Abstract
Species invasions and range shifts can lead to novel host–parasite communities, but we lack general rules on which new associations are likely to form. While many studies examine parasite sharing among host species, the directionality of transmission is typically overlooked, impeding our ability to derive principles of parasite acquisition. Consequently, we analysed parasite records from the non-native ranges of 11 carnivore and ungulate species. Using boosted regression trees, we modelled parasite acquisition within each zoogeographic realm of a focal host's non-native range, using a suite of predictors characterizing the parasites themselves and the host community in which they live. We found that higher parasite prevalence among established hosts increases the likelihood of acquisition, particularly for generalist parasites. Non-native host species are also more likely to acquire parasites from established host species to which they are closely related; however, the acquisition of several parasite groups is biased to phylogenetically specialist parasites, indicating potential costs of parasite generalism. Statistical models incorporating these features provide an accurate prediction of parasite acquisition, indicating that measurable host and parasite traits can be used to estimate the likelihood of new host–parasite associations forming. This work provides general rules to help anticipate novel host–parasite associations created by climate change and other anthropogenic influences.
Keywords: parasite, terrestrial mammal, invasive species, modelling, non-native
1. Introduction
Species invasions, introductions and range shifts can alter community composition and lead to novel host–parasite interactions [1]. Accelerating climate change and growing anthropogenic influences, such as land conversion and urbanization, will continue to perpetuate such community-level changes [2,3], thereby increasing the chances that species encounter novel parasites [4]. The spread of newly acquired parasites can lead to unexpected and catastrophic disease outbreaks, as seen for example with chytridiomycosis, a fungal pathogen that is a major threat to global amphibian biodiversity [5]. The effects extend to humans as well, such as through economic losses or direct morbidity and mortality from zoonotic pathogens [5,6].
While much progress has been made in understanding parasite sharing in general [7–12], studies including directional transmission are often specific case studies (e.g. [13–15]). Zoonotic transmission from animals to humans is a well-documented, special case of directional cross-species transmission, and has revealed that both host and parasite traits are important in understanding parasite acquisition [16–19]. However, in terms of behaviour and mobility, among other factors, humans are not representative of animals at large, calling for the broader study of parasite acquisition in other species and establishment of general rules governing cross-species transmission. In rare examples of directional transmission studied in natural host communities, host phylogeny and geographic range overlap were found to regulate cross-species transmission of bat rabies viruses [20], a finding echoed in comparative studies of primate parasites [10,21]. By contrast, over relatively short time periods, introduced populations of salmonid fish species attained similar levels of parasite diversity compared to their original range [22], indicating the high ecological, versus coevolutionary, potential for parasite acquisition. These observations invite a comparative study of parasite acquisition across species, which we present using a large dataset on terrestrial mammals [23] to determine the key factors controlling the acquisition of a taxonomically diverse set of parasites by several host species in their non-native ranges. Because such rules should not be specific to host and parasite species, we use 11 host species and 775 parasite species, and evaluate the ability of models developed for each host species to predict parasite acquisition by the other host species.
The establishment of general rules governing parasite acquisition by non-native hosts also promises to strengthen our understanding of related ecological theory. Parasite spillback theory [24] aims to quantify the increase in parasite pressure to a native community as a result of the introduction of a non-native host species competent for some set of the native parasites. Characterizing such competency in terms of host and parasite traits would aid in assessing which parasites in the novel community are most likely to be involved in this process, as well as determining how likely the non-native host is to acquire each of the parasites in the subset. Additionally, from the perspective of an invasive host moving from its native to non-native range, the enemy release hypothesis posits that the invasive host might enjoy a competitive advantage over native species by leaving behind natural enemies, including parasites [25–27]. The estimation of the likelihood of acquiring new parasite species in the non-native range allows for an assessment of how transient enemy release is likely to be. Finally, vacated niche theory suggests that parasite release creates an opportunity for a functionally similar parasite species to colonize a host population in the non-native range [28]. The study of the tendency for this to occur across invasive host species could allow the extent of the role of vacated niches in parasite acquisition to be established.
We expect both parasite and host community traits to drive patterns of parasite acquisition. Variation in parasite life histories might mean certain parasites have a greater propensity to jump the species barrier; such differences can be captured through, for example, parasite taxonomy (e.g. helminth versus virus) and transmission modes (e.g. close contact versus vector-borne). Parasite type will affect the production of genetic variation, among numerous other factors; parasites with higher genetic diversity and faster adaptation, such as viruses, might spread more easily to new hosts [29]. Meanwhile, transmission mode affects how, when and where interspecific transmission opportunities arise [30].
The degree of specialism or generalism exhibited by a parasite is also likely to affect its acquisition by new hosts. In theory, generalist parasites should have a greater chance of acquisition because they can infect a wider variety of hosts and thus are expected to jump between hosts more readily than specialists. In terms of mechanisms, generalist parasites have the intrinsic advantage of making use of a large set of often taxonomically distinct host species [31], which can help them attain large geographic ranges [32]. It can also protect them from extinction, as they are not reliant on a single host species which might itself experience local extinction or exhaustion of susceptible individuals [33]. However, while weak interspecific transmission can facilitate generalist parasite persistence in certain hosts [33], insufficient transmission between species can also lead to those same hosts losing local association with generalist parasites, which might, at the extreme, be deprived of cross-species transmission if some of the hosts become globally or locally extinct [34]. Further, generalist parasite strategies are likely to be associated with costs related to inefficient exploitation of hosts. These include the reduction of within-host replication as host range increases [35], the loss of fitness on the original host species as a parasite adapts to a new one [36], or maladaptive virulence, where the parasite is able to infect species phylogenetically distant from original hosts but causes harm to the new host, including mortality, limiting further transmission opportunities [37–39]. This ambiguity on the relative advantages of specialist and generalist parasite strategies, and the fact that they might manifest differently across parasite taxa, calls for consideration of parasite specificity as a predictor of acquisition by novel host species.
Host–parasite community composition is also likely to influence parasite acquisition. For example, parasites can exhibit different levels of infection prevalence across host species, where prevalence is defined as the proportion of sampled hosts that are infected. We expect this variation in force of infection to influence parasite spread to new hosts; that is, non-native hosts are more likely to acquire parasites that pre-exist at high prevalence in established host populations [40]. Parasites will also vary in the number of host species they infect within a given community. Under the common assumption that host competency and abundance are positively correlated [41], if the transmission is density-dependent and host community size increases with host species richness, the effect of host count on parasite acquisition is typically analogous to that of prevalence: contact and transmission opportunities for a parasite will scale positively with the number of host species it infects [42]. However, in cases with different competency–abundance relationships, frequency-dependent transmission or host communities at carrying capacities, opposite patterns are possible [42,43]. Consequently, host count has the potential to explain parasite acquisition via several latent mechanisms.
We can also consider the non-native host's ecological and evolutionary ‘compatibility’ with the community into which it enters. Host trait compatibility (defined as the similarity between hosts based on ecological traits) might facilitate parasite acquisition by a non-native host [44]; in other words, parasites acquired by the non-native host might be associated with established hosts that share more traits with the non-native host, such as diet [45,46]. Similarly, phylogenetic compatibility (measured as the phylogenetic distance between hosts) is also likely to affect parasite acquisition [46,47]; we expect that acquired parasites will be associated with hosts closely related to the non-native host. The phylogenetic distance between non-native and other hosts determines the size of the host jump required for parasite acquisition, which may modify the cost–benefit trade-off of being a generalist parasite.
Our goal in this study was to synthesize the theories discussed above in an exploration of parasite acquisition by terrestrial mammals that move into non-native ranges. We used host–parasite records from the recently published Global Mammal Parasite Database (GMPD) [23], which uses comprehensive literature review and error checking involving over 2700 publications to assemble our current understanding of host–parasite associations for wild carnivore, ungulate and primate host species. We modelled parasite acquisition as a function of both parasite and host community traits within the non-native range. Our results, which span 11 focal hosts and 3 zoogeographic realms, identify parasite prevalence, host phylogenetic compatibility and number of hosts per parasite as particularly influential factors. Overall, our analyses suggest that patterns of parasite acquisition are broadly generalizable and predictable, shaped by interacting effects of host and parasite characteristics.
2. Methods
As an overview, we collected parasite data for each zoogeographic realm of each focal host's non-native range as distinct dataframes. Each row of a dataframe corresponded to a parasite recorded in that portion of the host range (see table 1 ‘no. non-native parasites' column for sample sizes). To this, we added 15 predictors describing parasite and host community traits (table 2; electronic supplementary material). Here, host community traits refer to metrics that describe communities versus species, such as average relatedness between species, and not ecological interactions. We then created our response variable: a binary indicator of parasite acquisition status (whether or not each parasite was newly recorded in the focal host in the non-native range). These data were used to fit boosted regression tree (BRT) models; we ran 50 iterations per dataframe (i.e. 13 × 50 = 650 models). We assessed model performance using AUC and cross-validation and extracted relative influence values to summarize predictor importance.
Table 1.
Focal host names, realms, parasite data summary and AUC scores for the 13 clusters (focal host x realm combinations). Realms: N = Nearctic, P = Palearctic, S = Sino-Japanese. Sample sizes for modelling are given in the column ‘no. non-native parasites’. AUC scores are median values followed by minimum and maximum values in parentheses.
| cluster | host species | realm | host common name | host group | no. acquired parasites | no. non-native parasites | AUC score |
|---|---|---|---|---|---|---|---|
| 1 | Cervus elaphus | N | red deer | ungulate | 14 | 262 | 0.861 (0.748, 0.889) |
| 2 | Cervus nippon | N | sika deer | ungulate | 7 | 97 | 0.944 (0.911, 0.984) |
| 3 | Cervus nippon | P | sika deer | ungulate | 7 | 174 | 0.910 (0.841, 0.938) |
| 4 | Dama dama | N | fallow deer | ungulate | 13 | 142 | 0.923 (0.877, 0.924) |
| 5 | Dama dama | P | fallow deer | ungulate | 40 | 350 | 0.930 (0.891, 0.941) |
| 6 | Genetta genetta | P | common genet | carnivore | 13 | 191 | 0.937 (0.903, 0.959) |
| 7 | Lynx pardinus | P | Iberian lynx | carnivore | 5 | 143 | 0.554 (0.506, 0.701) |
| 8 | Neovison vison | P | American mink | carnivore | 28 | 344 | 0.915 (0.875, 0.932) |
| 9 | Nyctereutes procyonoides | P | raccoon dog | carnivore | 26 | 237 | 0.954 (0.945, 0.962) |
| 10 | Procyon lotor | S | common raccoon | carnivore | 11 | 102 | 0.834 (0.786, 0.859) |
| 11 | Rupicapra rupicapra | P | chamois | ungulate | 9 | 205 | 0.865 (0.773, 0.898) |
| 12 | Sus scrofa | N | wild boar | ungulate | 7 | 202 | 0.837 (0.760, 0.881) |
| 13 | Vulpes vulpes | N | red fox | carnivore | 13 | 319 | 0.816 (0.756, 0.862) |
Table 2.
Variables used to predict parasite acquisition. For trait.diff.mean and trait.diff.min, ‘host trait scores’ come from a principal component analysis of PanTHERIA, a database of mammal traits ([48]; see the electronic supplementary material for details).
| category | predictor | description |
|---|---|---|
| characteristics of parasites | ParType | parasite taxonomic group: arthropod, bacteria, helminth, protozoa or virus |
| close, nonclose, vector, intermediate | parasite transmission mode(s), each scored separately as a binary variable | |
| n.modes | parasite's number of transmission modes | |
| prev.mean, prev.max | mean and maximum parasite prevalence across infected hosts in the non-native range, excluding the focal host | |
| n.hosts | number of non-focal hosts in which the parasite was found in a focal host's non-native range | |
| all.hosts | number of known mammal hosts per parasite across all GMPD records, excluding the focal host | |
| z.score | standardized measurement of a parasite's phylogenetic range breadth; negative values indicate greater degree of host specialism | |
| characteristics of parasites’ hosts | PD.mean, PD.min | mean and minimum pairwise phylogenetic distances between a parasite's hosts (those tallied in n.hosts) and the relevant focal host; units are millions of years ago (Ma) |
| trait.diff.mean, trait.diff.min | mean and minimum difference in host trait scores between a parasite's hosts (those tallied in n.hosts) and the relevant focal host |
All data preparation, analysis and modelling were done in R, v. 3.5.1–3.6.1 [49].
(a). Data preparation
(i). Native versus non-native ranges
We obtained host–parasite associations and location data from the GMPD [23]. The GMPD contains over 24 000 records of parasites sampled from mammal hosts (carnivores, primates and ungulates), along with latitude–longitude data for most samples.
For our study, we focused only on terrestrial hosts scored in the GMPD as having parasite records in both native and non-native ranges (n = 33). Because accurate boundaries are needed to define range-specific host–parasite associations, we reclassified each host's GMPD records based on buffered IUCN native ranges ([50]; see the electronic supplementary material, text and figure S1 for details), with records outside these ranges classified as non-native. This resulted in 29 focal hosts (of our original 33) with GMPD records in both native and non-native ranges.
To translate non-native points into ecologically feasible non-native range areas, we overlapped GMPD records for our 29 focal hosts with The Nature Conservancy's Terrestrial Ecoregions map [51] (shapefiles from http://maps.tnc.org/gis_data.html). We defined non-native ranges as the sets of ecoregions in which non-native points were found. Ecoregions provide a meaningful way to approximate broad invasion areas, without relying on local buffering of individual sampling locations; using point-specific, localized areas would potentially lead to highly fragmented non-native ranges that miss nearby suitable areas. This approach has recently been assessed as viable in the field of host–parasite biogeography (e.g. the ‘ecoregion filling method’ of [52]). We deemed species distribution models (also known as ecological niche models) inappropriate for our study because invasive species are rarely in equilibrium with the environment, which violates assumptions of such models [53].
(ii). Non-native host–parasite communities
We analysed all terrestrial GMPD records to identify communities of hosts and parasites with which a focal host might interact in its non-native range. We first associated each GMPD record with an ecoregion (dropping those without latitude–longitude data) and then checked whether that ecoregion was found in each focal host's non-native ecoregion set.
Parasite classification. For the purposes of these analyses, we specifically wanted to identify parasites acquired by each focal host in its non-native range and compare those against parasites not acquired in the same range. We identified all unique GMPD parasites associated with each of our 29 focal hosts and created a binary classification in which acquired parasites were assigned 1 and not-acquired parasites were assigned 0. Using our previous classification of GMPD records as native or non-native, we assessed whether each parasite had been sampled from the host in its non-native, but not native, range. Parasites that met these criteria were flagged as having been acquired by the focal host in its non-native range. We then compared each focal host's set of acquired parasites to the full non-native parasite pool to identify those parasites present in the non-native range but not acquired by the focal host. Overall, many more parasites were not acquired than were (electronic supplementary material, figure S2).
(iii). Focal hosts
We excluded two focal hosts that had no GMPD records for other host species overlapping their non-native ranges, as we could not compare acquired parasites against others for these species. Seven more hosts were dropped because their acquired parasites were not sampled from any other mammal hosts in their non-native range. To ensure sufficient data for further range refinement (see below), we also excluded eight hosts whose non-native ranges contained fewer than 100 parasites and/or that had fewer than 5 acquired parasites. These removals left us with 12 focal hosts.
(iv). Refining non-native range definitions
Non-native ranges for several of our 12 focal hosts spanned multiple continents, which meant that grouping them into a single non-native community would make little ecological sense. To create more ecologically meaningful non-native ranges, we used Holt et al.'s [54] zoogeographic realms, which use both species distributions and phylogenetic relationships to demarcate geographic space.
We grouped each focal host's non-native ecoregions into realm-specific clusters, such that a cluster contained all non-native ecoregions for a single focal host within a single zoogeographic realm (table 1, columns 1–3). A focal host could have more than one associated cluster if its non-native ecoregions fell in more than one realm (figure 1).
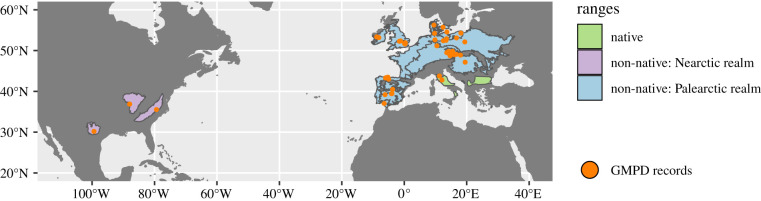
Figure 1.
Range map developed for example focal host Dama dama, whose non-native range consists of separate clusters in two zoogeographic realms. For map clarity, the unbuffered IUCN native range is shown. (Online version in colour.)
Splitting a focal host's non-native range into clusters meant parasite records were reduced. Thus, we re-checked parasite counts and dropped clusters with fewer than 50 parasites total and/or fewer than 5 acquired parasites, with the goal of minimizing negative effects of low sample sizes on model performance [55]. This left us with 13 clusters for 11 focal hosts and 3 zoogeographic realms (table 1).
(v). Predictors
For each cluster, we created a dataframe in which rows corresponded to parasites found in the cluster and added the binary classification of parasite acquisition status (1 = acquired, 0 = not acquired). This column was used as our response variable. We then collected 15 predictor variables to describe each cluster's non-native parasite community, both in terms of parasite characteristics and associated non-native hosts (table 2; see the electronic supplementary material for more detailed descriptions).
(b). Modelling
We used BRT as our modelling method due to its ability to accommodate many predictors of different types with missing data, and its flexibility in fitting complex responses and interactions [56]. We fitted our models in the ‘gbm’ package (version 2.1.5; [57]; see the electronic supplementary material for model details). We trained each model on all available data, in part due to limited records and in part because we were more interested in understanding patterns than producing predictions on withheld data; however, we did test the models on new data, as described below. To assess the stability of our modelling results, we fitted 50 models for each cluster.
(i). Model evaluation and validation
We calculated AUC scores for each of the 50 models per cluster, based on the internal cross-validation during model fitting. AUC (area under the curve of the receiver operating characteristic) is a threshold-independent performance measure for binary classification; an AUC of 1 indicates a perfect positive correlation between predictions and observations, 0 a perfect inverse correlation and 0.5 no correlation (random). The range of AUCs for each cluster allowed us to assess model stability across repetitions. We then identified each cluster's best model—the one with the highest AUC score (table 1)—and selected these models for further investigation.
This definition of ‘best models’ applies specifically to our models as tools to understand patterns in our data. We note, however, that this definition will not be appropriate or accurate in all circumstances. For example, models that receive good scores on internal validation metrics will not necessarily perform well when applied to new data (i.e. extrapolation [58]). As such, we tested extrapolative performance by validating each cluster's best model on the other 12 clusters' data and calculating AUC scores for each cross-cluster prediction. Aside from assessing extrapolative model performance, this ‘cross-cluster validation’ method helps us understand the degree to which rules of parasite acquisition are generalizable across host species and realms. To assess the consistency of our results, we repeated this process with models receiving the median or minimum AUC scores for each cluster; however, we found minimal differences in performance among these models, so we only present results from the best model cross-cluster validation.
(ii). Predictor assessment
We obtained relative influence values for all 15 predictors in the focal hosts' best models; these measures, which sum to 100 in a given model, account for how often a predictor is used for data classification in model fitting, weighted by the resultant improvement to the model [56]. We calculated the mean and variance of these values for each predictor across all 13 clusters, to assess consistency in predictor influence. We also created partial dependence plots (PDPs) for the most influential predictors, to examine cluster-specific predictor relationships with parasite acquisition. As interactions were included in our model, we further used Friedman's H statistic to test for the strength of two-way interactions among predictors [59]. Friedman's H ranges from 0 to 1 and represents the proportion of variance explained by an interaction [60]; naturally, interaction strength can be small in models already containing 15 potential main effects.
3. Results and discussion
Models for each cluster show fairly stable performance; maximum and minimum AUC scores differ by at most 0.195 (cluster 7) and on average by 0.09 (table 1; electronic supplementary material, figure S3a). This suggests that BRT is a robust modelling approach for our data and that we can make comparable statements for each cluster on the basis of any one of its models. Twelve clusters typically had AUCs greater than 0.8, indicating fairly strong positive correlations between predictions and observations (table 1; electronic supplementary material, figure S3a). The one remaining cluster (cluster 7: L. pardinus, P) still achieved AUCs greater than 0.5, or better-than-random performance (table 1; electronic supplementary material, figure S3a); we attribute the lower scores for this cluster to the fact that it contained the fewest acquired parasites (table 1).
To explore model performance across different focal hosts and realms, we conducted cross-cluster validation using each cluster's best model, again finding good model performance with most AUC values greater than 0.7. This analysis showed that internal evaluation scores (AUCs from model fitting) do not necessarily correspond with extrapolative predictive performance (electronic supplementary material, figure S3). We also observe that, broadly speaking, ‘shared host’ cross-validation (that is, models applied to data for the same host species) leads to the best results, followed by ‘shared realm’ and then ‘neither shared’ (electronic supplementary material, figure S3b, points).
Four clusters’ models under-performed relative to the others on cross-cluster validation: clusters 3 (C. nippon, P), 10 (P. lotor, S), 11 (R. rupicapra, P) and 13 (V. vulpes, N; electronic supplementary material, figure S3b). We note that these four clusters are among those with fewer acquired parasites (table 1). Cluster 10 is the only cluster unique in both host and realm; in addition, its PDPs (electronic supplementary material, figure S4b,c) show predictor relationships with parasite acquisition that differ from the general trends for other hosts, meaning this model might not have captured widely relevant patterns of acquisition. Interestingly, cluster 13 performed fairly well on clusters in the same zoogeographic realm; its overall performance was brought down by its application to clusters with which it shared neither host nor realm (electronic supplementary material, figure S3b, points). Variation aside, we can make reasonably accurate predictions across clusters; all models still perform well (electronic supplementary material, figure S3b). This suggests that common predictors drive parasite acquisition across focal hosts.
We found that both parasite and host community traits predict parasite acquisition. Across all clusters, the predictors with the highest mean relative influence were mean parasite prevalence (prev.mean, mean relative influence = 15.322), mean phylogenetic distance (PD.mean, mean relative influence = 11.989) and global mammal host count (all.hosts, mean relative influence = 11.374; electronic supplementary material, table S1). The importance of these three predictors demonstrates the value of incorporating data at multiple spatial scales, as they describe both range-specific and global factors that drive parasite acquisition.
These same three predictors were among those with the highest variance in relative influence. The partitioning of influence among predictors varied considerably across clusters, even though certain predictors were repeatedly among the most influential. We also saw variation across clusters in the specific relationship of each predictor with parasite acquisition, despite the consensus on predictor influence (electronic supplementary material, figure S4). This was particularly true for mean prevalence, where the relationship was highly variable at low prevalence values, before generally trending upwards at higher values (electronic supplementary material, figure S4a).
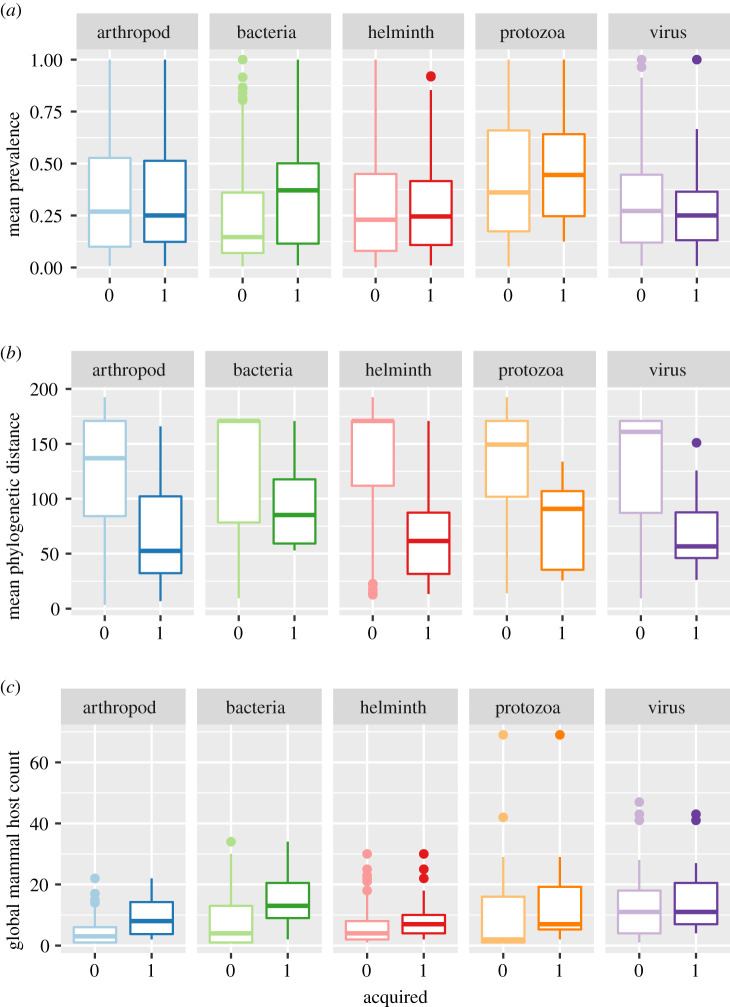
To look for broad-scale patterns in how our most influential predictors affect parasite acquisition, we examined the relationships in data pooled across all clusters, faceted by parasite type for insight into potential interactions (figure 2). Combining data from different communities obscures some of the specific detailed trends seen in the PDPs (electronic supplementary material, figure S4), but brings to light additional universal trends. These visualizations do not depict model output; rather, we are looking at patterns in the raw data.
Figure 2.
Box plots of parasite acquisition (1 = acquired, 0 = not acquired) for the most influential predictors: (a) mean parasite prevalence, (b) mean phylogenetic distance and (c) global mammal host count. Data pooled across all 13 clusters. Centre lines of boxes correspond to the median value; lower and upper hinges represent first and third quartiles (25th and 75th percentiles), respectively. Whiskers extend to the minimum and maximum values no farther than 1.5 × IQR from the lower and upper hinges, respectively (IQR = interquartile range, the distance between hinges). Additional outlying points are plotted individually. (Online version in colour.)
Mean prevalence, mean phylogenetic distance and global mammal host count show weak interactions with parasite type (mean Friedman's H across clusters = 0.062, 0.042, and 0.043, respectively); the importance of these predictors for parasite acquisition is not strongly dependent on the type of parasite. Across all parasite types, acquired parasites tend to be found at slightly higher prevalences, in established hosts more closely related to the non-native focal hosts (i.e. lower mean phylogenetic distance), and with higher global mammal host counts (figure 2). The bacteria group contains parasites which are acquired in spite of high mean phylogenetic distance between the focal host and other hosts in the community (figure 2b), perhaps because they tend to be less specialist than other parasite types [31]. Interestingly, bacteria also show some sensitivity to prevalence (figure 2a), whereby acquired parasites often pre-exist at relatively high prevalence in other hosts; in other parasite groups, the importance of having a high prevalence in established hosts might be outweighed by high parasite specificity. However, protozoa, which tend towards higher levels of specialism [31], also show a trend in which acquisition is associated with relatively high prevalence (figure 2a). Consequently, a case could also be made that costs of specialism, especially infrequent encounters between infected individuals and novel hosts, could be offset by high prevalence. Collectively, these nuanced effects of prevalence signal interesting areas for future study. Taken in combination with evidence for transmission between related host species being more likely, these results hint at variation in both the force of infection and susceptibility among species, which may support future studies characterizing variation in cross-species transmission from a mechanistic basis.
Meanwhile, viruses stand out as the only parasite taxon insensitive to global mammal host count (figure 2c). However, in line with other parasite taxa, they are strongly impacted by the phylogenetic relatedness between established hosts in a community and the non-native focal host (mean phylogenetic distance; figure 2b). Consequently, while the ability to infect a large number of species is broadly predictive of parasite acquisition, the distance of the species jump in the host community required for acquisition is more consistently informative across parasite taxa. Typically, viruses have larger numbers of hosts globally, which could contribute to their relative insensitivity to global mammal host count. Additionally, pooling viral subgroups (e.g. DNA versus RNA) could obscure separate trends, given the variation in viral ecology, transmission and evolution [61]. Overall, community-level characteristics are more predictive of virus acquisition and underscore the importance of considering interactions between community and parasite traits, as there may be exceptions to general rules or patterns driven by a minority of parasite species.
Given the consistent and distinct relationship between parasite acquisition and mean phylogenetic distance across parasite types (figure 2b), we investigated its potential interactions with other predictors. We expected that parasite specialism might modulate the importance of phylogenetic compatibility, and indeed, we found that mean phylogenetic distance interacts weakly with parasite specificity, as measured by phylogenetic z-score (mean Friedman's H across clusters = 0.067; electronic supplementary material, figure S5). Focal hosts that are more distantly related to parasites' host sets (101–200 Ma) show an overall tendency to acquire more generalist parasites, as evidenced by the z-scores for acquired parasites being more positive than those for not-acquired parasites. By contrast, when the focal host is more closely related to a parasite's host set (0–100 Ma), it is more likely to acquire specialists. This tendency suggests a potential fitness advantage to specialists over generalists at smaller phylogenetic distances. Such a trend could arise if phylogenetically specialist parasites have beneficial adaptations through infection of closely related hosts that offer no advantage in more distantly related hosts. Alternatively, this could be evidence of costs of generalism for the parasites, manifesting as a low probability of infection of hosts they are phylogenetically expected to be able to infect.
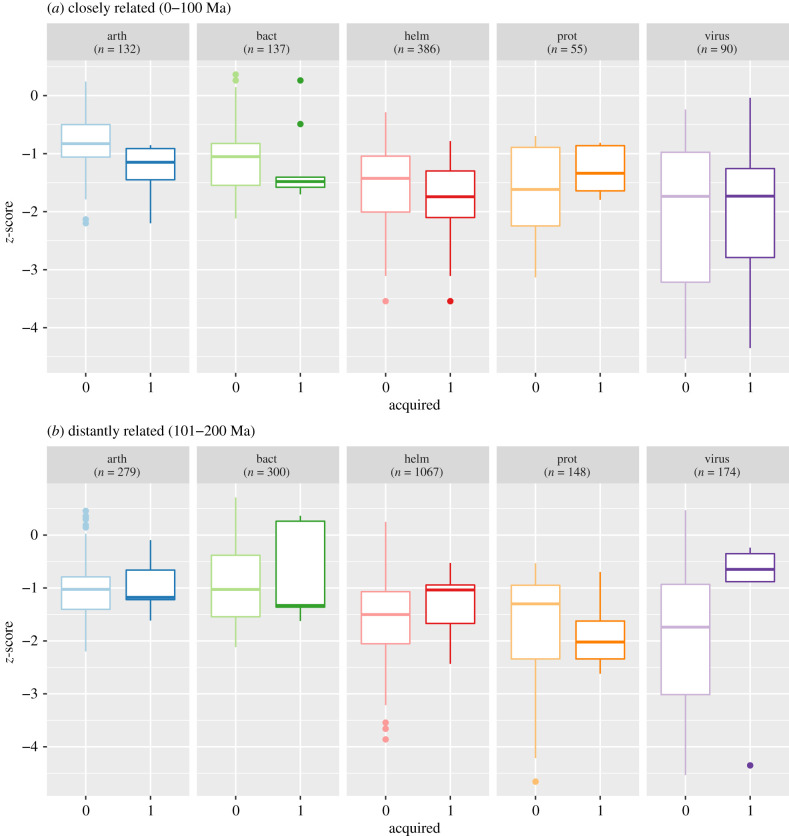
We can also break down each of the phylogenetic relatedness bins in the electronic supplementary material, figure S5 by parasite type (figure 3). While sample sizes render the strength of three-way interactions small, we observe that for smaller phylogenetic distances (figure 3a), acquired arthropods, bacteria and helminths tend towards greater levels of specialism than those not acquired. At greater phylogenetic distances (figure 3b), acquired helminths and viruses tend to be more generalist than those not acquired. Further research is required to clarify why generalist parasites of certain taxa are less likely to be acquired in this context; is this evidence of specialist fitness advantages or costs of generalizm? We note that such study should take into account the interaction seen here between parasite taxonomy and host phylogenetic relatedness, particularly evidenced by the virus group; at smaller phylogenetic distances viruses do not show a cost of generalism (figure 3a), but at larger distances, they show a pronounced cost of specialism (figure 3b).
Figure 3.
Parasite phylogenetic z-scores (z.score) by binned mean phylogenetic distances (Ma, PD.mean) and coloured by parasite taxonomic groups, summarized across all clusters. n denotes the number of parasite records per parasite type per phylogenetic distance bin. Parasite types: arth = arthropod, bact = bacteria, helm = helminth, prot = protozoa. Ma = millions of years ago. (Online version in colour.)
We acknowledge imperfections in our data and predictors. GMPD prevalence data, for example, is fairly noisy and includes multiple sampling methods. In addition, sampling effort is not evenly distributed across the GMPD—certain hosts, parasite types, geographic areas and time periods are more thoroughly sampled than others. Such variation will influence our definitions of non-native ranges, as well as our identification of acquired parasites. Some of our results could have been produced if, for example, researchers studying an invasive species are more likely to sample preferentially for locally prevalent parasites, or non-random, globally widespread pathogens [32]. While it is difficult to accurately predict the effects of such biases, the finding that global mammal host count is commonly a positive predictor of parasite acquisition might represent, in part, that parasite species commonly looked for, or easily detectable, are disproportionately represented among the acquired parasite species; the same could be said for the importance of parasite prevalence in predicting acquisition. Missing host–parasite associations would also affect the accuracy of estimated predictors capturing minimum ecological or phylogenetic distances, since a missing host could revise down the value of a predictor. To mitigate this effect, we used both minimum and mean distances, where the mean is less sensitive to a missing observation, and typically found mean distances to have higher predictive importance than minimum distances. However, the assembly of the underlying database is comprehensive in design with error-checking steps included [23]. We also note that our selection of predictors is, by necessity, far from exhaustive. Relevant information might also be provided by metrics quantifying the geographical opportunity for parasite acquisition (e.g. range overlap among host species) or environmental similarity between native and non-native ranges.
4. Conclusion
In conclusion, our work shows that parasite acquisition is non-random and predictable across multiple non-native terrestrial mammals occurring in several zoogeographic realms. We found parasite prevalence, host phylogenetic compatibility (phylogenetic distance) and global mammal host count to be particularly influential predictors. In addition, parasite taxonomy interacts with these predictors, which suggests that parasite and host community characteristics cannot necessarily be disentangled; each provides an important context in which to understand the influence of the other. Specifically, we identify both costs and benefits of generalist parasite infection strategies across host species that differ according to parasite group. While the number of host species considered is limited by data availability, our work greatly expands on previous case studies of directional parasite transmission; we show promising extrapolative performance of our models, even when predicting to different host species in distinct zoogeographic realms, supporting the idea that general rules for parasite acquisition apply broadly across host taxa and geographical locations. Our analytic framework may therefore provide a useful starting point from which to explore patterns of parasite acquisition for host species, locations, and contexts not yet studied. Further, this approach may even be extended to predict other types of symbiotic interactions likely to be altered by anthropogenic impacts, such as plant–fungal mutualisms [62], plant–pollinator associations [63] and host–parasitoid interactions [64].
Supplementary Material
Acknowledgements
We thank Patrick Stephens, John Vinson, David Vasquez, J. P. Schmidt and three anonymous reviewers for helpful comments on prior drafts.
Data accessibility
Data and reproducible code are available on figshare [65].
Authors' contributions
A.W.P. and A.M.S. conceived and designed the study; A.M.S. performed analyses; A.M.S. wrote the initial manuscript draft; A.M.S. and A.W.P. edited and approved the final manuscript.
Competing interests
The authors declare no conflict of interest.
Funding
This material is based upon work supported by the National Science Foundation under grant no. NSF DEB 1754255 and by the Macroecology of Infectious Disease Research Coordination Network (grant no. NSF DEB 1316223).
References
- 1.Telfer S, Bown K. 2012. The effects of invasion on parasite dynamics and communities. Funct. Ecol. 26, 1288-1299. ( 10.1111/j.1365-2435.2012.02049.x) [DOI] [Google Scholar]
- 2.Altizer S, Ostfeld RS, Johnson PTJ, Kutz S, Harvell CD. 2013. Climate change and infectious diseases: from evidence to a predictive framework. Science 341, 514-519. ( 10.1126/science.1239401) [DOI] [PubMed] [Google Scholar]
- 3.Carlson CJ, et al. 2017. Parasite biodiversity faces extinction and redistribution in a changing climate. Sci. Adv. 3, e1602422. ( 10.1126/sciadv.1602422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson CJ, Albery GF, Merow C, Trisos CH, Zipfel CM, Eskew EA, Olival KJ, Ross N, Bansal S. 2020. Climate change will drive novel cross-species viral transmission. bioRxiv. ( 10.1101/2020.01.24.918755) [DOI] [PubMed]
- 5.Karesh WB, Cook RA, Bennett EL, Newcomb J. 2005. Wildlife trade and global disease emergence. Emerg. Infect. Dis. 11, 1000-1002. ( 10.3201/eid1107.050194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham AA, Daszak P, Wood JLN. 2017. One health, emerging infectious diseases and wildlife: two decades of progress? Phil. Trans. R. Soc. B 372, 20160167. ( 10.1098/rstb.2016.0167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark NJ, Clegg SM, Sam K, Goulding W, Koane B, Wells K. 2018. Climate, host phylogeny and the connectivity of host communities govern regional parasite assembly. Divers. Distrib. 24, 13-23. ( 10.1111/ddi.12661) [DOI] [Google Scholar]
- 8.Cooper N, Griffin R, Franz M, Omotayo M, Nunn CL. 2012. Phylogenetic host specificity and understanding parasite sharing in primates. Ecol. Lett. 15, 1370-1377. ( 10.1111/j.1461-0248.2012.01858.x) [DOI] [PubMed] [Google Scholar]
- 9.Dallas TA, Han BA, Nunn CL, Park AW, Stephens PR, Drake JM. 2019. Host traits associated with species roles in parasite sharing networks. Oikos 128, 23-32. ( 10.1111/oik.05602) [DOI] [Google Scholar]
- 10.Davies JT, Pedersen AB. 2008. Phylogeny and geography predict pathogen community similarity in wild primates and humans. Proc. R. Soc. B Biol. Sci. 275, 1695-1701. ( 10.1098/rspb.2008.0284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrell MJ, Berrang-Ford L, Davies TJ. 2013. The study of parasite sharing for surveillance of zoonotic diseases. Environ. Res. Lett. 8, 015036. ( 10.1088/1748-9326/8/1/015036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang S, Bininda-Emonds ORP, Stephens PR, Gittleman JL, Altizer S. 2014. Phylogenetically related and ecologically similar carnivores harbour similar parasite assemblages. J. Anim. Ecol. 83, 671-680. ( 10.1111/1365-2656.12160) [DOI] [PubMed] [Google Scholar]
- 13.Allison AB, et al. 2012. Role of multiple hosts in the cross-species transmission and emergence of a pandemic parvovirus. J. Virol. 86, 865-872. ( 10.1128/JVI.06187-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heeney JL, Dalgleish AG, Weiss RA. 2006. Origins of HIV and the evolution of resistance to AIDS. Science 313, 462-466. ( 10.1126/science.1123016) [DOI] [PubMed] [Google Scholar]
- 15.Song D, Kang B, Lee C, Jung K, Ha G, Kang D, Park S, Park B, Oh J. 2008. Transmission of avian influenza virus (H3N2) to dogs. Emerg. Infect. Dis. 14, 741-746. ( 10.3201/eid1405.071471) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han BA, Schmidt JP, Bowden SE, Drake JM. 2015. Rodent reservoirs of future zoonotic diseases. Proc. Natl Acad. Sci. USA 112, 7039-7044. ( 10.1073/pnas.1501598112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olival KJ, Hosseini PR, Zambrana-Torrelio C, Ross N, Bogich TL, Daszak P. 2017. Host and viral traits predict zoonotic spillover from mammals. Nature 546, 646-650. ( 10.1038/nature22975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park AW. 2019. Phylogenetic aggregation increases zoonotic potential of mammalian viruses. Biol. Lett. 15, 20190668. ( 10.1098/rsbl.2019.0668) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plowright RK, Parrish CR, McCallum H, Hudson PJ, Ko AI, Graham AL, Lloyd-Smith JO. 2017. Pathways to zoonotic spillover. Nat. Rev. Microbiol. 15, 502-510. ( 10.1038/nrmicro.2017.45) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Streicker DG, Turmelle AS, Vonhof MJ, Kuzmin IV, McCracken GF, Rupprecht CE. 2010. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science 329, 676-679. ( 10.1126/science.1188836) [DOI] [PubMed] [Google Scholar]
- 21.Pedersen AB, Davies TJ. 2009. Cross-species pathogen transmission and disease emergence in primates. EcoHealth 6, 496-508. ( 10.1007/s10393-010-0284-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poulin R, Mouillot D. 2003. Host introductions and the geography of parasite taxonomic diversity. J. Biogeogr. 30, 837-845. ( 10.1046/j.1365-2699.2003.00868.x) [DOI] [Google Scholar]
- 23.Stephens PR, et al. 2017. Global mammal parasite database version 2.0. Ecology 98, 1476. ( 10.1002/ecy.1799) [DOI] [PubMed] [Google Scholar]
- 24.Kelly DW, Paterson RA, Townsend CR, Poulin R, Tompkins DM. 2009. Parasite spillback: a neglected concept in invasion ecology? Ecology 90, 2047-2056. ( 10.1890/08-1085.1) [DOI] [PubMed] [Google Scholar]
- 25.Torchin ME, Lafferty KD, Kuris AM. 2001. Release from parasites as natural enemies: increased performance of a globally introduced marine crab. Biol. Invasions 3, 333-345. ( 10.1023/A:1015855019360) [DOI] [Google Scholar]
- 26.Torchin ME, Lafferty KD, Dobson AP, McKenzie VJ, Kuris AM. 2003. Introduced species and their missing parasites. Nature 421, 628-630. ( 10.1038/nature01346) [DOI] [PubMed] [Google Scholar]
- 27.Wu J, Huang J, Han X, Gao X, He F, Jiang M, Jiang Z, Primack RB, Shen Z. 2004. The Three Gorges Dam: an ecological perspective. Front. Ecol. Environ. 2, 241-248. ( 10.1890/1540-9295(2004)002[0241:TTGDAE]2.0.CO;2) [DOI] [Google Scholar]
- 28.Lloyd-Smith JO. 2013. Vacated niches, competitive release and the community ecology of pathogen eradication. Phil. Trans. R. Soc. B 368, 20120150. ( 10.1098/rstb.2012.0150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woolhouse MEJ, Taylor LH, Haydon DT. 2001. Population biology of multihost pathogens. Science 292, 1109-1112. ( 10.1126/science.1059026) [DOI] [PubMed] [Google Scholar]
- 30.Borremans B, Faust C, Manlove KR, Sokolow SH, Lloyd-Smith JO. 2019. Cross-species pathogen spillover across ecosystem boundaries: mechanisms and theory. Phil. Trans. R. Soc. B 374, 20180344. ( 10.1098/rstb.2018.0344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park AW, et al. 2018. Characterizing the phylogenetic specialism–generalism spectrum of mammal parasites. Proc. R. Soc. B 285, 20172613. ( 10.1098/rspb.2017.2613) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Byers JE, Schmidt JP, Pappalardo P, Haas SE, Stephens PR. 2019. What factors explain the geographical range of mammalian parasites? Proc. R. Soc. B Biol. Sci. 286, 20190673. ( 10.1098/rspb.2019.0673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobson A. 2004. Population dynamics of pathogens with multiple host species. Am. Nat. 164, S64-S78. ( 10.1086/424681) [DOI] [PubMed] [Google Scholar]
- 34.Farrell MJ, Stephens PR, Berrang-Ford L, Gittleman JL, Davies TJ. 2015. The path to host extinction can lead to loss of generalist parasites. J. Anim. Ecol. 84, 978-984. ( 10.1111/1365-2656.12342) [DOI] [PubMed] [Google Scholar]
- 35.Poullain V, Gandon S, Brockhurst MA, Buckling A, Hochberg ME. 2008. The evolution of specificity in evolving and coevolving antagonistic interactions between a bacteria and its phage. Evolution 62, 1-11. ( 10.1111/j.1558-5646.2007.00260.x) [DOI] [PubMed] [Google Scholar]
- 36.Truyen U, Evermann JF, Vieler E, Parrish CR. 1996. Evolution of canine parvovirus involved loss and gain of feline host range. Virology 215, 186-189. ( 10.1006/viro.1996.0021) [DOI] [PubMed] [Google Scholar]
- 37.Farrell MJ, Davies TJ. 2019. Disease mortality in domesticated animals is predicted by host evolutionary relationships. Proc. Natl Acad. Sci. USA 116, 7911-7915. ( 10.1073/pnas.1817323116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guth S, Visher E, Boots M, Brook CE. 2019. Host phylogenetic distance drives trends in virus virulence and transmissibility across the animal–human interface. Phil. Trans. R. Soc. B 374, 20190296. ( 10.1098/rstb.2019.0296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longdon B, Hadfield JD, Day JP, Smith SCL, McGonigle JE, Cogni R, Cao C, Jiggins FM. 2015. The causes and consequences of changes in virulence following pathogen host shifts. PLoS Pathog. 11, e1004728. ( 10.1371/journal.ppat.1004728) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo Iacono G, et al. 2016. A unified framework for the infection dynamics of zoonotic spillover and spread. PLoS Negl. Trop. Dis. 10, e0004957. ( 10.1371/journal.pntd.0004957) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joseph MB, Mihaljevic JR, Orlofske SA, Paull SH. 2013. Does life history mediate changing disease risk when communities disassemble? Ecol. Lett. 16, 1405-1412. ( 10.1111/ele.12180) [DOI] [PubMed] [Google Scholar]
- 42.Mihaljevic JR, Joseph MB, Orlofske SA, Paull SH. 2014. The scaling of host density with richness affects the direction, shape, and detectability of diversity–disease relationships. PLoS ONE 9, e97812. ( 10.1371/journal.pone.0097812) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vinson JE, Park AW. 2019. Vector-borne parasite invasion in communities across space and time. Proc. R. Soc. B 286, 20192614. ( 10.1098/rspb.2019.2614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clark NJ, Clegg SM. 2017. Integrating phylogenetic and ecological distances reveals new insights into parasite host specificity. Mol. Ecol. 26, 3074-3086. ( 10.1111/mec.14101) [DOI] [PubMed] [Google Scholar]
- 45.Poulin R, Leung TLF. 2011. Body size, trophic level, and the use of fish as transmission routes by parasites. Oecologia 166, 731-738. ( 10.1007/s00442-011-1906-3) [DOI] [PubMed] [Google Scholar]
- 46.Stephens PR, Altizer S, Ezenwa VO, Gittleman JL, Moan E, Han B, Huang S, Pappalardo P. 2019. Parasite sharing in wild ungulates and their predators: effects of phylogeny, range overlap, and trophic links. J. Anim. Ecol. 88, 1017-1028. ( 10.1111/1365-2656.12987) [DOI] [PubMed] [Google Scholar]
- 47.Parker IM, Saunders M, Bontrager M, Weitz AP, Hendricks R, Magarey R, Suiter K, Gilbert GS. 2015. Phylogenetic structure and host abundance drive disease pressure in communities. Nature 520, 542-544. ( 10.1038/nature14372) [DOI] [PubMed] [Google Scholar]
- 48.Jones KE, et al. 2009. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648. ( 10.1890/08-1494.1) [DOI] [Google Scholar]
- 49.R Development Core Team. 2008. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. See http://www.r-project.org. [Google Scholar]
- 50.IUCN. 2016. The IUCN Red List of threatened species, version 2016-1. See https://www.iucnredlist.org/.
- 51.Olson DM, et al. 2001. Terrestrial ecoregions of the world: a new map of life on Earth: a new global map of terrestrial ecoregions provides an innovative tool for conserving biodiversity. BioScience 51, 933-938. ( 10.1641/0006-3568(2001)051[0933:TEOTWA]2.0.CO;2) [DOI] [Google Scholar]
- 52.Pappalardo P, Morales-Castilla I, Park AW, Huang S, Schmidt JP, Stephens PR. 2020. Comparing methods for mapping global parasite diversity. Glob. Ecol. Biogeogr. 29, 182-193. ( 10.1111/geb.13008) [DOI] [Google Scholar]
- 53.Elith J, Leathwick JR. 2009. Species distribution models: ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 40, 677-697. ( 10.1146/annurev.ecolsys.110308.120159) [DOI] [Google Scholar]
- 54.Holt BG, et al. 2013. An update of Wallace's zoogeographic regions of the world. Science 339, 74-78. ( 10.1126/science.1228282) [DOI] [PubMed] [Google Scholar]
- 55.Wisz MS, Hijmans RJ, Li J, Peterson AT, Graham CH, Guisan A. 2008. Effects of sample size on the performance of species distribution models. Divers. Distrib. 14, 763-773. ( 10.1111/j.1472-4642.2008.00482.x) [DOI] [Google Scholar]
- 56.Elith J, Leathwick JR, Hastie T. 2008. A working guide to boosted regression trees. J. Anim. Ecol. 77, 802-813. ( 10.1111/j.1365-2656.2008.01390.x) [DOI] [PubMed] [Google Scholar]
- 57.Greenwell B, Boehmke B, Cunningham J, GBM developers. 2019. gbm: generalized boosted regression models. See https://CRAN.R-project.org/package=gbm.
- 58.Bahn V, McGill BJ. 2013. Testing the predictive performance of distribution models. Oikos 122, 321-331. ( 10.1111/j.1600-0706.2012.00299.x) [DOI] [Google Scholar]
- 59.Friedman JH, Popescu BE. 2008. Predictive learning via rule ensembles. Ann. Appl. Stat. 2, 916-954. ( 10.1214/07-AOAS148) [DOI] [Google Scholar]
- 60.Fountain-Jones NM, Machado G, Carver S, Packer C, Recamonde-Mendoza M, Craft ME. 2019. How to make more from exposure data? An integrated machine learning pipeline to predict pathogen exposure. J. Anim. Ecol. 88, 1447-1461. ( 10.1111/1365-2656.13076) [DOI] [PubMed] [Google Scholar]
- 61.Albery GF, Eskew EA, Ross N, Olival KJ. 2020. Predicting the global mammalian viral sharing network using phylogeography. Nat. Commun. 11, 1-9. ( 10.1038/s41467-020-16153-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Desprez-Loustau M-L, Robin C, Buée M, Courtecuisse R, Garbaye J, Suffert F, Sache I, Rizzo DM. 2007. The fungal dimension of biological invasions. Trends Ecol. Evol. 22, 472-480. ( 10.1016/j.tree.2007.04.005) [DOI] [PubMed] [Google Scholar]
- 63.Morton EM, Rafferty NE. 2017. Plant–pollinator interactions under climate change: the use of spatial and temporal transplants. Appl. Plant Sci. 5, 1600133. ( 10.3732/apps.1600133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harvey JA. 2015. Conserving host–parasitoid interactions in a warming world. Curr. Opin. Insect Sci. 12, 79-85. ( 10.1016/j.cois.2015.09.001) [DOI] [Google Scholar]
- 65.Schatz AM, Park AW. 2021. Data and code for ‘Host and parasite traits predict cross-species parasite acquisition by introduced mammals’. figshare. Collection ( 10.6084/m9.figshare.c.5100305) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Schatz AM, Park AW. 2021. Data and code for ‘Host and parasite traits predict cross-species parasite acquisition by introduced mammals’. figshare. Collection ( 10.6084/m9.figshare.c.5100305) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data and reproducible code are available on figshare [65].