Abstract
Interferon (IFN) is a crucial first line of defense against viral infection. This cytokine induces the expression of several IFN-Stimulated Genes (ISGs), some of which act as restriction factors. Upon IFN stimulation, cells also express ISG15 and SUMO, two key ubiquitin-like (Ubl) modifiers that play important roles in the antiviral response. IFN itself increases the global cellular SUMOylation in a PML-dependent manner. Mass spectrometry-based proteomics enables the large-scale identification of Ubl protein conjugates to determine the sites of modification and the quantitative changes in protein abundance. Importantly, a key difference amongst SUMO paralogs is the ability of SUMO2/3 to form poly-SUMO chains that recruit SUMO ubiquitin ligases such RING finger protein RNF4 and RNF111, thus resulting in the proteasomal degradation of conjugated substrates. Crosstalk between poly-SUMOylation and ISG15 has been reported recently, where increased poly-SUMOylation in response to IFN enhances IFN-induced ISGylation, stabilizes several ISG products in a TRIM25-dependent fashion, and results in enhanced IFN-induced antiviral activities. This contribution will highlight the relevance of the global SUMO proteome and the crosstalk between SUMO, ubiquitin and ISG15 in controlling both the stability and function of specific restriction factors that mediate IFN antiviral defense.
Keywords: ubiquitin, SUMO, antiviral defense, interferon, restriction factors, ISG15, PML, TRIM25
Introduction
The establishment and maintenance of the innate antiviral response in cells is defined by the function of interferons (IFNs), important cytokines that were originally discovered by Isaacs and Lindenmann (1957). While they are mostly known for their antiviral activity, they also have important immunomodulatory, anti-proliferative and apoptotic activities (Borden et al., 2007; Chelbi-Alix and Wietzerbin, 2007). Based on the type of receptor through which they signal, human IFNs are classified into multiple type I species (including IFNα and IFNβ), one type II (IFNγ) and four members of type III species (IFNλ1-4) (Chelbi-Alix and Wietzerbin, 2007).
The innate immune system constitutes the first line of host defense during viral infection triggering IFN synthesis and establishing an antiviral state. IFNs are secreted and bind to their cognate receptors (IFNAR for IFNα/β, IFNGR for IFNγ, or IFNLR for IFNλ), and subsequently activate the JAK/STAT pathways to trigger the transcription of more than one hundred IFN-Stimulated Genes (ISGs) (Schneider et al., 2014). Some of these ISG products act as restriction factors blocking various steps of the viral life cycle while others are positive or negative regulators of the IFN pathway. The complex regulation of IFN activities is not only essential to ensure a strong antiviral response but is also required to elicit a return to cell homeostasis.
Briefly, type I and type III IFNs bind to their respective receptors and activate Janus kinase 1 (JAK1) and Tyrosine kinase 2 (TYK2), which phosphorylate Signal Transducer and Activator of Transcription proteins (STAT1 and STAT2) (Barrat et al., 2019). Phosphorylated STATs heterodimerize (pSTAT1:pSTAT2) or homodimerize (pSTAT2:pSTAT2) and form with the DNA binding protein IFN Regulatory Factor (IRF9), the IFN-Stimulated Gene Factor 3 (ISGF3) complex. ISGF3 translocates into the nucleus to induce ISGs containing in their promoters an IFN-Stimulated Response Element (ISRE). The interaction of type II IFN to its specific receptor induces the phosphorylation of STAT1 by JAK1 and JAK2, the homodimerization of pSTAT1, their migration to the nucleus to induce ISGs containing in their promoters a Gamma-Activated Sequence (GAS).
Interestingly, STAT1 SUMOylation inhibits IFN-induced STAT1 phosphorylation (pSTAT1) resulting in a decrease of IFNγ-induced transcription without affecting the activity of IFNα since pSTAT2 homodimers can compensate for the lack of pSTAT1 (Ungureanu et al., 2005; Maarifi et al., 2015). Accordingly, several reports indicate the existence of alternative STAT2 signaling pathways in response to type I IFN that are independent from STAT1 (Blaszczyk et al., 2015; Rengachari et al., 2018). Of note, either ISGylation or ubiquitylation can modify lysine residues of STAT1, suggesting a possible interplay between these ubiquitin-like (Ubl) proteins in the regulation of STAT1 activity. While STAT1 ISGylation maintains the levels of pSTAT1 and promotes STAT1 association with ProMyelocytic Leukemia (PML) nuclear bodies (NBs) (Fan et al., 2020), decreased levels of ISGylation lead to an increase in STAT1 poly-ubiquitylation, and its degradation by the proteasome (Ganesan et al., 2016).
Several ISG products act as restriction factors mediating the IFN-induced innate immune antiviral response. When the restriction factors are constitutively expressed, they mediate the intrinsic antiviral activity, which confer viral resistance in the absence of IFNs (Hannoun et al., 2016; Wang et al., 2017a). The restriction factors are also further induced upon viral infection or IFN treatment, and individually they can interfere with a particular stage of the viral life cycle (El Asmi et al., 2018). Interestingly, some restriction factors are conjugated to SUMO and their modifications are further enhanced in response to IFN and also required for their antiviral activity (Geoffroy and Chelbi-Alix, 2011; Marcos-Villar et al., 2013; Hannoun et al., 2016; El-Asmi et al., 2020a).
Interferon response is achieved in part through protein modifications of its key players. In particular, SUMOylation and ISGylation are important Ubls implicated in intrinsic and innate immunity regulating IFN production and IFN signaling as well as localization, stability and activity of many restriction factors (Maarifi et al., 2015; Perng and Lenschow, 2018; Dzimianski et al., 2019; El-Asmi et al., 2020b). Whereas the expression of ISG15 is directly induced by IFNs (Loeb and Haas, 1992), that of SUMO1/2/3 is enhanced by IFNs through a miRNA-based mechanism involving the Lin28/let-7 axis (Sahin et al., 2014).
SUMOylation emerged as a key regulator during IFN treatment or viral infection with a drastic change in the SUMO proteome (Domingues et al., 2015; Sloan et al., 2015; Maroui et al., 2018; El-Asmi et al., 2020a). The increase in IFN response of the level of cellular SUMOylation and the SUMOylation of several restriction factors contribute to the antiviral function of the host (Hannoun et al., 2016; El-Asmi et al., 2020a). In contrast, viruses have developed various strategies to usurp SUMO host pathway targeting each step of SUMOylation process for their own benefit and to the detriment of the host. SUMO either directly targets viral proteins or alters the expression and function of cellular proteins implicated in antiviral defense. Exploitation of the SUMO pathway by DNA and RNA viruses has been reported in several reviews (Everett et al., 2013; Wimmer and Schreiner, 2015; Lowrey et al., 2017; El Motiam et al., 2020).
Importantly, IFN was shown to enhance levels of cellular SUMOylation (Maroui et al., 2018), ubiquitylation (Seifert et al., 2010) and ISGylation (Harty et al., 2009). In addition, increased poly-SUMOylation further enhances ubiquitylation and ISGylation in reponse to IFN resulting in either dowregulation or upregulation of several ISG products (El-Asmi et al., 2020b).
In this review, we will focus on the global SUMO proteome analysis in response to IFN and the consequence of the interplay between SUMOylation, ubiquitylation and ISGylation on IFN-induced antiviral defense.
Proteome-Wide Analysis of Protein Sumoylation
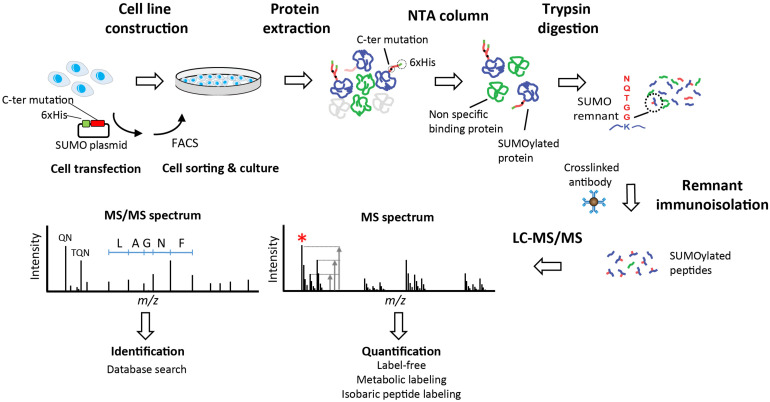
Different approaches including overexpression, knockdown, or knockout of either SUMO itself or the sole SUMO conjugating enzyme Ubc9 are used to understand the role of SUMOylation and to identify SUMO conjugates under different cell stimulation. However, the system-wide analysis of protein SUMOylation requires efficient affinity purification and sensitive mass spectrometry (MS) methods in view of the low abundance and rapid turnover of this modification. To identify and quantify protein SUMOylation in a global and site-specific manner, most methods rely on a two-step approach where SUMOylated proteins are first isolated by affinity purification, then digested with trypsin, and the modified peptides are subsequently purified by immunoaffinity precipitation (Figure 1) (Hendriks et al., 2014; Impens et al., 2014; Lamoliatte et al., 2014; Tammsalu et al., 2015). Exogenous expression of SUMO mutants containing a N-terminal poly-histidine tag is typically used to purify SUMO-modified proteins on Ni-NTA beads. In contrast to other Ubl such as ubiquitin, NEDD8 and ISG15, the tryptic digestion of SUMOylated proteins gives rise to large (26–32 amino acids) remnants that are difficult to sequence by MS. To facilitate peptide sequencing and SUMO peptide immunoprecipitation, an arginine residue is inserted near the C-terminus of SUMO to create smaller remnants upon tryptic digestion. Purified SUMOylated peptides can then by analyzed by MS to identify the protein substrates, the sites of modification and the type of SUMO chains appended to conjugates. Previous studies used SUMO remnant affinity purification in combination with quantitative proteomics to profile the changes in protein SUMOylation upon different cell stimuli including arsenic trioxide (As2O3) (Galisson et al., 2011; Rinfret Robert et al., 2020), proteasome inhibition (Hendriks et al., 2014; Lamoliatte et al., 2014), heat shock (Liebelt et al., 2019) and IFN (Maroui et al., 2018; El-Asmi et al., 2020a,b).
FIGURE 1.
Identification of SUMOylated proteins. Stable cell lines are generated using plasmids that facilitate the immunoisolation of conjugated substrates. Plasmid incorporate a 6xHis tag for the enrichment of SUMOylated proteins using immobilized metal affinity chromatography with Ni-NTA resin. Cells are lysed and proteins are extracted prior to purification of SUMOylated proteins on Ni-NTA beads. Proteins are digested with trypsin directly on beads and tryptic peptides with remnant SUMO are isolated by immunoaffinity using a specific antibody. Purified SUMO peptides are analyzed by LC-MS/MS to obtain a survey scan (MS spectrum) from which peptide ions are selected for MS/MS sequencing (red asterisk). The ratio of peptide ion intensities from the MS spectrum is used to determine the relative changes in abundance between conditions (Quantification). Different quantification strategies including label-free, metabolic labeling (e.g., stable isotope labeling in cell culture, SILAC) or isobaric peptide labeling can be used to determine changes in protein abundance. Fragment ions observed in the MS/MS spectrum are used to identify the peptide sequence and the cognate protein with one of several database search engines (Identification). Specific fragment ions associated with the SUMO remnant side chains are observed in the MS/MS spectrum (QN at m/z 243 and TQN at m/z 344) are used to confirm the identification of SUMOylated peptides.
It is noteworthy that ubiquitylated substrates display a diglycine remnant left on the side-chain lysine after trypsin digestion, and antibodies recognizing the corresponding diglycine motif are used for immunoaffinity purification (Xu et al., 2010; Kim et al., 2011). Building upon these technical advances, a sequential Ubl remnant immunoaffinity approach was introduced to identify substrates modified by both ubiquitin and SUMO (Lamoliatte et al., 2017). This latter approach enabled the identification of more than 10,000 SUMO sites and revealed important crosstalk between ubiquitin and SUMO including the SUMOylation of proteasome subunits, a modification required for the recruitment of proteasome to PML NBs.
PML, a Key Player in IFN Response and SUMO Pathway
ProMyelocytic leukemia protein, also known as TRIM19, is found in the nucleoplasm and assemble as PML NBs in the nuclear matrix. PML NBs are dynamic structures harboring a few permanent resident proteins (e.g., PML, Sp100, and SUMO) and numerous transient proteins depending on cell stimulation (i.e., stress, IFN, or viral infection) (van Damme et al., 2010; Geoffroy and Chelbi-Alix, 2011). PML NBs are implicated in various processus including stress response, apoptosis, protein degradation, viral infection and IFN response (Regad and Chelbi-Alix, 2001; Everett and Chelbi-Alix, 2007; Bernardi et al., 2008; Geoffroy and Chelbi-Alix, 2011; Nisole et al., 2013).
Several PML isoforms (PMLI to PMLVII) are generated from a single PML gene by alternative splicing. They share the same N-terminal region containing the RBCC/TRIM motif but differ in their C-terminus. The variability in their COOH-terminal region leads to the specific functions of some PML isoforms (Jensen et al., 2001; Maroui et al., 2011; Reichelt et al., 2011; Nisole et al., 2013).
ProMyelocytic leukemia protein emerges as a significant co-regulator of the IFN pathway. PML positively regulates virus-induced IFN synthesis (El Asmi et al., 2014) as well as type I and type II IFN signaling (El Bougrini et al., 2011; Kim and Ahn, 2015). Also, PML acts as a restriction factor in intrinsic and innate host antiviral defense both requiring its SUMOylation (Everett and Chelbi-Alix, 2007; Geoffroy and Chelbi-Alix, 2011; El Asmi et al., 2014). Key associations have been reported between PML NBs, IFN, antiviral defense and SUMO pathway.
PML and IFN Response
All IFNs directly induce the PML gene via the ISRE and the GAS motifs present in its promoter resulting in the increase of PML isoforms (Chelbi-Alix et al., 1995; Stadler et al., 1995). Analysis of PML knockout mice and their derived cell lines (PML−/−) shows that they are more susceptible to viral infections (Blondel et al., 2002; Bonilla et al., 2002; El McHichi et al., 2010). Also, PML−/− cells are defective in IFN signaling and in the induction of apoptosis induced by type I and type II IFNs (Wang et al., 1998; El Bougrini et al., 2011).
It is well established that several infections with DNA and RNA viruses result in PML degradation and/or PML NB disorganization (Everett and Maul, 1994; Chelbi-Alix and de The, 1999; Muller and Dejean, 1999; Everett and Chelbi-Alix, 2007; Tavalai and Stamminger, 2008; El McHichi et al., 2010; Geoffroy and Chelbi-Alix, 2011), suggesting that PML NB alteration could be a viral strategy to evade cellular resistance mechanisms. For instance, Herpes Simplex Virus 1 (HSV-1) encodes the E3 ubiquitin ligase ICP0 that targets PML and induces its proteasomal degradation (Boutell et al., 2003; Wang et al., 2012), whereas the IE1 of human cytomegalovirus (hCMV) disrupts PML NBs by inducing a loss of PML SUMOylation (Schilling et al., 2017).
The antiviral property of PML has been shown in human cells by its overexpression or its depletion. Indeed, expression of specific PML isoforms confers resistance to several DNA and RNA viruses (Everett and Chelbi-Alix, 2007) including HSV-1 (McNally et al., 2008; Cuchet et al., 2011), Varicella-Zoster Virus (VZV) (Reichelt et al., 2011), Adenovirus (Atwan et al., 2016), Human Foamy Virus (HFV) (Regad et al., 2001), Encephalomyocarditis virus (EMCV) (Maroui et al., 2011), Influenza A virus (IAV) (Chelbi-Alix et al., 1998), Vesicular Stomatitis Virus (VSV) (Chelbi-Alix et al., 1998; El Asmi et al., 2014), Dengue virus (DENV) (Giovannoni et al., 2015) or Rabies Virus (Blondel et al., 2010) by interacting with cellular or viral proteins inhibiting their functions (Regad et al., 2001; McNally et al., 2008; Maroui et al., 2011; Reichelt et al., 2011; El Asmi et al., 2014) or to poliovirus in a p53-dependent way by recruiting p53 within PML NBs and inducing apoptosis in infected cells (Pampin et al., 2006). Among all PML isoforms, PMLIV is implicated in innate immunity in addition to its intrinsic antiviral activity (El Asmi et al., 2014). Indeed, modification of PMLIV by SUMO positively regulates virus-induced IFNβ synthesis via the recruitment of the peptidylprolyl Cis/Trans Isomerase (PIN1) to PML NBs that results in a higher IRF3 phosphorylation (El Asmi et al., 2014). PML plays key roles in host antiviral defense and mediates the IFN-induced antiviral state since IFN has reduced antiviral activity in the absence of PML (Regad et al., 2001; Everett and Chelbi-Alix, 2007; Geoffroy and Chelbi-Alix, 2011; Maroui et al., 2011).
PML and SUMO Pathway
PML is subjected to multiple post-translational modifications, which include phosphorylation, acetylation, ubiquitylation, ISGylation, and SUMOylation (Geoffroy and Chelbi-Alix, 2011). PML localization is intimately linked to its SUMOylation. Within the nucleus, most of PML is expressed in the diffuse nuclear fraction of the nucleoplasm with only a small fraction in the matrix-associated NBs (Zhu et al., 1997; Porta et al., 2005; El McHichi et al., 2010). The transfer of PML from the nucleoplasm to PML NBs is associated with PML SUMOylation (Zhu et al., 1997; Muller et al., 1998; El McHichi et al., 2010, El-Asmi et al., 2019).
SUMO is the first identified PML NB-associated protein that interacts with PML (Boddy et al., 1996). SUMOylation has important consequences on PML functions as it can affect its localization, its stability, its ability to interact with other partners and its antiviral property (Everett and Chelbi-Alix, 2007; Geoffroy and Chelbi-Alix, 2011). Also, the non-covalent interactions of SUMO with PML via its SUMO-Interacting Motif (SIM) is required for further recruitment of PML NB partners (Shen et al., 2006; van Damme et al., 2010; Maroui et al., 2012). SUMOylation and the SIM in PML contribute substantially to body architecture and to multivalent interactions within PML NBs. In addition, PML exerts a SUMO E3 ligase activity (Chu and Yang, 2011) that facilitates SUMOylation of a large number of critical proteins by concentrating them into PML NBs where it serves as the scaffold.
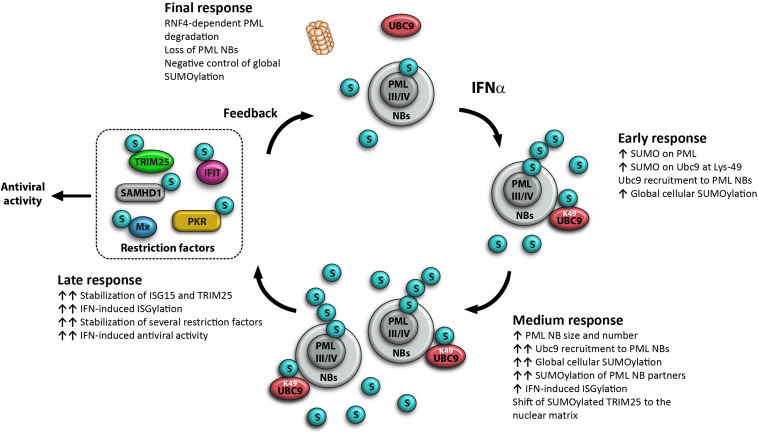
More recently, it has been shown that IFN enhances rapidly (45 min post-treatment) global cellular SUMOylation in an endogenous PML-dependent manner (Maroui et al., 2018). Also, IFN increases the SUMOylation of PML and of Ubc9, the only known SUMO E2-conjugating enzyme. The SUMOylation of Ubc9 at Lys-49 promotes its recruitment to PML NBs, and is associated with an increase in global SUMOylation within 16 h post-IFN treatment (Maroui et al., 2018) and in enhanced IFN-induced ISGylation (El-Asmi et al., 2020a). This process is followed by the stabilization of several restriction factors and an enhanced IFN-induced antiviral state (El-Asmi et al., 2020a). Later, IFN induces RNF4-dependent PML degradation with a loss of PML NBs (Maarifi et al., 2015) and a decrease of global cellular SUMOylation (Maroui et al., 2018), suggesting a feedback loop for the control of protein SUMOylation (Figure 2).
FIGURE 2.
Model for the key role of PML in IFN-increased global cellular SUMOylation. Early response at 45 min post-IFN treatment: IFN increases PML-dependent global cellular SUMOylation, with an enhancement of PML and Ubc9 SUMOylation; IFN induces PML-dependent Ubc9 shift to the nuclear matrix and the recruitment of SUMOylated Lys-49-Ubc9 to PML NBs; PMLIII and PMLIV are key players of the IFN-induced increase of cellular SUMOylation; Medium response at 16 h post-treatment: IFN increases the expression of PML isoforms, resulting in the increase of the number and the size of PML NBs, a further increase of Ubc9 recruitment to PML NBs, a maximum enhancement of global cellular SUMOylation, a positive regulation of IFN-induced ISGylation and a shift of SUMOylated TRIM25 to the nuclear matrix. Late response at 20 h: IFN promotes SUMOylation and a huge stabilization of several restriction factors resulting in enhanced IFN-induced antiviral activity. Final response at 24 h post-IFN treatment: IFN promotes RNF4-mediated proteasomal PML degradation with a loss of PML NBs and a decrease of global cellular SUMOylation, suggesting a negative control of global SUMOylation, this process is essential to ensure a return of the cell to homeostasis.
Interestingly, the expression of each of the human PML isoform (PMLI to PMLIV) into PML−/− cells reveals that increased SUMOylation in response to IFN is orchestrated by PMLIII and PMLIV isoforms (Maroui et al., 2018). The mechanism by which the isoform-specific protein sequences enhance global cellular SUMOylation is still unclear. Although PML isoforms may have related functions due to their common RBCC/TRIM domain, increasing evidences suggest that the variability of the C-terminal region confers specific functions of some PML isoforms (Maroui et al., 2011; Nisole et al., 2013; El Asmi et al., 2014). Importantly, PMLIV acts as a SUMO E3 ligase increasing the SUMOylation of various proteins (Chu and Yang, 2011) and among all PML isoforms, only the expression of PMLIII or PMLIV enhances overall SUMOylation in PML negative cells with a further increase upon IFN treatment (Maroui et al., 2018). As a SUMO ligase, PML interacts with Ubc9 via its RING finger motif (Chu and Yang, 2011), and PMLIII or PMLIV promotes the recruitment of SUMOylated Ubc9 within PML NBs. Indeed, the expression of PMLIII or PMLIV in PML−/− cells is able to recruit Ubc9 to PML NBs where both proteins colocalize (Maroui et al., 2018).
It is noteworthy that PML negative cells have a defect in IFN-induced apoptosis (Wang et al., 1998) and in increasing global cellular SUMOylation (Maroui et al., 2018). Many restriction factors and key regulators of IFN pathway are SUMOylated and require this modification for their functions (Hannoun et al., 2016). The expression of PMLIII or PMLIV in PML negative cells induces IFN-enhanced global cellular SUMOylation and could therefore restore IFN functions.
The SUMO, Ubiquitin, ISG15 Pathways and Protein Stability
In addition to ubiquitin, several Ubls are expressed following IFN stimulation, including SUMO and ISG15, which are key players of cellular antiviral defense (Hannoun et al., 2016; Maarifi et al., 2016; Dzimianski et al., 2019; El-Asmi et al., 2020b). Like ubiquitin, these Ubls can be conjugated to protein substrates via their own E1-E2-E3 enzymatic cascades. Ubc9, and UbcH8 are specific E2 conjugating enzymes for SUMOylation and ISGylation, respectively (Desterro et al., 1997; Johnson and Blobel, 1997; Kim et al., 2004). In some cases, a single Ubl is conjugated to a lysine residue while in other a long chain of polymerized Ubls is attached to an acceptor lysine. Ubiquitin and Ubls are involved in the regulation of a variety of cellular activities, including signal transduction, protein stability, intracellular trafficking and antiviral defense.
Several key players of IFN response are targets of ISGylation (Zhao et al., 2005) and/or SUMOylation (Maroui et al., 2018; El-Asmi et al., 2020a), and have been identified with different experimental global or specific approaches including affinity purification combined with MS. Many proteins implicated in IFN-induced antiviral defense can be targeted by both ISG15 and SUMO namely STAT1, RIG-I, IRF3, PKR, PML, IFIT1, MxA, TRIM25, p53 (Giannakopoulos et al., 2005; Zhao et al., 2005; Maroui et al., 2018; Perng and Lenschow, 2018; El-Asmi et al., 2020a). The consequences of ISGylation and SUMOylation on their functions are summarized in Table 1.
TABLE 1.
Effects of ISGylation and SUMOylation on key regulators of IFN pathway.
| Key regulators of IFN pathway | ISGylation | SUMOylation | References |
|
IFN production | |||
| IRF3 | Regulates positively IRF3 activation | Reduces virus-induced IFN synthesis | Kubota et al., 2008; Shi et al., 2010 |
| RIG-I | Reduces levels of virus-induced IFN promoter activity | Enhances type I IFN production | Kim et al., 2008; Mi et al., 2010 |
| MDA5 | Required for IFN synthesis | Regulates positively IFN synthesis | Fu et al., 2011; Kim et al., 2016; Liu et al., 2020 |
|
IFN signaling | |||
| STAT1 | Preserves STAT1 phosphorylation | Reduces STAT1 phosphorylation | Malakhov et al., 2003; Ungureanu et al., 2005; Maarifi et al., 2015 |
|
Restriction factors | |||
| MxA | The significance has not been evaluated | Increases protein stabilization and antiviral property | Zhao et al., 2005; Maarifi et al., 2016 |
| PML/TRIM19* | Enhances STAT1 association with PML NBs | Required for PML NB formation and functions Required for antiviral property | Everett and Chelbi-Alix, 2007; Shah et al., 2008; Geoffroy and Chelbi-Alix, 2011; Fan et al., 2020 |
| P53* | Promotes p53 degradation | Promotes cellular senescence | Zhao et al., 2005; Marcos-Villar et al., 2013; Huang et al., 2014 |
| IFIT1 | The significance has not been evaluated | Upregulates its protein level in response to IFN | Zhao et al., 2005; El-Asmi et al., 2020a |
| SAMHD1 | The significance has not been evaluated | Upregulates its protein level in response to IFN | El-Asmi et al., 2020a |
| PKR | Promotes PKR eIF2a activation in the absence of viral infection | Promotes PKR eIF2a activation in the absence of viral infection | Okumura et al., 2013; de la Cruz-Herrera et al., 2014; Maarifi et al., 2018 |
|
Ubl modifiers | |||
| Ubiquitin | Negatively regulates turnover of ubiquitinated proteins | Promotes Ubc9 localization within PML NBs | Fan et al., 2015; Maroui et al., 2018; McManus et al., 2018 |
| ISG15 | Poly-SUMOylation enhances IFN-induced ISG15 and ISGylation | El-Asmi et al., 2020a | |
| TRIM25 (E3 ligase) | Inhibits its own ISGylase activity | Upregulates its protein level in response to IFN | Zou et al., 2007; El-Asmi et al., 2020a |
| HERC5 (E3) | HERC5 is itself a target for ISGylation, but its functional significance is unknown. | Upregulates its protein level in response to IFN | Wong et al., 2006; El-Asmi et al., 2020a |
*All the keys regulators of IFN pathway indicated in the table are highly stabilized by SUMO3 in response to IFN except PML and p53, which are downregulated. PML and p53 are SUMOylated 1 h post IFN stimulation. PML recruits SUMOylated Ubc9 within PML NBs and promotes IFN-enhanced global SUMOylation. Later during IFN treatment PML is degraded suggesting a negative control of SUMOylation.
Interplay Between Poly-SUMO Chains and Ubiquitin Resulting in Protein Degradation
IFN Enhances the SUMOylation of Ubiquitin and Ubiquitin-Like Modifiers
The global identification of proteins conjugated to SUMO in response to IFNα has been performed using a novel proteomics approach based on SUMO remnant immunoaffinity purification (Galisson et al., 2011; Lamoliatte et al., 2014; Lamoliatte et al., 2017; Maroui et al., 2018; El-Asmi et al., 2020a). SUMO proteome analysis revealed that various SUMOylation sites are increased in response to IFNα in ISG products including several restriction factors (e.g., PML, ADAR1, Vimentin, SAMHD1, PKR, IFIT1, IRF1, and ISG20) and other regulators of IFN signaling or IFN production (e.g., STAT1, IRF1, IRF9, Tif1α/TRIM24, TRIM28, TRIM33, and TBK1) (Maroui et al., 2018; El-Asmi et al., 2020a). In addition to the three major SUMO sites on Lys-65, Lys-160 and Lys-490 (Kamitani et al., 1998), several other SUMO sites have been more recently identified on PML (Lys-209, Lys-226, Lys-337, Lys-380, Lys-394, Lys-400, Lys-401, Lys-476, Lys-478, Lys-487, Lys-497, and Lys-616) (El-Asmi et al., 2020a), but only Lys-65, Lys-380, and Lys-490 showed more than fourfold increase in SUMOylation upon IFNα stimulation (Maroui et al., 2018). Interestingly, most of the proteins from the SUMOylation machinery (e.g., Ubc9, SUMO1, SUMO2, SUMO3, PIAS1, PIAS2, RanBP2, TRIM28, and PML) display increased SUMOylation in response to IFNα (Maroui et al., 2018).
Recruitment of SUMOylated Ubc9 on PML NBs in Response to IFN
Ubc9 is SUMOylated at Lys-14, Lys-18, Lys-48, Lys-49, Lys-65, and Lys-154 and its SUMOylation at Lys-14 was shown to display enhanced binding to SIM-containing proteins (Knipscheer et al., 2008; Wieczorek et al., 2016; Maroui et al., 2018). Only SUMOylation at Lys-49 of Ubc9 is enhanced in the nucleus at an early time post-IFNα treatment (45 min), and further increase of SUMOylation at Lys-49 is found in the cytoplasm and the nucleus during prolonged IFNα treatment (Maroui et al., 2018). In untreated cells, Ubc9 is expressed in the cytoplasm and the nucleoplasm. Remarkably, IFNα induces PML-dependent Ubc9 transfer to the nuclear matrix where Ubc9 colocalizes with PML within PML NBs resulting in an enhancement of global cellular SUMOylation (Maroui et al., 2018). The SUMOylation of Ubc9 at Lys-49 promotes its localization to PML NBs (McManus et al., 2018). Altogether, these findings demonstrate that SUMOylated Ubc9 and PML are key players of IFN-enhanced global cellular SUMOylation.
Interplay Between Poly-SUMO Chains and Ubiquitin Resulting in Degradation of ISG Products
SUMOylation of proteins can alter their interaction properties and thereby their localization and/or stability thus providing a rapid and reversible way to control protein functions. SUMO is conjugated to lysine residues in target proteins through an isopeptide linkage catalyzed by SUMO-specific activating (E1), conjugating (E2), and ligating (E3) enzymes. Due to the expression of several SUMO paralogs, SUMO pathway analysis is more complicated than that of ubiquitin or ISG15. Mammalian cells can express four SUMO paralogs with SUMO1, SUMO2, and SUMO3 being conjugatable (SUMOylation) and ubiquitously expressed, while SUMO4 expression is restricted to renal, immune and pancreatic cells (Flotho and Melchior, 2013; Hay, 2013; Baczyk et al., 2017). Also, SUMO4 contains a proline residue close to the diglycine motif, which prevents its maturation and conjugation (Owerbach et al., 2005). In addition, SUMO via the SIMs is able to effect protein activity without being covalently attached (Song et al., 2004).
SUMO1 and SUMO2/3 (SUMO2 and SUMO3 have 97% peptide sequence similarity and are collectively referred to as SUMO2/3) modify both common and different substrates and growing evidences show that they may have distinct functions (Lallemand-Breitenbach et al., 2008; Tatham et al., 2008; Flotho and Melchior, 2013; Maarifi et al., 2015; Maarifi et al., 2018). SUMO2 and SUMO3 contain a lysine residue at position 11 (Lys-11) that can be used for self-conjugation and that is usually the site of poly-SUMO chains. In contrast, SUMO1 does not contain Lys-11 and therefore does not form poly-SUMO chains. However, alternatively linked non-canonical SUMO2/3 chains, mixed SUMO1-SUMO2/3 chains and SUMO2/3 chains capped with SUMO2, have been described (Sriramachandran and Dohmen, 2014; Gartner et al., 2018; Sriramachandran et al., 2019). It is well established that SUMO2/3 have the capability to efficiently form highly branched poly-SUMO chains that have the ability to recruit SUMO-targeted ubiquitin ligases (STUbls) such as the RING finger protein RNF4 and RNF111 (Sriramachandran and Dohmen, 2014; Kumar and Sabapathy, 2019). RNF4 harbors multiple SIMs in its N-terminus region that allow a strong interaction with SUMO leading to the ubiquitylation of poly-SUMO chains conjugated to substrates and resulting in the proteasomal degradation of SUMO2/3 conjugated proteins (Hay, 2013). The best-studied case is the SUMO-dependent PML degradation in cells treated with arsenic trioxide (As2O3) (Lallemand-Breitenbach et al., 2008; Tatham et al., 2008; Erker et al., 2013) or with IFN (Maarifi et al., 2015). More recently, RNF111 was reported to select proteins carrying SUMO1-capped SUMO2/3 hybrid conjugated and targets them for proteasomal degradation (Sriramachandran et al., 2019).
Treatment with IFNα results in a very rapid increase of global cellular SUMOylation (Maroui et al., 2018), which also increase the SUMOylation of endogenous PML and Sp100. This is followed by the increase of their mRNA and protein expression, and by their RNF4-dependent proteasomal degradation during prolonged IFN treatment (Maarifi et al., 2015). It should be noted that Sp100 is an ISG product permanently associated to PML NBs (Grotzinger et al., 1996). Therefore, the RNF4-dependent proteasomal degradation of PML and Sp100 proteins results in a loss of PML NBs. The expression of the restriction factors TRIM5α, p53 and Daxx is also dowregulated by SUMO3 in response to IFNα (El-Asmi et al., 2020a). Large-scale proteomic analyses revealed that in response to IFNα, SUMO3 expression results in the down-regulation of 586 proteins such as the restriction factor IFI16, which expression is restored upon RNF4 depletion (El-Asmi et al., 2020a) as previously shown for PML and Sp100 (Maarifi et al., 2015). The SUMOylation of PML and Sp100 is required for their degradation, suggesting that SUMOylation of other ISG products could also be necessary for their dowregulation.
SUMO and ISG15 Pathway in Response to IFN
IFN Stabilizes ISG15 and ISG15 Modifiers
ISG15 is expressed as a 17 kDa precursor protein that is processed into its mature 15 kDa form via protease cleavage to expose a carboxy-terminal LRLRGG motif (Potter et al., 1999). ISG15 exists as an unconjugated protein and is also covalently conjugated to lysine residues of several substrates through this motif by a process known as ISGylation (Loeb and Haas, 1992; Narasimhan et al., 1996). Similar to ubiquitin and SUMO, ISG15 can non-covalently bind to proteins and modulate their functions (Perng and Lenschow, 2018).
ISG15 was first identified from the study of type I IFN-treated cells (Korant et al., 1984; Haas et al., 1987). Whereas some Ubls are constitutively expressed in the host cell, ISG15 and the members of the enzymatic cascade mediating ISG15 conjugation, including the E1, E2, E3, and the deconjugating enzyme, are ISG products themselves (Loeb and Haas, 1992), suggesting their tight link to IFN-regulated cellular functions. The expression of the ISG15 gene is dependent on an ISRE found in its promoter region (Reich et al., 1987). The increase level of free ISG15 occurs within 2 h post-IFN treatment and is maximal by about 18 h, whereas increase of ISGylation is observable at least 12h post-IFN treatment. In addition, ISG15 is also induced by viral and bacterial infections (Yuan and Krug, 2001; Radoshevich et al., 2015), lipopolysaccharide (LPS) (Malakhova et al., 2002) and retinoic acid (Pitha-Rowe et al., 2004). Using MS, hundreds of host proteins have been identified as ISG15 targets upon IFN stimulation (Giannakopoulos et al., 2005; Zhao et al., 2005). Among these, the antiviral effectors, STAT1, IRF3, Retinoic acid Inducible Gene (RIG-I), double-stranded RNA (dsRNA)-dependent protein kinase (PKR), IFN-induced protein with tetratricopeptide repeats 1 (IFIT1), p53 and MxA (Giannakopoulos et al., 2005; Zhao et al., 2005; El-Asmi et al., 2019) (Table 2). Remarkably, the SUMO E3 ligase polycomb 2 (Pc2) and the SUMO protease SENP1 were also identified as ISG15 targets (Zhao et al., 2005), suggesting a possible crosstalk between the ISG15 and SUMO pathways. For a subset of these potential targets, modification has been validated, and the impact of ISGylation on their function has been investigated (Perng and Lenschow, 2018) (Table 1).
TABLE 2.
Antiviral properties of the restriction factors stabilized by SUMO3 in the presence of IFNα.
| Protein stability | SUMO3/wt cells | (SUMO3/wt cells) + IFN | Viral resistance |
|
IFN signaling | |||
| STAT1 | + | +++ | |
| STAT2 | +++ | ||
| IRF9 | +++ | ||
|
Restriction factors | |||
| MxA | + | +++ | VSV, IAV, VACV |
| MxB | +++ | HIV-1, HSV-1, HSV-2 | |
| GBP1 | +++ | VSV, IAV, DENV, KSHV | |
| GBP5 | +++ | IAV, RSV, HIV-1 | |
| SAMHD1 | + | +++ | HIV-1, HSV-1, HBV, VACV |
| IFITM1 | +++ | ZIKV, DENV | |
| IFITM2 | +++ | MLV, SARS-CoV-2 | |
| IFITM3 | +++ | MLV,VSV, IAV, IBV, DENV, SARS-CoV-2 | |
| IFIT1 | + | +++ | HBV, HPV, HCV |
| IFIT2 | +++ | VSV | |
| IFIT3 | +++ | HBV, HPV, HCV | |
| IFI44 | +++ | HIV-1, RSV | |
| IFI44L | +++ | HCV, RSV | |
| TRIM21 | +++ | CVB3, HBV | |
| Tetherin/BST2 | +++ | HIV-1, VSV, KSHV | |
| PKR | + | +++ | EMCV, VSV |
| ISG15 | +++ | IAV, IBV, Sindbis virus | |
Unlike ubiquitylation, ISGylation does not appear to directly target proteins for proteasome-mediated degradation (Liu et al., 2003). It has been reported that ISG15 can compete with ubiquitin for ubiquitin binding sites on a protein, thereby indirectly impairing the ubiquitin/proteasome pathway by interfering with protein degradation (Desai et al., 2006). In addition, ISG15-ubiquitin mixed chains have been identified and may negatively regulate the turnover of ubiquitylated proteins (Fan et al., 2015).
In response to IFN, SUMO3 expression results in a stabilization of ISG15 E2 conjugating enzyme Ube2L6/UbcH8 and the E3 ISG15 ligases TRIM25 and HERC5 (HECT domain and RCC1-like domain containing protein 5) (El-Asmi et al., 2020a) highlighting an unsuspected interplay between SUMOylation and ISGylation. Importantly, the upregulation of ISGylation in response to IFNα is observed in SUMO3- but not in SUMO1-expressing cells (El-Asmi et al., 2020a), thus providing a novel differential effect of SUMO1 and SUMO3 on the positive regulation of ISG products and IFNα-induced ISGylation.
Furthermore, Ni-NTA analysis revealed that IFNα drastically enhances global cellular ISGylation and ubiquitylation in cells overexpressing His-SUMO3, thus demonstrating that ISG15 may modify SUMO2/3 conjugated proteins (El-Asmi et al., 2020a) as previously shown for ubiquitin (Tatham et al., 2008). The crosstalk between SUMOylation and ISGylation is further demonstrated by Ubc9 depletion that results in a decrease of cellular SUMOylation and a decrease of IFN-induced cellular ISGylation (El-Asmi et al., 2020a).
TRIM25 a Key Player of ISG15 Modifiers
Several members of the tripartite motif-containing (TRIM) superfamily are expressed in response to IFNs and are implicated in various biological processes associated with innate immunity (Rajsbaum et al., 2008). This superfamily contains in its N-terminal region a TRIM (also named RBCC) motif that comprises a RING finger, one or two B-box domains and a Coiled-Coil domain in the amino-terminal region (Reymond et al., 2001; Ozato et al., 2008). The presence of a RING finger, which can mediate the conjugation of proteins with ubiquitin, SUMO or ISG15, contributes to the biological responses of TRIM proteins (Ozato et al., 2008).
TRIM25 (also named EFP, RNF147, and ZNF147) is an IFN-inducible E3 ligase by type I and type II IFNs (Nakasato et al., 2006; Martin-Vicente et al., 2017). TRIM25 participates in multiple cellular processes including the regulation of the antiviral innate immunity (Manokaran et al., 2015; Martin-Vicente et al., 2017; Chen et al., 2019; Lin et al., 2019). TRIM25 is involved in the RIG-I-mediated antiviral response by inducing the Lys-63-linked poly-ubiquitylation of RIG-I, and consequently the induction of type I IFN production (Gack et al., 2007). The importance of TRIM25 in RIG-I signaling was illustrated by reduced IFN production in TRIM25−/− mouse embryonic fibroblasts and a corresponding susceptibility to viral infection (Gack et al., 2007). In contrast, TRIM25 expression inhibits late-stage replication of HIV-1 and Murine Leukemia Virus (MLV) (Uchil et al., 2008). Remarkably, nuclear TRIM25 protein confers resistance to IAV independently of both its ubiquitin ligase activity and the IFN induction pathway (Meyerson et al., 2017). TRIM25 directly restricts viral RNA synthesis in infected cells by binding to nuclear viral ribonucleoproteins, demonstrating that TRIM25 exerts an intrinsic antiviral activity by interacting with a viral protein inhibiting its function.
TRIM25 acts as an E3 ligase that can catalyze both ubiquitylation and ISGylation (Nakasato et al., 2006; Zou and Zhang, 2006). In addition, increased expression of TRIM25 enhances ubiquitylation of multiple cellular proteins (Zheng et al., 2017). Also, TRIM25 can undergo auto ISGylation on Lys-117 residue, a modification that negatively regulates its ISG15 E3 ligase activity (Nakasato et al., 2006; Zou et al., 2007). In addition to be ISGylated and ubiquitylated (Nakasato et al., 2006), TRIM25 has been shown to be SUMOylated by Ni-NTA purification analysis, with a further increase of its modification upon IFN stimulation (El-Asmi et al., 2020a). However, the SUMO site of TRIM25 has not yet been identified.
The unmodified form of TRIM25 is found in the RIPA soluble fraction containing the cytoplasm and the nucleoplasm. Remarkably, IFNα enhances TRIM25 SUMOylation and shifts SUMO-modified TRIM25 to the nuclear matrix, which also contains most of the SUMOylated proteins and a small portion of ISGylated proteins (El-Asmi et al., 2020a). Whether SUMOylated TRIM25 is recruited within PML NBs colocalizing with PML is still unknown.
The E3 ISG15 ligases TRIM25 and HERC5 are stabilized by SUMO3 in IFNα-treated cells and their depletion expectedly impairs protein ISGylation, but only depletion of TRIM25 abrogates the stabilization of ISG products (El-Asmi et al., 2020a), demonstrating that TRIM25 is a key player in the SUMO3-mediated protein stabilization in response to IFN. Further experiments are required to determine the precise function of TRIM25 in this process.
Remarkably some members of TRIM protein family act as E3 ligases that can catalyze ubiquitylation, ISGylation and SUMOylation (Meroni and Diez-Roux, 2005; Nakasato et al., 2006; Zou and Zhang, 2006; Gillot et al., 2009; Chu and Yang, 2011). As seen for TRIM25, a single TRIM protein can have dual E3 activities. It is now of great interest to determine the regulation of the TRIM proteins’ SUMO E3 activity, its targets, and its interplay with the ubiquitin and ISG15 E3 activities.
Interplay Between Poly-SUMOylation and ISG15 Resulting in Protein Stabilization
SUMO3 but not SUMO1 stabilizes several ISG products in response to IFN. The large-scale proteomic analyses revealed that SUMO3 stabilizes 583 proteins in response to IFNα, among which are several proteins implicated in IFN production such as MAVS (also known as IPS-1/VISA/Cardif), TBK1, IRF3, RIG-I, and MDAR, in IFN signaling (STAT1, STAT2, and IRF9) and in antiviral defense such as SAMHD1, Tetherin/BST2, PKR, OAS3, SAMD9, TRIM21, ISG15 and members of GBP, IFITM, IFIT and IFI families (El-Asmi et al., 2020a).
The upregulation by SUMO3 of key players of IFN signaling STAT1, STAT2 and IRF9 in response to IFN (Table 2) could enhance ISG products independently of their SUMOylation since constitutive expression of ISGs can be mediated by the unphosphorylated ISGF3 (U-ISGF3) complex, consisting of IRF9 together with unphosphorylated STAT1 and STAT2 (Cheon et al., 2013; Sung et al., 2015; Wang et al., 2017b). While the transcription factor ISGF3 (IRF9 and tyrosine-phosphorylated STAT1 and STAT2) drives the first rapid response phase post-IFN treatment, the stabilization by SUMO3 of the U-ISGF3 (El-Asmi et al., 2020a) could drives a second prolonged response of ISG increase.
Ni-NTA analysis revealed that IFNα drastically enhances global cellular ISGylation in cells expressing His-SUMO3 (El-Asmi et al., 2020a), demonstrating that ISG15 may modify SUMO2/3 conjugated proteins as previously shown for ubiquitin (Tatham et al., 2008). In contrast, Ubc9 depletion decreases cellular SUMOylation and remarkably results in a decrease of global cellular ISGylation (El-Asmi et al., 2020a), further demonstrating the crosstalk between SUMOylation and ISGylation.
Significant progress has been made to understand the functional significance of protein ISGylation. Unlike ubiquitylation, ISGylation seems to counteract proteasome-dependent protein degradation via the conjugation of ISG15 to different E2 and E3 ubiquitin-conjugating enzymes (Okumura et al., 2008) or via the formation of mixed ubiquitin-ISG15 chains (Fan et al., 2015), thus resulting in a decrease of poly-ubiquitylated protein levels and protein protection from proteasomal degradation. Remarkably, many of the ISG products stabilized by SUMO3 in response to IFN (El-Asmi et al., 2020a) have been previously shown to be ISGylated such as MxA, GBP1, SAMHD1, PKR, IFIT1, IFIT3, UBE2L6, and HERC5 (Giannakopoulos et al., 2005; Zhao et al., 2005) (Table 1). The recent findings that the formation of the poly-SUMO chains results in enhanced IFNα-induced global ISGylation, and in a TRIM25-dependent stabilization of several ISG products highlight the consequences of the crosstalk between SUMOylation and ISGylation on protein stabilization. Further experiments are needed to identify SUMO-ISG15 mixed chains and to determine whether TRIM25 could act as a SUMO-ISG15 ligase.
SUMO3 Enhances IFN-Induced Antiviral Activities by Stabilizing Several Restriction Factors
Several restriction factors playing key roles in antiviral defense are stabilized by SUMO3 in response to IFN (El-Asmi et al., 2020a), they include SAMHD1, Tetherin/BST2, PKR, oligoadenylate synthetase (OAS)3, ISG15 and members of Mx, GBP, IFITM, IFIT, and IFI families (Al-Khatib et al., 2003; Goldstone et al., 2011; Hollenbaugh et al., 2013; Kane et al., 2013; Kim et al., 2013; Goujon et al., 2014; Krapp et al., 2016; Crameri et al., 2018). Several of these restriction factors are known to confer viral resistance by targeting different steps of viral replication (Blondel et al., 2015; El Asmi et al., 2018). For example, IFITM proteins inhibit entry (Weidner et al., 2010), MxA inhibits primary transcription (Staeheli and Pavlovic, 1991), IFIT, PKR and OAS inhibit viral translation (Balachandran et al., 2000; Terenzi et al., 2006; Kristiansen et al., 2011), and Tetherin prevents release of virions from the cell (Weidner et al., 2010).
Importantly, the increase in abundance of these restriction factors by SUMO3 in IFNα-treated cells is correlated with the increase of IFNα-induced anti-HIV-1 and anti-HSV-1 activities (El-Asmi et al., 2020a). In contrast, the loss of SUMOylation by Ubc9 depletion reduces the IFNα-mediated resistance to these viruses (Sahin et al., 2014). Taken together, these results demonstrate that the stabilization of these restriction factors by SUMO3 results in enhanced IFNα-induced antiviral defense. The antiviral activity of the most studied of these restriction factors is briefly described below and conveniently summarized in Table 2.
Mx Proteins (MxA and MxB)
Myxovirus resistance (Mx) proteins are known for their protection of mice from infection by IAV (Horisberger et al., 1983; Staeheli et al., 1986). They belong, like Guanylate Binging Proteins (GBP), to the guanosine triphosphatase (GTPase) subfamily. Humans have two Mx genes: MxA and MxB, which expression is strictly controlled by type I and type III IFNs. MxB shares 63% amino acid sequence identity with MxA. MxA (also known as MX1) accumulates in the cytoplasm and is partly associated with the plasma membrane as well as the smooth endoplasmic reticulum (Accola et al., 2002; Stertz et al., 2006). MxB (also known as MX2) exists in two isoforms, a long 78 kDa and a short 76 kDa molecule (Melen et al., 1996). The long 78 kDa MxB has a nuclear localization sequence (NLS) and is localized near the nuclear pores, whereas the 76 kDa isoform lacking the NLS is cytoplasmic.
MxA is SUMOylated at Lys-48 and interacts non-covalently with SUMO via two SIMs located in the GTPase binding domain that are required for its antiviral activity (Brantis-de-Carvalho et al., 2015). MxA is stabilized through its oligomerization in SUMO-expressing cells (Maarifi et al., 2016). MxA oligomerization capacity is important for its GTPase activity, and also for its interaction with SUMO and Ubc9 (Brantis-de-Carvalho et al., 2015). Accordingly, monomeric mutant MxA-L612K has a reduced interaction with SUMO and Ubc9 (Brantis-de-Carvalho et al., 2015), and is rapidly degraded in cells compared to wild-type MxA, demonstrating that the self-assembly of the MxA protein is critical for protein stability (Janzen et al., 2000).
Interestingly, the overexpression of each SUMO paralog confers resistance to VSV through MxA stabilization and MxA depletion in SUMO-expressing cells abrogates the anti-VSV effect of SUMO, thus signifying that MxA is a key mediator of SUMO-induced resistance to VSV (Maarifi et al., 2016). Although IFN production is highly reduced in VSV-infected SUMO-expressing cells, the intrinsic anti-VSV activity of SUMO mediated by MxA is still maintained. Further experiments are required to determine whether the replication process of other viral families can be inhibited by SUMO expression via the stabilization of MxA or other restriction factors. All SUMO paralogs stabilize MxA protein, however, SUMO3, but not SUMO1, promotes a higher increase of MxA protein level upon IFN stimulation compared to wild-type cells (El-Asmi et al., 2020a).
MxA inhibits a wide variety of RNA and DNA viruses. The overexpression of MxA protein inhibits the multiplication of several RNA viruses such as IAV, VSV, measles virus, and other viruses belonging to the family Bunyaviridae (Haller et al., 2007). MxA protein also confers resistance to a few DNA viruses like the vaccinia virus (VACV), the monkey poxvirus, and the African swine fever virus (Mitchell et al., 2013). In contrast to MxA, the level of MxB protein is not affected by SUMO alone, though SUMO3 promotes a significant increase in MxB protein level in response to IFNα compared to wild-type cells (El-Asmi et al., 2020a).
MxB was long considered non-antiviral, until the finding in 2013 that human immunodeficiency virus type 1 (HIV-1) is inhibited by MxB (Goujon et al., 2013; Kane et al., 2013; Liu et al., 2013). Since then, MxB was shown to restrict several members of the Flaviviridae, including Hepatitis C Virus (HCV), Japanese encephalitis virus and DENV (Yi et al., 2019) as well as several herpesviruses, including HSV-1, HSV-2, and Kaposi’s sarcoma-associated herpesvirus (KSHV) (Crameri et al., 2018). MxB restriction of HSV-1 and HSV-2 requires GTPase function, in contrast to restriction of lentiviruses.
Nuclear MxB was found by Ni-NTA analysis to be conjugated to SUMO3 with enhanced modification upon cell treatment with IFNα (El-Asmi et al., 2020a). Further investigations are needed to identify the MxB SUMO sites and to determine whether MxB SUMOylation is required for its antiviral activity. Both MxA and MxB proteins are stabilized by SUMO3 in response to IFNα (El-Asmi et al., 2020a) (Table 2), therefore their upregulation could contribute to the enhanced IFN-induced antiviral activity.
GBP Proteins (GBP1 and GBP5)
Guanylate-Binding Proteins (GBP) are a family of dynamin-related large GTPases, which are expressed in response to IFNs (Vestal and Jeyaratnam, 2011; Pilla-Moffett et al., 2016). Currently, 7 human GBPs have been identified. Recently, proteomic analysis revealed that SUMO3 upregulates the expression of the antiviral proteins GBP1 and GBP5 in response to IFNα (El-Asmi et al., 2020a).
GBP1 is abundantly expressed during the innate immune response (Vestal and Jeyaratnam, 2011; Pilla-Moffett et al., 2016). It is one of the large GTPases, with a relative molecular mass of 67 kDa, which can hydrolyze GTP to GDP and subsequently to GMP (Prakash et al., 2000). It is a large self-activating GTPase, and its dimerization is necessary for sufficient GTP-hydrolyzing activity (Chappie et al., 2010). The GTPase activity of GBP1 is required for its antiviral activity (Vestal, 2005). GBP1 exhibits antiviral activity against various RNA viruses such as VSV, DENV, IAV, classical swine fever virus, and HCV (Anderson et al., 1999; Itsui et al., 2009; Pan et al., 2012; Li et al., 2016). In addition, overexpression of GBP1 significantly inhibits Kaposi’s sarcoma-associated herpesvirus (KSHV) infection, while the knockdown of GBP1 promotes KSHV infection. The GTPase activity and dimerization of GBP1 are responsible for its anti-KSHV activity (Zou et al., 2017).
GBP5 overexpression confers resistance to IAV by increasing the expression of virus-induced IFN and ISGs, while knockdown of GBP5 has the opposite effect (Feng et al., 2017). Also, GBP5 inhibits respiratory syncytial virus (RSV) infection by reducing the cell-associated levels of the RSV small hydrophobic protein, which is a viroporin (Li et al., 2020) and reduces HIV-1 infectivity by interfering with maturation and virion incorporation of the envelope glycoprotein (Krapp et al., 2016; Hotter et al., 2017). However, whether GBP1 and GBP5 are SUMOylated and whether their modifications are required for their stability and/or antiviral property are still unknown.
SAMHD1
Sterile-α-motif and HD domain containing protein 1 (SAMHD1) regulates the cellular dNTP (2′-deoxynucleoside-5′-triphosphate) pool by catalyzing the hydrolysis of dNTP into 2′-deoxynucleoside and triphosphate products. By limiting the supply of dNTPs for viral DNA synthesis, SAMHD1 restricts the replication of several retroviruses, such as HIV-1, and some DNA viruses, vaccinia virus (VACV) and HSV-1, in dendritic and myeloid lineage cells and resting T-cells (Baldauf et al., 2012; Lahouassa et al., 2012; Gramberg et al., 2013; Hollenbaugh et al., 2013; Sze et al., 2013; Chen et al., 2014). SAMHD1 is SUMOylated at several lysine sites (Lys-43, Lys-148, Lys-294, Lys-304, Lys-332, Lys-467, Lys-469, Lys-492, Lys-523, Lys-544, Lys-595, and Lys-622) but only SUMOylation at the SAMHD1 consensus site of Lys-622 is enhanced in response to IFNα (El-Asmi et al., 2020a).
SAMHD1 protein is stabilized by SUMO3 alone with a much higher stabilization upon IFNα stimulation compared to wild-type cells (El-Asmi et al., 2020a). Also, it is interesting to note that SUMOylated SAMHD1 and MxB interact together in response to IFNα, suggesting that under certain conditions SUMOylated MxB and SAMHD1 can cooperate to confer viral resistance in IFN-treated cells. However, it is still unknown whether the SUMOylation of SAMHD1 is required for its antiviral activity.
Tetherin/BST2
Tetherin also know as bone marrow stromal antigen 2 (BST-2) is induced by type I IFN (Neil et al., 2008) and its protein level is stabilized by SUMO3 in response to this cytokine (El-Asmi et al., 2020a). The antiviral property of tetherin was demonstrated by its capacity to restrict the release of virions from HIV-1-infected cells (Neil et al., 2008). Since then, tetherin has been shown to potently block viral replication by inhibiting enveloped virus budding from the surface of infected cells (Swiecki et al., 2013; Berry et al., 2018). Tetherin restricts viruses from different families: arenaviruses (Lassa and Machupo) (Sakuma et al., 2009); herpesviruses (KSHV) (Mansouri et al., 2009); filoviruses (Ebola and Marburg) (Jouvenet et al., 2009; Sakuma et al., 2009); rhabdoviruses (VSV) (Weidner et al., 2010); paramyxoviruses (Nipah) (Radoshitzky et al., 2010) and flaviviruses (HCV) (Dafa-Berger et al., 2012).
IFITM Proteins (IFITM1, IFITM2, and IFITM3)
Humans have three IFN-induced transmembrane proteins (IFITM1, IFITM2, and IFITM3) (Siegrist et al., 2011). IFITM proteins are involved in the regulation of many activities, such as immune signal transduction and antiviral defense. It has been shown recently that the protein expression of these three IFITM members is stabilized by SUMO3 in response to IFNα (El-Asmi et al., 2020a).
IFITM proteins have been reported to suppress the entry of a wide range of viruses (Amini-Bavil-Olyaee et al., 2013), though other inhibitory mechanisms affecting entry have been proposed (Brass et al., 2009; Huang et al., 2011; Lu et al., 2011; Yu et al., 2015). IFITM1, IFITM2, and IFITM3 potently inhibit HIV-1 replication at least partially through interfering with virus entry (Lu et al., 2011). More recently, a novel mechanism by which IFITM proteins inhibit viral infection has been described. Indeed, IFITMs might also inhibit viral gene expression and viral protein synthesis (Lee et al., 2018; Liao et al., 2019). In addition, IFITM1/2/3 proteins confer resistance to different other viruses including IAV (Feeley et al., 2011), West Nile Virus (WNV), DENV (Jiang et al., 2010), VSV (Weidner et al., 2010), SARS Coronavirus (SARS-CoV), and Marburg virus (MARV) (Huang et al., 2011). The precise inhibitory mechanism of IFITMs on viral infection and replication still requires further exploration but their elevated expression by SUMO3 in response to IFN could enhance their antiviral response.
IFIT Proteins (IFT1, IFT2, and IFIT3)
IFN-induced proteins with tetratricopeptide repeats (IFITs) motifs play important roles in host innate immune response to viruses. There are four members in human IFIT family: IFIT1 (ISG56), IFIT2 (ISG54), IFIT3 (ISG60 or IFIT4) and IFIT5 (ISG58). The promoters of their genes, which are clustered, contain the ISREs and are induced by type I IFNs. IFIT1, IFIT2, and IFIT3 are stabilized by SUMO3 in response to IFNα with an increase of SUMOylation on four sites of IFIT1 (Lys-199, Lys-336, Lys-370, and Lys-407) (El-Asmi et al., 2020a).
IFITs are cytoplasmic proteins and do not have any known enzymatic roles. However, they are implicated in cellular functions by mediating protein–protein interactions and forming multiprotein complexes with cellular and viral proteins through their multiple tetratricopeptide repeats motifs (D’Andrea and Regan, 2003). IFIT1 and IFIT2 directly interact with eIF3, thus resulting in an inhibition of protein synthesis (Guo et al., 2000).
The IFIT family, especially IFIT1 and IFIT3, restrict DNA and RNA virus replication, such as hepatitis B virus (HBV), human papillomavirus (HPV), HCV, West Nile virus (WNV) (Saikia et al., 2010; Pichlmair et al., 2011; Raychoudhuri et al., 2011; Zhou et al., 2013). In addition, IFIT2 may limit replication of VSV in brain (Fensterl et al., 2014).
IFI Proteins
IFI44 has 444 amino acids, whereas IFI44L has 452 residues; the two proteins share 45% amino acid identity. Recently, it has been shown that the protein abundance of both IFI44 and IFI44L is enhanced by SUMO3 in response to IFNα (El-Asmi et al., 2020a). Overexpression of IFI44 has been shown to restrict Bunyamwera virus (Carlton-Smith and Elliott, 2012) and HIV-1 (Power et al., 2015) infection. IFI44L has weak antiviral activity against HCV infection (Schoggins et al., 2011). Recently, it has been reported that both IFI44 and IFI44L restrict replication of RSV (Busse et al., 2020). Indeed, overexpression of IFI44 or IFI44L is sufficient to restrict RSV infection at an early time post-infection. Knocking out these genes in human cells increases levels of infection, thus demonstrating a function for IFI44 and IFI44L in controlling RSV infection.
PKR
Double-stranded RNA (dsRNA)-dependent protein kinase (PKR) is a serine/threonine kinase that exerts its own phosphorylation and the phosphorylation of other substrates, the most studied being the α subunit of the protein synthesis initiation factor eIF-2α. This process results in a shut-off of protein translation and inhibition of virus replication (Garcia et al., 2007). PKR is ubiquitous and constitutively expressed. PKR is induced in an inactive form by IFN and activated by binding to viral dsRNA. In addition to being phosphorylated, PKR was also identified as a target of ISGylation on Lys-69 and Lys-159 by ISG15 (Okumura et al., 2013) and SUMOylation on Lys-60, Lys-150, and Lys-440 (de la Cruz-Herrera et al., 2014). More recently, PKR Lys-256 was also shown to be modified by SUMO (El-Asmi et al., 2020a). SUMOylation as well as non-covalent SUMO interaction are required for PKR-dsRNA binding, PKR dimerization, eIF-2α phosphorylation and anti-VSV activity (de la Cruz-Herrera et al., 2014, 2017).
Remarkably, SUMO1 and SUMO3 expression exert a differential effect on PKR activation SUMO1 expression alone results in PKR and eIF-2α phosphorylation, whereas SUMO3 reduces PKR and eIF-2α phosphorylation upon viral infection or dsRNA transfection (Maarifi et al., 2018). Furthermore, the higher SUMO1-induced PKR activation is correlated with an inhibition of EMCV. Importantly, SUMO1 by inducing PKR activation in the absence of viral infection and SUMO3 by counteracting both PKR activation and stability upon EMCV infection shed a new light on the differential effects of SUMO paralogs (Maarifi et al., 2018). It is interesting to note that PKR ISGylation at Lys-69 and Lys-159, both located in the ds-RNA binding motif, by ISG15 also triggers PKR and eIF-2α phosphorylation in the absence of viral infection (Okumura et al., 2013). Cells expressing human PKR confer partial resistance to EMCV that is correlated with PKR and eIF-2α phosphorylation (Meurs et al., 1992). Therefore the stabilization of PKR by SUMO3 in response to IFN (El-Asmi et al., 2020a) could enhance its antiviral function.
TRIM21
TRIM21 protein is an E3 ubiquitin ligase. The antiviral activity of TRIM21 varies among diverse viruses. Depletion of TRIM21 results in enhanced Coxsackievirus B3 (CVB3) replication, while its overexpression leads to increased IRF3 phosphorylation, increased IFNβ synthesis and reduced viral replication (Liu et al., 2018). More precisely, TRIM21 promotes IRF3 phosphorylation in infected cells by catalyzing the poly-ubiquitylation of MAVS, thereby enhancing type I IFN production. Interestingly, its implication in targeting viral proteins to regulate virus replication was recently shown (Mu et al., 2020). TRIM21 promotes the ubiquitylation and proteasomal degradation of HBV DNA polymerase using its RING finger domain, which has E3 ligase activity thus resulting in the restriction of HBV DNA replication (Mu et al., 2020).
ISG15
The antiviral actions of ISG15 are mediated by unconjugated ISG15 and/or by ISGylated host and viral proteins (Morales and Lenschow, 2013; Perng and Lenschow, 2018; Freitas et al., 2020). Mice lacking ISG15 or the ISG15 E1 enzyme, ubiquitin-activating enzyme E1 homolog (UBE1L; also known as UBA7), are more susceptible to Sindbis virus, IAV and IBV (Lai et al., 2009; Morales et al., 2015; Perng and Lenschow, 2018). Also, it has been reported that the ISGylation of both host and viral proteins and the non-covalent binding of ISG15 to host proteins can disrupt viral replication (Perng and Lenschow, 2018).
Several studies showed that ISG15 functions as a critical antiviral molecule against many viruses including RSV, IAV, IBV, Sindbis virus, HSV-1, and HCV (Okumura et al., 2006; Lenschow et al., 2007; Hsiang et al., 2009; Kim and Yoo, 2010; Morales and Lenschow, 2013; Gonzalez-Sanz et al., 2016; Hermann and Bogunovic, 2017; Perng and Lenschow, 2018). Conversely, some viruses induce viral specific proteins that can deconjugate ISG15 from its target proteins or prevent the generation of ISGylated proteins, thus abrogating the antiviral response (Yuan and Krug, 2001; Frias-Staheli et al., 2007; Guerra et al., 2008). In contrast, deletion of Ubiquitin Specific Peptidase 18 (USP18), the major ISG15 specific protease, which counteracts ISG15 conjugation, increases ISGylation in cells and renders cells more resistant to viral infection in response to IFN (Perng and Lenschow, 2018), suggesting that ISGylation is essential for host defense against viral infections.
Interestingly, it has been reported that protein ISGylation enhances condensation of STAT1 and STAT2, and their association with PML NBs (Banani et al., 2016), suggesting that ISGylation contribute with PML NBs to the formation of a favorable nuclear environment for the expression of specific genes. Therefore, SUMOylation–ISGylation network could play a central role in the stabilization of ISG products and in enhanced IFN-induced antiviral defense.
In conclusion, increased poly-SUMOylation upregulates IFN-induced ISGylation and stabilizes in TRIM25-dependent manner the level of several restriction factors known to inhibit various steps of virus cycle. Despite these mechanistic insights, cellular signaling by poly-SUMO chains is still incompletely understood. Therefore, further studies are needed to validate and identify the SUMOylation sites of several stabilized ISG products such as GBP1, GBP5, Tetherin/BST2 and members of IFIM and IFIT families. Also, some of the stabilized restriction factors such as TRIM25, MxB, and SAMHD1 have been shown to be SUMOylated (El-Asmi et al., 2020a) though it is unknown whether their SUMOylation is required for their antiviral activities. Remarkably, by increasing poly-SUMOylation, IFNα targets both SUMO and ISG15 pathways. Indeed, IFNα enhances SUMOylation of Ubc9 and proteins implicated in ubiquitin pathway (Maroui et al., 2018), and also stabilizes ISG15 and proteins implicated in ISG15 pathway (El-Asmi et al., 2020a).
Conclusion and Perspectives
ProMyelocytic leukemia protein, the organizer of PML NBs, is essential for IFN-enhanced global cellular SUMOylation and the SUMOylation of key players of IFN pathway. Its RNF4-dependent degradation later during IFN treatment suggests a negative regulation of SUMOylation and a return to cell homeostasis. Importantly, increased poly-SUMOylation in response to IFN upregulates both IFN-induced ISGylation and the expression of several restriction factors resulting in an enhanced IFN-induced antiviral state.
Multiple reports have demonstrated that SUMOylation and ISGylation are important for both intrinsic and innate immune antiviral responses. However, the biological significance of their cooperation in protein stability and antiviral defense is just emerging and necessitates further studies to clarify the role of the crosstalk between SUMOylation and ISGylation in intrinsic and innate immune responses. In this context, system-level identification of Ubl substrates using high sensitivity MS-based proteomic approaches is playing a key role to profile the changes in modification upon different cell stimuli and to identify acceptor sites.
The functions of poly-SUMO chains are growing. Recent findings showed that TRIM25 and RNF4, respectively, drive the SUMO3-dependent stabilization and destabilization of target proteins in response to IFNα. Whereas the ubiquitylation of poly-SUMO chains in proteasome-dependent protein degradation is well established, the ISGylation of poly-SUMO chains needs further experiments. UbcH8 and TRIM25 serve as E2- and E3-conjugating enzyme for both ubiquitin and ISG15, respectively. The identification of poly-SUMO-ISG15 chains and the determination whether TRIM25 and others Ubl conjugating enzyme could also act as a poly-SUMO-ISG15 ligase may play a key role in future discoveries of crosstalk between SUMOylation and ISGylation.
Further studies on the formation of hybrid chains ubiquitin-ISG15, SUMO-ubiquitin, SUMO-ISG15 and on the enzymes catalyzing their formation will confer an extra layer of complexity. The technological advances that led to the large-scale identification of Ubl substrates and their sites of modification provide important insights into Ubl functions. In particular, the identification of the acceptor sites, the sequence motifs, the types of branching, and the domains on which these modifications are located on target substrates in response to cell stimuli have significantly extend our understanding of Ubl regulation, The characterization of the interplay between SUMOylation, ubiquitylation and ISGylation pathways could help to identify antiviral targets, offering new opportunities to progress in antiviral defense mechanisms.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- CVB3
coxsackievirus B3
- Daxx
death domain associated protein
- DENV
dengue virus
- GBP
guanylate binding protein
- HIV-1
human immunodeficiency virus 1
- HCV
hepatitis C virus
- HSV-1
herpes simplex virus 1
- IAV
influenza A virus
- IBV
influenza B virus
- IFN
interferon
- IFI16
IFN γ inducible protein 16
- IFITM
interferon-induced transmembrane proteins
- IFIT
IFN-induced protein with tetratricopeptide repeats
- ISGs
IFN-stimulated genes
- KSHV
Kaposi’s sarcoma-associated herpesvirus
- MLV
murine leukemia virus
- Mx
myxovirus resistance protein
- PKR
double-stranded RNA-dependent protein kinase
- PML
ProMyelocytic leukemia protein
- RSV
respiratory syncytial virus
- SAMHD1
SAM domain and HD domain-containing
- SAMD9
Sterile Alpha Motif containing domain 9
- SUMO
Small Ubiquitin-related MOdifier
- STAT
signal transducer and activator of transcription
- TRIM
TRIpartite motif
- VACV
vaccinia virus
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- Ni-NTA
nickel-nitrilotriacetic acid
- Ubl
ubiquitin-like.
Footnotes
Funding. This work was funded by the Agence Nationale de Recherche sur le SIDA et les Hépatites Virales (ANRS ECTZ1994, MC-A) and the National Science and Engineering Research Council (NSERC 311598, PT). The Institute for Research in Immunology and Cancer (IRIC) receives infrastructure support from IRICoR and the Fonds de Recherche du Québec-Santé (FRQS). IRIC proteomics facility is a Genomics Technology platform funded in part by the Canadian Government through Genome Canada.
References
- Accola M. A., Huang B., Al Masri A., Mcniven M. A. (2002). The antiviral dynamin family member, MxA, tubulates lipids and localizes to the smooth endoplasmic reticulum. J. Biol. Chem. 277 21829–21835. 10.1074/jbc.m201641200 [DOI] [PubMed] [Google Scholar]
- Al-Khatib K., Williams B. R., Silverman R. H., Halford W., Carr D. J. (2003). The murine double-stranded RNA-dependent protein kinase PKR and the murine 2’,5’-oligoadenylate synthetase-dependent RNase L are required for IFN-beta-mediated resistance against herpes simplex virus type 1 in primary trigeminal ganglion culture. Virology 313 126–135. 10.1016/s0042-6822(03)00298-8 [DOI] [PubMed] [Google Scholar]
- Amini-Bavil-Olyaee S., Choi Y. J., Lee J. H., Shi M., Huang I. C., Farzan M., et al. (2013). The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe 13 452–464. 10.1016/j.chom.2013.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S. L., Carton J. M., Lou J., Xing L., Rubin B. Y. (1999). Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology 256 8–14. 10.1006/viro.1999.9614 [DOI] [PubMed] [Google Scholar]
- Atwan Z., Wright J., Woodman A., Leppard K. N. (2016). Promyelocytic leukemia protein isoform II inhibits infection by human adenovirus type 5 through effects on HSP70 and the interferon response. J. Gen. Virol. 97 1955–1967. 10.1099/jgv.0.000510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baczyk D., Audette M. C., Drewlo S., Levytska K., Kingdom J. C. (2017). SUMO-4: a novel functional candidate in the human placental protein SUMOylation machinery. PLoS One 12:e0178056. 10.1371/journal.pone.0178056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S., Roberts P. C., Brown L. E., Truong H., Pattnaik A. K., Archer D. R., et al. (2000). Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13 129–141. 10.1016/s1074-7613(00)00014-5 [DOI] [PubMed] [Google Scholar]
- Baldauf H. M., Pan X., Erikson E., Schmidt S., Daddacha W., Burggraf M., et al. (2012). SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat. Med. 18 1682–1687. 10.1038/nm.2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani S. F., Rice A. M., Peeples W. B., Lin Y., Jain S., Parker R., et al. (2016). Compositional control of phase-separated cellular bodies. Cell 166 651–663. 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrat F. J., Crow M. K., Ivashkiv L. B. (2019). Interferon target-gene expression and epigenomic signatures in health and disease. Nat. Immunol. 20 1574–1583. 10.1038/s41590-019-0466-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R., Papa A., Pandolfi P. P. (2008). Regulation of apoptosis by PML and the PML-NBs. Oncogene 27 6299–6312. 10.1038/onc.2008.305 [DOI] [PubMed] [Google Scholar]
- Berry K. N., Kober D. L., Su A., Brett T. J. (2018). Limiting respiratory viral infection by targeting antiviral and immunological functions of BST-2/tetherin: knowledge and gaps. Bioessays 40:e1800086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaszczyk K., Olejnik A., Nowicka H., Ozgyin L., Chen Y. L., Chmielewski S., et al. (2015). STAT2/IRF9 directs a prolonged ISGF3-like transcriptional response and antiviral activity in the absence of STAT1. Biochem. J. 466 511–524. 10.1042/bj20140644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel D., Kheddache S., Lahaye X., Dianoux L., Chelbi-Alix M. K. (2010). Resistance to rabies virus infection conferred by the PMLIV isoform. J. Virol. 84 10719–10726. 10.1128/jvi.01286-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel D., Maarifi G., Nisole S., Chelbi-Alix M. K. (2015). Resistance to Rhabdoviridae infection and subversion of antiviral responses. Viruses 7 3675–3702. 10.3390/v7072794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel D., Regad T., Poisson N., Pavie B., Harper F., Pandolfi P. P., et al. (2002). Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 21 7957–7970. 10.1038/sj.onc.1205931 [DOI] [PubMed] [Google Scholar]
- Boddy M. N., Howe K., Etkin L. D., Solomon E., Freemont P. S. (1996). PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene 13 971–982. [PubMed] [Google Scholar]
- Bonilla W. V., Pinschewer D. D., Klenerman P., Rousson V., Gaboli M., Pandolfi P. P., et al. (2002). Effects of promyelocytic leukemia protein on virus-host balance. J. Virol. 76 3810–3818. 10.1128/jvi.76.8.3810-3818.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden E. C., Sen G. C., Uze G., Silverman R. H., Ransohoff R. M., Foster G. R., et al. (2007). Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6 975–990. 10.1038/nrd2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutell C., Orr A., Everett R. D. (2003). PML residue lysine 160 is required for the degradation of PML induced by herpes simplex virus type 1 regulatory protein ICP0. J. Virol. 77 8686–8694. 10.1128/jvi.77.16.8686-8694.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantis-de-Carvalho C. E., Maarifi G., Goncalves Boldrin P. E., Zanelli C. F., Nisole S., Chelbi-Alix M. K., et al. (2015). MxA interacts with and is modified by the SUMOylation machinery. Exp. Cell Res. 330 151–163. 10.1016/j.yexcr.2014.10.020 [DOI] [PubMed] [Google Scholar]
- Brass A. L., Huang I. C., Benita Y., John S. P., Krishnan M. N., Feeley E. M., et al. (2009). The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139 1243–1254. 10.1016/j.cell.2009.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse D. C., Habgood-Coote D., Clare S., Brandt C., Bassano I., Kaforou M., et al. (2020). Interferon-induced protein 44 and interferon-induced protein 44-like restrict replication of Respiratory syncytial virus. J. Virol. 94:e00297-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton-Smith C., Elliott R. M. (2012). Viperin, MTAP44, and protein kinase R contribute to the interferon-induced inhibition of Bunyamwera Orthobunyavirus replication. J. Virol. 86 11548–11557. 10.1128/jvi.01773-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie J. S., Acharya S., Leonard M., Schmid S. L., Dyda F. (2010). G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature 465 435–440. 10.1038/nature09032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelbi-Alix M. K., de The H. (1999). Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18 935–941. 10.1038/sj.onc.1202366 [DOI] [PubMed] [Google Scholar]
- Chelbi-Alix M. K., Pelicano L., Quignon F., Koken M. H., Venturini L., Stadler M., et al. (1995). Induction of the PML protein by interferons in normal and APL cells. Leukemia 9 2027–2033. [PubMed] [Google Scholar]
- Chelbi-Alix M. K., Quignon F., Pelicano L., Koken M. H., De The H. (1998). Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J. Virol. 72 1043–1051. 10.1128/jvi.72.2.1043-1051.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelbi-Alix M. K., Wietzerbin J. (2007). Interferon, a growing cytokine family: 50 years of interferon research. Biochimie 89 713–718. 10.1016/j.biochi.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Chen S. T., Chen L., Lin D. S., Chen S. Y., Tsao Y. P., Guo H., et al. (2019). NLRP12 regulates anti-viral RIG-I activation via interaction with TRIM25. Cell Host Microbe 25 602–616.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhu M., Pan X., Zhu Y., Yan H., Jiang T., et al. (2014). Inhibition of Hepatitis B virus replication by SAMHD1. Biochem. Biophys. Res. Commun. 450 1462–1468. [DOI] [PubMed] [Google Scholar]
- Cheon H., Holvey-Bates E. G., Schoggins J. W., Forster S., Hertzog P., Imanaka N., et al. (2013). IFNbeta-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 32 2751–2763. 10.1038/emboj.2013.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y., Yang X. (2011). SUMO E3 ligase activity of TRIM proteins. Oncogene 30 1108–1116. 10.1038/onc.2010.462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crameri M., Bauer M., Caduff N., Walker R., Steiner F., Franzoso F. D., et al. (2018). MxB is an interferon-induced restriction factor of human herpesviruses. Nat. Commun. 9:1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuchet D., Sykes A., Nicolas A., Orr A., Murray J., Sirma H., et al. (2011). PML isoforms I and II participate in PML-dependent restriction of HSV-1 replication. J. Cell Sci. 124 280–291. 10.1242/jcs.075390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafa-Berger A., Kuzmina A., Fassler M., Yitzhak-Asraf H., Shemer-Avni Y., Taube R. (2012). Modulation of hepatitis C virus release by the interferon-induced protein BST-2/tetherin. Virology 428 98–111. 10.1016/j.virol.2012.03.011 [DOI] [PubMed] [Google Scholar]
- D’Andrea L. D., Regan L. (2003). TPR proteins: the versatile helix. Trends Biochem. Sci. 28 655–662. 10.1016/j.tibs.2003.10.007 [DOI] [PubMed] [Google Scholar]
- de la Cruz-Herrera C. F., Baz-Martinez M., Motiam A. E., Vidal S., Collado M., Vidal A., et al. (2017). Phosphorylable tyrosine residue 162 in the double-stranded RNA-dependent kinase PKR modulates its interaction with SUMO. Sci. Rep. 7:14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz-Herrera C. F., Campagna M., Garcia M. A., Marcos-Villar L., Lang V., Baz-Martinez M., et al. (2014). Activation of the double-stranded RNA-dependent protein kinase PKR by small ubiquitin-like modifier (SUMO). J. Biol. Chem. 289 26357–26367. 10.1074/jbc.m114.560961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai S. D., Haas A. L., Wood L. M., Tsai Y. C., Pestka S., Rubin E. H., et al. (2006). Elevated expression of ISG15 in tumor cells interferes with the ubiquitin/26S proteasome pathway. Cancer Res. 66 921–928. 10.1158/0008-5472.can-05-1123 [DOI] [PubMed] [Google Scholar]
- Desterro J. M., Thomson J., Hay R. T. (1997). Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 417 297–300. 10.1016/s0014-5793(97)01305-7 [DOI] [PubMed] [Google Scholar]
- Domingues P., Golebiowski F., Tatham M. H., Lopes A. M., Taggart A., Hay R. T., et al. (2015). Global reprogramming of host SUMOylation during influenza virus infection. Cell Rep. 13 1467–1480. 10.1016/j.celrep.2015.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzimianski J. V., Scholte F. E. M., Bergeron E., Pegan S. D. (2019). ISG15: it’s complicated. J. Mol. Biol. 431 4203–4216. 10.1016/j.jmb.2019.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Asmi F., Brantis-De-Carvalho C. E., Blondel D., Chelbi-Alix M. K. (2018). Rhabdoviruses, antiviral defense, and SUMO pathway. Viruses 10:686. 10.3390/v10120686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Asmi F., Maroui M. A., Dutrieux J., Blondel D., Nisole S., Chelbi-Alix M. K. (2014). Implication of PMLIV in both intrinsic and innate immunity. PLoS Pathog. 10:e1003975. 10.1371/journal.ppat.1003975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Bougrini J., Dianoux L., Chelbi-Alix M. K. (2011). PML positively regulates interferon gamma signaling. Biochimie 93 389–398. 10.1016/j.biochi.2010.11.005 [DOI] [PubMed] [Google Scholar]
- El McHichi B., Regad T., Maroui M. A., Rodriguez M. S., Aminev A., Gerbaud S., et al. (2010). SUMOylation promotes PML degradation during encephalomyocarditis virus infection. J. Virol. 84 11634–11645. 10.1128/jvi.01321-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Motiam A., Vidal S., Seoane R., Bouzaher Y. H., Gonzalez-Santamaria J., Rivas C. (2020). SUMO and cytoplasmic RNA viruses: from enemies to best friends. Adv. Exp. Med. Biol. 1233 263–277. 10.1007/978-3-030-38266-7_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Asmi F., El-Mchichi B., Maroui M. A., Dianoux L., Chelbi-Alix M. K. (2019). TGF-beta induces PML SUMOylation, degradation and PML nuclear body disruption. Cytokine 120 264–272. 10.1016/j.cyto.2019.05.008 [DOI] [PubMed] [Google Scholar]
- El-Asmi F., Mcmanus F. P., Brantis-de-Carvalho C. E., Valle-Casuso J. C., Thibault P., Chelbi-Alix M. K. (2020a). Cross-talk between SUMOylation and ISGylation in response to interferon. Cytokine 129:155025. 10.1016/j.cyto.2020.155025 [DOI] [PubMed] [Google Scholar]
- El-Asmi F., Mcmanus F. P., Thibault P., Chelbi-Alix M. K. (2020b). Interferon, restriction factors and SUMO pathways. Cytokine Growth Factor Rev. 55 37–47. 10.1016/j.cytogfr.2020.03.001 [DOI] [PubMed] [Google Scholar]
- Erker Y., Neyret-Kahn H., Seeler J. S., Dejean A., Atfi A., Levy L. (2013). Arkadia, a novel SUMO-targeted ubiquitin ligase involved in PML degradation. Mol. Cell. Biol. 33 2163–2177. 10.1128/mcb.01019-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R. D., Boutell C., Hale B. G. (2013). Interplay between viruses and host sumoylation pathways. Nat. Rev. Microbiol. 11 400–411. 10.1038/nrmicro3015 [DOI] [PubMed] [Google Scholar]
- Everett R. D., Chelbi-Alix M. K. (2007). PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89 819–830. 10.1016/j.biochi.2007.01.004 [DOI] [PubMed] [Google Scholar]
- Everett R. D., Maul G. G. (1994). HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13 5062–5069. 10.1002/j.1460-2075.1994.tb06835.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J. B., Arimoto K., Motamedchaboki K., Yan M., Wolf D. A., Zhang D. E. (2015). Identification and characterization of a novel ISG15-ubiquitin mixed chain and its role in regulating protein homeostasis. Sci. Rep. 5:12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J. B., Miyauchi S., Xu H. Z., Liu D., Kim L. J. Y., Burkart C., et al. (2020). Type I interferon regulates a coordinated gene network to enhance cytotoxic T cell-mediated tumor killing. Cancer Discov. 10 382–393. 10.1158/2159-8290.cd-19-0608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley E. M., Sims J. S., John S. P., Chin C. R., Pertel T., Chen L. M., et al. (2011). IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 7:e1002337. 10.1371/journal.ppat.1002337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Cao Z., Wang L., Wan Y., Peng N., Wang Q., et al. (2017). Inducible GBP5 mediates the antiviral response via interferon-related pathways during influenza A virus infection. J. Innate Immun. 9 419–435. 10.1159/000460294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensterl V., Wetzel J. L., Sen G. C. (2014). Interferon-induced protein Ifit2 protects mice from infection of the peripheral nervous system by vesicular stomatitis virus. J. Virol. 88 10303–10311. 10.1128/jvi.01341-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotho A., Melchior F. (2013). Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 82 357–385. 10.1146/annurev-biochem-061909-093311 [DOI] [PubMed] [Google Scholar]
- Freitas B. T., Scholte F. E. M., Bergeron E., Pegan S. D. (2020). How ISG15 combats viral infection. Virus Res. 286:198036. 10.1016/j.virusres.2020.198036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Staheli N., Giannakopoulos N. V., Kikkert M., Taylor S. L., Bridgen A., Paragas J., et al. (2007). Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2 404–416. 10.1016/j.chom.2007.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Xiong Y., Xu Y., Cheng G., Tang H. (2011). MDA5 is SUMOylated by PIAS2beta in the upregulation of type I interferon signaling. Mol. Immunol. 48 415–422. 10.1016/j.molimm.2010.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack M. U., Shin Y. C., Joo C. H., Urano T., Liang C., Sun L., et al. (2007). TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446 916–920. 10.1038/nature05732 [DOI] [PubMed] [Google Scholar]
- Galisson F., Mahrouche L., Courcelles M., Bonneil E., Meloche S., Chelbi-Alix M. K., et al. (2011). A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Mol. Cell. Proteomics 10:M110.004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan M., Poluektova L. Y., Tuma D. J., Kharbanda K. K., Osna N. A. (2016). Acetaldehyde disrupts interferon alpha signaling in hepatitis C virus-infected liver cells by up-regulating USP18. Alcohol. Clin. Exp. Res. 40 2329–2338. 10.1111/acer.13226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M. A., Meurs E. F., Esteban M. (2007). The dsRNA protein kinase PKR: virus and cell control. Biochimie 89 799–811. 10.1016/j.biochi.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Gartner A., Wagner K., Holper S., Kunz K., Rodriguez M. S., Muller S. (2018). Acetylation of SUMO2 at lysine 11 favors the formation of non-canonical SUMO chains. EMBO Rep. 19:e46117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy M. C., Chelbi-Alix M. K. (2011). Role of promyelocytic leukemia protein in host antiviral defense. J. Interferon Cytokine Res. 31 145–158. 10.1089/jir.2010.0111 [DOI] [PubMed] [Google Scholar]
- Giannakopoulos N. V., Luo J. K., Papov V., Zou W., Lenschow D. J., Jacobs B. S., et al. (2005). Proteomic identification of proteins conjugated to ISG15 in mouse and human cells. Biochem. Biophys. Res. Commun. 336 496–506. 10.1016/j.bbrc.2005.08.132 [DOI] [PubMed] [Google Scholar]
- Gillot I., Matthews C., Puel D., Vidal F., Lopez P. (2009). Ret Finger Protein: an E3 ubiquitin ligase juxtaposed to the XY body in meiosis. Int. J. Cell Biol. 2009:524858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni F., Damonte E. B., Garcia C. C. (2015). Cellular promyelocytic leukemia protein is an important dengue virus restriction factor. PLoS One 10:e0125690. 10.1371/journal.pone.0125690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone D. C., Ennis-Adeniran V., Hedden J. J., Groom H. C., Rice G. I., Christodoulou E., et al. (2011). HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480 379–382. 10.1038/nature10623 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Sanz R., Mata M., Bermejo-Martin J., Alvarez A., Cortijo J., Melero J. A., et al. (2016). ISG15 is upregulated in respiratory syncytial virus infection and reduces virus growth through protein ISGylation. J. Virol. 90 3428–3438. 10.1128/jvi.02695-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon C., Moncorge O., Bauby H., Doyle T., Barclay W. S., Malim M. H. (2014). Transfer of the amino-terminal nuclear envelope targeting domain of human MX2 converts MX1 into an HIV-1 resistance factor. J. Virol. 88 9017–9026. 10.1128/jvi.01269-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon C., Moncorge O., Bauby H., Doyle T., Ward C. C., Schaller T., et al. (2013). Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 502 559–562. 10.1038/nature12542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramberg T., Kahle T., Bloch N., Wittmann S., Mullers E., Daddacha W., et al. (2013). Restriction of diverse retroviruses by SAMHD1. Retrovirology 10:26. 10.1016/j.it.2011.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotzinger T., Jensen K., Will H. (1996). The interferon (IFN)-stimulated gene Sp100 promoter contains an IFN-gamma activation site and an imperfect IFN-stimulated response element which mediate type I IFN inducibility. J. Biol. Chem. 271 25253–25260. 10.1074/jbc.271.41.25253 [DOI] [PubMed] [Google Scholar]
- Guerra S., Caceres A., Knobeloch K. P., Horak I., Esteban M. (2008). Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog. 4:e1000096. 10.1371/journal.ppat.1000096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Hui D. J., Merrick W. C., Sen G. C. (2000). A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 19 6891–6899. 10.1093/emboj/19.24.6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A. L., Ahrens P., Bright P. M., Ankel H. (1987). Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J. Biol. Chem. 262 11315–11323. 10.1016/s0021-9258(18)60961-5 [DOI] [PubMed] [Google Scholar]
- Haller O., Stertz S., Kochs G. (2007). The Mx GTPase family of interferon-induced antiviral proteins. Microbes Infect. 9 1636–1643. 10.1016/j.micinf.2007.09.010 [DOI] [PubMed] [Google Scholar]
- Hannoun Z., Maarifi G., Chelbi-Alix M. K. (2016). The implication of SUMO in intrinsic and innate immunity. Cytokine Growth Factor Rev. 29 3–16. 10.1016/j.cytogfr.2016.04.003 [DOI] [PubMed] [Google Scholar]
- Harty R. N., Pitha P. M., Okumura A. (2009). Antiviral activity of innate immune protein ISG15. J. Innate Immun. 1 397–404. 10.1159/000226245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay R. T. (2013). Decoding the SUMO signal. Biochem. Soc. Trans. 41 463–473. 10.1042/bst20130015 [DOI] [PubMed] [Google Scholar]
- Hendriks I. A., D’Souza R. C., Yang B., Verlaan-De Vries M., Mann M., Vertegaal A. C. (2014). Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 21 927–936. 10.1038/nsmb.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann M., Bogunovic D. (2017). ISG15: in sickness and in health. Trends Immunol. 38 79–93. 10.1016/j.it.2016.11.001 [DOI] [PubMed] [Google Scholar]
- Hollenbaugh J. A., Gee P., Baker J., Daly M. B., Amie S. M., Tate J., et al. (2013). Host factor SAMHD1 restricts DNA viruses in non-dividing myeloid cells. PLoS Pathog. 9:e1003481. 10.1371/journal.ppat.1003481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horisberger M. A., Staeheli P., Haller O. (1983). Interferon induces a unique protein in mouse cells bearing a gene for resistance to influenza virus. Proc. Natl. Acad. Sci. U.S.A. 80 1910–1914. 10.1073/pnas.80.7.1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotter D., Sauter D., Kirchhoff F. (2017). Guanylate binding protein 5: impairing virion infectivity by targeting retroviral envelope glycoproteins. Small GTPases 8 31–37. 10.1080/21541248.2016.1189990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang T. Y., Zhao C., Krug R. M. (2009). Interferon-induced ISG15 conjugation inhibits influenza A virus gene expression and replication in human cells. J. Virol. 83 5971–5977. 10.1128/jvi.01667-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I. C., Bailey C. C., Weyer J. L., Radoshitzky S. R., Becker M. M., Chiang J. J., et al. (2011). Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 7:e1001258. 10.1371/journal.ppat.1001258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. F., Wee S., Gunaratne J., Lane D. P., Bulavin D. V. (2014). Isg15 controls p53 stability and functions. Cell Cycle 13 2200–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impens F., Radoshevich L., Cossart P., Ribet D. (2014). Mapping of SUMO sites and analysis of SUMOylation changes induced by external stimuli. Proc. Natl. Acad. Sci. U.S.A. 111 12432–12437. 10.1073/pnas.1413825111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs A., Lindenmann J. (1957). Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 147 258–267. [PubMed] [Google Scholar]
- Itsui Y., Sakamoto N., Kakinuma S., Nakagawa M., Sekine-Osajima Y., Tasaka-Fujita M., et al. (2009). Antiviral effects of the interferon-induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology 50 1727–1737. 10.1002/hep.23195 [DOI] [PubMed] [Google Scholar]
- Janzen C., Kochs G., Haller O. (2000). A monomeric GTPase-negative MxA mutant with antiviral activity. J. Virol. 74 8202–8206. 10.1128/jvi.74.17.8202-8206.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K., Shiels C., Freemont P. S. (2001). PML protein isoforms and the RBCC/TRIM motif. Oncogene 20 7223–7233. 10.1038/sj.onc.1204765 [DOI] [PubMed] [Google Scholar]
- Jiang D., Weidner J. M., Qing M., Pan X. B., Guo H., Xu C., et al. (2010). Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J. Virol. 84 8332–8341. 10.1128/jvi.02199-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. S., Blobel G. (1997). Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272 26799–26802. 10.1074/jbc.272.43.26799 [DOI] [PubMed] [Google Scholar]
- Jouvenet N., Neil S. J., Zhadina M., Zang T., Kratovac Z., Lee Y., et al. (2009). Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J. Virol. 83 1837–1844. 10.1128/jvi.02211-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani T., Kito K., Nguyen H. P., Wada H., Fukuda-Kamitani T., Yeh E. T. (1998). Identification of three major sentrinization sites in PML. J. Biol. Chem. 273 26675–26682. 10.1074/jbc.273.41.26675 [DOI] [PubMed] [Google Scholar]
- Kane M., Yadav S. S., Bitzegeio J., Kutluay S. B., Zang T., Wilson S. J., et al. (2013). MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502 563–566. 10.1038/nature12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. T., White T. E., Brandariz-Nunez A., Diaz-Griffero F., Weitzman M. D. (2013). SAMHD1 restricts herpes simplex virus 1 in macrophages by limiting DNA replication. J. Virol. 87 12949–12956. 10.1128/jvi.02291-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. I., Giannakopoulos N. V., Virgin H. W., Zhang D. E. (2004). Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol. Cell. Biol. 24 9592–9600. 10.1128/mcb.24.21.9592-9600.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. J., Hwang S. Y., Imaizumi T., Yoo J. Y. (2008). Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J. Virol. 82 1474–1483. 10.1128/jvi.01650-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. J., Yoo J. Y. (2010). Inhibition of hepatitis C virus replication by IFN-mediated ISGylation of HCV-NS5A. J. Immunol. 185 4311–4318. 10.4049/jimmunol.1000098 [DOI] [PubMed] [Google Scholar]
- Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., et al. (2011). Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44 325–340. 10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. E., Ahn J. H. (2015). Positive role of promyelocytic leukemia protein in type I interferon response and its regulation by human cytomegalovirus. PLoS Pathog. 11:e1004785. 10.1371/journal.ppat.1004785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Kim E. T., Kim Y. E., Lee M. K., Kwon K. M., Kim K. I., et al. (2016). Consecutive inhibition of ISG15 expression and ISGylation by cytomegalovirus regulators. PLoS Pathog. 12:e1005850. 10.1371/journal.ppat.1005850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P., Flotho A., Klug H., Olsen J. V., Van Dijk W. J., Fish A., et al. (2008). Ubc9 sumoylation regulates SUMO target discrimination. Mol. Cell 31 371–382. 10.1016/j.molcel.2008.05.022 [DOI] [PubMed] [Google Scholar]
- Korant B. D., Blomstrom D. C., Jonak G. J., Knight E., Jr. (1984). Interferon-induced proteins. Purification and characterization of a 15,000-dalton protein from human and bovine cells induced by interferon. J. Biol. Chem. 259 14835–14839. 10.1016/s0021-9258(17)42679-2 [DOI] [PubMed] [Google Scholar]
- Krapp C., Hotter D., Gawanbacht A., Mclaren P. J., Kluge S. F., Sturzel C. M., et al. (2016). Guanylate Binding Protein (GBP) 5 is an interferon-inducible inhibitor of HIV-1 infectivity. Cell Host Microbe 19 504–514. 10.1016/j.chom.2016.02.019 [DOI] [PubMed] [Google Scholar]
- Kristiansen H., Gad H. H., Eskildsen-Larsen S., Despres P., Hartmann R. (2011). The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J. Interferon Cytokine Res. 31 41–47. 10.1089/jir.2010.0107 [DOI] [PubMed] [Google Scholar]
- Kubota T., Matsuoka M., Chang T. H., Tailor P., Sasaki T., Tashiro M., et al. (2008). Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J. Biol. Chem. 283 25660–25670. 10.1074/jbc.m804479200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Sabapathy K. (2019). RNF4-A paradigm for SUMOylation-mediated ubiquitination. Proteomics 19:e1900185. [DOI] [PubMed] [Google Scholar]
- Lahouassa H., Daddacha W., Hofmann H., Ayinde D., Logue E. C., Dragin L., et al. (2012). SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13 223–228. 10.1038/ni.2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C., Struckhoff J. J., Schneider J., Martinez-Sobrido L., Wolff T., Garcia-Sastre A., et al. (2009). Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and non-mouse-adapted influenza B virus infection. J. Virol. 83 1147–1151. 10.1128/jvi.00105-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., et al. (2008). Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 10 547–555. 10.1038/ncb1717 [DOI] [PubMed] [Google Scholar]
- Lamoliatte F., Caron D., Durette C., Mahrouche L., Maroui M. A., Caron-Lizotte O., et al. (2014). Large-scale analysis of lysine SUMOylation by SUMO remnant immunoaffinity profiling. Nat. Commun. 5:5409. [DOI] [PubMed] [Google Scholar]
- Lamoliatte F., Mcmanus F. P., Maarifi G., Chelbi-Alix M. K., Thibault P. (2017). Uncovering the SUMOylation and ubiquitylation crosstalk in human cells using sequential peptide immunopurification. Nat. Commun. 8:14109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. J., Fu R. M., Liang C., Sloan R. D. (2018). IFITM proteins inhibit HIV-1 protein synthesis. Sci. Rep. 8:14551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenschow D. J., Lai C., Frias-Staheli N., Giannakopoulos N. V., Lutz A., Wolff T., et al. (2007). IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. U.S.A. 104 1371–1376. 10.1073/pnas.0607038104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. F., Yu J., Li Y., Wang J., Li S., Zhang L., et al. (2016). Guanylate-Binding Protein 1, an interferon-induced GTPase, exerts an antiviral activity against classical swine fever virus depending on its GTPase activity. J. Virol. 90 4412–4426. 10.1128/jvi.02718-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Qu X., Liu X., Huan C., Wang H., Zhao Z., et al. (2020). GBP5 is an interferon-induced inhibitor of respiratory Syncytial Virus. J. Virol. 94: e01407-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Goraya M. U., Yuan X., Zhang B., Chiu S. H., Chen J. L. (2019). Functional involvement of interferon-inducible transmembrane proteins in antiviral immunity. Front. Microbiol. 10:1097. 10.3389/fmicb.2019.01097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebelt F., Sebastian R. M., Moore C. L., Mulder M. P. C., Ovaa H., Shoulders M. D., et al. (2019). SUMOylation and the HSF1-regulated chaperone network converge to promote proteostasis in response to heat shock. Cell Rep. 26 236–249.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Jiang M., Liu L., Yang Z., Ma Z., Liu S., et al. (2019). The long noncoding RNA Lnczc3h7a promotes a TRIM25-mediated RIG-I antiviral innate immune response. Nat. Immunol. 20 812–823. 10.1038/s41590-019-0379-0 [DOI] [PubMed] [Google Scholar]
- Liu G., Lee J. H., Parker Z. M., Acharya D., Chiang J. J., Van Gent M., et al. (2020). ISG15-dependent activation of the RNA sensor MDA5 and its antagonism by the SARS-CoV-2 papain-like protease. bioRxiv [Preprint]. 10.1101/2020.10.26.356048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Li M., Song Y., Xu W. (2018). TRIM21 restricts coxsackievirus B3 replication, cardiac and pancreatic injury via interacting with MAVS and positively regulating IRF3-mediated type-I interferon production. Front. Immunol. 9:2479. 10.3389/fimmu.2018.02479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Li X. L., Hassel B. A. (2003). Proteasomes modulate conjugation to the ubiquitin-like protein, ISG15. J. Biol. Chem. 278 1594–1602. 10.1074/jbc.m208123200 [DOI] [PubMed] [Google Scholar]
- Liu Z., Pan Q., Ding S., Qian J., Xu F., Zhou J., et al. (2013). The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe 14 398–410. 10.1016/j.chom.2013.08.015 [DOI] [PubMed] [Google Scholar]
- Loeb K. R., Haas A. L. (1992). The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J. Biol. Chem. 267 7806–7813. 10.1016/s0021-9258(18)42585-9 [DOI] [PubMed] [Google Scholar]
- Lowrey A. J., Cramblet W., Bentz G. L. (2017). Viral manipulation of the cellular sumoylation machinery. Cell Commun. Signal. 15:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Pan Q., Rong L., He W., Liu S. L., Liang C. (2011). The IFITM proteins inhibit HIV-1 infection. J. Virol. 85 2126–2137. 10.1128/jvi.01531-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maarifi G., El Asmi F., Maroui M. A., Dianoux L., Chelbi-Alix M. K. (2018). Differential effects of SUMO1 and SUMO3 on PKR activation and stability. Sci. Rep. 8:1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maarifi G., Hannoun Z., Geoffroy M. C., El Asmi F., Zarrouk K., Nisole S., et al. (2016). MxA mediates SUMO-induced resistance to vesicular stomatitis virus. J. Virol. 90 6598–6610. 10.1128/jvi.00722-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maarifi G., Maroui M. A., Dutrieux J., Dianoux L., Nisole S., Chelbi-Alix M. K. (2015). Small ubiquitin-like modifier alters IFN response. J. Immunol. 195 2312–2324. 10.4049/jimmunol.1500035 [DOI] [PubMed] [Google Scholar]
- Malakhov M. P., Kim K. I., Malakhova O. A., Jacobs B. S., Borden E. C., Zhang D. E. (2003). High-throughput immunoblotting. Ubiquitiin-like protein ISG15 modifies key regulators of signal transduction. J. Biol. Chem. 278 16608–16613. [DOI] [PubMed] [Google Scholar]
- Malakhova O., Malakhov M., Hetherington C., Zhang D. E. (2002). Lipopolysaccharide activates the expression of ISG15-specific protease UBP43 via interferon regulatory factor 3. J. Biol. Chem. 277 14703–14711. 10.1074/jbc.m111527200 [DOI] [PubMed] [Google Scholar]
- Manokaran G., Finol E., Wang C., Gunaratne J., Bahl J., Ong E. Z., et al. (2015). Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350 217–221. 10.1126/science.aab3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri M., Viswanathan K., Douglas J. L., Hines J., Gustin J., Moses A. V., et al. (2009). Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 83 9672–9681. 10.1128/jvi.00597-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos-Villar L., Perez-Giron J. V., Vilas J. M., Soto A., de la Cruz-Hererra C. F., Lang V., et al. (2013). SUMOylation of p53 mediates interferon activities. Cell Cycle 12 2809–2816. 10.4161/cc.25868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroui M. A., Kheddache-Atmane S., El Asmi F., Dianoux L., Aubry M., Chelbi-Alix M. K. (2012). Requirement of PML SUMO interacting motif for RNF4- or arsenic trioxide-induced degradation of nuclear PML isoforms. PLoS One 7:e44949. 10.1371/journal.pone.0044949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroui M. A., Maarifi G., Mcmanus F. P., Lamoliatte F., Thibault P., Chelbi-Alix M. K. (2018). Promyelocytic leukemia protein (PML) requirement for interferon-induced global cellular SUMOylation. Mol. Cell. Proteomics 17 1196–1208. 10.1074/mcp.ra117.000447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroui M. A., Pampin M., Chelbi-Alix M. K. (2011). Promyelocytic leukemia isoform IV confers resistance to encephalomyocarditis virus via the sequestration of 3D polymerase in nuclear bodies. J. Virol. 85 13164–13173. 10.1128/jvi.05808-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Vicente M., Medrano L. M., Resino S., Garcia-Sastre A., Martinez I. (2017). TRIM25 in the regulation of the antiviral innate immunity. Front. Immunol. 8:1187. 10.3389/fimmu.2017.01187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus F. P., Bourdeau V., Acevedo M., Lopes-Paciencia S., Mignacca L., Lamoliatte F., et al. (2018). Quantitative SUMO proteomics reveals the modulation of several PML nuclear body associated proteins and an anti-senescence function of UBC9. Sci. Rep. 8:7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally B. A., Trgovcich J., Maul G. G., Liu Y., Zheng P. (2008). A role for cytoplasmic PML in cellular resistance to viral infection. PLoS One 3:e2277. 10.1371/journal.pone.0002277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melen K., Keskinen P., Ronni T., Sareneva T., Lounatmaa K., Julkunen I. (1996). Human MxB protein, an interferon-alpha-inducible GTPase, contains a nuclear targeting signal and is localized in the heterochromatin region beneath the nuclear envelope. J. Biol. Chem. 271 23478–23486. 10.1074/jbc.271.38.23478 [DOI] [PubMed] [Google Scholar]
- Meroni G., Diez-Roux G. (2005). TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays 27 1147–1157. 10.1002/bies.20304 [DOI] [PubMed] [Google Scholar]
- Meurs E. F., Watanabe Y., Kadereit S., Barber G. N., Katze M. G., Chong K., et al. (1992). Constitutive expression of human double-stranded RNA-activated p68 kinase in murine cells mediates phosphorylation of eukaryotic initiation factor 2 and partial resistance to encephalomyocarditis virus growth. J. Virol. 66 5805–5814. 10.1128/jvi.66.10.5805-5814.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson N. R., Zhou L., Guo Y. R., Zhao C., Tao Y. J., Krug R. M., et al. (2017). Nuclear TRIM25 specifically targets influenza virus ribonucleoproteins to block the onset of RNA chain elongation. Cell Host Microbe 22 627–638.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi Z., Fu J., Xiong Y., Tang H. (2010). SUMOylation of RIG-I positively regulates the type I interferon signaling. Protein Cell 1 275–283. 10.1007/s13238-010-0030-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. S., Emerman M., Malik H. S. (2013). An evolutionary perspective on the broad antiviral specificity of MxA. Curr. Opin. Microbiol. 16 493–499. 10.1016/j.mib.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales D. J., Lenschow D. J. (2013). The antiviral activities of ISG15. J. Mol. Biol. 425 4995–5008. 10.1016/j.jmb.2013.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales D. J., Monte K., Sun L., Struckhoff J. J., Agapov E., Holtzman M. J., et al. (2015). Novel mode of ISG15-mediated protection against influenza A virus and Sendai virus in mice. J. Virol. 89 337–349. 10.1128/jvi.02110-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu T., Zhao X., Zhu Y., Fan H., Tang H. (2020). The E3 ubiquitin ligase TRIM21 promotes HBV DNA polymerase degradation. Viruses 12:346. 10.3390/v12030346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S., Dejean A. (1999). Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73 5137–5143. 10.1128/jvi.73.6.5137-5143.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S., Matunis M. J., Dejean A. (1998). Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17 61–70. 10.1093/emboj/17.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasato N., Ikeda K., Urano T., Horie-Inoue K., Takeda S., Inoue S. (2006). A ubiquitin E3 ligase Efp is up-regulated by interferons and conjugated with ISG15. Biochem. Biophys. Res. Commun. 351 540–546. 10.1016/j.bbrc.2006.10.061 [DOI] [PubMed] [Google Scholar]
- Narasimhan J., Potter J. L., Haas A. L. (1996). Conjugation of the 15-kDa interferon-induced ubiquitin homolog is distinct from that of ubiquitin. J. Biol. Chem. 271 324–330. 10.1074/jbc.271.1.324 [DOI] [PubMed] [Google Scholar]
- Neil S. J., Zang T., Bieniasz P. D. (2008). Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451 425–430. 10.1038/nature06553 [DOI] [PubMed] [Google Scholar]
- Nisole S., Maroui M. A., Mascle X. H., Aubry M., Chelbi-Alix M. K. (2013). Differential roles of PML isoforms. Front. Oncol. 3:125. 10.3389/fonc.2013.00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura A., Lu G., Pitha-Rowe I., Pitha P. M. (2006). Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc. Natl. Acad. Sci. U.S.A. 103 1440–1445. 10.1073/pnas.0510518103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura A., Pitha P. M., Harty R. N. (2008). ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc. Natl. Acad. Sci. U.S.A. 105 3974–3979. 10.1073/pnas.0710629105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura F., Okumura A. J., Uematsu K., Hatakeyama S., Zhang D. E., Kamura T. (2013). Activation of double-stranded RNA-activated protein kinase (PKR) by interferon-stimulated gene 15 (ISG15) modification down-regulates protein translation. J. Biol. Chem. 288 2839–2847. 10.1074/jbc.m112.401851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owerbach D., Mckay E. M., Yeh E. T., Gabbay K. H., Bohren K. M. (2005). A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem. Biophys. Res. Commun. 337 517–520. 10.1016/j.bbrc.2005.09.090 [DOI] [PubMed] [Google Scholar]
- Ozato K., Shin D. M., Chang T. H., Morse H. C., III (2008). TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 8 849–860. 10.1038/nri2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampin M., Simonin Y., Blondel B., Percherancier Y., Chelbi-Alix M. K. (2006). Cross talk between PML and p53 during poliovirus infection: implications for antiviral defense. J. Virol. 80 8582–8592. 10.1128/jvi.00031-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W., Zuo X., Feng T., Shi X., Dai J. (2012). Guanylate-binding protein 1 participates in cellular antiviral response to dengue virus. Virol. J. 9:292. 10.1186/1743-422x-9-292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng Y. C., Lenschow D. J. (2018). ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 16 423–439. 10.1038/s41579-018-0020-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A., Lassnig C., Eberle C. A., Gorna M. W., Baumann C. L., Burkard T. R., et al. (2011). IFIT1 is an antiviral protein that recognizes 5’-triphosphate RNA. Nat. Immunol. 12 624–630. 10.1038/ni.2048 [DOI] [PubMed] [Google Scholar]
- Pilla-Moffett D., Barber M. F., Taylor G. A., Coers J. (2016). Interferon-inducible GTPases in host resistance, inflammation and disease. J. Mol. Biol. 428 3495–3513. 10.1016/j.jmb.2016.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitha-Rowe I., Hassel B. A., Dmitrovsky E. (2004). Involvement of UBE1L in ISG15 conjugation during retinoid-induced differentiation of acute promyelocytic leukemia. J. Biol. Chem. 279 18178–18187. 10.1074/jbc.m309259200 [DOI] [PubMed] [Google Scholar]
- Porta C., Hadj-Slimane R., Nejmeddine M., Pampin M., Tovey M. G., Espert L., et al. (2005). Interferons alpha and gamma induce p53-dependent and p53-independent apoptosis, respectively. Oncogene 24 605–615. 10.1038/sj.onc.1208204 [DOI] [PubMed] [Google Scholar]
- Potter J. L., Narasimhan J., Mende-Mueller L., Haas A. L. (1999). Precursor processing of pro-ISG15/UCRP, an interferon-beta-induced ubiquitin-like protein. J. Biol. Chem. 274 25061–25068. 10.1074/jbc.274.35.25061 [DOI] [PubMed] [Google Scholar]
- Power D., Santoso N., Dieringer M., Yu J., Huang H., Simpson S., et al. (2015). IFI44 suppresses HIV-1 LTR promoter activity and facilitates its latency. Virology 481 142–150. 10.1016/j.virol.2015.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash B., Praefcke G. J., Renault L., Wittinghofer A., Herrmann C. (2000). Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature 403 567–571. 10.1038/35000617 [DOI] [PubMed] [Google Scholar]
- Radoshevich L., Impens F., Ribet D., Quereda J. J., Nam Tham T., Nahori M. A., et al. (2015). ISG15 counteracts Listeria monocytogenes infection. Elife 4:e06848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoshitzky S. R., Dong L., Chi X., Clester J. C., Retterer C., Spurgers K., et al. (2010). Infectious Lassa virus, but not filoviruses, is restricted by BST-2/tetherin. J. Virol. 84 10569–10580. 10.1128/jvi.00103-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R., Stoye J. P., O’garra A. (2008). Type I interferon-dependent and -independent expression of tripartite motif proteins in immune cells. Eur. J. Immunol. 38 619–630. 10.1002/eji.200737916 [DOI] [PubMed] [Google Scholar]
- Raychoudhuri A., Shrivastava S., Steele R., Kim H., Ray R., Ray R. B. (2011). ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J. Virol. 85 12881–12889. 10.1128/jvi.05633-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad T., Chelbi-Alix M. K. (2001). Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20 7274–7286. 10.1038/sj.onc.1204854 [DOI] [PubMed] [Google Scholar]
- Regad T., Saib A., Lallemand-Breitenbach V., Pandolfi P. P., de The H., Chelbi-Alix M. K. (2001). PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator. EMBO J. 20 3495–3505. 10.1093/emboj/20.13.3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich N., Evans B., Levy D., Fahey D., Knight E., Jr., Darnell J. E., Jr. (1987). Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc. Natl. Acad. Sci. U.S.A. 84 6394–6398. 10.1073/pnas.84.18.6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt M., Wang L., Sommer M., Perrino J., Nour A. M., Sen N., et al. (2011). Entrapment of viral capsids in nuclear PML cages is an intrinsic antiviral host defense against varicella-zoster virus. PLoS Pathog. 7:e1001266. 10.1371/journal.ppat.1001266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengachari S., Groiss S., Devos J. M., Caron E., Grandvaux N., Panne D. (2018). Structural basis of STAT2 recognition by IRF9 reveals molecular insights into ISGF3 function. Proc. Natl. Acad. Sci. U.S.A. 115 E601–E609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., et al. (2001). The tripartite motif family identifies cell compartments. EMBO J. 20 2140–2151. 10.1093/emboj/20.9.2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinfret Robert C., Mcmanus F. P., Lamoliatte F., Thibault P. (2020). Interplay of ubiquitin-like modifiers following arsenic trioxide treatment. J. Proteome Res. 19 1999–2010. 10.1021/acs.jproteome.9b00807 [DOI] [PubMed] [Google Scholar]
- Sahin U., Ferhi O., Carnec X., Zamborlini A., Peres L., Jollivet F., et al. (2014). Interferon controls SUMO availability via the Lin28 and let-7 axis to impede virus replication. Nat. Commun. 5:4187. [DOI] [PubMed] [Google Scholar]
- Saikia P., Fensterl V., Sen G. C. (2010). The inhibitory action of P56 on select functions of E1 mediates interferon’s effect on human papillomavirus DNA replication. J. Virol. 84 13036–13039. 10.1128/jvi.01194-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T., Noda T., Urata S., Kawaoka Y., Yasuda J. (2009). Inhibition of Lassa and Marburg virus production by tetherin. J. Virol. 83 2382–2385. 10.1128/jvi.01607-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling E. M., Scherer M., Reuter N., Schweininger J., Muller Y. A., Stamminger T. (2017). The human cytomegalovirus IE1 protein antagonizes PML nuclear body-mediated intrinsic immunity via the inhibition of PML de novo SUMOylation. J. Virol. 91:e02049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W. M., Chevillotte M. D., Rice C. M. (2014). Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32 513–545. 10.1146/annurev-immunol-032713-120231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J. W., Wilson S. J., Panis M., Murphy M. Y., Jones C. T., Bieniasz P., et al. (2011). A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472 481–485. 10.1038/nature09907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert U., Bialy L. P., Ebstein F., Bech-Otschir D., Voigt A., Schroter F., et al. (2010). Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell 142 613–624. 10.1016/j.cell.2010.07.036 [DOI] [PubMed] [Google Scholar]
- Shah S. J., Blumen S., Pitha-Rowe I., Kitareewan S., Freemantle S. J., Feng Q., et al. (2008). UBE1L represses PML/RAR{alpha} by targeting the PML domain for ISG15ylation. Mol. Cancer Ther. 7 905–914. 10.1158/1535-7163.mct-07-0515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen T. H., Lin H. K., Scaglioni P. P., Yung T. M., Pandolfi P. P. (2006). The mechanisms of PML-nuclear body formation. Mol. Cell 24 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H. X., Yang K., Liu X., Liu X. Y., Wei B., Shan Y. F., et al. (2010). Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol. Cell. Biol. 30 2424–2436. 10.1128/mcb.01466-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist F., Ebeling M., Certa U. (2011). The small interferon-induced transmembrane genes and proteins. J. Interferon Cytokine Res. 31 183–197. 10.1089/jir.2010.0112 [DOI] [PubMed] [Google Scholar]
- Sloan E., Tatham M. H., Groslambert M., Glass M., Orr A., Hay R. T., et al. (2015). Analysis of the SUMO2 proteome during HSV-1 infection. PLoS Pathog. 11:e1005059. 10.1371/journal.ppat.1005059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., Chen Y. (2004). Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 101 14373–14378. 10.1073/pnas.0403498101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriramachandran A. M., Dohmen R. J. (2014). SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta 1843 75–85. 10.1016/j.bbamcr.2013.08.022 [DOI] [PubMed] [Google Scholar]
- Sriramachandran A. M., Meyer-Teschendorf K., Pabst S., Ulrich H. D., Gehring N. H., Hofmann K., et al. (2019). Arkadia/RNF111 is a SUMO-targeted ubiquitin ligase with preference for substrates marked with SUMO1-capped SUMO2/3 chain. Nat. Commun. 10:3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler M., Chelbi-Alix M. K., Koken M. H., Venturini L., Lee C., Saib A., et al. (1995). Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene 11 2565–2573. [PubMed] [Google Scholar]
- Staeheli P., Haller O., Boll W., Lindenmann J., Weissmann C. (1986). Mx protein: constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell 44 147–158. 10.1016/0092-8674(86)90493-9 [DOI] [PubMed] [Google Scholar]
- Staeheli P., Pavlovic J. (1991). Inhibition of vesicular stomatitis virus mRNA synthesis by human MxA protein. J. Virol. 65 4498–4501. 10.1128/jvi.65.8.4498-4501.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz S., Reichelt M., Krijnse-Locker J., Mackenzie J., Simpson J. C., Haller O., et al. (2006). Interferon-induced, antiviral human MxA protein localizes to a distinct subcompartment of the smooth endoplasmic reticulum. J. Interferon Cytokine Res. 26 650–660. 10.1089/jir.2006.26.650 [DOI] [PubMed] [Google Scholar]
- Sung P. S., Cheon H., Cho C. H., Hong S. H., Park D. Y., Seo H. I., et al. (2015). Roles of unphosphorylated ISGF3 in HCV infection and interferon responsiveness. Proc. Natl. Acad. Sci. U.S.A. 112 10443–10448. 10.1073/pnas.1513341112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M., Omattage N. S., Brett T. J. (2013). BST-2/tetherin: structural biology, viral antagonism, and immunobiology of a potent host antiviral factor. Mol. Immunol. 54 132–139. 10.1016/j.molimm.2012.11.008 [DOI] [PubMed] [Google Scholar]
- Sze A., Belgnaoui S. M., Olagnier D., Lin R., Hiscott J., Van Grevenynghe J. (2013). Host restriction factor SAMHD1 limits human T cell leukemia virus type 1 infection of monocytes via STING-mediated apoptosis. Cell Host Microbe 14 422–434. 10.1016/j.chom.2013.09.009 [DOI] [PubMed] [Google Scholar]
- Tammsalu T., Matic I., Jaffray E. G., Ibrahim A. F., Tatham M. H., Hay R. T. (2015). Proteome-wide identification of SUMO modification sites by mass spectrometry. Nat. Protoc. 10 1374–1388. 10.1038/nprot.2015.095 [DOI] [PubMed] [Google Scholar]
- Tatham M. H., Geoffroy M. C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., et al. (2008). RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10 538–546. 10.1038/ncb1716 [DOI] [PubMed] [Google Scholar]
- Tavalai N., Stamminger T. (2008). New insights into the role of the subnuclear structure ND10 for viral infection. Biochim. Biophys. Acta 1783 2207–2221. 10.1016/j.bbamcr.2008.08.004 [DOI] [PubMed] [Google Scholar]
- Terenzi F., Hui D. J., Merrick W. C., Sen G. C. (2006). Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J. Biol. Chem. 281 34064–34071. 10.1074/jbc.m605771200 [DOI] [PubMed] [Google Scholar]
- Uchil P. D., Quinlan B. D., Chan W. T., Luna J. M., Mothes W. (2008). TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 4:e16. 10.1371/journal.ppat.0040016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungureanu D., Vanhatupa S., Gronholm J., Palvimo J. J., Silvennoinen O. (2005). SUMO-1 conjugation selectively modulates STAT1-mediated gene responses. Blood 106 224–226. 10.1182/blood-2004-11-4514 [DOI] [PubMed] [Google Scholar]
- van Damme E., Laukens K., Dang T. H., Van Ostade X. (2010). A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int. J. Biol. Sci. 6 51–67. 10.7150/ijbs.6.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestal D. J. (2005). The guanylate-binding proteins (GBPs): proinflammatory cytokine-induced members of the dynamin superfamily with unique GTPase activity. J. Interferon Cytokine Res. 25 435–443. 10.1089/jir.2005.25.435 [DOI] [PubMed] [Google Scholar]
- Vestal D. J., Jeyaratnam J. A. (2011). The guanylate-binding proteins: emerging insights into the biochemical properties and functions of this family of large interferon-induced guanosine triphosphatase. J. Interferon Cytokine Res. 31 89–97. 10.1089/jir.2010.0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Long J., Zheng C. F. (2012). The potential link between PML NBs and ICP0 in regulating lytic and latent infection of HSV-1. Protein Cell 3 372–382. 10.1007/s13238-012-2021-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu L., Su J., Peppelenbosch M. P., Pan Q. (2017a). Transcriptional regulation of antiviral interferon-stimulated genes. Trends Microbiol. 25 573–584. 10.1016/j.tim.2017.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Yin Y., Xu L., Su J., Huang F., Wang Y., et al. (2017b). Unphosphorylated ISGF3 drives constitutive expression of interferon-stimulated genes to protect against viral infections. Sci. Signal. 10:eaah4248. 10.1126/scisignal.aah4248 [DOI] [PubMed] [Google Scholar]
- Wang Z. G., Ruggero D., Ronchetti S., Zhong S., Gaboli M., Rivi R., et al. (1998). PML is essential for multiple apoptotic pathways. Nat. Genet. 20 266–272. 10.1038/3073 [DOI] [PubMed] [Google Scholar]
- Weidner J. M., Jiang D., Pan X. B., Chang J., Block T. M., Guo J. T. (2010). Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J. Virol. 84 12646–12657. 10.1128/jvi.01328-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek E., Kedracka-Krok S., Soltys K., Jankowska U., Holubowicz R., Seliga J., et al. (2016). Is transthyretin a regulator of Ubc9 SUMOylation? PLoS One 11:e0160536. 10.1371/journal.pone.0160536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer P., Schreiner S. (2015). Viral mimicry to usurp ubiquitin and SUMO host pathways. Viruses 7 4854–4872. 10.3390/v7092849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J. J., Pung Y. F., Sze N. S., Chin K. C. (2006). HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc. Natl. Acad. Sci. U.S.A. 103 10735–10740. 10.1073/pnas.0600397103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Paige J. S., Jaffrey S. R. (2010). Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 28 868–873. 10.1038/nbt.1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi D. R., An N., Liu Z. L., Xu F. W., Raniga K., Li Q. J., et al. (2019). Human MxB inhibits the replication of hepatitis C Virus. J. Virol. 93:e01285-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Li M., Wilkins J., Ding S., Swartz T. H., Esposito A. M., et al. (2015). IFITM proteins restrict HIV-1 infection by antagonizing the envelope glycoprotein. Cell Rep. 13 145–156. 10.1016/j.celrep.2015.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W., Krug R. M. (2001). Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20 362–371. 10.1093/emboj/20.3.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Denison C., Huibregtse J. M., Gygi S., Krug R. M. (2005). Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. U.S.A. 102 10200–10205. 10.1073/pnas.0504754102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Wang X., Tu F., Wang Q., Fan Z., Gao G. (2017). TRIM25 is required for the antiviral activity of Zinc Finger antiviral protein. J. Virol. 91:e00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Michal J. J., Zhang L., Ding B., Lunney J. K., Liu B., et al. (2013). Interferon induced IFIT family genes in host antiviral defense. Int. J. Biol. Sci. 9 200–208. 10.7150/ijbs.5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Koken M. H., Quignon F., Chelbi-Alix M. K., Degos L., Wang Z. Y., et al. (1997). Arsenic-induced PML targeting onto nuclear bodies: implications for the treatment of acute promyelocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 94 3978–3983. 10.1073/pnas.94.8.3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W., Wang J., Zhang D. E. (2007). Negative regulation of ISG15 E3 ligase EFP through its autoISGylation. Biochem. Biophys. Res. Commun. 354 321–327. 10.1016/j.bbrc.2006.12.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W., Zhang D. E. (2006). The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J. Biol. Chem. 281 3989–3994. 10.1074/jbc.m510787200 [DOI] [PubMed] [Google Scholar]
- Zou Z., Meng Z., Ma C., Liang D., Sun R., Lan K. (2017). Guanylate-binding protein 1 inhibits nuclear delivery of Kaposi’s sarcoma-associated herpesvirus virions by disrupting formation of actin filament. J. Virol. 91:e00632-17. [DOI] [PMC free article] [PubMed] [Google Scholar]