Abstract
Purpose
The ratio of mitochondrial DNA to genomic DNA (mtDNA/gDNA) in embryo culture medium as a predictor of embryonic development is a new method of noninvasive embryo screening. However, current tests based on this concept have proven inconsistent. The aim of this study was to define the predictive value of the ratio of mtDNA/gDNA for embryonic developmental potential.
Materials and Methods
We used digital PCR to measure mtDNA/gDNA ratios in day 3 culture media of 223 embryos from 56 patients. We compared the relationship between the predictive value of mtDNA/gDNA ratio and each of embryo fragmentation, embryo morphological grade, and blastocyst formation.
Results
mtDNA/gDNA ratio decreased significantly with a decrease in embryo rating: 22.54 (44.66); 31.25 (36.97) and 46.33 (57.11); Grades A vs C, P = 0.006; B vs C, P = 0.015. mtDNA/gDNA ratio increased overall with an increase in embryo fragment content but did not differ significantly between high-, -medium, and poor-quality embryos. Interestingly, this trend differed from that of the unformed blastocysts. mtDNA/gDNA ratio of cleavage stage embryos forming blastocysts was lower (P=0.005). Trends of mtDNA/gDNA ratio differed according to inner cell mass (ICM) and trophectoderm (TE) levels, but not significantly. mtDNA/gDNA ratio in day 3 culture medium was not significantly improved over morphological scores.
Conclusion
We hereby show the correlation of mtDNA/gDNA ratio in the culture medium of developing embryos. The correlation between the mtDNA/gDNA ratio and early embryonic development was controversial. Furthermore, an increase in mtDNA/gDNA ratio might indicate reduced development potential, but the difference remains insufficient for application as a clinical predictor.
Keywords: mtDNA/gDNA, embryo, digital PCR, blastocyst
Plain Language Summary
Noninvasive embryo screening technology is a new method to evaluate the contents of metabolites and nutrients in embryo culture medium. During the development of embryo, the contents of mitochondrion in the blastomere decreased gradually, and the contents of cell-free mtDNA in the corresponding embryo medium increased. It has been studied that the ratio of mtDNA/gDNA in embryo culture medium can be used to predict embryo development, but its predictive value is controversial, while the positive detection rate of gDNA is low, and a more accurate technology is to be found for verification. In this study, we used digital PCR system to explore the relationship between cell-free mtDNA in culture medium and the development potential of embryo. The mtDNA/gDNA ratio is associated with the cleavage stage embryo rating, fragmentation, and developmental state, and correlates with the blastocyst rating. According to our research, shortcomings are still associated with using only this indicator to predict embryonic development potential.
Introduction
New techniques to screen for the highest quality embryos for transfer can reduce transfer embryo numbers, improve pregnancy outcomes, and reduce the risk of multiple births. This topic has become a focus of assisted reproductive technology (ART) research. Selective single blastocyst transplantation can improve pregnancy rates and reduce the risk of multiple pregnancies.1 Embryos during the cleavage and blastocyst phases have been rated during in vitro fertilization (IVF) using morphological scoring methods. Some of these screening metrics have been applied to predict developmental potential including embryonic development speed, degrees of uneven blastomere cleavage, fragment content, inner cell mass (ICM), and trophectoderm (TE) levels.2–6 However, that the best quality embryos will be selected for transfer is not guaranteed. Because morphology can appear normal, morphological screening cannot detect genetic defects, nor can it completely ensure optimal embryo development potential. Traditional observation at a fixed time point will miss some important parameters and key events during embryonic development. These parameters are all related to embryonic development potential, and to achieve standardization by relying on subjective differences in artificial observation scores is difficult.7–9
Therefore, new methods are needed to screen embryos to ensure that blastocysts of the best quality are selected for transfer. Chromosomal abnormalities in human embryos are mainly caused by chromosomal segregation errors during stage 1 meiosis. This is usually related to maternal age and is also an important cause of early miscarriage and chromosomal diseases.10 Preimplantation genetic screening (PGS) can eliminate embryos with abnormal chromosomes, but this technique has several disadvantages: embryo biopsy remains invasive, little is known about the safety of children born after embryo biopsy, trained experienced embryologists are required to conduct technical operations, and specialized equipment is required. These increase costs and are time-consuming for patients undergoing IVF assistance.11 Based on the limitations of blastocyst TE biopsy technology, new non-invasive embryo selection technologies are urgently required.
Mitochondria are important cytoplasmic organelles. They function not only as sites of ATP synthesis, but also for the regulation of calcium homeostasis and redox balance.12,13 Mitochondria provide various intermediate metabolites and mediate many signal transduction pathways. Mitochondria are sensitive to the environment and induce apoptosis during injury.14 The mitochondria of oocytes have profound significance for the reproductive process, as they support fertilization and embryo growth before implantation. During the early stages of embryonic development, the zygotic genome is not activated, and the mitochondria do not replicate. All energy-consuming activities of early embryos depend mainly on mitochondria stored by oocytes. Oocyte mitochondrial status and egg apoptosis are closely associated with embryonic developmental arrest.15,16 Therefore, the quality of mitochondria in mature oocytes is an important indicator during fertility screening.17 Mitochondrial genes are transferred to offspring. Mitochondria are the only organelles in human cells that contain maternally inherited, extranuclear genetic material.18 Sperm mitochondrial DNA (mtDNA) in the fertilized egg is quickly recognized and degraded by ubiquitin hydrolase in the cytoplasm.19 Thus, mtDNA in offspring normally originates from the mother. If mutations carried by oocytes escape reproductive screening, they might result in deleterious effects such as increased organ oxidative phosphorylation requirements after birth.18 Therefore, oocyte mtDNA has a significant and extensive impact on the health of offspring.
Embryos can reportedly be screened by detecting mtDNA and genomic (g) DNA based on embryo culture medium;20–22 However others have suggested the opposite.23 We explored the relationship between the mtDNA/gDNA ratio and embryonic developmental potential by measuring levels of cell-free DNA in embryonic culture medium using digital polymerase chain reaction (dPCR) technology that has better detection accuracy than real-time quantitative (RT-q PCR). Digital PCR is a new method of measuring DNA that independently divides PCR reaction units. The terminal dPCR measures fluorescence signals and uses a positive partition number determined from the total target concentration calculated according to Poisson distribution and the proportion of the positive partition number.24 Compared with traditional RT- qPCR, dPCR does not require the inclusion of a standard or creation of a standard curve, and directly determines copy numbers. Digital PCR also has several other advantages,24 such as better sensitivity and precision, although it uses the same primers and probes as qPCR. Real-time-qPCR cannot distinguish < 2-fold differences in gene expression or copy number variations. Conventional methods also cannot identify alleles with frequencies < 1% because they also detect highly abundant common alleles with similar sequences.25 Here, we measured cell-free DNA in embryo culture medium using dPCR, and applied mtDNA/gDNA ratios to predict embryonic development potential. This technology improved detection accuracy and positive detection rates.
Materials and Methods
Patient Selection
This study included patients aged < 35 years from whom 5–15 eggs were harvested, and those who conceived at our center for the first time. We recruited 56 pairs of patients who were unable to conceive due to male infertility, from the Department of Reproductive Medicine of Xiamen Maternity and Child Health Hospital. We collected 223 embryos from these patients to prevent the influence of sperm and granulosa cells on culture medium. All selected patients had undergone fertilization via intracytoplasmic sperm injection (ICSI). The selected patients used their own eggs and sperm. We excluded patients who donated eggs and sperm, and who underwent cycles of implementation with frozen eggs and/or sperm, and without surgically recovered semen.
Ethical Approval
All patients provided written, informed consent to the use of embryonic culture medium for this study. The ethics committee at Xiamen Maternity and Child Health Hospital approved this study (Approval No. KY-2020-064), which proceeded in accordance with the Declaration of Helsinki (2013).
Embryo Culture and Media Collection
After ICSI fertilization, embryos were cultured to the blastula stage in Sydney IVF Cleavage Medium (Cook Medical Inc., Bloomington, IN, USA) then transferred to blastocyst culture medium for embryo exchange on day 3. The cultures were placed in MINC® Benchtop Incubators (Cook Medical Inc.) and only 2PN-derived embryos were collected. We sampled 20 μL of CM culture media per embryo in DNase and RNase-free sterile PCR tubes and froze them at −40°C. Unused culture medium was the negative control.
DNA Purification
Free DNA in culture medium was purified using QIAamp Circulating Nucleic Acid kit (Qiagen, Hilden, Germany) as described by the manufacturer, mixed with 30 μL of elution buffer, then quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific Inc., Waltham, MA, USA).
Digital PCR Quantitative Detection
We established a digital PCR detection system with reference to primers in the literature. We then optimized and amplified the D-loop region of the mitochondrial (NC_012920.1) gene, and β-actin (NG_007992.1) was the internal reference gene. Table 1 shows the primers and probes.
Table 1.
Primer and Probe Sequences
| Name | Sequences (5ʹ to 3ʹ) |
|---|---|
| MTF | TGACCACCATCCTCCGTGAAAT |
| MTR | ATCGTGATGTCTTATTTAAGGGGAA |
| MT-TZ | FAM-CAAGAGTGCTACTCTCCTCGCT-MGB |
| Act-F2 | GCCTCGCTGTCCACCTTCCA |
| Act-R3 | TTTTGTCAAGAAAGGGTGTAACGCA |
| Act-TZ2 | VIC-AAGCAGGAGTATGACGAGTCCG-MGB |
We used a QX200 droplet dPCR system (Bio-Rad Laboratories Inc., Hercules, CA, USA). We found mtDNA in 223 samples, (Average: 219.80, n = 223). We did not detect gDNA in 25 samples (Average: 8.04, n = 198), and the minimum detection value was 1.8. Therefore, the number of copies of gDNA that were not detected were counted as 1.8.
Reaction Mixture
The primers and probes in this paper were diluted to 10μM (10μmol/L). Reaction mixtures (20 μL) contained 10 μL ddPCR Supermix for Probes (No dUTP) (Bio-Rad Laboratories Inc.), forward and reverse primers (1.8 µL) for the target and β-actin genes, the target gene probe (0.4 µL), the β-actin gene probe (0.4 µL), and 2 µL of template.
Preparation of Microdroplets
Reaction mixtures (20 µL) were added to 8 holes in the middle row of a DG8 cartridge (20 μL X control buffer doubled for ~8 samples). About 70 µL of microdrop-generated (DG) oil was added to the bottom row of 8 holes in the cartridges, and all holes were filled. The gasket was covered and all holes on both sides were secured. The holder was placed above the QX200 microdroplet generator to produce microdroplets for 2 minutes in the top row of the cartridge. These were then transferred to 96-well plates, covered with a membrane and heat-sealed.
PCR Amplification
The PCR amplification conditions were 95°C for 10 min, followed by 40 cycles of 94°C for 30 s, 60°C for 60 s, 98°C for 10 min, then the reactions were cooled at 4°C.
Embryo Scores
We modified the described morphological scoring method6 as follows: grade 1, 4 cells on day 2, 8 cells on day 3, uniform size, regular shape, uniform arrangement, uniform cytoplasm, no vacuoles, no polynuclear phenomenon, and ≤5% fragments; grade 2, 3 ~ 5 cells on day 2 (mostly 4 cells), 7 ~ 9 cells on day 3 (mostly 8 cells). The blastomeres were uniform or approximately uniform, the number was equal or approximately equal, the cytoplasm was uniform, large vacuoles and multinucleation were absent, and the fraction accounted for 10% ~20%; grade 3, 2–6 cells on day 2, 6–12 cells on day 3. Blastomeres were uniform/uneven, the number was equal/unequal, and the cytoplasm contained more vacuoles, no polynuclear phenomenon, fragments accounted for 20% ~30%; grade 4, > 6 embryonic cells or no cleavage on day 2, < 6 or >12 embryonic cells on day 3, blastomere was not uniform, cytoplasm was not uniform and contained many vacuoles, multicore phenomenon, fragment content >30%. Current scoring rules indicated very few grade 1 embryos. Therefore, we combined grades 1 and 2 embryos and classified them as Grade A and considered them as being of high quality. Embryos classified as Grade B were usable, and equivalent to grade 3. Embryos classified Grade C were not usable, and equivalent to embryo grade 4. Blastocysts were scored with reference to the Istanbul consensus.26 We grouped blastocysts according to the ICM and TE ratings: group A was rated as high-quality, with ICM and TE ratings of AA, AB, AB, BA, and BB; group B was rated as medium with ICM and TE rating of BC, and group C was unusable, containing early (stages 1, 2, and 3) blastocysts on day 6, and the ICM and TE rating of CC.
Statistical Analysis
All data were statistically analyzed using SPSS 20 (IBM Corp., Armonk, NY, USA). Variables that were not normally distributed were analyzed using Mann–Whitney and Kruskal–Wallis tests, and qualitative variables were analyzed using chi-square tests. Optimal cut-off values and the validity of specific variables were determined from receiver operator characteristics (ROC) curves. Quantitative variables are described as medians with interquartile ranges (Q1-Q3), and qualitative variables are described as numbers (n) with ratios (%). Values with P < 0.05 were considered statistically significant.
Results
Free DNA was quantified by dPCR in culture medium of 223 embryos and five negative controls. Figure 1 shows significantly increased mtDNA/gDNA ratios in day 3 culture medium (33.33 [42.54] and 5.00 [3.90], P = 0.001).
Figure 1.
mtDNA/gDNA ratio in culture medium (n=223) and negative control medium (n=5). D3 was experimental group, NC was the control groups that identically processed but did not contain embryo. One black dot or square represents one sample. Data are presented as the median with interquartile ranges (Q1-Q3). (**P <0.01).
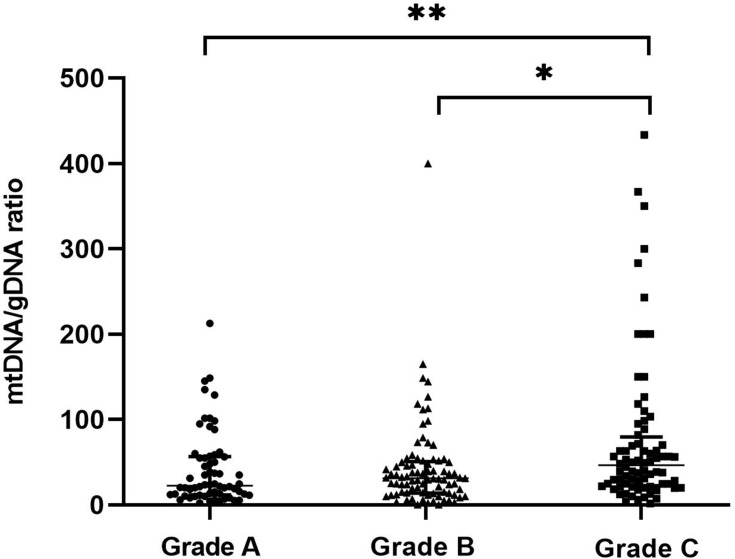
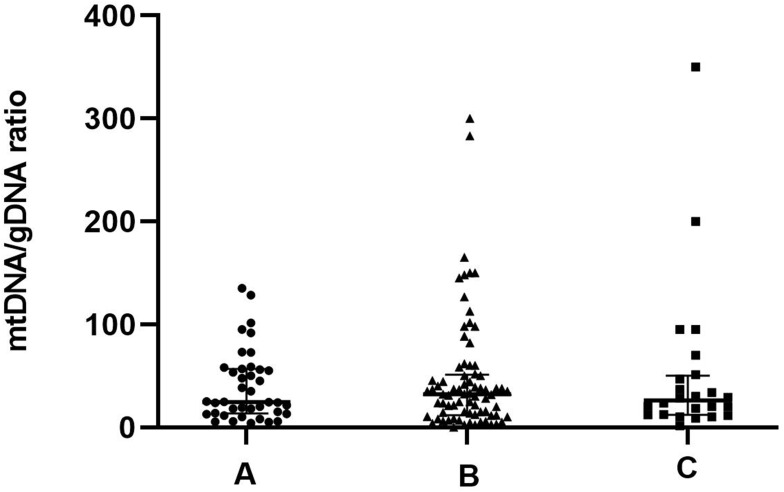
Morphological criteria remain the predominant factors when selecting embryos for uterine transfer during IVF. Therefore, relationships between variations in mtDNA quantities and aspects of embryonic morphology are important to understand. We examined a subset of 223 embryos for which morphological data were available (Figure 2). The results revealed three groups of embryonic morphological quality: grades A, B and C were considered the highest, medium, and poorest quality (n = 62, 85 and 76), respectively. The ratios of mtDNA and gDNA copy numbers in the embryo culture medium were measured by dPCR. The mtDNA/gDNA ratios were 22.54 (44.66), 31.25 (36.97), and 46.33 (57.11) for Grades A, B, and C, respectively, and the results are shown as medians with interquartile ranges (IQR; Q1-Q3). The ratios of mtDNA/gDNA detected in the culture medium did not significantly differ between Grades A and B (P = 1.000), but significantly differed between grades A and C (P = 0.006), and between grades B and C (P = 0.015). These findings revealed that the copy numbers of mtDNA in the medium were associated with embryonic morphological criteria, especially in comparisons of poor- and high-quality embryos. As the embryo quality rating decreased, the number of mtDNA copies in the medium tended to increase, but the absence of a statistical difference between Grades A and B indicated that poor-quality embryos are more likely to release more free mtDNA into the medium.
Figure 2.
mtDNA/gDNA ratio in embryo culture medium for different cleavage stages on day 3. The high-quality embryo group is categorized as Grade A (n = 62), the medium-quality group is Grade B (n = 85), and the poor-quality embryo group is Grade C (n = 76). One black dot, triangle, or square represents one sample. Data are presented as the median with interquartile ranges (Q1–Q3). (**P <0.01, *P<0.05).
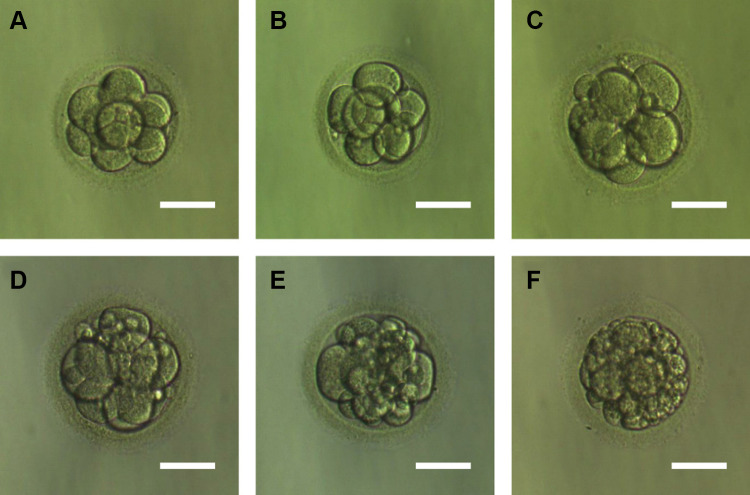
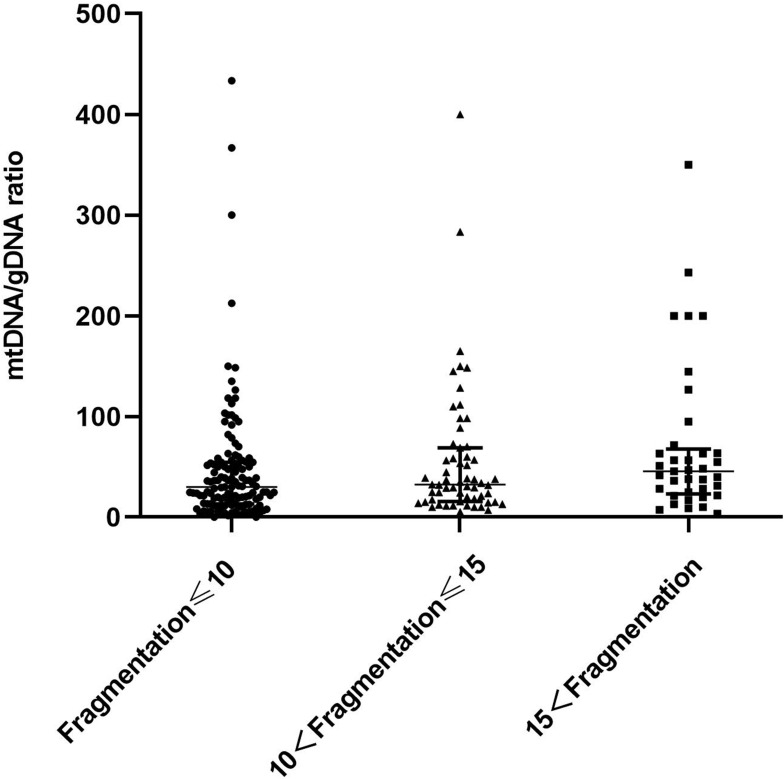
Figure 3 shows embryos with different fragment contents. We investigated relationships between embryo fragmentation and mtDNA/gDNA ratios (Figure 4). The embryos were assigned to groups according to embryo fragment contents of <10% (n = 127), 10–15% (n = 59), and >15% (n = 37).
Figure 3.
Embryos with different fragments content. (A) Day 3 embryos developing showed fragmentation less than 5%. (B) Day 3 embryos developing showed 5–10% fragmentation. (C) Day 3 embryos developing showed 10–15% fragmentation. (D) Day 3 embryos developing showed 15–20% fragmentation. (E) Day 3 embryos developing showed 20–25% fragmentation. (F) Day 3 embryos developing showed more than 25% fragmentation. Scale bar, 60 μm.
Figure 4.
Difference in mtDNA/gDNA ratio in the culture medium of embryos with different fragment content at day 3. The mtDNA/gDNA ratio corresponding to different proportions of embryo fragments in the embryo culture medium were divided into three groups: Fragmentation of embryos in the first group is less than 10% (n = 127), the fragment content of embryos in the second group is between 10% and 15% (n = 59), and in the third group, embryo fragmentation>15% (n = 37). One black dot, black triangle, or black square represents one sample. Data are presented as the median with interquartile ranges (Q1–Q3).
The results showed that that as the proportion of embryo fragments increased, the mtDNA/gDNA ratios in the embryo culture medium also increased from 30.00 (41.33) to 32.40 (53.18), to 45.63 (44.34), respectively. However, the mtDNA/gDNA ratios did not significantly differ among the three groups (P = 0.582, F ≤ 10 vs 10 < F ≤ 15; P = 0.153, F ≤ 10 vs 15 < F; P = 0.763, 10 < F ≤ 15 vs 15 < F.
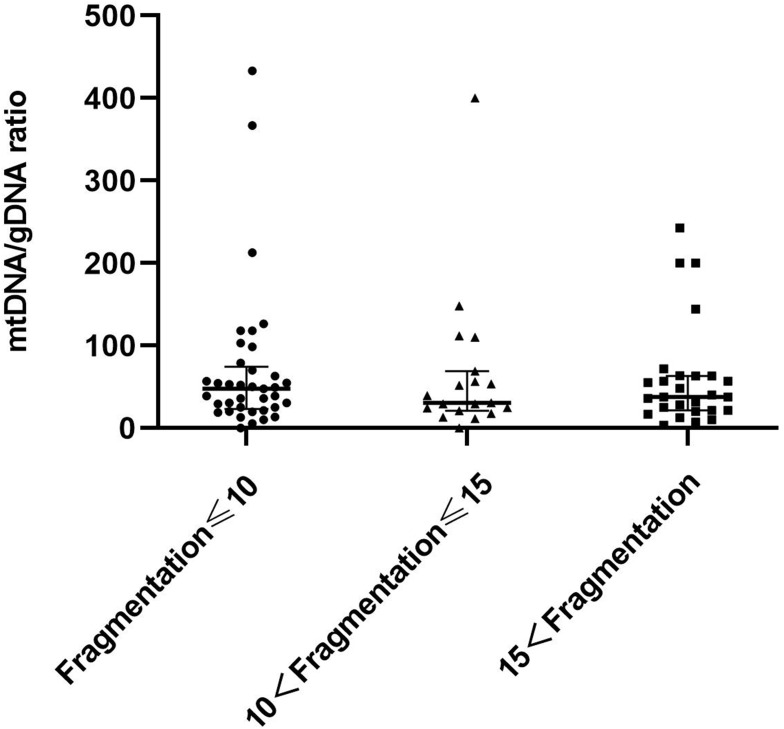
We further investigated whether the mtDNA/gDNA ratio in the group that did not form blastocysts was consistent with above findings, by analyzing 82 non-embryogenic embryos based on fragment contents (Figure 5). Fragmentation of embryos in the first, second and third groups were ≤10% (n = 37), 10–15% (n = 18) and >15% (n = 27). The mtDNA/gDNA ratio was higher in the medium of embryos that did not form blastocysts and had fewer fragments than those with more fragments. However, the three groups overall did not significantly differ 47 (51.23), 34.97 (55.60) and 37.78 (41.90), respectively (P = 0.946).
Figure 5.
Group comparison of cleavage stage embryo fragments without blastocyst formation. Fragmentation of embryos in the first group was ≤10% (n = 37), the fragment content of embryos in the second group between 10% and 15% (n = 18), fragmentation of embryos in the third group was >15% (n = 27). One black dot, black triangle, or black square represents one sample. Data are presented as the median with interquartile ranges (Q1–Q3).
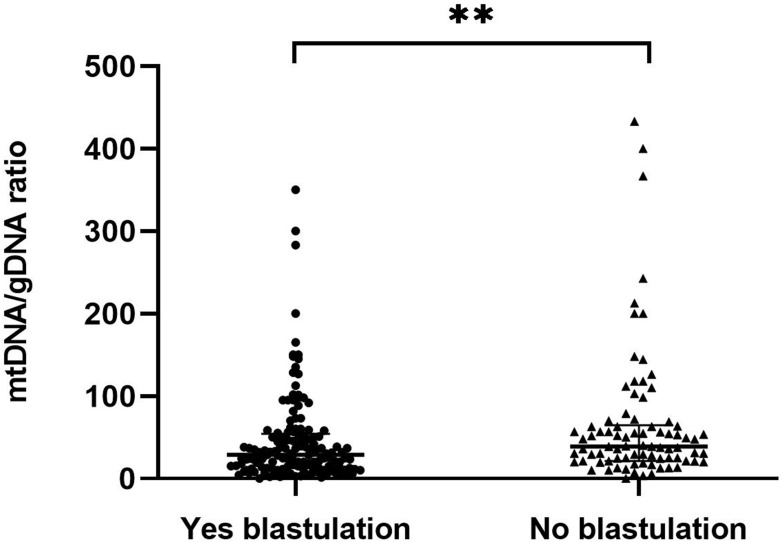
Embryos were classified based on whether they formed blastocysts (n = 141) or not (n = 82) (Figure 6). The mtDNA/gDNA ratios were 29.09 (41.80) and 39.15 (43.39), respectively (P=0.005). Therefore, the ratio of mtDNA/gDNA was lower in the culture medium of cleavage stage embryos that formed blastocysts.
Figure 6.
Comparison of mtDNA/gDNA in embryo culture medium of groups with different blastulation outcomes. Embryos were grouped according to whether they grew to the blastocyst stage or not. One black dot or black triangle represents one sample. Data are presented as the median with interquartile ranges (Q1–Q3). (**P <0.01).
Embryos were then classified based on their ICM and TE morphology (Figure 7) as high- (Group A, n = 41), medium- (Group B, n = 76), and poor- (Group C, n = 24) quality blastocysts. The average ratios of mtDNA/gDNA in the culture media did not significantly differ (24.50 [43.56], 32.20 [39.26], and 26.03 [37.85], P = 0.965).
Figure 7.
Blastocysts divided into high-quality, non-high-quality, and unusable blastocyst groups based on classification of blastocyst ICM and TE. There were 41 high-quality blastocysts are divided into group A(AA/AB/BA/BB), 76 medium-quality blastocysts are divided into group B(BC), 24 poor-quality blastocysts are divided into group C (stage 1/stage 2/stage 3/CC). One black dot, black triangle, or black square represents one sample. Data are presented as the median with interquartile ranges (Q1–Q3).
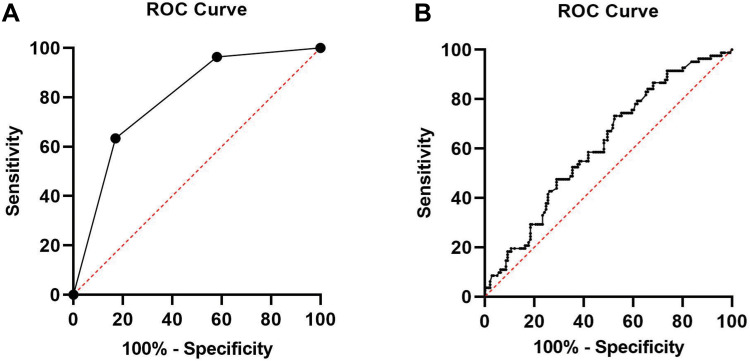
The predictive performance of day 3 morphological grades and mtDNA/gDNA ratios in embryo culture media for blastocyst formation was analyzed using ROC curves (Figure 8).
Figure 8.
ROC curve of morphological grade and the mtDNA/gDNA ratio: (A) day 3 morphological grade group undergoing IVF: area under the curve (AUC)= 0.793 [0.734–0.853], cutoff value= 3.50 with sensitivity=63.4% and specificity= 83.0%. (B) mtDNA/gDNA ratio group undergoing IVF: area under the curve (AUC)= 0.614 [0.539–0.689], cutoff value= 24.83 with sensitivity= 73.2%and specificity= 47.5%.
Univariate comparisons revealed significant differences in the numbers of embryonic cells, % fragmentation, morphology grade on D3 and mtDNA/gDNA ratios between the groups that formed and did not form blastocysts (Table 2).
Table 2.
Univariate Comparison of Exposure Factors Between Blastocyst Formation Group and the Non-Blastocyst Formation Group
| Blastocyst Formation Group (n=141) | Non-Blastocyst Formation Group (n=82) | Z/χ2 | P | |
|---|---|---|---|---|
| Age | 30.00(5.00) | 29.0(5.00) | −0.721 | 0.471 |
| Number of embryonic cells ** | ||||
| n≤6 | 45(31.91%) | 60(73.17%) | 35.42 | <0.001 |
| 6<n≤9 | 87(61.70%) | 20(24.39%) | ||
| 9<n | 9(6.38%) | 2(2.44%) | ||
| % fragmentation** | ||||
| F≤10 | 90(63.83%) | 37(45.12%) | 25.038 | <0.001 |
| 10<F≤15 | 41(29.08%) | 18(21.95%) | ||
| 15<F | 10(7.09%) | 27(32.93%) | ||
| Morphological grade** | ||||
| Grade A | 59(41.84%) | 3(3.66%) | 60.85 | <0.001 |
| Grade B | 58(41.13%) | 27(32.93%) | ||
| Grade C | 24(17.02%) | 52(63.41%) | ||
| The mtDNA/gDNA ratio** | 29.09(41.8) | 39.15(43.39) | 4462.50 | 0.005 |
Note: **Statistically significant differences, with P < 0.01.
Logistic regression analysis significantly associated only day 3 embryo morphology grade and % fragmentation with blastocyst development. The possibility of developing into a blastocyst was reduced as the morphological quality of the embryo decreased (P < 0.0001). The mtDNA/gDNA ratio in culture medium was not associated with blastocyst formation (Table 3).
Table 3.
The Logistic Regression Analysis
| B | S.E, | Sig. | Exp (B) | ||
|---|---|---|---|---|---|
| Step 1a | n≤6 | 0.502 | |||
| 6<n≤9 | −0.386 | 0.492 | 0.432 | 0.680 | |
| 9<n | 0.505 | 0.885 | 0.568 | 1.658 | |
| GradeA** | 0.000 | ||||
| GradeB VS GradeA** | −2.509 | 0.687 | 0.000 | 0.081 | |
| GradeC VS GradeA** | −3.921 | 0.829 | 0.000 | 0.020 | |
| F≤10 | 0.081 | ||||
| 10<F≤15 VS F≤10 | 0.527 | 0.396 | 0.184 | 1.694 | |
| 15<F VS F≤10 | −0.607 | 0.476 | 0.202 | 0.545 | |
| The mtDNA/gDNA ratio | −0.001 | 0.002 | 0.554 | 0.999 | |
| Constant | 3.352 | 0.772 | 0.000 | 28.570 | |
Notes: aVariable (s) entered on step 1: number of embryonic cells, morphology grade, % fragmentation and the mtDNA/gDNA ratio. **Statistically significant differences, with P < 0.01.
We further analyzed the embryo parameters affecting embryo blastocyst formation. We compared the number of embryonic cells, morphology grade on D3, % fragmentation, and mtDNA/gDNA ratios in culture medium among the groups using a generalized estimating equation model (Table 4).
Table 4.
Generalized Estimating Equation Model Compare Embryological Parameters Between Groups
| Parameter | B | S.E, | Wald Chi-Square | Sig. | Exp (B) |
|---|---|---|---|---|---|
| n≤6 | −0.672 | 0.8169 | 0.677 | 0.411 | 0.511 |
| 6<n≤9 | −0.931 | 0.7850 | 1.408 | 0.235 | 0.394 |
| 9<n | 0a | 1 | |||
| Grade=A** | 3.714 | 0.7365 | 25.432 | 0.000 | 41.033 |
| Grade=B** | 1.302 | 0.4276 | 9.276 | 0.002 | 3.678 |
| Grade=C | 0a | 1 | |||
| F≤10 | 0.583 | 0.4625 | 1.591 | 0.207 | 1.792 |
| 10<F≤15 | 1.109 | 0.5959 | 3.462 | 0.063 | 3.030 |
| 15<F | 0a | 1 | |||
| mtDNA/gDNA=0–15.79 | 0.576 | 0.5392 | 1.140 | 0.286 | 1.778 |
| mtDNA/gDNA=15.80–33.33 | −0.114 | 0.3811 | 0.089 | 0.765 | 0.892 |
| mtDNA/gDNA=33.34–58.33 | −0.163 | 0.4315 | 0.143 | 0.705 | 0.850 |
| mtDNA/gDNA=58.33–433.33 | 0a | 1 |
Notes: Dependent: Whether blastocyst is formed. Model: (Intercept), number of cells, D3 morphology grade, % fragmentation and the mtDNA/gDNA ratio. aThis parameter is redundant, so it is set to zero. **Statistically significant differences, with P < 0.01.
Discussion
Non-invasive embryo selection techniques have focused for the past decade on embryonic oxygen consumption, and metabolites and nutrients in embryo culture media, such as glucose, lactic acid, pyruvate, amino acids, pyruvate and glucose intake, and amino acid metabolism. However, such screening techniques do not have substantial proven value for clinical applications, and prospective randomized controlled trials have not found that relative morphological screening methods have obvious advantages.27 Therefore, a simple, reliable, non-invasive, embryo screening technology is needed that can predict embryo developmental potential. This should result in the best quality blastocysts being identified and selected for single blastocyst transfer to the uterus, and consequently increase pregnancy rates and reduce multiple birth rates.21,22,28
Commercial embryo culture media might be contaminated with small, or negligible amounts of DNA, but reported analytical details remain scant.20,29 Levels of DNA in medium exposed to embryos are generally thought to be much higher. This indicates that embryonic DNA enters the medium. The present findings showed a significantly lower mtDNA/gDNA ratio in the blank control than in the embryo culture medium. Therefore, we could disregard the small amount of DNA already in the medium.
Some studies have found that the mtDNA/gDNA ratio in embryo culture medium can predict embryo development potential, and this notion has recently attracted interest. The implantation potential of blastocysts can be evaluated by detecting the mitochondrial content in the TE of embryos.30 However, the source of DNA in the culture medium should not be ignored, as it could be contaminated by maternal granulosa cells. Additional precautions such as more thorough cumulus cell denudation should be taken to reduce genetic contamination.31
Here, we used dPCR technology to detect the copy number of free DNA in culture medium. This strategy improved the sensitivity of detection and greatly improved the detection rate of gDNA. The mtDNA/gDNA ratios varied among the morphological scores. Grades A and B did not significantly differ, whereas Grades A and C and B and C did (P < 0.001 and P < 0.05, respectively). We identified a positive correlation between the ratio of mtDNA/gDNA and the morphological rating of embryonic development in day 3 media. Therefore, the mtDNA/gDNA ratio tended to increase as the embryo morphological rating decreases, especially in unusable embryos.
The mtDNA/gDNA ratio in culture medium correlated with embryo fragmentation. This result is consistent with that of previous reports.20 The mtDNA/gDNA ratio increased as that of embryo fragments increased, but the difference did not reach statistical significance. This also indicated that an increase in embryo fragmentation might lead to increased mtDNA release. Embryos with fewer fragments have a lower mtDNA/gDNA ratio in culture medium, and they are more likely to develop to the blastocyst stage, which is consistent with previous reports describing fragments as unfavorable factors in embryo development.32,33 Trends of mtDNA/gDNA and fragment ratios in embryos that had not formed blastocysts were inconsistent with previous findings, which also suggested that although fragments can retard blastocyst formation and development, other factors might also cause embryos to release more mtDNA into culture medium. An embryo could have mild or extensive fragmentation involving the entire blastomere during development. Extensive fragmentation is usually associated with genetic abnormalities, namely aneuploidy and mosaicism,34,35 and fragmentation is in general, a manifestation of embryonic defects. Cytoskeleton and spindle defects can also cause chromosomal aberrations and fragmentation. The postulated origin of embryo fragmentation is that it is a function of apoptosis.36,37 Our results of mtDNA/gDNA ratios and whether the embryo can develop to the blastocyst stage are inconsistent with previous findings. The mtDNA/gDNA ratio in culture medium reportedly correlates with fragmentation; the amounts of cell-free DNA appeared larger in spent media of embryos with fair and poor morphological score compared with those of high-grade embryos, although the difference was only statistically significant for dsDNA (P = 0.0452). The ratio was lower in patients aged ≤ 35, than in those aged > 35 years (P < 0.05).20 But other studies have shown different results, they found that a high mtDNA/gDNA ratio in spent medium was associated with successful implantation outcome (P = 0.0452) of good-quality embryos.21 Which seems to be the opposite of the previous and fragment results. It is hard to explain why embryos with more fragments are more likely to be implanted. This might have been due to the selection of different populations. With the multiple annealing and looping-based amplification cycles method of next-generation sequencing for whole genome amplification, a study reported that the mt/gDNA ratio of medium from embryos that reached blastulation with successful pregnancy showed a decreasing trend, but the differences were not statistically significant.23 Based on previous research, we reduced the impact of individual differences and the influence of age by selecting standard patients in our center seeking assistance with pregnancy. We also used digital PCR technology, which provided better detection sensitivity and more accurate results.
We further analyzed the relationship between blastocyst ratings and the mtDNA/gDNA ratio of cleavage stage blastocysts and found no relationship between them. The small sample size might also account for the results differing between the present and previous studies.23 We classified blastocysts as high-, -medium, and poor-quality according to the ICM ratings and TE as an integrity factor. The ICM and TE were separately analyzed a previous study.23
We further investigated the relationship between the mtDNA/gDNA ratio and embryo development, by subdividing the unformed blastocysts according to embryo fragmentation. We found a tendency towards increased embryo fragmentation, and a decreased mtDNA/gDNA ratio in the group that did not form blastocysts. However, the tendency did not reach statistical significance. This might have been due to the fact that among the 82 embryos, blastocysts did not form due to fragments, and other factors might have affected blastocyst formation. Other factors will cause embryos with fewer fragments to consume more mitochondria and increase the content of mtDNA in the medium. However, this needs to be addressed in a larger patient cohort.
Although the non-invasive detection of embryos has good application prospects, the use of the mtDNA/gDNA ratio in culture medium for prediction requires more parameters. Logistic regression analysis significantly associated only the morphology grade of day 3 embryos with the blastocyst formation (P < 0.001). The same conclusion was obtained when comparing embryological parameters between groups using a generalized estimating equation model; when the embryo was Grade A, P < 0.001 and the odds ratio was 41.033. A comparison of the blastulation effect of the embryo morphological rating and the mtDNA/gDNA ratio as the area under the ROC curve showed that the embryo morphological rating has obvious advantages.
We used dPCR to assess the relationship more accurately between the mitochondrial genome content in culture medium and embryonic developmental potenFn a larger study cohort and increasing the proportion of single blastocyst transplants in the data population.
Conclusion
The mtDNA/gDNA ratio is associated with the cleavage stage embryo rating, fragmentation, and developmental state, and correlates with the blastocyst rating. However, shortcomings are still associated with using only this indicator to predict embryonic development potential.
Funding Statement
This work was supported by Natural Science Foundation of Fujian Province (2018D0021) and Xiamen Science and Technology Bureau Science and Technology Benefits People Program (3502Z20184045).
Abbreviations
AUC, area under the curve; ICSI, intracytoplasmic sperm injection; SPSS, Statistical Package of Social Science; ROC, receiver operator characteristic curve; dPCR, digital polymerase chain reaction.
Disclosure
The authors declare no competing interests in this work.
References
- 1.Papanikolaou EG, Kolibianakis EM, Tournaye H, et al. Live birth rates after transfer of equal number of blastocysts or cleavage-stage embryos in IVF. A systematic review and meta-analysis. Hum Reprod. 2008;23(1):91–99. doi: 10.1093/humrep/dem339 [DOI] [PubMed] [Google Scholar]
- 2.Hardarson T, Hanson C, Sjögren A, Lundin K. Human embryos with unevenly sized blastomeres have lower pregnancy and implantation rates: indications for aneuploidy and multinucleation. Hum Reprod. 2001;16(2):313–318. doi: 10.1093/humrep/16.2.313 [DOI] [PubMed] [Google Scholar]
- 3.Giorgetti C, Terriou P, Auquier P, et al. Embryo score to predict implantation after in-vitro fertilization: based on 957 single embryo transfers. Hum Reprod. 1995;10(9):2427–2431. doi: 10.1093/oxfordjournals.humrep.a136312 [DOI] [PubMed] [Google Scholar]
- 4.Ziebe S, Petersen K, Lindenberg S, Andersen AG, Gabrielsen A, Andersen AN. Embryo morphology or cleavage stage: how to select the best embryos for transfer after in-vitro fertilization. Hum Reprod. 1997;12(7):1545–1549. doi: 10.1093/humrep/12.7.1545 [DOI] [PubMed] [Google Scholar]
- 5.Roux C, Joanne C, Agnani G, Fromm M, Clavequin MC, Bresson JL. Morphometric parameters of living human in-vitro fertilization embryos; importance of the asynchronous division process. Hum Reprod. 1995;10(5):1201–1207. doi: 10.1093/oxfordjournals.humrep.a136119 [DOI] [PubMed] [Google Scholar]
- 6.Scott L, Finn A, O’Leary T, McLellan S, Hill J. Morphologic parameters of early cleavage-stage embryos that correlate with fetal development and delivery: prospective and applied data for increased pregnancy rates. Hum Reprod. 2007;22(1):230–240. doi: 10.1093/humrep/del358 [DOI] [PubMed] [Google Scholar]
- 7.Ohata K, Ezoe K, Miki T, et al. Blastomere movement post first cell division correlates with embryonic compaction and subsequent blastocyst formation. Reprod Biol Endocrinol. 2019;17(1):44. doi: 10.1186/s12958-019-0488-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaninovic N, Irani M, Meseguer M. Assessment of embryo morphology and developmental dynamics by time-lapse microscopy: is there a relation to implantation and ploidy? Fertil Steril. 2017;108(5):722–729. doi: 10.1016/j.fertnstert.2017.10.002 [DOI] [PubMed] [Google Scholar]
- 9.Gallego RD, Remohí J, Meseguer M. Time-lapse imaging: the state of the art. Biol Reprod. 2019;101(6):1146–1154. doi: 10.1093/biolre/ioz035 [DOI] [PubMed] [Google Scholar]
- 10.Munné S. Chromosome abnormalities and their relationship to morphology and development of human embryos. Reprod Biomed Online. 2006;12(2):234–253. doi: 10.1016/s1472-6483(10)60866-8 [DOI] [PubMed] [Google Scholar]
- 11.Xu J, Fang R, Chen L, et al. Noninvasive chromosome screening of human embryos by genome sequencing of embryo culture medium for in vitro fertilization. Proc Natl Acad Sci U S A. 2016;113(42):11907–11912. doi: 10.1073/pnas.1613294113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motta PM, Nottola SA, Makabe S, Heyn R. Mitochondrial morphology in human fetal and adult female germ cells. Hum Reprod. 2000;15(Suppl 2):129–147. doi: 10.1093/humrep/15.suppl_2.129 [DOI] [PubMed] [Google Scholar]
- 13.Sathananthan AH, Trounson AO. Mitochondrial morphology during preimplantational human embryogenesis. Hum Reprod. 2000;15(Suppl 2):148–159. doi: 10.1093/humrep/15.suppl_2.148 [DOI] [PubMed] [Google Scholar]
- 14.Dumollard R, Duchen M, Carroll J. The role of mitochondrial function in the oocyte and embryo. Curr Top Dev Biol. 2007;77:21–49. doi: 10.1016/S0070-2153(06)77002-8 [DOI] [PubMed] [Google Scholar]
- 15.Perez GI, Trbovich AM, Gosden RG, Tilly JL. Mitochondria and the death of oocytes. Nature. 2000;403(6769):500–501. doi: 10.1038/35000651 [DOI] [PubMed] [Google Scholar]
- 16.Thouas GA, Trounson AO, Wolvetang EJ, Jones GM. Mitochondrial dysfunction in mouse oocytes results in preimplantation embryo arrest in vitro. Biol Reprod. 2004;71(6):1936–1942. doi: 10.1095/biolreprod.104.033589 [DOI] [PubMed] [Google Scholar]
- 17.Van Blerkom J. Mitochondrial function in the human oocyte and embryo and their role in developmental competence. Mitochondrion. 2011;11(5):797–813. doi: 10.1016/j.mito.2010.09.012 [DOI] [PubMed] [Google Scholar]
- 18.Shoubridge EA, Wai T. Mitochondrial DNA and the mammalian oocyte. Curr Top Dev Biol. 2007;77:87–111. doi: 10.1016/S0070-2153(06)77004-1 [DOI] [PubMed] [Google Scholar]
- 19.Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G. Ubiquitin tag for sperm mitochondria. Nature. 1999;402(6760):371–372. doi: 10.1038/46466 [DOI] [PubMed] [Google Scholar]
- 20.Stigliani S, Anserini P, Venturini PL, Scaruffi P. Mitochondrial DNA content in embryo culture medium is significantly associated with human embryo fragmentation. Hum Reprod. 2013;28(10):2652–2660. doi: 10.1093/humrep/det314 [DOI] [PubMed] [Google Scholar]
- 21.Stigliani S, Persico L, Lagazio C, Anserini P, Venturini PL, Scaruffi P. Mitochondrial DNA in day 3 embryo culture medium is a novel, non-invasive biomarker of blastocyst potential and implantation outcome. Mol Hum Reprod. 2014;20(12):1238–1246. doi: 10.1093/molehr/gau086 [DOI] [PubMed] [Google Scholar]
- 22.Stigliani S, Orlando G, Massarotti C, et al. Non-invasive mitochondrial DNA quantification on day 3 predicts blastocyst development: a prospective, blinded, multi-centric study. Mol Hum Reprod. 2019;25(9):527–537. doi: 10.1093/molehr/gaz032 [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Sun Y, Dong X, et al. Mitochondrial DNA and genomic DNA ratio in embryo culture medium is not a reliable predictor for in vitro fertilization outcome. Sci Rep. 2019;9(1):5378. doi: 10.1038/s41598-019-41801-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quan P-L, Sauzade M, Brouzes E. dPCR: a technology review. Sensors (Basel). 2018;18(4):1271. doi: 10.3390/s18041271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manoj P. Droplet digital PCR technology promises new applications and research areas. Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27(1):742–746. doi: 10.3109/19401736.2014.913168 [DOI] [PubMed] [Google Scholar]
- 26.Balaban B, Brison D, Calderon G; Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology. The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum Reprod. 2011;26(6):1270–1283. doi: 10.1093/humrep/der037 [DOI] [PubMed] [Google Scholar]
- 27.Hardarson T, Ahlström A, Rogberg L, et al. Non-invasive metabolomic profiling of day 2 and 5 embryo culture medium: a prospective randomized trial. Hum Reprod. 2012;27(1):89–96. doi: 10.1093/humrep/der373 [DOI] [PubMed] [Google Scholar]
- 28.Sayed GA, Al-Sawaf HA, Al-Sawaf AH, Saeid M, Maged A, Ibrahim IH. Mitochondrial DNA in fresh versus frozen embryo culture media of polycystic ovarian syndrome patients undergoing invitro fertilization: a possible predictive marker of a successful pregnancy. Pharmgenomics Pers Med. 2021;14:27–38. doi: 10.2147/PGPM.S284064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Assou S, Aït-Ahmed O, El Messaoudi S, Thierry AR, Hamamah S. Non-invasive pre-implantation genetic diagnosis of X-linked disorders. Med Hypotheses. 2014;83(4):506–8. doi: 10.1016/j.mehy.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 30.Ravichandran K, McCaffrey C, Grifo J, et al. Mitochondrial DNA quantification as a tool for embryo viability assessment: retrospective analysis of data from single euploid blastocyst transfers. Hum Reprod. 2017;32(6):1282–1292. doi: 10.1093/humrep/dex070 [DOI] [PubMed] [Google Scholar]
- 31.Hammond ER, McGillivray BC, Wicker SM, et al. Characterizing nuclear and mitochondrial DNA in spent embryo culture media: genetic contamination identified. Fertil Steril. 2017;107(1):220–228.e5. doi: 10.1016/j.fertnstert.2016.10.015 [DOI] [PubMed] [Google Scholar]
- 32.Fujimoto VY, Browne RW, Bloom MS, Sakkas D, Alikani M. Pathogenesis, developmental consequences, and clinical correlations of human embryo fragmentation. Fertil Steril. 2011;95(4):1197–1204. doi: 10.1016/j.fertnstert.2010.11.033 [DOI] [PubMed] [Google Scholar]
- 33.Maintenance of the inner cell mass in human blastocysts from fragmented embryos - PubMed. Available from: https://pubmed.ncbi.nlm.nih.gov/12606492/. Accessed March13, 2021. [DOI] [PubMed]
- 34.Sathananthan H, Bongso A, Ng SC, Ho J, Mok H, Ratnam S. Ultrastructure of preimplantation human embryos co-cultured with human ampullary cells. Hum Reprod. 1990;5(3):309–318. doi: 10.1093/oxfordjournals.humrep.a137096 [DOI] [PubMed] [Google Scholar]
- 35.Ebner T, Yaman C, Moser M, Sommergruber M, Pölz W, Tews G. Embryo fragmentation in vitro and its impact on treatment and pregnancy outcome. Fertil Steril. 2001;76(2):281–285. doi: 10.1016/s0015-0282(01)01904-5 [DOI] [PubMed] [Google Scholar]
- 36.Dawson KJ, Conaghan J, Ostera GR, Winston RM, Hardy K. Delaying transfer to the third day post-insemination, to select non-arrested embryos, increases development to the fetal heart stage. Hum Reprod. 1995;10(1):177–182. doi: 10.1093/humrep/10.1.177 [DOI] [PubMed] [Google Scholar]
- 37.Chi H-J, Koo J-J, Choi S-Y, Jeong H-J, Roh S-I. Fragmentation of embryos is associated with both necrosis and apoptosis. Fertil Steril. 2011;96(1):187–192. doi: 10.1016/j.fertnstert.2011.04.020 [DOI] [PubMed] [Google Scholar]