ABSTRACT
Shiga toxin-producing Escherichia coli, a foodborne bacterial pathogen, has been linked to a broad spectrum of clinical outcomes ranging from asymptomatic carriage to fatal hemolytic uremic syndrome (HUS). Here, we collected clinical data and STEC strains from HUS patients from 1994 through 2018, whole-genome sequencing was performed to molecularly characterize HUS-associated STEC strains, statistical analysis was conducted to identify bacterial genetic factors associated with severe outcomes in HUS patients. O157:H7 was the most predominant serotype (57%) among 54 HUS-associated STEC strains, followed by O121:H19 (19%) and O26:H11 (7%). Notably, some non-predominant serotypes such as O59:H17 (2%) and O109:H21 (2%) also caused HUS. All O157:H7 strains with one exception belonged to clade 8. During follow-up at a median of 4 years, 41% of the patients had renal sequelae. Fifty-nine virulence genes were found to be statistically associated with severe renal sequelae, these genes encoded type II and type III secretion system effectors, chaperones, and other factors. Notably, virulence genes associated with severe clinical outcomes were significantly more prevalent in O157:H7 strains. In contrast, genes related to mild symptoms were evenly distributed across all serotypes. The whole-genome phylogeny indicated high genomic diversity among HUS-STEC strains. No distinct cluster was found between HUS and non-HUS STEC strains. The current study showed that O157:H7 remains the main cause of STEC-associated HUS, despite the rising importance of other non-O157 serotypes. Besides, O157:H7 is associated with severe renal sequelae in the follow-up, which could be a risk factor for long-term prognosis in HUS patients.
KEYWORDS: Shiga toxin-producing Escherichia coli, hemolytic uremic syndrome, clinical outcomes, O157:H7, virulence genes, whole-genome sequencing
Introduction
Shiga toxin-producing E. coli (STEC) infections can cause a spectrum of clinical manifestations ranging from asymptomatic carriage to the life-threatening complication hemolytic uremic syndrome (HUS), which is characterized by microangiopathic hemolytic anemia, thrombocytopenia, and acute kidney injury (AKI) [1,2]. Most HUS cases are caused by STEC infection, whereas atypical HUS (aHUS) is associated with gene mutations or autoantibodies, which results in complement dysregulation [3]. It is estimated that 6–25% of patients with STEC-related hemorrhagic colitis develop HUS, among which, up to 50% of patients need dialysis during the acute phase [4]. STEC associated-HUS has been described in all age groups but most susceptible to very young children below five years and the elderly [5]. Serotype O157:H7 has been implicated worldwide as the major cause of post-diarrhea HUS either during STEC outbreaks or in sporadic cases [1]. Previous study showed that patients with HUS was significantly more likely to be infected with O157:H7 strains belonging to clade 8 lineage, which might acquire critical factors leading to more severe disease [6]. It is estimated that up to 15% of STEC-infected children below ten years developed HUS [1]. Nevertheless, non-O157:H7 STEC strains, especially those of serogroups O26, O103, O111, and O145, are variably present as causes of HUS and diarrhea throughout Europe [7–9] and other countries [10,11]. In 2011, an E. coli O104: H4 outbreak resulted in 4,321 confirmed cases, 852 HUS, and 54 deaths in Europe [12–14], highlighting the clinical importance of non-O157 serotypes.
The pathophysiology of HUS is thought to be due to the production of bacteriophage-encoded Shiga toxin (Stx), which causes endothelial damage and activates the complement cascade, leading to thrombotic microangiopathy that principally affects the kidney. It may also involve the central nervous system (CNS), heart, liver, pancreas, and hematopoietic cells [3,4,15]. Stx comprises two immunologically distinct toxin types, Stx1 and Stx2, which share 56% amino acid sequence similarity [16]. Strains producing Stx2, especially Stx2a, Stx2c, or Stx2d subtypes, show a stronger association with the development of HUS than other subtypes [17,18]. Aside from stx, additional virulence factors also contribute to the pathological process. For instance, eae-encoded intimin, is an important “effacement/adherence” factor in intestinal colonization [19,20], which can induce a strong antibody response in HUS patients [21]. Several other potential adherence factors, such as ToxB, Saa, Iha, Efa1, and Paa, have also been described in HUS-STEC strains [22,23]. However, there is no single or combination of genetic markers that can predict the potential of STEC strains to cause HUS, as various bacterial and host-related factors contribute to the pathogenicity. The correlation between bacterial genetic factors and clinical symptoms is not fully understood.
The prognosis of HUS has improved over time due to the advances in intensive care and dialysis. However, HUS in children is still a significant cause of mortality, which varies between 1% and 5% of cases [24]. In the acute phase, nearly 50% of the STEC-HUS patients need dialysis [25]. In the long term, 15–30% develop proteinuria, 5–15% hypertension, 9–18% decreased kidney function, and 3% even end-stage renal disease [25]. Prolonged dialysis and the long duration of oligo-anuria have been described as the most significant risk factors for an unfavorable long-term outcome. Besides, the presence of neurological symptoms, high white blood cell counts, high hematocrit levels, have also been suggested as risk factors for poor prognosis [25–27]. However, there is no curative treatment for STEC-HUS yet. Antibiotic therapy during the acute gut STEC infection is considered contraindicated as it boosts the release of Stx and stimulates the absorption of toxin, which ultimately increases the risk of HUS [28].
In Sweden, human STEC O157 cases have been mandatory to report since 1996 and all serotypes since July 2004 [29]. Most STEC infections were sporadic cases or smaller family outbreaks, although a few community outbreaks have been described [30–32]. In 2005, a large O157 outbreak occurred on the west coast of Sweden, 135 cases were recorded totally, including 11 cases of HUS [29]. However, limited study had described molecular characteristic of HUS-causing STECs of all serotypes in Sweden so far. Comprehensive analysis is needed to unveil the entire molecular picture of HUS-STEC strains, and to identify bacterial genetic factors associated with HUS clinical outcomes. In the current study, we report STEC associated-HUS cases treated in Gothenburg and Stockholm in Sweden from 1994 through 2018. By whole-genome sequencing-based molecular characterization and comparative genomic analysis, we attempt to ascertain molecular features of highly pathogenic HUS-associated STEC strains, and to identify bacterial genetic markers for severe outcomes in HUS patients.
Materials and Methods
Case definition, collection of isolates and clinical data
HUS was characterized by three primary symptoms: thrombocytopenia, microangiopathic hemolytic anemia, and acute kidney injury [33]. The HUS-associated STEC isolates were collected from routine STEC diagnostics. Clinical data were collected by reviewing the patients’ medical records. The metadata are shown in Table S1. Two isolates were originated from the same patient who sought medical treatment in two cities.
We investigated six clinical parameters during the acute phase of HUS. For each parameter, we divided patients into two groups: severe and mild. Hemoglobin concentration (Hb) ≤65 g/L, leukocytes (LPK) >20 × 109/L, duration of dialysis (dialysdur) >10 days, anuria >1 day, any kind of complication, and antihypertensive treatment (btdur) for more than one day, were considered as severe (Table 1).
Table 1.
Clinical outcomes of 54 patients with STEC-HUS
| Clinical parameter | Severe | Mild | No. of severe/all patients* (%) |
|---|---|---|---|
| HUS episode | |||
| Hemoglobin (g/L) | ≤65 | >65 | 17/54 (31.48) |
| Leukocytes (×109/L) | >20 | ≤20 | 28/54 (51.85) |
| Duration of anuria (day) | ≥1 | < 1 | 29/53 (54.72) |
| Any kind of complication during the HUS episode | Yes | No | 23/54 (42.59) |
| Duration of dialysis (days) | ≥10 | <10 | 26/53 (49.06) |
| Duration of antihypertensive treatment (day) | ≥1 | <1 | 17/50 (34.00) |
| Follow-up | |||
| Estimated glomerular filtration rate (ml/min/m2 body surface area) at follow-up | <90 | ≥90 | 6/42 (14.29) |
| Hypertension or antihypertensive treatment at follow-up | Yes | No | 4/43 (9.30) |
| Albuminuria at follow-up | Yes | No | 14/40 (35.00) |
| Some renal sequelae at follow-up | Yes | No | 18/44 (40.91) |
*Number of patients with available corresponding clinical information.
The duration of the follow-up was continued 4 years after the acute phase of the disease. We examined three follow-up parameters: creatinine, cystatin C and estimated glomerular filtration rate (eGFR) [34]. eGFR < 90 ml/min/m2 (body surface area) was defined as decreased and therefore a sign of severe sequelae. The presence of hypertension or antihypertensive treatment (HT) and albuminuria (Alb) at follow-up were also defined as signs of severe sequelae. Renal sequelae (any kind of kidney-related abnormality at follow-up) was defined as the presence of at least one of the above parameters.
DNA extraction and whole-genome sequencing
Bacterial DNA was obtained from overnight cultures on blood agar plates by EZ1 DNA Tissue Kit on EZ1 instrument (Qiagen, Hilden, Germany) according to the manufacturer’s instructions for Gram-negative bacteria. The library was prepared using Nextera DNA Flex Library Prep kit (Illumina). For the 33 isolates from Gothenburg, library was pair-end (2×100 bp) sequenced to average 2 M read pairs on HiSeq 2500 (Illumina) platform at SciLifeLab (Stockholm, Sweden). For the 21 isolates from Stockholm, library was pair-end (2×150 bp) sequenced to minimum 3 M read pairs on NovaSeq 6000 (Illumina) at SciLifeLab. Base-calling and demultiplexing were done using bcl2fastq v2.20.0.422, without allowing any mismatch in the index sequence. The sequencing reads were de novo assembled with SKESA (version 2.3.0) [35]. The draft genome sequences were annotated with Prokka (version 1.14.5) [36].
Determination of stx subtypes, virulence factors, serotypes, sequence types, and O157:H7 clade 8 strains
The stx subtypes of STEC isolates were determined by ABRicae version 0.8.10 (https://github.com/tseemann/abricate) with default parameter. Briefly, an in-house stx subtyping database was created with ABRicate by including representative nucleotide sequences of all identified stx1 and stx2 subtypes. The assemblies were then used to search against the stx subtyping database. The assemblies were compared against VFDB database (http://www.mgc.ac.cn/VFs/) for virulence factors using ABRICATE 0.8.10 using the following parameters: coverage ≥75%, identity ≥80%. Serotype was determined by comparing assemblies to the SerotypeFinder database (DTU, Denmark) (http://www.genomicepidemiology.org/) using BLAST+ v2.2.3. Multilocus sequence typing (MLST) was conducted in silico using the on-line tool provided by the Warwick E. coli MLST scheme website (https://enterobase.warwick.ac.uk/species/ecoli/allele_st_search).
The clade 8 status of O157:H7 strains was determined using an in silico-PCR approach, which was based on the study from Manning et al. [6], an SNP in the ECs2357 gene was used for clade 8 identification in the in silico-PCR approach as described previously [37].
Whole-genome phylogenetic analysis
To assess the phylogenetic relationship of HUS-STEC strains in this study, an ad hoc whole-genome multilocus typing (wgMLST) analysis was performed using fast-GeP (version 1.0.2) [38]. The draft genomes of 54 HUS-STEC strains were analyzed with draft genomes of 30 non-HUS STEC strains from Sweden that were processed using same pipeline as recently described [39]. The 30 non-HUS STEC representative strains included 10 STEC isolates from bloody diarrheal (BD) patients, 10 from non-bloody diarrheal (NBD) patients, and 10 from individuals without diarrhea (ND) (Table S1). The complete genome sequence of strain Sakai (Acc. NC_002695) was used as a reference. The whole-genome phylogeny was inferred from the concatenated sequences of the loci shared by all genome sequences, which was found in the ad hoc wgMLST analysis. All the regions with elevated densities of base substitutions were eliminated and phylogenetic relationships were generated by Gubbins (version 2.4.1) [40].
Ethics statement
The study was approved by the regional ethics committees in Gothenburg (2015/335-15) and Stockholm (2020–02338), Sweden, respectively.
Statistical Analysis
χ2 and Fisher’s exact test were used to analyze if specific virulence genes of HUS-STEC isolates were significantly related to clinical outcomes of HUS patients, and to determine the associations between bacterial genetic factors of HUS-STEC isolates, using Statistica12 (StatSoft, Inc. Tibco). A p-value below 0.05 was regarded as statistically significant.
Data deposition
GenBank accession numbers of the 54 HUS-STEC genomes in this study are shown in Table S1.
Results
Molecular characterization of the HUS-associated STEC strains
Fifty-four HUS-STEC strains were included in the study, among which, O157:H7 was the most predominant serotype accounting for 57% (31/54) of strains. Five of “big six” non-O157 serogroups [41] were identified, namely, O121 (10/54; 19%), O26 (4/54; 7%), O111 (2/54; 4%), O103 (2/54; 4%), O145 (1/54; 2%). Notably, some non-predominant serotypes such as O165:H25 (2/54; 4%), O59:H17 (1/54; 2%), and O109:H21 (1/54; 2%) were also identified among HUS-STEC strains (Figure 1a). Five different stx subtypes were identified, the combination of stx2a and stx2c was the most common subtype, which was present in 52% (28/54) of all strains, and 87% (27/31) of O157:H7 strains, followed by stx2a (19/54; 35%), stx2c (3/54; 6%), stx1a (2/54; 4%), stx1a+stx2a (2/54; 4%) (Figure 1b). Ten MLST sequence types (STs) were assigned in 54 HUS-STEC strains. Isolates with the same serotype (O157:H7, O121: H19, O165: H25) were assigned as same ST (i.e., ST11, ST655, ST119, respectively) (Table S2). Thirty out of 31 O157:H7 strains belonged to clade 8 strains.
Figure 1.
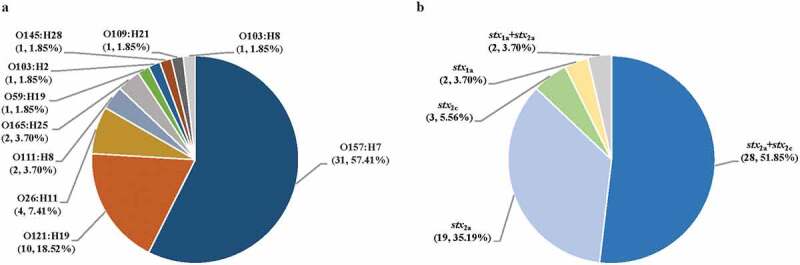
Distribution of serotypes (a) and stx subtypes (b) in 54 HUS-STEC strains The number and percentage of strains belonging to corresponding serotypes and stx subtypes is shown in parentheses (number, percentage)
Serotype O157:H7 was found to be associated with stx2a+stx2c (p < 0.01). Correspondingly, stx2a+stx2c was associated with ST11 (p < 0.01). O165: H25 serotype showed a link with ST119 (p = 0.001).
Clinical characterization of HUS patients
Twenty-nine out of 53 (55%) HUS patients had anuria for more than one day; this data was unavailable for one HUS patient. Twenty-six out of 53 (49%) patients needed dialysis for more than 10 days, this information was missing for one patient. Twenty-eight (28/54; 52%) patients had leukocytosis (leukocytes >20 ×109/L). Twenty-three (23/54; 43%) patients developed complications during the HUS episode. Follow-up data was available for 44 out of 54 patients, among which, 18 (41%) developed renal sequelae. Detailed information of the hemoglobin levels, the presence of hypertension, duration of antihypertensive treatment, either eGFR were presented in Table 1. The prevalence of renal sequelae at follow-up, either eGFR <90 mL/min/m2, hypertension and albuminuria, was significantly associated with O157:H7 serotype, the presence of stx2a+stx2c, and ST11 (Table 2).
Table 2.
Prevalence of Renal sequelaea in correlation to serotype, stx subtype and MLST
| Serotype/stx/MLST | No. of isolates (%) |
p-value | |
|---|---|---|---|
| no Renal sequelaea (n = 26) | Renal sequelae (n = 18) | ||
| O157:H7 | 12 (46.15) | 14 (77.78) | 0.036* |
| stx2a+stx2c | 9 (34.62) | 13 (72.22) | 0.014* |
| MLST11 | 12 (46.15) | 14 (77.78) | 0.036* |
aRenal sequelae: Any kind of kidney-related abnormality at follow-up (eGFR/HT/Alb).
* Statistically significant difference.
Virulence factors in correlation to clinical symptoms
We identified 588 virulence factors (VFs) in 54 HUS-STEC isolates, among which 59 genes were statistically associated with the prevalence of renal sequelae at follow-up, while cfaA gene was negatively associated with kidney-related pathology. These virulence genes can be classified into several groups based on their functions: type III secretion system effectors (espX2, espX3, espY2, nleB2, nleC), type II secretion proteins (etpD, etpE, etpG, etpH, etpI, etpJ, etpK, etpL, etpM, etpN), chaperones (cesAB, cesL, escE, escG), hypothetical proteins and other factors (Table S3).
The presence of virulence genes cadA, ecpA, ecpD, ecpR, and pixH were negatively associated with antihypertensive treatment (Table S4). nleG2-2 gene was statistically significantly associated with a mild clinical outcome with no hypertension or antihypertensive treatment at follow-up (p = 0.019). espK and nleC were associated with mild anemia (Hb > 65 g/L), with a p-value of 0.021 and 0.014, respectively. espM1 and pixH were associated with high leukocytes level above 20×109/L (p = 0.047). Interestingly, espX2, espY1 and espY4 were associated with decreased eGFR (<90 ml/min/m2) at follow-up (p = 0.029), while nleH2 was related to normal eGFR (≥90 ml/min/m2) (p = 0.049). nleG homologs nleG-3, nleG2-3, and nleG5-1, as well as nleC, were associated with the presence of albuminuria at follow-up (Table S5). Virulence factors related to renal sequelae at the follow-up were significantly more prevalent in O157:H7 strains. In contrast, those associated with mild symptoms such as no hypertension or antihypertensive treatment and higher hemoglobin were relatively evenly distributed in strains of all serotypes (Figure 2).
Figure 2.
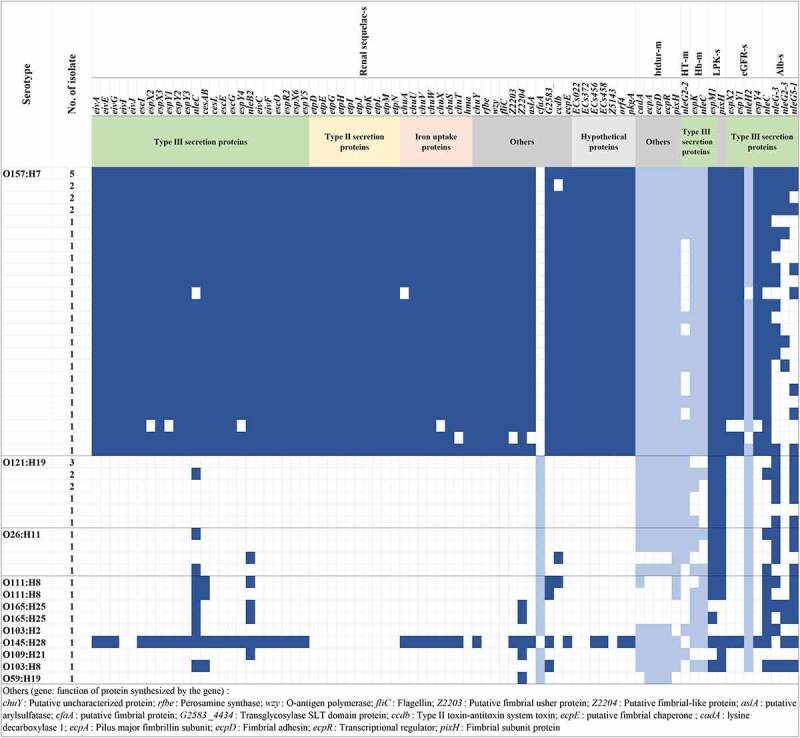
Distribution of virulence factor genes significantly related to clinical symptoms in different serotypes Virulence factor genes significantly associated with different clinical symptoms are shown on the top. The functions of genes are indicated below the genes. Serotypes, number of isolate with the same genes spectrum are indicated on the left. Virulence factor genes associated with severe (s) clinical symptoms are shown in dark blue, those related to mild (m) symptoms are shown in light blue, blank box means the virulence factor gene was absent. Renal sequelae: any kind of kidney-related abnormality at follow up (eGFR/HT/Alb); btdur: duration of antihypertensive treatment; HT: hypertension or antihypertensive treatment at follow up; Hb: hemoglobin; LPK: leukocytes; eGFR: estimated glomerular filtration rate; Alb: Albuminuria at follow-up
Phylogenomic comparison of HUS associated STEC and non-HUS STEC
The whole-genome phylogeny was used to accurately determine relationships of HUS-STEC strains, together with reference non-HUS STEC strains. In total, 2,846 shared loci were found in 85 STEC genomes used in this study, comprising of 54 HUS-STEC strains, 30 non-HUS strains and one strain Sakai as reference. HUS-STEC isolates scattered throughout the phylogenetic tree, with most strains being clustered in O157:H7 clade. Surprisingly, all O157:H7 strains with one exception belonged to highly virulent clade 8 (Figure 3). No distinct cluster was observed among strains from non-HUS including bloody or non-bloody diarrheal cases, and individuals without diarrhea. Isolates with the same serotype were grouped together (Figure 3).
Figure 3.
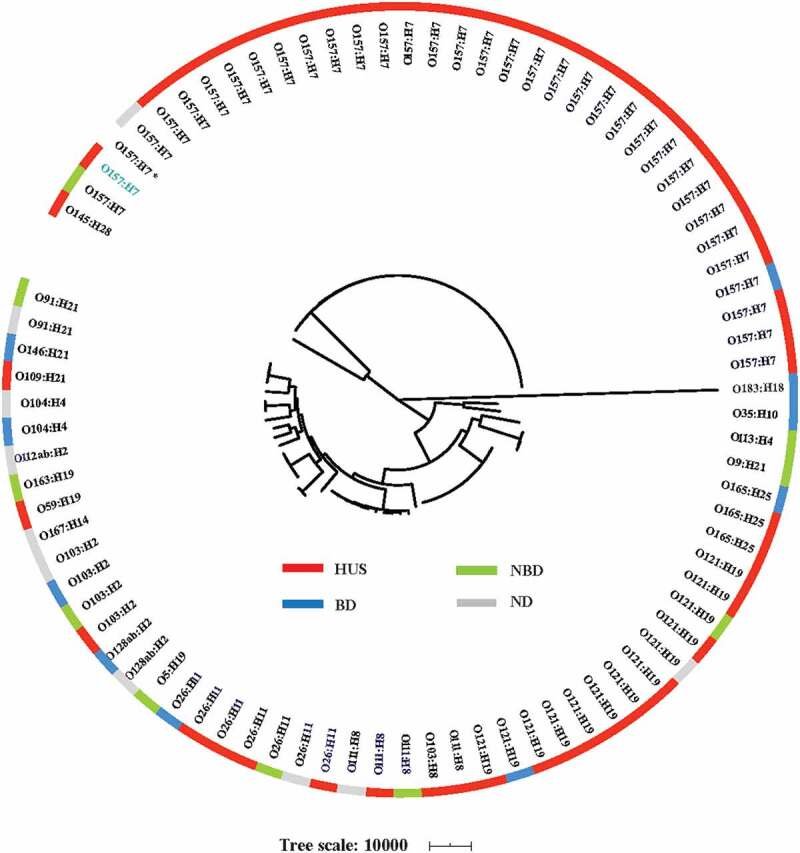
Whole-genome phylogeny of STEC strains Circular representation of the Gubbins tree generated from the concatenated sequences of the shared loci that found in the wgMLST analysis. Gubbins tree was annotated with strain classification using iTOL. The outer circle shows the clinical trait as indicated. One non-clade 8 O157 strain is shown in blue. Reference O157:H7 strain Sakai is indicated with an asterisk. HUS: hemolytic uremic syndrome; BD: bloody diarrhea; NBD: non-bloody diarrhea; ND: no diarrhea
Discussion
This is the first genomic study on clinical HUS-associated STEC isolates systematically collected over a long period in Sweden. The incidence of HUS among STEC-infected individuals is increasing, with current estimation of 5 to 15 cases per 100 STEC-patients, or even higher [42–44]. A microbiologic criterion released by Europe legislation for specific STEC serogroups, highlighted O157, O26, O103, O111, O145 and O104 as the most common cause for HUS in European Union [45]. An earlier study analyzed a large collection of HUS-associated STEC isolates in Germany, demonstrating that O157:H7/H− (67.7%) were the most predominant serotypes among 524 STEC isolates from HUS patients [46]. Consistently, our study showed that O157:H7 was the main STEC serotype (57%) associated with HUS cases in Sweden, which was also in line with a recent study indicating that O157:H7 caused the majority of HUS cases in Finland [26]. Although the proportion of non-O157 STEC infection (mainly caused by O26, O45, O103, O111, O121, and O145 serogroups) has increased in recent years [47,48], O157:H7 still remains as the predominant serotype to cause severe clinical outcomes such as HUS. Among non-O157 serotypes, O121:H19 was the most common serotype (19%) in this study, which was different from the German study [46], where O26:H11/H− (42.6%) was the most predominant serotype among 169 non-O157 HUS-associated STEC strains, followed by O145:H28/H – (18.9%), O111:H8/H – (8.3%), and O103:H2/H – (8.3%). These results indicated that O157:H7 remains as main cause of HUS globally, while the predominant non-O157 STEC serotypes causing severe diseases varied among different countries. It has been reported that Stx genotype rather than the amount of Stx or the cytotoxicity correlates with the appearance of the HUS, i.e., the stx2a and stx2c were associated with high virulence and the ability to cause HUS [18]. In line with earlier studies, we observed that stx subtype stx2a+stx2c (52%) and MLST type ST11 (57%) accounted for the largest proportion among 54 HUS-STEC strains in the present study, which all belonged to O157:H7 serotype, with one exception. Among the HUSEC collection in Germany, stx2a is the most predominant stx subtype [46], similar with a Finnish study, indicating that stx2a was one risk factor for HUS in STEC-positive children [26]. Notably, we found that O157:H7, stx2a+stx2c, and ST11 were associated with renal sequelae at follow-up, indicating that O157:H7 is highly associated with development of long-term sequelae.
STEC pathogenesis is complex as various bacterial and host factors interact with each other, and contribute jointly to disease progression. The association between bacterial factors and disease severity has not been well defined in HUS cases, as most studies mainly focus on several key virulence factors. In this study, we attempt to identify the discriminative genetic features of STEC strains associated with severe outcomes of HUS. We found that espM1 and pixH were associated with higher leukocytes level in acute phase, espX2, espY1 and espY4 were associated with decreased eGFR at follow-up, and nleC, nleG-3, nleG2-3, as well as nleG5-1 were associated with follow-up albuminuria. Approximately sixty virulence factors, including chuA, eivA, etpD, espX and their family genes, as well as nleB2 and nleC, were found to be associated with renal sequelae at follow-up. The functions of some virulence factor genes remain unclear. Notably, some virulence genes related to severe outcomes play a significant role in the STEC pathogenesis. For instance, nleB, the encoded protein can suppress host cell death and inflammation [49], which was shown to be highly associated with enhanced virulence for humans [50]; nleC, which could repress NF-κB and MAPK signaling [51]; espM, which inhibits pedestal formation and disrupts the architecture of a polarized epithelial monolayer [52]. Most of the unfavorable-outcome related-virulence genes were only found in O157:H7 strains, while those associated with better outcome were distributed among all serotypes, indicating that O157:H7 pose higher risk for severe renal sequelae of HUS. Hence, this study suggests that a subset of the genes associated with severe outcomes could be included in the STEC diagnostics, in addition to key virulence genes, e.g., stx, eae. Further studies are warranted to detect the expression level and function of these genes in correlation to disease severity.
The whole-genome phylogeny indicated high diversity of STEC strains. HUS-STEC strains scattered throughout the phylogenetic tree, with most strains being clustered in O157:H7 clade. O157:H7 strains were classified into nine different clades, among which, clade 8 strains were reported to produce higher level of Stx2 [53,54], which might partly explain why these strains cause severe disease and HUS. We found that all O157 strains with one exception in this study belonged to clade 8, consistent with a previous study indicated that HUS patients were more frequently associated with strains from clade 8 [6]. However, our study could not clearly distinguish between HUS-associated and non-HUS STEC strains on the genomic level, HUS-STEC strains scattered throughout the O157 clade and non-O157 clades on the whole genome phylogenetic tree. This was in agreement with an earlier Norwegian study [55]. The present study implies that other factors than bacterial genetic factors, for instance, genes expression level, host-related factors, and bacteria-host interactions might play a joint role in STEC pathogenesis, which is valuable future work.
In conclusion, the current study confirms that O157:H7 serotype is the main cause of HUS, while the predominate non-O157 STEC serotypes associated with severe disease vary among countries. stx subtype stx2a+stx2c and several other virulence factors genes were found to be associated with severe clinical outcomes. In addition, rapid diagnostic targeting of these genes directly on the fecal samples may contribute to improved care and infection control measures ultimately leading to less patients suffering from severe STEC disease.
Supplementary Material
Funding Statement
This work was supported by the Scandinavian Society for Antimicrobial Chemotherapy Foundation under grant[SLS884041]; National Natural Science Foundation of China under grant [81701977];and the Natural Science Foundations of Guangdong Province under grant [2019A1515111004 and 2021A1515011240]. All funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Phillip CAGW, Tarr I.. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. LANCET. 2005;365(9464):1073–1086. [DOI] [PubMed] [Google Scholar]
- [2].Picard C, Burtey S, Bornet C, et al. Pathophysiology and treatment of typical and atypical hemolytic uremic syndrome. Pathol Biol. 2015;63(3):136–143. [DOI] [PubMed] [Google Scholar]
- [3].Jokiranta TS. HUS and atypical HUS. BLOOD. 2017;129(21):2847–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bryan A, Youngster I, McAdam AJ. Shiga toxin producing Escherichia coli. CLIN LAB MED. 2015;35(2):247–272. [DOI] [PubMed] [Google Scholar]
- [5].Smith JL, Fratamico PM, Gunther NT. Shiga toxin-producing Escherichia coli. Adv Appl Microbiol. 2014;86:145–197. [DOI] [PubMed] [Google Scholar]
- [6].Manning SD, Motiwala AS, Springman AC, et al. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc Natl Acad Sci U S A. 2008;105(12):4868–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Beutin L, Krause G, Zimmermann S, et al. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J CLIN MICROBIOL. 2004;42(3):1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Messens W, Bolton D, Frankel G, et al. Defining pathogenic verocytotoxin-producing Escherichia coli (VTEC) from cases of human infection in the European Union, 2007–2010. EPIDEMIOL INFECT. 2015;143(8):1652–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tozzi AE, Caprioli A, Minelli F, et al. Shiga Toxin–Producing Escherichia coli infections associated with hemolytic uremic syndrome, Italy, 1988–2000. EMERG INFECT DIS. 2003;9(1):106–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Elliott EJ, Robins-Browne RM, O’Loughlin EV, et al. Nationwide study of haemolytic uraemic syndrome: clinical, microbiological, and epidemiological features. ARCH DIS CHILD. 2001;85(2):125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Brooks JT, Sowers EG, Wells JG, et al. Non-O157 Shiga Toxin–Producing Escherichia coli infections in the United States, 1983–2002. J INFECT DIS. 2005;192(8):1422–1429. [DOI] [PubMed] [Google Scholar]
- [12].Buchholz U, Bernard H, Werber D, et al. German Outbreak of Escherichia coli O104:H4 Associated with Sprouts. NEW ENGL J MED. 2011;365(19):1763–1770. [DOI] [PubMed] [Google Scholar]
- [13].Frank C, Faber MS, Askar M, et al. Large and ongoing outbreak of haemolytic uraemic syndrome, Germany, May 2011. Euro Surveill. 2011;16(21):19878. [PubMed] [Google Scholar]
- [14].Karch H, Denamur E, Dobrindt U, et al. The enemy within us: lessons from the 2011 European Escherichia coli O104:H4 outbreak. EMBO MOL MED. 2012;4(9):841–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Trachtman H, Austin C, Lewinski M, et al. Renal and neurological involvement in typical Shiga toxin-associated HUS. Nat Rev Nephrol. 2012;8(11):658–669. [DOI] [PubMed] [Google Scholar]
- [16].Bai X, Fu S, Zhang J, et al. Identification and pathogenomic analysis of an Escherichia coli strain producing a novel Shiga toxin 2 subtype. SCI REP-UK. 2018;8(1):6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kawano K, Okada M, Haga T, et al. Relationship between pathogenicity for humans and stx genotype in Shiga toxin-producing Escherichia coli serotype O157. EUR J CLIN MICROBIOL. 2008;27(3):227–232. [DOI] [PubMed] [Google Scholar]
- [18].Orth D, Grif K, Khan AB, et al. The Shiga toxin genotype rather than the amount of Shiga toxin or the cytotoxicity of Shiga toxin in vitro correlates with the appearance of the hemolytic uremic syndrome. DIAGN MICR INFEC DIS. 2007;59(3):235–242. [DOI] [PubMed] [Google Scholar]
- [19].McDaniel TK, Jarvis KG, Donnenberg MS, et al. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A. 1995;92(5):1664–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nguyen Y, Sperandio V. Enterohemorrhagic E. coli (EHEC) pathogenesis. FRONT CELL INFECT MI. 2012;2:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. NAT REV MICROBIOL. 2004;2(2):123–140. [DOI] [PubMed] [Google Scholar]
- [22].Baranzoni GM, Fratamico PM, Gangiredla J, et al. Characterization of Shiga toxin subtypes and virulence genes in porcine Shiga toxin-producing Escherichia coli. FRONT MICROBIOL. 2016;7. DOI: 10.3389/fmicb.2016.00574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].McWilliams BD, Torres AG. Enterohemorrhagic Escherichia coli Adhesins. Microbiol Spectr. 2014;2(3). DOI: 10.1128/microbiolspec.EHEC-0003-2013 [DOI] [PubMed] [Google Scholar]
- [24].Bitzan M, Lapeyraque A, Postinfectious hemolytic uremic syndrome, Europe PMC, 2020, pp. 653–731.
- [25].Spinale JM, Ruebner RL, Copelovitch L, et al. Long-term outcomes of Shiga toxin hemolytic uremic syndrome. PEDIATR NEPHROL. 2013;28(11):2097–2105. [DOI] [PubMed] [Google Scholar]
- [26].Ylinen E, Salmenlinna S, Halkilahti J, et al. Hemolytic uremic syndrome caused by Shiga toxin–producing Escherichia coli in children: incidence, risk factors, and clinical outcome. PEDIATR NEPHROL. 2020;35(9):1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].VanSickle JS, Srivastava T, Alon US. Risk factors for short- and long-term outcomes in children with STEC-HUS/D+ HUS: a single-center experience. Glob Pediatr Health. 2018;5:1881692X–2333794X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wong CS, Jelacic S, Habeeb RL, et al. The risk of the Hemolytic–uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 Infections. N Engl J Med. 2000;342(26):1930–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Soderstrom A, Osterberg P, Lindqvist A, et al. A large Escherichia coli O157 outbreak in Sweden associated with locally produced lettuce. FOODBORNE PATHOG DIS. 2008;5(3):339–349. [DOI] [PubMed] [Google Scholar]
- [30].Sartz L, De B, Hjertqvist JONG,M, et al. An outbreak of Escherichia coli O157:H7 infection in southern Sweden associated with consumption of fermented sausage; aspects of sausage production that increase the risk of contamination. EPIDEMIOL INFECT. 2008;136(3):370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Welinder-olsson C, Stenqvist K, Badenfors M, et al. EHEC outbreak among staff at a children’s hospital – use of PCR for verocytotoxin detection and PFGE for epidemiological investigation. EPIDEMIOL INFECT. 2004;132(1):43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ziese T, Anderson Y, De Jong B, et al. Outbreak of Escherichia coli O157 in Sweden. EUROSURVEILLANCE. 1996;1(1):2S–135S. [DOI] [PubMed] [Google Scholar]
- [33].Noris M, Remuzzi G. Hemolytic Uremic Syndrome. J AM SOC NEPHROL. 2005;16:1035–1050. [DOI] [PubMed] [Google Scholar]
- [34].Björk J, Nyman U, Berg U, et al. Validation of standardized creatinine and cystatin C GFR estimating equations in a large multicentre European cohort of children. PEDIATR NEPHROL. 2019;34(6):1087–1098. [DOI] [PubMed] [Google Scholar]
- [35].Souvorov A, Agarwala R, Lipman DJ. SKESA: strategic k-mer extension for scrupulous assemblies. GENOME BIOL. 2018;19(1). DOI: 10.1186/s13059-018-1540-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Seemann T. Prokka: rapid prokaryotic genome annotation. BIOINFORMATICS. 2014;30(14):2068–2069. [DOI] [PubMed] [Google Scholar]
- [37].Soderlund R, Jernberg C, Ivarsson S, et al. Molecular typing of Escherichia coli O157:H7 Isolates from Swedish Cattle and Human Cases: population Dynamics and Virulence. J CLIN MICROBIOL. 2014;52(11):3906–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhang J, Xiong Y, Rogers L, et al. Genome-by-genome approach for fast bacterial genealogical relationship evaluation. BIOINFORMATICS. 2018;34(17):3025–3027. [DOI] [PubMed] [Google Scholar]
- [39].Hua Y, Bai X, Zhang J, et al. Molecular characteristics of eae-positive clinical Shiga toxin-producing Escherichia coli in Sweden. Emerg Microbes Infect. 2020;9(1):2562–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Croucher NJ, Page AJ, Connor TR, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. NUCLEIC ACIDS RES. 2015;43(3):e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yoon SC, Windham WR, Ladely S, et al. Differentiation of big-six non-O157 Shiga-toxin producing Escherichia coli (STEC) on spread plates of mixed cultures using hyperspectral imaging. J FOOD MEAS CHARACT. 2013;7(2):47–59. [Google Scholar]
- [42].López EL, Contrini MM, Glatstein E, et al. An Epidemiologic Surveillance of Shiga-like Toxin-producing Escherichia coli Infection in Argentinean Children. Pediatr Infect Dis J. 2012;31(1):20–24. [DOI] [PubMed] [Google Scholar]
- [43].Keithlin J, Sargeant J, Thomas MK, et al. Chronic Sequelae of E. coli O157: systematic Review and Meta-analysis of the Proportion of E. coli O157 Cases That Develop Chronic Sequelae. FOODBORNE PATHOG DIS. 2014;11(2):79–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gould LH, Demma L, Jones TF, et al. Hemolytic uremic syndrome and death in persons with Escherichia coli O157:H7 infection, foodborne diseases active surveillance network sites, 2000–2006. CLIN INFECT DIS. 2009;49(10):1480–1485. [DOI] [PubMed] [Google Scholar]
- [45].Delannoy S, Beutin L, Fach P. Discrimination of Enterohemorrhagic Escherichia coli (EHEC) from Non-EHEC Strains Based on Detection of Various Combinations of Type III Effector Genes. J CLIN MICROBIOL. 2013;51(10):3257–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mellmann A, Bielaszewska M, Köck R, et al. Analysis of collection of hemolytic uremic syndrome–associated enterohemorrhagic Escherichia coli. EMERG INFECT DIS. 2008;14(8):1287–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bosilevac JM, Koohmaraie M. Prevalence and characterization of non-O157 Shiga toxin-producing Escherichia coli Isolates from commercial ground beef in the United States. APPL ENVIRON MICROB. 2011;77(6):2103–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Monaghan Á, Byrne B, Fanning S, et al. Serotypes and virulence profiles of non-O157 Shiga toxin-producing Escherichia coli Isolates from bovine farms. APPL ENVIRON MICROB. 2011;77(24):8662–8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lu Q, Li S, Shao F. Weet talk: protein glycosylation in bacterial interaction with the host. TRENDS MICROBIOL. 2015;23(10):630–641. [DOI] [PubMed] [Google Scholar]
- [50].Bugarel M, Beutin L, Fach P. Low-density macroarray targeting non-locus of enterocyte effacement effectors (nle Genes) and major virulence factors of Shiga toxin-producing Escherichia coli (STEC): a new approach for molecular risk assessment of STEC Isolates. APPL ENVIRON MICROB. 2010;76(1):203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Litvak Y, Sharon S, Hyams M, et al. Epithelial cells detect functional type III secretion system of enteropathogenic Escherichia coli through a novel NF-κB signaling pathway. PLOS PATHOG. 2017;13(7):e1006472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Simovitch M, Sason H, Cohen S, et al. EspM inhibits pedestal formation by enterohaemorrhagic Escherichia coli and enteropathogenic E. coli and disrupts the architecture of a polarized epithelial monolayer. CELL MICROBIOL. 2010;12(4):489–505. [DOI] [PubMed] [Google Scholar]
- [53].Amigo N, Mercado E, Bentancor A, et al. Clade 8 and Clade 6 Strains of Escherichia coli O157:H7 from Cattle in Argentina have Hypervirulent-Like Phenotypes. PLOS ONE. 2015;10(6):e127710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Iyoda S, Manning SD, Seto K, et al., Phylogenetic clades 6 and 8 of enterohemorrhagic Escherichia coli O157: h7With Particular stx Subtypes are more frequently found in isolates from hemolytic uremic syndrome patients than from asymptomatic carriers, OPEN FORUM INFECT DI 1 (2014). [DOI] [PMC free article] [PubMed]
- [55].Haugum K, Johansen J, Gabrielsen C, et al. Comparative genomics to delineate pathogenic potential in non-O157 Shiga toxin-producing Escherichia coli (STEC) from patients with and without haemolytic uremic syndrome (HUS) in Norway. PLOS ONE. 2014;9(10):e111788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


