation. Nucleic. Acids. Res. 2011; 39:5935–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Chan K.L., Palmai-Pallag T., Ying S., Hickson I.D.. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 2009; 11:753–760. [DOI] [PubMed] [Google Scholar]
- 208. Harrigan J.A., Belotserkovskaya R., Coates J., Dimitrova D.S., Polo S.E., Bradshaw C.R., Fraser P., Jackson S.P.. Replication stress induces 53BP1-containing OPT domains in G1 cells. J. Cell Biol. 2011; 193:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Lukas C., Savic V., Bekker-Jensen S., Doil C., Neumann B., Pedersen R.S., Grøhfte M., Chan K.L., Hickson I.D., Bartek J.et al.. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 2011; 13:243–253. [DOI] [PubMed] [Google Scholar]
- 210. Pedersen R.T., Kruse T., Nilsson J., Oestergaard V.H., Lisby M.. TopBP1 is required at mitosis to reduce transmission of DNA damage to G1 daughter cells. J. Cell Biol. 2015; 210:565–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Spies J., Lukas C., Somyajit K., Rask M.B., Lukas J., Neelsen K.J.. 53BP1 nuclear bodies enforce replication timing at under-replicated DNA to limit heritable DNA damage. Nat. Cell Biol. 2019; 21:487–497. [DOI] [PubMed] [Google Scholar]
- 212. Lips E.H., Mulder L., Oonk A., Van Der Kolk L.E., Hogervorst F.B.L., Imholz A.L.T., Wesseling J., Rodenhuis S., Nederlof P.M.. Triple-negative breast cancer: BRCAness and concordance of clinical features with BRCA1-mutation carriers. Br. J. Cancer. 2013; 108:2172–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Nguyen L., Martens J., Hoeck A., van and Cuppen E.. Pan-cancer landscape of homologous recombination deficiency. Nat. Commun. 2020; 11:5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Staaf J., Glodzik D., Bosch A., Vallon-Christersson J., Reuterswärd C., Häkkinen J., Degasperi A., Amarante T.D., Saal L.H., Hegardt C.et al.. Whole-genome sequencing of triple-negative breast cancers in a population-based clinical study. Nat. Med. 2019; 25:1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Gulhan D.C., Lee J.J.K., Melloni G.E.M., Cortés-Ciriano I., Park P.J. Detecting the mutational signature of homologous recombination deficiency in clinical samples. Nat. Genet. 2019; 51:912–919. [DOI] [PubMed] [Google Scholar]
- 216. Ceccaldi R., Liu J.C., Amunugama R., Hajdu I., Primack B., Petalcorin M.I.R., O’Connor K.W., Konstantinopoulos P.A., Elledge S.J., Boulton S.J.et al.. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature. 2015; 518:258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Lok B.H., Carley A.C., Tchang B., Powell S.N.. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene. 2013; 32:3552–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Kok Y.P., Llobet S.G., Schoonen P.M., Everts M., Bhattacharya A., Fehrmann R.S.N., Tempel N. Van Den, Vugt M.A.T.M. Van. Overexpression of Cyclin E1 or Cdc25A leads to replication stress, mitotic aberrancies, and increased sensitivity to replication checkpoint inhibitors. Oncogenesis. 2020; 9:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Dreyer S.B., Upstill-Goddard R., Paulus-Hock V., Paris C., Lampraki E.-M., Dray E., Serrels B., Caligiuri G., Rebus S., Plenker D.et al.. Targeting DNA damage response and replication stress in pancreatic cancer. Gastroenterology. 2020; 160:362–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Polak P., Kim J., Braunstein L.Z., Tiao G., Karlic R., Rosebrock D., Livitz D., Kübler K., Mouw K.W., Haradhvala N.W.et al.. Mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat. Genet. 2017; 49:1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]