I read with interest the Personal View by Rosanna Peeling and colleagues1 on the performance of rapid antigen detection tests (Ag-RDTs) across different SARS-CoV-2 prevalence settings. The authors elegantly show that the negative predictive value (NPV) increases with decreasing disease prevalence and conclude that “for asymptomatic individuals in low prevalence settings, for travel, return to schools, workplaces, and mass gatherings, Ag-RDTs with high negative predictive values can be used with confidence to rule out infection”.1 However, the clinical interpretation of NPVs requires attention.
Independent evaluation of different Ag-RDTs has shown that their sensitivity ranges between 70% and 90% (lower confidence limits 50–80%) in symptomatic individuals,2 but it deteriorates remarkably (<50%) in asymptomatic close contacts,3 in those with low nasopharyngeal viral loads,2 and in paediatric patients.4 By contrast, the NPV is excellent (>97%) in all instances,2, 3, 4 which has led most investigators to conclude that a negative Ag-RDT might reliably rule out the infection in low-prevalence settings.1, 4
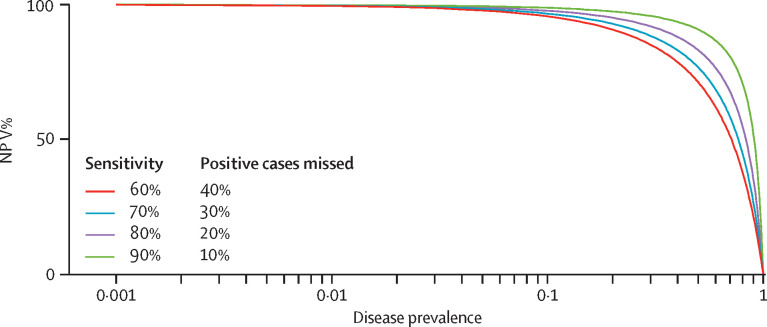
Predictive values are inherently dependent on disease prevalence and, as such, they can be misleading. When the probability of the disease is low, the NPV of any diagnostic test is high, irrespective of its sensitivity (figure ). Assuming a disease prevalence of 2·5%, an Ag-RDT with 80% sensitivity and 97% specificity would result in a NPV of 99·5%, whereas one with 50% sensitivity (same specificity) would yield a NPV of 98·7%. Although both NPVs are excellent, the second test would miss five out of ten infected cases (the false-negative rate is equivalent to 100 minus the sensitivity, which equals 50%).
Figure.
NPV in relation to disease prevalence.
NPV=negative predictive value.
Current mathematical models suggest that the SARS-CoV-2 pandemic is driven by early and asymptomatic viral transmission and that prompt identification of low-risk and asymptomatic individuals has the strongest effect in controlling viral spread.5 Thus, if our goal is to ensure SARS-CoV-2-free environments (eg, workplaces, schools, gatherings) by allowing those who test negative to resume their usual activities, false-negative results should not be tolerable; to achieve this goal, screening tools with the highest possible sensitivity are required, since sensitivity is the only parameter that reflects the rate of cases who erroneously test negative, irrespective of the disease prevalence.
Because most of the currently available Ag-RDTs have a considerable false-negative rate,2, 3, 4 health-care professionals should be aware that a single negative test cannot conclusively rule out SARS-CoV-2 infection; this is particularly true in low-prevalence settings, where the typically excellent NPV of Ag-RDTs is misleading.
I report non-financial support from ELPEN and grants from University of Patras, outside the submitted work.
References
- 1.Peeling RW, Olliaro PL, Boeras DI, Fongwen N. Scaling up COVID-19 rapid antigen tests: promises and challenges. Lancet Infect Dis. 2021 doi: 10.1016/S1473-3099(21)00048-7. published online Feb 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.FIND FIND evaluations of SARS-CoV-2 assays. https://www.finddx.org/covid-19/sarscov2-eval/
- 3.Torres I, Poujois S, Albert E, Colomina J, Navarro D. Evaluation of a rapid antigen test (Panbio COVID-19 Ag rapid test device) for SARS-CoV-2 detection in asymptomatic close contacts of COVID-19 patients. Clin Microbiol Infect. 2021 doi: 10.1016/j.cmi.2020.12.022. published online Jan 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albert E, Torres I, Bueno F, et al. Field evaluation of a rapid antigen test (Panbio COVID-19 Ag rapid test device) for COVID-19 diagnosis in primary healthcare centres. Clin Microbiol Infect. 2021;27:472.e7–472.e10. doi: 10.1016/j.cmi.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCombs A, Kadelka C. A model-based evaluation of the efficacy of COVID-19 social distancing, testing and hospital triage policies. PLoS Comput Biol. 2020;16 doi: 10.1371/journal.pcbi.1008388. [DOI] [PMC free article] [PubMed] [Google Scholar]