Abstract
The majority of regulated protein degradation in eukaryotes is accomplished by the 26S proteasome, the large proteolytic complex responsible for removing regulatory proteins and damaged proteins. Proteins are targeted to the proteasome by ubiquitination, and degradation is initiated at a disordered region within the protein. The ability of the proteasome to precisely select which proteins to break down is necessary for cellular functioning. Recent studies reveal the subtle mechanisms of substrate recognition by the proteasome – diverse ubiquitin chains can act as potent proteasome targeting signals, ubiquitin receptors function uniquely and cooperatively, and modification of initiation regions modulate degradation. Here, we summarize recent findings illuminating the nature of substrate recognition by the proteasome.
Keywords: Proteasome, UPS, protein degradation, specificity of degradation
Introduction
Protein destruction is required for maintaining cellular homeostasis. In eukaryotes, the majority of regulated protein degradation is carried out by the ubiquitin proteasome system (UPS), an essential pathway that regulates diverse cellular processes such as cell cycle progression, signal transduction, DNA repair, and protein quality control [1]. At the center of the UPS is a marcromolecular proteolytic complex called the 26S proteasome. Proteins are typically targeted to the proteasome by the covalent attachment of one or more ubiquitin moieties or polyubiquitin chains to lysine residues within target proteins by the sequential action of E1, E2, and E3 enzymes. Ubiquitin chains are formed by conjugation of the C-terminus of one ubiquitin to one of seven lysine residues (K6, K11, K27, K29, K33, K48, K63) or the N-terminus of another ubiquitin (M1) [2]. The ubiquitin chains are then recognized by the proteasome, and once a protein is bound to the proteasome, degradation is initiated at a stretch of unstructured amino acids within the substrate.
Many studies within the past decade have contributed to the structural, mechanistic, and kinetic understanding of the proteasome. High-resolution cryo-electron microscopy (cryoEM) structures of substrate-free and substrate-processing proteasomes have identified several conformational states demonstrating its dynamic nature and substrate processing mechanism [3–10]. Elegant biochemical studies have elucidated events triggering the conformational switches, drawing a kinetic map of substrate processing [11]. Cryo-electron tomography (cryo-ET) has investigated proteasome localization patterns in cells to reveal the functional importance of proteasome compartmentalization. Cryo-ET has also shown how the proteasome responds to neuronal protein aggregates and shed light on the relationship between the UPS and disease pathology [12–14].
To maintain the integrity of the proteome the proteasome must select precisely and process its substrates from the vast number of proteins in the cell. Incongruously, the degradation signal ubiquitin is also used as a signal in many non-proteolytic cellular processes. Substrate selectivity is achieved in part through a combination of ubiquitin receptor binding preferences, initiation region recognition preferences, and proteasome architecture. Here, we highlight recent structural, biochemical, and cell-based studies that have yielded insights into the mechanisms of substrate recognition by the proteasome.
Structure and the dynamic nature of the proteasome
The proteasome is a 2.5 MDa machine composed of at least 33 unique subunits organized into two main subcomplexes: the 20S core particle (CP) and the 19S regulatory particle (RP) (Figure 1a). The CP contains the proteolytic activity of the proteasome and is a barrel-shaped structure that consists of four stacked rings: two inner heteroheptameric rings of β subunits (β1–7) flanked by an outer heteroheptameric ring of α subunits (α1–7) on each side [15]. The proteolytic sites are sequestered within the CP and located the inner surface of β1, β2, and β5 with caspase-like, trypsin-like, and chymotrypsin-like activity, respectively [16]. The six proteolytic sites allow the proteasome to process almost any protein into peptides between 3–12 residues in length [1]. Access to the inner chamber is tightly restricted by the N-termini of the α subunits, which form a narrow gate that can be opened by the 19S RP [8].
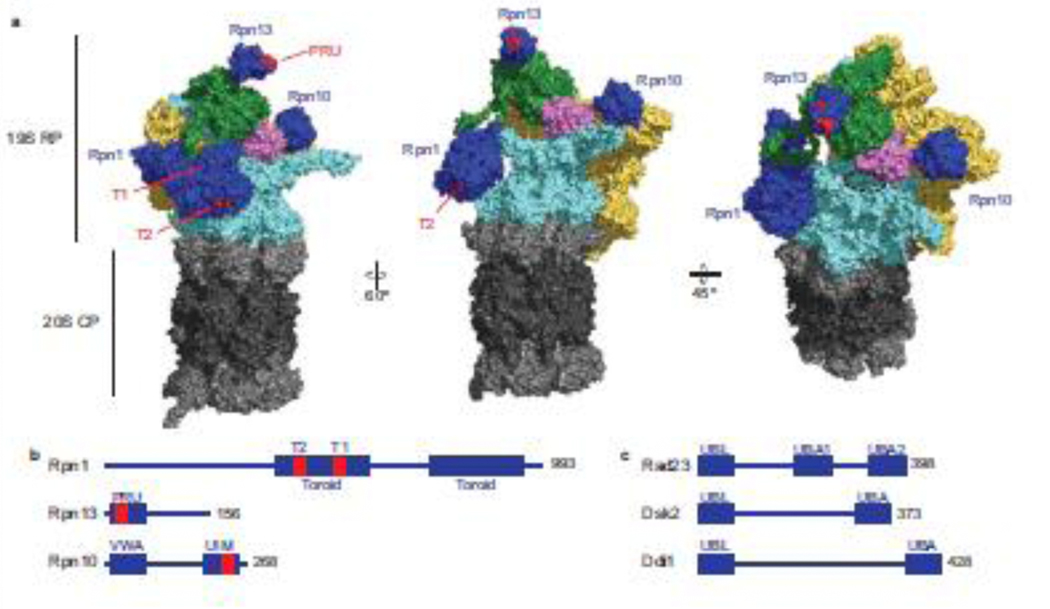
Figure 1: Architecture of the 26S proteasome and ubiquitin receptors.
(a) CryoEM surface structure of the substrate-free yeast 26S proteasome (s1 state) depicted in three different orientations (PDB: 6FVT). The 20S CP is shown in grey: α subunits in light grey and β subunits in dark grey. The 19S RP is shown in multiple colors. Ubiquitin receptors Rpn1, Rpn13, and Rpn10 are shown in blue. Residues within the T1 and T2 sites of Rpn1 and the PRU domain of Rpn13 critical for ubiquitin binding are highlighted in red. The UIM domain of Rpn10 is not resolved, therefore only the VWA domain of Rpn10 is shown. Rpn2 is shown in green, the non-ATPase lid subunits are shown in yellow, and the DUB Rpn11 is shown in pink. The heterohexameric ring of ATPases (Rpt1–6) that form the translocation channel is shown in cyan. The entrance to the substrate translocation channel is indicated by a dashed black circle. (b) Schematic of yeast proteasomal ubiquitin receptors Rpn10, Rpn13, and Rpn1 with location of the essential ubiquitin-binding residues indicated in red. (c) Schematic of yeast UBL-UBA shuttle receptor proteins Rad23, Dsk2, and Ddi1.
The 19S RP is made of at least 19 distinct subunits and caps one or both ends of the CP. The RP recognizes proteasomal substrates and transfers them to the CP for destruction. Two subcomplexes, the base and lid, comprise the RP [17,18]. A heterohexameric ring of AAA+ ATPases (Rpt1–6) forms the entrance to the substrate translocation channel. The ATPases use power generated from repeated cycles of ATP hydrolysis to mechanically unfold the substrate while translocating them into the CP. Five of the ATPases contain hydrophobic-tyrosine-X (HbYX) or related motifs at their C-terminus that insert into inter-subunit pockets between the α subunits to induce CP gate opening and to facilitate substrate translocation from the RP to the CP [6,19].
Subunits Rpn10, Rpn1, and Rpn13 serve as ubiquitin receptors (Figure 1a,b). Rpn10 is located the closest to the translocation channel. It binds ubiquitin through its C-terminal ubiquitin-interacting motif (UIM), which is tethered via a flexible linker to an N-terminal von Willebrand factor type A (VWA) domain tightly docked on the proteasome [20,21]. Rpn1 is on the opposite side of the translocation channel relative to the VWA domain of Rpn10 and binds ubiquitin through its T1 site. Rpn1 can also bind the ubiquitin-like (UBL) domain of deubiquitinating enzyme (DUB) Ubp6 through its T2 site. T1 and T2 are located within one of the toroidal domains of Rpn1 and face away from the translocation channel [22]. Rpn13 is located at the top of the RP and binds ubiquitin through its Pleckstrin-like receptor for ubiquitin (PRU) domain [9,20,23,24].
Binding to the proteasome is not sufficient for degradation. Degradation only occurs when the proteasome can physically engage a disordered region within the substrate (i.e., initiation region) to initiate degradation [1,25]. When a substrate is bound to the ubiquitin receptors, its initiation region enters the translocation channel and is engaged by loops within the ATPases (i.e., the conserved pore-1 loops) [5]. During substrate translocation, ubiquitin encounters the essential DUB Rpn11, which is directly adjacent to the entrance of the translocation channel [3]. Ubiquitin binding to Rpn11 triggers en bloc removal of the remaining ubiquitin chain from the substrate [5,7,26,27]. This mechanism of degradation-coupled deubiquitination prevents premature ubiquitin removal and substrate escape [27].
High-resolution cryoEM structures of the proteasome have identified at least six conformational states (s1-s6) corresponding to various stages of substrate processing with major conformational changes occurring between states [3–10]. In the substrate-free state, there is an equilibrium between s1 and s2 conformations, though s1 is presumed to be the substrate-accepting conformation since Rpn11 blocks initiation region access to the pore-1 loops in the s2 state. In the s1 state, Rpn11 is offset from the entrance to the translocation channel permitting initiation region entry. However, the translocation channels of the RP and the CP are not coaxially aligned preventing access to the CP. The substrate-processing state is represented by s3, s4, and s6 conformations. In these states Rpn11 is shifted above the entrance to the translocation channel and translocation channels within the RP and CP are coaxially aligned to create a continuous path to the proteolytic sites of the CP from the RP [3].
The expanding diversity of ubiquitin modifications in proteasome targeting
Ubiquitin is used as a signal in many cellular pathways and the configuration of the ubiquitin moieties is often linked to the fate of the modified protein. The canonical degradation targeting signal has long been defined as a K48-linked ubiquitin chain of at least 4 molecules [28] (Figure 2b) though more recent studies show that other ubiquitin modifications can also target proteins to the proteasome. K11-linked chains are associated with proteasomal degradation during mitosis yet fail to trigger efficient degradation in vitro [29,30]. This may be because the chains assembled by the APC/C ubiquitination complex during mitosis typically consist of K11 and K48 linkages. K63-linked chains are generally associated with membrane trafficking and other non-proteolytic processes but can also target proteins for degradation in vitro and in some cases in vivo [29,31].
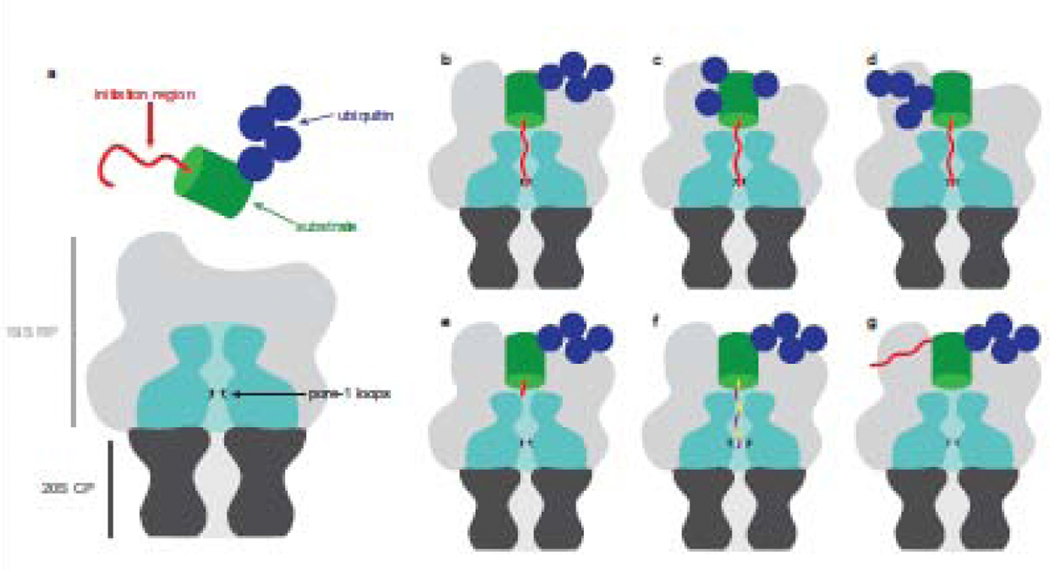
Figure 2: Substrate recognition is mediated by ubiquitin and its engagement is determined by the initiation region.
(a) Schematic representation of the proteasome and its substrate. The 20S CP is shown in dark grey and the ATPases of the 19S RP are shown in cyan. The pore-1 loops of the ATPases are sequestered inside the translocation channel and depicted in black. Proteins (green) are typically targeted to the proteasome through the attachment of ubiquitin (blue) and degradation is initiated at a disordered region within the substrate (red). Targeting can be accomplished by diverse ubiquitin modifications such as homotypic chains (b), multi-monoubiquitination (c), and branched chains (d). Effective engagement of a bound substrate requires the initiation region be sufficient length (e), the appropriate composition of amino acids (f), and suitably located relative to the ubiquitin modification in order to access the translocation channel (g).
Ubiquitin can form chains with homotypic linkages where ubiquitin moieties are connected through the same Lys or chains with heterotypic linkages where ubiquitin moieties are linked through different Lys within a single chain. These chains can be mixed (i.e., different linkage types but each ubiquitin is connected to only one other ubiquitin) or branched (i.e., one ubiquitin is connected to multiple ubiquitin moieties). Branched chains including K48/K63, K29/K48, and K11/K48 can target proteins for proteasomal degradation [32,33] (Figure 2d). Branched K11/K48 chains enhance degradation of mitotic proteins compared to homogeneous K11 or K48 chains [30,34]. A recent study using NMR, X-ray crystallography, and small angle neutron scattering (SANS) solved the structure of branched K11/K48 tri-ubiquitin discovering a novel interdomain interface between distal ubiquitin moieties [35]. Branched K11/K48 tri-ubiquitin binds with enhanced affinity to Rpn1, suggesting that branched chains may improve degradation through more productive proteasome binding. Since branched K11/K48 chains are synthesized during mitosis to rapidly remove cell-cycle regulator proteins and ensure timely progression through the cell cycle, it makes physiological sense that these chains would be highly efficient degradation targeting signals.
Ubiquitin can be linked to form complex chains but it can also take contrastingly simple forms. Proteins can be decorated with a single ubiquitin or multiple single ubiquitin moieties. Mono- and multi-monoubiquitination were associated with nonproteolytic function, but are now known to be effective degradation signals also [29,36,37] (Figure 2c). Multi-monoubiquitination of a natural protein is sufficient to target it for degradation in a reconstituted system [38]. Likewise, a model substrate containing two ubiquitin domains designed to mimic a multi-monoubiquitinated substrate is degraded by purified proteasome [29].
Substrate recognition through the ubiquitin receptors
Many types of ubiquitin modifications can target substrates for degradation (Figure 2b,c,d), and the three ubiquitin receptors of the proteasome must recognize these numerous and topologically diverse signals. Indeed, the 19S RP appears to function as a versatile recognition platform [29]. A biochemical study measuring the degradation of defined model substrates in presence of purified proteasomes in which individual receptors were mutated to abrogate ubiquitin binding showed that receptors can collaborate to recognize substrates with varied ubiquitin length and topology [29]. Degradation of K48-linked chains is primarily mediated by Rpn10, since proteasome in which Rpn10 was the only functional receptor degraded substrates as efficiently as wildtype proteasome. Substrates with K63-linked chains and multiple K48-linked chains are targeted cooperatively between Rpn10 and Rpn1 or Rpn10 and Rpn13. The UIM of Rpn10 is tethered to its proteasome-docked VWA domain by a long flexible linker, perhaps permitting substrates to explore wider conformational space compared to the proteasome docked ubiquitin binding sites of Rpn1 or Rpn13. Rpn10 is a critical receptor for ubiquitinated substrates perhaps due in part to the orientation of bound substrates being less conformationally restricted increasing the likelihood of engagement of the initiation region. Proximity of Rpn10 to Rpn11 may also contribute to degradation of substrate directed through Rpn10 [39].
The structures of various ubiquitin chains bound to ubiquitin receptors informs our understanding of how receptors recognize ubiquitinated substrates. Receptors display both overlapping and unique linkage binding preferences, strengthening the idea that the 19S is capable of recognizing diverse substrates [35,40,41]. Using NMR, the structures of free K48-linked di-ubiquitin and of K48-linked di-ubiquitin bound to hRpn13 in association with the proteasome structural subunit hRpn2 were recently solved [42]. This work discovered a previously unidentified conformation of K48-linked di-ubiquitin proposed to be the preferred form for receptor binding and revealed highly dynamic interactions between hRpn2:hRpn13 and bound ubiquitin chains. The dynamic nature of these interactions may assist in orienting substrates for engagement and deubiquitination by allowing receptors to bind different locations within ubiquitin chains or transferring ubiquitin chains between receptors [42].
The three ubiquitin receptors of the proteasome can also bind the UBL domains of UBL-UBA proteins, which are thought to act as shuttling substrate receptors by binding to the proteasome through their UBL domain and to ubiquitinated proteins through their ubiquitin-associating (UBA) domain(s) [22–24,43,44] (Figure 1c). Isothermal titration calorimetry (ITC) and NMR-based studies have measured tight affinities of UBL-UBA proteins for ubiquitin receptors and elucidated the mechanism of binding [45,46]. Biochemical and cell-based work has shown model substrates containing a UBL domain are efficiently degraded by the proteasome [29,44,47]. Though some UBL-UBA proteins play a role in the degradation of select natural proteins, their mechanism of action remains unclear [48].
Substrate engagement is determined by initiation regions
Proteasome localization of a substrate is not sufficient to cause degradation. Degradation requires the presence within the substrate protein of a disordered region of appropriate length, composition, and location [1]. Several studies suggest that C-terminal initiation regions must be 20–30 amino acids in length to allow efficient degradation, which is consistent with structural studies of the proteasome [25,44,49–53] (Figure 2e). The pore-1 loops, responsible for grabbing disordered regions and engaging substrates, are arranged in a spiral staircase lining the translocation channel and are located approximately 30–40 Angstroms from its entrance in substrate-free proteasomes [8]. Substrates whose initiation regions are located internally must be much longer than terminally located ones, presumably because internal disordered regions must enter and exit the translocation channel before the flanking domains can be unfolded [54].
In vivo and in vitro studies using model substrates with different disordered regions demonstrated that nonpolar, hydrophobic, stiff, and complex amino acid compositions result in more effective degradation. Conversely, polar, acidic, highly flexible, and compositionally biased sequences are largely avoided [1,50,51,55] (Figure 2f). For instance, the E2 Cdc34 escapes proteasomal degradation despite ubiquitination because its disordered region, though sufficiently long, is acidic and has a biased amino acid composition [51,54]. The proteasome’s preferences for initiation region sequences are likely a reflection of the interaction between residues within the translocation channel and the initiation region. The internal surface of the translocation channel surrounding the pore-1 loops is largely negatively charged, possibly repelling acidic disordered regions from engagement [56]. An explanation for the preference of hydrophobic sequences could be that the pore-1 loops, which contain conserved Tyr residues, and the translocation channel, which is enriched in Tyr residues, interact more effectively with similarly hydrophobic residues [56]. Disordered regions enriched in residues with small side chains (i.e., Gly, Ser) are structurally flexible, which could prevent interactions between the pore loops and disordered region required for gripping the substrate and generating mechanical force necessary to unfold folded domains [8].
Location of the initiation region within a substrate also modulates degradation: it must be neither too far nor too close to a ubiquitin modification [49] (Figure 2g). Substrate binding sites on Rpn1, Rpn13, and Rpn10 and the pore-1 loops are at fairly well-defined locations on the proteasome particle and this likely restricts the arrangements of ubiquitin modification and disordered region within a substrate that result in simultaneous binding of ubiquitin and initiation region. Presumably both components of the degradation signal have to bind the proteasome at the same time for efficient degradation [5,7].
A recent study investigated how the proteasome avoids self-destruction even though many subunits have disordered regions. Proteasomal subunits Rpn10 and Rpn3 contain disordered regions that can reach the translocation channel yet escape degradation because these regions are compositionally biased [57]. Degradation of these subunits can be induced if the disordered region is replaced with an effective initiation region. On the other hand, Rpn2 and Rpn13 are far from the translocation channel preventing the proteasome from engaging their disordered regions. Considering the intrinsically disordered regions in some proteasomal subunits play significant regulatory roles through post-translational modification or protein interaction, sequence bias and proteasome architecture may have evolved to maintain proteasome stability and function [57,58].
While most degradation is ubiquitin-mediated, some proteins are degraded independent of ubiquitination. These ubiquitin-independent substrates are recognized by the proteasome directly at their initiation region or indirectly through binding partners serving as degradation adaptors. Well studied examples of such proteins include ornithine decarboxylase (ODC), the proteasome transcription factor Rpn4, and p53 [59].
Preparing substrates for the proteasome that lack initiation regions
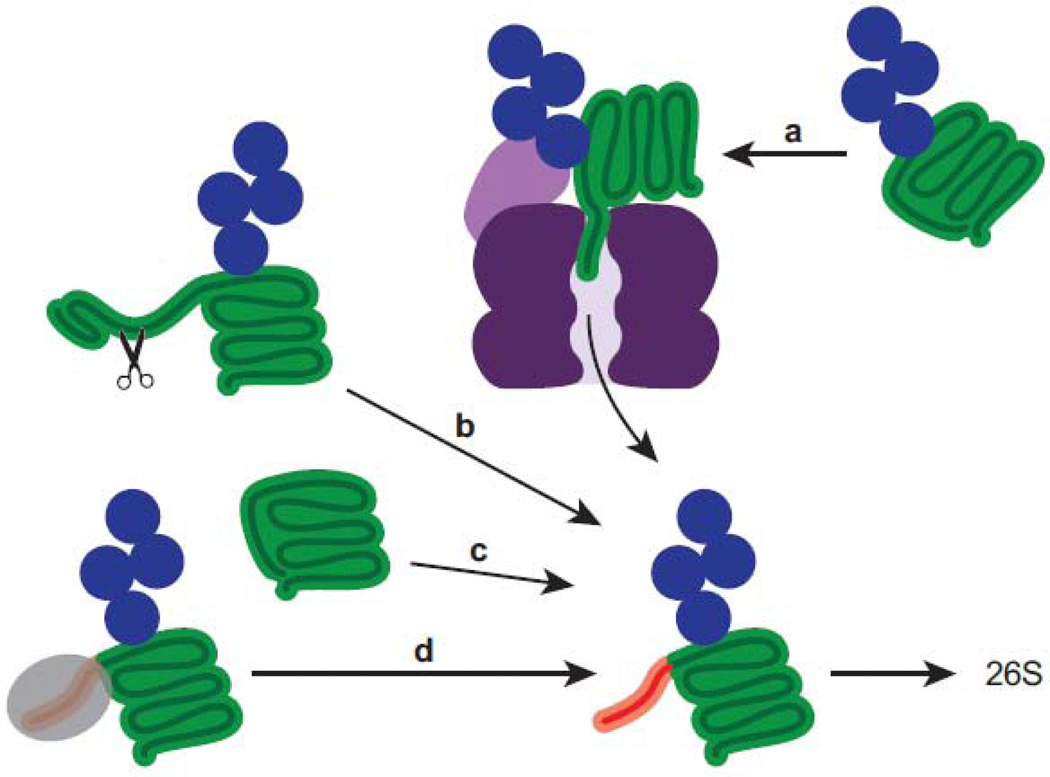
An initiation region is required for effective substrate engagement by the proteasomal ATPases, but some proteins lack an obvious unstructured region [60]. The proteasome must be able to degrade a wide array of proteins, including those lacking effective initiation regions. Various mechanisms have been identified that solve this problem. Cdc48 is an ATPase complex that unfolds ubiquitinated proteins and has been proposed to prepare substrates for the proteasome by generating initiation regions through unfolding [61,62] (Figure 3a). It was recently demonstrated using an in vitro reconstituted system that Cdc48 prepares substrates for the proteasome by partially unfolding well-folded ubiquitinated proteins that lack an initiation region [63]. These findings represent the first direct evidence of collaboration between Cdc48 and the proteasome in protein degradation. Site-specific ubiquitination can locally destabilize folded structures and likewise generate unstructured regions competent for engagement by the proteasome [60,64,65] (Figure 3c). Site-specific cleavage by proteases can reveal an initiation region that was previously inaccessible to the proteasome (Figure 3b), as was recently demonstrated [66]. Degradation of retinoblastoma protein (Rb), a tumor suppressor, is induced by the E7 protein of human papilloma virus (HPV) to promote cell cycle progression. Proteasomal degradation of Rb requires cleavage by the protease calpain-1, mediated by E7, which exposes a region competent for proteasome engagement [66]. The disordered region at the C-terminus of native Rb is negatively charged and not engaged by the proteasome. Initiation regions may also be inaccessible to the proteasome due to a binding partner, whose dissociation would then free the initiation region permitting proteasomal engagement (Figure 3d). For instance, yeast protein Mdy2 binds the proteasome through its UBL domain and can be degraded but is protected from degradation through binding of Get4 to the initiation region [44].
Figure 3: Substrates can be processed or modified to reveal initiation regions.
An unstructured region (red) is required for degradation since it allows the proteasome to physically engage the substrate and initiate degradation. Proteins that lack an initiation region can be processed or modified in order to reveal unstructured regions that the proteasome can efficiently engage. (a) Well-folded proteins can be processed by the unfoldase Cdc48 (cross section shown in shades of purple) to generate an initiation region. (b) Cleavage by proteases can reveal an accessible initiation region. (c) Site-specific ubiquitination may locally destabilize a protein thereby generating an initiation region. (d) Removal of a binding partner (grey) can liberate an accessible initiation region.
Conclusions and future directions
Work over the past several years has greatly expanded our knowledge of proteasome structure and function. Recent studies demonstrate that substrate recognition by the proteasome can be achieved by a variety of mechanisms including ubiquitin modification, cooperation between ubiquitin receptors, initiation region, and overall architecture of the proteasome. These findings suggest a synergistic relationship between ubiquitin and initiation region that confers specificity and a means of regulating the substrate recognition step of proteasomal degradation. However, it remains poorly characterized how ubiquitin and initiation region collaborate in a cellular environment to target natural proteins for degradation. Can a weak initiation region be compensated for by a strong ubiquitin modification and vice versa? Future studies will refine and reveal mechanistic and physiological processes of substrate recognition, leading to a deeper understanding of protein degradation by the UPS.
Acknowledgments
Funding Sources
This work was supported by R01GM124501 and R01GM135264 from NIGMS and grant F-1817 from the Welch Foundation.
Footnotes
Conflicts of interest statement
Andreas Matouschek is a paid consultant of Kymera Therapeutics.
AM is a paid consultant of Kymera Therapeutics. None of the other authors have conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- [1].Tomita T, Matouschek A, Substrate selection by the proteasome through initiation regions., Protein Science : A Publication of the Protein Society. 28 (2019) 1222–1232. 10.1002/pro.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Komander D, Rape M, The ubiquitin code., Annual Review of Biochemistry. 81 (2012) 203–229. 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- [3].Matyskiela ME, Lander GC, Martin A, Conformational switching of the 26S proteasome enables substrate degradation., Nature Structural & Molecular Biology. 20 (2013) 781–788. 10.1038/nsmb.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Unverdorben P, Beck F, Śledź P, Schweitzer A, Pfeifer G, Plitzko JM, Baumeister W, Förster F, Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome., Proceedings of the National Academy of Sciences of the United States of America. 111 (2014) 5544–5549. 10.1073/pnas.1403409111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].de la Peña AH, Goodall EA, Gates SN, Lander GC, Martin A, Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation., Science (New York, N.Y.). 301 (2018) eaav0725. 10.1126/science.aav0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Eisele MR, Reed RG, Rudack T, Schweitzer A, Beck F, Nagy I, Pfeifer G, Plitzko JM, Baumeister W, Tomko RJ, Sakata E, Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating., Cell Reports. 24 (2018) 1301–1315.e5. 10.1016/j.celrep.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dong Y, Zhang S, Wu Z, Li X, Wang WL, Zhu Y, Stoilova-McPhie S, Lu Y, Finley D, Mao Y, Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome, Nature. 565 (2018) 49–55. 10.1038/s41586-018-0736-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bard JAM, Goodall EA, Greene ER, Jonsson E, Dong KC, Martin A, Structure and Function of the 26S Proteasome., Annual Review of Biochemistry. 87 (2018) 697–724. 10.1146/annurev-biochem-062917-011931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wehmer M, Rudack T, Beck F, Aufderheide A, Pfeifer G, Plitzko JM, Förster F, Schulten K, Baumeister W, Sakata E, Structural insights into the functional cycle of the ATPase module of the 26S proteasome., Proceedings of the National Academy of Sciences of the United States of America. 114 (2017) 1305–1310. 10.1073/pnas.1621129114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Greene ER, Goodall EA, de la Peña AH, Matyskiela ME, Lander GC, Martin A, Specific lid-base contacts in the 26s proteasome control the conformational switching required for substrate degradation., ELife. 8 (2019) 8626. 10.7554/elife.49806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bard JAM, Bashore C, Dong KC, Martin A, The 26S Proteasome Utilizes a Kinetic Gateway to Prioritize Substrate Degradation., Cell. 177 (2019) 286–298.e15. 10.1016/j.cell.2019.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Guo Q, Lehmer C, Martínez-Sánchez A, Rudack T, Beck F, Hartmann H, Pérez-Berlanga M, Frottin F, Hipp MS, Hartl FU, Edbauer D, Baumeister W, Fernández-Busnadiego R, In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment., Cell. 172 (2018) 696–705.e12. 10.1016/j.cell.2017.12.030.**In this study neurons expressing aggregates of poly-GA, a mutant protein linked to ALS/FTD, were analyzed by cryo-electron tomography. The authors revealed that poly-GA aggregates form ribbon-like structures that heavily recruit proteasomes. They found many of the proteasomes interacting with the aggregates were functionally impaired, suggesting stalled degradation.
- [13].Albert S, Schaffer M, Beck F, Mosalaganti S, Asano S, Thomas HF, Plitzko JM, Beck M, Baumeister W, Engel BD, Proteasomes tether to two distinct sites at the nuclear pore complex., Proceedings of the National Academy of Sciences of the United States of America. 114 (2017) 13726–13731. 10.1073/pnas.1716305114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Asano S, Fukuda Y, Beck F, Aufderheide A, Förster F, Danev R, Baumeister W, Proteasomes. A molecular census of 26S proteasomes in intact neurons., Science (New York, N.Y.). 347 (2015) 439–442. 10.1126/science.1261197. [DOI] [PubMed] [Google Scholar]
- [15].Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik HD, Huber R, Structure of 20S proteasome from yeast at 2.4 A resolution., Nature. 386 (1997) 463–471. 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- [16].Tanaka K, The proteasome: Overview of structure and functions, Proc Jpn Acad Ser B Phys Biological Sci. 85 (2009) 12–36. 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D, A subcomplex of the proteasome regulatory particle required for ubiquitinconjugate degradation and related to the COP9-signalosome and eIF3., Cell. 94 (1998) 615–623. [DOI] [PubMed] [Google Scholar]
- [18].Glickman MH, Rubin DM, Fried VA, Finley D, The regulatory particle of the Saccharomyces cerevisiae proteasome., Molecular and Cellular Biology. 18 (1998) 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rabl J, Smith DM, Yu Y, Chang S-C, Goldberg AL, Cheng Y, Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases., Molecular Cell. 30 (2008) 360–368. 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sakata E, Bohn S, Mihalache O, Kiss P, Beck F, Nagy I, Nickell S, Tanaka K, Saeki Y, Förster F, Baumeister W, Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy., P Natl Acad Sci Usa. 109 (2012) 1479–84. 10.1073/pnas.1119394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Deveraux Q, Ustrell V, Pickart CM, Rechsteiner M, A 26 S protease subunit that binds ubiquitin conjugates., The Journal of Biological Chemistry. 269 (1994) 7059–7061. [PubMed] [Google Scholar]
- [22].Shi Y, Chen X, Elsasser S, Stocks BB, Tian G, Lee B, Shi Y, Zhang N, de Poot SAH, Tuebing F, Sun S, Vannoy J, Tarasov SG, Engen JR, Finley D, Walters KJ, Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome., Science (New York, N.Y.). 351 (2016) aad9421–aad9421. 10.1126/science.aad9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I, Proteasome subunit Rpn13 is a novel ubiquitin receptor, Nature. 453 (2008) 481–488. 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M, Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction., Nature. 453 (2008) 548–552. 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A, An unstructured initiation site is required for efficient proteasome-mediated degradation., Nature Structural & Molecular Biology. 11 (2004) 830–837. 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- [26].Worden EJ, Padovani C, Martin A, Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation., Nature Structural & Molecular Biology. 21 (2014) 220–227. 10.1038/nsmb.2771. [DOI] [PubMed] [Google Scholar]
- [27].Worden EJ, Dong KC, Martin A, An AAA Motor-Driven Mechanical Switch in Rpn11 Controls Deubiquitination at the 26S Proteasome., Molecular Cell. 67 (2017) 799–811.e8. 10.1016/j.molcel.2017.07.023. [DOI] [PubMed] [Google Scholar]
- [28].Thrower JS, Hoffman L, Rechsteiner M, Pickart CM, Recognition of the polyubiquitin proteolytic signal., The EMBO Journal. 19 (2000) 94–102. 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Martinez-Fonts K, Davis C, Tomita T, Elsasser S, Nager AR, Shi Y, Finley D, Matouschek A, The proteasome 19S cap and its ubiquitin receptors provide a versatile recognition platform for substrates., Nature Communications. 11 (2020) 477–16. 10.1038/s41467-019-13906-8.**The authors characterize degradation of model substrates with well-defined ubiquitin modifications by purified proteasome in which individual receptors have been mutated to abrogate ubiquitin binding. The study determines the three ubiquitin receptors of the proteasome enable recognition of substrates with different orientation and diverse ubiquitin chain topologies with some receptors functioning as the primary receptor for certain proteins.
- [30].Grice GL, Lobb IT, Weekes MP, Gygi SP, Antrobus R, Nathan JA, The Proteasome Distinguishes between Heterotypic and Homotypic Lysine-11-Linked Polyubiquitin Chains., Cell Reports. 12 (2015) 545–553. 10.1016/j.celrep.2015.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Saeki Y, Kudo T, Sone T, Kikuchi Y, Yokosawa H, Toh-e A, Tanaka K, Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome., The EMBO Journal. 28 (2009) 359–371. 10.1038/emboj.2008.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ohtake F, Tsuchiya H, Saeki Y, Tanaka K, K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains., P Natl Acad Sci Usa. 115 (2018) E1401–E1408. 10.1073/pnas.1716673115.*The authors characterize in detail an example of an endogenous protein whose proteasomal degradation is regulated by K48/K63 branched chains. A combination of cell-based and biochemical approaches revealed that attachment of K63 chains to tumor suppressor TXNIP by the E3 ligase ITCH triggers the assembly of K48/K63 branched chains by the E3 ligase UBR5, leading to its degradation by the proteasome.
- [33].Liu C, Liu W, Ye Y, Li W, Ufd2p synthesizes branched ubiquitin chains to promote the degradation of substrates modified with atypical chains., Nature Communications. 8 (2017) 14274. 10.1038/ncomms14274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Meyer H-J, Rape M, Enhanced protein degradation by branched ubiquitin chains., Cell. 157 (2014) 910–921. 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Boughton AJ, Krueger S, Fushman D, Branching via K11 and K48 Bestows Ubiquitin Chains with a Unique Interdomain Interface and Enhanced Affinity for Proteasomal Subunit Rpn1, Structure. 28 (2020) 29–43.e6. 10.1016/j.str.2019.10.008.**The authors used a combination of X-ray crystallography, NMR, and SANS to solve the structure of branched K11/K48 tri-ubiquitin and discovered a novel hydrophobic interface between distal ubiquitin molecules. They demonstrate that chain branching has a minimal effect on deubiquitination activity or binding to proteasome-associated UBL-UBA receptors. Instead, branching enhances affinity for the proteasomal ubiquitin receptor Rpn1.
- [36].Braten O, Livneh I, Ziv T, Admon A, Kehat I, Caspi LH, Gonen H, Bercovich B, Godzik A, Jahandideh S, Jaroszewski L, Sommer T, Kwon YT, Guharoy M, Tompa P, Ciechanover A, Numerous proteins with unique characteristics are degraded by the 26S proteasome following monoubiquitination., Proceedings of the National Academy of Sciences of the United States of America. 113 (2016) E4639–47. 10.1073/pnas.1608644113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sun H, Mali SM, Singh SK, Meledin R, Brik A, Kwon YT, Kravtsova-Ivantsiv Y, Bercovich B, Ciechanover A, Diverse fate of ubiquitin chain moieties: The proximal is degraded with the target, and the distal protects the proximal from removal and recycles, Proc National Acad Sci. 116 (2019) 7805–7812. 10.1073/pnas.1822148116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Dimova NV, Hathaway NA, Lee B, Kirkpatrick DS, Berkowitz ML, Gygi SP, Finley D, King RW, APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1., Nature Cell Biology. 14 (2012) 168–176. 10.1038/ncb2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chen X, Dorris Z, Shi D, Huang RK, Khant H, Fox T, de Val N, Williams D, Zhang P, Walters KJ, Cryo-EM Reveals Unanchored M1-Ubiquitin Chain Binding at hRpn11 of the 26S Proteasome., Struct Lond Engl 1993. (2020). 10.1016/j.str.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhang N, Wang Q, Ehlinger A, Randles L, Lary JW, Kang Y, Haririnia A, Storaska AJ, Cole JL, Fushman D, Walters KJ, Structure of the S5a:K48-Linked Diubiquitin Complex and Its Interactions with Rpn13, Molecular Cell. 35 (2009) 280–290. 10.1016/j.molcel.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Riedinger C, Boehringer J, Trempe J-F, Lowe ED, Brown NR, Gehring K, Noble MEM, Gordon C, Endicott JA, Structure of Rpn10 and its interactions with polyubiquitin chains and the proteasome subunit Rpn12., The Journal of Biological Chemistry. 285 (2010) 33992–34003. 10.1074/jbc.m110.134510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lu X, Ebelle DL, Matsuo H, Walters KJ, An Extended Conformation for K48 Ubiquitin Chains Revealed by the hRpn2:Rpn13:K48-Diubiquitin Structure, Structure. 28 (2020) 495–506.e3. 10.1016/j.str.2020.02.007.**The authors used NMR and X-ray crystallography to solve the structure of free K48-linked di-ubiquitin and of K48-linked di-ubiquitin bound to hRpn2 in association with hRpn13. The authors identified a previously undetermined extended conformation of K48-linked di-ubiquitin, which is selected for by hRpn2:hRpn13. They found the receptor and chain to interact dynamically and propose that the nature of these interactions aids in preparing substrates for deubiquitination and engagement.
- [43].Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Müller B, Feng MT, Tübing F, Dittmar GAG, Finley D, Proteasome subunit Rpn1 binds ubiquitin-like protein domains., Nature Cell Biology. 4 (2002) 725–730. 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- [44].Yu H, Kago G, Yellman CM, Matouschek A, Ubiquitin-like domains can target to the proteasome but proteolysis requires a disordered region, The EMBO Journal. 35 (2016) 1522–1536. 10.15252/embj.201593147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chen X, Randles L, Shi K, Tarasov SG, Aihara H, Walters KJ, Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome., Structure (London, England : 1993). 24 (2016) 1257–1270. 10.1016/j.str.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chen X, Ebelle DL, Wright BJ, Sridharan V, Hooper E, Walters KJ, Structure of hRpn10 Bound to UBQLN2 UBL Illustrates Basis for Complementarity between Shuttle Factors and Substrates at the Proteasome, J Mol Biol. 431 (2019) 939–955. 10.1016/j.jmb.2019.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Collins GA, Goldberg AL, Proteins containing ubiquitin-like (Ubl) domains not only bind to 26S proteasomes but also induce their activation., P Natl Acad Sci Usa. 117 (2020) 4664–4674. 10.1073/pnas.1915534117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Verma R, Oania R, Graumann J, Deshaies RJ, Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system., Cell. 118 (2004) 99–110. 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- [49].Inobe T, Fishbain S, Prakash S, Matouschek A, Defining the geometry of the two-component proteasome degron., Nature Chemical Biology. 7 (2011) 161–167. 10.1038/nchembio.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yu H, Gautam AKS, Wilmington SR, Wylie D, Martinez-Fonts K, Kago G, Warburton M, Chavali S, Inobe T, Finkelstein IJ, Babu MM, Matouschek A, Conserved Sequence Preferences Contribute to Substrate Recognition by the Proteasome., The Journal of Biological Chemistry. 291 (2016) 14526–14539. 10.1074/jbc.m116.727578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Fishbain S, Inobe T, Israeli E, Chavali S, Yu H, Kago G, Babu MM, Matouschek A, Sequence composition of disordered regions fine-tunes protein half-life., Nature Structural & Molecular Biology. 22 (2015) 214–221. 10.1038/nsmb.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].van der Lee R, Lang B, Kruse K, Gsponer J, de Groot NS, Huynen MA, Matouschek A, Fuxreiter M, Babu MM, Intrinsically disordered segments affect protein half-life in the cell and during evolution., Cell Reports. 8 (2014) 1832–1844. 10.1016/j.celrep.2014.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Takeuchi J, Chen H, Coffino P, Proteasome substrate degradation requires association plus extended peptide, Embo J. 26 (2006) 123–131. 10.1038/sj.emboj.7601476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Fishbain S, Prakash S, Herrig A, Elsasser S, Matouschek A, Rad23 escapes degradation because it lacks a proteasome initiation region, Nature Communications. 2 (2011) 192. 10.1038/ncomms1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kraut DA, Israeli E, Schrader EK, Patil A, Nakai K, Nanavati D, Inobe T, Matouschek A, Sequence- and species-dependence of proteasomal processivity., ACS Chemical Biology. 7 (2012) 1444–1453. 10.1021/cb3001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Chen S, Wu J, Lu Y, Ma Y-B, Lee B, Yu Z, Ouyang Q, Finley DJ, Kirschner MW, Mao Y, Structural basis for dynamic regulation of the human 26S proteasome, Proceedings of the National Academy of Sciences. (2016) 201614614. 10.1073/pnas.1614614113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Gautam AKS, Yu H, Yellman C, Elcock A, Matouschek A, Design principles that protect the proteasome from self-destruction, (2020). 10.21203/rs.3.rs-38559/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kors S, Geijtenbeek K, Reits E, Schipper-Krom S, Regulation of Proteasome Activity by (Post-)transcriptional Mechanisms., Frontiers Mol Biosci. 6 (2019) 48. 10.3389/fmolb.2019.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Erales J, Coffino P, Ubiquitin-independent proteasomal degradation., Biochim Biophys Acta. 1843 (2013) 216–21. 10.1016/j.bbamcr.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hagai T, Azia A, Tóth-Petróczy Á, Levy Y, Intrinsic Disorder in Ubiquitination Substrates, J Mol Biol. 412 (2011) 319–324. 10.1016/j.jmb.2011.07.024. [DOI] [PubMed] [Google Scholar]
- [61].Twomey EC, Ji Z, Wales TE, Bodnar NO, Ficarro SB, Marto JA, Engen JR, Rapoport TA, Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding., Sci New York N Y. 365 (2019) eaax1033. 10.1126/science.aax1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Barthelme D, Sauer RT, Origin and Functional Evolution of the Cdc48/p97/VCP AAA+ Protein Unfolding and Remodeling Machine., J Mol Biol. 428 (2015) 1861–9. 10.1016/j.jmb.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Olszewski MM, Williams C, Dong KC, Martin A, The Cdc48 unfoldase prepares well-folded protein substrates for degradation by the 26S proteasome., Commun Biology. 2 (2019) 29. 10.1038/s42003-019-0283-z.*The authors demonstrate directly that Cdc48 prepares substrates for proteasomal degradation by unfolding tightly folded proteins to generate disordered regions for engagement by the proteasome. Using an in vitro reconstituted system, they show that Cdc48 and the proteasome collaborate to degrade ubiquitinated proteins lacking an initiation region. The authors highlight the importance of Cdc48 in regulating substrate recognition by the proteasome.
- [64].Carroll EC, Greene ER, Martin A, Marqusee S, Site-specific ubiquitination affects protein energetics and proteasomal degradation, Nat Chem Biol. 16 (2020) 866–875. 10.1038/s41589-020-0556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hagai T, Levy Y, Ubiquitin not only serves as a tag but also assists degradation by inducing protein unfolding., Proceedings of the National Academy of Sciences of the United States of America. 107 (2010) 2001–2006. 10.1073/pnas.0912335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Tomita T, Huibregtse JM, Matouschek A, A masked initiation region in retinoblastoma protein regulates its proteasomal degradation., Nature Communications. 11 (2020) 2019–8. 10.1038/s41467-020-16003-3.*Using a biochemical and cell-based approach, the authors show that the proteasome cannot efficiently degrade full-length retinoblastoma protein (Rb). Instead, Rb cleavage by calpain-1 is necessary to first expose an initiation region suitable for proteasome engagement. Their findings describe a physiological mechanism used to control proteasomal substrate recognition through regulating access to initiation regions.