B-cell prolymphocytic leukemia (B-PLL) is a very rare disease; it accounts for <1% of all chronic B-cell leukemias. According to the World Health Organization’s definition, B-PLL is diagnosed when peripheral blood (PB) prolymphocytes account for more than 55% of lymphoid cells in a de novo context. B-PLL generally occurs in elderly people presenting B symptoms, a rapidly rise in the lymphocyte count, massive splenomegaly but little or no lymphadenopathy. There are no specific genetic abnormalities; B-PLL has genomic similarities with other chronic B-cell malignancies but displays well-defined combinations of alterations. The karyotype is frequently complex. MYC aberrations resulting from mutually exclusive translocations or gains are observed in about 75% of cases. These translocations place the MYC gene under the control of an enhancer (usually immunoglobulin genes IGH, IGK, or IGL enhancers) and lead to MYC overexpression. Deletions of the short arm of chromosome 17 including the TP53 gene (del(TP53)) are also frequent. TP53, MYD88, BCOR, MYC, SF3B1, SETD2, CHD2, CXCR4, and BCLAF1 are the most frequently mutated genes in B-PLL. We recently reported on 3 subgroups in which the prognosis depended on the MYC and TP53 status. Patients with both a MYC aberration and del(TP53) belong to the high-risk subgroup and have a short mean overall survival time.1
Burkitt lymphoma (BL) is an aggressive mature B-cell lymphoma that occurs in adults and children. BL is subdivided into a sporadic subtype (often diagnosed in developed countries, accounting for ~1% of adult lymphomas and ~30% of pediatric lymphomas), the Epstein-Barr-virus-associated endemic subtype, and an HIV-associated subtype. This lymphoma comprises medium-sized monomorphic B-cells with round nuclei, finely clumped chromatin, and deeply basophilic cytoplasm that usually contains lipid vacuoles, numerous mitoses, and tingible body macrophages with a “starry sky” appearance. Although BL characteristic morphology and immunophenotype often enable a rapid diagnosis, testing for genomic aberrations is needed to differentiate BL from other high-grade B-cell neoplasms. Although MYC rearrangements are not specific for BL, they are considered as a hallmark feature and are found in almost all cases. The typical t(8;14)(q24;q32) rearrangement (MYC-IGH) occurs in 80% of cases. Rearrangements involving the light chain loci IGL t(2;8) or IGK t(8;22) are less frequent.2 MYC is also the most frequently mutated gene in BL (in 70% of cases). Mutations in the transcription factor 3 (TCF3) gene or its negative regulator ID3 have been reported in about 70% of sporadic subtypes. The other frequently mutated genes are CCND3, TP53, RHOA, SMARCA4, and ARID1A.3–5
Here, we describe a case of concomitant B-PLL and BL. Cytogenetic and molecular analyses revealed a common origin, with the acquisition of additional genetic lesions in the BL clone.
A 46-year-old woman with an unremarkable medical history presented with hyperleukocytosis and thrombocytopenia but no splenomegaly or lymphadenopathy. The white blood cell count was 14.1 × 109/L with 79% lymphocytes, the hemoglobin level was 96 g/L, and the platelet count was 25 × 109/L. In a blood smear examination prolymphocytes accounted for 72% of lymphoid cells. Flow cytometry of PB cells revealed a CD5+CD23-CD79b−FMC7+IgMλweak clonal B-lymphocyte population. The karyotype (K) was 46,XX,t(8;22)(q24;q11)[2]/46,XX[34]. fluorescence in situ hybridization (FISH) analyses confirmed the t(8;22) with MYC rearrangement in 52% of the nuclei and revealed a cryptic del(TP53) in 71% of the nuclei. Cohybridization with MYC and TP53 FISH probes showed that 55% of the cells harbored both abnormalities, and 18% had a del(TP53) only; hence, the MYC translocation had occurred after the del(TP53). Our diagnosis was de novo B-PLL. A bone marrow (BM) aspirate showed a massive infiltration by BL cells (accounting for 87% of the BM cells). The clonal BM B-cells’ immunophenotype was CD5-CD10+bcl2-IgMλhigh. The K was 46,X,-X,t(8;22)(q24;q11),der(13)t(7;13)(q21;q34),+20[19]/46,XX[1], and a FISH analysis detected a MYC rearrangement and a del(TP53) in 59% and 84% of the nuclei, respectively. IGHV sequencing in PB and BM samples showed that both displayed the same VH3-21/DH3-10/JH6 recombination, with full sequence identity. The sequences contained somatic mutations (96.9% homology with the germline counterparts). The patient was diagnosed with medullar BL clonally related to B-PLL. After treatment with a cyclophosphamide/oncovin/adriamycin/prednisolone/methotrexate regimen, the patient achieved a complete response in the BM but prolymphocytic cells persisted in the PB. She relapsed 5 months later, with massive BL cell invasion of the BM. An allogeneic BM transplant was performed but she died 1 month later, following BL relapse.6
To further investigate the clonal relationship between the B-PLL and BL cells, we performed whole exome sequencing (WES) on DNA extracted from sorted CD19+CD5+ PB tumor cells, BM cells, and sorted nontumor CD3+ PB cells (considered to be germinal controls) sampled at the time of diagnosis. Somatic coding mutations were confirmed by polymerase chain reaction-based targeted deep resequencing (See Supplemental Digital Content, http://links.lww.com/HS/A154). In the PB sample, 19 genes were mutated and the variant allele frequency (VAF) ranged from 29.9% to 95.83%; these included TP53 (c.T824A, p.L275Q, VAF: 95.83%); CHD2 (c.4160_4178del, p.P1387Rfs*13, VAF: 47.06%), and SETD2 (c.2628_2629insAG, p.G878Qfs*14, VAF: 34.88%). The same 19 mutations were present in the BM sample, with similar VAFs. Twenty-seven additional mutations were detected in the BM, including TCF3 (c.G1663C, p.E555Q, VAF: 40.53%) and CCND3 (c.T875A, p.L292Q, VAF: 37.25%) (Supplemental Digital Content, Table 1, http://links.lww.com/HS/A155). The copy number aberration analysis from WES data confirmed the chromosomal abnormalities observed by K/FISH, and detected cryptic 17q gain and 14q loss in the PB, and 17q gain and 11q loss in the BM (Figure 1; Supplemental Digital Content, Table 2, http://links.lww.com/HS/A154).
Figure 1.
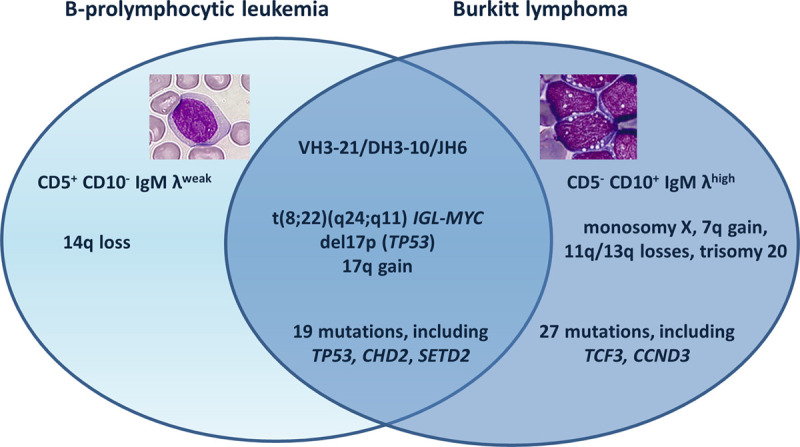
Venn diagram summarizing the biological data.
This case provided an unusual illustration of a dual B-cell neoplasm, with B-PLL in the PB and BL in the BM. The B-PLL cells harbored a translocation that deregulates MYC expression and had biallelic inactivation of TP53 (by deletion and mutation); these are the 2 most prevalent abnormalities in B-PLL and, when combined, confer a poor prognosis. Mutations in CHD2 and SETD2 (involved in chromatin remodeling) are also frequent in B-PLL.1 The medullar BL cells carried the same somatic mutations, chromosomal abnormalities and VDJ recombination (using the IGHV3-21 gene) as the B-PLL cells but also had other genetic lesions. IGHV3 is the predominant subgroup in both BL and B-PLL cells.1,7 Our results demonstrate that the B-PLL and BL cells had the same clonal origin and suggest strongly that the BL developed from the B-PLL (Figure 2).
Figure 2.
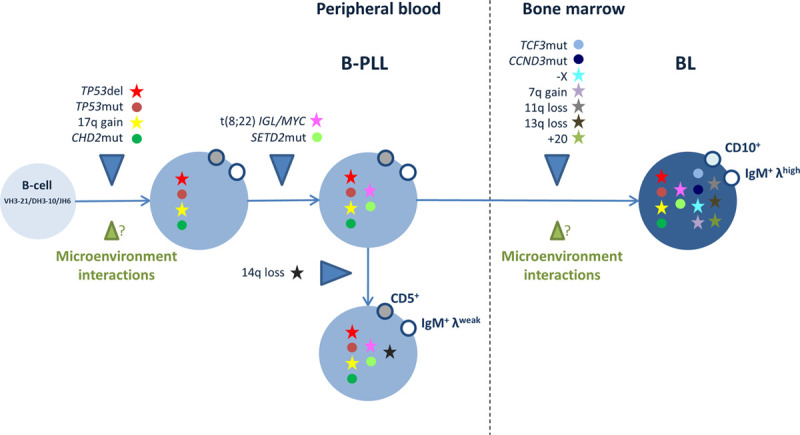
Hypothetical model of the development of B-PLL and BL in the case described here. The illustration depicts the putative sequential acquisition of chromosomal abnormalities and gene mutations, and the possible role of interactions with the microenvironment. B-PLL = B-cell prolymphocytic leukemia; BL = Burkitt lymphoma.
The transformation of a chronic B lymphoproliferative disease into an aggressive lymphoma is well known in chronic lymphocytic leukemia (CLL, as Richter’s syndrome [RS]), follicular lymphoma (FL), and marginal zone B-cell lymphoma of mucosa-associated lymphoid tissue lymphomas (MALT) but has not been described previously in B-PLL. Our case is unique in this respect. In CLL, RS occurs in 2%–8% of patients. Most frequently, clonally related (80%) or unrelated (20%) diffuse large B-cell lymphoma (DLBCL) develops, whereas transformation to classical Hodgkin lymphoma is rare.8 Transformation to BL is very unusual in CLL; only a few cases have been reported.9–11 Transformation is linked to the acquisition of additional chromosomal abnormalities and somatic mutations. In DLBCL-type RS, genetic lesions typically affect the TP53, NOTCH1, MYC, and CDKN2A genes. The MYC network is deregulated in ~70% of samples.8 MYC pathway deregulation is also considered to be a key event in transformation of MALT (40%–80% of cases) and FL (~40%) to DLBCL, and is often associated with TP53 aberrations.12,13 In our case, transformation to BL was not due to MYC deregulation or TP53 inactivation alone because these aberrations were already present in the B-PLL cells. Moreover, MYC deregulation is known to be insufficient for BL oncogenesis. Additional genetic lesions cooperate with MYC to generate human BL.14 The additional mutations observed in our patient’s BM included mutations in the TCF3 and CCND3 genes, both of which are frequently mutated in de novo BL. The L292Q CCND3 missense mutation, novel in BL, affects a conserved residue in the proline (P), glutamic acid (E), serine (S), and threonine (T) (PEST) domain, which has a role in the protein’s degradation. The great majority of the CCND3 mutations observed in BL and other B-lymphoid neoplasms target the PEST domain and result in the intracellular accumulation of cyclin D3 and deregulation of the cell cycle.5 The E555Q TCF3 mutation, already identified in BL,5 affects the basic helix–loop–helix domain. Gain-of-function monoallelic mutations in TCF3 and biallelic inactivating mutations in the ID3 gene (encoding TCF3’s inhibitor) activate B-cell receptor signaling, and thus sustain BL cell survival by engaging the phosphoinositide-3-kinase pathway. These mutations are essentially absent in other mature B-cell malignancies, suggesting that the TCF3/ID3 module has a determining role in the pathogenesis of BL. Indeed, it has been shown that TCF3 contributes to the BL phenotype by enforcing a germinal center-derived transcriptional program; it controls a centroblast-restricted gene expression signature that is “inherited” by BL cells and is intensified in cases with TCF3/ID3 aberrations.5 Hence, in the present case, the acquisition of the TCF3 and CCND3 mutations may have contributed strongly to the development of BL.
We described a unique chemotherapy-refractory case of de novo high-risk B-PLL with concomitant, clonally related BL. Deregulation of MYC is a nonspecific, oncogenic event shared by B-PLL and BL (and other lymphoid malignancies). It is necessary for tumor transformation but does not fully explain the phenotype, which is probably dictated by specific combinations of genetic and epigenetic abnormalities. Clonal evolution, cell migration, disease progression, and drug resistance may all be influenced by the tumor microenvironment.15 This case further confirms the crucial role of TCF3 and CCND3 in BL lymphomagenesis.
Disclosures
The authors have no conflicts of interest to disclose.
Sources of funding
This investigation was funded by grants from GEFLUC (to EC), the Association Laurette Fugain (ALF 14/08, to FN-K), the Institut Thématique Multi-Organisme Cancer, the Institut National du Cancer and Roche Diagnostics. This work was supported by the Cancer United Research Associating Medicine, University and Society (CURAMUS) «INCA-DGOS-Inserm_12560».
Supplementary Material
Footnotes
Supplemental digital content is available for this article.
References
- 1.Chapiro E, Pramil E, Diop M, et al. the Groupe Francophone de Cytogénétique Hématologique (GFCH); the French Innovative Leukemia Organization (FILO). Genetic characterization of B-cell prolymphocytic leukemia: a prognostic model involving MYC and TP53. Blood. 2019; 134:1821–1831 [DOI] [PubMed] [Google Scholar]
- 2.Dalla-Favera R, Bregni M, Erikson J, et al. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 1982; 79:7824–7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Love C, Sun Z, Jima D, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012; 44:1321–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richter J, Schlesner M, Hoffmann S, et al. ICGC MMML-Seq Project. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet. 2012; 44:1316–1320 [DOI] [PubMed] [Google Scholar]
- 5.Schmitz R, Young RM, Ceribelli M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012; 490:116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen-Khac F, Davi F, Receveur A, et al. Burkitt-type acute leukemia in a patient with B-prolymphocytic leukemia: evidence for a common origin. Cancer Genet Cytogenet. 2005; 159:74–78 [DOI] [PubMed] [Google Scholar]
- 7.Baptista MJ, Calpe E, Fernandez E, et al. Analysis of the IGHV region in Burkitt’s lymphomas supports a germinal center origin and a role for superantigens in lymphomagenesis. Leuk Res. 2014; 38:509–515 [DOI] [PubMed] [Google Scholar]
- 8.Rossi D, Spina V, Gaidano G. Biology and treatment of Richter syndrome. Blood. 2018; 131:2761–2772 [DOI] [PubMed] [Google Scholar]
- 9.Asou N, Osato M, Horikawa K, et al. Burkitt’s type acute lymphoblastic transformation associated with t(8;14) in a case of B cell chronic lymphocytic leukemia. Leukemia. 1997; 11:1986–1988 [PubMed] [Google Scholar]
- 10.Mohamed AN, Compean R, Dan ME, et al. Clonal evolution of chronic lymphocytic leukemia to acute lymphoblastic leukemia. Cancer Genet Cytogenet. 1996; 86:143–146 [DOI] [PubMed] [Google Scholar]
- 11.Torelli UL, Torelli GM, Emilia G, et al. Simultaneously increased expression of the c-myc and mu chain genes in the acute blastic transformation of a chronic lymphocytic leukaemia. Br J Haematol. 1987; 65:165–170 [DOI] [PubMed] [Google Scholar]
- 12.Maeshima AM, Taniguchi H, Toyoda K, et al. Clinicopathological features of histological transformation from extranodal marginal zone B-cell lymphoma of mucosa-associated lymphoid tissue to diffuse large B-cell lymphoma: an analysis of 467 patients. Br J Haematol. 2016; 174:923–931 [DOI] [PubMed] [Google Scholar]
- 13.Pasqualucci L, Khiabanian H, Fangazio M, et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014; 6:130–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitz R, Ceribelli M, Pittaluga S, et al. Oncogenic mechanisms in Burkitt lymphoma. Cold Spring Harb Perspect Med. 2014; 4:a014282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mangolini M, Ringshausen I. Bone marrow stromal cells drive key hallmarks of B cell malignancies. Int J Mol Sci. 2020; 21:E1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


