Abstract
Intervertebral disc degeneration and resulting low back pain arises from the programmed apoptosis of nucleus pulposus cells (NPCs). Recent studies show that hypoxia-inducible factor-1α plays a vital role in the etiology and pathogenesis of disc degeneration. However, the underlying mechanism of HIF-1α in NPCs is unclear. The present study identified 994 significant differentially expressed miRNAs by analyzing microarray data downloaded from the Gene Expression Omnibus database. MicroRNA(miR)-32-5p expression was 2.81-fold upregulated in NPCs compared with that of the healthy control tissues (P<0.05). A total of 331 significant differentially expressed mRNAs were identified, and PTEN was downregulated in NPCs of non-degenerative disc tissues from young patients. miR-32-5p was predicted to target the PTEN 3'-untranslated region (UTR). To confirm these results, in-vitro experiments investigating the molecular function of miR-32-5p and PTEN were performed. Furthermore, hypoxia induced miR-32-5p and PTEN expression. HIF-1α inhibited NPC proliferation and promoted cell apoptosis by regulating miR-32-5p and PTEN. miR-32-5p promoted NPC proliferation and decreased cell apoptosis. Next, it was verified whether miR-32-5p targeted the PTEN 3'-UTR using dual-luciferase reporter assays. Finally, it was observed that PI3K/AKT/mTOR signaling pathway was upregulated by a miR-32-5p mimic, which improved cell proliferation and decreased apoptosis. Importantly, PTEN was downregulated in these experiments; and inhibition of miR-32-5p had the opposite effect. Overall, these results demonstrate that HIF-1α regulates cell proliferation and apoptosis by controlling the miR-32-5p/PTEN/PI3K/AKT/mTOR axis in NPCs.
Keywords: hypoxia-inducible factor-1α, microRNA-32-5p, PTEN, nucleus pulposus cells, PI3K/AKT/mTOR signaling pathway
Introduction
Intervertebral disc degeneration is a leading cause of chronic low back pain, which imposes a substantial clinical and socioeconomic burden (1). Low proliferative viability and increased apoptosis of human nucleus pulposus cells (NPCs) are two common features of intervertebral disc degeneration. Notably, nucleus pulposus therapies are being developed to treat intervertebral disc degeneration, and clinical trials are advancing (2).
Hypoxia-inducible factor-1α (HIF-1α) is a crucial player in the progression of nucleus pulposus growth. A previous study revealed that HIF-1α regulates metabolic flux by coordinating glycolysis and mitochondrial TCA cycle interactions, thereby controlling the overall biosynthetic capacity of NPCs (3). Moreover, it was previously observed that the alteration of HIF-1α concentration affects NPC proliferation and apoptosis. Therefore, recombinant HIF-1α protein may be a potential preventive medicine and therapeutic application.
Phosphatase and tensin homolog (PTEN) has been extensively studied in various diseases, such as neoplastic and orthopedic diseases (4,5). PTEN regulates intervertebral disc degeneration, which is an orthopedic disease. Liu et al (6) found that PTEN affects NPC proliferation during intervertebral disc degeneration (6). Notably, PTEN inhibitors such as VO-OHpic are being studied as a potential therapy for intervertebral disc erosion (7). Indeed, PTEN is a critical factor in the development of degenerating intervertebral discs.
MicroRNA(miR)-32-5p is a potential diagnostic target in cancer (8). Previous studies reported that the long non-coding RNA (lncRNA) SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4(9). Wu et al (10) found that miR-32 regulates PTEN expression and promotes growth, migration and invasion in colorectal carcinoma cells. However, the involvement of miR-32-5p in intervertebral disc degeneration has not been investigated. Further, the mechanism underlying HIF-1α regulation by the miR-32-5p/PTEN axis has not yet been reported. In the present study, the differential expression of miR-32-5p and PTEN was investigated by analyzing microarray data. The influence of HIF-1α and the role of miR-32-5p or PTEN on NPCs in vitro was also studied. Overall, the findings suggest that the maintenance of the miR-32-5p/PTEN axis is critical for HIF-1α expression in NPCs during intervertebral disc homeostasis.
Materials and methods
Differentially expressed mRNA analysis by raw microarray data mining
Raw microarray data (GSE34095_RAW.tar) was downloaded from the NCBI Gene Expression Omnibus database (ncbi.nlm.nih.gov/geo). Three samples were from NPCs of young patients with non-degenerative disc tissues (GSM841717, GSM841719 and GSM841721), and three samples were taken from NPCs from elderly patients with disc degeneration (GSM841718, GSM841720 and GSM841722). An Affymetrix Human Genome U133A Array (cat. no. GPL96; Affymetrix; Thermo Fisher Scientific, Inc.) was analyzed using the Affymetrix Transcriptome Analysis Console (version 1.3). Differentially expressed mRNA was identified by |fold change (FC)|>1.5 and P<0.05.
Differentially expressed miRNA data mining using online GEO2R analysis
The GSE116726 dataset contained 3 NPC samples from healthy control tissues (GSM3259525, GSM3259526 and GSM3259527) and 3 samples of NPCs from patients with degenerative intervertebral disc (GSM3259528, GSM3259529 and GSM3259530). The differentially expressed miRNA were screened using the DataSet analysis tool (GEO2R; ncbi.nlm.nih.gov/geo/geo2r). Differentially expressed miRNA were identified by|FC|>2 and P<0.05, shown in Table I.
Table I.
Differentially expressed miRNA data mining using online GEO2R analysis (|logFC|>2 and P<0.05).
| miRNA_ID | adj.P.Val | P-value | -lgP | logFC |
|---|---|---|---|---|
| hsa-miR-32-5p | 2.37x10-17 | 1.17x10-20 | 19.93181414 | -2.81 |
| hsa-miR-187-3p | 2.37x10-17 | 1.85x10-20 | 19.73282827 | 2.52 |
| hsa-miR-2682-3p | 3.64x10-17 | 4.91x10-20 | 19.30891851 | 2.01 |
| hsa-miR-141-3p | 7.80x10-14 | 4.55x10-16 | 15.3419886 | 2.52 |
| hsa-miR-223-5p | 8.89x10-14 | 5.53x10-16 | 15.25727487 | 2.41 |
| hsa-miR-1273h-3p | 1.35x10-13 | 1.21x10-15 | 14.91721463 | 2 |
| hsa-miR-130b-5p | 1.35x10-13 | 1.21x10-15 | 14.91721463 | 2 |
| hsa-miR-15a-3p | 1.35x10-13 | 1.21x10-15 | 14.91721463 | 2 |
| hsa-miR-1911-5p | 1.35x10-13 | 1.21x10-15 | 14.91721463 | 2 |
| hsa-miR-27a-5p | 1.35x10-13 | 1.21x10-15 | 14.91721463 | 2 |
| hsa-miR-339-5p | 1.35x10-13 | 1.21x10-15 | 14.91721463 | 2 |
| hsa-miR-379-3p | 1.35x10-13 | 1.21x10-15 | 14.91721463 | 2 |
| hsa-miR-4715-5p | 1.42x10-13 | 1.82x10-15 | 14.73992861 | 2.01 |
| hsa-miR-492 | 1.42x10-13 | 1.82x10-15 | 14.73992861 | 2.01 |
| hsa-miR-302c-5p | 3.38x10-13 | 6.84x10-15 | 14.1649439 | 2.01 |
| hsa-miR-3164 | 4.48x10-13 | 1.09x10-14 | 13.9625735 | 2.01 |
| hsa-miR-516a-3p | 4.48x10-13 | 1.09x10-14 | 13.9625735 | 2.01 |
| hsa-miR-1288-5p | 4.48x10-13 | 1.10x10-14 | 13.95860731 | 2.01 |
| hsa-miR-4482-3p | 9.23x10-13 | 3.18x10-14 | 13.49757288 | 2.01 |
| hsa-miR-127-5p | 3.44x10-9 | 1.31x10-10 | 9.882728704 | 2.4 |
| hsa-miR-3606-3p | 8.20x10-9 | 3.19x10-10 | 9.496209317 | 2 |
| hsa-miR-373-3p | 5.31x10-8 | 2.40x10-9 | 8.619788758 | 2.01 |
| hsa-miR-597-3p | 1.68x10-7 | 8.50x10-9 | 8.070581074 | 2.15 |
| hsa-miR-4434 | 4.44x10-7 | 2.75x10-8 | 7.560667306 | 2 |
| hsa-miR-518b | 4.52x10-7 | 2.83x10-8 | 7.548213564 | 2.04 |
| hsa-miR-6810-3p | 4.84x10-7 | 3.09x10-8 | 7.510041521 | 2 |
| hsa-miR-4697-3p | 5.64x10-7 | 3.67x10-8 | 7.435333936 | 2.01 |
| hsa-miR-6827-3p | 5.64x10-7 | 3.68x10-8 | 7.434152181 | 2.02 |
| hsa-miR-7846-3p | 6.16x10-7 | 4.05x10-8 | 7.392544977 | 2.22 |
| hsa-miR-6728-3p | 8.10x10-7 | 5.64x10-8 | 7.248720896 | 2.03 |
| hsa-miR-15b-3p | 8.41x10-7 | 5.92x10-8 | 7.227678293 | 2.1 |
| hsa-miR-6759-3p | 9.89x10-7 | 7.12x10-8 | 7.147520006 | 2.02 |
| hsa-miR-410-5p | 1.08x10-6 | 8.05x10-8 | 7.09420412 | -2.49 |
| hsa-miR-376a-5p | 1.08x10-6 | 8.05x10-8 | 7.09420412 | -2.5 |
| hsa-miR-6799-3p | 1.08x10-6 | 8.06x10-8 | 7.093664958 | 2.03 |
| hsa-miR-6511b-3p | 1.08x10-6 | 8.10x10-8 | 7.091514981 | 2.02 |
| hsa-miR-6801-3p | 1.17x10-6 | 8.94x10-8 | 7.048662481 | 2.03 |
| hsa-miR-21-5p | 1.28x10-6 | 9.90x10-8 | 7.004364805 | 2.35 |
| hsa-miR-518e-5p | 1.59x10-6 | 1.28x10-7 | 6.89279003 | 2.03 |
| hsa-miR-627-5p | 1.64x10-6 | 1.34x10-7 | 6.872895202 | 2.02 |
| hsa-miR-378a-5p | 1.99x10-6 | 1.69x10-7 | 6.772113295 | -2.46 |
| hsa-miR-486-5p | 2.64x10-6 | 2.43x10-7 | 6.614393726 | -2.77 |
| hsa-miR-6830-5p | 7.26x10-6 | 8.67x10-7 | 6.061980903 | 2.02 |
| hsa-miR-338-3p | 8.41x10-6 | 1.03x10-6 | 5.987162775 | 2.41 |
| hsa-miR-146a-5p | 8.71x10-6 | 1.07x10-6 | 5.970616222 | 2.38 |
| hsa-miR-455-5p | 8.91x10-6 | 1.10x10-6 | 5.958607315 | 2.52 |
| hsa-miR-520b | 1.00x10-5 | 1.26x10-6 | 5.899629455 | 2.22 |
| hsa-miR-6507-3p | 1.69x10-5 | 2.32x10-6 | 5.634512015 | 2.12 |
| hsa-miR-1268b | 2.27x10-5 | 3.31x10-6 | 5.480172006 | 2.01 |
| hsa-miR-563 | 2.34x10-5 | 3.50x10-6 | 5.455931956 | 2.37 |
| hsa-miR-29c-3p | 3.28x10-5 | 5.21x10-6 | 5.283162277 | 2.58 |
| hsa-miR-192-5p | 4.48x10-5 | 7.67x10-6 | 5.115204636 | 2.37 |
| hsa-miR-634 | 6.42x10-5 | 1.21x10-5 | 4.91721463 | 2.01 |
| hsa-miR-34a-5p | 6.46x10-5 | 1.23x10-5 | 4.910094889 | 2.46 |
| hsa-miR-518a-5p | 7.81x10-5 | 1.57x10-5 | 4.804100348 | 2.33 |
| hsa-miR-610 | 7.86x10-5 | 1.59x10-5 | 4.798602876 | 2.44 |
| hsa-miR-590-5p | 8.15x10-5 | 1.66x10-5 | 4.779891912 | -2.34 |
| hsa-miR-139-5p | 3.78x10-4 | 1.02x10-4 | 3.991399828 | 2.42 |
| hsa-miR-1306-3p | 4.07x10-4 | 1.11x10-4 | 3.954677021 | 2.06 |
| hsa-miR-125b-1-3p | 1.89x10-3 | 6.30x10-4 | 3.200659451 | 2.25 |
| hsa-miR-204-5p | 7.03x10-3 | 2.69x10-3 | 2.57024772 | -2.26 |
miRNA/miR, microRNA; FC, fold change.
Target gene prediction
The TargetScan database (targetscan.org) was used to predict microRNA target genes. Base-pairing and minimum free energy for miR-138 binding to the target sequences were predicted using the RNAhybrid software (version 2.1; bibiserv.techfak.uni-bielefeld.de/rnahybrid).
Cell culture
Human nucleus pulposus cells (HNPCs) were purchased from ScienCell Research Laboratories, Inc. Cells were maintained in Nucleus Pulposus Cell Medium (ScienCell Research Laboratories, Inc.) under constant oxygenated culture conditions (37˚C; 20% oxygen; 5% carbon dioxide; 75% nitrogen) or hypoxic culture conditions (37˚C; 1% oxygen; 5% carbon dioxide; 94% nitrogen). Hypoxic cell culture conditions were generated by a hypoxia incubator (HF100; Heal Force). Cells were grown in constant oxygen or hypoxic conditions for 24-72 h, according to the experimental requirements. Recombinant HIF-1α protein (0,0.5,1,2,5 µg/ml; cat. no. H00003091-P01; Abnova,) was added at the beginning of the normoxic or hypoxic experiments and maintained at 37˚C for 24 h.
Transfection
The miR-32-5p mimics or inhibitor, and their negative controls (scramble), as well as the PTEN small interfering (si)RNA or PTEN overexpression plasmid (ov-PTEN), and their negative controls (scrambled) were synthesized by Ribobio Co., Ltd. According to the manufacturer's instructions, 2x105 human nucleus pulposus cells (HNPC) were seeded in a 6-well plate and incubated overnight at 37˚C with 5% CO2. Then, HNPC cells were transfected with miRNA mimics or inhibitors (100 nM), si-PTEN or PTEN overexpression plasmid (50 nM) at 37˚C using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Negative control (NC) mimic was used as the negative control of miR-32-5p mimic, and NC inhibitor was used as the negative control of miR-32-5p inhibitor. Si-NC (100 nM) was used as the negative control of si-PTEN, and ov-NC (100 nM) was used as the negative control of ov-PTEN. After 4 h, the mixture was replaced with a fresh medium containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). After 24 h, the cells were collected and centrifuged at 300 x g for 5 min at 37˚C. Then, TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was added for reverse transcription-quantitative PCR (RT-qPCR) detection. Sequences of si-PTEN, miRNA mimics or inhibitors, including the negative controls (scramble), were as follows: si-PTEN, 5'-CAATATTGATGATGTAGTAAG-3'; si-NC, 5'-UUCUCCGAACGUGUCAGUTT-3'; miR-32-5p mimics, 5'-UAUUGCACAUUACUAAGUUGCA-3'; miR-32-5p mimics negative control, 5'-CUAAUUAUCCUGAAGAUCUUAG-3'; miR-32-5p inhibitor, 5'-TGCAACTTAGTAATGTGCAATA-3'; and miR-32-5p inhibitor negative control, 5'-CAGUACUUUUGUGUAGUACAA-3'.
RT-qPCR
According to the manufacturer's instructions, total RNA was extracted from HNPCs using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The extracted RNA was reverse transcribed into cDNA using the PrimeScript RT reagent kit (Invitrogen; Thermo fisher Scientific, Inc.) in accordance with the manufacturers protocol. qPCR reactions were performed using SYBR Premix ExTaq II kits (Takara Biotechnology Co., Ltd.) on a 7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The PCR thermocycling conditions were as follows: 95˚C for 5 min, followed by 40 cycles of 95˚C for 15 sec and 60˚C for 1 min. U6 was used to normalize miR expression and β-actin to normalize PTEN expression. The following primers were used: miRNA-32-5p forward, 5'-ACACTCCAGCTGGGTATTGCACATTACTAA-3'; miRNA-32-5p reverse, 5'-CTCAACTGGTGTCGTGGA-3'; U6 forward, 5'-CTCGCTTCGGCAGCACA-3'; U6 reverse, 5'-AACGCTTCACGAATTTGCGT-3'; PTEN forward, 5'-TGAGTTCCCTCAGCCGTTACCT-3'; PTEN reverse, 5'-GAGGTTTCCTCTGGTCCTGGTA-3'; β-actin forward, 5'-AGCGAGCATCCCCCAAAGTT-3'; β-actin reverse, 5'-GGGCACGAAGGCTCATCATT-3'. Relative miR-32-5p and PTEN expression levels were calculated using the 2-ΔΔCq method (11). All PCR experiments were performed in triplicate for each sample, and all experiments were repeated three times.
Western blotting
Cells were harvested and lysed with ice-cold lysis buffer (Beyotime Institute of Biotechnology) and the protein concentration was determined using BCA protein assay kit (Nanjing KeyGen Biotech Co., Ltd.). Denatured proteins (20 µg) were separated by sodium dodecyl sulfate-polyacrylamide 10% gel electrophoresis and transferred to polyvinylidene fluoride membranes (EMD Millipore). The membrane was blocked with 5% BSA Blocking Buffer (cat. no. SW3015; Beijing Solarbio Science & Technology Co., Ltd.) and incubated with PTEN primary antibodies (1:1,000; cat. no. ab267787; Abcam) at 4˚C overnight Next, the membranes were incubated with secondary antibody Goat Anti-Rabbit (1:10,000; cat. no. ab205718; Abcam) for 2 h at 25˚C. The bound proteins were visualized by enhanced chemiluminescence (Thermo Fisher Scientific, Inc.) and detected using an imaging system (DNR Bio-Imaging Systems Ltd., Ma'ale Hahamisha). β-actin was used as a loading control. Densitometric analysis was performed using ImageJ software (version 1.8.0; National Institutes of Health).
Cell proliferation assay
For cell proliferation assays, single-cell suspensions were prepared by trypsinization, and cells were seeded onto 6-well plates with a density of 500 cells per well. After 2 weeks of culture, the cells were digested with trypsin. Then, 10 µl cell suspension was mixed with 10 µl phenolic blue dye and added to a counting plate. The cells were incubated at room temperature for 3 min, observed, and counted using light microscope (100x; Olympus; Tokyo, Japan). The remaining cells were then inoculated in 96-well plates at a density of 3x103 cells/well and cultured for 24-72 h. Optical density values were measured every 24 h using a Cell Counting Kit-8 (cat. no. CA1210; Solarbio), according to the manufacturer's instructions.
Cell apoptosis assay
Apoptosis assays were performed using an Annexin V-FITC Apoptosis Detection kit (Nanjing KeyGen Biotech Co., Ltd.) according to the manufacturer's instructions. Cells (1x106 cells/ml) were harvested and washed twice with cold PBS and resuspended in 500 µl binding buffer. Next, the cells were incubated with 5 µl annexin V-FITC and 5 µl propidium iodide in the dark for 15 min at 25˚C. Apoptosis was assessed by flow cytometry (BD Biosciences) and using flowjo software (version 10.0.6; FlowJo LLC) to analysis. Each experiment was repeated three times.
Luciferase reporter assay
The PTEN 3'-UTR was cloned into the psi-CHECK2 vector (Promega Corporation). Mutant reporters (binding site mutations in PTEN) were then generated. Luciferase reporter assays were performed by transfecting the mutated promoter reporter plasmid and the psi-CHECK2-PTEN vector into human 293T cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, luciferase activity was detected using a dual luciferase assay system (Promega Corporation), according to the manufacturer's instructions. The ratio of firefly to Renilla was used to normalize the firefly luciferase values and each experiment was repeated three times.
Statistical analysis
All data are expressed as the mean ± standard deviation (SD), and were analyzed using the SPSS version 19.0 statistical analysis package (SPSS, Inc.). For independent two-group paired analyses, Student's t-tests were used (unpaired). All experimental groups were compared with a control group for the multiple comparisons, which were analyzed using oneway analysis of variance (ANOVA) followed by Dunnett's post-hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Selection of miR-32-5p and PTEN as candidate targets
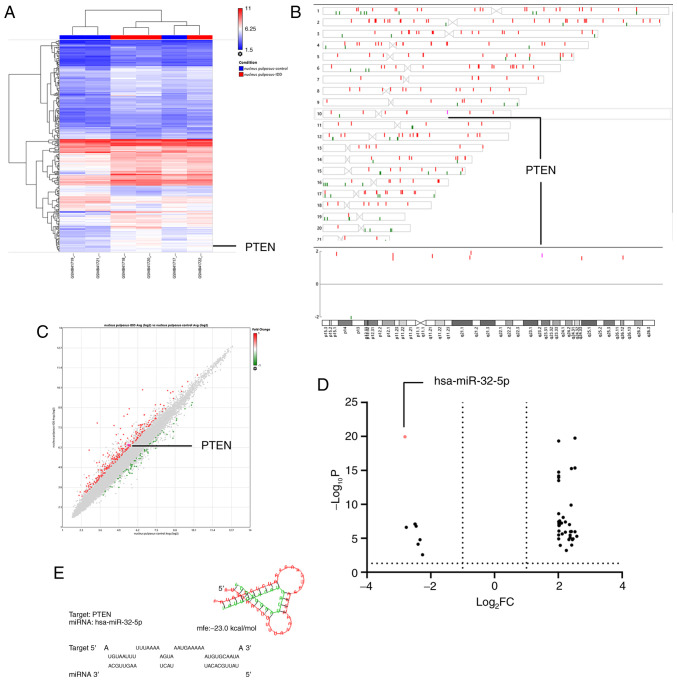
The raw microarray data mining results showed a negative association between miRNA-32-5p and intervertebral disc degeneration. In contrast, PTEN on chromosome 10 (q-arm) was upregulated in NPCs of degenerative disc tissues from elderly patients (GSE34095; Fig. 1A-C). miRNA-32-5p was downregulated in NPCs of tissues from patients with intervertebral disc degeneration, according to the online analysis on GEO2R of GSE116726 (Fig. 1D). The PTEN gene sequence was examined and putative miRNA-32-5p binding sites were identified at position 2859-2866 of the PTEN 3'-UTR. Therefore, it was hypothesized that miR-32-5p interacts with PTEN. Thus, miR-32-5p and PTEN were chosen for subsequent experiments.
Figure 1.
Selection of miR-32-5p and PTEN for subsequent experiments. (A) Heatmap of hierarchical clustering |FC|>1.5 and P<0.05. (B) Differentially expressed genes on chromosome 10. (C) Scatter plot showing 331 differentially expressed genes |FC|>1.5 and P<0.05. (D) Volcano plot showing 93 differentially expressed miRNAs |log2FC|>2 and P<0.05. (E) RNAHybrid prediction analysis demonstrated that miR-32-5p targets the PTEN 3'-untranslated region. FC, fold change; miR, microRNA.
Hypoxia induces miR-32-5p and PTEN expression
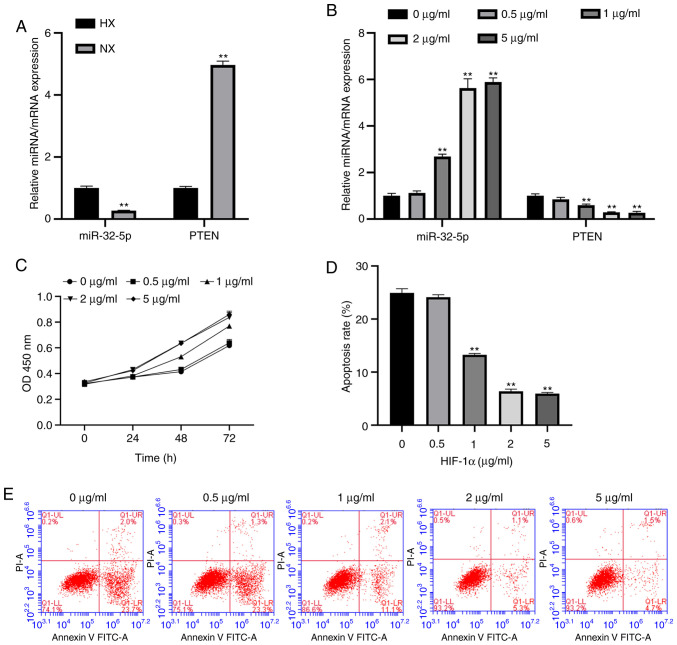
To evaluate the association between miR-32-5p, PTEN and the NPC oxemic status, the expression levels of miR-32-5p and PTEN were measured under hypoxic and normoxic conditions using RT-qPCR. The results suggest that miR-32-5p expression increases during hypoxia compared with the normoxia group, while PTEN expression decreases (Fig. 2A).
Figure 2.
Hypoxia and HIF-1α increase miR-32-5p and decrease PTEN expression in NPC (A) RT-qPCR analysis of miR-32-5p and PTEN in NPCs cultured under HX or NX conditions. miR-32-5p expression was significantly increased while PTEN expression was decreased in HX compared with NX. (B) RT-qPCR analysis of miR-32-5p or PTEN expression in NPCs cultured with different concentrations of HIF-α. miR-32-5p expression was significantly increased and PTEN was significantly decreased by high HIF-1α concentrations. (C) Cell Counting Kit-8 assays confirmed that HIF-1α promoted NPC proliferation. (D and E) Flow cytometry analysis demonstrated that HIF-1α inhibited cell apoptosis. **P<0.01. HIF-1α, hypoxia-inducible factor-1α; miR, microRNA; NPC, nucleus pulposus cells; RT-qPCR, reverse transcription-quantitative PCR; HX, hypoxic; NX, normoxic.
HIF-1α promotes NPC proliferation, inhibits apoptosis and alters miR-32-5p and PTEN expression
A previous study reported that HIF-1α is involved in the regulation of hypoxic stress responses (3). To further investigate whether recombinant HIF-1α protein controls hypoxic regulation, the expression levels of miR-32-5p and PTEN were examined following HIF-1α treatment. It was found that HIF-1α increased miR-32-5p expression and decreased PTEN expression (Fig. 2B). In addition, HIF-1α promoted NPCs proliferation and inhibited apoptosis (Fig. 2C-E). The optimal concentration of HIF-1 was 2 µg/ml; this concentration will be used as the experimental concentration for the present study.
Hsa-miR-32-5p mimics or inhibitor and PTEN overexpression or inhibition plasmids were successfully constructed
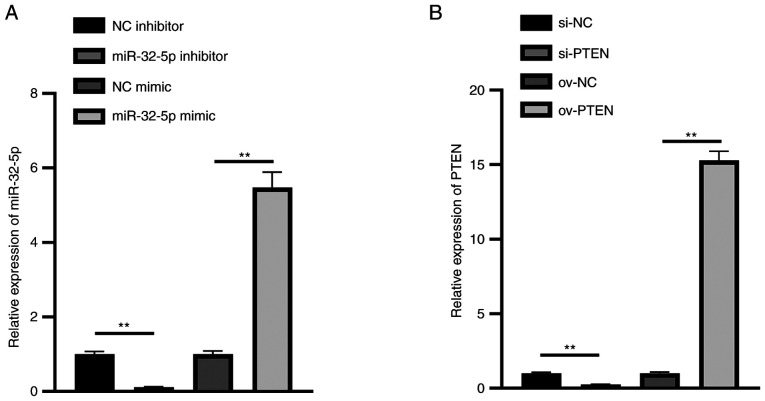
To validate Hsa-miR-32-5p mimics or inhibitor and PTEN overexpression or inhibition plasmids, these RNAs were transfected into NPCs. The results show that the expression of miR-32-5p in the miR-32-5p mimics group was significantly upregulated, while that in the miR-32-5p inhibitor group was significantly downregulated (Fig. 3A). Meanwhile, the expression of PTEN in the ov-PTEN group was significantly upregulated, while that in the si-PTEN group was significantly downregulated (Fig. 3B). These results illustrate that hsa-miR-32-5p mimics or inhibitor and PTEN overexpression or inhibition plasmids were successfully constructed.
Figure 3.
Verification of miR-32-5p mimics or inhibitor and si-PTEN or ov-PTEN plasmids. Reverse transcription-quantitative PCR analysis of (A) miR-32-5p mimics or inhibitor, and (B) si-PTEN or ov-PTEN in nucleus pulposus cells. **P<0.01. miR, microRNA; si-, small interfering RNA; ov-, overexpression plasmid; NC, negative control.
miR-32-5p promotes NPC proliferation and decreases apoptosis
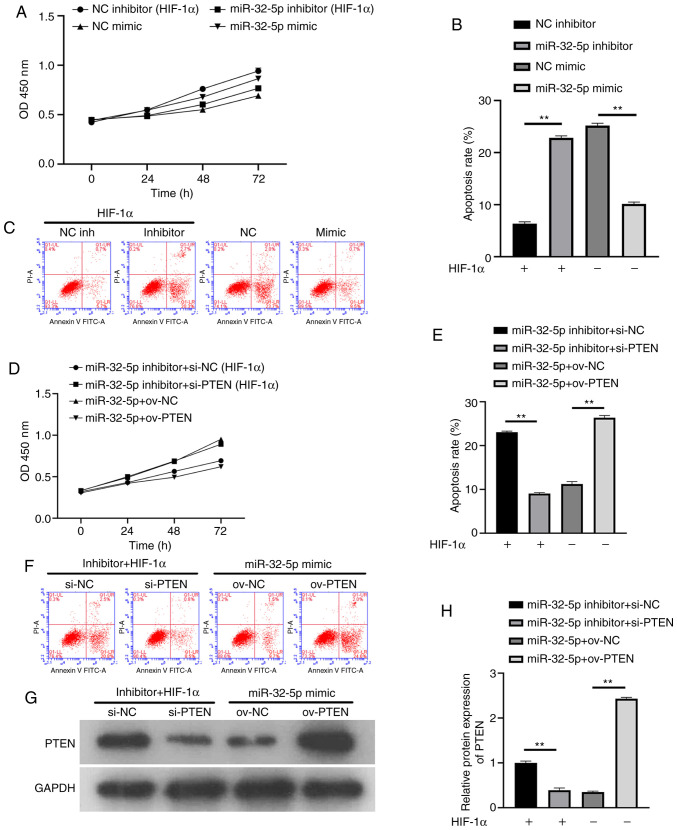
To further confirm that NPC proliferation and apoptosis are dependent on miR-32-5p or PTEN, the effects of miRNA or PTEN inhibition and overexpression on cell proliferation were investigated. The results showed that miR-32-5p inhibitor, after the addition of recombinant HIF-1α protein, inhibited cell proliferation and increased apoptosis. In the absence of HIF-1α, miR-32-5p overexpression promoted cell proliferation and inhibited apoptosis (Fig. 4A-C). Following the addition of recombinant HIF-1α protein, decreased PTEN levels increased cell proliferation and inhibited apoptosis. In the absence of HIF-1α, PTEN overexpression inhibited cell proliferation and promoted apoptosis (Fig. 4D-F). To ensure the success of the transfection experiment, western blotting was used to verify the PTEN expression (Fig. 4G and H). The results suggest that miR-32-5p or PTEN can induce NPC proliferation and apoptosis. At the same time, a significant negative association was demonstrated between miR-32-5p and PTEN (P<0.01).
Figure 4.
Effects of miR-32-5p or PTEN on nucleus pulposus cells proliferation and apoptosis. (A-C) miR-32-5p promoted cell proliferation and inhibited apoptosis. (D-F) PTEN inhibited cell proliferation and promoted apoptosis. (G and H) PTEN expression was analyzed by western blotting. **P<0.01. miR, microRNA; NC, negative control; HIF-1α, hypoxia-inducible factor-1α; si-, small interfering RNA; ov-, overexpression plasmid.
PTEN is a direct target of miR-32-5p
The function of miR-32-5p in NPCs was validated using PTEN as a target. A single putative miR-32-5p recognition site was predicted within the PTEN 3'-UTR sequence. The mechanism through which miR-32-5p modulates PTEN expression was examined by introducing the PTEN 3'-UTR containing one miR-32-5p binding site downstream of a luciferase reporter (Fig. 5A). The luciferase activity of the WT PTEN 3'-UTR construct in transfected cells was significantly lower compared with that of cells transfected with the control construct. A luciferase reporter containing a mutant miR-32-5p binding site at the PTEN 3'-UTR was also constructed. The luciferase activity of the mutant PTEN 3'-UTR construct was not suppressed upon transfection with miR-32-5p mimics. These results indicate that miR-32-5p selectively binds to PTEN mRNAs and that the single recognition element identified in the PTEN 3'-UTR mRNA is sufficient for miR-32-5p activity (Fig. 5B). Thus, miR-32-5p directly targets PTEN.
Figure 5.
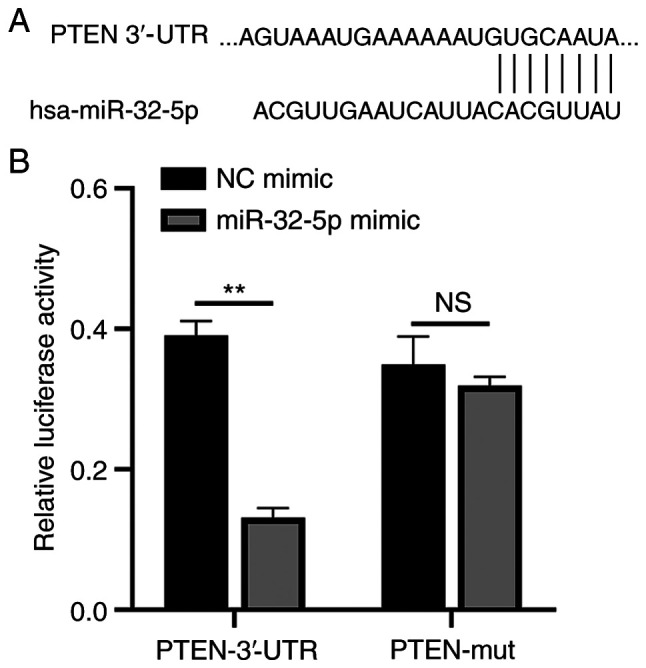
miR-32-5-p directly combined with PTEN 3'-UTR. (A) The TargetScan database predicted binding sites for miR-32-5p and PTEN. (B) Fluorescence expression of WT/Mut mimic group and NC mimic group were detected by dual luciferase at OD450 nm wavelength. All experiments were repeated three times. **P<0.01; NS, not significant; miR, microRNA; UTR, untranslated region; WT, wild-type; Mut, mutant; NC, negative control.
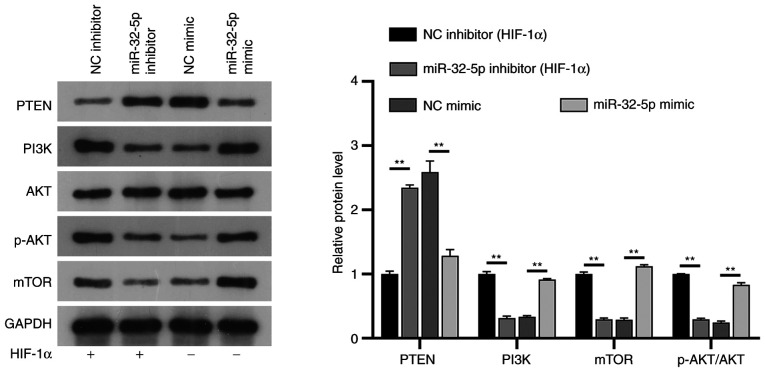
The mechanism of miR-32-5p/PTEN/PI3K/AKT/mTOR interaction in NPCs
PTEN is a well-known inhibitor of the PI3K/AKT/mTOR pathway in several diseases, such as autism, spinal cord injury or prostate cancer. To further understand the potential mechanisms of miR-32-5p and PTEN in NPCs, their association with the PI3K/AKT/mTOR signaling pathway was investigated. The results showed that under constant oxygen conditions, following the addition of HIF-1α, mir-32-5p mimic could upregulate the expressions of PI3K, phosphorylated (p)-AKT and mTOR, while the expression of PTEN was downregulated, and the expression of AKT did not change significantly (P>0.05). Following the addition of HIF-1α in the miR-32-5p inhibitor group, PTEN expression was upregulated, PI3K/p-AKT/mTOR expression was downregulated, and AKT expression was not significantly changed compared with the NC inhibitor group (P>0.05). Notably, the expression of PI3K/AKT/mTOR was significantly downregulated and PTEN was upregulated in the NC mimic group compared with the NC inhibitor group, confirming the inhibitory effect of HIF-1α on the PI3K/AKT/mTOR pathway. HIF-1α regulates the PI3K/AKT/mTOR signaling pathway through miR-32-5p/PTEN (Fig. 6).
Figure 6.
miR-32-5p regulates PI3K/AKT/mTOR through PTEN. Western blotting verified the effect of transfection with mimic and inhibitor on the protein expression of PTEN/PI3K/AKT/p-AKT/mTOR in NPC. **P<0.01. miR, microRNA; p-, phosphorylated; NC, negative control; HIF-1α, hypoxia-inducible factor-1α.
Discussion
The present study demonstrated for the first time that miR-32-5p and PTEN induce NPC proliferation and apoptosis, and that hypoxia induces miR-32-5p and PTEN expression. Programmed NPC apoptosis leads to intervertebral disc degeneration (12); this previous finding combined with the present results indicates that miR-32-5p is a key factor in the degeneration of intervertebral discs. Several studies have focused on the preclinical development of a microRNA-based therapy for intervertebral disc degeneration (1,13), and the present results corroborate those findings. The second major finding of the present study is that HIF-1α controls the miR-32-5p/PTEN axis to regulate cell proliferation and apoptosis in NPCs. Together, in the hypoxic intervertebral disc, HIF-1α, miR-32-5p and PTEN are crucial for NPC proliferation.
It was observed that PTEN was upregulated in the nucleus pulposus from elderly patient with degenerative disc tissues, which was consistent with a previous study (7). Interestingly, PTEN, a tumor suppressor that is frequently mutated in several cancer types, such as breast cancer and stomach cancer, was downregulated, which generally inhibits tumorigenesis (14). Thus, the question was what the major cause of high PTEN expression in NPCs from healthy patients is? The role of hypoxia was investigated, which showed that hypoxia induces PTEN expression. Therefore, PTEN can have different biological functions under different conditions or in different diseases.
The present study also showed a significant negative association between miR-32-5p and PTEN expression. Based on the dual role of PTEN as an inhibitor in the human body, miR-32-5p-based therapy holds potential in intervertebral disc degeneration treatment. Emerging evidence indicates that miR-32-5p may suppress cancer metastasis (15). Indeed, Wang et al (8) found that miR-32-5p suppresses clear cell renal cell carcinoma metastasis, and Li et al (16) found that HIF-1α is a key responder to tumor hypoxia. The results of the present study clearly suggest that miR-32-5p promotes NPC proliferation and decreases apoptosis. Further, the present results suggest that miR-32-5p has similar function in intervertebral disc degeneration and in cancer. In the process of studying PTEN, it was observed that several articles have reported that PTEN and PI3K/AKT/mTOR signaling pathway are closely associated in different diseases. Therefore, it was investigated whether the proliferation and apoptosis of NPC is influenced by miR-32-5p/PTEN through the regulation of the PI3K/AKT/mTOR signaling pathway, which was confirmed to be the case in the present study.
Although HIF-1α is widely studied in various diseases as a transcription factor that regulates other genes in the nucleus (17,18), almost no literature reports its function as a recombinant protein in vitro. Stegen et al (19) found that HIF-1α controls collagen synthesis and modification in chondrocytes. Notably, HIF-1α is a therapeutic target for hepatocellular carcinoma in the nucleus (20). Chen and Steidl (21) demonstrated that inhibition of HIF1α signaling is effective for myelodysplastic syndromes therapy. In vitro recombinant HIF-1α regulates cell proliferation and apoptosis in NPCs, similar to its role in the nucleus. To the best of our knowledge, this is the first study to investigate HIF-1α as a recombinant protein.
There are limitations to the present study. Firstly, other miRNAs and mRNAs and associated signaling pathways were not analyzed in this study, despite the bioinformatics analysis. In addition, this study was only conducted at the cellular level, and further verification in animal models is needed in the future.
In summary, the present study findings support that HIF-1α, as a recombinant protein, controls the miR-32-5p/PTEN/PI3K/AKT/mTOR axis to regulate cell proliferation and apoptosis.
Acknowledgements
Not applicable.
Funding Statement
Funding: This study was supported by Natural Science Foundation of Hainan Province (grant no. 819QN350).
Availability of data and materials
The microarray data (GSE34095, GSE116726) referenced in the study are available in a public repository from the GEO website (http://www.ncbi.nlm.nih.gov/geo). The authors declare that all the other data supporting the findings of this study are available within the article and its Supplementary Information files and from the corresponding authors on reasonable request.
Authors' contributions
DZ, ML, JC and GW conceived and designed the study, and developed the methodology. JC, WC and BW performed the experiments and collected the data. DZ, ML, JC, WC, JL, YF and YL analyzed the data. ML, YF, JL, YL, BW and GW interpreted the data. DZ drafted the manuscript. All authors read and approved the final manuscript. DZ and GW confirm the authenticity of all the raw data.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ji ML, Jiang H, Zhang XJ, Shi PL, Li C, Wu H, Wu XT, Wang YT, Wang C, Lu J. Preclinical development of a microRNA-based therapy for intervertebral disc degeneration. Nat Commun. 2018;9(5051) doi: 10.1038/s41467-018-07360-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sloan SR Jr, Wipplinger C, Kirnaz S, Navarro-Ramirez R, Schmidt F, McCloskey D, Pannellini T, Schiavinato A, Härtl R, Bonassar LJ. Combined nucleus pulposus augmentation and annulus fibrosus repair prevents acute intervertebral disc degeneration after discectomy. Sci Transl Med. 2020;12(eaay2380) doi: 10.1126/scitranslmed.aay2380. [DOI] [PubMed] [Google Scholar]
- 3.Madhu V, Boneski PK, Silagi E, Qiu Y, Kurland I, Guntur AR, Shapiro IM, Risbud MV. Hypoxic regulation of mitochondrial metabolism and mitophagy in nucleus pulposus cells is dependent on HIF-1α-BNIP3 Axis. J Bone Miner Res. 2020;35:1504–1524. doi: 10.1002/jbmr.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamid AA, Gray KP, Shaw G, MacConaill LE, Evan C, Bernard B, Loda M, Corcoran NM, Van Allen EM, Choudhury AD, Sweeney CJ. Compound genomic alterations of TP53, PTEN, and RB1 tumor suppressors in localized and metastatic prostate cancer. Eur Urol. 2019;76:89–97. doi: 10.1016/j.eururo.2018.11.045. [DOI] [PubMed] [Google Scholar]
- 5.Meng J, Zhang W, Wang C, Zhang W, Zhou C, Jiang G, Hong J, Yan S, Yan W. Catalpol suppresses osteoclastogenesis and attenuates osteoclast-derived bone resorption by modulating PTEN activity. Biochem Pharmacol. 2020;171(113715) doi: 10.1016/j.bcp.2019.113715. [DOI] [PubMed] [Google Scholar]
- 6.Liu H, Huang X, Liu X, Xiao S, Zhang Y, Xiang T, Shen X, Wang G, Sheng B. miR-21 promotes human nucleus pulposus cell proliferation through PTEN/AKT signaling. Int J Mol Sci. 2014;15:4007–4018. doi: 10.3390/ijms15034007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Y, Guo W, Chen KW, Xiao ZM. VO-OHpic attenuates intervertebral disc degeneration via PTEN/Akt pathway. Eur Rev Med Pharmacol Sci. 2020;24:2811–2819. doi: 10.26355/eurrev_202003_20642. [DOI] [PubMed] [Google Scholar]
- 8.Wang M, Sun Y, Xu J, Lu J, Wang K, Yang DR, Yang G, Li G, Chang C. Preclinical studies using miR-32-5p to suppress clear cell renal cell carcinoma metastasis via altering the miR-32-5p/TR4/HGF/Met signaling. Int J Cancer. 2018;143:100–112. doi: 10.1002/ijc.31289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao L, Han T, Li Y, Sun J, Zhang S, Liu Y, Shan B, Zheng D, Shi J. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2017;31:893–903. doi: 10.1096/fj.201600994R. [DOI] [PubMed] [Google Scholar]
- 10.Wu W, Yang J, Feng X, Wang H, Ye S, Yang P, Tan W, Wei G, Zhou Y. MicroRNA-32 (miR-32) regulates phosphatase and tensin homologue (PTEN) expression and promotes growth, migration, and invasion in colorectal carcinoma cells. Mol Cancer. 2013;12(30) doi: 10.1186/1476-4598-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Sakai D, Nakamura Y, Nakai T, Mishima T, Kato S, Grad S, Alini M, Risbud MV, Chan D, Cheah KS, et al. Exhaustion of nucleus pulposus progenitor cells with ageing and degeneration of the intervertebral disc. Nat Commun. 2012;3(1264) doi: 10.1038/ncomms2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith LJ, Silverman L, Sakai D, Le Maitre CL, Mauck RL, Malhotra NR, Lotz JC, Buckley CT. Advancing cell therapies for intervertebral disc regeneration from the lab to the clinic: Recommendations of the ORS spine section. JOR Spine. 2018;1(e1036) doi: 10.1002/jsp2.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, et al. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2016;6:202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang D, Ma M, Zhou W, Yang B, Xiao C. Inhibition of miR-32 activity promoted EMT induced by PM2.5 exposure through the modulation of the Smad1-mediated signaling pathways in lung cancer cells. Chemosphere. 2017;184:289–298. doi: 10.1016/j.chemosphere.2017.05.152. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Jia Y, Wang Y. Targeting HIF-1α signaling pathway for gastric cancer treatment. Pharmazie. 2019;74:3–7. doi: 10.1691/ph.2019.8674. [DOI] [PubMed] [Google Scholar]
- 17.Ling S, Shan Q, Zhan Q, Ye Q, Liu P, Xu S, He X, Ma J, Xiang J, Jiang G, et al. USP22 promotes hypoxia-induced hepatocellular carcinoma stemness by a HIF1α/USP22 positive feedback loop upon TP53 inactivation. Gut. 2020;69:1322–1334. doi: 10.1136/gutjnl-2019-319616. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Zhang X, Chen K, Cheng Y, Liu S, Xia M, Chen Y, Zhu H, Li Z, Cao X. CCR7 Chemokine receptor-inducible inc-Dpf3 restrains dendritic cell migration by inhibiting HIF-1α-mediated glycolysis. Immunity. 2019;50:600–615.e15. doi: 10.1016/j.immuni.2019.01.021. [DOI] [PubMed] [Google Scholar]
- 19.Stegen S, Laperre K, Eelen G, Rinaldi G, Fraisl P, Torrekens S, Van Looveren R, Loopmans S, Bultynck G, Vinckier S, et al. HIF-1α metabolically controls collagen synthesis and modification in chondrocytes. Nature. 2019;565:511–515. doi: 10.1038/s41586-019-0874-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu FQ, Fang T, Yu LX, Lv GS, Lv HW, Liang D, Li T, Wang CZ, Tan YX, Ding J, et al. ADRB2 signaling promotes HCC progression and sorafenib resistance by inhibiting autophagic degradation of HIF1α. J Hepatol. 2016;65:314–324. doi: 10.1016/j.jhep.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Steidl U. Inhibition of HIF1α Signaling: A grand slam for MDS therapy? Cancer Discov. 2018;8:1355–1357. doi: 10.1158/2159-8290.CD-18-1061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The microarray data (GSE34095, GSE116726) referenced in the study are available in a public repository from the GEO website (http://www.ncbi.nlm.nih.gov/geo). The authors declare that all the other data supporting the findings of this study are available within the article and its Supplementary Information files and from the corresponding authors on reasonable request.