Abstract
During a large variety of common pathogens, E. coli, P. aeruginosa, MRSA, MRCNS, V. parahaemolyticus, L. monocytogenes and Salmonella are the leading pathogens responsible for large number of human infections and diseases. In this study, a high flux screening based on nucleic acid isothermal amplification technique has been developed. For the 8 common pathogens, species-specific targets had been selected and analyzed for their unique specificity. After optimization, separate LAMP reaction assays had been bioprocessed and integrated into one systematic detection platform, including 8 strips (PCR tubes) and 96-well plates. Eight standard strains verified for the accuracy. Application of the established high flux screening platform was used for detection for 48 samples in 4 different 96-well plates, with 2 groups of 2 operators using double-blind procedure. The accuracy of 100% was obtained, with the total time consumption as 66–75 min (for 12 samples detection on 8 different pathogens). As concluded, through the bioprocess of the systematic platform based on LAMP technique, it’s been demonstrated to be capable of simultaneous detection of 8 pathogens, with high sensitivity, specificity, rapidity and convenience.
Keywords: Nucleic acid, Isothermal amplification, LAMP, High flux screening, Detection
Introduction
Pathogens mediated infectious diseases in human beings and animals remain a major concern in public health [1]. During a large variety of common pathogens, E. coli, P. aeruginosa, S. aureus (especially methicillin-resistant Staphylococcus aureus, MRSA), coagulase-negative staphylococci (MRCNS), V. parahaemolyticus, L. monocytogenes and Salmonella are the leading pathogens responsible for large number of human infections and diseases [2–15]. However, bacterial identification of such pathogens requires up to several days [16]. For example, using “golden standard” culturing takes up 3–7 days depending on the results [17]. The required time length has posed a major concern for clinicians. Early diagnosis of such pathogens is significantly important for further therapeutic treatment and thus substantially influences the prognosis of the infections and diseases [18–20].
In recent years, PCR and Q-PCR have been well studied and documented to be a promising technique for rapid detection and bacterial identification [21–25]. However, regular PCR requires laborious results determination process, such as electrophoresis or hybridization. (The former takes several hours and the latter takes up to 36 h.) For Q-PCR, expensive equipment and reagents are required, which poses an obstacle during its broad application [26, 27]. Since 2000, a novel nucleic acid amplification method has been developed and established, named loop-mediated isothermal amplification (LAMP) [28, 29]. For the past 2 decades, LAMP has been developed for the detection of various microorganisms and other genes [30, 31]. However, the clinical application of LAMP assays, especially platform for high flux screening based on LAMP, has been rarely reported [32–34]. Advantages of LAMP include high sensitivity, specificity, rapidity, simpliness in operation and labor, convenience and expense [35]. However, lack of the bioprocessing and systematic integration of this technique into applicable platforms has been the biggest obstacle during the application of this technique [36–38]. As a consequence, development and application of rapid detection on such pathogens as well as their virulent factors for high flux screening application are of utmost importance and necessity [39–41].
In this study, a high flux screening of pathogens, including E. coli, P. aeruginosa, MRSA, MRCNS, V. parahaemolyticus, L. monocytogenes and Salmonella, has been developed based on the loop-mediated isothermal amplification methodology.
Materials and methods
Bacterial strains
Nine standard strains were used to establish the high flux screening assays in this study, including E. coli C600 ATCC25922, Pseudomonas aeruginosa: ATCC27853, MRSA 85/2082, MSSA ATCC14458, MRCNS ATCC29887, MSCNS ATCC27844, Listeria monocytogenes: ATCC19118, Salmonella enterica subsp. enterica: ATCC29629, Vibrio parahaemolyticus: ATCC27969. The storage, inoculation, culturing and incubation have been conducted as described previously.
Design on the systematic integration of detection
Specific targets were selected for different pathogens, with the detailed information as follows. For E. coli, rfbE (the specific O-antigen) was selected. For P. aeruginosa, oprI was selected. For MRSA, MSSA, MRCNS, MSCNS, mecA, femA and 16SrRNA (specific for the genus of Staphylococci) were selected. For Salmonella, invA was selected. For V. parahaemolyticus, tlh (thermolable haemolysin, considered to be a species-specific marker for V. parahaemolyticus) gene was selected. For L. monocytogenes, hlyA was selected. In our previous studies, the primer sets for each of the targets had been separately designed, and optimized for LAMP reaction. Such primer sets were also selected in this study, and additionally, new primer sets for each targets had been designed using Primer Explorer V4 [42–44]. Optimal parameters were selected by each set of primers. Principles for the systematic integration of the high flux screening platform included: firstly, unique temperature is required for the reaction occurred for each primers set. Secondly, unique reaction time is also required for each target. The DNA samples of E. coli, Pseudomonas aeruginosa, MRSA, MSSA, MRCNS, MSCNS, L. monocytogenes, Salmonella, V. parahaemolyticus were isolated using the DNA extraction kit. DNA quality and concentration had been confirmed with NanoDrop before further detection. LAMP reaction was performed under different temperatures (59 °C to 66 °C), reaction time (0, 15 min, 30 min, 45 min, 60 min, 75 min, 90 min), concentrations of betaine (0.3 M, 0.4 M, 0.5 M, 0.6 M and 0.7 M) and ratios of calcein and Mn2 + (1:20, 1:16, 1:12, 1:8, 1:4, 1:2) [45]. The results determination was performed by observation by naked eye and SYBR Green I, electrophoresis [46, 47]. At last, 8 primer sets were selected for the high flux screening platform (Table 1).
Table 1.
The primers used for each separate LAMP reaction
| Gene | Sequence (5′-3′) | Size | References |
|---|---|---|---|
| femA | 6 | ||
| F3 | ATGCTGGTGGTACATCAA | 18 | |
| B3 | TGGTTTAATAAAGTCACCAACAT | 23 | |
| FIP | GGTCAATGCCATGATTTAATGCATAGCATTCCGTCATTTTGCC | 43 | |
| BIP | CAGAAGATGCTGAAGATGCTGGTCAATAATTTCAGCATTGTAACC | 45 | |
| LF | AATCATTTCCCATTGCACT | 22 | |
| LB | TGTAGTTAAATTCAA | 15 | |
| mecA | 6 | ||
| F3 | AAGATGGCAAAGATATTCAACT | 22 | |
| B3 | AGGTTCTTTTTTATCTTCGGTTA | 23 | |
| FIP | GTGGATAGCAGTACCTGAGCCTTGATGCTAAAGTTCAAAAGAGT | 44 | |
| BIP | CCTCAAACAGGTGAATTATTAGCACCTTCGTTACTCATGCCATAC | 45 | |
| LF | TAATCATTTTTCATGTTG | 18 | |
| LB | TGTAAGCACACCTTCATATGACGT | 24 | |
| rfbE | 2 | ||
| F3 | AACAGTCTTGTACAAGTCCA | 20 | |
| B3 | GGTGCTTTTGATATTTTTCCG | 21 | |
| FIP | CTCTCTTTCCTCTGCGGTCC-GATGTTTTTCACACTTATTGGAT | 43 | |
| BIP | TAAGGAATCACCTTGCAGATAAACT-AGTACATTGGCATCGTGT | 43 | |
| LF | CCAGAGTTAAGATTGAT | 17 | |
| LB | CGAAACAAGGCCAGTTTTTTACC | 23 | |
| 16S rRNA | 6 | ||
| F3 | CGTGGGGATCAAACAGGATT | 20 | |
| B3 | CATGCTCCACCGCTTGTG | 18 | |
| FIP | TAGCTGCAGCACTAAGGGGC-CCACGCCGTAAACGATGAG | 39 | |
| BIP | ACGCATTAAGCACTCCGCCT-GGGTCCCCGTCAATTCCT | 38 | |
| LF | GGAAACCCCCTAACACT | 17 | |
| LB | GGGGAGTACGACCGCAAGGT | 20 | |
| invA | 1 | ||
| F3 | TCAACAATGCGGGGATCTG | 19 | |
| B3 | GAAGCGTA CTGGAAAGGGAA | 21 | |
| FIP | ACRCGCCATGGTATGGATTTGTGACCATCACCAATGGTCAGC | 41 | |
| BIP | ATGATGCCGGCAATAGCGTCAAGCCAGCTTTACGGTTCCT | 40 | |
| LF | TCCGCTCTGTCTACTTATACCAT | 23 | |
| LB | TGATAAACTTCATCGCACCGTCAA | 25 | |
| oprI | 7 | ||
| F3 | CTGGCTGCTGTTCTGG | 16 | |
| B3 | CGCTCGTTAGCCTCGT | 16 | |
| FIP | CTGCGTCTTCGGTAGCGGGGTTGCAGCAGCCACT | 34 | |
| BIP | TCAGGCTCGCGCTGACGA-AGTCTGCTGAGCTTTCTGAG | 38 | |
| LF | TCTTTGGCTTCGAGCAGACT | 20 | |
| LB | GCCTATCGCAAGGCTGACGAA | 21 | |
| hlyA | 8 | ||
| F3 | GGAGGMTACGTTGCTCAA | 18 | |
| B3 | AAGCTAAACCAGTGCATTC | 19 | |
| FIP | TCGCTCCAGTTTTTATGTTGAACAC-CTTGGGATGAARTAAATTATGATCC | 50 | |
| BIP | AGCAAGCTAGCTCATTTCACAT-AGCGTAAACATTAATATTTCTCGC | 46 | |
| LF | ACTTCCATTKCTTTA | 15 | |
| LB | CGTCCATCTATTTGCCAGGTAAC | 24 | |
| tlh | 9 | ||
| F3 | CGCTGACAATCGCTTCTCAT | 20 | |
| B3 | GTTCTTCGCTTTGGCAATGT | 20 | |
| FIP | CTGTCACCGAGTGCAACCACTTAACCACACGATCTGGAGCA | 41 | |
| BIP | GCATCACAATGGCGCTTCCCACCGTTGGAGAAGTGACCTA | 40 | |
| LF | GTTGATTTGATCTGGCTGCATTG | 23 | |
| LB | AACCCGAACAGCTGGTTCT | 19 |
Bioprocessing the separate LAMP assays into high flux screening platform
As mentioned above, 8 primers sets specifically targeted for rfbE, oprI, mecA, femA, 16S rRNA for staphylococci, invA, tlh and hlyA have been included [48]. As LAMP reactions were concerned, 65 °C was selected due to the efficiency, stability and reproducibility of this methodology. Also, reaction time as 45 min was selected as at this time point sufficient reaction products for results determination as well as minimal amounts of amplicons were both achieved [49–51]. For the concentration of betaine, insignificant difference was found, and thus 0.3 M was used for the consideration of minimal expense. For the ratio between calcein and Mn2+, 1:4 was found to be optimal and thus selected. In addition, calcein was selected to replace SYBR Green I for the color change for results determination as calcein is capable of preload in the reaction volume but SYBR Green I [52]. For application, 8 strips (PCR tubes) were employed for the detection of standard strains, and 96-well plates were employed for the detection of different samples. Different sets of primers as well as reaction volume (25 micro liters were used, with 0.3 M betaine and 1:4 of calcein and Mn2+) were prepared at each tube of 8 strips or 96-well plates (Fig. 1). Both 8 strips and 96-well plates could be stored under −20 °C. For standard strains detection including E. coli, Pseudomonas aeruginosa, MRSA, MSSA, MRCNS, MSCNS, L. monocytogenes, Salmonella, V. parahaemolyticus, 8 strips were used for 8 targets, and template DNA was loaded, followed by reaction on waterbath at 65 °C for 45 min [53–55]. Color change was determined for test results.
Fig. 1.
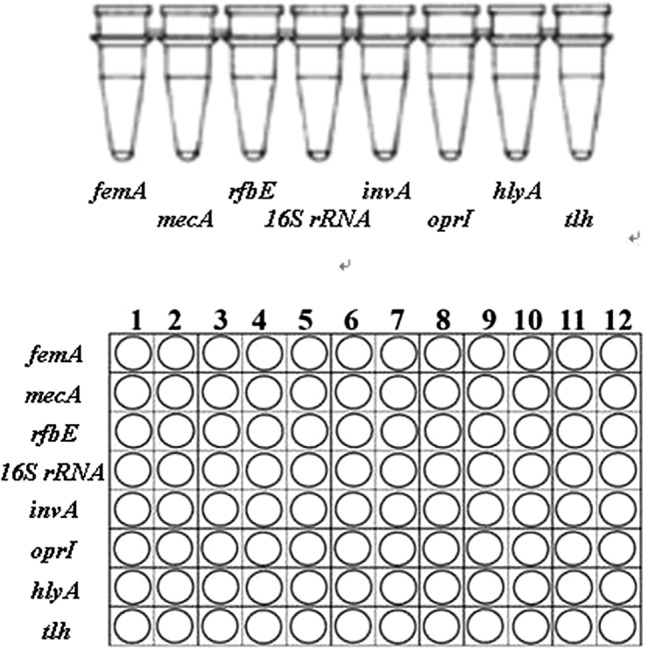
8-tube strips and PCR well plates for the biosystem platform
Application of the high flux screening platform
In this study, a common type of food sample, cake was employed to verify the applicability of this high flux screening platform. Sample artificially contamination was performed as described previously. A total of 16 strains other than standard strains were used in the application study, including 2 E. coli, 2 P. aeruginosa, 2 MRSA, 2 MSSA, 2 MRCNS, 2 L. monocytogenes, 2 Salmonella, 2 V. parahaemolyticus strains. A total of four 96-well plates were used. For each 96-well plate, there are 12 panels, and for each panel, 8 detected targets were included. Experiments were performed using a double-blind method, and the demonstration is as follows [56–58]. Firstly, 2 separate operators (Group A) randomly selected different number of strains (from 1 to 8) for each of the 12 panels and conducted DNA extraction without informing the strain selection. Two other operators (Group B) further performed the detection using the 96-well plate. After the detection, 2 groups of operators compared the strains selection and detection results. Then, the same procedure was conducted vice versa, with Group B conducted strains selection and Group A conducted detection [59]. For a single 96-well plate, a total of 12 samples were detected for 8 pathogens at one reaction. Simple and rapid DNA extraction was performed as described previously, followed by loading of 5 micro liters of template DNA (with the 96-well plates placed on ice). LAMP was proceeded on waterbath at 65 °C for 45 min [60]. Color change was determined for test results. Besides, regular PCR detection was also performed as control.
Results
Development of separate LAMP platform and their integration
According to the development of LAMP assays, positive results had been obtained from DNA amplification of standard strains, with color change from orange to green by either SYBR Green I or calcein, as well as typical ladder bands pattern from electrophoresis [61–63]. Optimization of LAMP reaction was also performed, in details as follows. For reaction processed under different temperatures ranging from 59 °C to 66 °C, insignificant difference was observed between 63 °C and 65 °C, and 65 °C was selected due to the efficiency, stability (according to previous studies), applicability (significantly higher number of reported LAMP reactions had used this temperature) and reproducibility of this methodology. For reaction time ranging from 0, 15 min, 30 min, 45 min, 60 min, 75 min, 90 min, the first time point for positive results to occur was found to be 30 min, and sufficient amplicons were obtained since 45 min. As a consequence, 45 min was selected. For concentrations of betaine (0.3 M, 0.4 M, 0.5 M, 0.6 M and 0.7 M), insignificant difference was found, and thus 0.3 M was used for the consideration of minimal expense (Fig. 2). In this study, calcein was selected to replace SYBR Green I for the color change for results determination as calcein is capable of preload in the reaction volume but SYBR Green I [64, 65]. For different ratios of calcein and Mn2 + (1:20, 1:16, 1:12, 1:8, 1:4, 1:2), 1:4 was found to be optimal and thus selected (Fig. 3).
Fig. 2.
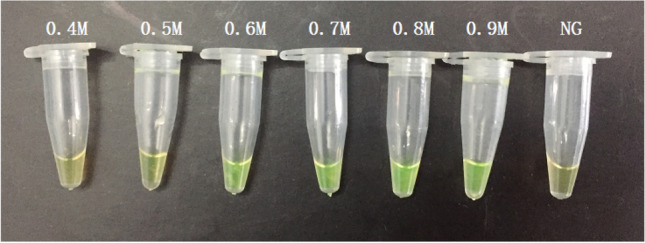
Effect of different concentration of betain on the LAMP reaction
Fig. 3.
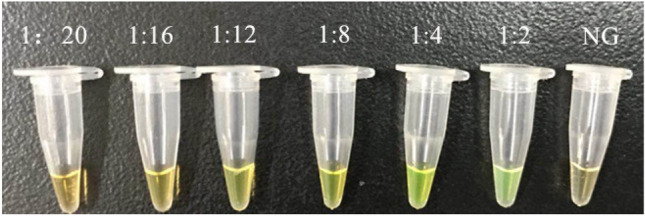
Optimization of the concentration ratio of calcein and Mn2+ (C(calcein) and C(Mn2+) is 1:20, 1:16, 1:12, 1:8, 1:4, 1:2, respectively; NG, negative control)
Verification of the high flux screening platform
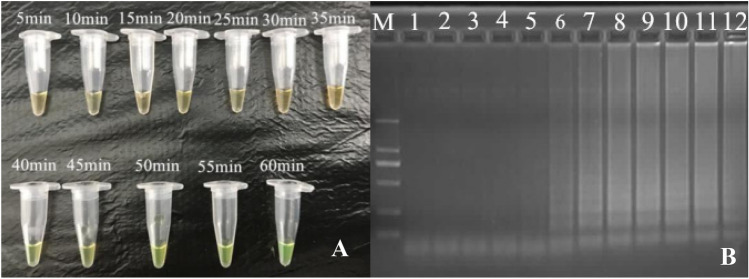
For verification of the high flux screening platform on standard strains including E. coli, Pseudomonas aeruginosa, MRSA, MSSA, MRCNS, MSCNS, L. monocytogenes, Salmonella, V. parahaemolyticus, 8 strips (PCR tubes) were employed with different sets of primers, reaction volume (25 micro liters were used, with 0.3 M betaine and 1:4 of calcein and Mn2 +) preloaded at each tube of 8 strips. After loading of template DNA for each standard strain, reaction was proceeded in waterbath at 65 °C for 45 min [66]. As shown, color change from orange to green was observed for positive results, and 100% of specificity was obtained in this study (Fig. 4a). From template DNA loading to results determination, 50 min was required (Fig. 4b).
Fig. 4.
a Visual detection by calcein with fluorescence in LAMP amplified product, from 40 min onwards to 60 min and orange color in 5-35 min. b Agarose gel electrophoresis of LAMP amplification products at different time interval (5–60 min), Lane M: 2000 bp ladder marker, 1–12 are 5 min, 10 min, 15 min, 20 min, 25 min, 30 min, 35 min, 40 min, 45 min, 50 min, 55 min, 60 min)
Application of the high flux screening platform
According to the results, for the first round as Group A for strains selection and Group B for detection, Operator 1 had selected 1 E. coli, 1 P. aeruginosa, 2 MRSA, 1 MSSA, 1 MRCNS, 2 L. monocytogenes, 2 Salmonella, 2 V. parahaemolyticus strains. As shown by the detection, all of the selected strains had been diagnosed by Operator 1 from Group B. For Operator 2, 2 E. coli, 1 P. aeruginosa, 2 MRSA, 2 MSSA, 2 MRCNS, 1 L. monocytogenes, 1 Salmonella, 1 V. parahaemolyticus strains were selected, and all strains were correctly detected by Operator 2 from Group B [67–69]. For the second round as Group B for strains selection and Group A for detection, Operator 1 had selected 2 E. coli, 2 P. aeruginosa, 2 MRSA, 2 MSSA, 2 MRCNS, 2 L. monocytogenes, 0 Salmonella, 0 V. parahaemolyticus strains. As shown by the detection, all of the selected strains had been diagnosed by Operator 1 from Group A. For Operator 2, 1 E. coli, 1 P. aeruginosa, 1 MRSA, 1 MSSA, 2 MRCNS, 2 L. monocytogenes, 2 Salmonella, 2 V. parahaemolyticus strains were selected, and all strains were correctly detected by Operator 2 from Group A [70]. In summary, 100% of accuracy was obtained by both groups for 2 rounds of experiments (Fig. 5). As rapidity was concerned, 10–15, 8–10, 45 and 3–5 min are required for DNA extraction, template DNA loading, LAMP reaction and results determination, respectively [71, 72]. In summary, the total time consumption is 66–75 min, for simultaneous detection of 12 samples for 8 different pathogens.
Fig. 5.
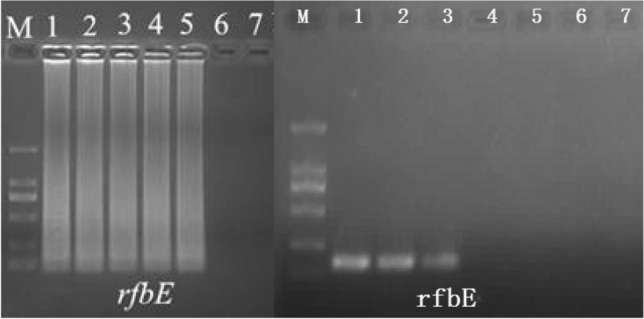
Example of electrophoresis of rfbE
Discussion
In this study, a high flux screening based on nucleic acid isothermal amplification technique has been developed. Firstly, 8 common pathogens were selected and 8 species-specific targets had been selected and analyzed for their unique specificity. Then, 8 different sets of primers for LAMP reaction had been further designed and optimized to obtain unique reaction temperature and time. Furtherly, the 8 detection assays had been integrated into a biosystem panel for isothermal detection, including 8 strips (PCR tubes) and 96-well plates, with 8 standard strains verified for the accuracy [73–76]. At last, application of the established high flux screening platform was used for detection for 48 samples in 4 different 96-well plates, with 2 groups of 2 operators using double-blind procedure. The accuracy of 100% was obtained, with the total time consumption as 66–75 min (for 12 samples detection on 8 different pathogens) [77–79]. As concluded, through the bioprocess of the systematic platform based on LAMP technique, it’s been demonstrated to be capable of simultaneous detection of 8 pathogens, with high sensitivity, specificity, rapidity and convenience.
Acknowledgements
This study was supported by Grants from the Science, Technology and Innovation Commission of Shenzhen municipality (JCYJ20170306113900538; JCYJ20170306140020487), and the Sanming Project of Medicine in Shenzhen (SZSM201812090).
Compliance with ethical standards
Conflict of interest
The authors declare that there are no conflicts of interest.
Footnotes
Parallel First Author: Huamin Zhong, Hongwei Deng.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pal M, Kerorsa GB, Marami LM, et al. Epidemiology, pathogenicity, animal infections, antibiotic resistance, public health significance, and economic impact of staphylococcus aureus: a comprehensive review. Am J Public Health Res. 2020;8(1):14–21. [Google Scholar]
- 2.Paquette SJ, Reuter T. Properties of an antimicrobial molecule produced by an Escherichia coli champion. Antibiotics. 2019;9(1):6. doi: 10.3390/antibiotics9010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis SJ, Crossman LC, McGrath CJ, et al. Identification and characterisation of enteroaggregative Escherichia coli subtypes associated with human disease. Sci Rep. 2020;10:7475. doi: 10.1038/s41598-020-64424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eklöf J, Sørensen R, Ingebrigtsen TS, et al. Pseudomonas aeruginosa and risk of death and exacerbations in patients with chronic obstructive pulmonary disease: an observational cohort study of 22 053 patients. Clin Microbiol Infect. 2020;26(2):227–234. doi: 10.1016/j.cmi.2019.06.011. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs DM, Ochsbalcom HM, Noyes K, et al. Impact of Pseudomonas aeruginosa isolation on mortality and outcomes in an outpatient chronic obstructive pulmonary disease cohort. Open Forum Infect Dis. 2020;7(1):546. doi: 10.1093/ofid/ofz546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isaac S, Yi M, Valerie A, et al. Trends in incidence of methicillin-resistant Staphylococcus aureus bloodstream infections differ by strain type and healthcare exposure, United States, 2005–2013. Clin Infect Dis. 2019;1:1. doi: 10.1093/cid/ciz158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliveira D, Borges A, Simoes M, et al. Staphylococcus aureus toxins and their molecular activity in infectious diseases. Toxins. 2018;10(6):252. doi: 10.3390/toxins10060252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parlet CP, Brown MM, Horswill AR, et al. Commensal Staphylococci Influence Staphylococcus aureus Skin Colonization and Disease. Trends Microbiol. 2019;27(6):497–507. doi: 10.1016/j.tim.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee LH, Mutalib NSA, Law WF, et al. Discovery on Antibiotic Resistance Patterns of Vibrio parahaemolyticus in Selangor Reveals Carbapenemase Producing Vibrio parahaemolyticus in Marine and Freshwater Fish. Front Microbiol. 2018;9:2513. doi: 10.3389/fmicb.2018.02513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Meng H, Gu D, Li Y, Jia M. Molecular mechanisms of Vibrio parahaemolyticus pathogenesis. Microbiol Res. 2019;1(222):43–51. doi: 10.1016/j.micres.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Fontana C, Favaro M. Coagulase-positive and coagulase-negative staphylococci in human disease[M]//Pet-To-Man Travelling Staphylococci. 2018
- 12.Fulvio M, Cristina EDF, Barbara DM. Chapter 4–Coagulase-Positive and Coagulase-Negative Staphylococci Animal Diseases. Pet-To-Man Travelling Staphylococci. 2018: 43-50,
- 13.Bahrami A, Baboli ZM, Schimmel K, Jafari SM, Williams L. Efficiency of novel processing technologies for the control of Listeria monocytogenes in food products. Trends Food Sci Technol. 2020;1(96):61–78. [Google Scholar]
- 14.Gast, Richard K. and Robert E. Porter, Jr., 2020. Salmonella infections. Pages 719-753 in Diseases of Poultry, 14th edition. D. E. Swayne, ed. John Wiley and Sons, Hoboken, New Jersey.
- 15.Tanner JR, Kingsley RA. Evolution of Salmonella within Hosts. Trends Microbiol. 2018;26(12):986–998. doi: 10.1016/j.tim.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balestra GM, Misaghi IJ. Increasing the efficiency of the plate counting method for estimating bacterial diversity. J Microbiol Methods. 1997;30(2):111–117. [Google Scholar]
- 17.Zhao X, Wang L, Chu J, Li Y, Li Y, Xu Z, et al. Development and application of a rapid and simple loop-mediated isothermal amplification method for food-borne Salmonella detection. Food Sci Biotechnol. 2010;19(6):1655–1659. [Google Scholar]
- 18.Zhao X, Li Y, Wang L, You L, Xu Z, Li L, et al. Development and application of a loop-mediated isothermal amplification method on rapid detection Escherichia coli O157 strains from food samples. Mol Biol Rep. 2010;37(5):2183–2188. doi: 10.1007/s11033-009-9700-6. [DOI] [PubMed] [Google Scholar]
- 19.Zhao X, Li M, Xu Z. Detection of foodborne pathogens by surface enhanced raman spectroscopy. Front Microbiol. 2018;12(9):1236. doi: 10.3389/fmicb.2018.01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Deng Y, Peters BM, Li L, Li B, Chen L, Xu Z, Shirtliff ME. Transcriptomic analysis on the formation of the viable putative non-culturable state of beer-spoilage Lactobacillus acetotolerans. Sci Rep. 2016;6(1):1. doi: 10.1038/srep36753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Deng Y, Li L, Li B, Li Y, Zhou S, et al. Discovery and control of culturable and viable but non-culturable cells of a distinctive Lactobacillus harbinensis strain from spoiled beer. Sci Rep. 2018;8:11446. doi: 10.1038/s41598-018-28949-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Z, Li L, Chu J, Peters BM, Harris ML, Li B, Shi L, Shirliff ME. Development and application of loop-mediated isothermal amplification assays on rapid detection of various types of staphylococci strains. Food Res Int. 2012;47:166–173. doi: 10.1016/j.foodres.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao X, Wang L, Li Y, Xu Z, Li L, He X, Liu Y, Wang J, Yang L. Development and application of a loop-mediated isothermal amplification method on rapid detection of Pseudomonas aeruginosa strains. World J Microbiol Biotechnol. 2011;27(1):181–184. [Google Scholar]
- 24.Garibyan L, Avashia N. Research techniques made simple: polymerase chain reaction (PCR) J Investigative Dermatol. 2013;133(3):e6. doi: 10.1038/jid.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bustin SA. Why the need for qPCR publication guidelines?—The case for MIQE. Methods. 2010;50(4):217–226. doi: 10.1016/j.ymeth.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Li Y, Chu J, Xu Z, Zhong Q. Development and application of a simple loop-mediated isothermal amplification method on rapid detection of Listeria monocytogenes strains. Mol Biol Rep. 2012;39:445–449. doi: 10.1007/s11033-011-0757-7. [DOI] [PubMed] [Google Scholar]
- 27.Zhao X, Li Y, Chu J, Wang L, Shirtliff ME, He X, Liu Y, Wang J, Xu Z, Li L. Rapid detection of Vibrio parahaemolyticus strains and virulent factors by loop-mediated isothermal amplification assays. Food Sci Biotechnol. 2010;19:1191–1197. [Google Scholar]
- 28.Lin Shiqi, Li Lin, Li Bing, Zhao Xihong, Lin Chii-wann, Deng Yang, Zhenbo Xu. Development and evaluation of quantitative detection of N-epsilon-carboxymethyl-lysine in Staphylococcus aureus biofilm by LC-MS method. Basic Clin Pharmacol Toxicol. 2016;118:33. [Google Scholar]
- 29.Notomi T, Okayama H, Masubuchi HO, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28(12) [DOI] [PMC free article] [PubMed]
- 30.Miao Jian, Peters Brian M, Li Lin, Li Bing, Zhao Xihong, Zhenbo Xu, Shirtliff Mark E. Evaluation of ERIC-PCR for fingerprinting methicillin-resistant staphylococcus aureus strains. Basic Clin Pharmacol Toxicol. 2016;118(S1):33. [Google Scholar]
- 31.Becherer L, Borst N, Bakheit M, Frischmann S, Zengerle R, von Stetten F. Loop-mediated isothermal amplification (LAMP)–review and classification of methods for sequence-specific detection. Anal Methods. 2020;12(6):717–746. [Google Scholar]
- 32.Kimanya ME, Mamiro PRS, Van Camp J, Devlieghere F, Opsomer A, Kolsteren P, et al. Growth of Staphylococcus aureus and Bacillus cereus during germination and drying of finger millet and kidney beans. Int J Food Sci Technol. 2003;38(2):119–125. [Google Scholar]
- 33.Liu L, Xu R, Li L, Li B, Zhang X, Zhang X, et al. Correlation and in vitro mechanism of bactericidal activity on E coli with whey protein isolate during ultrasonic treatment. Microbial Pathogenesis. 2018;115:154–158. doi: 10.1016/j.micpath.2017.12.062. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Zhou R, Li L, Peters BM, Li B, Lin C, et al. Viable but non-culturable state and toxin gene expression of enterohemorrhagic Escherichia coli 0157 under cryopreservation. Res Microbiol. 2017;168(3):188–193. doi: 10.1016/j.resmic.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Deng Y, Liu J, Peters BM, Chen L, Miao J, Li B, et al. Antimicrobial resistance investigation on Staphylococcus strains in a Local Hospital in Guangzhou, China, 2001-2010. Microbial Drug Resist. 2015;21(1):102–104. doi: 10.1089/mdr.2014.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong N, Gui Z, Xu L, Huang J, Hu K, Gao Y, Zhang X, Xu Z, Su J, Li B. Solvent-free enzymatic synthesis of 1, 3-Diacylglycerols by direct esterification of glycerol with saturated fatty acids. Lipids Health Dis. 2013;12(1):1–7. doi: 10.1186/1476-511X-12-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen D, Wen S, Feng D, Xu R, Liu J, Peters BM, Su D, Lin Y, Yang L, Xu Z, Shirtliff ME. Microbial virulence, molecular epidemiology and pathogenic factors of fluoroquinolone-resistant Haemophilus influenzae infections in Guangzhou, China. Annals Clin Microbiol Antimicrobial. 2018;17(1):41. doi: 10.1186/s12941-018-0290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bao X, Yang L, Chen L, Li B, Li L, Li Y, et al. Analysis on pathogenic and virulent characteristics of the Cronobacter sakazakii strain BAA-894 by whole genome sequencing and its demonstration in basic biology science. Microb Pathog. 2017;109:280–286. doi: 10.1016/j.micpath.2017.05.030. [DOI] [PubMed] [Google Scholar]
- 39.Bao X, Yang L, Chen L, Li B, Li L, Li Y, et al. Virulent and pathogenic features on the Cronobacter sakazakii polymyxin resistant pmr mutant strain s-3. Microb Pathog. 2017;110:359–364. doi: 10.1016/j.micpath.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 40.Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG., Jr Staphylococcus aureus Infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28(3):603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Z, Xie J, Soteyome T, Peters BM, Shirtliff ME, Liu J, Harro JM. Polymicrobial interaction and biofilms between Staphylococcus aureus and Pseudomonas aeruginosa: an underestimated concern in food safety. Current Opinion Food Sci. 2019;1(26):57–64. [Google Scholar]
- 42.Miao Jian, Lin Shiqi, Soteyome Thanapop, Peters Brian M, Li Yanmei, Chen Huishan, Jianyu Su, Li Lin, Li Bing, Zhenbo Xu, Shirtliff Mark E, Harro Janette M. Biofilm formation of Staphylococcus aureus under food heat processing conditions-first report on CML production within biofilm. Sci Rep. 2019;9:1313. doi: 10.1038/s41598-018-35558-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Junyan, Xie Jinhong, Yang Ling, Chen Dingqiang, Peters Brian M, Zhenbo Xu, Shirtliff Mark E. Identification of the KPC plasmid pCT-KPC334: new insights on the evolution pathway of the epidemic plasmids harboring fosA3-blaKPC-2 genes. Int J Antimicrob Agents. 2018;52:510–512. doi: 10.1016/j.ijantimicag.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 44.Liu Junyan, Yang Ling, Chen Dingqiang, Peters Brian M, Li Lin, Li Bing, Zhenbo Xu, Shirtliff Mark E. Complete sequence of pBM413, a novel multi-drug-resistance megaplasmid carrying qnrVC6 and blaIMP-45 from Pseudomonas aeruginosa. Int J Antimicrob Agents. 2018;51:145–150. doi: 10.1016/j.ijantimicag.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Li L, Li B, Peters B, Deng Y, Zhenbo X, Shirtliff M. The viable but nonculturable state induction and genomic analyses of Lactobacillus casei BM-LC14617, a beer-spoilage bacterium. Microbiol Open. 2017;6(5):e506. doi: 10.1002/mbo3.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Junyan, Li Lin, Zhou Lizhen, Li Bing, Zhenbo Xu. Effect of ultrasound treatment conditions on Saccharomyces cerecisiae by response surface methodology. Microb Pathog. 2017;111:497–502. doi: 10.1016/j.micpath.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 47.Liu Junyan, Li Lin, Li Bing, Peters Brian M, Deng Yang, Zhenbo Xu, Shirtliff Mark E. Study on spoilage capability and VBNC state formation and recovery of Lactobacillus plantarum. Microb Pathog. 2017;110:257–261. doi: 10.1016/j.micpath.2017.06.044. [DOI] [PubMed] [Google Scholar]
- 48.Xie Jinhong, Yang Ling, Peters Brian M, Chen Lequn, Chen Dingqiang, Li Bing, Li Lin, Guangchao Yu, Zhenbo Xu, Shirtliff Mark E. A 16-year retrospective surveillance report on the pathogenic features and antimicrobial susceptibility of Pseudomonas aeruginosa isolated from Guangzhou representative of Southern China. Microb Pathog. 2017;110:37–41. doi: 10.1016/j.micpath.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 49.Liu Junyan, Deng Yang, Soteyome Thanapop, Li Yanyan, Jianyu Su, Li Lin, Li Bing, Shirtliff Mark E, Zhenbo Xu, Peters Brian M. Induction and recovery of the viable but nonculturable state of hop-resistance Lactobacillus brevis. Front Microbiol. 2018;9:2076. doi: 10.3389/fmicb.2018.02076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Junyan, Yang Ling, Hou Yuchao, Soteyome Thanapop, Zeng Bingbing, Jianyu Su, Li Lin, Li Bing, Chen Dingqiang, Li Yanyan, Aiwu Wu, Shirtliff Mark E, Harro Janette M, Zhenbo Xu, Peters Brian M. Transcriptomics study on Staphylococcus aureus biofilm under low concentration of ampicillin. Front Microbiol. 2018;9:2413. doi: 10.3389/fmicb.2018.02413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao Xihong, Zhixue Yu, Zhenbo Xu. Study the features of 57 confirmed CRISPR Loci in 38 Strains of Staphylococcus aureus. Front Microbiol. 2018;9:1591. doi: 10.3389/fmicb.2018.01591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Junyan, Li Lin, Peters Brian M, Li Bing, Chen Dingqiang, Zhenbo Xu, Shirtliff Mark E. Complete genome sequence and bioinformatics analyses of Bacillus thuringiensis strain BM-BT15426. Microb Pathog. 2017;108:55–60. doi: 10.1016/j.micpath.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 53.Liu Junyan, Li Lin, Li Bing, Peters Brian M, Zhenbo Xu, Shirtliff Mark E. First study on the formation and resuscitation of viable but nonculturable state and beer spoilage capability of Lactobacillus lindneri. Microb Pathog. 2017;107:219–224. doi: 10.1016/j.micpath.2017.03.043. [DOI] [PubMed] [Google Scholar]
- 54.Xie Jinhong, Peters Brian M, Li Bing, Li Lin, Guangchao Yu, Zhenbo Xu, Shirtliff Mark E. Clinical features and antimicrobial resistance profiles of important Enterobacteriaceae pathogens in Guangzhou representative of Southern China, 2001-2015. Microb Pathog. 2017;107:206–211. doi: 10.1016/j.micpath.2017.03.038. [DOI] [PubMed] [Google Scholar]
- 55.Zhenbo Xu, Xie Jinhong, Liu Junyan, Ji Lili, Soteyome Thanapop, Peters Brian M, Chen Dingqiang, Li Bing, Li Lin, Shirtliff Mark E. Whole-genome resequencing of Bacillus cereus and expression of genes functioning in sodium chloride stress. Microb Pathog. 2017;104:248–253. doi: 10.1016/j.micpath.2017.01.040. [DOI] [PubMed] [Google Scholar]
- 56.Miao Jian, Liang Yanrui, Chen Lequn, Wang Wenxin, Wang Jingwen, Li Bing, Li Lin, Chen Dingqiang, Zhenbo Xu. Formation and development of Staphylococcus biofilm: with focus on food safety. J Food Saf. 2017;7:e12358. [Google Scholar]
- 57.Miao J, Chen L, Wang J, Wang W, Chen D, Li L, Li B, Deng Y, Xu Z. Current methodologies on genotyping for nosocomial pathogen methicillin-resistant Staphylococcus aureus (MRSA) Microbial Pathog. 2017;1(107):17–28. doi: 10.1016/j.micpath.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 58.Bao X, Jia X, Chen L, Peters BM, Lin CW, Chen D, Li L, Li B, Li Y, Xu Z, Shirtliff ME. Effect of polymyxin resistance (pmr) on biofilm formation of Cronobacter sakazakii. Microbial Pathog. 2017;1(106):16–19. doi: 10.1016/j.micpath.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 59.Zhenbo Xu, Xie Jinhong, Peters Brian M, Li Bing, Li Lin, Guangchao Yu, Shirtliff Mark E. Longitudinal surveillance on antibiogram of important gram-positive pathogens in Southern China, 2001 to 2015. Microb Pathog. 2017;103:80–86. doi: 10.1016/j.micpath.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 60.Liu Junyan, Chen Dingqiang, Peters Brian M, Li Lin, Li Bing, Zhenbo Xu, Shirliff Mark E. Staphylococcal chromosomal cassettes mec (SCCmec): a mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb Pathog. 2016;101:56–67. doi: 10.1016/j.micpath.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 61.Guangchao Yu, Wen Wangrong, Peters Brian M, Liu Junyan, Ye Congxiu, Che Yuchuan, Liu Juzhen, Cao Kaiyuan, Zhenbo Xu, Shirtliff Mark E. First report of novel genetic array aacA4-bla(IMP-25)-oxa30-catB3 and identification of novel metallo-beta-lactamase gene bla(IMP25): a Retrospective Study of antibiotic resistance surveillance on Psuedomonas aeruginosa in Guangzhou of South China, 2003-2007. Microb Pathog. 2016;95:62–67. doi: 10.1016/j.micpath.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 62.Liu Junyan, Li Lin, Peters Brian M, Li Bing, Deng Yang, Zhenbo Xu, Shirtliff Mark E. Draft genome sequence and annotation of Lactobacillus acetotolerans BM-LA14527, a beer-spoilage bacteria. FEMS Microbiol Lett. 2016;363:18. doi: 10.1093/femsle/fnw201. [DOI] [PubMed] [Google Scholar]
- 63.Zhenbo Xu, Hou Yuchao, Peters Brian M, Chen Dingqiang, Li Bing, Li Lin. Chromogenic Media for MRSA Diagnostics. Mol Biol Rep. 2016;43:1205–1212. doi: 10.1007/s11033-016-4062-3. [DOI] [PubMed] [Google Scholar]
- 64.Zhenbo Xu, Liang Yanrui, Lin Shiqi, Chen Dingqiang, Li Bing, Li Lin, Deng Yang. Crystal violet and XTT assays on Staphylococcus aureus biofilm quantification. Curr Microbiol. 2016;73:474–482. doi: 10.1007/s00284-016-1081-1. [DOI] [PubMed] [Google Scholar]
- 65.Deng Y, Liu J, Li L, Fang H, Tu J, Li B, Liu J, Li H, Xu Z. Reduction and restoration of culturability of beer-stressed and low-temperature-stressed Lactobacillus acetotolerans strain 2011-2018. Int J Food Microbiol. 2015;3(206):96–101. doi: 10.1016/j.ijfoodmicro.2015.04.046. [DOI] [PubMed] [Google Scholar]
- 66.Xu Z, Gui Z, Zhao X, Zhang Y, He X, Li W, Yang L, Li L, Li B, Su J, Wang J. Expression and purification of gp41-gp36 fusion protein and application in serological screening assay of HIV-1 and HIV-2. Afr J Microbiol Res. 2012;6(33):6295–6299. [Google Scholar]
- 67.Zhenbo Xu, Li Lin, Shi Lei, Shirliff Mark E. Class 1 integron in staphylococci. Mol Biol Rep. 2011;38:5261–5279. doi: 10.1007/s11033-011-0676-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhenbo Xu, Lin Li, Xihong Zhao, Jing Chu, Bing Li, Lei Shi, Jianyu Su. Mark E. Shirtliff. Development and application of a novel multiplex polymerase chain reaction (PCR) assay for rapid detection of various types of staphylococci strains. Afr J Microbiol Res. 2011. 5:1869-1873
- 69.Xu Z, Li L, Shirtliff ME, Peters BM, Peng Y, Alam MJ, Yamasaki S, Shi L. First report of class 2 integron in clinical Enterococcus faecalis and class 1 integron in Enterococcus faecium in South China. Diagnostic Microbio Infect Dis. 2010;68(3):315–317. doi: 10.1016/j.diagmicrobio.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 70.Xu Z, Li L, Alam MJ, Zhang L, Yamasaki S, Shi L. First confirmation of integron-bearing methicillin-resistant Staphylococcus aureus. Curr Microbiol. 2008;57(3):264–268. doi: 10.1007/s00284-008-9187-8. [DOI] [PubMed] [Google Scholar]
- 71.Zhenbo Xu, Shi Lei, Zhang Chao, Zhang Liyan, Li Xinhui, Cao Yicheng, Li Lin, Yamasaki Shinji. Nosocomial infection caused by class 1 integron-carrying Staphylococcus aureus in a hospital in South China. Clin Microbiol Infect. 2007;13:980–984. doi: 10.1111/j.1469-0691.2007.01782.x. [DOI] [PubMed] [Google Scholar]
- 72.Zhenbo Xu, Luo Yuting, Soteyome Thanapop, Lin Chii-Wann, Xingyong Xu, Mao Yuzhu, Jianyu Su, Liu Junyan. Rapid detection of food-borne Escherichia coli O157:H7 with visual inspection by crossing priming amplification (CPA) Food Anal Methods. 2020;13:474–481. [Google Scholar]
- 73.Xu Z, Li L, Shirtliff ME, Alam MJ, Yamasaki S, Shi L. Occurrence and characteristics of class 1 and 2 integrons in Pseudomonas aeruginosa isolates from patients in southern China. J Clin Microbiol. 2009;47(1):230–234. doi: 10.1128/JCM.02027-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu Z, Hou Y, Qin D, Liu X, Li B, Li L, et al. Evaluation of current methodologies for rapid identification of methicillin-resistant Staphylococcus aureus strains. Basic Clin Pharmacol Toxicol. 2016;118:33–39. [Google Scholar]
- 75.Xu Z, Shi L, Alam MJ, Li L, Yamasaki S. Integron-bearing methicillin-resistant coagulase-negative staphylococci in South China, 2001-2004. FEMS Microbiol Lett. 2008;278(2):223–230. doi: 10.1111/j.1574-6968.2007.00994.x. [DOI] [PubMed] [Google Scholar]
- 76.Xu Z, Li L, Shirtliff ME, Peters BM, Li B, Peng Y, et al. Resistance class 1 integron in clinical methicillin-resistant Staphylococcus aureus strains in southern China, 2001–2006. Clin Microbiol Infect. 2011;17(5):714–718. doi: 10.1111/j.1469-0691.2010.03379.x. [DOI] [PubMed] [Google Scholar]
- 77.You Z, Xu Z, Shirtliff ME, Yu G, Zhao X, Shi L, Li B, Su J, Li L. Methicillin-resistance Staphylococcus aureus detection by an improved rapid PCR assay. Afr J Microbiol Res. 2012;6(6):7131–7133. [Google Scholar]
- 78.Miao J, Wang W, Xu W, Su J, Li L, Li B, et al. The fingerprint mapping and genotyping systems application on methicillin-resistant Staphylococcus aureus. Microb Pathog. 2018;125:246–251. doi: 10.1016/j.micpath.2018.09.031. [DOI] [PubMed] [Google Scholar]
- 79.Miao J, Chen L, Wang J, Wang W, Chen D, Li L, et al. Evaluation and application of molecular genotyping on nosocomial pathogen-methicillin-resistant Staphylococcus aureus isolates in Guangzhou representative of Southern China. Microb Pathog. 2017;107:397–403. doi: 10.1016/j.micpath.2017.04.016. [DOI] [PubMed] [Google Scholar]