Abstract
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a public health emergency. The most common symptoms of COVID-19 are fever, cough, and fatigue. While most patients with COVID-19 present with mild illness, some patients develop pneumonia, an important risk factor for mortality, at early stage of viral infection, putting these patients at increased risk of death. So far, little has been known about differences in the T cell repertoires between COVID-19 patients with and without pneumonia during SARS-CoV-2 infection. Herein, we aimed to investigate T cell receptor (TCR) repertoire profiles and patient-specific SARS-CoV-2-associated TCR clusters between COVID-19 patients with mild disease (no sign of pneumonia) and pneumonia. The TCR sequencing was conducted to characterize the peripheral TCR repertoire profile and diversity. The TCR clustering and CDR3 annotation were exploited to further discover groups of patient-specific TCR clonotypes with potential SARS-CoV-2 antigen specificities. Our study indicated a slight decrease in the TCR repertoire diversity and a skewed CDR3 length usage in patients with pneumonia compared to those with mild disease. The SARS-CoV-2-associated TCR clusters enriched in patients with mild disease exhibited significantly higher TCR generation probabilities and most of which were highly shared among patients, compared with those from pneumonia patients. Importantly, using similarity network-based clustering followed by the sequence conservation analysis, we found different patterns of CDR3 sequence motifs between mild disease- and pneumonia-specific SARS-CoV-2-associated public TCR clusters. Our results showed that characteristics of overall TCR repertoire and SARS-CoV-2-associated TCR clusters/clonotypes were divergent between COVID-19 patients with mild disease and patients with pneumonia. These findings provide important insights into the correlation between the TCR repertoire and disease severity in COVID-19 patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10875-021-01045-z.
Keywords: COVID-19, T cell receptor, TCR repertoire, TCR sequencing, SARS-CoV-2
Introduction
Beginning in December 2019, the newly emergent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) rapidly became a pandemic (coronavirus disease 2019; COVID-19). SARS-CoV-2 has so far infected more than one hundred million people and killed over two million people worldwide. The clinical manifestations of patients with SARS-CoV-2 infection vary considerably, with 70–80% of patients either asymptomatic or experiencing moderate symptoms, including fever, fatigue, dry cough, sore throat, dyspnea, myalgia, and impaired taste and smell. About 15% of confirmed symptomatic cases have severe respiratory distress and shock, and these patients may rapidly progress into acute respiratory distress syndrome, which requires intensive care. The known risk factors for developing respiratory failure and acute respiratory distress syndrome include old age, lymphopenia, high lactate dehydrogenase (LDH), and high D-dimer [1]. Moreover, the viral load of SARS-CoV-2 was reported to be predictive of COVID-19 mortality [2].
To quantify the high rate of COVID-19 transmission, several studies have estimated the basic reproduction number of SARS-CoV-2 at approximately 3.28, while the World Health Organization (WHO) has estimated it to be approximately 1.95 [3]. The precise interval during which a patient infected with SARS-CoV-2 can transmit infection to others is uncertain, but it has been suggested that infected patients were most contagious during the early stage of illness, when the level of viral RNA in the upper airway appears to be highest [4]. He et al. reported that the mean serial interval between the onset of symptoms among 77 transmission pairs was 5.8 days, suggesting that patients became infectious 2.3 days prior to symptom onset and infectiousness peaked 0.7 days before symptom onset and then declined over the course of 7 days [5]. Recent studies showed that some patients experience prolonged periods of detectable viral RNA in the respiratory tract [6]; however, detectable viral RNA does not always indicate the presence of infectious virus. In addition, infectious virus was only detected in stored respiratory specimens that had a high concentration of viral RNA (RT-PCR positive at cycle threshold ([Ct] < 24)] [7], suggesting that the virus became less infectious with prolonged storage. According to guidance from the United States Centers for Disease Control and Prevention (CDC), if viral RNA is still detectable in upper respiratory specimens 3 days after clinical recovery, the RNA concentrations are generally below the levels at which replication-competent virus can be reliably isolated [8].
The clinical relevance of dynamic changes in the immune landscape is still unclear in SARS-CoV-2 infection, especially with regard to T cells. As with any virus infection, the adaptive immune response plays a central role in clearing SARS-CoV-2. Thus, the T cell receptor (TCR) repertoire is a key factor in viral clearance, since it describes the ability of CD8+ T cells to recognize various viral antigens expressed on major histocompatibility class I (MHC I) molecules in infected cells and dendritic cells [9]. After activation by antigen recognition, T cells undergo clonal expansion, during which activated T cells rapidly proliferate to generate large numbers with identical TCRs to eliminate virus-infected cells [10]. Most of the TCRs on circulating T cells are alpha and beta subunit heterodimers, and the specificity for an antigen is shaped by VDJ recombination [11]. This process can generate up to 1015 unique TCRs from the same heterodimer [12]. The TCR repertoire dynamically changes in response to acute and chronic infections, and exposure to antigen triggers clonal expansion that skews the TCR repertoire to favor T cells that have specificity for the antigen [13]. Such dynamic changes in TCR repertoire in antiviral immunity have been demonstrated by a longitudinal study of yellow fever immunization in human [14]. Moreover, TCR analysis of influenza-positive individuals revealed several T cell clonotypes with the same recurrent TCR motifs [15]. Therefore, we postulated that unique and dynamic changes of the TCR repertoire occur in response to SARS-CoV-2 infection. In this study, we performed TCR sequencing and analysis on peripheral blood lymphocytes of SARS-CoV-2-infected patients and correlated features and characteristics of their TCR repertoire with disease severity.
Materials and Methods
Patient Enrollment
A total of nine COVID-19 patients diagnosed with SARS-CoV-2 infection by quantitative reverse transcription polymerase chain reaction (RT-qPCR) were enrolled in Taipei Medical University-Shuang Ho Hospital, Ministry of Health and Welfare, Taiwan. The patient classification for COVID-19 was performed according to Interim Guidelines for Clinical Management of SARS-CoV-2 Infection (5th edition, March 26, 2020), published by Taiwan Centers for Disease Control (Taiwan CDC). Based on criteria of the guidelines, three and six of recruited patients were assigned to “mild disease” and “pneumonia” groups, respectively. Both mild disease and pneumonia groups of patients displayed common symptoms of COVID-19, including fever, fatigue, and cough; however, only pneumonia patients were diagnosed with pneumonia throughout the course of the disease. According to Taiwan CDC regulation, all individuals testing positive for SARS-CoV-2 were isolated in hospital until the RT-PCR tests for SARS-CoV-2 showed three consecutive negative results. Blood samples were collected from patients during hospitalization. All COVID-19 patients of the study were recovered and discharged after three consecutive negative results of RT-qPCR test for SARS-CoV-2. Six (67%) of the patients are male in our cohort, with an average age of 48.8 years old (24–73). The study was approved by Taipei Medical University Joint Internal Review Board (TMU-JIRB: N202004076). All patients were provided with and signed informed consents.
Sample Processing and TCRβ Library Preparation
RNA was extracted from COVID-19 patients’ peripheral blood mononuclear cells (PBMCs) using a Trizol-based method (Thermo Fisher Scientific, MA, USA). Library preparation of T cell receptor beta chain (TCRβ) repertoire for high-throughput sequencing (HTS) was performed as previously described [16]. Purified RNA sample was first subjected to template-switching reverse transcription polymerase chain reaction (RT-PCR) using SMARTer PCR cDNA Synthesis Kit (Takara Bio USA, CA, USA) and a modified cDNA synthesis (CDS) oligo (5′-CGG GGT ACG ATG AGA CAC CAT TTT TTT TTT TTT TTT TTT TVN-3′). Full-length cDNAs synthesized from whole mRNA transcripts were obtained and subsequently amplified using Q5 High-Fidelity 2X Master Mix (New England Biolabs, MA, USA) with a self-designed CDS oligo-specific primer (5′-CGG GGT ACG ATG AGA CAC CA-3′). Following first-strand cDNA synthesis and amplification, TCRβ fragments were enriched using Q5 High-Fidelity 2X Master Mix with self-designed TRBC1/TRBC2-specific primer (5′-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG DVH DVT CTG ATG GCT CAA ACA CAG C-3′) and template-switching oligo-specific primer (5′-GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGC AGT GGT ATC AAC GCA GAG TAC-3′). Size selection was performed to extract TCRβ cDNA fragments with sizes between 300 and 1000 base pairs (bp) by Pippin DNA Size Selection System (Sage Science, MA, USA). Illumina index and adaptor sequences were finally added to enriched TCRβ fragments using KAPA HiFi HotStart ReadyMix (Roche, Basel, Switzerland) and Nextera XT Index Kit (Illumina, CA, USA). AMPure XP reagent kit (Beckman Coulter, CA, USA) was used for purification of PCR products throughout the TCR library preparation. The final product of TCRβ library was ready for subsequent HTS analysis.
TCRβ Sequencing and Data Preprocessing
TCRβ libraries constructed from different patients’ PBMC samples were pooled and sequenced using MiSeq Reagent Kit v3 with 2 × 300 bp read length (Illumina, CA, USA). The demultiplexed sequencing files corresponding to different patient’s TCRβ libraries were generated in a fastq format. The raw sequencing data was subjected to adaptor trimming and quality filtering of raw reads using Cutadapt [17] and Trimmomatic (LEADING:15 TRAILING:15 SLIDINGWINDOW:4:20 MINLEN:50) [18]. Filtered reads were subsequently aligned to variable (V), diversity (D), joining (J), and constant (C) gene segments of T cell receptor beta locus (TRB) for clonotype assembly of complementarity-determining region 3 (CDR3) nucleic acid sequences by MiXCR software (v 3.0.12) [19]. Furthermore, clonotypes with the same CDR3 amino acid sequences were collapsed, and the abundancy of each TCRβ clonotype was recalculated. Finally, the TCRβ repertoire profile was constructed for each COVID-19 patient and used for following TCRβ repertoire analysis. The rarefaction curve of TCRβ repertoire was generated using VDJtools [20] for each subject (Fig. S1).
TCRβ Repertoire Diversity Profile and Index
To establish the diversity profile of each individual’s TCRβ repertoire, we calculated Hill’s numbers (αD), a series of diversity indices, according to the equation as follows:
where n is the total number of clonotypes and pi means the frequency of the ith clonotype in TCRβ repertoire. The α value indicated different orders of the repertoire diversity, representing distinct extents of weighting on abundant clonotypes in TCRβ repertoire. In the study, we computed Hill’s numbers of order α values from 0 to 5 for each TCRβ repertoire of COVID-19 patients and transformed Hill’s number into Rényi entropy (αH) for visualization, according to the equation as follows:
The means and the 95% confident intervals (CIs) of Rényi entropies at different α values were calculated for mild disease and pneumonia groups. Accordingly, the patient group-specific diversity profile curve was built using a locally estimated scatterplot smoothing (LOESS) function deployed in R software. In addition, several common diversity indices, including Shannon entropy (Shannon index), inverse Simpson index, and Pielou’s evenness index, were calculated for comparing differences in TCRβ repertoire diversities between patients with mild disease and pneumonia. Equations of the diversity indices are shown as follows:
Principal Component Analysis and Calculation of Repertoire Dissimilarity Index Calculation for V Gene, J Gene, and CDR3 Length Profiles
The gene usage of TRBV and TRBJ was measured by calculating occurrences of each V and J gene segment of unique TCRβ clonotypes in each patient’s repertoire. The CDR3 length usage was estimated by frequencies of CDR3 amino acid lengths of unique TCRβ clonotypes within the TCRβ repertoire of each patient. The count matrices based on usages of V and J gene and CDR3 length were generated and subjected to principal component analysis (PCA) and repertoire dissimilarity index (RDI) calculation. PCA was performed using a prcomp() R function with default parameters. RDI were computed between pairwise combinations of intragroup patients with mild disease and pneumonia using a rdi() function of rdi R package based on an analytic method as previously described [21, 22]. The CDR3 length distribution for comparison was constructed by counting numbers of each CDR3 length from all TCRβ repertoires of patients in the same group. The statistical difference between the CDR3 length distribution of different patient groups was determined using a Kolmogorov–Smirnov (KS) test.
Clustering, Generation Probabilities (Pgen) Estimation, Annotation, and Assignment of TCRβ Clonotypes
To group TCRβ clonotypes with a high possibility of having the same or similar antigen specificity, we analyzed pooled TCRβ repertoires from all of the nine COVID-19 patients using a reported clustering tool—GLIPH2 (grouping of lymphocyte interaction by paratope hotspots version 2) algorithm [23, 24]. A reference TCRβ dataset of combined naïve CD4+ and CD8+ repertoires and default parameters recommended by the GLIPH2 developer (http://50.255.35.37:8080/) were adopted. The GLIPH2 analysis identified both global- and motif-based TCRβ clusters based on CDR3 sequence similarity (hamming distance of same-length CDR3) and motif enrichment of TCRβ clonotypes. Based on the GLIPH2 clustering results, we further predicted theoretical generation possibilities (Pgen) of CDR3 amino acid sequences of clustered TCRβ clonotypes using an established OLGA (Optimized Likelihood estimate of immunoGlobulin Amino-acid sequences) algorithm [25].
Following the clustering analysis by GLIPH2, we next annotated clustered TCRβ clonotypes with potential antigen specificities against SARS-CoV-2 epitopes by matching TRBV gene segments and CDR3 amino acid sequences of clustered clonotypes to that of known SARS-CoV-2-specific clonotypes of a combined COVID-19 TCR dataset. Herein, two released TCR datasets from VDJdb [26] and ImmuneCODE [27] were downloaded and merged as a reference dataset, and a TCR annotation tool, VDJmatch [28], was used to annotate each global-clustered TCRβ clonotype with single or multiple SARS-CoV-2 epitope specificity according to matching rules as follows: (1) The V gene segment of querying TCRβ clonotype was matched to that of SARS-CoV-2-specific TCRβ clonotypes in the reference dataset. (2) CDR3 amino acid sequences were matched between querying and reference SARS-CoV-2-specific TCRβ clonotypes with one or less substitution. For each motif-based cluster, the enriched CDR3 motif amino acid sequence, determined by GLIPH2 analysis, of querying TCRβ clonotype was searched in substrings spanning IMGT (the international ImMunoGeneTics information system)-defined positions 107–116 within CDR3 amino acid sequences of SARS-CoV-2-specific TCRβ clonotypes in the reference dataset. Accordingly, motif-clustered clonotypes with enriched CDR3 motif sequences discovered in the reference dataset were annotated with corresponding SARS-CoV-2 antigen specificities. Finally, those of global- or motif-based TCRβ clusters containing at least one SARS-CoV-2-annotated TCRβ clonotype were assigned potential SARS-CoV-2-specific (or SARS-CoV-2-associated) TCRβ clusters.
We next examined the enrichment of TCRβ clusters in either mild disease, pneumonia, or both groups of patients. For TCRβ clusters with all TCRβ clonotypes from the identical group of patients, the corresponding mild disease- or pneumonia-specific cluster was assigned. For those of clusters with TCRβ clonotypes contributed from both mild disease and pneumonia patients, absolute differences of median frequencies between TCRβ clonotypes from patients with mild disease and pneumonia were computed with 9999 permutations for cluster assignment. In this case, all TCRβ clonotypes within each cluster were randomly assigned to mild disease or pneumonia group, and the median frequency difference between reassigned clonotypes of mild disease and pneumonia groups was then computed. We repeated the permutation step 9999 times for each TCRβ cluster with clonotypes from both mild disease and pneumonia patients and then calculated the probability (as the p value) of that original median frequency difference was larger than/smaller than/equal to median frequency differences from 9999 permutation plus one original estimation. Based on the permutation test, TCRβ clusters with significant difference (p value < 0.05) between median frequencies of clustered TCRβ clonotype from different patient groups were classified as either mild disease- or pneumonia-specific clusters; otherwise, they were assigned shared-specific clusters. Furthermore, the publicity of each mild disease-/pneumonia-/shared-specific cluster was determined by the number of mild disease/pneumonia/all patients contributing clustered TCRβ clonotypes to the cluster. The low, medium, and high levels of sharing were used to represent the extent of publicity of each TCRβ cluster based on the number of patients contributing clustered clonotypes to the mild disease- (clustered clonotypes from 2 or 3 of patients with mild disease as medium or high level of sharing), pneumonia- (clustered clonotypes from 2, 3–4, or 5–6 of patients with pneumonia as low, medium, or high level of sharing), or shared-specific (clustered clonotypes from 2–3, 4–6, or 7–9 of all patients as low, medium, or high level of sharing) cluster. For those of TCRβ clusters, where all of clustered clonotypes derived from the same patient, they were assigned private.
Network Analysis of TCRβ Clonotypes
To establish public SARS-CoV-2-associated TCR networks, we selected TCRβ clonotypes from mild disease- and pneumonia-specific TCRβ clusters that were annotated with potential SARS-CoV-2 antigen specificity and determined as high level of sharing. TCRβ clonotypes with identical amino acid sequence but from different TCRβ clusters or patients were collapsed and displayed as one vertex on the network graph. All TCRβ clonotypes within the same cluster were linked together. In addition, the hamming distance between TCRβ clonotypes from different GLIPH2 clusters was calculated, and those with same-length and highly similar CDR3 sequences (hamming distance equal to or less than one) were connected or overlapped. We further re-grouped TCRβ clonotypes for mild disease- and pneumonia-specific TCR similarity network using a clusters() function of igraph R package. Accordingly, TCRβ clonotypes from different GLIPH2-defined clusters but linked together based on high inter-group similarity were re-clustered in the new TCRβ cluster.
Visualization of Sequence Conservation of TCRβ Clonotypes
To visualize CDR3 sequence conservation of re-clustered TCRβ clonotypes, the logo plotting was performed using a ggseqlogo R package. For TCRβ clonotypes from motif-based clusters with diverse length of CDR3 sequences, their CDR3 amino acid sequences were transformed into a gapped sequence format according to the IMGT numbering system (http://www.imgt.org/). The length of transformed sequences was determined by the maximum CDR3 length of TCRβ clonotype for each motif-based TCRβ cluster. A gapped alignment was then performed for logo plotting of motif-based TCRβ clusters. In addition, the consensus region of GLIPH2-identifying enriched motif sequence for each motif-based cluster was determined by searching the most frequently observed positions of the start and end of the motif sequence among motif-clustered TCRβ clonotypes. For global-based clusters, the diversity of each IMGT-defined CDR3 position was evaluated by Shannon entropy. CDR3 sequences of each clonotypes were transformed into gapped sequences with specific length, which was determined by the maximal CDR3 length of all global-based re-clustered clonotypes, as described above. The Shannon index of each CDR3 position among TCRβ clonotypes of each global-based cluster was calculated, and the mean of Shannon indices of each CDR3 position across 104 to 118 among clusters from the same patient-specific type was then evaluated.
Statistical Analysis
All of TCRβ repertoire analysis and statistical calculation were performed using R software. All figures were generated using the ggplot2 [29] and ggpubr R package. Wilcoxon rank sum test was used for the comparison of TCRβ repertoire diversity, RDI, and cumulative TCRβ clonal proportion. T-test was used for comparison of cluster Pgen and frequencies. Chi-square test was used for comparison of distribution of sharing levels among clusters. KS test was used for comparison of CDR3 length distribution. Kendall rank correlation was used to test associations between biological/biochemical measurements and the repertoire diversity index. Examining the difference in age and sex between patient groups was performed using t-test and Fisher’s exact test. The statistical significance was defined by a p value less than 0.05, and p values < 0.05, < 0.01, or < 0.001 were denoted by “*,” “**,” or “***.”
Results
Clinical Characteristics and TCRβ Repertoire Profiles of COVID-19 Patients with Mild Disease and Pneumonia
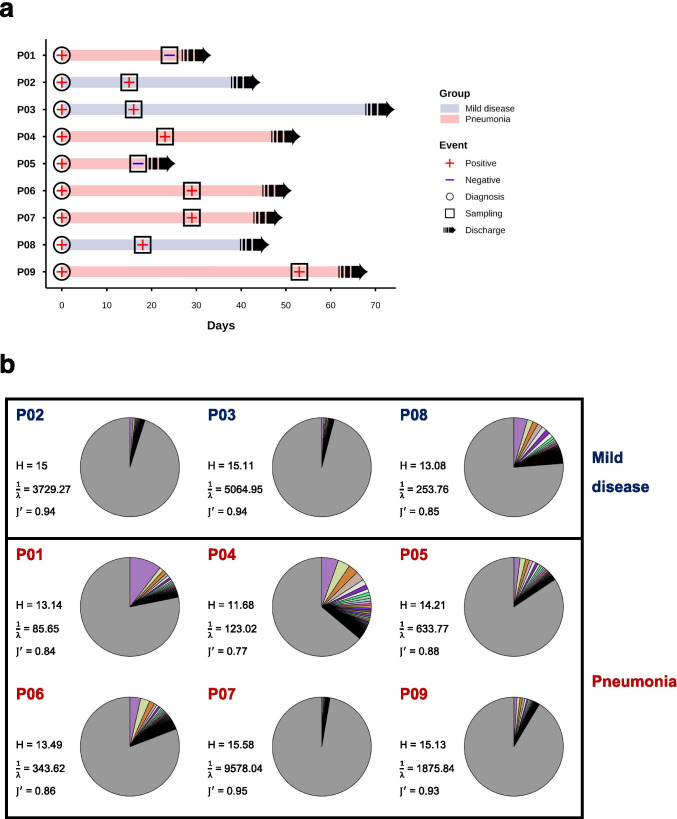
In the study, we recruited nine COVID-19 patients, who were diagnosed with SARS-CoV-2 infection confirmed by polymerase chain reaction (PCR) test. These patients were categorized into mild disease (n = 3) and pneumonia (n = 6) groups according to pneumonia severity, scored by 0 (no pneumonia), 1 (mild pneumonia), and 2 (severe pneumonia) at time of diagnosis (Table 1 and Fig. S2). Most of patients showed no symptom of pneumonia at time of sample collection except P04, who still had mild pneumonia at days 23 from diagnosis (Table 1). No statistically significant difference in age or sex was observed between the two groups of patients (Table S1). All patients were hospitalized, recovered, and then discharged in 22 ~ 71 days, and their convalescent peripheral blood samples were collected at days 15 ~ 53 after diagnosis for T cell receptor (TCR) sequencing (Fig. 1a). Profiling of TCR beta chain (TCRβ) repertoire showed that the overall frequencies of abundant TCRβ clonotypes (> 0.1%) were relatively higher in patients with pneumonia compared with those with mild disease (Fig. 1b and Fig. S3a,c). In addition, potentially higher cumulative proportions of TCRβ clonotypes with medium- to hyper- (> 0.01%) and hyper-abundant (> 1%) clone sizes were observed in patients with pneumonia, relative to those with mild disease (Fig. S3b,d), although not statistical significance was revealed. These results showed that pneumonia patients had more expanded TCRβ clonotypes than patients with mild disease in the convalescent stage.
Table 1.
Characteristics of COVID-19 patients
| Subject | Age | Sex | Pneumonia severity† | PCR test | Day of sampling‡ | Group§ | ||
|---|---|---|---|---|---|---|---|---|
| Diagnosis | Sampling | Diagnosis | Sampling | |||||
| P01 | 48 | Male | 1 | 0 | Positive | Negative | 24 | Pneumonia |
| P02 | 24 | Male | 0 | 0 | Positive | Positive | 15 | Mild disease |
| P03 | 71 | Female | 0 | 0 | Positive | Positive | 16 | Mild disease |
| P04 | 62 | Male | 2 | 1 | Positive | Positive | 23 | Pneumonia |
| P05 | 46 | Female | 1 | 0 | Positive | Negative | 17 | Pneumonia |
| P06 | 73 | Male | 1 | 0 | Positive | Positive | 29 | Pneumonia |
| P07 | 30 | Male | 1 | 0 | Positive | Positive | 29 | Pneumonia |
| P08 | 54 | Male | 0 | 0 | Positive | Positive | 18 | Mild disease |
| P09 | 31 | Female | 1 | 0 | Positive | Positive | 53 | Pneumonia |
†The pneumonia severity was denoted by scores of 0 (none), 1 (mild), and 2 (severe)
‡Duration of days from diagnosis to sampling
§All patients were classified into pneumonia (pneumonia severity score of diagnosis > 0) or mild disease (pneumonia severity score of diagnosis = 0) group
Fig. 1.
Disease course and TCRβ repertoire profiles in COVID-19 patients. a A swimmer’s plot illustrated an overview of disease progression in three of patients with mild disease (blue bar) and six of patients with pneumonia (red bar) COVID-19 patients. All patients were recorded from day 0 after diagnosis (symbol of “◯”) with SARS-CoV-2 infection based on RT-PCR test (symbol of “ + ” and “-” for positive and negative results, respectively). Blood samples were collected (symbol of “□”) from patients during the convalescent phase. All patients were discharged (symbol of “➟”) after 3 ~ 10 weeks of hospitalization. b Dominant TCRβ clonotypes with abundances larger than 0.1% were illustrated with different colors for TCRβ repertoire profiles of patients with mild disease (colored by blue) and pneumonia (colored by red) in pie charts. TCRβ repertoire diversity were estimated by calculating Shannon index (), inverse Simpson index () and Pielou’s evenness index ()
Differences in TCRβ Repertoire Diversities Between COVID-19 Patients with Mild Disease and Pneumonia
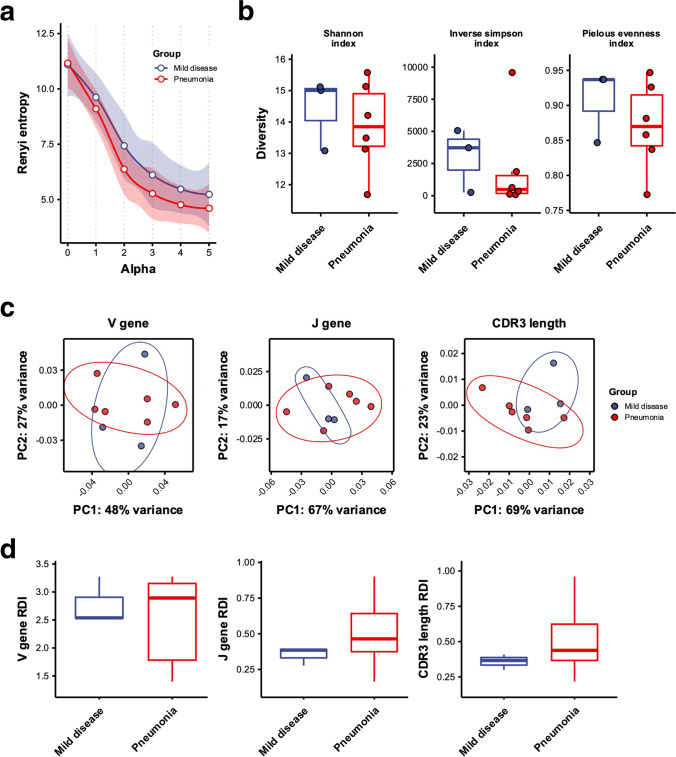
Based on the observed difference in clonotype distribution between mild disease and pneumonia groups described above, we next compared characteristics of TCRβ repertoire between two groups of COVID-19 patients. The diversity profiles based on Rényi entropy (αH) showed that the average αH of pneumonia patients was lower than those of patients with mild disease when α > 0, which tended to weight abundant clonotypes when calculating αH (Fig. 2a). Common diversity indices, including Shannon index (), inverse Simpson index (), and Pielou’s evenness index (), between two groups of patients also revealed that TCRβ repertoire diversity was slightly reduced in pneumonia patients (Fig. 2b). Such results suggested that during convalescent phase, pneumonia patients may have lower peripheral T cell repertoire diversity compared with individuals with mild disease.
Fig. 2.
Characterization of TCRβ repertoire diversities in COVID-19 patients with mild disease and pneumonia. a The diversity profile showed TCRβ repertoire diversities based on average Rényi entropies (y-axis) calculated with alpha values (x-axis) from 0 to 5 in mild disease (blue points) and pneumonia (red points) groups of patients. The Rényi entropy was surrogated by Shannon index of TCRβ repertoire when alpha value was equal to 1. The LOESS function was used to perform curve fitting and estimation of 95% confidence interval (CI) for TCRβ repertoire diversity profiles of mild disease (blue line and shading) and pneumonia (red line and shading) patients. b Comparison of TCRβ repertoire diversity between patients with mild disease (colored blue) and pneumonia (colored red) was evaluated by Shannon indices (left panel), inverse Simpson indices (middle panel) and Pielou’s evenness indices (right panel) of two groups of patients. The difference between groups was calculated using Wilcoxon rank sum test. c Principal component analysis (PCA) for TRBV (left panel), TRBJ (middle panel), and CDR3 length (right panel) usage of TCRβ repertoire in patients with mild disease (colored blue) and pneumonia (colored red) patients was illustrated. The 95% CI was shown by colored ellipses for mild disease (blue) and pneumonia (red) groups. d Comparison between intragroup repertoire similarity index (RDI) values for TRBV (left panel), TRBJ (middle panel), and CDR3 length (right panel) usage of TCRβ repertoire of patients with mild disease (colored blue) and pneumonia (colored red) patients was illustrated. The difference between groups was calculated using Wilcoxon rank sum test. p Values larger than 0.05 were considered to be not statistically significant and were not shown
To explore whether combinatorial and junctional diversities between two groups of patients were also divergent, we next examined variable gene usage and complementarity-determining region 3 (CDR3) length distribution of COVID-19 patients’ TCRβ repertoire. The variable gene usage was determined by frequencies of TRBV or TRBJ genes used by each unique TCRβ clonotype, while CDR3 length usage/distribution was evaluated by lengths of CDR3 amino acid sequences of each unique clonotype. Profiling of TCRβ variable gene usage revealed no obvious difference in V and J gene usage between patients with mild disease and pneumonia (Fig. S4a,b). The principal component analysis (PCA) also showed that both V and J gene usage were not biased by patient group; however, the usage of CDR3 lengths was skewed and distinct between patients with mild disease and pneumonia (Fig. 2c). A two-sample KS test indicated a significant difference in CDR3 length distribution between patients with mild disease and pneumonia (Fig. S4c). Compared to individuals with mild disease, pneumonia patients had higher frequencies of longer CDR3 lengths. We further calculated repertoire dissimilarity indices (RDI) [21, 22] for V and J gene usage and CDR3 length distribution of peripheral TCRβ repertoire and compared intragroup dissimilarities between mild disease and pneumonia groups. Comparison of intragroup RDI showed that dissimilarities of V, J, and CDR3 length usage among pneumonia patients was potentially higher than that among patients with mild disease, suggesting that patterns of combinatorial and junctional diversities were more distinct between pneumonia patients (Fig. 2d). In addition, we examined whether TCRβ repertoire diversity was correlated with clinical features of COVID-19 patients. However, no significant difference was observed between Pielou’s evenness index and any biochemical measurement in our COVID-19 patient cohort (Fig. S5).
Divergent Characteristics of SARS-CoV-2 Associated TCRβ Clusters Between COVID-19 Patients with Mild Disease and Pneumonia
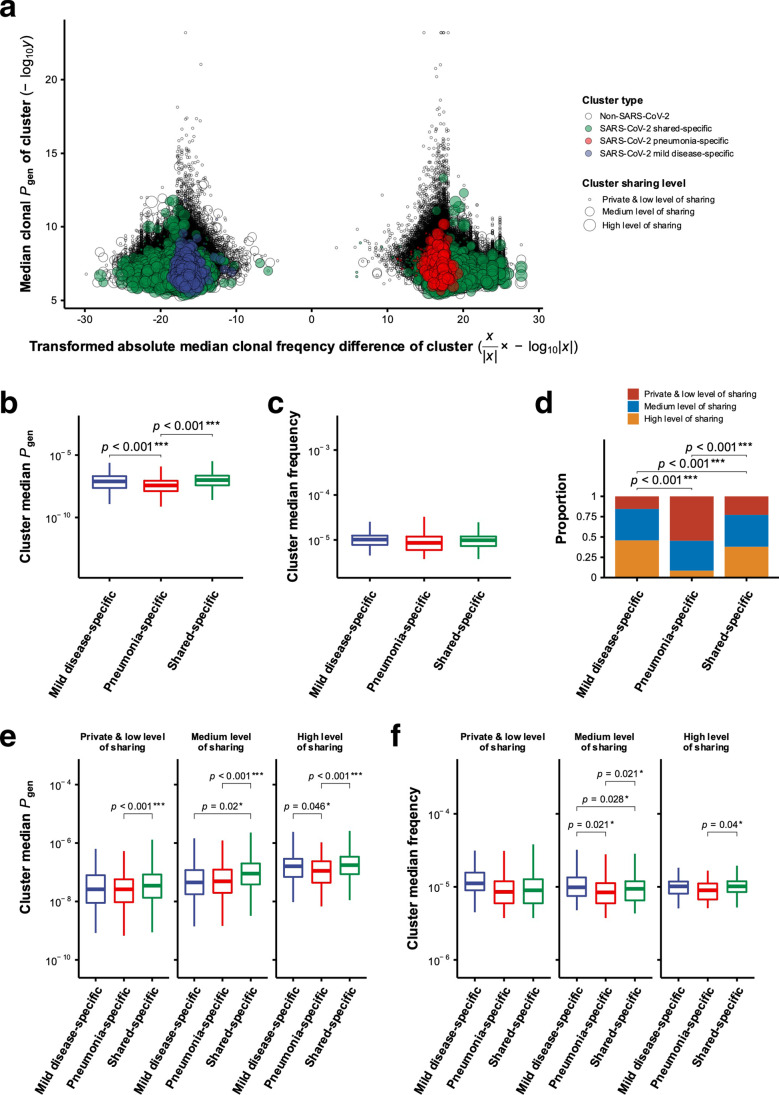
The overall characteristics of TCRβ repertoire in COVID-19 patients suggested that patients with mild disease and pneumonia expressed distinct T cell repertoire dynamics during SARS-CoV-2 infection. To further discover and analyze TCRβ clonotypes potentially specific to SARS-CoV-2 antigens in COVID-19 patients, we next performed TCR clustering and annotation for grouping and predicting antigen specificities of clonotypes, respectively. The GLIPH2 (grouping of lymphocyte interaction by paratope hotspots version 2) [24] algorithm was used to cluster TCRβ clonotypes with high probabilities of shared antigen specificities in pooled TCRβ repertoires from all of the nine patients with mild disease and pneumonia. Prediction of SARS-CoV-2-specific TCRβ clonotypes was performed by matching TCRβ CDR3 or enriched motif sequences against reported SARS-CoV-2 specific TCRs from two recently announced COVID-19 TCR datasets collected in VDJdb and ImmuneCODE [26, 27, 30]. We next calculated absolute difference of median frequencies between TCRβ clonotypes from patients with mild disease and pneumonia for each TCRβ cluster and determined whether the cluster was enriched with clonotypes from either mild disease, pneumonia, or both groups of patients based on exclusivity of patient group and permutation test of clustered TCRβ clonotypes. In addition, predicted generation probabilities (Pgen) of TCRβ clonotypes were computed using the OLGA (Optimized Likelihood estimate of immunoGlobulin Amino-acid sequences) [25] algorithm in order to interrogate whether the selection pattern of TCRβ clonotypes/clusters between patients with mild disease and pneumonia was different. We integrated above information and identified potential SARS-CoV-2-specific (SARS-CoV-2-associated) TCRβ clusters that were enriched with clonotypes from either mild disease, pneumonia, or both groups of COVID-19 patients (defined as mild disease-, pneumonia-, and shared-specific clusters) and exhibited diverse cluster median Pgen values, frequencies, and levels of sharing (Fig. 3a). The statistical analysis showed that the overall median Pgen of SARS-CoV-2-associated pneumonia-specific clusters was significantly lower than that of SARS-CoV-2-associated mild disease- and shared-specific clusters (means of cluster median Pgen of SARS-CoV-2-associated mild disease-, pneumonia-, and shared-specific clusters: 1.72 × 10–7, 7.95 × 10–8, and 1.89 × 10–7), while no significant difference in overall median Pgen was observed between the latter two types of SARS-CoV-2-associated clusters (Fig. 3b). Such results suggested that some TCRβ clonotypes with higher Pgen and potential SARS-CoV-2 specificity may be public and either exclusively or widely present in COVID-19 patients with mild disease. In contrast, overall median frequencies between SARS-CoV-2-associated mild disease- and pneumonia- and shared-specific TCRβ clusters were not significantly different (Fig. 3c). Further investigating the distribution of sharing level of SARS-CoV-2-associated TCRβ clusters revealed that mild disease-specific clusters were mostly public (clustered clonotypes from all individuals with mild disease), whereas pneumonia-specific clusters were mainly private (clustered clonotypes from one or two pneumonia patient(s)) (Fig. 3d).
Fig. 3.
Characterization of SARS-CoV-2-associated TCRβ clusters enriched with TCRβ clonotypes from mild disease group, pneumonia group, and both groups of patients. a The distribution of transformed absolute difference of cluster median TCRβ frequencies (x-axis) and cluster median TCRβ generation probabilities (Pgen) (y-axis) of GLIPH2-identifying clusters was shown. Each circle represented one TCRβ cluster. The color and size of circles represented different patient-specific types and levels of sharing for TCRβ clusters, respectively. Only mild disease-, pneumonia-, and shared-specific TCRβ clusters with potential SARS-CoV-2 specificities were colored by blue, red, and green, respectively. The sharing level was denoted by private and low level of sharing, medium level of sharing, and high level of sharing. The value X for x-axis represented the absolute difference between median frequencies of clustered clonotypes contributed from pneumonia and mild disease groups of patients for each cluster. The value Y for y-axis indicated the median generation probability of all clonotypes within each cluster. b, c Comparison of cluster median Pgen and frequencies between SARS-CoV-2-associated mild disease- and pneumonia- and shared-specific TCRβ clusters was performed. Two-sided p values were shown from t-test. d Proportions of private and low level of sharing, medium level of sharing, and high level of sharing were compared between SARS-CoV-2-associated mild disease- and pneumonia- and shared-specific TCRβ clusters. p Values were shown from chi-square test. e, f Comparison of cluster median Pgen and frequencies between SARS-CoV-2-associated mild disease- and pneumonia- and shared-specific clusters, grouped by private and low level of sharing, medium level of sharing, and high level of sharing, was performed. Two-sided p values were shown from t-test. p Values larger than 0.05 were considered to be not statistically significant and were not shown
Since our findings showed that the distribution of sharing level was divergent between different patient group-specific TCRβ clusters, we next separated all SARS-CoV-2-associated TCRβ clusters into three groups (private and low level of sharing, medium level of sharing and high level of sharing) based on the number of patients contributing clonotypes to the cluster and examined differences in median Pgen and frequency between mild disease- and pneumonia- and shared-specific clusters at different levels of sharing. Our results showed that among SARS-CoV-2-associated TCRβ clusters at high level of sharing, the overall median Pgen of SARS-CoV-2-associated mild disease-specific clusters was significantly higher than that of SARS-CoV-2-associated pneumonia-specific clusters (Fig. 3e). In addition, the overall median Pgen of SARS-CoV-2-associated shared-specific clusters was significantly higher than that of SARS-CoV-2-associated pneumonia-specific clusters at any level of sharing, while no significant difference of overall median Pgen was observed between SARS-CoV-2-associated mild disease- and shared-specific clusters at low level (including private clusters) or high level of sharing (Fig. 3e). Of note, a significant difference of overall median Pgen between mild disease- and shared-specific clusters at medium level of sharing was detected (Fig. 3e). Such results supported our finding of that the cluster median Pgen was significantly lower in SARS-CoV-2-associated pneumonia-specific clusters, especially in those at high level of sharing, relative to mild disease-specific ones. When comparing clonal frequencies between clusters within different groups of sharing level, we observed a significant difference in cluster median frequencies between SARS-CoV-2-associated mild disease- and pneumonia- and shared-specific clusters at medium level of sharing (Fig. 3f). At high level of sharing group, shared-specific clusters had significantly higher cluster median frequencies than pneumonia-specific clusters; however, no significant difference in the cluster median frequency was observed between mild disease- and pneumonia-specific clusters (Fig. 3f). These findings suggested that SARS-CoV-2-associated TCRβ clonotypes/clusters may exhibit different characteristics, which are correlated with VDJ recombination and TCR clonal expansion, between COVID-19 patients with mild disease and pneumonia.
Distinct Patterns of Clustered TCRβ CDR3 Similarity Network and Sequence Conservation Between COVID-19 Patients with Mild Disease and Pneumonia
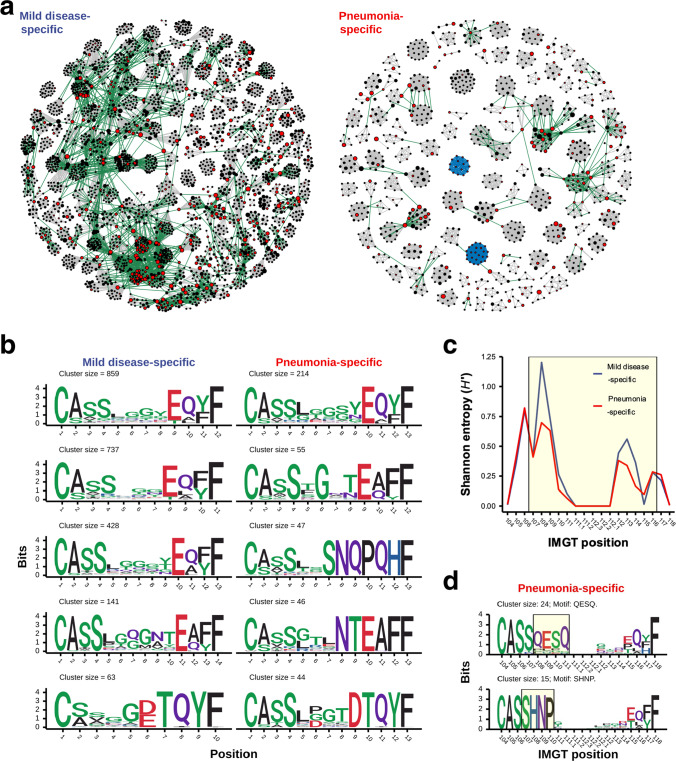
Our clustering analysis revealed distinct characteristics between potential SARS-CoV-2-specific TCRβ clusters enriched and highly shared by COVID-19 patients with mild disease and pneumonia. We next constructed similarity networks for TCRβ clonotypes from public SARS-CoV-2-associated mild disease- and pneumonia-specific clusters. To address this, TCRβ clonotypes from SARS-CoV-2-associated clusters at high level of sharing were selected, and those with high global similarity (hamming distance of same-length TCRβ CDR3 sequences equal to and less than one) or clustered by GLIPH2 algorism (global- or motif-based prediction) were connected to depict unweighted similarity networks of TCRβ clonotypes from mild disease- and pneumonia-specific TCRβ clusters (Fig. 4a). The clonotype network of public SARS-CoV-2-associated mild disease-specific TCRβ clusters exhibited a larger extent of cross-group relationship (edges colored green) between clonotypes from different global-based clusters, compared with the clonotype network of public SARS-CoV-2-associated pneumonia-specific clusters (Fig. 4a). Such result suggested that TCRβ clonotypes from different global-based SARS-CoV-2-associated clusters still exhibit similar characteristics, although they were considered to possess divergent antigen specificities based on GLIPH2 clustering algorithm. We presumed that TCRβ clonotypes of connected TCRβ clusters may still have high probability of sharing SARS-CoV-2 antigen specificity. We thus combined these connected clusters for each public mild disease- and pneumonia-specific cluster network. The CDR3 sequence conservations of the top five largest merged clusters were characterized (Fig. 4b). Compared to public SARS-CoV-2-associated pneumonia-specific TCRβ clusters, TCRβ clonotypes from mild disease-specific clusters showed highly diverse sequences within CDR3 central region (Fig. 4c), where the primary contact site of CDR3 motif for antigen recognition was reported [23]. In addition, we observed that motif-based SARS-CoV-2-associated TCRβ clusters (edges colored blue) were exclusive to the public pneumonia-specific cluster similarity network (Fig. 4a). These motif-based pneumonia-specific clusters revealed a shared pattern of SARS-CoV-2-associated CDR3 motif position (Fig. 4d). Taken together, our results revealed several discrepancies between TCRβ clonotypes from public SARS-CoV-2-associated mild disease- and pneumonia-specific TCRβ clusters, suggesting that patients with mild disease and pneumonia have underwent distinct T cell response during SARS-CoV-2 viral infection and therefore exhibited different characteristics of public TCR clonotypes/clusters in the convalescent stage.
Fig. 4.
The similarity network and CDR3 sequence conservation of TCRβ clonotypes of public SARS-CoV-2-associated mild disease- and pneumonia-specific clusters. a Network graphs illustrated similarities between TCRβ clonotypes from SARS-CoV-2-associated mild disease- (left panel) and pneumonia-specific (right panel) clusters with high level of sharing. Each vertex indicated a TCRβ clonotype with unique CDR3 amino acid sequence. Edges represented high similarities between TCRβ clonotype based on global- (grey line) and motif-based (blue line) clustering by GLIPH2 algorithm as well as calculated small hamming distance (equal to one) between cross-cluster clonotypes with same CDR3 sequence length (green line). Vertex size indicated the frequency of clonotypes among all patients. SARS-CoV-2-annotated TCRβ clonotypes were colored by red. b Logo plots illustrated CDR3 amino acid sequences of top 5 largest SARS-CoV-2-associated mild disease- (left panel) and pneumonia-specific (right panel) TCRβ clusters from single and merged cross-group GLIPH2 global-based TCRβ clusters in (a). c The mean of Shannon entropy of each CDR3 amino acid position among all public SARS-CoV-2-associated mild disease- (blue line) and pneumonia-specific (red line) GLIPH2 global-based TCRβ clusters were shown. The CDR3 region with high structural contact probabilities (107–116) was highlighted by the yellow box. d Sequence logos of annotated TCRβ clonotypes from SARS-CoV-2-associated pneumonia-specific GLIPH2 motif-based TCRβ clusters in (a) were shown. The label on x-axis represented positions of CDR3 amino acid defined by IMGT. The consensus region of enriched motif sequence in each motif-based TCRβ cluster was highlighted by the yellow box
Discussion
Here, we applied an immunosequencing approach to profile peripheral TCRβ repertoires of COVID-19 patients with mild disease and pneumonia in the convalescent stage. Results suggested a distinct diversity and revealed a skewed CDR3 length usage in TCRβ repertoires of pneumonia patients compared with individuals with mild disease. Furthermore, we found that the overall generation probabilities (Pgen) and the dominant level of sharing were significantly different between mild disease- and pneumonia-specific TCRβ clusters. Specifically, divergent patterns of CDR3 sequence diversity and motif usage between public mild disease- and pneumonia-specific TCRβ clusters were identified. Consistent with previous studies that indicated a decreased TCR repertoire diversity in COVID-19 patients compared with healthy individuals [31, 32], our findings revealed a slight reduction in TCRβ repertoire diversity in pneumonia patients. Such findings are comparable to a recent report showing that the single-cell TCR repertoire diversity is decreased in patients with severe COVID-19 compared with those classified as mild severity [33]. In addition, we further found that TCRβ repertoire of pneumonia patients contained longer lengths of CDR3 sequences compared to patients with mild disease. Such phenomenon was previously observed in severe COVID-19 patients’ B cell receptor (BCR) repertoires, where the length of CDR3 nucleic acid sequence of BCR clonotypes with top copy numbers was significantly longer, compared with patients with moderate severity or healthy individuals [34]. Taken together, we suggested that compared to COVID-19 patients with mild illness, patients with pneumonia should possess stronger and prolonged T cell responses with a larger extent of TCR clonal expansion (reflected by the slight decrease in repertoire diversity) and a distinct CDR3 feature (reflected by the skewed CDR3 length) throughout SARS-CoV-2 infection. Such inference has been supported by previous researches showing that severe COVID-19 patients exhibit increased T cell activation markers and IFN-γ–producing T cell populations, while asymptomatic/mild patients seemed to have a weaker or limited immune response to SARS-CoV-2 infection [35, 36].
In this study, a framework for TCR clustering, annotation, and classification was used to identify groups of TCRβ clonotypes with probably high similarity in antigen specificity against known SARS-CoV-2 epitopes. We determined SARS-CoV-2-associated mild disease-, pneumonia-, and shared-specific clusters based on enrichment of clustered clonotypes in each patient group. Then, we estimated the theoretical Pgen of TCRβ clonotype for each SARS-CoV-2-associated cluster. We found that the overall Pgen was significantly higher in mild disease- and shared-specific clusters, relative to pneumonia-specific clusters. The findings suggested that such clustered SARS-CoV-2-associated TCRβ clonotypes were public TCRs and probably generated via a convergent VDJ recombination [25, 37]. Indeed, we observed a skewed distribution of publicity in SARS-CoV-2-associated mild disease-specific TCRβ clusters, most of which were highly shared across patients with mild disease. On the contrary, most of SARS-CoV-2-associated pneumonia-specific TCRβ clusters were private or lowly shared among pneumonia patients. In addition, the sequence conservation analysis of public SARS-CoV-2-associated TCRβ clusters revealed distinct patterns of CDR3 sequence motifs between mild disease- and pneumonia-specific clusters. These results indicated that COVID-19 patients are very likely to share similar T cell immune responses; however, individuals with different symptom status could develop divergent T cell antigen recognition or expansion strategies, which are reflected by their TCR repertoires. On the other hand, the compositions and dynamics of individual TCR repertoires might associate with disease symptoms and severity of COVID-19 [31].
There are some limitations worth noting. First, we were only able to analyze a small cohort of COVID-19 patients, which might influence the statistical power. Second, the time point of sampling for patients with mild disease (15–18 days) and pneumonia (17–53 days) was different. Such factors might affect the profiling of TCR repertoire in different patients, since SAS-CoV-2-specific TCR clonotypes are dynamic throughout the course of viral infection [38, 39]. Nevertheless, TCR repertoire status of COVID-19 patients in our study should still predominantly reflect a stable SARS-CoV-2-responding T cell immunity as they were in the convalescent stage at the time of sample collection. Moreover, the unavailability of patients’ HLA typing data limited the analysis for TCR clustering and annotation of SARS-CoV-2 specificity. Inclusion of HLA typing data from COVID-19 patients would be helpful for the discovery and following validation of SARS-CoV-2-specific TCR clonotypes. In addition, functional validation for clustered SARS-CoV-2-associated TCRβ clonotypes is very important. Experiments such as MHC-tetramer staining [40] or yeast-display screening [41, 42] coupled with immunosequencing will allow us to determine whether these TCRβ clonotypes possess antigen specificities for SARS-CoV-2 epitopes in future studies. Finally, our TCR annotation analysis was based on known SARS-CoV-2 epitope-specific TCRs from two public datasets [27, 30], and therefore, SARS-CoV-2-unannotated clustered TCRβ clonotypes and the unknown SARS-CoV-2 antigen specificities may have been discarded. Those non-SARS-CoV-2-annotated TCRβ clusters with simultaneous high level of sharing and low overall Pgen might contain novel SARS-CoV-2-specific TCRβ clonotypes that could be of great interest, as previously suggested [31]. A larger SARS-CoV-2-specific TCR dataset would capture more TCRβ clonotypes/clusters with SARS-CoV-2 antigen specificities.
In summary, our study revealed that COVID-19 patients with mild disease and pneumonia exhibited distinct features of TCRβ repertoire and SARS-CoV-2-associated TCRβ clonotypes/clusters, suggesting a divergence in T cell response between mild disease and pneumonia patients infected with SARS-CoV-2. These findings provided a better understanding of the role of T cell immunity in SARS-CoV-2 infection.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Taipei Medical University-Shuang Ho Hospital for patient recruitment and sample collections and Ministry of Science and Technology, Taiwan, for supporting the study.
Authors’ Contributions
K.Y.L. and W.C.C. initiated, designed and supervised the study. P.H.F. and K.Y.L. enrolled patients and collected blood samples as well as clinical data. C.M.C. and T.H.W. performed the experiments. C.M.C. performed data analysis. C.M.C. P.H.F., T.H.W., H.A., K.Y.L. and W.C.C. interpreted data and results of the study. C.M.C., P.H.F., K.Y.L., and W.C.C. wrote the manuscript. C.M.C., P.H.F., H.A., K.Y.L., and W.C.C. revised the manuscript.
Funding
This work was supported by the grants from Ministry of Science and Technology, Taiwan (MOST-108–2314-B-038–027, MOST-109–2314-B-038–131 and MOST-109–2628-B-038–012), and Taipei Medical University, Taiwan (12310-106079, Yusuke Nakamura Chair Professorship).
Data Availability
The data that support the findings of this study are available on reasonable request from the corresponding authors.
Code Availability
R codes for analysis of the study are available from the corresponding authors on reasonable request.
Declarations
Ethics Approval
The study was conducted with approval from the Taipei Medical University Joint Institutional Review Board (TMUJIRB) [IRB number: N202004076].
Consent to Participate
Informed consent was obtained from all participants recruited in the study.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Che-Mai Chang and Po-Hao Feng contributed equally to this work.
Corresponding authors: Kang-Yun Lee and Wei-Chiao Chang.
Contributor Information
Kang-Yun Lee, Email: leekangyun@tmu.edu.tw.
Wei-Chiao Chang, Email: wcc@tmu.edu.tw.
References
- 1.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan. China Jama. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pujadas E, Chaudhry F, McBride R, Richter F, Zhao S, Wajnberg A, et al. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir Med. 2020;8(9):e70. doi: 10.1016/s2213-2600(20)30354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Gayle AA, Wilder-Smith A, Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. 2020;27(2). 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed]
- 4.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26(5):672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 6.Xiao AT, Tong YX, Zhang S. Profile of RT-PCR for SARS-CoV-2: a preliminary study from 56 COVID-19 patients. Clin Infect Dis. 2020. 10.1093/cid/ciaa460. [DOI] [PMC free article] [PubMed]
- 7.Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, et al. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perera R, Tso E, Tsang OTY, Tsang DNC, Fung K, Leung YWY, et al. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis. 2020;26(11):2701–2704. doi: 10.3201/eid2611.203219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang-Huau TL, Gueguen P, Goudot C, Durand M, Bohec M, Baulande S, et al. Human in vivo-generated monocyte-derived dendritic cells and macrophages cross-present antigens through a vacuolar pathway. Nat Commun. 2018;9(1):2570. doi: 10.1038/s41467-018-04985-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet. 2020;395(10223):497–506. doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dash P, Fiore-Gartland AJ, Hertz T, Wang GC, Sharma S, Souquette A, et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature. 2017;547(7661):89–93. doi: 10.1038/nature22383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikolich-Zugich J, Slifka MK, Messaoudi I. The many important facets of T-cell repertoire diversity. Nat Rev Immunol. 2004;4(2):123–132. doi: 10.1038/nri1292. [DOI] [PubMed] [Google Scholar]
- 13.Trautmann L, Rimbert M, Echasserieau K, Saulquin X, Neveu B, Dechanet J, et al. Selection of T cell clones expressing high-affinity public TCRs within Human cytomegalovirus-specific CD8 T cell responses. J Immunol. 2005;175(9):6123–6132. doi: 10.4049/jimmunol.175.9.6123. [DOI] [PubMed] [Google Scholar]
- 14.Pogorelyy MV, Minervina AA, Touzel MP, Sycheva AL, Komech EA, Kovalenko EI, et al. Precise tracking of vaccine-responding T cell clones reveals convergent and personalized response in identical twins. ProcNatlAcadSci U S A. 2018;115(50):12704–12709. doi: 10.1073/pnas.1809642115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sant S, Grzelak L, Wang Z, Pizzolla A, Koutsakos M, Crowe J, et al. Single-cell approach to influenza-specific CD8(+) T cell receptor repertoires across different age groups, tissues, and following influenza virus infection. Front Immunol. 2018;9:1453. doi: 10.3389/fimmu.2018.01453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang CM, Liao YM, Lan GY, Chang WC, Yen Y. Using T-cell repertoire profiles as predictor in a primary mucosal melanoma. ClinTransl Med. 2020;10(4):e136. doi: 10.1002/ctm2.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):3. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 18.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12(5):380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 20.Shugay M, Bagaev DV, Turchaninova MA, Bolotin DA, Britanova OV, Putintseva EV, et al. VDJtools: Unifying post-analysis of T cell receptor repertoires. PLoS Comput Biol. 2015;11(11):e1004503. doi: 10.1371/journal.pcbi.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubelt F, Bolen CR, McGuire HM, Vander Heiden JA, Gadala-Maria D, Levin M, et al. Individual heritable differences result in unique cell lymphocyte receptor repertoires of naïve and antigen-experienced cells. Nat Commun. 2016;7:11112. doi: 10.1038/ncomms11112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolen CR, Rubelt F, Vander Heiden JA, Davis MM. The repertoire dissimilarity index as a method to compare lymphocyte receptor repertoires. BMC Bioinformatics. 2017;18(1):155. doi: 10.1186/s12859-017-1556-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F, et al. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547(7661):94–98. doi: 10.1038/nature22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang H, Wang C, Rubelt F, Scriba TJ, Davis MM. Analyzing the mycobacterium tuberculosis immune response by T-cell receptor clustering with GLIPH2 and genome-wide antigen screening. Nat Biotechnol. 2020;38(10):1194–1202. doi: 10.1038/s41587-020-0505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sethna Z, Elhanati Y, Callan CG, Walczak AM, Mora T. OLGA: fast computation of generation probabilities of B- and T-cell receptor amino acid sequences and motifs. Bioinformatics. 2019;35(17):2974–2981. doi: 10.1093/bioinformatics/btz035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bagaev DV, Vroomans RMA, Samir J, Stervbo U, Rius C, Dolton G, et al. VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Res. 2020;48(D1):D1057–D1062. doi: 10.1093/nar/gkz874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snyder TM, Gittelman RM, Klinger M, May DH, Osborne EJ, Taniguchi R et al. Magnitude and dynamics of the T-cell response to SARS-CoV-2 infection at both individual and population levels. medRxiv. 2020. 10.1101/2020.07.31.20165647.
- 28.Pogorelyy MV, Shugay M. A Framework for annotation of antigen specificities in high-throughput T-cell repertoire sequencing studies. Front Immunol. 2019;10:2159. doi: 10.3389/fimmu.2019.02159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2016. [Google Scholar]
- 30.Shomuradova AS, Vagida MS, Sheetikov SA, Zornikova KV, Kiryukhin D, Titov A, et al. SARS-CoV-2 epitopes are recognized by a public and diverse repertoire of human T cell receptors. Immunity. 2020;53(6):1245–57.e5. doi: 10.1016/j.immuni.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schultheiß C, Paschold L, Simnica D, Mohme M, Willscher E, von Wenserski L, et al. Next-generation sequencing of T and B cell receptor repertoires from COVID-19 patients showed signatures associated with severity of disease. Immunity. 2020;53(2):442–55.e4. doi: 10.1016/j.immuni.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang JY, Wang XM, Xing X, Xu Z, Zhang C, Song JW, et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat Immunol. 2020;21(9):1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- 33.Zhang F, Gan R, Zhen Z, Hu X, Li X, Zhou F, et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal Transduct Target Ther. 2020;5(1):156. doi: 10.1038/s41392-020-00263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuri-Cervantes L, Pampena MB, Meng W, Rosenfeld AM, Ittner CAG, Weisman AR et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci Immunol. 2020;5(49). 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed]
- 35.Wang F, Hou H, Luo Y, Tang G, Wu S, Huang M et al. The laboratory tests and host immunity of COVID-19 patients with different severity of illness. JCI Insight. 2020;5(10). 10.1172/jci.insight.137799. [DOI] [PMC free article] [PubMed]
- 36.Long QX, Tang XJ, Shi QL, Li Q, Deng HJ, Yuan J, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26(8):1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 37.Elhanati Y, Sethna Z, Callan CG, Jr, Mora T, Walczak AM. Predicting the spectrum of TCR repertoire sharing with a data-driven model of recombination. Immunol Rev. 2018;284(1):167–179. doi: 10.1111/imr.12665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minervina AA, Komech EA, Titov A, Bensouda Koraichi M, Rosati E, Mamedov IZ et al. Longitudinal high-throughput TCR repertoire profiling reveals the dynamics of T-cell memory formation after mild COVID-19 infection. Elife. 2021;10. 10.7554/eLife.63502. [DOI] [PMC free article] [PubMed]
- 39.Niu X, Li S, Li P, Pan W, Wang Q, Feng Y, et al. Longitudinal analysis of T and B cell receptor repertoire transcripts reveal dynamic immune response in COVID-19 patients. Front Immunol. 2020;11:582010. doi: 10.3389/fimmu.2020.582010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramachandiran V, Grigoriev V, Lan L, Ravkov E, Mertens SA, Altman JD. A robust method for production of MHC tetramers with small molecule fluorophores. J Immunol Methods. 2007;319(1–2):13–20. doi: 10.1016/j.jim.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Birnbaum ME, Mendoza JL, Sethi DK, Dong S, Glanville J, Dobbins J, et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 2014;157(5):1073–1087. doi: 10.1016/j.cell.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gee MH, Han A, Lofgren SM, Beausang JF, Mendoza JL, Birnbaum ME, et al. Antigen identification for orphan T cell receptors expressed on tumor-infiltrating lymphocytes. Cell. 2018;172(3):549–63.e16. doi: 10.1016/j.cell.2017.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available on reasonable request from the corresponding authors.
R codes for analysis of the study are available from the corresponding authors on reasonable request.