Abstract
The spread of SARS-CoV-2 is a global concern that has taken a toll on entire human health. Researchers across the globe have been working in devising the strategies to combat this dreadful disease. Studies focused on genetic variability help design effective drugs and vaccines. Considering this, the present study entails the information regarding the genome-wide mutations detected in the 108 SARS CoV-2 genomes worldwide. We identified a few hypervariable regions localized in orf1ab, spike, and nucleocapsid gene. These nucleotide polymorphisms demonstrated their effect on both codon usage as well as amino acid usage pattern. Altogether the present study provides valuable information that would be helpful to ongoing research on 2019-nCoV vaccine development.
Keywords: Mutations, Hypervariable region, Amino acids, Genetic variations, S-protein
1. Introduction
The novel coronavirus of 2019 (2019-nCoV/SARS CoV-2), has affected nearly every corner of the world, affecting human health and causing enormous human loss (Seah and Agrawal, 2020). Originating in Hubei province, China, it causes severe respiratory problems in mammals (Lorusso et al., 2020; Pal et al., 2020). This unprecedented epidemic has already been declared an international public health emergency by the World Health Organization (WHO). At the time of writing (12 June 2020), the confirmed cases of this rapidly spreading coronavirus globally exceeded 12.6 million. Currently, there is no specific treatment for nCoV (Lu, 2020). Developing a new drug or vaccine is a major challenge; therefore, other antiviral drugs are fast-tracked for testing against nCoV (Li et al., 2020). Recent studies have shown that nCoV is mutating and evolving frequently (Zhang et al., 2020). Worldwide efforts were made to sequence the entire SARS CoV-2 genome to study genetic variation and its evolutionary origin (Ceraolo and Giorgi, 2020). Moreover, docking studies simultaneously screening antagonist against SARS-COV-2 coronaviruses (Yu et al., 2020).
Genome-level analysis will provide information on host responses and may improve vaccine development. SARS CoV-2 is an RNA virus with higher rate of mutation that may have facilitated the widespread of this virus across different weather conditions (Pachetti et al., 2020). Information regarding the composition of amino acids and the codon usage pattern may help gain more insight into gene expression or other selection pressures. In this study, genetic variations of SARS CoV-2 were analyzed based on available data. Here, we reported the emerging genetic variants that might have developed over time in humans since the virus became pandemic. To assess the genetic variation, one hundred and sixty four complete genome sequences of SARS CoV-2 were retrieved from GISAID database (Shu and McCauley, 2017). The genome sequence of Wuhan 2019-nCoV was used as a reference to identify the genetic diversity and mutations between the 2019-nCoV sequences from different countries. The hypervariable 2019-nCoV genomic region undergoing mutations were assessed for their tendency to alter the amino acid sequence in the respective polypeptide sequences.
2. Materials and methods
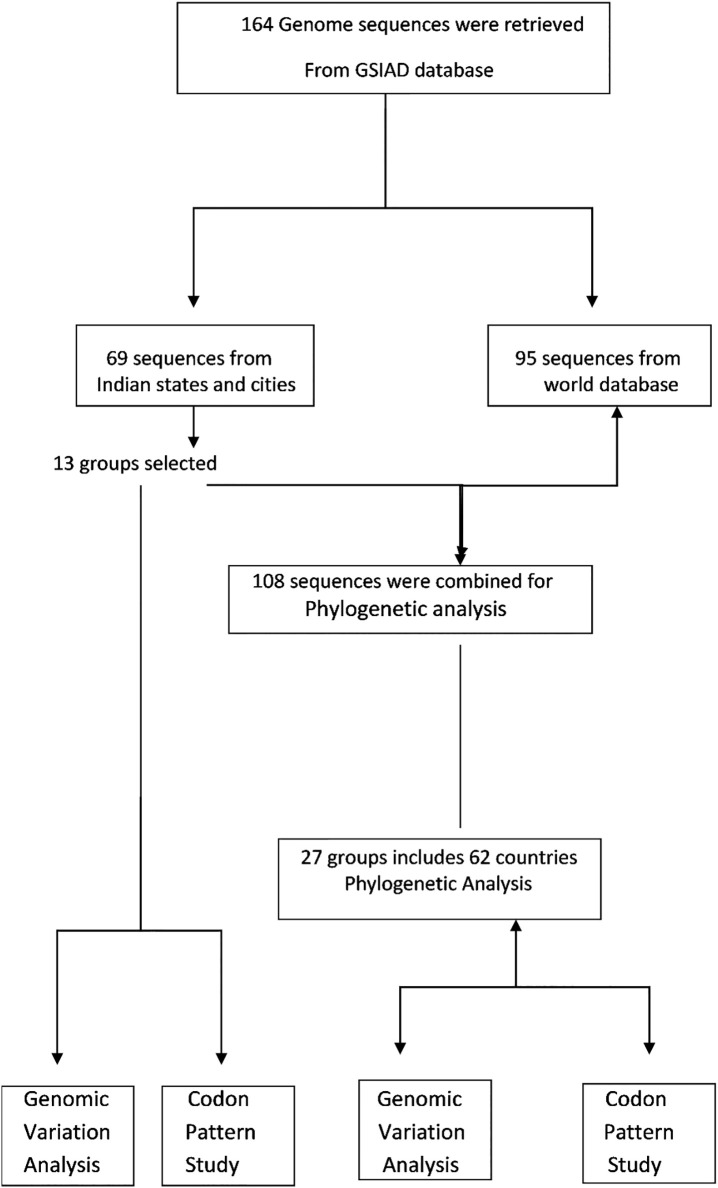
SARS Cov-2 genomic sequences were retrieved from GISAID database (Global Initiative on Sharing Avian Influenza Data) (Shu and McCauley, 2017). As of December 2019, GISAID became established as a database for the coronavirus. A total of 164 genomic sequences were collected for the present analysis, comprising 69 sequences from Indian states (randomly selected) and 95 genomic sequences from different continents, including 62 countries (Data was retrieved on 4th May 2020). The RefSeq genome (NC_045512) from Wuhan was used as the reference genome. The pipeline of the workflow is shown in Fig. 1 . Here, we masked the initial 64 nucleotides and the sequences exceeding the position 29686 while performing multiple sequence alignment. ClustalW software was used for sequence alignment (Guo and Sun, 2000). The aligned sequences were used to create phylogenetic tree using MEGA X software (Kumar et al., 2018). The sequence data were analyzed using neighbor-joining method. To assess the robustness of individual nodes, the bootstrap method was employed to test the phylogeny with 1000 replicates (Soltis and Soltis, 2003). Sequence handling and data processing were carried out in Bioedit software (Hall et al., 2011). We only considered the mutations present in at least 3 different genomes in a given position for amino acid substitution analysis. An in-house python script was developed to extract the amino acid substitutions in each position. AutoDock 4.2 software was used to dock the protein structures with their potential ligands (Rizvi et al., 2013). Codon usage pattern of individual genomic sequence was determined based on RSCU analysis. INCA 2.1 software was used for codon usage as well as amino acid usage bias analysis (Supek and Vlahoviček, 2004).
Fig. 1.
Flow chart representing the overall work done from retrieval to analysis of genomic sequences.
3. Results
3.1. Phylogenetic analysis
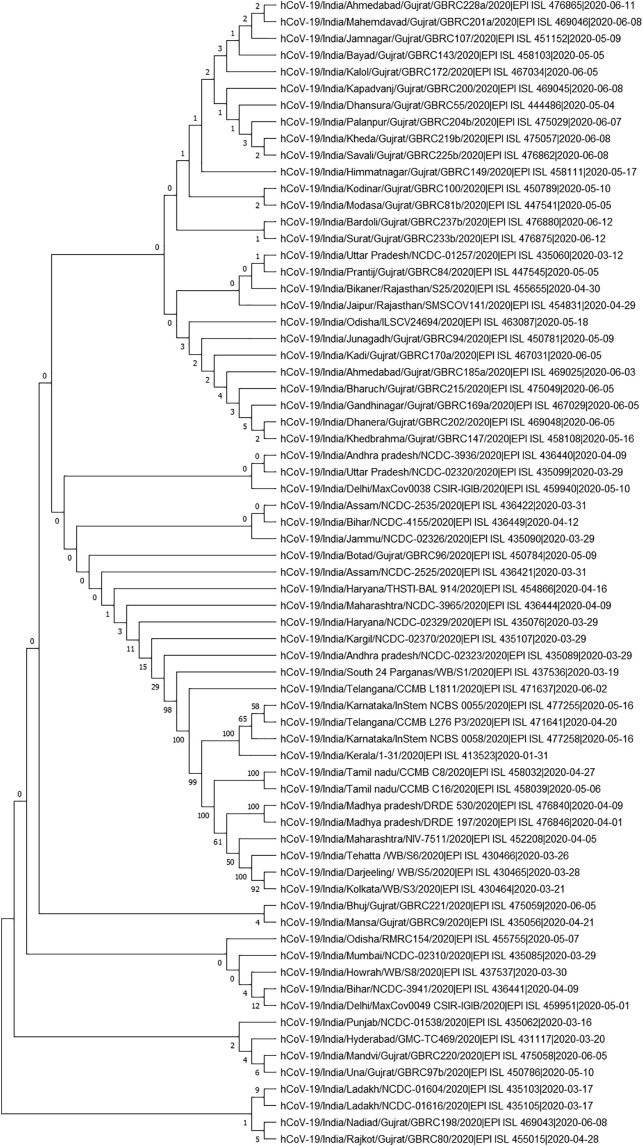
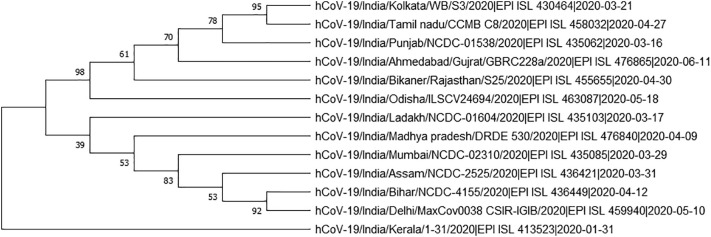
GSAID database was used to fetch 164 SARS CoV-2 genomic sequences which comprised of 69 genomic sequences from India and 95 from other countries of the World. Genomic sequences from different Indian states have been phylogenetically analyzed to gain insight into the genetic variation prevalent in India (Fig. 2) Based on the phylogenetic tree we grouped the SARS CoV-2 genome sequences of Indian states into 13 major groups (Fig. 3). These 13 representative groups (states) of India were then phylogenetically analyzed along with SARS CoV-2 genomes (95 coronavirus sequences) obtained from different countries around the world. Further, on the basis of phylogenetic analysis, 27 major groups were created which included 62 countries (Fig. 4 & supplementary file 1).
Fig. 2.
Phylogenetic tree of 69 SARS CoV-2 genomes representing different cities/states of India.
Fig. 3.
Phylogenetic tree of SARS CoV-2 genome sampled from 13 selected Indian states.
Fig. 4.
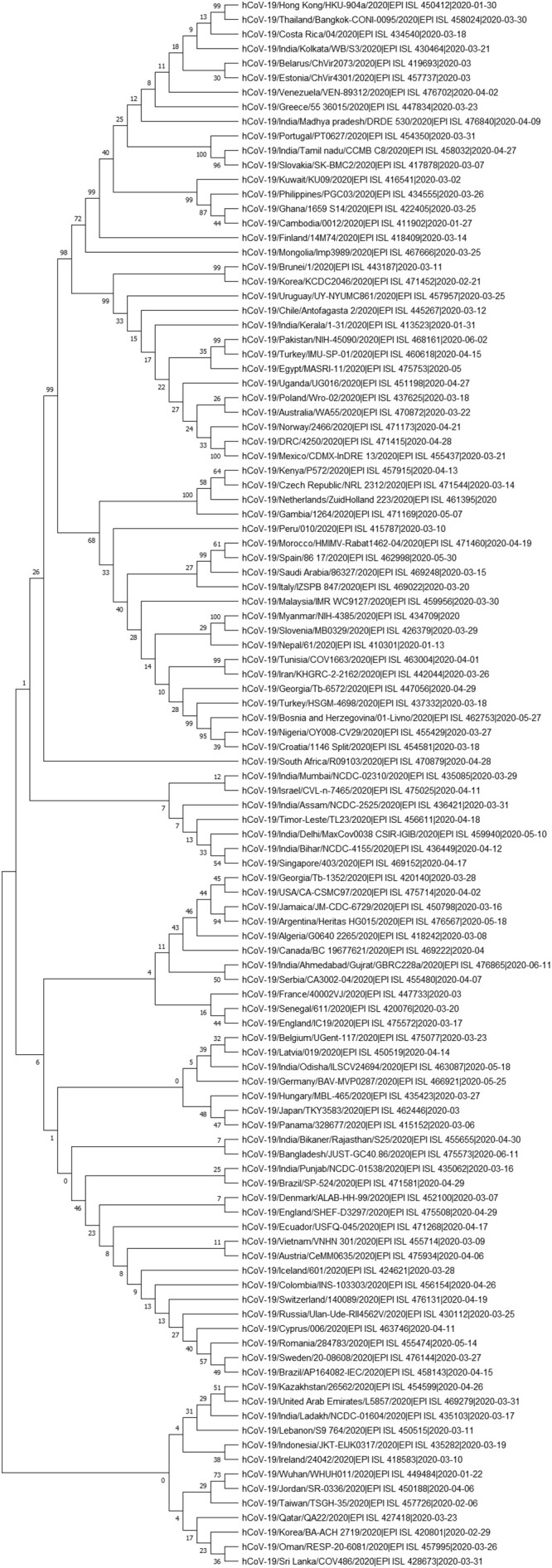
Phylogenetic tree constructed using 108 (95 global and 13 Indian states) SARS CoV-2 genomes (method: Maximum-likelihood).
3.2. Analysis of genetic variation across the world
To investigate the genetic variation in the novel coronavirus, we compared the SARS-CoV-2 genomic sequences from 62 countries with the Wuhan SARS-CoV-2 (Wuhan SARS-CoV-2 was used as reference genome). Here we observed sequence conservation of 99.9% similar to previous findings (Lu et al., 2020). In comparison to the reference sequence, 83 single nucleotide polymorphisms (SNPs) were observed at various genomic locations in the coronavirus genome obtained from different countries. Among 83 SNPs, maximum mutations were detected in orf1ab gene (37) followed by genomic region (26). The changes in the nucleotide have altered 32 amino acids whereas 25 mutations were recorded as synonymous mutations (Table 1).
Table 1.
Single Nucleotide Polymorphism and amino acid changes identified in the entire Coronavirus genome (World Data).
| Genes | Total SNP | SNP led to change in amino acid | Synonymous mutations |
|---|---|---|---|
| orf1ab | 37 | 16 | 21 |
| spike | 8 | 4 | 4 |
| Orf3a | 4 | 4 | 0 |
| Memberane | 3 | 3 | 0 |
| Orf8 | 3 | 3 | 0 |
| Nucleocapsid | 2 | 2 | 0 |
| Genomic | 26 | ||
| Total | 83 | 32 | 25 |
The description of genetic variation occurred in SARS CoV-2 genome with respect to each country (Selected for the present study) has been provided in supplementary file 2. In addition, gene-wise mutational information for the selected countries have also been given (supplementary file 3). For the present study, we focused on those regions which showed nucleotide variations in at least more than 3 genomic sequences analyzed and are thus considered as hypervariable regions (Table 2). Hypervariable regions at 1068 and 11092 genomic position falls under the orf1ab gene (Fig. 5). At the amino acid level the mutation at 1068 (C → T mutation), and 11092 (G → T mutation) resulted in alteration of amino acid from threonine (ACT) to isoleucine (ATT) and asparagine (TTG) to phenylalanine (TTT) respectively. While at 3046 position, we observed a synonymous mutation. Another hypervariable region was located at genomic position 23412, this area encodes spike protein. Here we observed G instead of A, which leads to a shift in the encoded protein sequence i.e. aspartate (GAT) to glycine (GGT). In addition, variation at genomic position 25572 was localized in the gene orf3a. At this location, G → T mutation has resulted in glutamine (CAG) to histidine (CAT) substitution. Likewise, nucleotide (C → T) alteration was also observed in the nucleocapsid gene at the genomic position 28230 which resulted in a change in the amino acid i.e. proline (CCC) to glutamate (CTC).
Table 2.
Hypervariable regions identified from the selected SARS CoV-2 genome sequences (World Data).
| Genomic position | Gene position | Nucleotide in reference | Change nucleotide | Amino acid and codon | Change codon and amino acid | Countries involved |
|---|---|---|---|---|---|---|
| 250 | Genomic | C | T | NA | NA | Belarus, Estonia, Slovakia, Tamil Nadu, Portugal, Ghana, Philippines, Cambodia, Pakistan, Turkey, Egypt, Mexico, Norway, Kenya, Czech Republic, Netherlands, Morocco, Spain, Saudi Arabia, Italy, Bosnia, Nigeria, Croatia, Georgia, USA, Jamaica, Argentina, Serbia, Ahmedabad (India) |
| 1068 | orf1ab | C | T | ACC (Threonine) | ATC (Isoleucine) | Bosnia, Croatia, Nigeria, USA, Georgia, Jamaica, Argentina |
| 3046 | orf1ab | C | T | TTC and Phenylalanine | TTT and Phenylalanine | Belarus, Estonia, Slovakia, Tamil Nadu, Portugal, Ghana, Philippines, Cambodia, Pakistan, Turkey, Egypt, Mexico, Norway, Kenya, Czech Republic, Netherlands, Morocco, Spain, Saudi Arabia, Italy, Bosnia, Nigeria, Croatia, Georgia, USA, Jamaica, Argentina, Serbia, Ahmedabad (India) |
| 11092 | orf1ab | G | T | TTG (Asparagine) | TTT (Phenylalanine) | Pakistan, Turkey, Egypt, Delhi (India), Bihar (India), Singapore, UAE, Kazakhstan |
| 23412 | Spike | A | G | GAT (Aspartate) | GGT (Glycine) | Belarus, Estonia, Slovakia, Tamil Nadu, Portugal, Ghana, Philippines, Cambodia, Pakistan, Turkey, Egypt, Mexico, Norway, Kenya, Czech Republic, Netherlands, Morocco, Spain, Saudi Arabia, Italy, Bosnia, Nigeria, Croatia, Georgia, USA, Jamaica, Argentina, Serbia, Ahmedabad (India) |
| 25572 | orf3a | G | T | CAG (Glutamine) | CAT (Histidie) | Belarus, Estonia, Slovakia, Tamil Nadu, Portugal, Ghana, Philippines, Cambodia, Pakistan, Turkey, Egypt, Mexico, Norway, Kenya, Czech Republic, Netherlands, Morocco, Spain, Saudi Arabia, Italy, Bosnia, Nigeria, Croatia, Georgia, USA, Jamaica, Argentina, Serbia, Ahmedabad (India) |
| 28320 | Nucleocapsid | C | T | CCC (Proline) | CTC (Glutamate) | Pakistan, Turkey, Egypt, Israel, Mumbai (India), Delhi (India), Bihar (India), Singapore |
Fig. 5.
Multiple sequence alignment of SARS CoV-2 genomes (65 countries including 5 Indian states) representing the hypervariable regions across the entire genome. (Mutations at 1068 bp, 3046 bp and 11,092 bp genomic positions are localized in gene orf1ab), (Mutations at 23412 bp and 25,572 bp are localized in spike gene), (Mutations at 28320 bp are localized in Nucleocapsid (N) gene).
3.3. Analysis of genetic variation across Indian states
Genomic sequences of SARS CoV-2 obtained from different Indian states were also compared with the reference genome (Wuhan) to detect the genetic variation. A total of 47 SNPs were detected in 2019 nCoV-2 genomes obtained from Indian states. The implication of the nucleotide polymorphism has led to alterations in 21 amino acid while 16 mutations were recorded as synonymous (Table 3). State wise (selected states) description of genetic variations across the genome has been given in supplementary file 4. Additionally, the number of mutations occurred in all the selected states with respect to their gene location has also been tabulated in supplementary file 5.
Table 3.
Single Nucleotide Polymorphism and subsequent amino acid changes identified in the SARS CoV-2 genome sequences retrieved from selected states of India.
| Genes | Total SNP | SNP led to chane in amino acid | Synonymous mutations |
|---|---|---|---|
| orf1ab | 24 | 14 | 10 |
| spike | 8 | 4 | 4 |
| Orf3a | 2 | 1 | 1 |
| Memberane | 1 | 0 | 1 |
| Orf8 | 1 | 1 | 0 |
| Nucleocapsid | 1 | 1 | 0 |
| Genomic | 10 | ||
| Total | 47 | 21 | 16 |
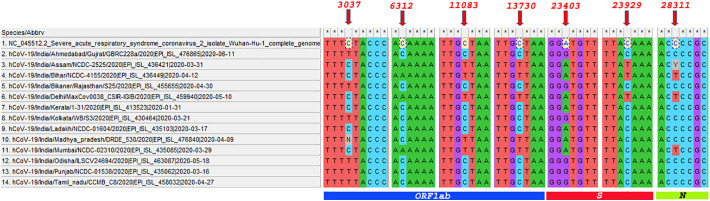
For the present investigation, we have described the mutations that took place at hypervariable regions involving 3 or more than 3 SARS CoV-2 genomes of Indian states (Table 4). We found four hypervariable genomic regions at positions 3037, 6312, 11083, and 13730 corresponding to orf1ab gene. The alteration in the nucleotide at position 3037 resulted in synonymous mutation (Fig. 6). While at position 6312 replacement of nucleotide led to the transition in amino acid from threonine (ACA) to lysine (AAA). Similarly, at 1083 genomic position, there was a substitution of leucine (TTG) by phenylalanine (TTT) due to G → T mutation. At 13730 position, C → T mutation caused the alanine (GCT) to be changed to valine (GTT) in the polypeptide chain. Genetic variability was also identified in the genomic region of the spike gene at 23403 and 23929 locations. At 23403, non-synonymous mutation (GGA by GGT, i.e. AG mutation) was found forcing aspartate to alter in the final protein sequence by glycine. Likewise, amino acid modification from methionine (TAC) to tyrosine (TAT) occurred at 23929 position due to C → T mutation. Non-synonymous mutation was also observed in the nucleocapsid gene at 28311 genomic position that causes C → T mutation and results in the replacement of amino acid from glutamate (CTC) to proline (CCC).
Table 4.
Hypervariable regions identified from SARS CoV-2 genome sequences retrieved from selected states of India.
| Genomic position | Gene position | Nucleotide in reference | Change nucleotide | Amino acid and codon | Change codon and amino acid | States involved |
|---|---|---|---|---|---|---|
| 241 | Genomic | C | T | NA | NA | Ahmedabad, Rajasthan, Kolkatta, Odisha, Punjab, Tamil Nadu |
| 3037 | orf1ab | C | T | TTC (Phenylalanine) | TTT (Phenylalanine) | Ahmedabad, Rajasthan, Kolkata, Odisha, Punjab, Tamil Nadu |
| 6312 | orf1ab | C | A | ACA Threonine | AAA and Lysine | Assam, Bihar, Delhi, Mumbai |
| 11083 | orf1ab | G | T | TTG (Leucine) | TTT (Phenylalanine) | Assam, Bihar, Delhi, Ladakh, Mumbai |
| 13730 | orf1ab | C | T | GCT (Alanine) | GTT (Valine) | Assam, Bihar, Delhi, MP, Mumbai |
| 23403 | Spike | GAT (Aspartic acid) | GGT (Glycine) | Ahmedabad, Rajasthan, Kolkata, Odisha, Punjab Tamil nadu | ||
| 23929 | Spike | C | T | TAC (Tyrosine) | TAT (Tyrosine) | Assam, Bihar, Delhi, |
| 28311 | Nucleocapsid | C | T | CCC (Proline) | CTC (Glutamate) | Bihar, Delhi, Mumbai |
Fig. 6.
Multiple sequence alignment of SARS CoV-2 genomes from 13 selected states of India representing the hypervariable regions across the entire genome (Mutations at 3037 bp, bp and 6312 bp, 11,083 bp, 13,730 bp genomic positions are localized in gene orf1ab), (Mutations at 23403 bp and 23,929 bp are localized in spike gene), (Mutation at 28311 bp are localized in Nucleocapsid (N) gene). Initially we retrieved two sequences from each Indian states, which was grouped into 13 states based on their sequence similarity.
3.4. Worldwide: codon usage and amino acid usage bias
Genome of SARS CoV-2 sequenced from 62 countries (27 groups based on phylogeny) were analyzed for codon usage trend and amino acid composition. Based on the similarity in the pattern of codon usage and amino acid composition among 27 groups, these were further assembled into 3 sub-groups A, B, and C. Groups 1 to 4 formed sub-group A, Groups 5–7 clustered into sub-group B and Groups 8 onward constituted sub-group C. We observed that coronavirus genome in all the 3 sub-groups showed a similar pattern in codon usage with respect to 7 amino acids namely Cys, Glu, Phe, Ileu, Lys, Leu, and Gln with preference towards U/A ending codons. However, differences in the codon usage pattern was observed for 11 amino acids among the 3 sub-groups. SARS CoV-2 genome of sub-group A showed a tendency to use U-ending codons for 10/11 amino acids except for arginine (AGG). Genomes from sub-group C also preferred to use U-ending codons for 8/11 amino acids except Pro, Arg and Thr that tends to use A-ending codons. Whereas the coronavirus genomes of sub-group B displayed a distinct pattern of codon usage by choosing A and C ending codons. Moreover, uniqueness in the use of G-ending codon for valine was also detected in the genomes of sub-group B. The data has been provided in (supplementary file 6). Amino acid composition analysis revealed that leucine was predominantly present among all the three sub-groups though differences were observed in the usage frequency. Sub-group B showed maximum frequency (0.16) as compared to sub-group A (0.12) and sub-group C (0.08) (supplementary file 7).
3.5. Indian States: codon usage and amino acid usage bias
Coronavirus genomes from 13 selected Indian states (Based on phylogeny) have been studied for their variability in codon usage. Diversity in the usage of synonymous codons was observed in the genomes sequenced from different states. We noticed that nine states (Ahmedabad, Assam, Bihar, Rajasthan, Delhi, Ladakh, Mumbai, Odisha and Punjab) exhibited identical trends in codon usage. Genome sequence from the states Madhya Pradesh (MP) and Tamil Nadu (TN) showed a similar trend, while Kerala and Kolkata states showed a distinct pattern compared to other states. We also observed that SARS CoV-2 genomes from all states favors U-ending codons to encode majority of the amino acids except Leu, Lys, Gln, Arg, Ser, and Thr, which preferred to be coded by A-ending codons. Biasness in codon selection was observed with respect to three amino acids Ser, Tyr and Val in the genome from different states Genomeic sequences from Kerala, Kolkata, MP and TN prefer to use UCA codon to encode serine, while genomes from other states tend to use AGU codon. Interestingly, genomes sequenced from Kerala showed a unique pattern in tyrosine (UAC) and valine (GUG) codon selection as compared to other strains isolated from India (supplementary file 8).
Compositional analysis of amino acids showed the dominance of leucine in all the 2019-nCoV genomes analyzed. However, variability in the frequency of leucine usage was observed among the genomes retrieved from different states. Kerala showed the highest frequency (0.16) followed by Kolkata, MP, TN (0.12) and rest of the states (0.08). It has been observed that 6-fold and 4-fold degenerative amino acids were more common compared to 2-fold amino acids. Glu, Asp and Pro were among the lowest frequency amino acids in all the coronavirus genomes (supplementary file 9).
4. Discussion
Rapid evolution and spread of the corona virus across the world intrigued us to understand the genetic variation in the SARS CoV-2 genome. Our findings also revealed the existence of high substitution rates in 2019-nCoV genomes sequenced from different parts of the world. Moreover, phylogenetic analysis further confirmed the migratory context of 2019-nCoV and its spread. The hypervariable regions identified in the SARS CoV-2 genes (orf1ab, S and nucleocapsid N gene) also supports the constant evolution of this virus across the world. In orf1ab gene, we found two hypervariable amino acid changing region (1068 and 11092 genomic locations) worldwide and three hypervariable regions (6312, 11083, and 13730 genomic locations) in Indian states when compared with the Wuhan strain. Single nucleotide polymorphisms were observed highest in Orf1ab. The mutations reported in the Orf1ab gene will be of interest to investigate the clinical function of this protein in Covid-19.
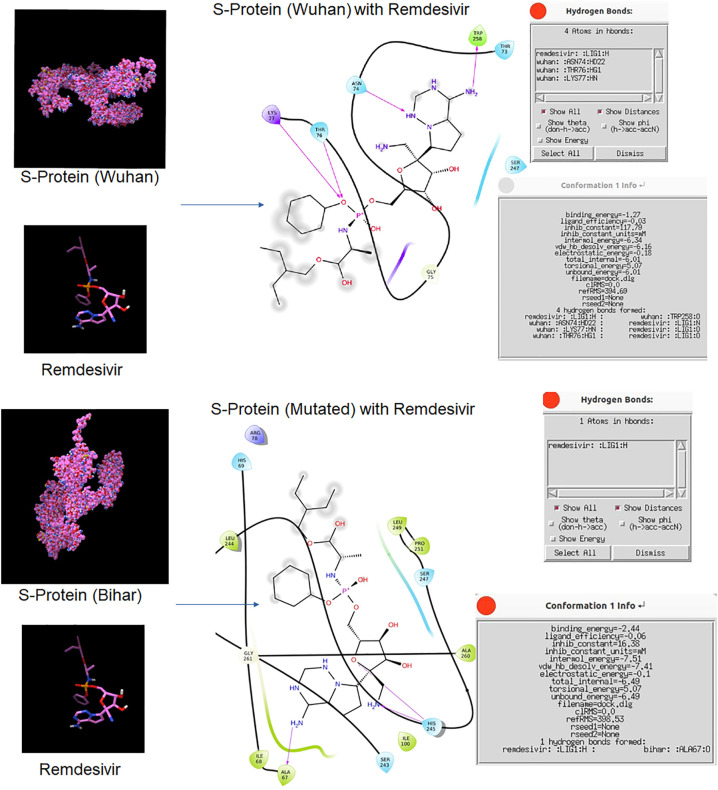
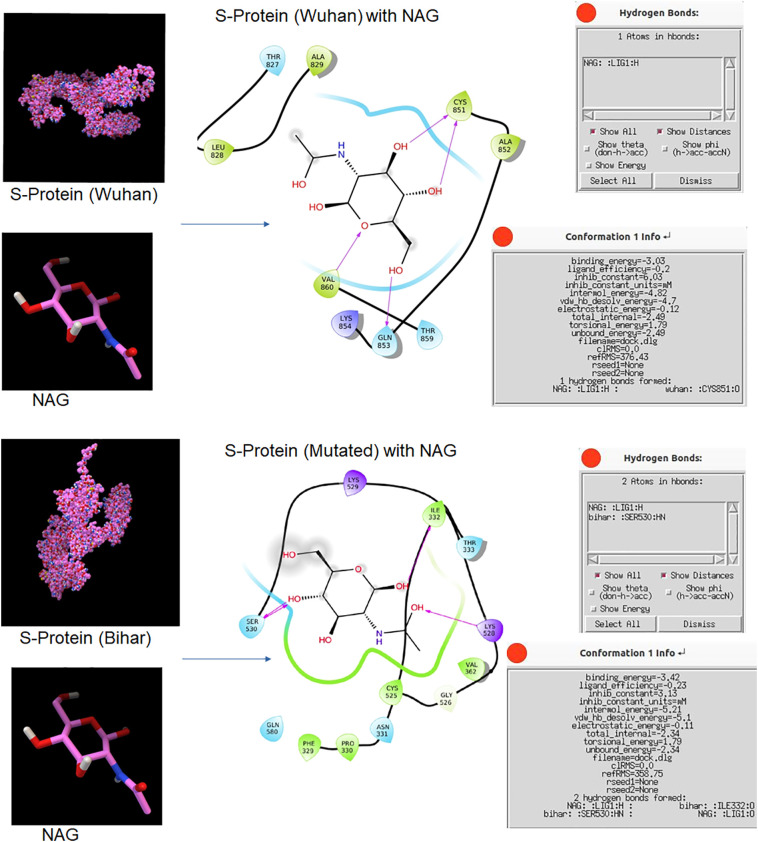
In addition Nucleocapsid N gene also showed a hypervariable region where proline was replaced by glutamate due to mutation. The N protein plays a key role in virion assembly (McBride et al., 2014). Therefore, this hypervariable region might have influenced its assembly efficiency and thus pathogenicity. Similarly, we observed a hypervariable region in S protein where (due to mutation) aspartate was replaced by glycine. Across the Indian states, we found another non-synonymous mutation in S protein where methionine was replaced by tyrosine. Non-synonymous mutation affects the overall conformation of proteins. It has been reported earlier that S protein and ACE2 genes play a key role in the establishment of the pathogenesis of coronavirus (Koyama et al., 2020; Phan, 2020). The non-synonymous mutation in spike protein (particularly aspartate to glycine) has been implicated with increase transmissibility of the virus by enhancing its affinity towards ACE-2 receptors (Korber et al., 2020; Raghav et al., 2020; Zhang et al., 2020). In addition to its increased infectivity it has been implied for causing high viral load (Korber et al., 2020). Furthermore, we observed altered binding confirmation when we docked the mutated S protein with remdesivir and NAG compared to the Wuhan S protein (Fig. 7, Fig. 8 ). This altered amino acid might have made a contribution towards the pathogenesis and transmission of 2019-nCoV.
Fig. 7.
Docked structure of S protein with Remdesivir.
Fig. 8.
Docked structure of S protein with NAG.
Since the COVID-19 outbreak in Dec 2019, plethora of studies have reported the presence of SNPs in different genomic locations. Here we observed 37 SNPs at different genomic locations encoding ORF1ab gene. Genome wide comparison showed that Denmark, England and Ahmedabad (India) has the maximum mutation in ORF1ab gene compared to all the countries analyzed. Banerjee and colleagues reported 14 SNPs in the ORF1ab gene (Banerjee et al., 2020). Similarly, Ceraolo and Giorgi (2020) revealed the presence of one hypervariable region in ORF1ab. Previous research on S protein isolated from India showed the presence of single mutation when compared with the Wuhan strain (Saha et al., 2020). We observed, 2 noteworthy mutation in the genomic locations corresponding to S protein in SARS CoV-2 genome from Tunisia and Iran. Similar findings were observed for the following Indian states namely Ahmedabad, Bihar, Rajasthan, Kolkata and Punjab. In addition to the mutation in the coding region we also detected 26 mutations in different genomic locations of SARS-CoV-2.
Furthermore, genome-wide codon usage comparison between Indian sates revealed the bias in codon selection to encode three amino acids, namely serine, tyrosine, and valine. Likewise, we observed eleven amino acids namely, Ala, Arg, Asn, Asp, Gly, His, Pro, Ser, Thr, Tyr, and Val showed variation in codon selection between world genomes divided into three sub-groups. These findings, which reflect substantial differences in the use of codon between the 2019-nCoV sequences, will be useful for assessing the adaptation, evolution of a host-virus and are therefore of interest to vaccine design strategies.
In conclusion, our analysis confirms high variability among 2019-nCoV sequenced genomes, highlighting hypervariable positions within three key protein-coding regions. Such variability in proteins might have affected the patient's clinical outcomes because the viral genome that infects them is slightly different. Nevertheless, there is a chance that these mutational variants might have modulated the disease's spread. Our results provide positive light on the prospect of developing 2019-nCoV therapy for patients from different locations.
The following are the supplementary data related to this article.
Assemblage of SARS-CoV-2 genomes obtained from 62 countries into 27 groups based on phylogenetic analysis. RSCU score of individual genomes were presented.
Analysis of genetic variation prevalent in SARS-CoV-2 genome sequences retrieved from selected countries across the world.
Gene wise mutational information on SARS-CoV-2 genome sequences retrieved from selected countries across the world.
Analysis of single nucleotide polymorphism prevalent in SARS-CoV-2 genome sequences retrieved from selected states of India.
Gene wise mutational information on SARS-CoV-2 genome sequences retrieved from selected states of India.
Comparative codon usage pattern analysis of SARS-CoV-2 genome sequences between three sub-groups A, B and C (Sub-group A, Sub-group B and Sub-group C are constituted from 27 groups based on similar codon usage pattern).
Comparative amino acid pattern analysis of SARS-CoV-2 genome sequences between three sub-groups A, B and C (Sub-group A, Sub-group B and Sub-group C are constituted from 27 groups based on similar codon usage pattern).
Codon usage pattern analysis of SARS-CoV-2 genome sequences obtained from 13 selected states of India.
Amino acid pattern analysis of SARS-CoV-2 genome sequences retrieved from 13 selected states of India.
CRediT authorship contribution statement
All the authors carried out the analysis, RS and PP conceived the idea. SG prepared display items. SG and RS wrote the research article. PP edited the final manuscript. All the authors reviewed the manuscript before submission.
Declaration of competing interest
None declared.
Acknowledgements
Authors are thankful to Negenome Bio Solutions Pvt. Ltd. for their support. Authors are also grateful to the IIIM Jammu for providing the necessary research facilities to carry out this work.
References
- Banerjee S., Seal S., Dey R., Bhattacharjee P. Mutational spectra of SARS-CoV-2 orf1ab polyprotein and signature mutations in the United States of America. J. Med. Virol. 2020;93(3):1428–1435. doi: 10.1002/jmv.26417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceraolo C., Giorgi F.M. Genomic variance of the 2019-nCoV coronavirus. J. Med. Virol. 2020;92:522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Sun M. ClustalW—a software for multiple sequence alignment of protein and nucleic acid sequence. Biotechnol. Lett. 2000;11:146–149. [Google Scholar]
- Hall T., Biosciences I., Carlsbad C. BioEdit: an important software for molecular biology. GERF Bull. Biosci. 2011;2:60–61. [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., et al. Tracking Changes in SARS-CoV-2 Spike: evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020;98 doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang C., Liu L., Gu M. Existing bitter medicines for fighting 2019-nCoV-associated infectious diseases. FASEB J. 2020;34:6008–6016. doi: 10.1096/fj.202000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorusso A., Calistri P., Petrini A., Savini G., Decaro N. Novel coronavirus (SARS-CoV-2) epidemic: a veterinary perspective. Vet. Ital. 2020;56:5–10. doi: 10.12834/VetIt.2173.11599.1. [DOI] [PubMed] [Google Scholar]
- Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Biosci. Trends. 2020;14:69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- Lu R., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., Van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachetti M., et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18:1–9. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M., Berhanu G., Desalegn C., Kandi V. Severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2): an update. Cureus. 2020;12 doi: 10.7759/cureus.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020;81 doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghav S., Ghosh A., Turuk J., et al. Analysis of Indian SARS-CoV-2 genomes reveals prevalence of D614G mutation in spike protein predicting an increase in interaction with TMPRSS2 and virus infectivity. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.594928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi S.M.D., Shakil S., Haneef M. A simple click by click protocol to perform docking: AutoDock 4.2 made easy for non-bioinformaticians. EXCLI J. 2013;12:831. [PMC free article] [PubMed] [Google Scholar]
- Saha P., Banerjee A.K., Tripathi P.P., Srivastava A.K., Ray U. A virus that has gone viral: amino acid mutation in S protein of Indian isolate of coronavirus COVID-19 might impact receptor binding, and thus, infectivity. Biosci. Rep. 2020;40 doi: 10.1042/BSR20201312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seah I., Agrawal R. Can the coronavirus disease 2019 (COVID-19) affect the eyes? A review of coronaviruses and ocular implications in humans and animals. Ocul. Immunol. Inflamm. 2020;28:391–395. doi: 10.1080/09273948.2020.1738501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data–from vision to reality. Eurosurveillance. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis P.S., Soltis D.E. Applying the bootstrap in phylogeny reconstruction. Stat. Sci. 2003:256–267. [Google Scholar]
- Supek F., Vlahoviček K. INCA: synonymous codon usage analysis and clustering by means of self-organizing map. Bioinformatics. 2004;20:2329–2330. doi: 10.1093/bioinformatics/bth238. [DOI] [PubMed] [Google Scholar]
- Yu R., Chen L., Lan R., Shen R., Li P. Computational screening of antagonist against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Int. J. Antimicrob. Agents. 2020;56(2) doi: 10.1016/j.ijantimicag.2020.106012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11(1):1–9. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Shen F.-M., Chen F., Lin Z. Origin and evolution of the 2019 novel coronavirus. Clin. Infect. Dis. 2020;7(15):882–883. doi: 10.1093/cid/ciaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Assemblage of SARS-CoV-2 genomes obtained from 62 countries into 27 groups based on phylogenetic analysis. RSCU score of individual genomes were presented.
Analysis of genetic variation prevalent in SARS-CoV-2 genome sequences retrieved from selected countries across the world.
Gene wise mutational information on SARS-CoV-2 genome sequences retrieved from selected countries across the world.
Analysis of single nucleotide polymorphism prevalent in SARS-CoV-2 genome sequences retrieved from selected states of India.
Gene wise mutational information on SARS-CoV-2 genome sequences retrieved from selected states of India.
Comparative codon usage pattern analysis of SARS-CoV-2 genome sequences between three sub-groups A, B and C (Sub-group A, Sub-group B and Sub-group C are constituted from 27 groups based on similar codon usage pattern).
Comparative amino acid pattern analysis of SARS-CoV-2 genome sequences between three sub-groups A, B and C (Sub-group A, Sub-group B and Sub-group C are constituted from 27 groups based on similar codon usage pattern).
Codon usage pattern analysis of SARS-CoV-2 genome sequences obtained from 13 selected states of India.
Amino acid pattern analysis of SARS-CoV-2 genome sequences retrieved from 13 selected states of India.