Abstract
Many archaea express histones, which organize the genome and play a key role in gene regulation. The structure and function of archaeal histone–DNA complexes remain however largely unclear. Recent studies show formation of hypernucleosomes consisting of DNA wrapped around an ‘endless’ histone-protein core. However, if and how such a hypernucleosome structure assembles on a long DNA substrate and which interactions provide for its stability, remains unclear. Here, we describe micromanipulation studies of complexes of the histones HMfA and HMfB with DNA. Our experiments show hypernucleosome assembly which results from cooperative binding of histones to DNA, facilitated by weak stacking interactions between neighboring histone dimers. Furthermore, rotational force spectroscopy demonstrates that the HMfB–DNA complex has a left-handed chirality, but that torque can drive it in a right-handed conformation. The structure of the hypernucleosome thus depends on stacking interactions, torque, and force. In vivo, such modulation of the archaeal hypernucleosome structure may play an important role in transcription regulation in response to environmental changes.
INTRODUCTION
Dynamic genome organization is a prerequisite for a compact yet active genome throughout all domains of life. Current views on evolution agree that eukaryotes are part of the branch of Archaea, which are single-cellular organisms that share many cellular mechanisms with Eukaryotes (1–4). Eukaryotes use histones, which wrap DNA into nucleosomes to compact and functionally organize their genomes. These histones have N-terminal tails that can be post-translationally modified, which changes their physico-chemical properties, and is key in defining the functional chromatin state (5). Most archaeal species express rudimentary homologues of eukaryotic histones with the characteristic histone fold (6–8), but lack the N-terminal tail (9–13). In addition to histones, Archaea express other small architectural proteins, nucleoid-associated proteins (NAPs). These NAPs are also involved in genome organization and may complement or compete with histones to regulate genes (14,15).
The prototypical archaeal histones are HMfA and HMfB from Methanothermus fervidus (16). HMfA and HMfB are homologues of eukaryotic histones in terms of sequence and structure (13). HMfA and HMfB share 84% sequence identity. These proteins are expressed at varying ratios as a function of growth phase (17). Although estimates of absolute expression levels of individual histones are lacking, HMfA is prevalent during exponential growth (1.5× the amount of HMfB), and present in equal amounts as HMfB during stationary growth (17). This might imply that these proteins have evolved different DNA binding properties—and correspondingly distinct functions—in the cell. HMfA and HMfB are capable of forming homo- and heterodimers, which subsequently can form larger structures (17–20). The binding of HMf dimers to DNA results in bends in DNA at low protein concentrations, and has been reported to yield beads-on-a-string nucleosome-like structures at higher protein to DNA ratios (16). HMf proteins bind preferentially to intrinsically curved DNA sequences in vitro (21). There is currently no evidence of the existence of specific DNA sequence signatures that enhance histone binding in vivo, although studies in Methanothermobacter thermautotrophicus and Thermococcus kodakarensis indicate that repetitive motifs of AT and GC base pair steps favor histone positioning (22). These motifs facilitate the distortion needed for DNA wrapping. Systematic evolution of ligands by exponential enrichment (SELEX) experiments have yielded two high-affinity DNA binding sequences for HMfB in vitro (23). Binding of HMf proteins to this sequence yields nucleosome-like structures, in which 60 bp of DNA is proposed to be wrapped around a tetrameric protein core composed of two interacting HMf dimers (24). Here, we will refer to the DNA compaction mode of HMf proteins as wrapping, even when the DNA is wrapped less than one turn.
The histone tetramer model is supported by micrococcal nuclease (MNase) digestion studies of chromatin from Haloferax volcanii, which yield undigested DNA fragments 60 bp in size (22,25). However, the histone tetramer model has been challenged by the results of similar studies in T. kodakarensis, which yield DNA fragments ranging from 30 to 450 bp in 30 bp increments (26). The sizes of these DNA fragments were interpreted as reflecting the existence of histone multimers of varying sizes bound along the genome. Similar results were obtained when HMfA and HMfB proteins were heterologously expressed in E. coli. These studies also revealed a mild generic repressive effect of HMf multimer formation on transcription (27). A most noteworthy recent crystallography study of HMfB on 90 base pair DNA fragments supports the formation of a multimeric histone filament established by interactions between adjacent histone dimers and of wrapping of DNA fragments around an endless protein core, creating a quasi-continuous superhelix (Figure 1A) (28). This structure is referred to as the hypernucleosome (29). The hypernucleosome is left-handed like the eukaryotic nucleosome, although it has been reported that archaeal histones can accommodate both left- and right-handed wrapping configurations (20,30,31). It was proposed that hypernucleosomes are not just held together by interactions between adjacent dimers, but also by stacking interactions between the layers of dimers within the hypernucleosome (Figure 1B and C) (29). The disparate findings from H. volcanii and T. kodakarensis, as well as the crystal structure of HMfB-DNA complexes, indicate that molecular insights into the role of histones in archaeal chromatin organization are important for understanding archaeal genome compaction.
Figure 1.
The hypernucleosome and Methanothermus fervidus histones HMfB and HMfA. (A) The HMfB hypernucleosome structure as described by Mattiroli et al. (PDB: 5T5K (28)). Figure taken from Henneman et al. 2018 with permission (29). (B) Hypernucleosome interfaces of HMfB (left; water-refined and starting from the 5T5K structure) and HMfA (right; water-refined and starting from the homology model). Top panels show the stacking interface, bottom panels show the tetramer interface. Interacting residues are labeled; hydrogen bonds are shown by dashed lines. Important residue differences between HMfA and HMfB are colored in orange. The closely packed residues G16 in the stacking interface are shown in yellow spheres (Cα only). (C) Sequence alignment of HMfA and HMfB. Conserved residues are shown in black boxes, conservative changes in grey and non-conservative changes in white boxes. Residues are numbered according to HMfB sequence. The two sequences are 84% identical and have 94% similarity. Residues directly contacting the DNA are indicated with diamonds (◊), residues forming the tetramerization interface are marked with asterisks (*) and stacking residues are indicated with triangles (▵) for HMfA (top) and HMfB (bottom). Differences in interacting residues between HMfA and HMfB are indicated with red symbols. Important residue differences between HMfA and HMfB are colored in orange and G16 residues are shown in yellow, matching panel B.
In this study, we provide direct evidence of hypernucleosome formation and its capability of transitioning in handedness. HMfA and HMfB homodimers exhibit subtle differences in amino acid composition, resulting in different stacking energies in the hypernucleosomes. This results in different stabilities of the hypernucleosome, which may be the key mechanism of transcription regulation in Archaea.
MATERIALS AND METHODS
DNA substrate preparation
For the Tethered Particle Motion (TPM) DNA substrate, a random sequence of 50% GC content (CGGCGCAAATTCGTGACCAGTTGCATCAGCTGCGTGAGCTGTTTATCGCAGCATCGTAACAGGATAGTGAAGAAGACT) was cloned into pBR322, as described previously (32), resulting in plasmid pRD121. We used PCR to generate and amplify a 685 base pairs linear substrate (sequence in supplemental information) containing the cloned sequence, using digoxygenin- and biotin-labeled oligonucleotides (33).
For magnetic tweezers a 3646 base pairs DNA fragment based on plasmid pFW01 (pBPCYC1(wt)/3 derivative (34); sequence in supplemental information) was digested with BsaI and BseYI (New England Biolabs). The fragment was isolated using agarose gel electrophoresis and purified using a Promega Wizard® SV Gel and PCR Clean-Up System. It was subsequently labeled with digoxigenin and biotin at either end by a Klenow reaction.
For the torsionally constrained construct, pUC18-based plasmid pTB01, containing 12 repeats of the Widom 601 sequence separated by 25 bp of linker DNA (35) (sequence in supplemental information), was digested with BsaI and BseYI. The 4092 base pairs digestion product was isolated using agarose gel electrophoresis and purified using a Promega Wizard® SV Gel and PCR Clean-Up System. Subsequently, a 650 bp handle containing multiple digoxigenin- or biotin-modified bases was ligated on each end. Details of both protocols are provided elsewhere (36). The 4092 bp substrate, based on plasmid pTB01, qualitatively behaved the same as the 3646 bp pFW01-based substrate. We did not observe different features of the hypernucleosome on this nucleosome reconstitution template.
Protein preparation
HMfA and HMfB were kindly provided by John Reeve and Kathleen Sandman. HMfAK31A E35A and HMfBD14A K30A E34A were synthesized by GeneArt (Thermo Fisher Scientific, MA) and cloned into pET30b vectors using Gibson assembly. This resulted in plasmids pRD323 and pRD324, expressing HMfAK31A E35A and HMfBD14A K30A E34A respectively. These proteins were expressed and purified as described in detail in the supplementary information.
Tethered particle motion
The tethered particle motion experiments were carried out as described previously (37) with minor modifications. Briefly, the flow-cell was washed with 100 μl experimental buffer (50 mM Tris–HCl, pH 7 and 75 mM KCl), followed by flushing in 100 μl diluted protein in experimental buffer. This was incubated for 10 min before washing with again 100 μl protein dilution. The flow cell was sealed with nail polish, dried and placed in the holder. To stabilize the temperature at 25°C, the flow cell was left in the holder for 5 min before the first measurement. For each flow cell more than 300 beads were measured and for each concentration the measurement was done in triplicate. We used an anisotropic ratio cut-off of 1.3 and a standard deviation cut-off of 8% to select single-tethered beads. The end-to-end distance of the DNA substrate was determined by taking the average of the extremities (2.5% largest values) of the deflection of 25 beads in the xy plane at 50 nM of HMfB and subtracting the radius of the bead (Supplementary Figure S1). The end-to-end distance was obtained by triangular calculation.
Force spectroscopy and rotational spectroscopy
Details of the force spectroscopy experiments and the multiplexed magnetic tweezers setup have been described elsewhere (36,38,39). Briefly, flow cells were flushed with isopropanol and subsequently incubated with a solution of 10 μg/ml anti-digoxigenin (Sigma Aldrich; 11333089001) in PBS for 2 h at 4°C. After that, the flow cell surface was blocked using 2% BSA in PBS and incubated for another 2 h at 4°C. HMfA and HMfB were diluted to 100 nM in measurement buffer (10 mM HEPES, 100 mM KCl, 10 mM NaN3, 20mM MgCl2, 0.1% Tween20, 0.2% BSA). HMfAK31A E35A and HMfBD14A K30A E34A were diluted to 3 μM and 200 nM respectively. Subsequently, 50 μl of the histone solution was mixed with 1 pM DNA substrate and 0.2 μl paramagnetic M270 beads (Dynabeads;Thermo Fisher Scientific, USA). After this sample was loaded into the flow cell, the flow cell was left in the holder for 15 min before the first measurement to stabilize the temperature. Force is exerted by a magnet positioned above the flow cell. In each pulling experiment, the force was increased exponentially from 0.01 to 40 pN in 70 seconds by lowering the magnet toward the flow cell, and decreased at the same speed. In measurements involving rotation, the magnet was rotated with a speed of two turns per second at a force of 0.1 pN before pulling.
Quantitative modeling of stretching the hypernucleosome
To interpret the force-extension behavior of the hypernucleosomes, we developed a statistical mechanics model. HMfA and HMfB dimers bound to the DNA were distributed over three conformations, representing different levels of compaction. In the lowest force regime, HMfA and HMfB homodimers assembled into a hypernucleosome. The extension of the hypernucleosome was modeled by a freely-jointed chain (FJC) (40,41) (Equation 1)
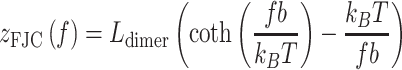 |
(1) |
where 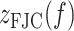 is the extension per dimer,
is the extension per dimer, 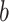 the Kuhn length that characterizes the tether flexibility,
the Kuhn length that characterizes the tether flexibility, 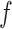 the force,
the force,  Boltzmann's constant,
Boltzmann's constant, 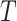 the temperature, and
the temperature, and 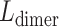 the height of a single dimer (when fully wrapped), which was fixed at 4 nm. The FJC was chosen for both its initial linear increase of extension with force, as observed experimentally, and its asymptote at high force. For low forces
the height of a single dimer (when fully wrapped), which was fixed at 4 nm. The FJC was chosen for both its initial linear increase of extension with force, as observed experimentally, and its asymptote at high force. For low forces 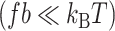 , the FJC describes a Hookean spring with linear extension, and its stiffness
, the FJC describes a Hookean spring with linear extension, and its stiffness  of the stacked fiber is inversely proportional to the effective Kuhn length (Equation 2) (40).
of the stacked fiber is inversely proportional to the effective Kuhn length (Equation 2) (40).
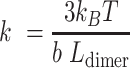 |
(2) |
Note that only the linear part of the FJC, i.e. up to an extension up to 2 nm per dimer, mattered, as the dimers unstacked at larger forces.
The second force regime is characterized as a beads-on-a-string structure. In this structure, DNA wraps onto non-interacting HMfA or HMfB homodimers resulting in moderate compaction of the fiber. The beads-on-a-string structure was modeled as an extensible worm-like chain (WLC) (42,43) (Equation 3).
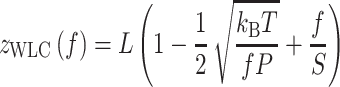 |
(3) |
in which is the extension of an HMf dimer-DNA complex,
is the extension of an HMf dimer-DNA complex, 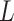 is the contour length of the DNA substrate,
is the contour length of the DNA substrate, 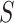 the stretch modulus of DNA and
the stretch modulus of DNA and 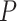 the persistence length. Since the footprint of an HMfB homodimer is 30 bp (44), each transition of a dimer from the hypernucleosome into a beads-on-a-string conformation reduces the contour length of the FJC by
the persistence length. Since the footprint of an HMfB homodimer is 30 bp (44), each transition of a dimer from the hypernucleosome into a beads-on-a-string conformation reduces the contour length of the FJC by 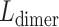 and increases the contour length of the WLC with 30 bp. Moreover, each HMf dimer bends the DNA, which reduces the persistence length of DNA to an apparent persistence length
and increases the contour length of the WLC with 30 bp. Moreover, each HMf dimer bends the DNA, which reduces the persistence length of DNA to an apparent persistence length 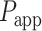 (45) (Equation 4).
(45) (Equation 4).
 |
(4) |
in which 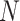 is the number of dimers in the beads-on-a-string conformation and
is the number of dimers in the beads-on-a-string conformation and  is the DNA deflection angle induced by the dimer. The model fitted the data best using a deflection angle of ∼15°, which corresponds roughly to the structure depicted in Figure 3C.
is the DNA deflection angle induced by the dimer. The model fitted the data best using a deflection angle of ∼15°, which corresponds roughly to the structure depicted in Figure 3C.
Figure 3.

Force spectroscopy experiments on the hypernucleosome reveal stronger stacking in HMfB tethers than in HMfA tethers. (A) Force spectroscopy on a HMfB-DNA complex (blue dots) at 100 nM of HMfB reveals three levels of compaction. A bare DNA molecule is shown in yellow dots. We fitted different parts of the curve to a freely-jointed chain (FJC) (I), kinked worm-like chain (WLC) (II) and WLC (III). Fixed parameters per stacked dimer in regime I: force independent length = 0.5 nm, FJC contour length = 4 nm, length of wrapped DNA = 30 bp. Fixed parameters for DNA in regime III: contour length = 3646 bp, DNA persistence length = 50 nm, DNA stretch modulus = 900 pN. The release curves overlapped with the stretch curves, which indicates that HMfB-DNA the stretch-release cycle was in equilibrium. (B) Comparison between HMfA hypernucleosomes (red dots) and HMfB hypernucleosomes (blue dots). Each curve shows a pulling trace in color and a refolding curve in grey, typically largely obscured by overlap with the pulling curve. (C) Structural models of three states of the HMfB-DNA complex corresponding to the model fit in A, illustrating a 7-fold compaction of the hypernucleosome compared to bare DNA. (D–H) Histogram of fit parameters to Equation (10) for HMfA and HMfB complexes. Statistics are shown in Table 1.
At high forces, the HMfA and HMfB homodimers appeared to remain bound, but did not bend the DNA. Each transition therefore reduced the number of kinks in the DNA, resulting in an increase of the apparent persistence length to that of bare DNA, for which we used  nm and S = 900 pN (41,46,47).
nm and S = 900 pN (41,46,47).
A large number of fiber states 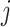 can be defined, reflecting different distributions of each of the three dimer conformations
can be defined, reflecting different distributions of each of the three dimer conformations 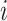 , representing the hypernucleosome, the beads-on-a-string, or the straight conformation. The total extension
, representing the hypernucleosome, the beads-on-a-string, or the straight conformation. The total extension of the tether is a linear combination of the extension of each dimer conformation
of the tether is a linear combination of the extension of each dimer conformation  multiplied by the number of dimers
multiplied by the number of dimers 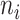 in conformation
in conformation 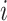 (Equation 5).
(Equation 5).
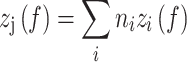 |
(5) |
The free energy per dimer in the hypernucleosome is given by:
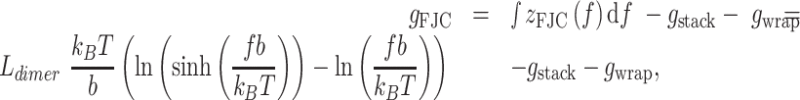 |
(6) |
using a protein-protein interaction energy 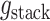 and a protein-DNA interaction energy
and a protein-DNA interaction energy 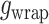 . The free energy per dimer in the wrapped and the straight conformation is given by:
. The free energy per dimer in the wrapped and the straight conformation is given by:
 |
(7) |
with 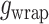 = 0 for the straight conformation. In addition, the work
= 0 for the straight conformation. In addition, the work 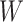 done by the bead corresponds to:
done by the bead corresponds to:
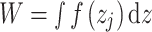 |
(8) |
The total free energy 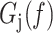 of a particular tether is the work by the bead minus the sum of the free energy contributions of each dimer:
of a particular tether is the work by the bead minus the sum of the free energy contributions of each dimer:
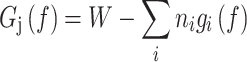 |
(9) |
Finally, the force dependent extension of the HMf-DNA complex was calculated as the Boltzmann-weighted mean extension:
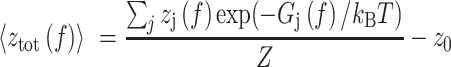 |
(10) |
with partition function 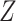 =
= 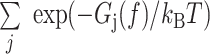 .
.
z0 reflects the relative position of the DNA tether on the bead, which can be offset from the bottom due to the anisotropy of the magnetic moment of the bead. Equation 10 was fitted to the force-extension curve in the force range between 0.1 and 55 pN to extract the number of dimers, the stiffness of the hypernucleosome k, the deflection angle α, the stacking energy 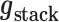 , the wrapping energy
, the wrapping energy 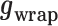 and bead offset z0 from each curve.
and bead offset z0 from each curve.
RESULTS
Both HMfA and HMfB cooperatively compact DNA
To determine the degree of DNA compaction by HMfA and HMfB, we performed Tethered Particle Motion experiments (TPM) (33). Here, the root mean squared displacement (RMS) of a bead connected to a surface-attached DNA tether provides a quantitative readout of DNA conformation (Supplementary Figure S1A). We investigated the binding of HMfA and HMfB to a 685 bp DNA molecule containing a random, naturally occurring sequence (see Materials and Methods). We observed an abrupt and drastic reduction in RMS of the DNA tether over a narrow concentration range during titration of both proteins, which indicates that DNA compaction by both HMfA and HMfB is highly cooperative. Such cooperativity is due to either direct protein-protein interactions or facilitated protein-DNA binding through structural effects of adjacently bound HMfA or HMfB proteins (20,48) (Figure 2).
Figure 2.

Methanothermus fervidus histone proteins HMfA and HMfB cooperatively compact DNA in Tethered Particle Motion experiments. Root mean square displacement (RMS) excursion of the bead for HMfA and HMfB on a DNA substrate is shown as a function of protein concentration. Dashed lines are to guide the eye. Error bars indicating the standard error of the data points (N ≥ 100) are small and mostly hidden behind the data points. In grey, saturation levels of the bacterial NAP HU and archaeal NAPs Sso10a1 and Cren7—which are known to organize and compact DNA—are indicated, as reported by Driessen et al., Driessen et al. and Driessen et al. (32,49,52).
As a dimer, HMf proteins bend DNA to an angle that is similar to those induced by other DNA-bending proteins, such as the bacterial NAP HU and the archaeal NAPs Cren7 and Sso10a1 (16,32,49–52). These proteins reduce the RMS of a 685 bp DNA substrate from 150 to 110–120 nm at saturating protein concentrations (32,52). A much stronger reduction in RMS down to 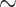 70 nm was observed for HMfB and
70 nm was observed for HMfB and 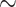 75 nm for HMfA. The extent of this change cannot be explained by DNA bending induced by individual HMf dimers. The high degree of DNA compaction together with the pronounced binding cooperativity suggests adjacent packing of HMfB (and HMfA) proteins that together wrap DNA into multiple turns. By determining the maximum deflection of the bead in TPM, we found an end-to-end distance of 24 ± 4.7 nm for the HMfB–DNA complex at saturating protein concentrations for this specific DNA substrate (Supplementary Figure S1). This number agrees with the theoretical end-to-end distance of the hypernucleosomal structure observed in the crystallography studies (28). Both HMf proteins bind to DNA in a cooperative manner, but HMfB shows a more pronounced cooperativity than HMfA. The reason for the differential cooperativity remains unclear, but differences in amino acid composition of the C-terminus may underlie this effect (Figure 1B and C).
75 nm for HMfA. The extent of this change cannot be explained by DNA bending induced by individual HMf dimers. The high degree of DNA compaction together with the pronounced binding cooperativity suggests adjacent packing of HMfB (and HMfA) proteins that together wrap DNA into multiple turns. By determining the maximum deflection of the bead in TPM, we found an end-to-end distance of 24 ± 4.7 nm for the HMfB–DNA complex at saturating protein concentrations for this specific DNA substrate (Supplementary Figure S1). This number agrees with the theoretical end-to-end distance of the hypernucleosomal structure observed in the crystallography studies (28). Both HMf proteins bind to DNA in a cooperative manner, but HMfB shows a more pronounced cooperativity than HMfA. The reason for the differential cooperativity remains unclear, but differences in amino acid composition of the C-terminus may underlie this effect (Figure 1B and C).
HMfB organizes long DNA tethers into endless hypernucleosomes
The mechanical stability of the hypernucleosome was further investigated by force spectroscopy. HMfB hypernucleosomes were reconstituted in vitro on a torsionally unconstrained DNA construct, at 100 nM HMfB, well above the KD. Hence, we expect full coverage of the DNA. Force spectroscopy revealed characteristic reversible force-extension curves (Figure 3A and B). Binding of HMfB resulted in a large compaction of the DNA. The absence of hysteresis indicates that the manipulation of the HMfB–DNA complex occurs in thermodynamic equilibrium: stretch-release cycles could be repeated multiple times without qualitative changes in the data. The force extension curve of the HMfB-DNA complex showed three regimes of extension (Figure 3A). In regime I, at forces below 1 pN, the force-extension curve featured a small extension and a high stiffness. In regime II, between 2 and 10 pN, the tether is longer and appears to be less stiff than in regime I. In regime III, at forces above 20 pN, the extension of the complex follows that of bare DNA, which is stiffer than the complex in regime II. Most of the force extension curve of the HMfB-coated DNA could be captured in these three regimes, and we interpret the intermediate parts as transitions between these regimes.
We modeled the two most extended structures, in force regime II and III, with kinked and regular worm-like chains (WLCs). In regime III the complex approached the force-extension curve of bare DNA with a persistence length of 50 nm. For regime II, we fitted the same contour length and a persistence length of approximately 4 nm, suggesting a much more flexible tether. The most compacted structure, regime I, could not be modeled by a WLC and shows a linear extension between 0.1 and 2 pN. We used the linear part of a freely-jointed chain (FJC) to describe this regime of the force-extension curve. Note that this choice of the mechanical response in regime I does not imply that the tether behaves as a random-walk polymer. Rather, we assume a more general Hookean extension (like that of a FJC in the low force regime) and do not apply the FJC beyond this low force regime. The FJC model has the numerical advantage that it results in prohibitively large stretching energies for large extensions, which facilitates the statistical physics model of the transitions between the regimes.
The mechanical properties of the complex suggest three different structures of the tether. Stacking of HMfB dimers into a hypernucleosome would create a relatively stiff structure that can only be stretched to a limited extent without breaking the protein-protein stacking interactions. Based on the crystal structure of the HMfB hypernucleosome (28), we expect an approximate height of 4 nm per dimer for the most condensed structure (Figure 3C, I). When the stacking interactions are broken due to excessive force, individual dimers may remain bound to the DNA. Such a structure would have a much larger extension per HMfB dimer than the stacked hypernucleosome. If all DNA-protein contacts remain intact, the DNA follows a highly curved trajectory (Figure 3C, II). Such curved DNA results in a decrease of the apparent persistence length (45,53), which is consistent with the observed force-extension relation in regime II. Increasing the force further breaks protein-DNA interactions to yield an unperturbed DNA trajectory (Figure 3C, III), which would have similar mechanical properties as bare DNA. Indeed, at forces larger than 20 pN the force-extension curve overlaps with that of DNA. Thus, the experimental force-extension curves suggest a two-step transition from a hypernucleosome into a fully stretched DNA tether.
For a more quantitative analysis we developed a statistical mechanics model that includes the transitions between the three structures. The entire force-extension curve was fitted to the statistical physics model described in the materials and methods section. We fitted the parameters number of dimers, stiffness, stacking energy, wrapping energy, deflection angle and a global offset (which is a priori unknown for magnetic tweezers due to differences in the location where the tether attaches to the bead). All DNA mechanical parameters were fixed to validated numbers. Release curves fully overlapped with the stretch curves. The absence of hysteresis in all curves indicates that all transitions are in thermodynamic equilibrium, allowing an equilibrium model to describe the transitions between the states. In analogy with the forced unfolding of eukaryotic chromatin (47), we modeled the entire force-extension curve as a linear combination of the extension of individual dimer-DNA complexes in one of three states supplemented by a small fraction of bare DNA (see Materials and Methods for model details). For every dimer that binds to the DNA tether, the extension changes and the free energy is reduced by a binding and/or stacking energy. For each dimer in state II, we fitted a deflection angle of ∼15 degrees, resulting in a reduction of the persistence length as described by Kulić and Schiessel (45).
The force-induced transitions between all three states of the HMfB-coated DNA tether were adequately captured by the model (Figure 3B). The first transition (I to II) at 2 pN yields a stacking energy of 3.44 ± 0.02 kBT, which indicates that the stacked hypernucleosome is easily disrupted by force. Such unstacking yielded a lengthening of the tether of 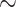 500 nm. Unwrapping of the DNA from the dimers at 10 pN takes more energy, 5.23 ± 0.07 kBT, and increases the extension of the tether by
500 nm. Unwrapping of the DNA from the dimers at 10 pN takes more energy, 5.23 ± 0.07 kBT, and increases the extension of the tether by 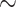 200 nm. These transitions appear as gradual changes in extension, likely because of the non-cooperative behavior of the large number of dimers, the small changes in extension per dimer and the fast kinetics.
200 nm. These transitions appear as gradual changes in extension, likely because of the non-cooperative behavior of the large number of dimers, the small changes in extension per dimer and the fast kinetics.
For multiple tethers, we fitted narrowly distributed parameters (Figure 3D–H). For HMfB tethers we obtained an average of 109 ± 8 dimers per tether. A footprint of 30 base pairs per HMfB dimer would allow 121 dimers to bind this DNA molecule. It therefore appears that the DNA molecule is not fully saturated, implicating defects in stacking of the hypernucleosome (Figure 3C–I). Interestingly, the fitted number of dimers did not change in multiple stretch and release curves of the same hypernucleosome, suggesting that the dimers remain bound to and positioned on the DNA, even at high forces. The data, nor the model, can differentiate between multiple stretches of hypernucleosomes with intermitted stacking defects or one extended hypernucleosome in combination with a patch of bare DNA. Given the small stacking energy and the conserved number of dimers in successive pulling experiments, it is most likely that stacking defects are distributed over the tether, yielding an average size of the hypernucleosome of ∼100 dimers.
TPM experiments showed that HMfB compacts DNA slightly more than HMfA (Figure 2). In force spectroscopy, the force-extension curves for HMfA-DNA complexes were similar to those for HMfB-DNA complexes (Figure 3B), although the transitions between stacked and unstacked states occurred at lower forces, i.e. at 0.5 rather than 2 pN and the stiffness of an HMfA tether was smaller than that of HMfB (Figure 3B and E). We found a smaller stacking energy but a similar dimer occupancy for the HMfA-DNA complex compared to the HMfB-DNA complex. Also, the wrapping energy was similar (Figure 3G). The deflection angle appeared to be larger for HMfA. All fitting parameters are summarised in Table 1. The high similarity in the force-extension curves highlights the universality of the hypernucleosome structure, whereas the difference in stacking stiffness, stacking energy and deflection angle clearly distinguishes the mechanical properties of the protein-DNA filaments formed by the two homologues. Note that the statistical mechanics model is not detailed enough to pinpoint the residues that are responsible for this interaction. The stacking energy therefore reflects the net difference in free energy between a condensed and a more extended organization. Indeed, this low value implies that these dimer-dimer interactions are unstable, making the HMfA hypernucleosome a dynamic rather than static structure. This may be biologically relevant as differences in mechanical stability may modulate the balance between gene compaction and accessibility in vivo.
Table 1.
Fit results of Equation 10 to force-extension curves of HMfA (N = 82) and HMfB (N = 56) complexes. Values represent mean ± standard deviation
| HMfA | HMfB | |
|---|---|---|
| N dimers (–) | 105 ± 11 | 109 ± 8 |
| Stiffness (pN/nm) | 0.45 ± 0.11 | 0.83 ± 0.24 |
| Stacking energy (kBT) | 1.4 ± 0.4 | 3.5 ± 0.6 |
| Wrapping energy (kBT) | 5.7 ± 1.1 | 5.0 ± 1.1 |
| Deflection angle (°) | 14.9 ± 1.7 | 11.1 ± 1.0 |
Stacking interactions mediate hypernucleosome formation
The basis for stacking of HMfA and HMfB homodimers lies in their primary structure. We analyzed both histones in terms of interaction residues (Figure 1C). Using water refinement with HADDOCK (54) on the HMfB hypernucleosome crystal structure and modeling the HMfA hypernucleosome structure based on that same crystal structure, we were able to identify the possible stacking interactions within the hypernucleosome. For HMfB, we determined interactions between R48-D14, K30-E61 and E34-R65. For HMfA, we found interactions between K31-E62 and E35-K62. In order to investigate whether these interactions indeed mediate hypernucleosome formation, we expressed and purified HMfAK31A E35A and HMfBD14A K30A E34A. We performed similar force spectroscopy experiments as described above, but at higher protein concentrations, which reflects a lower affinity of the mutants, to achieve a similar density on the DNA. The resulting fit parameters are listed and compared to those of the wild type proteins in Table 2. We found that the DNA-protein complex with HMfAK31A E35A indeed unfolds at a lower force than that with wt HMfA (Figure 4A). Strikingly, the second unfolding plateau is largely absent, suggesting that the mutations not only affect the stacking between dimers, but also the wrapping of DNA. This is reflected in the reduced wrapping energies and a reduced deflection angle of the mutant complexes. When comparing HMfBD14A K30A E34A to wt HMfB, the effect is more pronounced (Figure 4B). The DNA-protein complex with HMfBD14A K30A E34A unfolds at lower force than with wt HMfB, meaning that the interactions within the hypernucleosome are destabilized. In addition to changes in the stacking and wrapping energies we also found a reduced stiffness of the tethers. This may be caused by a larger number of stacking defects, as a consequence of the reduced interaction strength between mutant dimers. Overall, force-extension data confirm the crucial role of residues K31 and E35 in HMfA and residues D14, K30 and E34 in HMfB in both mediating stacking interactions and stabilizing DNA wrapping.
Table 2.
Fit results of Equation (10) to the force-extension curves of representative HMfA and HMfB complexes and their stacking mutants shown in Figure 4A and B. Values represent mean ± standard error of fit
| HMfA | HMfB | |||
|---|---|---|---|---|
| wt | Mutant | wt | Mutant | |
| N dimers | 108.4 ± 0.5 | 114.70 ± 0.30 | 107.3 ± 0.4 | 108.4 ± 0.4 |
| Stiffness (pN/nm) | 0.424 ± 0.011 | 0.312 ± 0.004 | 0.771 ± 0.010 | 0.254 ± 0.004 |
| Stacking energy (kBT) | 1.811 ± 0.013 | 1.183 ± 0.007 | 3.440 ± 0.020 | 1.067 ± 0.009 |
| Wrapping energy (kBT) | 4.18 ± 0.05 | 0.686 ± 0.009 | 5.23 ± 0.07 | 0.638 ± 0.009 |
| Deflection angle (°) | 10.33 ± 0.04 | 6.73 ± 0.06 | 10.93 ± 0.05 | 6.95 ± 0.08 |
Figure 4.

Mutations in the stacking interface of HMfA and HMfB destabilize the hypernucleosome. (A) DNA–protein complexes of HMfAK31AE35A show similar compaction as wt HMfA below 0.5 pN, but unfold at lower force. The pink curve shows the wt data. Note that HMfAK31A E34A does not feature a second unfolding plateau. Grey circles show the refolding curve, which overlaps with the unfolding data. Solid lines show fits to Equation (10). The mutant features a reduced stiffness, stacking energy, wrapping energy and deflection angle, suggesting that the mutations not only affect stacking but also wrapping of DNA (B) DNA-protein complexes of HMfBD14A K30A E34A show similar stretching behavior as the HMfA mutant, but unfold at slightly lower force. A representative curve of wt HMfB is plotted in light blue. The fits show the same trend as for HMfA, but the mutations have a stronger effect, yielding similar mechanical properties for both mutants. Fit parameters are listed in Table 2.
The hypernucleosome forms a left-handed structure on DNA
Wrapping DNA into a hypernucleosome imposes a distinct chirality. To reveal handedness and torsional stiffness of the hypernucleosome, we used rotational force spectroscopy on a torsionally constrained HMfB-DNA complexes. As opposed to the experiments described in the previous section, the hypernucleosome was unable to release torsional stress by swiveling around the attachment point in this experiment. The resulting torque could stabilize the HMfB-DNA hypernucleosome when stretched.
We monitored the extension of the hypernucleosome with force, when applying negative (Figure 5A) and positive (Figure 5B) twist. The force plateau that represents the unstacking transition shifted up to 10 pN for negatively twisted complexes. As more force is required to unstack the hypernucleosome, the energy for the unstacking transition is increased. Large amounts of negative twist compacted the HMfB-DNA complex by buckling the hypernucleosome at forces below 1 pN. The second transition, from wrapped to unbent DNA, appeared unaffected by negative twist. Positive twist, on the other hand, only marginally extended the range of the unstacking transition to 2–10 pN. Such behavior is characteristic of a left-handed structure, and in agreement with the HMfB-DNA co-crystal structure (28). Strikingly, the bent DNA was stabilized by positive twist, but not by negative twist. Thus, it appears that unstacked HMfB dimers can better accommodate positive twist than bare DNA. In accordance, the rupture force for HMfB dimers increased when overtwisting the DNA.
Figure 5.

Stretching a torsionally constrained HMfB–DNA complex reveals left-handedness of the hypernucleosome, and shows increased affinity of HMfB dimers for positively twisted DNA. (A) Negative twist as a result of 5–25 rotations increases the unstacking force of the hypernucleosome. In addition, excess negative twist causes the HMfB-DNA complex to buckle, resulting in a dramatic decrease of its extension. Both observations point to the left-handedness of the hypernucleosome. The unwrapping transition appears largely unaffected, resulting in a continuous decondensation of the tether around 10 pN. (B) Positive twist as a result of 5–25 rotations recovers the unstacking plateau. Strikingly, positive twist increases the rupture force of the unwrapping transition of HMfB dimers. An increasing force is necessary to reverse the bending effect of HMfB dimers on DNA. At very low forces the beads appear to stick to the cover slip for certain curves (−5, −20 and +20 rotations, especially).
Unstacked HMfB–DNA complexes impose a right-handed DNA structure
The unstacking transition of a torsionally constrained hypernucleosome features an increased and more gradual force plateau compared to a torsionally unconstrained hypernucleosome (Supplementary Figure S2). As a result, the unstacking transition merges with the transition that we attribute to unwrapping DNA from the HMfB dimers (regime II to III in Figure 3A). Since HMfB dimers appeared to remain bound to the DNA, the unwrapping of one dimer would create additional torsional stress, which impedes unwrapping of the next dimer, resulting in an anti-cooperative transition that manifests itself by an increased force range in which unfolding takes place.
When maintaining a fixed force, and monitoring the extension as a function of twist, we were able to determine the handedness of the unstacked HMfB-DNA complex (Figure 6). At 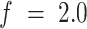 pN, both negative and positive twist induced a reduction in extension due to buckling of the hypernucleosome structure. Note that the presence of HMfB-induced bends favored plectonemes over DNA melting, which is apparent from the decrease in extension. In bare DNA, a twist-independent extension would be observed for negative twist at this force. Such asymmetry in twist response in HMfB-decorated tethers occurred only at
pN, both negative and positive twist induced a reduction in extension due to buckling of the hypernucleosome structure. Note that the presence of HMfB-induced bends favored plectonemes over DNA melting, which is apparent from the decrease in extension. In bare DNA, a twist-independent extension would be observed for negative twist at this force. Such asymmetry in twist response in HMfB-decorated tethers occurred only at 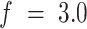 pN. At this force, negative twist helped to unwrap the HMfB dimers, increasing the length of the tether. Similar to bare DNA, positive twist induced supercoils in the tether, which leads to reduction of the extension. This implies that the unstacked HMfB–DNA complex formed a right-handed superhelix and that the HMfB dimer DNA complexes were stabilized by positive torque. The force required to completely unwrap the DNA also depended on the applied twist. In this intermediate force regime, HMfB dimers appeared to have a higher wrapping affinity for positively twisted DNA than for negatively twisted or relaxed DNA. This observation was supported by the release curves (Supplementary Figure S3), which showed that positively twisted complexes more readily restack into hypernucleosomes than relaxed or negatively twisted tethers.
pN. At this force, negative twist helped to unwrap the HMfB dimers, increasing the length of the tether. Similar to bare DNA, positive twist induced supercoils in the tether, which leads to reduction of the extension. This implies that the unstacked HMfB–DNA complex formed a right-handed superhelix and that the HMfB dimer DNA complexes were stabilized by positive torque. The force required to completely unwrap the DNA also depended on the applied twist. In this intermediate force regime, HMfB dimers appeared to have a higher wrapping affinity for positively twisted DNA than for negatively twisted or relaxed DNA. This observation was supported by the release curves (Supplementary Figure S3), which showed that positively twisted complexes more readily restack into hypernucleosomes than relaxed or negatively twisted tethers.
Figure 6.

Torque stabilizes the hypernucleosome and reveals a right-handed chirality of unstacked HMfB-DNA complexes. A torsionally constrained HMfB hypernucleosome was compacted by twisting the molecule under constant force. At 2.0 pN, both positive and negative twist reduced the extension, presumably by buckling. At higher forces, negative twist caused the HMfB dimers to unwrap from the DNA, which induced stretching of the HMfB–DNA complex. These measurements suggest that the unstacked HMfB–DNA complex is right-handed. As a reference, we included a twist-extension curve of bare DNA at 1.0 pN. Note that the slope for the HMfB–DNA complex is much smaller than that of bare DNA.
The slope of the curve beyond the buckling transitions, as shown in Figure 6, reveals more details of the twisted complex. Bare plectonemic DNA reduces its extension by 40–100 nm per rotation, depending on force, as the size of the loop at the tip of the plectoneme decreases with force (55). For unstacked HMfB-DNA complexes we measured a reduction of the extension of 16 nm per rotation, much lower than bare DNA, which was independent of force. This is consistent with a fixed HMfB-induced bend at the tip of the plectoneme that defines the tip curvature, and thus the geometry of the plectoneme.
The structural origin of the change in handedness upon unstacking may be found in the relaxation of the constraints imposed by the hypernucleosome. Since separate HMf dimers are free to optimize their conformation, it could be that the dimer rearranges relative to the structure found in the hypernucleosome. Such a change was also proposed for eukaryotic centromeric nucleosomes, that can be forced into right-handed heterotypic tetrasomes (hemisomes) (38) and for canonical tetrasomes under torsional stress (39). In both cases, it was implied that this change in handedness may have functional consequences in genome regulation. HMf may use similar mechanisms to function.
DISCUSSION
The hypernucleosome structure may be a key feature of archaeal chromosome compaction. Here we characterized the structural and mechanical properties of single DNA tethers containing such hypernucleosomes. We found that hypernucleosomes composed of stacked M. fervidus histone homodimers HMfA and HMfB compact DNA much stronger than similar DNA-bending proteins. Both HMfA–DNA and HMfB–DNA complexes featured a two-step unfolding mechanism that could be captured quantitatively in the transitions between a hypernucleosome, an array of wrapped dimers that kink the DNA trajectory, and a stretched DNA molecule to which dimers remain bound. HMfB featured a higher stacking energy than HMfA, resulting in a larger occupancy as well as a higher stability of the hypernucleosome against force.
There is an interesting interplay between stiffness and stacking energy of the fiber. On a global scale, reducing either will make the genome more pliable and will facilitate dynamic higher order (re-) organization of the genome. On a local scale the effect is quite different. A lower stacking energy will increase the number of defects, allowing other factors to bind the vacant DNA. A lower stiffness on the other hand retains DNA occupancy by HMf proteins. We observed different stiffness and stacking energies for the two HMf variants, suggesting that this local modulation of genome organization is key to different functional roles of the two proteins.
The stiffness of hypernucleosomes is a bit larger than that of stacked eukaryotic nucleosomes (47). This may be explained by the different nature of the stacking interactions. Interactions between eukaryotic nucleosomes are known to be mediated by flexible histone tails. Structural data suggest that the archaeal hypernucleosome on the other hand is mediated by stacking interactions between the globular histones that form the hypernucleosome (28). Stacking energies of HMfA and HMfB dimers ( 2 kBT) are much lower than the 17 kBT that we reported for eukaryotic nucleosome stacking (56). In contrast, the unwrapping of DNA from the HMfA and HMfB dimers starts at similar forces as the unwrapping of eukaryotic nucleosomes. However, in hypernucleosomes this happened reversibly, whereas nucleosome unwrapping is generally not in equilibrium.
2 kBT) are much lower than the 17 kBT that we reported for eukaryotic nucleosome stacking (56). In contrast, the unwrapping of DNA from the HMfA and HMfB dimers starts at similar forces as the unwrapping of eukaryotic nucleosomes. However, in hypernucleosomes this happened reversibly, whereas nucleosome unwrapping is generally not in equilibrium.
The equilibrium conditions made it possible to capture the entire force-extension curve in a statistical physics model. This worked better for the unstacking transition than for the unwrapping transition. It should be noted though that modelling of the wrapped dimers as kinks in the DNA trajectory might be somewhat naive, as the high occupancy of the DNA yields dimer-dimer distances that are much smaller than the typical deflection length for which the Kulić and Schiessel model is valid (45). Moreover, this model assumes a flat one-dimensional kink, while the super-helical structure of the unfolded hypernucleosome may be more complex. A deflection angle of ∼15°, which matched the curves best, is rather small for the 30 base pair footprint of a HMfA or HMfB dimer. Nevertheless, the resulting force dependent reduction of the end-to-end distance matched quantitatively with the force-extension curve. We resolved a clear transition from a flexible (regime II) to a stiffer WLC (regime III), using the same number of dimers as in the unstacking transition at low force, indicating that this unfolding intermediate can be effectively modeled as an array of wrapped dimers.
In the crystal structure, the hypernucleosome compacts DNA in a left-handed manner, which resembles DNA wrapping by eukaryotic histones. In accordance, we found that negative supercoiling stabilized this most compact regime of the HMfB–DNA complex. However, after breaking the stacking by force, a right-handed HMfB-DNA structure was observed. Such a right-handed HMfB-DNA complex has been described previously as a functional form, responding to environmental cues in vitro (17,31). Depending on conditions, force and torque, a right-handed HMfB–DNA complex may have a biological function in Archaea.
The archaeal genome is also organized by other NAPs. The components of the organizational machinery however differ per phylum, class and order. Histones are found in almost every branch of the archaeal domain. Currently, evidence in support of hypernucleosome formation has only been reported for M. fervidus using x-ray crystallography, and for T. kodakarensis showing in vivo effects on transcription and MNase digestion results (26,28). It is likely though that histones from other species also assemble into hypernucleosomes (29). Initiation of the hypernucleosome is probably stochastic, and initial complex formation at multiple sites may lead to frustration when two expanding hypernucleosomes meet. Therefore, long stretches of hypernucleosome will probably be rare in the presence of other factors that regulate chromatin structure (57). Ultimately, long straight hypernucleosomes might clash with physical boundaries of the cell. Such large and highly ordered structures should be visible with microscopic techniques, but empirical evidence for such large filaments is currently lacking (26).
M. fervidus histones form homo- and heterodimers, and, based on sequence similarities, histones from other species may be able to do so as well. The majority of archaeal chromosomes contains genes coding for two or more different histones. This may provide Archaea with a means to regulate gene expression via changing the length and stability of the hypernucleosome in response to growth phase or environmental cues (17). For Archaea, transcription regulation may follow a variety of mechanisms (58). For instance, heterodimers may bind to DNA and interact with other heterodimers via one multimerization site, while other sites remain unbound. Homodimers may form hypernucleosomes, which can be interrupted by other homodimers incapable of certain interactions. Histone variants proposed to be capable of perturbing hypernucleosome growth have been referred to as ‘capstones’ (59). Furthermore, sequence specificity in certain histone variants may direct histones to specific genomic targets. The specialization of the eukaryotic histone and emergence of the N-terminal tail have diverted the chromatin organization away from the hypernucleosome, eventually allowing a wide range of chromatin factors to access the nucleosome and histone surface to regulate chromatin biology. (29). The current methods of biological analysis at the single-molecule level allow for detailed, mechanistic measurements that have the potential to shed light on the evolutionary path of genome compaction and transcription regulation.
DATA AVAILABILITY
The datasets generated during the current study are available from the 4TU repository (https://data.4tu.nl/) under DOI: https://doi.org/10.4121/13265459.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kathleen Sandman and John Reeve for providing wild type HMfA and HMfB. We thank Helmut Schiessel for fruitful discussions and guidance during the development of the statistical mechanics model.
Author contributions: R.T.D., J.v.N., H.v.I. and R.A.v.d.V. conceived the research. B.H., T.B., A.M.E., G.A.J.T.K and N.C.S.K. carried out the experiments, and B.H., J.v.N. and T.B. analyzed the data. T.B. and J.v.N. developed the quantitative model. M.T. provided technical support. The manuscript was written by B.H., T.B., A.M.E, C.v.E, R.T.D, J.v.N. and H.v.I.
Contributor Information
Bram Henneman, Leiden Institute of Chemistry, Leiden University, Einsteinweg 55, 2333CC Leiden, The Netherlands.
Thomas B Brouwer, Biological and Soft Matter Physics, Huygens-Kamerlingh Onnes Laboratory, Leiden University, Niels Bohrweg 2, 2333CA Leiden, The Netherlands.
Amanda M Erkelens, Leiden Institute of Chemistry, Leiden University, Einsteinweg 55, 2333CC Leiden, The Netherlands.
Gert-Jan Kuijntjes, Biological and Soft Matter Physics, Huygens-Kamerlingh Onnes Laboratory, Leiden University, Niels Bohrweg 2, 2333CA Leiden, The Netherlands.
Clara van Emmerik, Bijvoet Center for Biomolecular Research, Utrecht University, Padualaan 8, 3584CH Utrecht, The Netherlands.
Ramon A van der Valk, Leiden Institute of Chemistry, Leiden University, Einsteinweg 55, 2333CC Leiden, The Netherlands.
Monika Timmer, Leiden Institute of Chemistry, Leiden University, Einsteinweg 55, 2333CC Leiden, The Netherlands.
Nancy C S Kirolos, Leiden Institute of Chemistry, Leiden University, Einsteinweg 55, 2333CC Leiden, The Netherlands.
Hugo van Ingen, Bijvoet Center for Biomolecular Research, Utrecht University, Padualaan 8, 3584CH Utrecht, The Netherlands.
John van Noort, Biological and Soft Matter Physics, Huygens-Kamerlingh Onnes Laboratory, Leiden University, Niels Bohrweg 2, 2333CA Leiden, The Netherlands.
Remus T Dame, Leiden Institute of Chemistry, Leiden University, Einsteinweg 55, 2333CC Leiden, The Netherlands; Centre for Microbial Cell Biology, Leiden University, Einsteinweg 55, 2333CC Leiden, The Netherlands.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Netherlands Organization for Scientific Research [VICI 016.160.613 to R.T.D., VICI 680.47.616 to J.v.N., VIDI 723.013.010 to H.v.I.]; FOM Foundation for Fundamental Research on Matter program ‘Crowd management: The physics of genome processing in complex environments’ [to R.T.D.]; Human Frontiers Science program [RGP0014/2014 to R.T.D.]. Funding for open access charge: Leiden University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Williams T.A., Foster P.G., Cox C.J., Embley T.M.. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2013; 504:231–236. [DOI] [PubMed] [Google Scholar]
- 2. Zaremba-Niedzwiedzka K., Caceres E.F., Saw J.H., Backstrom D., Juzokaite L., Vancaester E., Seitz K.W., Anantharaman K., Starnawski P., Kjeldsen K.U.et al.. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017; 541:353–358. [DOI] [PubMed] [Google Scholar]
- 3. Eme L., Spang A., Lombard J., Stairs C.W., Ettema T.J.G.. Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 2017; 15:711–723. [DOI] [PubMed] [Google Scholar]
- 4. Cox C.J., Foster P.G., Hirt R.P., Harris S.R., Embley T.M.. The archaebacterial origin of eukaryotes. PNAS. 2008; 105:20356–20361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bowman G.D., Poirier M.G.. Post-translational modifications of histones that influence nucleosome dynamics. Chem. Rev. 2015; 115:2274–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sandman K., Reeve J.N.. Archaeal chromatin proteins: different structures but common function. Curr. Opin. Microbiol. 2005; 8:656–661. [DOI] [PubMed] [Google Scholar]
- 7. Peeters E., Driessen R.P., Werner F., Dame R.T.. The interplay between nucleoid organization and transcription in archaeal genomes. Nat. Rev. Microbiol. 2015; 13:333–341. [DOI] [PubMed] [Google Scholar]
- 8. Luijsterburg M.S., White M.F., van Driel R., Dame R.T.. The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2008; 43:393–418. [DOI] [PubMed] [Google Scholar]
- 9. Becker E.A., Seitzer P.M., Tritt A., Larsen D., Krusor M., Yao A.I., Wu D., Madern D., Eisen J.A., Darling A.E.et al.. Phylogenetically driven sequencing of extremely halophilic archaea reveals strategies for static and dynamic osmo-response. PLos Genet. 2014; 10:e1004784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bult C.J., White O., Olsen G.J., Zhou L., Fleischmann R.D., Sutton G.G., Blake J.A., FitzGerald L.M., Clayton R.A., Gocayne J.D.et al.. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996; 273:1058–1073. [DOI] [PubMed] [Google Scholar]
- 11. Darcy T.J., Sandman K., Reeve J.N.. Methanobacterium formicicum, a mesophilic methanogen, contains three HFo histones. J. Bacteriol. 1995; 177:858–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tabassum R., Sandman K.M., Reeve J.N.. HMt, a histone-related protein from Methanobacterium thermoautotrophicum delta H. J. Bacteriol. 1992; 174:7890–7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Henneman B., Dame R.T.. Archaeal histones: dynamic and versatile genome architects. Aims Microbiol. 2015; 1:72–81. [Google Scholar]
- 14. Fukui T., Atomi H., Kanai T., Matsumi R., Fujiwara S., Imanaka T.. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 2005; 15:352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anderson I., Djao O.D., Misra M., Chertkov O., Nolan M., Lucas S., Lapidus A., Del Rio T.G., Tice H., Cheng J.F.et al.. Complete genome sequence of Methanothermus fervidus type strain (V24S). Stand Genomic Sci. 2010; 3:315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sandman K., Krzycki J.A., Dobrinski B., Lurz R., Reeve J.N.. HMf, a DNA-binding protein isolated from the hyperthermophilic archaeon Methanothermus fervidus, is most closely related to histones. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:5788–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sandman K., Grayling R.A., Dobrinski B., Lurz R., Reeve J.N.. Growth-phase-dependent synthesis of histones in the archaeon methanothermus-Fervidus. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:12624–12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Musgrave D.R., Sandman K.M., Reeve J.N.. DNA binding by the archaeal histone HMf results in positive supercoiling. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:10397–10401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grayling R.A., Sandman K., Reeve J.N.. DNA stability and DNA binding proteins. Adv. Protein Chem. 1996; 48:437–467. [DOI] [PubMed] [Google Scholar]
- 20. Marc F., Sandman K., Lurz R., Reeve J.N.. Archaeal histone tetramerization determines DNA affinity and the direction of DNA supercoiling. J. Biol. Chem. 2002; 277:30879–30886. [DOI] [PubMed] [Google Scholar]
- 21. Howard M.T., Sandman K., Reeve J.N., Griffith J.D.. HMf, a histone-related protein from the hyperthermophilic archaeon Methanothermus fervidus, binds preferentially to DNA containing phased tracts of adenines. J. Bacteriol. 1992; 174:7864–7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nalabothula N., Xi L., Bhattacharyya S., Widom J., Wang J.P., Reeve J.N., Santangelo T.J., Fondufe-Mittendorf Y.N.. Archaeal nucleosome positioning in vivo and in vitro is directed by primary sequence motifs. BMC Genomics. 2013; 14:391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bailey K.A., Pereira S.L., Widom J., Reeve J.N.. Archaeal histone selection of nucleosome positioning sequences and the procaryotic origin of histone-dependent genome evolution. J. Mol. Biol. 2000; 303:25–34. [DOI] [PubMed] [Google Scholar]
- 24. Bailey K.A., Marc F., Sandman K., Reeve J.N.. Both DNA and histone fold sequences contribute to archaeal nucleosome stability. J. Biol. Chem. 2002; 277:9293–9301. [DOI] [PubMed] [Google Scholar]
- 25. Ammar R., Torti D., Tsui K., Gebbia M., Durbic T., Bader G.D., Giaever G., Nislow C.. Chromatin is an ancient innovation conserved between Archaea and Eukarya. Elife. 2012; 1:e00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maruyama H., Harwood J.C., Moore K.M., Paszkiewicz K., Durley S.C., Fukushima H., Atomi H., Takeyasu K., Kent N.A.. An alternative beads-on-a-string chromatin architecture in Thermococcus kodakarensis. EMBO Rep. 2013; 14:711–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rojec M., Hocher A., Stevens K.M., Merkenschlager M., Warnecke T.. Chromatinization of Escherichia coli with archaeal histones. Elife. 2019; 8:e52542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mattiroli F., Bhattacharyya S., Dyer P.N., White A.E., Sandman K., Burkhart B.W., Byrne K.R., Lee T., Ahn N.G., Santangelo T.J.et al.. Structure of histone-based chromatin in Archaea. Science. 2017; 357:609–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Henneman B., van Emmerik C., van Ingen H., Dame R.T.. Structure and function of archaeal histones. PLos Genet. 2018; 14:e1007582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Efremov A.K., Qu Y., Maruyama H., Lim C.J., Takeyasu K., Yan J.. Transcriptional repressor TrmBL2 from Thermococcus kodakarensis forms filamentous nucleoprotein structures and competes with histones for DNA binding in a Salt- and DNA Supercoiling-dependent manner. J. Biol. Chem. 2015; 290:15770–15784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Musgrave D., Forterre P., Slesarev A.. Negative constrained DNA supercoiling in archaeal nucleosomes. Mol. Microbiol. 2000; 35:341–349. [DOI] [PubMed] [Google Scholar]
- 32. Driessen R.P., Lin S.N., Waterreus W.J., van der Meulen A.L., van der Valk R.A., Laurens N., Moolenaar G.F., Pannu N.S., Wuite G.J., Goosen N.et al.. Diverse architectural properties of Sso10a proteins: Evidence for a role in chromatin compaction and organization. Sci. Rep. 2016; 6:29422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van der Valk R.A., Laurens N., Dame R.T.. Tethered particle motion analysis of the DNA binding properties of architectural proteins. Methods Mol. Biol. 2017; 1624:127–143. [DOI] [PubMed] [Google Scholar]
- 34. Pollock W.B., Rosell F.I., Twitchett M.B., Dumont M.E., Mauk A.G.. Bacterial expression of a mitochondrial cytochrome c. Trimethylation of lys72 in yeast iso-1-cytochrome c and the alkaline conformational transition. Biochemistry. 1998; 37:6124–6131. [DOI] [PubMed] [Google Scholar]
- 35. Lowary P.T., Widom J.. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998; 276:19–42. [DOI] [PubMed] [Google Scholar]
- 36. Brouwer T.B., Kaczmarczyk A., Pham C., van Noort J.. Unraveling DNA organization with single-molecule force spectroscopy using magnetic tweezers. Methods Mol. Biol. 2018; 1837:317–349. [DOI] [PubMed] [Google Scholar]
- 37. Henneman B., Heinsman J., Battjes J., Dame R.T.. Quantitation of DNA-binding affinity using tethered particle motion. Methods Mol. Biol. 2018; 1837:257–275. [DOI] [PubMed] [Google Scholar]
- 38. Henikoff S., Furuyama T.. The unconventional structure of centromeric nucleosomes. Chromosoma. 2012; 121:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vlijm R., Lee M., Lipfert J., Lusser A., Dekker C., Dekker N.H.. Nucleosome assembly dynamics involve spontaneous fluctuations in the handedness of tetrasomes. Cell Rep. 2015; 10:216–225. [DOI] [PubMed] [Google Scholar]
- 40. Strick T.R., Allemand J.F., Croquette V., Bensimon D.. Physical approaches to the study of DNA. J. Stat. Phys. 1998; 93:647–672. [Google Scholar]
- 41. Strick T., Allemand J.F., Croquette V., Bensimon D.. Twisting and stretching single DNA molecules. Prog. Biophys Mol. Bio. 2000; 74:115–140. [DOI] [PubMed] [Google Scholar]
- 42. Meng H., Andresen K., van Noort J.. Quantitative analysis of single-molecule force spectroscopy on folded chromatin fibers. Nucleic. Acids. Res. 2015; 43:3578–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marko J.F., Siggia E.D.. Stretching DNA. Macromolecules. 1995; 28:8759–8770. [Google Scholar]
- 44. Pereira S.L., Grayling R.A., Lurz R., Reeve J.N.. Archaeal nucleosomes. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:12633–12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kulic I.M., Mohrbach H., Thaokar R., Schiessel H.. Equation of state of looped DNA. Phys. Rev. E. 2007; 75:011913. [DOI] [PubMed] [Google Scholar]
- 46. Bustamante C., Bryant Z., Smith S.B.. Ten years of tension: single-molecule DNA mechanics. Nature. 2003; 421:423–427. [DOI] [PubMed] [Google Scholar]
- 47. Meng H., Andresen K., van Noort J.. Quantitative analysis of single-molecule force spectroscopy on folded chromatin fibers. Nucleic Acids Res. 2015; 43:3578–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tomschik M., Karymov M.A., Zlatanova J., Leuba S.H.. The archaeal histone-fold protein HMf organizes DNA into bona fide chromatin fibers. Structure. 2001; 9:1201–1211. [DOI] [PubMed] [Google Scholar]
- 49. Driessen R.P., Meng H., Suresh G., Shahapure R., Lanzani G., Priyakumar U.D., White M.F., Schiessel H., van Noort J., Dame R.T.. Crenarchaeal chromatin proteins Cren7 and Sul7 compact DNA by inducing rigid bends. Nucleic Acids Res. 2013; 41:196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Feng Y., Yao H., Wang J.. Crystal structure of the crenarchaeal conserved chromatin protein Cren7 and double-stranded DNA complex. Protein Sci. 2010; 19:1253–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen L., Chen L.R., Zhou X.E., Wang Y., Kahsai M.A., Clark A.T., Edmondson S.P., Liu Z.J., Rose J.P., Wang B.C.et al.. The hyperthermophile protein Sso10a is a dimer of winged helix DNA-binding domains linked by an antiparallel coiled coil rod. J. Mol. Biol. 2004; 341:73–91. [DOI] [PubMed] [Google Scholar]
- 52. Driessen R.P., Sitters G., Laurens N., Moolenaar G.F., Wuite G.J., Goosen N., Dame R.T.. Effect of temperature on the intrinsic flexibility of DNA and its interaction with architectural proteins. Biochemistry. 2014; 53:6430–6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. van Noort J., Verbrugge S., Goosen N., Dekker C., Dame R.T.. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:6969–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. van Zundert G.C.P., Rodrigues J., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S.J., van Dijk M., de Vries S.J., Bonvin A.. The HADDOCK2.2 web server: User-Friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016; 428:720–725. [DOI] [PubMed] [Google Scholar]
- 55. Lipfert J., Klijnhout S., Dekker N.H.. Torsional sensing of small-molecule binding using magnetic tweezers. Nucleic Acids Res. 2010; 38:7122–7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kaczmarczyk A., Allahverdi A., Brouwer T.B., Nordenskiold L., Dekker N.H., van Noort J.. Single-molecule force spectroscopy on histone H4 tail-cross-linked chromatin reveals fiber folding. J. Biol. Chem. 2017; 292:17506–17513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sanders T.J., Lammers M., Marshall C.J., Walker J.E., Lynch E.R., Santangelo T.J.. TFS and Spt4/5 accelerate transcription through archaeal histone-based chromatin. Mol. Microbiol. 2018; 111:784–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nishida H., Oshima T.. Archaeal histone distribution is associated with archaeal genome base composition. J. Gen. Appl. Microbiol. 2017; 63:28–35. [DOI] [PubMed] [Google Scholar]
- 59. Stevens K.M., Swadling J.B., Hocher A., Bang C., Gribaldo S., Schmitz R.A., Warnecke T.. Histone variants in archaea and the evolution of combinatorial chromatin complexity. Proc. Natl. Acad. Sci. U.S.A. 2020; doi:10.1073/pnas.2007056117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available from the 4TU repository (https://data.4tu.nl/) under DOI: https://doi.org/10.4121/13265459.



