Abstract
RING E3s comprise the largest family of ubiquitin (UB) and ubiquitin-like protein (UBL) ligases. RING E3s typically promote UB or UBL transfer from the active site of an associated E2 enzyme to a distally-recruited substrate. Many RING E3s – including the cullin-RING ligase family - are multifunctional, interacting with various E2s (or other E3s) to target distinct proteins, transfer different UBLs, or to initially modify substrates with UB or subsequently elongate UB chains. Here we consider recent structures of cullin-RING ligases, and their partner E2 enzymes, representing ligation reactions. The studies collectively reveal multimodal mechanisms – interactions between ancillary E2 or E3 domains, post-translational modifications, or auxiliary binding partners - directing cullin-RING E3-E2 enzyme active sites to modify their specific targets.
Keywords: Ubiquitin, NEDD8, cullin-RING ligase, RING E3
Introduction
Virtually all eukaryotic intracellular processes depend on regulation by post-translational modifications with ubiquitin (UB) and ubiquitin-like proteins (UBLs) such as NEDD8 or SUMO (referred to here in general terms as UB/UBL)[1,2]. The isopeptide linkage of a UB/UBL to a targeted lysine(s) can alter the fate of a modified protein in myriad ways, ranging from determining protein interaction partners to directing proteasomal or autophagic degradation[3]. Regulation is achieved by cascades of E1 activating, E2 conjugating, and E3 ligase enzymes. E3 ligases, often together with partner E2 enzymes, are responsible for matching specific protein targets with particular UB/UBL modifications[1]. Substrates are recruited to E3 substrate receptor domains, while UB and UBLs are transferred from distinctive active sites determined by the class of E3. The vast majority of E3 ligases – estimated at 600 in humans – are classified by a hallmark RING domain.
RING domains themselves lack ubiquitin transferase activity. Rather, RING domains partner with other ubiquitin carrying enzymes, typically E2s, where the active site cysteine is linked to the C-terminus of UB or a UBL by a covalent but reactive thioester bond. Such E2~UB/UBL intermediates are relatively stable on their own (here “~” refers to reactive but covalent thioester linkage between enzyme and UB/UBL, or to stabilized mimics of such bonds). NMR studies provided an explanation: the E2 and UB/UBL are flexibly tethered about the thioester bond between them. The capacity for E2 and UB/UBL to adopt infinite different orientations relative to each other reduces propensity for proper alignment of the E2~UB/UBL active site[4]. However, E3 ligase RING domains typically corral their partner E2~UB/UBL intermediates into a distinct architecture referred to as the ‘closed conformation[4–15] (Figure 1a). The RING domain binds both E2 and its linked UB/UBL such that the otherwise flexible UB/UBL C-terminal tail folds back, wrapping around and packing against the E2 domain, supported by contacts between the UB/UBL Ile44-centered hydrophobic patch and E2’s long central helix. Typically, a RING linchpin residue inserted into the interface between the E2 and UB stabilizes the 3-way interaction. An E3 “non-RING priming element” may further buttress this activated RING-E2~UB/UBL conformation[11,16]. As such, RING domains stimulate UB/UBL transfer from an E2 to a suitably placed nucleophile, such as lysine in a target distally recruited via the E3’s substrate receptor domain. In addition, many RING E3s are multifunctional (Figure 1b, c), interacting with various E2s (or other E3s) to modify distinct targets, transfer UB or various UBLs, and/or separately initiate and elongate UB chain formation.
Figure 1. Context determines specificity of structurally similar RING-E2~UB/UBL modules.
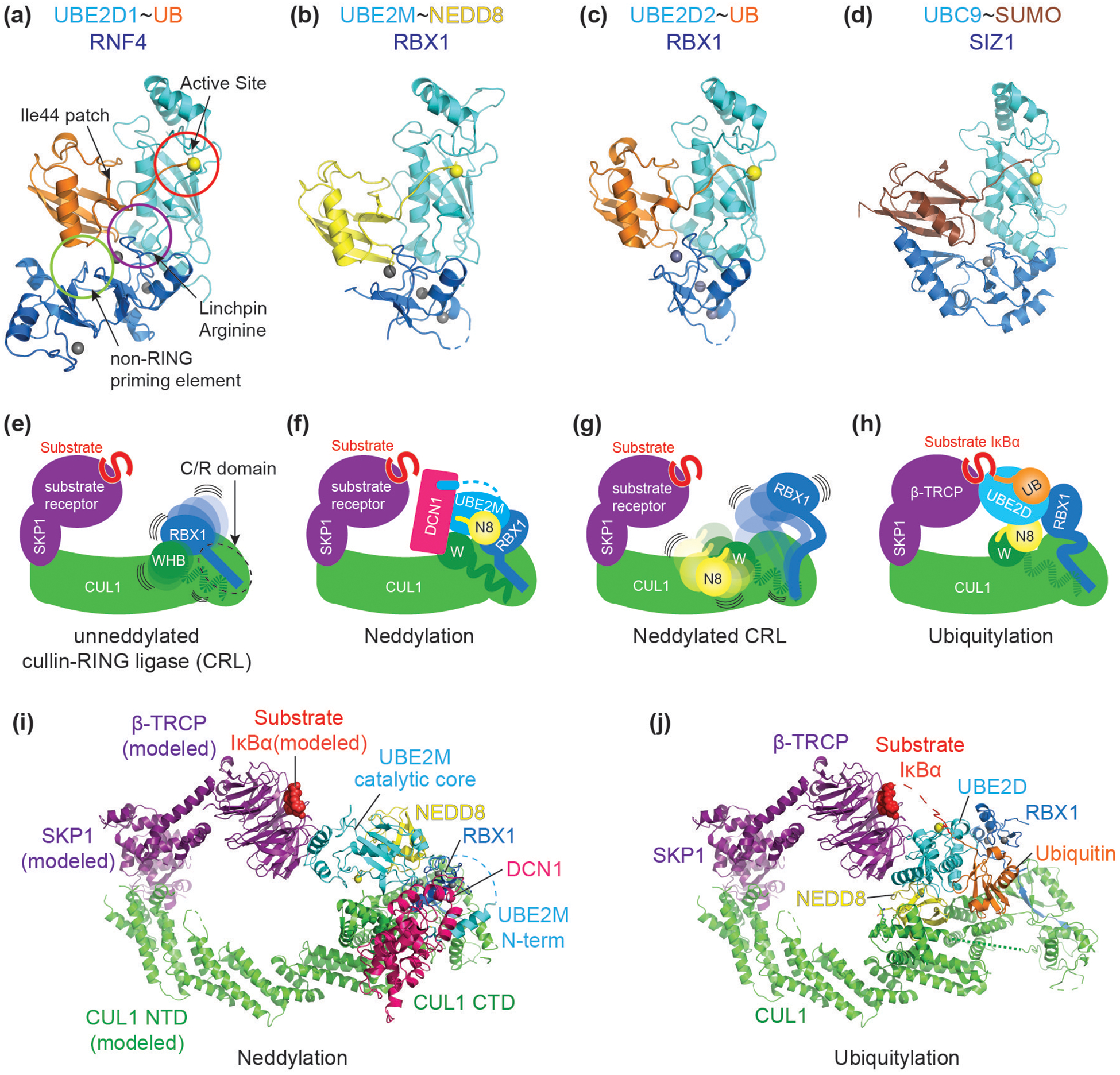
(a) A common closed conformation across RING E3-E2~UB/UBL assemblies (“~” refers to thioester linkage between E2 catalytic cysteine and UB/UBL C-terminus). In the closed conformation, ubiquitin’s Ile44 hydrophobic interacts with the E2. This orientation is reinforced through interaction with a RING E3 linchpin (often an arginine, purple circle) and non-RING priming element (light green circle). The closed conformation stabilizes the position of the UB/UBL C-terminal tail, thereby activating the E2~ubiquitin active site (red circle) for catalysis. Highlighted in yellow sphere is the catalytic residue of E2 throughout Figure 1. Representative example shown is RNF4-UBE2D1~UB (PDB ID: 4AP4).
(b) Cullin associated RBX1’s RING domain activates multiple E2~UBL conjugates. RBX1 promotes closed conformation of UBE2M~NEDD8 (PDB ID: 4P5O).
(c) RBX1’s RING domain interacts with and promotes the closed conformation of UBE2D~UB (PDB ID: 6TTU).
(d) SIZ1 promotes UBC9~SUMO closed conformation (PDB ID: 5JNE).
(e) Schematic of a unneddylated cullin-RING ligase (CRL) bound to a SKP1-Fbox-substrate complex with its N-terminus on the left side (EMDB ID: 10582). On the right side, RBX1’s catalytic RING domain is flexibly tethered to RBX1’s N-terminal strand, embedded within the cullin forming a “C/R” domain (dotted circle). CUL1’s WHB domain is flexibly tethered to its connecting helix-29.
(f) Schematic of a cullin-RING ligase neddylation intermediate (PDB ID: 4P5O). RBX1 RING facilitates closed conformation of UBE2M~NEDD8, and with the participation of a co-E3 ligase DCN1, promotes transfer of NEDD8 to CUL1’s Lys720 located within the WHB domain.
(g) Schematic of a neddylated cullin-RING ligase (EMDB ID: 10583). Once NEDD8 is isopeptide linked to CUL1’s WHB domain, RBX1’s RING domain is further liberated. NEDD8, the linked WHB domain, and RBX1’s RING domain sample multiple orientations with enhanced flexibility.
(h) Schematic of a cullin-RING ligase intermediate during ubiquitylation (PDB ID: 6TTU). RBX1 promotes closed conformation of UBE2D~UB.
(i) Crystal structure of CUL1-RBX1-DCN1-UBE2M-NEDD8 representing an intermediate for neddylation (PDB ID: 4P5O. CUL1’s N-terminal domain (NTD) and SKP1-β-TRCP-IκBα are modeled (from PDB ID: 6TTU).
(j) Cryo-EM structure of CUL1-RBX1-SKP1-β-TRCP-IκBα-UBE2D-Ubiquitin representing an intermediate for ubiquitylation (PDB ID: 6TTU).
Over the past two decades, many structural studies have revealed how E3 ligase substrate receptor domains uniquely recruit their specific protein targets for ubiquitylation[for examples see 17–21]. There are also many structures of E3 RING domains bound to stable mimics of their E2~UB/UBL partners. These latter structures are similar for different E3s, E2s, and UBLs (Figure 1a–d). Nonetheless, few high-resolution structures have shown how RING-E2~UB/UBL modules target specific sites within distally recruited targets. Structures of two distinct such intermediates of cullin-RING E3 ligases (CRLs) offer insights into how a single RING E3 partners with different E2~UB/UBL intermediates to modify distinct targets. CRLs account for nearly half of all E3 ligases, and are responsible for around 20% of all proteasomal degradation. Therefore, there is considerable interest in understanding the underlying structural and biochemical mechanisms that drive CRL-mediated ubiquitylation.
CRLs are multiprotein complexes, with overall similar modular architectures first defined for the “CRL1” (for CUL1-RBX1), aka “SCF” (SKP1-CUL1-F-box protein), family[22] (Figure 1e). The amino-terminal end of a banana-shaped cullin protein (CUL1 in a CRL1 complex) assembles with a swappable substrate receptor (a SKP1 adaptor/F-box protein receptor complex for a CRL1 [23]). In humans, nearly 70 F-box proteins are thought to serve as interchangeable receptors recruiting distinct cohorts of substrates. The C-terminal end of CUL1-RBX1 – the cullin-RING core -contains multiple domains. CUL1 and RBX1 come together in a structurally stable “C/R” domain, which continues into the dynamically connected C-terminal domains of CUL1 (the so-called cullin “WHB” domain) and RBX1’s E3 ligase RING domain (Figure 1e). RBX1’s RING domain functions with at least five distinct families of E2s (UBE2M, UBE2F, UBE2D, UBE2G, and UBE2R) and one E3 (ARIH1)[24–26]. First, RBX1 employs UBE2M, or in some cases UBE2F, to ligate the UBL NEDD8 to a specific lysine (720 in CUL1). This post-translational modification in which NEDD8 modifies the cullin, termed neddylation, has been shown to regulate CRL assembly in vivo[27], and to biochemically enhance CRL-catalyzed ubiquitylation reactions[28]. Neddylated CRL1 complexes employ the other abovementioned partner E2 and E3 enzymes to transfer UB to recruited substrates or to generate polyUB chains.
Here we review the structures representing RBX1 RING-catalyzed transfer of NEDD8 from UBE2M to CUL1[12] (Figure 1f,i), and in the context of a neddylated CRL1 complex, UB transfer from UBE2D to an F-box protein-bound substrate[29] (Figure 1h,j). We also highlight other recent structures showing how E2 domains N- or C-terminal of the catalytic core can also influence ubiquitylation reactions by CRL or CRL-like E3s[30,31]. Together, the studies have defined principles governing RING E2~UB/UBL targeting specificity, through conformational changes choreographed by auxiliary proteins, ancillary domains, and substrate-assisted catalysis.
Multimodal allosteric activation of RING-E2~NEDD8 for cullin neddylation
Neddylation had been known to involve RBX1’s RING domain binding and activating UBE2M~NEDD8, such that the active site is aligned with CUL1’s Lys720 in its WHB domain. But it was not possible to model the reaction because docking RING-E2~UB structures (as proxies for E2~NEDD8) onto early structures of CUL1-RBX1 showed massive gaps between where NEDD8 starts on the E2 and where it becomes linked to CUL1. The keys to capturing a structure representing the transition state were mutationally making a stabilized isosteric oxyester-bonded mimic of the UBE2M~NEDD8 intermediate, substituting CUL1’s acceptor Lys720 to arginine, and discovering that a co-E3, DCN1, accelerates the reaction with UBE2M that has been co-translationally modified by N-terminal acetylation[32,33].
The structure of the stabilized DCN1-CUL1-RBX1-UBE2M~NEDD8 complex showed how a RING-activated E2~UB/UBL active site is coordinated with a target lysine through multiple complementary mechanisms[12]. (1) The substrate, CUL1, is “recruited” by the sheer existence of the C/R domain constitutive to all CRL complexes, which flexibly tethers both the CUL1 WHB domain harboring the neddylation site and RBX1’s RING domain. (2) RBX1’s RING domain binds and activates UBE2M~NEDD8, stabilizing the closed conformation through a distinctive non-canonical linchpin, (Figure 2a) and promoting NEDD8’s Ile44 hydrophobic patch to contact the central helix of UBE2M (Figure 2b). This specific conformation locks the NEDD8 C-terminal tail, resulting in an RBX1-UBE2M~NEDD8 unit ready to search for and modify the target lysine. (3) The E2 UBE2M is recruited not only by the RING, but also by an auxiliary interaction with DCN1 via a ‘Dual E3’ mechanism[32]. The acetylated N-terminus of UBE2M is embraced by a DCN1 hydrophobic pocket(Figure 2c)[33]. DCN1 also binds CUL1’s WHB domain distal from the neddylation site. The multisite interactions avidly stabilize the complex between CUL1-RBX1 and UBE2M~NEDD8, and restrain the relative orientations of the flexibly tethered RBX1 RING-UBE2M~NEDD8 unit and the targeted CUL1 WHB domain. (4) The orientation of RBX1’s RING is further guided by specific residues of the UBL. When linked to UBE2M, NEDD8 interacts with RBX1 residues in the flexible linker between the C/R and RING domains. Here, RBX1’s own linker serves as the non-RING priming element for the closed UBE2M~NEDD8 configuration, while NEDD8 reciprocally steers the RING-UBE2M~NEDD8 unit through alignment of two negatively charged side-chains (glutamates in NEDD8) around a tryptophan in RBX1 (Figure 2d). (5) The substrate, CUL1’s WHB domain, also assists catalysis in three ways. First, there is shape complementarity between the activated UBE2M~NEDD8 intermediate and targeted surface of CUL1’s well-folded, globular WHB domain (Figure 2f). Second, the acceptor lysine is presented with accuracy, guided by neighboring residues including CUL1’s Tyr774 and its C-terminus (Figure 2g). Proper presentation of the CUL1’s Lys720 not only specifies it as the target, but also enhances intrinsic reactivity of UBE2M~NEDD8 independently of NEDD8 transfer to CUL1. Altogether, these components and their associated conformational rearrangements (Figure 2e, h) contribute to create a highly specific architecture where all factors work together to optimally orient the RBX1-UBE2M~NEDD8 towards its target lysine in CUL1.
Figure 2. Structure of CUL1-RBX1 intermediate during neddylation.
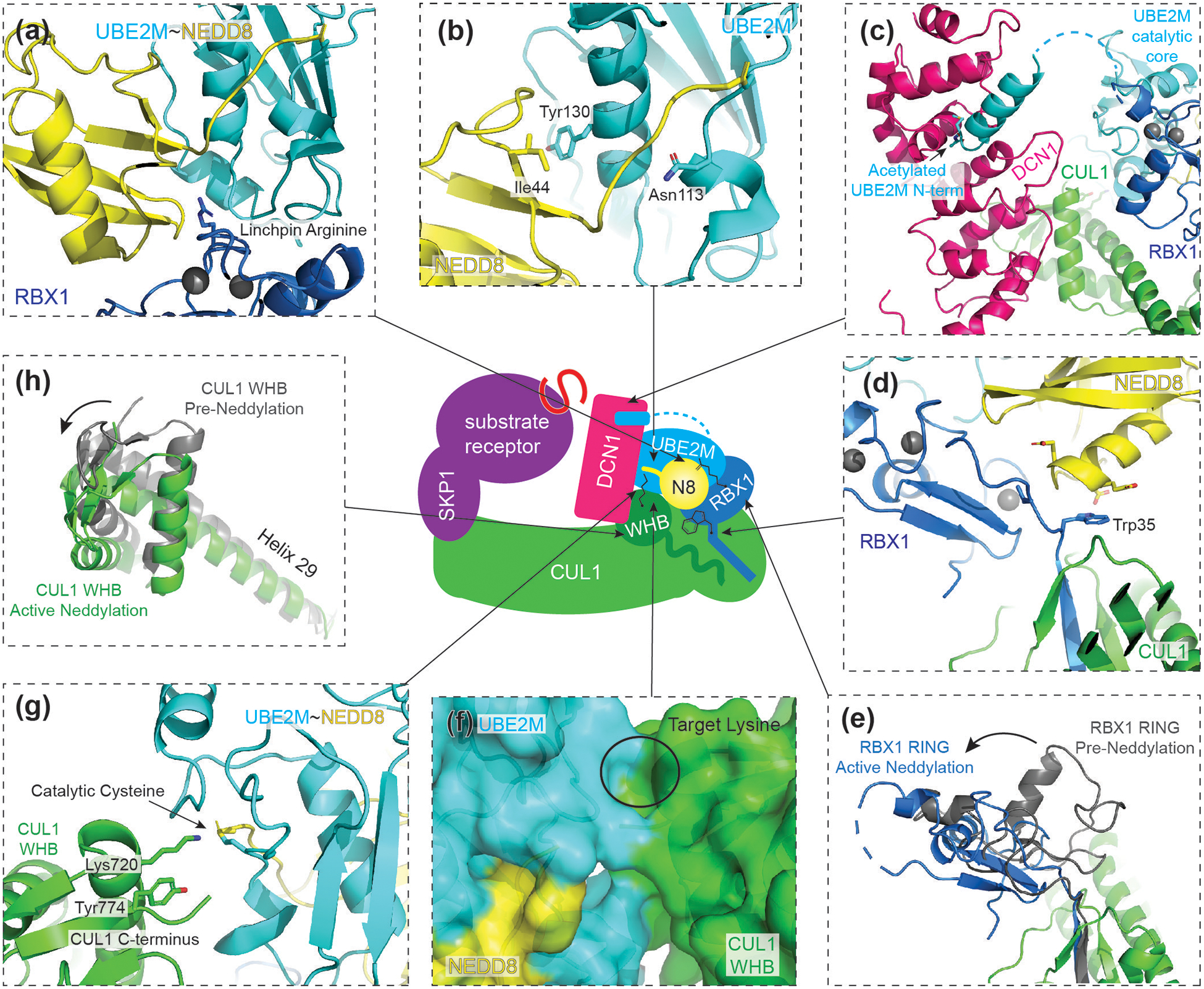
Central cartoon represents the CUL1 neddylation intermediate structure (PDB ID: 4P5O), with arrows indicating locations of structural details.
(a) RBX1 displays a linchpin arginine in a non-conical location, which promotes closing of the UBE2M~NEDD8 conjugate.
(b) When activated, NEDD8’s Ile44 hydrophobic patch interacts with UBE2M’s central helix. The C-terminal tail of NEDD8 is locked and primed for catalysis by Asn113 of UBE2M.
(c) UBE2M~NEDD8 recruitment to CRL is facilitated by both RING E3 RBX1 and co-E3 DCN1. UBE2M’s acetylated N-terminus engages a hydrophobic pocket of DCN1 while its catalytic domain interacts with the RBX1 RING domain.
(d) Unique residues of NEDD8 orient RBX1. NEDD8 steers UBE2M~NEDD8 by aligning two charged residues unique to NEDD8 to RBX1’s Trp35. Other residues in between RBX1’s cullin-binding strand and catalytic RING domain also function as non-RING priming elements.
(e) Comparison of RBX1’s RING position before (grey) and during active neddylation (blue). During neddylation, RBX1 is reoriented, positioning UBE2M~NEDD8 for its active site to align with CUL1’s target lysine.
(f) CUL1’s WHB domain and UBE2M present complementary surfaces to buttress the active conformation.
(g) CUL1’s acceptor Lys720 is guided by neighboring residues of CUL1, including Tyr774 and CUL1’s C-terminus. The acceptor lysine itself intrinsically activates UBE2M~NEDD8.
(h) Comparison of CUL1’s WHB domain before (grey) and during active neddylation (green). CUL1’s WHB domain and its connecting helix 29 translocate during neddylation.
Multimodal allosteric activation of RING-E2~NEDD8 for substrate ubiquitylation
Once CUL1’s Lys720 is modified by NEDD8, UBE2M disengages and the NEDD8 modified CUL1-RBX1 acquires further flexibility and capabilities (Figure 1g)[34]. The RING domain and CUL1 WHB domain and its linked NEDD8 adopt multiple conformations, which crosslinking data suggest differ from those preferred by an unneddylated CRL[35]. Even with this in mind, it had not been possible to accurately model the reaction whereby UB is transferred from the RBX1 RING-bound E2 UBE2D to a substrate recruited to an F-box protein. All prior models based on crystal structures showed a ~50Å gap between the E2 active site and the recruited substrate (Figure 3).
Figure 3. Structure of neddylated CRL1 intermediate during ubiquitylation.
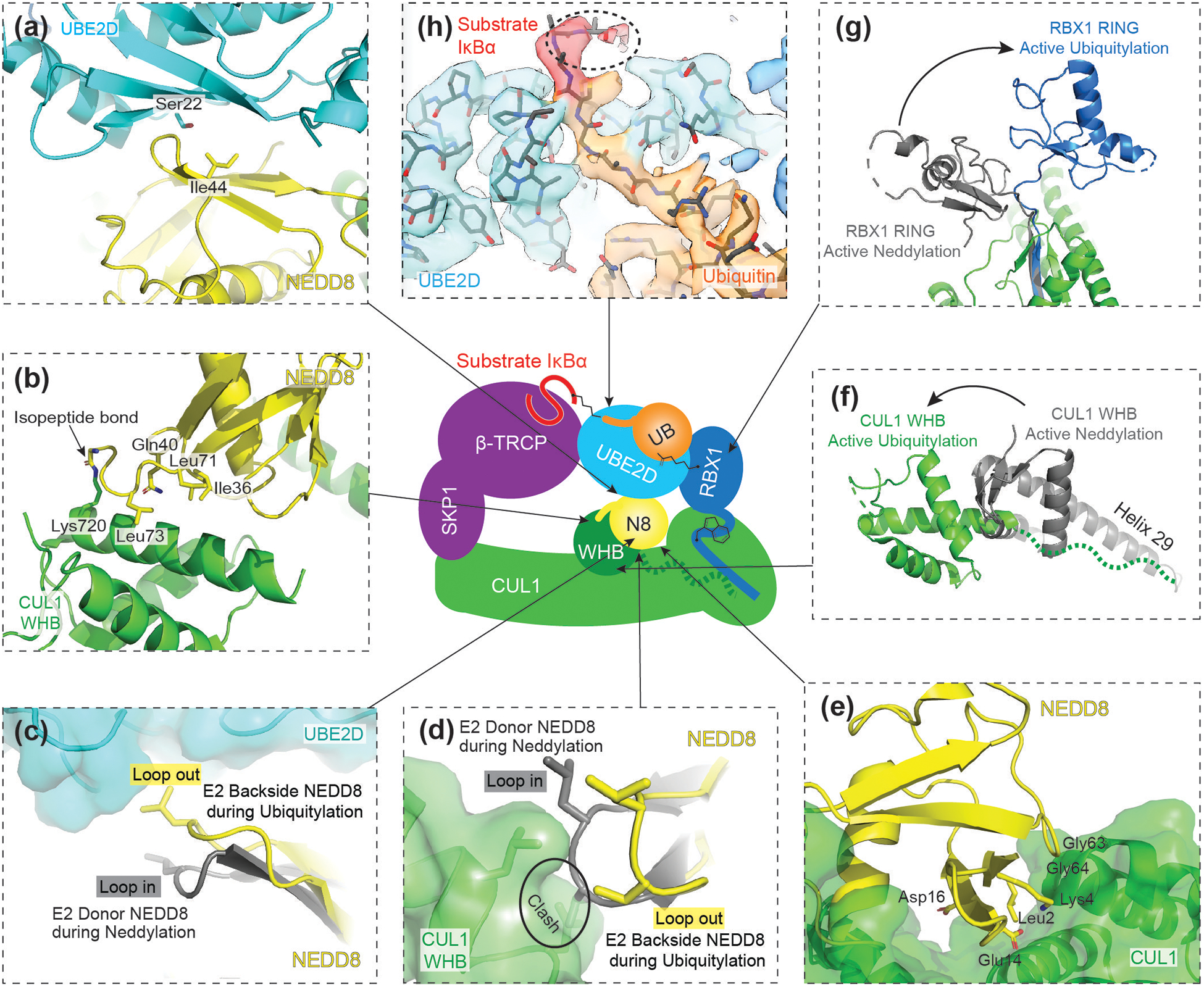
Central cartoon represents neddylated CRL1β-TRCP ubiquitylation intermediate structure, where arrows indicate locations of structural details.
(a) NEDD8’s Ile44 hydrophobic patch interacts with UBE2D’s backside, opposite of the active site, during ubiquitylation. This interaction encompasses several residues of UBE2D’s backside including Ser22.
(b) During ubiquitylation, NEDD8 and its isopeptide-linked CUL1 WHB interact via an interface that involves NEDD8’s Ile36 hydrophobic patch.
(c) To bind the backside of UBE2D, NEDD8 adopts a “loop-out” (shown in yellow) conformation. The “loop-in” (shown in grey) conformation of NEDD8, which is typically observed in the E2 active site bound donor position, is incompatible with UBE2D backside binding.
(d) The “loop-out” (yellow) conformation of NEDD8 is also required to interact with the CUL1 WHB domain.
(e) Residues unique to NEDD8 make direct contacts with CUL1.
(f) Comparison of CUL1’s WHB domain position during neddylation (grey) and ubiquitylation (green). CUL1’s WHB domain translocates to a position beyond the length of helix requiring remodeling to accommodate the location of the WHB linked NEDD8.
(g) Comparison of RBX1’s RING domain during active neddylation (grey) and active ubiquitylation (blue). During ubiquitylation, the RING domain adopts a unique orientation accommodating the position of UBE2D~UB.
(h) Cryo-EM density of UBE2D~UB active site. The density for the substrate IκBα is shown in red. Highlighted in dotted circle is the density of backbone atoms of IκBα, showing how a target lysine could potentially be guided by neighboring residues of the substrate.
This conundrum was resolved and the gap closed in a recent cryo EM structure representing ubiquitylation of a substrate (IκBα) recruited to the adaptor/F-box protein receptor complex SKP1/β-TRCP[29]. Compared to the neddylation intermediate, the RBX1 RING-E2~UB/UBL intermediate is strikingly rearranged, with UBE2D~UB oriented towards its β-TRCP-bound substrate. For this ubiquitylation, NEDD8 steers the flexibly tethered RBX1 RING-E2~UB unit, which resembles prior RING-E2~UB/UBL structures including that with UBE2M~NEDD8. However, NEDD8 is essentially encircled, with three NEDD8 surfaces mediating interactions that contribute to orienting and activating UBE2D~UB for substrate ubiquitylation. First, NEDD8’s Ile44-centered hydrophobic patch interacts with the ‘backside’ of UBE2D, opposite the active site (Figure 3a). Although numerous E2s interact with partner proteins (including UB) through backside interactions[36–39], it was questionable if NEDD8 could participate in homologous interactions – based on prior CRL1 structures, it was impossible to make a model where NEDD8 could simultaneously be linked to CUL1’s Lys720 and bound to UBE2D’s backside. However, the structure representing substrate ubiquitylation shows distinct rearrangement of both CUL1’s WHB (Figure 3f) and RBX1’s RING (Figure 3g) domains. The second interface involves CUL1’s WHB domain and the Ile36 hydrophobic patch of its isopeptide-linked NEDD8 (Figure 3b). Interestingly, these two interactions both depend on NEDD8 adopting a specific conformation termed ‘Loop-out’, which differs from the ‘Loop-in’ conformation required for the neddylation reaction (Figure 3c, d). Thus, it seems that NEDD8 interactions with UBE2D’s backside, and with its linked CUL1 WHB domain would be mutually stabilizing in part because either interface would sculpt NEDD8 to favor the other. Finally, other residues unique to NEDD8 make direct contact with CUL1 (Figure 3e), and swapping these residues for those in UB severely hampered ubiquitylation activity, providing further insight as to why NEDD8 uniquely activates CRL ubiquitylation.
Altogether, NEDD8 is spatially restricted by the three interfaces which coordinately orient the RING-UBE2D~UB assembly. In order to accommodate the location of NEDD8, the CUL1 WHB domain projects further than would be possible for a helix connecting to the C/R domain (Figure 3f). Cryo-EM data suggests this helix is remodeled into a dynamic loop to accommodate the repositioning of CUL1’s WHB domain for its linked NEDD8 to orchestrate ubiquitylation. In addition, the structure of the active site hints that the substrate could potentially guide the target lysine for ubiquitylation, much like during neddylation (Figure 3h). Despite differing from UBE2M’s interface with its well-folded substrate, UBE2D’s active site embraces its intrinsically disordered substrate in such a way that the peptide backbone is positioned much like the cullin residues that guide the targeted lysine.
E2 elements allosterically regulating other CRL-catalyzed ubiquitylation reactions
The drivers of conformational regulation of UB/UBL transfer are not limited to the E3, UBL, and substrate. E2 conjugating enzymes also participate in interactions steering their own activities. Such interactions are often mediated by structural appendages beyond the catalytic core domain conserved across E2s. Two recent examples of this are for the E2s UBE2R and UBE2S[30,31]. Neddylated CRL1s employ E2s in the UBE2R family to build polyUB chains, through transfer of a UB from UBE2R’s catalytic cysteine to Lys48 on a UB previously linked to an F-box protein-bound substrate. This polyubiquitylation reaction was known to depend on the UBE2R~UB intermediate forming the closed conformation even without binding to RBX1’s RING domain. Yet it was unclear how this conformation was specified. A recent crystal structure showed a role for residues in a C-terminal extension beyond UBE2R’s catalytic core domain[30]. C-terminal extension elements traverse the backside of UBE2R, and ultimately serve a linchpin-like role by inserting between the E2 core domain and UB to promote a closed conformation for UBE2R~UB (Figure 4a, b). It seems likely that in the context of a neddylated CRL, RBX1’s RING domain would further buttress the UBE2R~UB assembly, and potentially promote remodeling of UBE2R loop insertions to promote polyubiquitylation. Future studies will be required to visualize other elements explaining the incredible speed by which UBE2R forms polyUB chains on CRL substrates.
Figure 4.
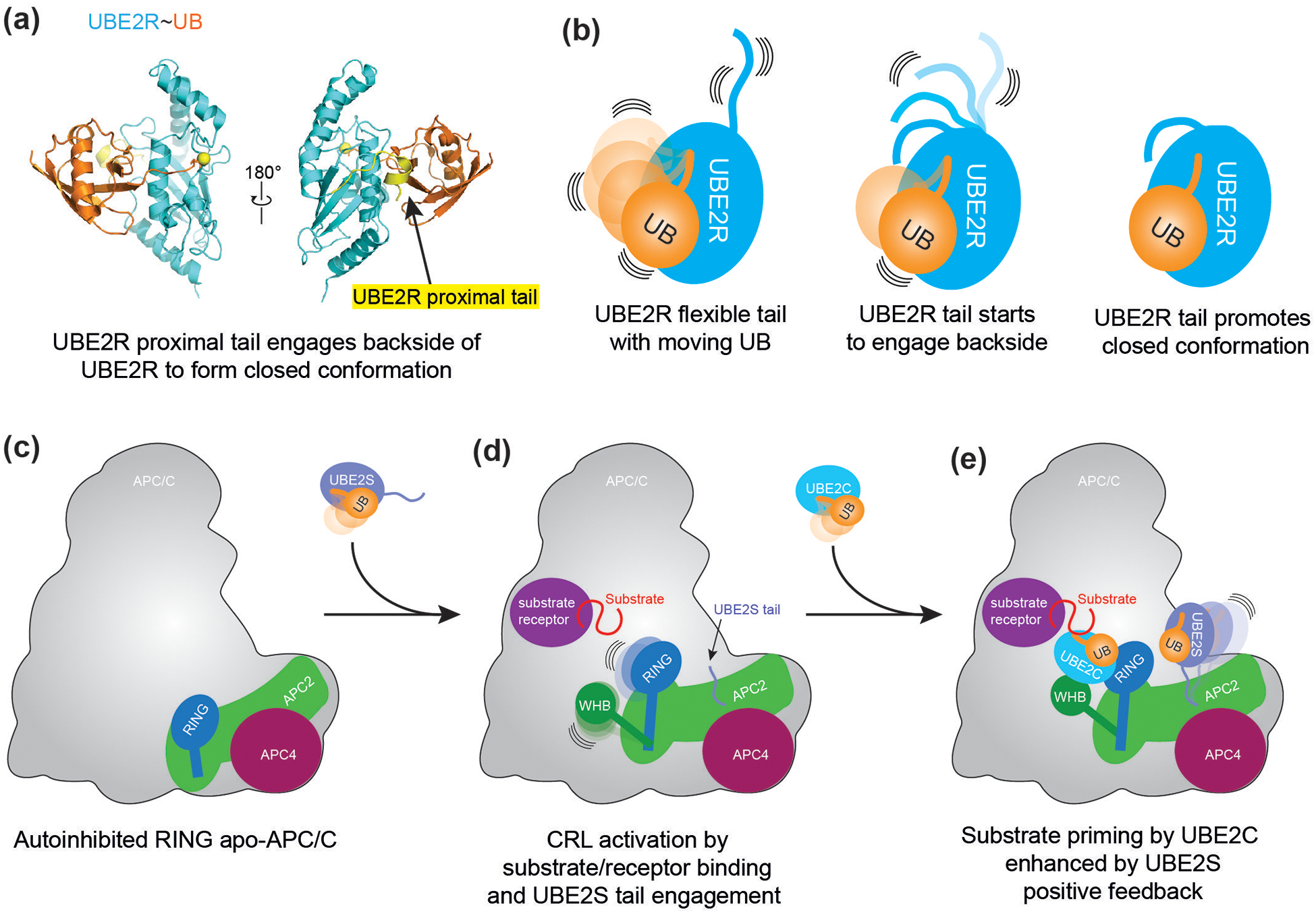
(a) Crystal structure of UBE2R~UB. Highlighted in yellow are residues of UBE2R’s C-terminal extension comprising the proximal tail. CC0651, an inhibitor of UBE2R, is not depicted for simplification. Yellow sphere connected to Ubiquitin’s (orange) C-terminus indicates catalytic residue of UBE2R.
(b) Schematic of the role of UBE2R C-terminal extension in stabilizing the closed conformation of the UBE2R~UB intermediated.
(c) Schematic of APC11 RING domain in autoinhibited, apo-APC/C.
(d) Upon substrate receptor/substrate recruitment to APC/C, the CRL unit of APC/C (APC2-APC11) shifts upward, and the RING domain is relieved from autoinhibition. The WHB domain of APC2 is liberated to engage UBE2C, while the C-terminal tail of UBE2S can interact with a separate region of APC2 and APC4.
(e) APC/C substrates are primed by UBE2C. The priming reaction can be allosterically activated by the C-terminal tail of the chain elongating E2, UBE2S. Acceleration of priming by UBE2S enhances the formation of UBE2S’s target substrate for polyubiquitylation.
A different multiprotein E3 ligase, Anaphase-Promoting Complex/Cyclosome (APC/C), relies on an embedded dedicated cullin-RING core (APC2-APC11) to mediate ubiquitylation[40,41]. Unlike other cullins, APC2 is not neddylated, and thus APC/C must be activated by other means, including conformational rearrangement mediated by the binding of a substrate receptor subunit (Figure 4c, d). Like conventional CRLs, APC/C employs multiple E2 partners for different reactions: UBE2C to transfer UB to acceptor-bound substrates, and subsequently UBE2S in a non-standard mechanism that does not involve RING-E2 interactions to mediate polyubiquitylation. A recent study showed UBE2S’s C-terminal extension binds the cullin subunit APC2, to allosterically activate substrate ubiquitylation by the canonical APC11 RING-UBE2C~UB unit[31]. Because UBE2C-modified proteins in turn become substrates of UBE2S, this mechanism enables UBE2S to accelerate formation of its own substrate for UB chain elongation (Figure 4e).
Outlook
The modular cullin-RING ligase system includes 6 different cullins (CUL1, 2, 3, 4A, 4B, and 5) paired with either RBX1 or RBX2. Each CUL-RBX complex interacts with dozens of receptors, or adaptor/receptor complexes. While the numerous cullin-receptor combinations determine the regulated recruitment of thousands of substrates, their ubiquitylation ultimately depends on common elements, RBX1’s RING (or RBX2 in CUL5-based CRL5s) and NEDD8 covalently-linked to the cullin activating various UB carrying enzymes - several E2s or ARIH1-family RBR E3s. In order to compensate for the structural and functional diversity of CRL substrates, the common catalytic elements seem to have evolved as highly dynamic contortionists, with specificity determined by conformational regulation. It seems likely that future studies of other CRL complexes with different UB carrying enzymes will reveal even more remarkable mechanisms enabling UB transfer to various substrates, and their modification by assorted UB chains. Moreover, the capacity for CRL redirection by pathogenic viruses to degrade and thereby avert host defense factors [42]- and by small molecules [43–46](e.g. PROTAC or molecular glue) to ubiquitylate therapeutically-important proteins at will - likely depends on conformational plasticity of the E3 ligase elements.
Beyond CRLs, it is becoming clear that common general principles determine activities and specificities of RING E3s in general. Various RING E3s - forming homologous assemblies with E2~NEDD8, E2~SUMO, or E2~UB intermediates - are activated and directed to their specific targets through remarkably diverse combinations of avid multisite interactions and allosteric modulation[11,12,14,29,47–50]. The few that have been studied structurally to date have shown conformational regulation depending on E2 or E3 appendages, UBL identity, co-E3s, post-translational modifications, and even metabolite binding. Furthermore, a sufficient number of elements mediating an avid, stable RING E3-E2-UBL-substrate assembly can even overcome suboptimal target lysine presentation to the active site to achieve catalysis[14]. We anticipate many future studies representing active E3 ligases will show remarkable, multifarious coordination of seemingly similar catalytic elements determining targeting specificity.
Acknowledgements
We thank all our colleagues in the UB/UBL field, and all past and present Schulman lab members for inspiring discoveries. We apologize to those whose research we could not cite here. This work was supported by NIH R01CA247365, P30CA021765, St. Jude Children’s Research Hospital, ALSAC, the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 789016-NEDD8Activate), and the Leibniz Prize (to B.A.S.) from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation - SCHU 3196/1-1), and the Max Planck Society.
Footnotes
Conflict of interest
The authors declare no competing interest.
References and Recommended reading
Papers of particular interest, published within a period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Cappadocia L, Lima CD: Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism. Chem Rev 2018, 118:889–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K: Nedd8 on cullin: building an expressway to protein destruction. Oncogene 2004, 23:1985–1997. [DOI] [PubMed] [Google Scholar]
- 3.Komander D, Rape M: The ubiquitin code. Annu Rev Biochem 2012, 81:203–229. [DOI] [PubMed] [Google Scholar]
- 4.•.Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, Klevit RE: Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol Cell 2012, 47:933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]; A NMR-based study and structural model defining a key hydrogen bond between a conserved side chain within E3s and carbonyl of E2s for stabilizing the closed E2~UB conformation.
- 5.Hamilton KS, Ellison MJ, Barber KR, Williams RS, Huzil JT, McKenna S, Ptak C, Glover M, Shaw GS: Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure 2001, 9:897–904. [DOI] [PubMed] [Google Scholar]
- 6.Reverter D, Lima CD: Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 2005, 435:687–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M: The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 2011, 144:769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ: Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol Cell 2011, 42:75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.•.Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT: Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 2012, 489:115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first structure of a RING E3 bound to a UB-loaded E2 defining the structural and biochemical determinants for RING E3 priming of catalysis
- 10.•.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT: BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol 2012, 19:876–883. [DOI] [PMC free article] [PubMed] [Google Scholar]; Structural and biochemical characterization of BIRC7-UBCH5B~UB forming the closed conformation for catalysis.
- 11.•.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT: Essentiality of a non-RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nat Struct Mol Biol 2013, 20:982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies a phosphorylated residue of CBL-B outside of its RING domain as an essential component for activiating UB transfer, demonstrating how monomeric E3s use regions outside of their RING domains to stabilize E2~UB conjugates.
- 12.••.Scott DC, Sviderskiy VO, Monda JK, Lydeard JR, Cho SE, Harper JW, Schulman BA: Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell 2014, 157:1671–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study describes the crystal structure of a trapped NEDD8 ligation intermediate, providing detailed insight into the structural mechanisms governing activation and lysine selectivity of UBL transfer.
- 13.Koliopoulos MG, Esposito D, Christodoulou E, Taylor IA, Rittinger K: Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J 2016, 35:1204–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.•.Streich FC Jr., Lima CD: Capturing a substrate in an activated RING E3/E2-SUMO complex. Nature 2016, 536:304–308. [DOI] [PMC free article] [PubMed] [Google Scholar]; An elegant combination of biochemical traps and engineered proteins to structurally illuminate a SUMO transfer intermediate.
- 15.••.Branigan E, Carlos Penedo J, Hay RT: Ubiquitin transfer by a RING E3 ligase occurs from a closed E2~ubiquitin conformation. Nat Commun 2020, 11:2846. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, Branigan et. al. utilize a single molecule FRET approach to experimentally demonstrate that ubiquitin transfer occurs from the closed E2~UB conformation. These results confirm that the active form of the E2~UB conjugate is the closed form.
- 16.Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL: Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ 2008, 15:841–848. [DOI] [PubMed] [Google Scholar]
- 17.Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, Miller DJ, Walden H, Duda DM, Seyedin SN et al. : Insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell 2009, 36:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Zapien D, Ruiz FX, Poirson J, Mitschler A, Ramirez J, Forster A, Cousido-Siah A, Masson M, Vande Pol S, Podjarny A, et al. : Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature 2016, 529:541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rusnac DV, Lin HC, Canzani D, Tien KX, Hinds TR, Tsue AF, Bush MF, Yen HS, Zheng N: Recognition of the Diglycine C-End Degron by CRL2(KLHDC2) Ubiquitin Ligase. Mol Cell 2018, 72:813–822 e814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kung WW, Ramachandran S, Makukhin N, Bruno E, Ciulli A: Structural insights into substrate recognition by the SOCS2 E3 ubiquitin ligase. Nat Commun 2019, 10:2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F: Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 2003, 112:243–256. [DOI] [PubMed] [Google Scholar]
- 22.Rusnac DV, Zheng N: Structural Biology of CRL Ubiquitin Ligases. Adv Exp Med Biol 2020, 1217:9–31. [DOI] [PubMed] [Google Scholar]
- 23.•.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. : Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 2002, 416:703–709. [DOI] [PubMed] [Google Scholar]; First structure of a cullin-RING ligase complex.
- 24.Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, et al. : E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell 2009, 33:483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott DC, Rhee DY, Duda DM, Kelsall IR, Olszewski JL, Paulo JA, de Jong A, Ovaa H, Alpi AF, Harper JW, et al. : Two Distinct Types of E3 Ligases Work in Unison to Regulate Substrate Ubiquitylation. Cell 2016, 166:1198–1214 e1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill S, Reichermeier K, Scott DC, Samentar L, Coulombe-Huntington J, Izzi L, Tang X, Ibarra R, Bertomeu T, Moradian A, et al. : Robust cullin-RING ligase function is established by a multiplicity of poly-ubiquitylation pathways. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierce NW, Lee JE, Liu X, Sweredoski MJ, Graham RL, Larimore EA, Rome M, Zheng N, Clurman BE, Hess S, et al. : Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell 2013, 153:206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu K, Chen A, Pan ZQ: Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J Biol Chem 2000, 275:32317–32324. [DOI] [PubMed] [Google Scholar]
- 29.••.Baek K, Krist DT, Prabu JR, Hill S, Klugel M, Neumaier LM, von Gronau S, Kleiger G, Schulman BA: NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly. Nature 2020, 578:461–466. [DOI] [PMC free article] [PubMed] [Google Scholar]; First structure of a NEDD8 activated cullin-RING ligase transfer intermediate. The structure defines structural and biochemical principles underlying the NEDD8 dependent activation of UB transfer by CULLIN-RINGs.
- 30.••.Williams KM, Qie S, Atkison JH, Salazar-Arango S, Alan Diehl J, Olsen SK: Structural insights into E1 recognition and the ubiquitin-conjugating activity of the E2 enzyme Cdc34. Nat Commun 2019, 10:3296. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, Williams et. al. determine the structure of a CDC34~UB thioester mimetic revealing contacts between the C-terminal extension of CDC34 and UB which stabilize the closed E2~UB conformation in the absence of E3.
- 31.••.Martinez-Chacin RC, Bodrug T, Bolhuis DL, Kedziora KM, Bonacci T, Ordureau A, Gibbs ME, Weissmann F, Qiao R, Grant GD, et al. : Ubiquitin chain-elongating enzyme UBE2S activates the RING E3 ligase APC/C for substrate priming. Nat Struct Mol Biol 2020, 27:550–560. [DOI] [PMC free article] [PubMed] [Google Scholar]; A sophisticated biochemical study demonstrating that the tail of UBE2S activates APC/C for priming by UBCH10. These results reveal that E2 appendages can themselves directly activate E3 activity towards other E2 conjugating enzymes. In this case, the activation serves to accelerate production of its own substrate.
- 32.Scott DC, Monda JK, Grace CR, Duda DM, Kriwacki RW, Kurz T, Schulman BA: A dual E3 mechanism for Rub1 ligation to Cdc53. Mol Cell 2010, 39:784–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA: N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 2011, 334:674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA: Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 2008, 134:995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu C, Mao H, Novitsky EJ, Tang X, Rychnovsky SD, Zheng N, Huang L: Gln40 deamidation blocks structural reconfiguration and activation of SCF ubiquitin ligase complex by Nedd8. Nat Commun 2015, 6:10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hibbert RG, Huang A, Boelens R, Sixma TK: E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc Natl Acad Sci U S A 2011, 108:5590–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metzger MB, Liang YH, Das R, Mariano J, Li S, Li J, Kostova Z, Byrd RA, Ji X, Weissman AM: A structurally unique E2-binding domain activates ubiquitination by the ERAD E2, Ubc7p, through multiple mechanisms. Mol Cell 2013, 50:516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S, Liang YH, Mariano J, Metzger MB, Stringer DK, Hristova VA, Li J, Randazzo PA, Tsai YC, Ji X, et al. : Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B. J Biol Chem 2015, 290:30225–30239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buetow L, Gabrielsen M, Anthony NG, Dou H, Patel A, Aitkenhead H, Sibbet GJ, Smith BO, Huang DT: Activation of a primed RING E3-E2-ubiquitin complex by non-covalent ubiquitin. Mol Cell 2015, 58:297–310. [DOI] [PubMed] [Google Scholar]
- 40.Brown NG, VanderLinden R, Watson ER, Qiao R, Grace CR, Yamaguchi M, Weissmann F, Frye JJ, Dube P, Ei Cho S, et al. : RING E3 mechanism for ubiquitin ligation to a disordered substrate visualized for human anaphase-promoting complex. Proc Natl Acad Sci U S A 2015, 112:5272–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watson ER, Brown NG, Peters JM, Stark H, Schulman BA: Posing the APC/C E3 Ubiquitin Ligase to Orchestrate Cell Division. Trends Cell Biol 2019, 29:117–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Tan X: Viral Manipulations of the Cullin-RING Ubiquitin Ligases. Adv Exp Med Biol 2020, 1217:99–110. [DOI] [PubMed] [Google Scholar]
- 43.Bulatov E, Ciulli A: Targeting Cullin-RING E3 ubiquitin ligases for drug discovery: structure, assembly and small-molecule modulation. Biochem J 2015, 467:365–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petzold G, Fischer ES, Thoma NH: Structural basis of lenalidomide-induced CK1alpha degradation by the CRL4(CRBN) ubiquitin ligase. Nature 2016, 532:127–130. [DOI] [PubMed] [Google Scholar]
- 45.Verma R, Mohl D, Deshaies RJ: Harnessing the Power of Proteolysis for Targeted Protein Inactivation. Mol Cell 2020, 77:446–460. [DOI] [PubMed] [Google Scholar]
- 46.Burslem GM, Crews CM: Proteolysis-Targeting Chimeras as Therapeutics and Tools for Biological Discovery. Cell 2020, 181:102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mattiroli F, Uckelmann M, Sahtoe DD, van Dijk WJ, Sixma TK: The nucleosome acidic patch plays a critical role in RNF168-dependent ubiquitination of histone H2A. Nat Commun 2014, 5:3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGinty RK, Henrici RC, Tan S: Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 2014, 514:591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DaRosa PA, Wang Z, Jiang X, Pruneda JN, Cong F, Klevit RE, Xu W: Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal. Nature 2015, 517:223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magnussen HM, Ahmed SF, Sibbet GJ, Hristova VA, Nomura K, Hock AK, Archibald LJ, Jamieson AG, Fushman D, Vousden KH, et al. : Structural basis for DNA damage-induced phosphoregulation of MDM2 RING domain. Nat Commun 2020, 11:2094. [DOI] [PMC free article] [PubMed] [Google Scholar]


