Abstract
Esophageal squamous cell carcinoma (ESCC) is a common malignant tumour type of the digestive system. Long non-coding RNA (lncRNA) c-Myc upregulated (MYU), also known as VPS9 domain-containing 1 antisense 1, was recently discovered. However, the expression of lncRNA MYU in ESCC and its role in tumour progression have remained elusive. In the present study, the expression of lncRNA MYU, Ki-67 and the epithelial-mesenchymal transition-related proteins E-cadherin and Vimentin in ESCC tissues was detected by reverse transcription-quantitative PCR. The expression of Ki-67, E-cadherin and Vimentin in ESCC tissues was also detected by immunohistochemistry. A small interfering RNA plasmid was employed to establish a TE-2 cell line with knockdown on lncRNA MYU. The results indicated that the expression of lncRNA MYU was higher in ESCC tissues than in normal adjacent tissues and that upregulation of lncRNA MYU was a potential biomarker for poor prognosis. The results also suggested that the expression levels of lncRNA MYU were correlated with the histological grade, lymph node metastasis and TNM stage (P<0.05). Silencing of lncRNA MYU expression inhibited the proliferation, migration and invasion, while the expression of lncRNA MYU increased as cell proliferation increased. In addition, the mRNA expression of Vimentin and Ki-67 was decreased in TE-2 cells after lncRNA MYU was knocked down, while E-cadherin mRNA expression was elevated. In conclusion, the present results indicated that lncRNA MYU may regulate the proliferation, migration and invasion of ESCC cells, and may serve as a prognostic biomarker for ESCC.
Keywords: long non-coding RNA MYU, esophageal squamous cell carcinoma, prognosis, proliferation, invasion
Introduction
Esophageal cancer is the fourth most common cancer type in China and nearly half of all newly diagnosed cases of esophageal cancer and associated deaths worldwide occur in China (1,2). The most common histological subtypes are esophageal squamous cell carcinoma (ESCC) and adenocarcinoma; however, in China, >90% of esophageal cancer cases are squamous cell carcinoma (3).
Long non-coding RNAs (lncRNAs) are a class of RNA molecules that are >200 nucleotides long and do not have any protein-coding capacity (4,5). In recent years, an increasing number of studies suggested that lncRNAs have important roles in regulating biological behaviours, including cell proliferation, differentiation, migration and invasion (6-8). They are able to affect biological activities by regulating mechanisms associated with gene expression, chromatin modification and cell metabolism (9,10). lncRNAs are also closely associated with tumour development and influence the prognosis of patients with tumours (11,12). Abnormal expression of lncRNAs has been reported in ESCC (13-15).
Studies on ESCC have indicated that lncRNAs may have a role in suppressing or promoting cancer-associated genes during the development of esophageal cancer. Previous studies have also indicated that certain lncRNAs may be potential markers for ESCC tumour diagnosis, treatment and estimation of prognosis (16,17). It was also reported that the expression level of lncRNA lymphoid enhancer binding factor 1 (LEF1)-antisense 1 (AS1) in ESCC tissues is significantly increased and that LEF1-AS1 is associated with lymph node metastasis, TNM stage and poor prognosis in patients with ESCC (16). It has also been suggested that lnc-ABC A12-3 expression is increased in ESCC and that this lncRNA is able to promote the migration, invasion and proliferation of tumour cells by regulating fibronectin 1(17). lncRNA cancer susceptibility 2 is able to negatively regulate microRNA-181a by inhibiting the Akt pathway in ESCC cells, thereby increasing the sensitivity of ESCC cells to cisplatin (18).
c-Myc upregulated lncRNA (MYU), also known as VPS9 domain-containing 1 (VPS9D1)-AS1, is a recently discovered lncRNA that was produced from the reverse strand of the VPS9D1 gene. To date, the expression and clinical significance of lncRNA MYU in ESCC has not been systematically reported in the literature. Thus, the present study explored the expression of lncRNA MYU in ESCC and its role in tumour development and progression. The effect of lncRNA MYU expression on the growth, proliferation and migration of various gastric cancer cell lines was also investigated.
Materials and methods
Patients and tissue specimens
Tissue specimens were collected from 112 patients with ESCC who underwent radical resection between January 2015 and December 2017 at the Affiliated Hospital of Nantong University (Nantong, China). None of the patients recruited for the study received any radiotherapy or chemotherapy prior to surgery. The ages of the patients ranged from 33 to 78 years, with a median age of 68 years, and there were 71 males and 41 females. The detailed clinicopathological parameters of the patients are provided in Table I. Tumor tissue samples were collected with written informed consent from the patients. The present study was approved by the Ethics Committee of the Affiliated Hospital of Nantong University (Nantong, China).
Table I.
lncRNA MYU expression in esophageal squamous cell carcinoma.
| lncRNA MYU | |||||
|---|---|---|---|---|---|
| Clinicopathological features | Number of patients | High expression | Low expression | χ2 value | P-value |
| Age (years) | 0.169 | 0.681 | |||
| <60 | 34 | 18 | 16 | ||
| ≥60 | 78 | 38 | 40 | ||
| Sex | 1.885 | 0.170 | |||
| Male | 71 | 32 | 39 | ||
| Female | 41 | 24 | 17 | ||
| Histological grade | 4.390 | 0.036 | |||
| High + moderate | 63 | 26 | 37 | ||
| Low | 49 | 30 | 19 | ||
| Vascular tumour thrombus | 0.728 | 0.393 | |||
| Present | 30 | 17 | 13 | ||
| Absent | 82 | 39 | 43 | ||
| T classification | 5.659 | 0.059 | |||
| T1 | 29 | 9 | 20 | ||
| T2 | 36 | 20 | 16 | ||
| T3 | 47 | 27 | 20 | ||
| Lymph node metastasis | 6.196 | 0.013 | |||
| Present | 47 | 30 | 17 | ||
| Absent | 65 | 26 | 39 | ||
| TNM stage | 5.486 | 0.019 | |||
| I/II | 70 | 29 | 41 | ||
| III/IV | 42 | 27 | 15 | ||
Statistical analyses were performed by using Pearson's χ2 test. lncRNA, long non-coding RNA; MYU, c-Myc upregulated lncRNA.
A total of four ESCC cell lines (EC9706, KYSE150, ECA109 and TE-2 cells) and normal human esophageal epithelial cells (HEECs) were obtained from the Shanghai Institute of Cell Research, Chinese Academy of Sciences. All cells were cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) medium supplemented with 10% foetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and maintained under standard conditions (37˚C, 5% CO2).
Antibodies and reagents
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Inc.), a green fluorescent protein (GFP)-tagged MYU-specific small interfering RNA (siRNA) plasmid (Shanghai Genechem Co., Ltd.), a Ki-67 antibody (cat. no. MA5-15690), an E-cadherin antibody (cat. no. MA5-15711) and a Vimentin antibody (cat. no. MA5-35320; all from Invitrogen; Thermo Fisher Scientific, Inc.), and Lipofectamine™ 2000 (Santa Cruz Biotechnology) were used in the present study.
Immunohistochemistry
Tissue specimens were fixed with 4% formaldehyde, routinely embedded in paraffin, sectioned, dewaxed and hydrated. Subsequently, endogenous peroxidase activity was blocked. The samples were then incubated with primary antibodies [Ki-67 (1:500 dilution), E-cadherin (1:200; dilution) and Vimentin (1:200 dilution)] overnight at 4˚C, followed by incubation with the corresponding secondary antibodies: Goat anti-rabbit (1:2,000; cat. no. ab207995; Abcam) and goat anti-mouse (1:1,000, cat. no. ab6788; Abcam) at room temperature for 30 min. The antibodies were visualized with diaminobenzidine and finally, the samples were counterstained with haematoxylin.
The immunostaining intensity was evaluated according to immunoreactivity scores, which were determined based on staining intensity and the percentage of positive cells. Five high-power visual fields were observed in each tissue, and 1x103 cells were counted for statistical analysis. The staining intensity score was multiplied by the score indicating the percentage of positive cells. The staining intensity of tumour cells was scored using the following criteria: Negative, 0; weak, 1; moderate, 2; and strong, 3. The scoring criteria for the percentage of positive cells were as follows: <5%, 0; 5-25%, 1; 26-50%, 2; 51-75%, 3; and >75%, 4.
Cell transfection
A lncRNA MYU siRNA and control siRNA were chemically synthesized by Shanghai GenePharma, Co., Ltd. The sequences of the lncRNA MYU-specific siRNAs were as follows: 5'-CCCUGCAAGCCAUGGGUAA-3' (siRNA-1) and 5'-CAAGAAGGCUGGUCACAGU-3' (siRNA-2). The sequence of the lncRNA MYU control siRNA was as follows: 5'-AAUUCUCCGAACGUGUCACGU-3' (siRNA control). Plasmid transfection was performed using Lipofectamine™ 2000 in accordance with the manufacturer's instructions. ESCC TE-2 cells were seeded into 24-well plates (2x105 cells/well) and transfected with the lncRNA MYU siRNA or control siRNA at 37˚C for 20 min. After incubation for 48 h, ESCC TE-2 cells were collected for the subsequent experiments.
RNA extraction and reverse transcription-quantitative (RT-qPCR)
Total RNA was extracted from tissues and cultured cells with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RT was conducted using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Inc.). The specific primer sequences for fluorescence qPCR were as follows: MYU forward, 5'-ATGGGTAACCAGGGGTCAAG-3' and reverse, 5'-AGTAACAGTGGTAGAGCCGAC-3'; E-cadherin forward, 5'-CGAGAGCTACACGTTCACGG-3' and reverse, 5'-GGGTGTCGAGGGAAAAATAGG-3'; Vimentin forward, 5'-GACGCCATCAACACCGAGTT-3' and reverse, 5'-CTTTGTCGTTGGTTAGCTGGT-3'; GAPDH forward, 5'-AATGGAGCCGTTAGGAAA-3' and reverse, 5'-GCGCCATACGACAATC-3'. The thermocycling conditions were as follows: Initial step at 94˚C for 40 sec, followed by 30 cycles of 94˚C for 10 sec and 65˚C for 30 sec. An ABI 7500 qPCR instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for relative quantitative analysis. GAPDH was used as the internal reference gene and the relative expression level of lncRNA MYU was calculated. Each sample was tested at least three times.
Cell Counting Kit-8 (CCK-8) assay
To analyse the growth of the ESCC cell lines, the CCK-8 assay (Dojindo Laboratories) was performed according to the manufacturer's protocol. ESCC cells transfected with lncRNA MYU siRNA or control siRNA were cultured in 96-well plates overnight. The cells were incubated for another 2 h and then detected at a wavelength of 450 nm using a microplate reader (Bio-Rad Laboratories). GraphPad Prism 8 software (GraphPad Software, Inc.) was used to plot the cell growth curve. The experiments were performed at least three times.
Cell cycle analysis
BD cell cycle reagent (BD Biosciences) was used according to the manufacturer's protocol. ESCC cells were washed with cold PBS and then fixed in 70% ethanol. After fixation, the cells were stained with propidium iodide. Finally, the cell suspensions were detected and analysed with a flow cytometer (BD Biosciences) and evaluated with ModFit LT 4.1 software (Verity Software House).
Wound-healing assay
ESCC cells transfected with lncRNA MYU siRNA or control siRNA were seeded in 6-well plates. After the cells reached confluence in a single layer, the scratch test was performed. A line-shaped scratch was performed, and the cells were rinsed with phosphate buffer to remove the detached cells, and then fresh medium without serum was added. Images of the scraped area with migrated cells were captured at 0 and 48 h after scratching.
Transwell assay
Serum-deprived ESCC cells were inoculated into the upper chamber of a Transwell chamber (Corning Inc.) according to the manufacturer's protocol and culture medium was added to the lower chamber as a chemoattractant while the serum was not contained in the upper chamber. A total of 1x105 cells/well were seeded in serum-free medium in the upper chamber with a membrane with 8-µm width filter pore that was pre-coated with Matrigel (BD Biosciences). After incubation for 48 h at 37˚C, the medium was aspirated and the cells that had invaded to the lower chamber were fixed in 4% formaldehyde for 30 min at room temperature. Subsequently, the cells were stained with 0.1% crystal violet for 10 min at room temperature and the cells on the lower surface of the membrane were counted under a microscope.
Statistical analysis
Each biological experiment was performed three times. Statistical significance was determined by Student's t-test, the χ2 test or one-way ANOVA with Tukey's post-hoc test as appropriate. Survival analysis was performed using the Kaplan-Meier method and the log-rank test. All statistical analyses were performed using SPSS 25.0 software (IBM Corp.). P<0.05 was considered to indicate statistical significance.
Results
lncRNA MYU is increased in ESCC tissues
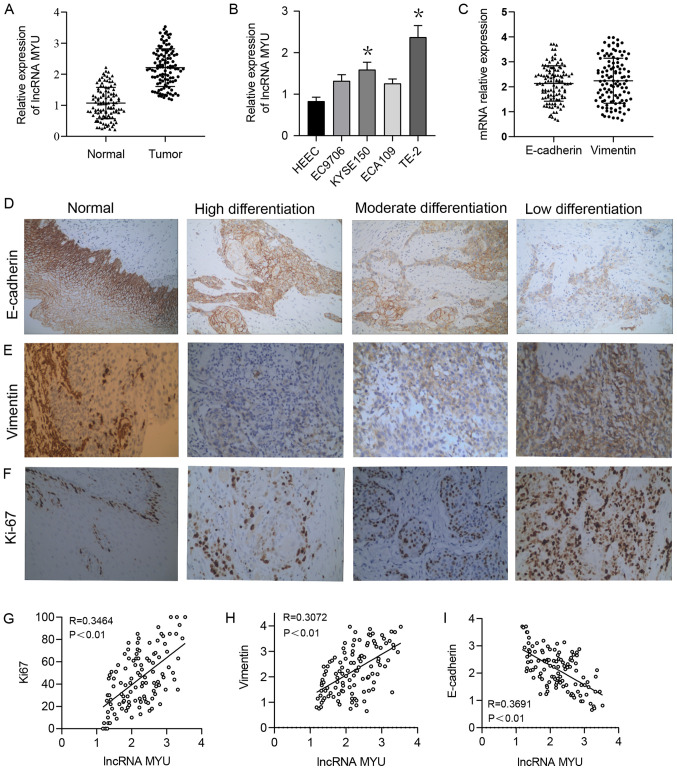
The RT-qPCR results suggested that the relative expression of lncRNA MYU in ESCC tissues was significantly higher than that in normal adjacent tissues, with relative expression scores of 2.2122±0.606 and 1.0712±0.504, respectively (P<0.0001; Fig. 1A). The median relative expression of lncRNA MYU in ESCC tumour tissue was 2.195, and based on this, low MYU expression was defined as an expression score of <2.195 and high MYU expression was defined as an expression score ≥2.195.
Figure 1.
Expression of lncRNA MYU, E-cadherin, Vimentin and Ki-67 in ESCC tissues. (A) The relative expression of lncRNA MYU was increased in ESCC tissues compared with that in normal tissues. GAPDH was used as a loading control. (B) Reverse transcription-quantitative PCR analysis of lncRNA MYU expression in 3 ESCC cell lines and HEECs. The relative expression level of lncRNA MYU was significantly upregulated in TE-2 cells. *P<0.05 vs. HEEC. (C) Relative mRNA expression of E-cadherin and Vimentin in ESCC tissues. GAPDH was used as a loading control. (D-F) Representative images of immunohistochemical staining for (D) E-cadherin, (E) Vimentin and (F) Ki-67 in ESCC tissues (magnification, x200). (G-I) Pearson correlation analysis of correlations between the relative expression of lncRNA MYU and that of (G) Ki-67, (H) Vimentin and (I) E-cadherin in ESCC tissues (P<0.01). ESCC, esophageal squamous cell carcinoma; HEECs, human esophageal epithelial cells; lncRNA, long non-coding RNA; MYU, c-Myc upregulated lncRNA.
In addition, lncRNA MYU expression was examined in 3 human ESCC cell lines (EC9706, KYSE150, ECA109 and TE-2) and HEECs by RT-qPCR. As lncRNA MYU expression was significantly higher in TE-2 cells than in the other cells (Fig. 1B), this cell line was selected for the subsequent experiments.
Expression of lncRNA MYU is associated with the clinicopathological features of ESCC
Statistical analysis revealed that the relative expression level of lncRNA MYU in ESCC tissue was significantly associated with the histological grade (P=0.036), lymph node metastasis (P=0.013) and TNM stage (P=0.019), but was not associated with the patients' age or sex, vascular tumour thrombus or T classification (P>0.05; Table I).
Expression of lncRNA MYU is correlated with Ki-67, Vimentin and E-cadherin expression
The expression of Ki-67 and the epithelial-mesenchymal transition (EMT)-related proteins E-cadherin and Vimentin in ESCC samples were then examined by immunohistochemistry (Fig. 1C-F). The Spearman rank correlation coefficient indicated that lncRNA MYU expression was positively correlated with the expression of the proliferation-associated proteins Ki-67 and the EMT-related proteins Vimentin (Fig. 1G and H, respectively) and negatively correlated with the expression of the EMT-related protein E-cadherin (Fig. 1I) in ESCC tumour tissues.
Expression of lncRNA MYU is associated with poor prognosis of patients with ESCC
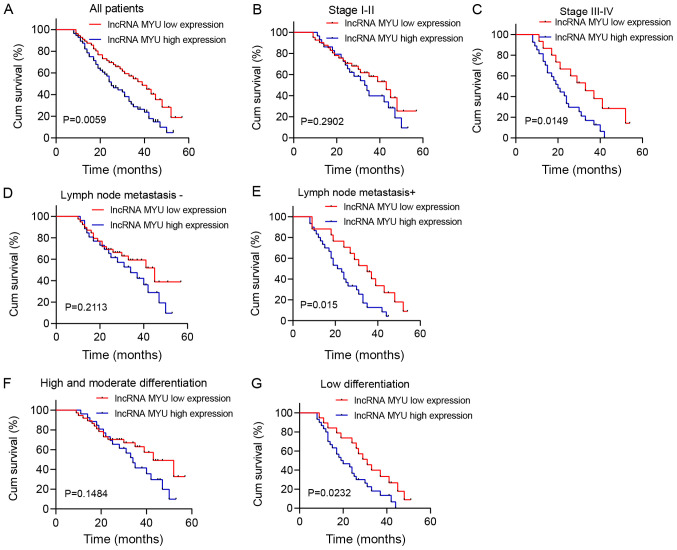
Survival analysis was performed by drawing Kaplan-Meier plots (Fig. 2). It was revealed that patients with high lncRNA MYU expression in their ESCC tissues had shorter overall survival (OS) (median survival of 25 months) than patients with low lncRNA MYU expression (median survival of 39 months; P=0.0059; Fig. 2A).
Figure 2.
OS curves of 112 cases based on relative lncRNA expression of MYU. (A) All patients; (B) stage I/II; (C) stage III/IV; (D) no lymph node metastasis; (E) lymph node metastasis; (F) high and moderate degree of differentiation; (G) poor differentiation. In A, C, E and G, patients with high lncRNA MYU expression had shorter OS than those with low lncRNA MYU expression (log-rank test, P<0.05). OS, overall survival; lncRNA, long non-coding RNA; MYU, c-Myc upregulated lncRNA; Cum, cumulative; m, months.
Further subgroup analysis suggested that in patients with stage III/IV, lymph node metastasis and low degree of differentiation, the OS of the high lncRNA MYU expression group was significantly shorter than that of the low lncRNA MYU expression group (P<0.05; Fig. 2C, E and G). However, OS was not associated with MYU expression in patients with stage I/II, lymph node metastasis-negative status and a high/moderate degree of differentiation (P>0.05; Fig. 2B, D and F).
lncRNA MYU promotes the proliferation of TE-2 cells
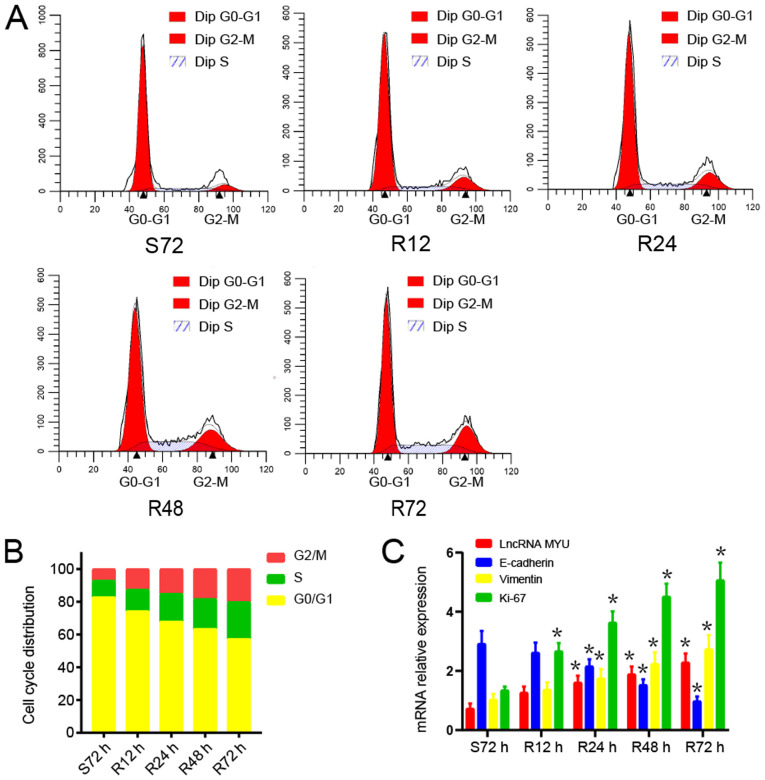
TE-2 cells were used for the serum starvation-release experiment. After TE-2 cells were subjected to serum starvation for 72 h, the serum supply was restored.
The changes in the cell cycle distribution after serum starvation and restoration were analysed by flow cytometry. After 72 h of serum starvation, TE-2 cells were arrested in G0/G1 phase. Once serum was restored, TE-2 cells were released from G0/G1 phase and gradually entered the S and G2/M phases (Fig. 3A and B).
Figure 3.
Expression of lncRNA MYU promotes the proliferation of TE-2 cells. (A and B) Cells were synchronized in the G0/G1 phase and progressed through the cell cycle when serum was added. (A) Representative cell cycle profiles obtained by flow cytometry and (B) quantified proportions of cells in the different phases of the cell cycle at various experimental time-points. (C) Bar graph indicating the expression of lncRNA MYU, E-cadherin, Vimentin and Ki-67 relative to that of GAPDH in proliferating TE-2 cells. Results for the time-points S72 h, R12 h, R24 h, R48 h and R72 h are provided, with S indicating serum starvation and R serum release. *P<0.05 vs. respective S72h groups. lncRNA, long non-coding RNA; MYU, c-Myc upregulated lncRNA.
The mRNA expression of lncRNA MYU, E-cadherin, Vimentin and Ki-67 during the serum starvation-release experiment was then examined. After serum stimulation, the expression of lncRNA MYU, Vimentin and Ki-67 mRNA in TE-2 cells gradually increased as the cell proliferation increased, while E-cadherin mRNA expression declined gradually (Fig. 3C). These results indicated that lncRNA MYU may promote the proliferation of tumour cells and be related to tumour metastasis.
Knockdown of lncRNA MYU inhibits the proliferation, migration and invasion of TE-2 cells
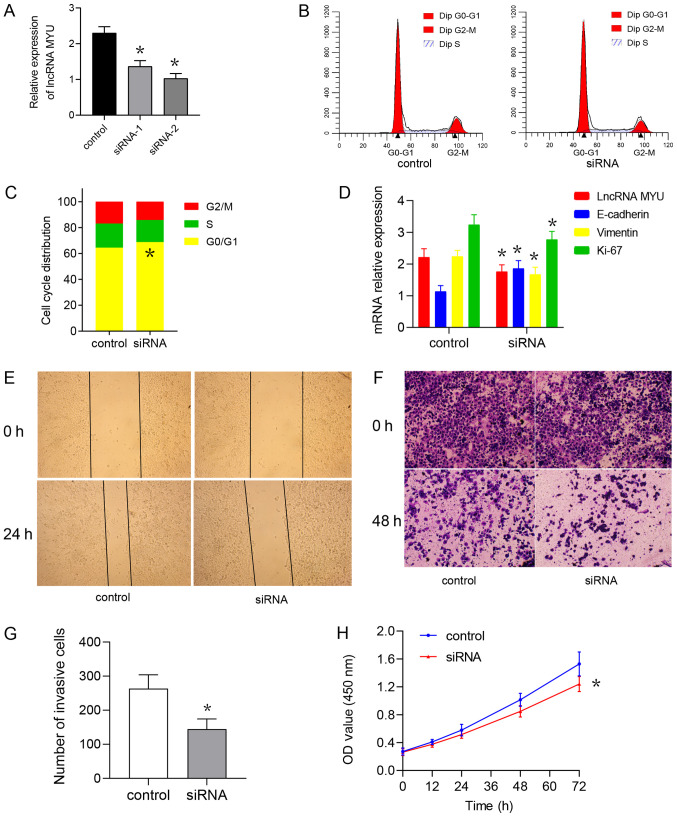
TE-2 cells were transfected with the lncRNA MYU siRNA plasmid to interfere with the expression of lncRNA MYU. RT-qPCR was used to identify the MYU siRNA with the highest knockdown efficiency and the results suggested that the MYU siRNA2-transfected group had the lowest lncRNA MYU expression and the greatest knockdown with good transfection efficiency (Fig. 4A). The flow cytometry results also indicated that the cell cycle was inhibited and that the proportion of cells in G1/G0 phase was increased in the MYU siRNA2-transfected group as compared with that in the MYU siRNA control-transfected group (Fig. 4B and C). It was also observed that after lncRNA MYU was knocked down, the mRNA expression of Vimentin and Ki-67 was decreased in TE-2 cells, while E-cadherin mRNA expression was elevated (Fig. 4D).
Figure 4.
lncRNA MYU knockdown inhibits the proliferation of TE-2 cells. (A) The relative expression level of lncRNA MYU in esophageal squamous cell carcinoma cells transfected with control siRNA or lncRNA MYU siRNA was analysed by reverse transcription-quantitative PCR. *P<0.05 vs. control. (B) Cell cycle analysis was performed after TE-2 cells were transfected with control siRNA or lncRNA MYU siRNA-2. (C) Representative cell cycle profiles obtained by flow cytometry and quantified proportions of cells in the different phases of the cell cycle. *P<0.05 vs. control. (D) Bar graph presenting the expression of lncRNA MYU, E-cadherin, Vimentin and Ki-67 relative to that of GAPDH in TE-2 cells transfected with control siRNA or lncRNA MYU siRNA-2. *P<0.05 vs. control. (E) A wound-healing assay was used to evaluate the migration ability of TE-2 cells transfected with control siRNA or lncRNA MYU siRNA-2. (F) The invasiveness of TE-2 cells transfected with control siRNA or lncRNA MYU siRNA-2 was evaluated using a Transwell assay. (G) Bar graph presenting the number of invasive TE-2 cells transfected with control siRNA or lncRNA MYU siRNA. *P<0.05 vs. control. (H) A Cell Counting Kit-8 assay was performed in TE-2 cells transfected with control siRNA or lncRNA MYU siRNA-2. *P<0.05 vs. control. lncRNA, long non-coding RNA; MYU, c-Myc upregulated lncRNA; siRNA, small interfering RNA; OD, optical density.
The wound-healing assay suggested that at 48 h after scratching, the control group exhibited significantly faster healing than the experimental group (Fig. 4E), indicating that interference with lncRNA MYU expression is able to inhibit the migration ability of TE-2 cells. The Transwell invasion test indicated that after the expression of lncRNA MYU was knocked down, the number of cells observed under the microscope in the experimental group und was significantly lower than that observed in the control group (Fig. 4F and G).
The CCK-8 cell proliferation assay indicated that the absorbance of the MYU siRNA group at 0, 24, 48, 72 and 96 h was lower than that of the control group at these time-points, suggesting that interference with lncRNA MYU expression inhibited the proliferation of TE-2 cells (Fig. 4H).
The results demonstrated that interference with the expression of lncRNA MYU may inhibit the proliferation, migration and invasion ability of TE-2 cells.
Mechanistically, lncRNA MYU may influence migration and invasion of ESCC cells by regulating the expression of the EMT-associated proteins E-cadherin and Vimentin.
Discussion
Despite continuous advancements of surgery and other treatments in recent years, the prognosis of esophageal cancer has not significantly improved (19). Studies have indicated that abnormal expression of lncRNAs is closely associated with the occurrence and development of esophageal cancer. Certain lncRNAs may become effective markers and therapeutic targets for the diagnosis and prognostic monitoring of esophageal cancer.
It has been reported that MYU is highly expressed in colorectal cancer and is associated with poor patient prognosis (20). MYU may stabilize the transcription of cyclin-dependent kinase 6 by combining with heterogeneous nuclear ribonucleoprotein K, thereby promoting colon cancer progression (21). A study on prostate cancer indicated that MYU is significantly upregulated in tumour tissue (22). Furthermore, MYU expression was reported to be significantly increased in squamous cell carcinoma of the lung and to be associated with the number of lymph node metastases and poor prognosis (23). The present study suggested that the expression level of lncRNA MYU in ESCC tumour tissues was significantly higher than that in normal adjacent tissues and that the expression of lncRNA MYU was associated with lymph node metastasis and TNM stage. Furthermore, survival analysis indicated that the OS of patients with high expression of lncRNA MYU was significantly shorter than that of patients with low expression. In addition, it was demonstrated that MYU expression was correlated with the expression of the proteins Ki-67, Vimentin and E-cadherin, which were assessed by immunohistochemical analysis. These results indicated that MYU has an important role in the progression and metastasis of ESCC.
Interfering with MYU expression is able to inhibit the growth and migration of cancer cells in prostate cancer and there is no correlation between MYU expression and VPS9D1 expression (24). MYU may also be transported outside of cells through exosomes to promote the proliferation and migration of adjacent cells. The present study further investigated the effect of MYU on the biological behaviour of ESCC cell lines. The serum starvation and release assay suggested that, as cell proliferation increased, the expression of MYU also increased accordingly, while the mRNA expression of E-cadherin decreased. Thus, this result suggested that MYU might play a role in cell cycle regulation (22). However, due to the limitations of the serum starvation and release assay, the specific role of MYU in ESCC cell proliferation still needs to be studied further. After MYU expression was knocked down, the proliferation, migration and invasion of esophageal cancer cells were inhibited, while the mRNA expression of E-cadherin was increased. MYU may regulate the proliferation, migration and invasion of ESCC cells, but the associated regulatory mechanisms require to be further studied and discussed.
In conclusion, the results of the present study suggested that an increase in the expression levels of lncRNA MYU may be an indicator of poor prognosis in patients with ESCC and may have value for determining the prognosis of patients with ESCC and serve as a potential therapeutic target.
Acknowledgements
Not applicable.
Funding Statement
Funding: The present study was supported by the Science and Technology Project of Nantong (grant no. JC2018154).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SG and LQ conceived and designed the study. JM, HS, YL and SZ were responsible for collecting patient data and conducted the data analysis. YL, SG and GM performed the experiments and analyzed the test data. SG, LQ and GM wrote and revised the manuscript and gave final approval for submission. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Affiliated Hospital of Nantong University (Nantong, China). The patients provided written informed consent for the use of their tumour tissues.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Zhang S, Zeng H, Xia C, Zuo T, Yang Z, Zou X, He J. Cancer incidence and mortality in China, 2013. Cancer Lett. 2017;401:63–71. doi: 10.1016/j.canlet.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 3.Liang H, Fan JH, Qiao YL. Epidemiology, etiology, and prevention of esophageal squamous cell carcinoma in China. Cancer Biol Med. 2017;14:33–41. doi: 10.20892/j.issn.2095-3941.2016.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez CA, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109:2093–2100. doi: 10.1111/cas.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 6.Chi Y, Wang D, Wang J, Yu W, Yang J. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;8(1015) doi: 10.3390/cells8091015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunch H. Gene regulation of mammalian long non-coding RNA. Mol Genet Genomics. 2018;293:1–15. doi: 10.1007/s00438-017-1370-9. [DOI] [PubMed] [Google Scholar]
- 8.Palmieri G, Paliogiannis P, Sini MC, Manca A, Palomba G, Doneddu V, Tanda F, Pascale MR, Cossu A. Long non-coding RNA CASC2 in human cancer. Crit Rev Oncol Hematol. 2017;111:31–38. doi: 10.1016/j.critrevonc.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12:433–446. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 11.Chen B, Li Y, He Y, Xue C, Xu F. The emerging roles of long non-coding RNA in gallbladder cancer tumorigenesis. Cancer Biomark. 2018;22:359–366. doi: 10.3233/CBM-170979. [DOI] [PubMed] [Google Scholar]
- 12.Xie H, Ma B, Gao Q, Zhan H, Liu Y, Chen Z, Ye S, Li J, Yao L, Huang W. Long non-coding RNA CRNDE in cancer prognosis: Review and meta-analysis. Clin Chim Acta. 2018;485:262–271. doi: 10.1016/j.cca.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Ma J, Xiao Y, Tian B, Chen S, Zhang B, Wu J, Wu Z, Li X, Tang J, Yang D, et al. Genome-wide analyses of long non-coding RNA expression profiles and functional network analysis in esophageal squamous cell carcinoma. Sci Rep. 2019;9(9162) doi: 10.1038/s41598-019-45493-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Li R, Ding X, Zhang K, Qin W. Upregulation of long non-coding RNA SNHG6 promote esophageal squamous cell carcinoma cell malignancy and its diagnostic value. Am J Transl Res. 2019;11:1084–1091. [PMC free article] [PubMed] [Google Scholar]
- 15.Razavi M, Ghorbian S. Up-regulation of long non-coding RNA-PCAT-1 promotes invasion and metastasis in esophageal squamous cell carcinoma. EXCLI J. 2019;18:422–428. doi: 10.17179/excli2018-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zong MZ, Feng WT, Du N, Yu XJ, Yu WY. Upregulation of long noncoding RNA LEF1-AS1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Eur Rev Med Pharmacol Sci. 2019;23:7929–7934. doi: 10.26355/eurrev_201909_19007. [DOI] [PubMed] [Google Scholar]
- 17.Ma J, Xiao Y, Tian B, Chen S, Zhang B, Wu J, Wu Z, Li X, Tang J, Yang D, et al. Long noncoding RNA lnc-ABCA12-3 promotes cell migration, invasion, and proliferation by regulating fibronectin 1 in esophageal squamous cell carcinoma. J Cell Biochem. 2020;121:1374–1387. doi: 10.1002/jcb.29373. [DOI] [PubMed] [Google Scholar]
- 18.Zhu D, Yu Y, Qi Y, Wu K, Liu D, Yang Y, Zhang C, Zhao S. Long non-coding RNA CASC2 enhances the antitumor activity of cisplatin through suppressing the Akt pathway by inhibition of miR-181a in esophageal squamous cell carcinoma cells. Front Oncol. 2019;9(350) doi: 10.3389/fonc.2019.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou H, Meng Z, Zhao X, Ding G, Sun M, Wang W, Wang Y. Survival of esophageal cancer in China: A pooled analysis on hospital-based studies from 2000 to 2018. Front Oncol. 2019;9(548) doi: 10.3389/fonc.2019.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L, Xu L, Wang Q, Wang M, An G. Dysregulation of long non-coding RNA profiles in human colorectal cancer and its association with overall survival. Oncol Lett. 2016;12:4068–4074. doi: 10.3892/ol.2016.5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawasaki Y, Komiya M, Matsumura K, Negishi L, Suda S, Okuno M, Yokota N, Osada T, Nagashima T, Hiyoshi M, et al. MYU, a target lncRNA for Wnt/c-Myc signaling, mediates induction of CDK6 to promote cell cycle progression. Cell Rep. 2016;16:2554–2564. doi: 10.1016/j.celrep.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Yang X, Li R, Wang L, Gu Y, Zhao Y, Huang KH, Cheng T, Yuan Y, Gao S. Long non-coding RNA MYU promotes prostate cancer proliferation by mediating the miR-184/c-Myc axis. Oncol Rep. 2018;40:2814–2825. doi: 10.3892/or.2018.6661. [DOI] [PubMed] [Google Scholar]
- 23.Tan J, Yang L. Long noncoding RNA VPS9D1-AS1 overexpression predicts a poor prognosis in non-small cell lung cancer. Biomed Pharmacother. 2018;106:1600–1606. doi: 10.1016/j.biopha.2018.07.113. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Chen Q, Wang X, Li W, Yu G, Zhu Z, Zhang W. ZEB1 activated-VPS9D1-AS1 promotes the tumorigenesis and progression of prostate cancer by sponging miR-4739 to upregulate MEF2D. Biomed Pharmacother. 2020;122(109557) doi: 10.1016/j.biopha.2019.109557. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.