Abstract
The expression levels of microRNA (miR)-221-3p and miR-222-3p in thyroid cancer have been found to be upregulated compared with those in normal tissues. The present study aimed to determine the effects and potential underlying mechanisms of miR-221-3p and miR-222-3p on the regulation of radioactive iodine (131I) uptake and radiosensitivity of thyroid cancer cells. The potential regulatory target genes of miR-221-3p and miR-222-3p were predicted by bioinformatics analysis, and reverse transcription-quantitative polymerase chain reaction was used to verify miR-221-3p, miR-222-3p and target gene expression levels in thyroid cancer tissues and cell lines. Overexpression of miR-221-3p or miR-222-3p in cell models was performed using lentivirus infection. Knockdown of miR-221-3p and miR-222-3p in cells was achieved using oligonucleotide inhibitor transfection. Western blotting was used to analyze the expression levels of target proteins. In addition, the effects of miR-221-3p and miR-222-3p on the radiosensitivity of thyroid cancer cells were verified using a colony formation assay. The results of the present study revealed that the expression levels of miR-221-3p and miR-222-3p were significantly upregulated, while the expression levels of suppressor of cytokine signaling 3 (SOCS3) were downregulated in thyroid cancer tissues. Furthermore, miR-221-3p and miR-222-3p overexpression downregulated the expression levels of SOCS3, E-cadherin and solute carrier family 5 member 5 (NIS), and upregulated the expression levels of phosphorylated STAT3 and vimentin. Following the overexpression of miR-221-3p or miR-222-3p in the FTC133 and TPC1 cell lines, their radiosensitivity was suppressed. In conclusion, the findings of the present study suggested that miR-221-3p and miR-222-3p may downregulate the expression levels of NIS and promote radioresistance. The potential mechanism was hypothesized to be associated with the miR-221-3p and miR-222-3p targeting of the SOCS3 gene, which may subsequently activate the STAT3 signaling pathway.
Keywords: differentiated thyroid cancer, microRNA-221, microRNA-222, suppressor of cytokine signaling 3, solute carrier family 5 member 5, radiosensitivity
Introduction
The incidence of thyroid cancer has significantly increased in recent years (1). Currently, ≥90% of patients with thyroid cancer are diagnosed with differentiated thyroid cancer (DTC), which mainly includes papillary thyroid cancer (PTC) and follicular thyroid cancer (FTC) (1). Radioactive iodine (131I) can be taken up by DTC cells and emit β-rays, which exert a radiotherapeutic effect on DTC cells. Patients who are resistant to 131I therapy, due to the inability to take up 131I or the occurrence of radioresistance in distant lesions, have a poor prognosis and shorter survival (2-4). The uptake of 131I by metastatic DTC is facilitated by a series of iodine metabolism genes, among which the solute carrier family 5 member 5 (NIS) protein has been reported to serve a key role in 131I uptake and treatment (3). However, the current understanding of radioresistance following 131I treatment in DTC remains limited.
MicroRNAs (miRNAs/miRs) are a class of mononucleotide, small non-coding RNAs that combine with the 3'-untranslated region (3'-UTR) of target genes to suppress target gene expression and have a wide range of biological functions (5). Notably, miR-221 and miR-222 share the same promoter, have a highly homologous sequence and share the same seed site. The expression levels of miR-221 and miR-222 in bladder cancer (6), breast cancer (7) and other types of tumor tissues or cell lines were found to be significantly upregulated compared with those in normal tissues (8). In addition, miR-221 and miR-222 were demonstrated to play important roles in regulating tumor cell functions, such as invasion, metastasis, epithelial-to-mesenchymal transition (EMT), proliferation and resistance to treatment, by targeting target genes, such as suppressor of cytokine signaling 3 (SOCS3), transcriptional repressor GATA binding 1 and p27 (8,9). It has also been demonstrated that the expression levels of miR-221 and miR-222 were significantly upregulated in DTC tissues compared with those in normal thyroid tissues (10-12). Furthermore, the expression levels of miR-221 and miR-222 in the blood of patients with thyroid cancer were found to be associated with the progression of DTC (13,14). A recent large-scale meta-analysis revealed that the expression levels of miR-221 and miR-222 could predict poor overall survival in patients with cancer. In particular, miR-222 exhibited a significant predictive value for secondary outcomes, including disease-free and recurrence-free survival (15).
Signal transducer and activator of transcription 3 (STAT3) regulates a variety of cell functions, including EMT, through its downstream gene signaling pathways. Treatment of thyroid cancer cells with the STAT3 inhibitor, cucurbitacin I, was shown to upregulate the expression levels of thyroid-specific genes and significantly enhance 131I uptake; furthermore, cucurbitacin I treatment enhanced the sensitivity of thyroid cancer cells to radiation and chemotherapy (16). SOCS3 is a cytokine-inducible negative regulator of cytokine signaling, which can bind to Janus kinase (JAK)2 and inhibit the activity of the JAK/STAT signaling pathway (9). The genetic silencing of the miR-221/miR-222 cluster was shown to attenuate angiogenesis in glioblastoma by inactivating the JAK/STAT signaling pathway via the upregulation of SOCS3(9). This finding suggested that miR-221 and miR-222 may activate the STAT3 signaling pathway by downregulating the expression levels of their target gene, SOCS3. miR-221 and miR-222 were also found to be expressed at high levels in DTC, which suggested that miR-221 and miR-222 may be involved in the regulation of thyroid carcinoma resistance to 131I treatment. However, the role and corresponding underlying mechanism of miR-221 and miR-222 during the 131I treatment of DTC remain unclear. Therefore, the present study aimed to investigate the role and potential mechanism underlying the upregulated expression levels of miR-221-3p and miR-222-3p in the resistance of thyroid cancer to 131I therapy.
Materials and methods
Cell lines and culture
Nthy-ori-3, K1 and BCPAP cell lines were purchased from the Cell Resource Center, Shanghai Institutes for Biological Sciences of the Chinese Academy of Sciences. FTC133 and TPC1 cells were purchased from Nanjing Branch Bai Biotechnology Co., Ltd. The human thyroid cancer cell lines FTC133, TPC1 and K1 were cultured at 37˚C in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher, Shanghai, China), in a humidified incubator with 5% CO2 at 37˚C. The Nthy-ori 3-1 human thyroid follicular epithelial cells and the BCPAP human thyroid cancer cell line were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 1% non-essential amino acids (Gibco; Thermo Fisher Scientific, Inc.), 1% sodium pyruvate (100 mM) solution (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 g/ml streptomycin. The BCPAP and K1 cell lines were authenticated by comparing DNA short tandem repeats data with the ATCC, DSMZ, JCRB RIKEN and EXPASY databases, and no multiple alleles were found in these cell lines. The BCPAP cell line was authenticated by Applied Biological Materials, Inc. The K1 cell line was authenticated by Jianlian Gene Technology (Beijing) Co., Ltd. In experiments analyzing the expression levels of NIS, FTC133, TPC1 and BCPAP cells were supplemented with 10 mU/ml thyroid-stimulating hormone, bovine pituitary (cat. no. 609385-5; Merck KGaA).
Cell transfection
BCPAP cells were transfected with 100 nM scrambled oligonucleotide [negative control (NC)] or 50 nM miR-221-3p inhibitor and 50 nM miR-222-3p inhibitor (Invitrogen; Thermo Fisher Scientific, Inc.) to construct transient knockdown miR-221-3p and miR-222-3p cell lines. The sequences were as follows: miR-221-3p inhibitor, 5'-GAA ACCCAGCAGACAAUGUAGCU-3' (lot no. AS0299J7); miR-222-3p inhibitor, 5'-ACCCAGUAGCCAGAUGUA GCU-3' (lot no. AS0299J8); and NC, 5'-CAGUACUUUUGU GUAGUACAA-3', lot no. AS026V49). Cell transfection was performed using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) in 6-well plates. A total of 8 µl Lipofectamine® 3000 and 100 nM inhibitor or NC were mixed in 2 ml RPMI-1640 medium and placed at room temperature for 20 min. The cells were cultured at 37˚C with 5% CO2 in the mixture for 6 h. Then the cells were cultured in RPMI-1640 medium supplemented with 10% FBS. Total RNA was extracted at 24 h or 48 h and protein was extracted at 48 h after transfection with inhibitors or NC.
Patient studies
A total of 15 tissue samples from patients with PTC were obtained retrospectively under strict anonymity from the tissue bank of Zhejiang Cancer Hospital (Hangzhou, China). The patients, including 8 women and 7 men, with a median age of 43 years (range, 27-72 years), were treated for PTC in our hospital from January 2018 to December 2018. Postoperative samples with other pathological types, patients with a history of other tumors or aged <18 years were excluded. All samples were fresh-frozen in liquid nitrogen following surgery and stored at -80˚C. Frozen tissue samples were homogenized using TissueRuptor II (Qiagen, Inc.) prior to RNA extraction. The experimental protocols were approved by the Ethics Committee of Zhejiang Cancer Hospital (approval no. IRB-2020-337). Informed consent forms were signed by all the patients prior to surgery.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from tissues and cell lines using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Total RNA concentration was determined using a NanoDrop spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.). For miRNA expression, total RNA (1 µg) was reverse-transcribed into cDNA at 42˚C for 60 min and 70˚C for 10 min and qPCR was subsequently performed using a Bulge-Loop miRNA RT-qPCR Starter kit (Guangzhou RiboBio Co., Ltd.) on an ABI StepOnePlus quantitative PCR instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.) with 2 µl cDNA as the template. Bulge-loop miRNA RT-qPCR Primer sets (one RT primer and a pair of qPCR primers for each gene) specific for miR-221-3p, miR-222-3p and U6 were designed by Guangzhou RiboBio Co., Ltd. Due to the patented technology, the company refused to disclose the sequence of primers. The following thermocycling conditions were used for the qPCR: Initial denaturation at 95˚C for 10 min; followed by 40 amplification cycles at 95˚C for 10 sec, 60˚C for 20 sec and 70˚C for 20 sec, at a ramp-rate of 1.6˚C/sec.
For mRNA expression, total RNA was reverse-transcribed into cDNA using a M-MLV Reverse Transcriptase for qPCR (Promega Corporation). qPCR was subsequently performed using PowerUp™ SYBR™-Green Master Mix (Thermo Fisher Scientific, Inc.) on an ABI StepOnePlus quantitative PCR instrument. The primer sequences for SOCS3 and β-actin (Shanghai GenePharma Co., Ltd.) were as follows: SOCS3 forward, 5'-TCGCCACCTACTGAACCCT-3' and reverse, 5'-GGTCCAGGAACTCCCGAAT-3'; and β-actin forward, 5'-AGCGGGAAATCGTGCGTG-3' and reverse, 5'-GGTCCA GGAACTCCCGAAT-3'. The following thermocycling conditions were used for the qPCR: Initial denaturation at 95˚C for 5 min, followed by 40 amplification cycles at 95˚C for 5 sec and 60˚C for 31 sec, with a ramp-rate of 1.6˚C/sec. The mRNA and miRNA expression levels were calculated using the 2-ΔΔCq method (17). miRNA expression levels were normalized to U6 snRNA and mRNA expression levels were normalized to GAPDH.
Lentivirus infection
A total of 1x106 FTC133 or TPC1 cells were seeded into 6-well plates using DMEM medium supplemented with 10% FBS overnight. FTC133 or TPC1 cells were then infected with lentivirus vectors (MOI=1.0). Green fluorescent protein (GFP)-miR-221-3p (HmiR0369), GFP-miR-222-3p (HmiR0370) and GFP lentiviral vectors (pEZX-MR03, CmiR0001) (all from GeneCopoeia, Inc.). The cells were re-infected with the lentivirus vectors (MOI=1.0) 72 h after the first infection. The pEZX-MR03-GFP vector was used as the NC. GFP expression was visualized under an inverted fluorescence microscope (Olympus Corporation) at a magnification of x200. The subsequent experiments were performed 72 h after the last infection. During the subsequent experiments, cell lines were cultured and passaged for no more than 3 generations.
Western blotting
Total protein was extracted from cells using RIPA lysis buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate and 0.1% SDS] supplemented with a protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA). Total protein was quantified using the bicinchoninic acid (BCA) protein assay kit (Beyotime Institute of Biotechnology) and 30 µg protein/lane was separated via 10% SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were subsequently blocked with 5% non-fat dry milk for 1 h at room temperature and incubated with the following primary antibodies at 4˚C overnight: Anti-SOCS3 (1:500; rabbit; cat. no. ab16030; Abcam), anti-phosphorylated (p)-STAT3 (1:1,000; rabbit; cat. no. ab76315; Abcam), anti-vimentin (1:1,000; rabbit; cat. no. ab8069; Abcam), anti-E-cadherin (1:500; rabbit; cat. no. ab133597; Abcam), anti-NIS (1:500; rabbit; cat. no. ab83816; Abcam) and anti-GAPDH (1:2,000; mouse; cat. no. sc365062; Santa Cruz Biotechnology, Inc.). Following primary antibody incubation, the membranes were washed and incubated with a goat anti-mouse (1:2,000; cat. no. sc2005; Santa Cruz Biotechnology, Inc.) or anti-rabbit (1:2,000; cat. no. sc2004; Santa Cruz Biotechnology, Inc.) antibody for 1 h at room temperature. Protein bands were visualized using ECL (cat. no. sc-2048; Santa Cruz Biotechnology, Inc.). Densitometric analysis was performed using ImageJ software, version 7.0 (National Institutes of Health). The relative protein expression levels were expressed as a ratio of the gray value of the target protein band: GAPDH protein band.
Bioinformatics analysis
miRNA sequences and annotation were acquired form the miRBase database (18). miRNA target prediction software, TargetScanHuman v.7.1 was used to identify miR-221-3p and miR-222-3p (MIMAT0000278 and MIMAT0000279) putative targets (19). Microarray data were extracted from the Oncomine database (20). Analysis of differential gene expression was performed using the R statistical computing package (http://www.r-project.org). All datasets were logarithmically transformed and centered on the median per array, and the standard deviations were normalized to one for each array (20).
Colony formation assay
Cells were seeded into six-well plates at different densities (400, 1,000, 2,000, 3,000 or 4,000 cells/well) and incubated for 24 h. Cells were subsequently treated with different X-ray doses (Siemens AG; 200 cGy/min). Following 12 days of incubation, the cells were fixed with 10% formaldehyde at room temperature for 10 min, stained with 1.0% crystal violet solution at room temperature for 10 min and then the number of colonies (>50 cells) was counted under an inverted microscope (magnification, x200). Surviving fractions were estimated using the following equation: (Colony number)/(cell number seeded x plating efficiency) x 100% (21). Each experiment was independently repeated in triplicate.
Statistical analysis
Statistical analysis was performed using SPSS software, version 22.0 (IBM Corp.). The significance of the differences between groups were determined by paired or unpaired Student's t-test (for comparison between two groups) or one way ANOVA followed by Bonferroni's post hoc test. Histograms were used to check for the distribution of the data. The data in this study exhibited normal distribution. The experiments were performed in triplicate. Data are presented as the mean ± SD. P<0.05 was considered to indicate a statistically significant difference.
Results
SOCS3 is a potential target gene of miR-221-3p and miR-222-3p in thyroid cancer
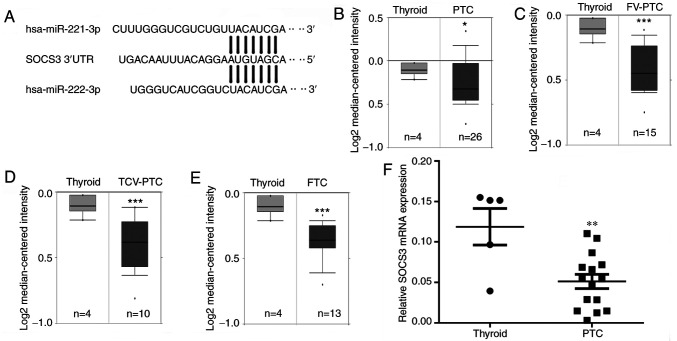
miR-221-3p and miR-222-3p target genes were predicted using TargetScan (Fig. 1A). Microarray data was extracted from the Oncomine database. Genes expressed at low levels in thyroid cancer were screened as potential target genes. The mRNA expression levels of SOCS3 were downregulated in different subtypes of DTC, including PTC, follicular variant PTC (FV-PTC), tall-cell variant PTC (TCV-PTC) and FTC (*P<0.05, ***P<0.001; Fig. 1B-E). The expression levels of the STAT3 inhibitor, SOCS3, were of particular interest, as STAT3 not only promotes EMT, but also participates in the regulation of iodine uptake (16). Thus, RT-qPCR was used to analyze the differences in SOCS3 gene expression between 15 thyroid cancer and 5 normal thyroid tissues; the results further confirmed that the expression levels of SOCS3 were significantly downregulated in thyroid cancer specimens (**P<0.01; Fig. 1F).
Figure 1.
SOCS3 is a potential target gene of miR-221-3p and miR-222-3p. (A) Complementary sequence of miR-221-3p and miR-222-3p to the seed region in the SOCS3 3'- UTR. mRNA expression levels of SOCS3 in (B) PTC, (C) FV-PTC, (D) TCV-PTC and (E) FTC tissues obtained from the Oncomine database. (F) Reverse transcription-quantitative polymerase chain reaction was used to analyze the differences in SOCS3 mRNA expression levels between thyroid cancer and normal thyroid tissues. The results are presented as the mean ± SD of three independent experiments. The relative mRNA levels were normalized to β-actin. *P<0.05, **P<0.01, ***P<0.001 vs. normal thyroid tissue. SOCS3, suppressor of cytokine signaling 3; miR, microRNA; PTC, papillary thyroid cancer; FV, follicular variant; TCV, tall-cell variant; FTC, follicular thyroid cancer; UTR, untranslated region.
Expression levels of miR-221-3p and miR-222-3p in thyroid cancer cell lines
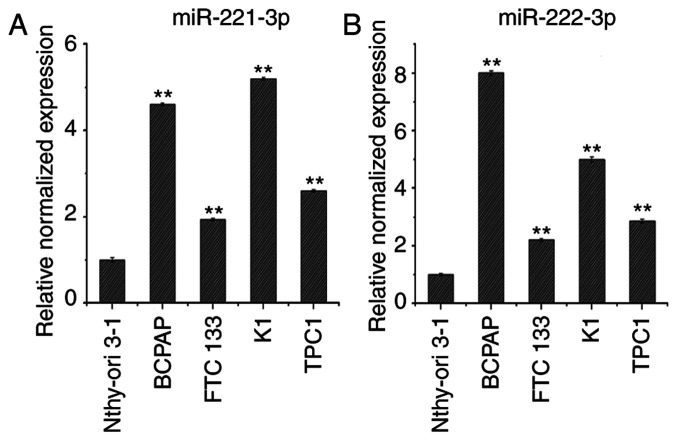
RT-qPCR analysis was used to determine miR-221-3p and miR-222-3p expression levels in the thyroid cancer cell lines BCPAP, FTC133, K1 and TPC1, compared with Nthy-ori 3-1 cells. As shown in Fig. 2A and B, the expression levels of miR-221-3p and miR-222-3p were upregulated in BCPAP, FTC133, K1 and TPC1 cells compared with Nthy-ori3-1 cells. Of note, FTC133 and TPC1 cells exhibited lower expression levels of miR-221-3p and miR-222-3p compared with K1 and BCPAP cells; therefore, the BCPAP cell line was selected to establish knockdown cell models and FTC133 and TPC1 cell lines were selected to establish overexpression cell models.
Figure 2.
Expression levels of miR-221-3p and miR-222-3p in thyroid cancer cell lines. Reverse transcription-quantitative polymerase chain reaction analysis of (A) miR-221-3p and (B) miR-222-3p expression levels in thyroid cancer cell lines and normal thyroid cells. n=3; **P<0.01 vs. Nthy-ori 3-1. miR, microRNA; DTC, differentiated thyroid cancer.
SOCS3 expression levels in thyroid cancer cells are downregulated by miR-221-3p and miR-222-3p
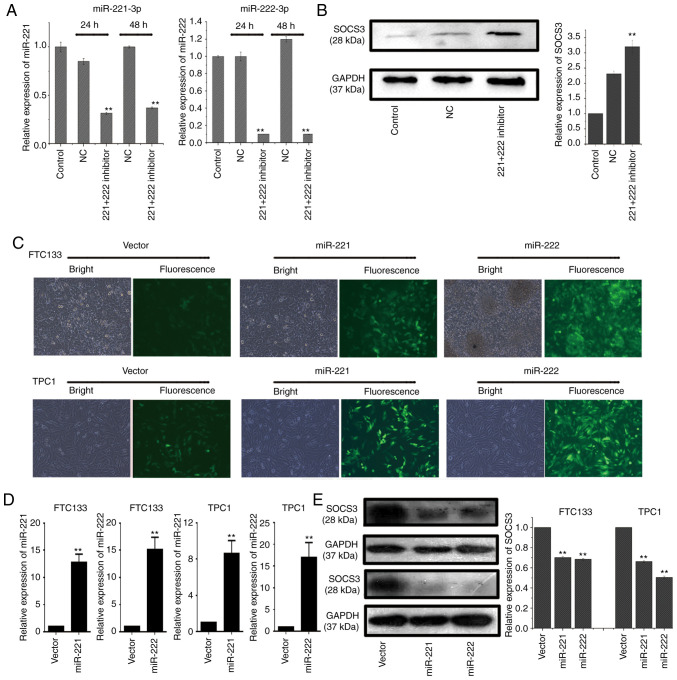
BCPAP cells were transfected with a miR-221-3p inhibitor, miR-222-3p inhibitor or NC, and total RNA was extracted at 24 and 48 h post-transfection. RT-qPCR analysis revealed that the co-transfection with the miR-221-3p and miR-222-3p inhibitors could effectively downregulate the expression levels of miR-221-3p and miR-222-3p compared with the NC group at 24 and 48 h post-transfection in BCPAP cells (Fig. 3A). In addition, compared with the NC group, co-transfection of BCPAP cells with miR-221-3p and miR-222-3p inhibitors upregulated the protein expression levels of SOCS3 (Fig. 3B).
Figure 3.
Downregulation of SOCS3 expression by miR-221-3p and miR-222-3p in thyroid cancer cell lines. (A) RT-qPCR was used to analyze endogenous miR-221-3p and miR-222-3p expression levels in BCPAP cells following co-transfection with miR-221-3p and miR-222-3p inhibitors for 24 or 48 h. Each reaction was performed in triplicate. **P<0.01 vs. NC. (B) BCPAP cells were co-transfected with a miR-221-3p and miR-222-3p inhibitor and western blotting was used to analyze the expression levels of SOCS3. Relative expression levels of SOCS3/GAPDH were analyzed by ImageJ software. Each band was measured in triplicate. **P<0.01 vs. control and NC. (C) GFP, GFP-miR-221-3p or GFP-miR-222-3p lentiviral vectors were infected into FTC133 and TPC1 cells and visualized using a fluorescence microscope. (magnification, x200). (D) RT-qPCR was used to analyze the endogenous expression levels of miR-221-3p or miR-222-3p in FTC133 and TPC1 cells following transfection with lentiviral vectors. Each reaction was performed in triplicate. GFP lentiviral vectors served as the NC. **P<0.01 vs. vector. (E) FTC133 (upper two bands) and TPC1 (lower two bands) cells were infected with GFP-miR-221-3p, GFP-miR-222-3p or GFP lentiviral vectors, and western blotting was used to analyze the expression levels of SOCS3. Grouping of FTC133 and TPC1 images from different parts of the same gel. Relative expression levels of SOCS3/GAPDH were analyzed using ImageJ software. Each band was measured in triplicate. **P<0.01 vs. vector. SOCS3, suppressor of cytokine signaling 3; miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; GFP, green fluorescent protein; NC, negative control; vector, GFP lentiviral vector.
FTC133 and TPC1 cells were subsequently infected with lentiviral vectors to construct cell models overexpressing miR-221-3p and miR-222-3p. The fluorescence of the lentiviral vector was observed under a fluorescence microscope at 72 h post-transfection, and the results demonstrated that the lentiviral vectors were successfully infected into the cells (Fig. 3C). RT-qPCR analysis also revealed that the infection with miR-221-3p and miR-222-3p lentiviral vectors markedly upregulated the expression levels of miR-221-3p and miR-222-3p at 72 h post-infection (Fig. 3D). The western blotting results also demonstrated that the protein expression levels of SOCS3 were significantly downregulated following overexpression of miR-221-3p or miR-222-3p (Fig. 3E).
miR-221-3p and miR-222-3p regulate the expression levels of p-STAT3, EMT-related markers and NIS in thyroid cancer cells
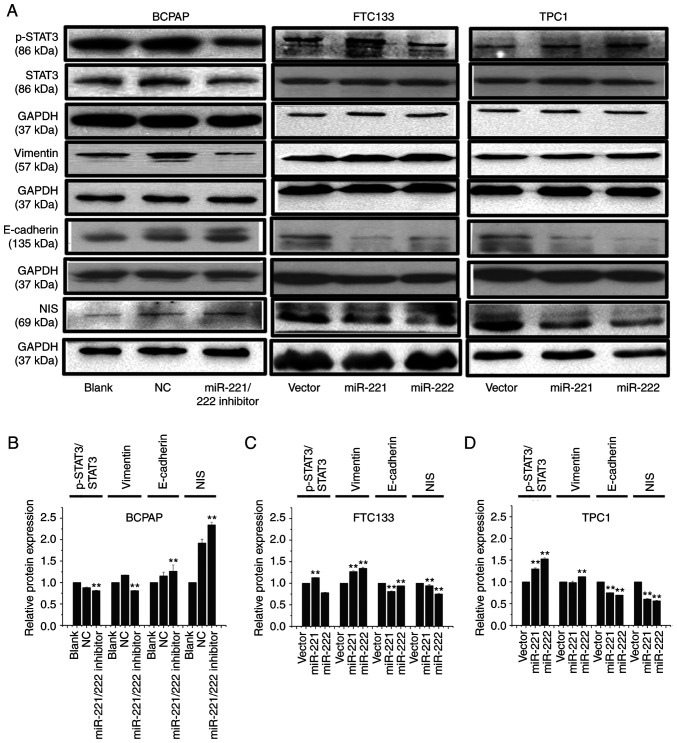
The western blotting results revealed that, following inhibition of miR-221-3p and miR-222-3p expression in BCPAP cells, the expression levels of p-STAT3 and vimentin were downregulated, while the expression levels of E-cadherin and NIS were upregulated (Fig. 4A and B). Conversely, following overexpression of miR-221-3p or miR-222-3p in FTC133 and TPC1 cells, the expression levels of p-STAT3 and vimentin were upregulated, while the expression levels of E-cadherin and NIS were significantly downregulated (Fig. 4A, C and D).
Figure 4.
miR-221-3p and miR-222-3p upregulate the protein expression levels of p-STAT3 and vimentin and downregulate the expression of NIS and E-cadherin. (A) BCPAP cells were transfected with miR-221-3p, miR-222-3p inhibitor and NC. FTC133 and TPC1 cells were infected with GFP-miR-221-3p, GFP-miR-222-3p or GFP lentiviral vectors. Western blotting was used to analyze the expression levels of p-STAT3, STAT3, vimentin, E-cadherin and NIS. Grouping of BCPAP images from different gels than those of FTC133 and TPC1. Grouping of FTC133 and TPC1 images from different parts of the same gel. Bands of p-STAT3 and STAT3 from different gels. (B-D) Relative expression levels of p-STAT3/STAT3, vimentin/GAPDH, E-cadherin/GAPDH and NIS/GAPDH were analyzed using ImageJ 7.0 software in each cell model. Each band was measured in triplicate. **P<0.01 vs. NC or vector. miR, microRNA; p-, phosphorylated; STAT, signal transducer and activator of transcription; NIS, solute carrier family 5 member 5; GFP, green fluorescent protein; NC, negative control; vector, GFP lentiviral vector.
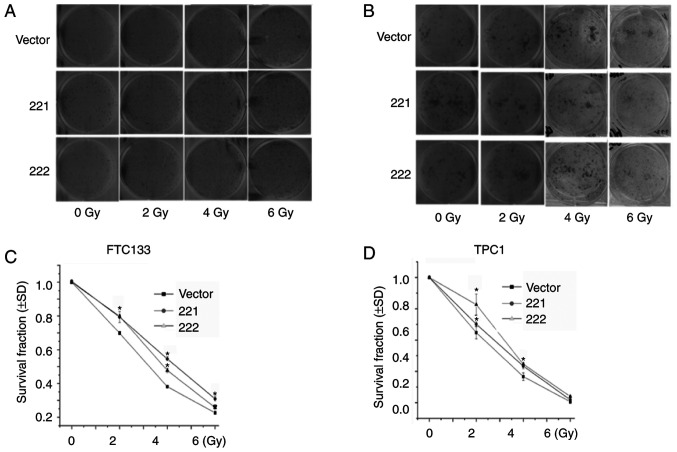
miR-221-3p and miR-222-3p reduce radiosensitivity in thyroid cancer cells
It was previously reported that EMT was associated with decreased cancer sensitivity to radiation therapy (22-25). Therefore, a colony formation assay was used to determine the role of miR-221-3p and miR-222-3p in radiosensitivity. FTC133 (Fig. 5A and C) and TPC1 (Fig. 5B and D) cells were infected with lentivirus vectors, treated with different doses of X-ray irradiation and cultured for 12 days. The results demonstrated that the overexpression of miR-221-3p or miR-222-3p significantly increased the surviving fraction of FTC133 (at 2, 4 and 6 Gy; Fig. 5C) and TPC1 (at 2 and 4 Gy; Fig. 5D) cells. These results indicated that overexpression of miR-221-3p or miR-222-3p may increase radioresistance in DTC cell lines.
Figure 5.
miR-221-3p and miR-222-3p regulate the radiosensitivity of thyroid cancer cells. FTC133 and TPC1 cells were infected with GFP-miR-221-3p, GFP-miR-222-3p or GFP lentiviral vectors. Radiation sensitivity was evaluated using colony formation assay. Representative images of the plates for the colony formation assay of (A) FTC133 and (B) TCP1 cells. The effect of miR-221-3p or miR-222-3p on radiation-treated (C) FTC133 and (D) TPC1 cells was assessed by survival curves. n=3; *P<0.05 vs. vector. miR, microRNA; GFP, green fluorescent protein; vector, GFP lentiviral vector.
Discussion
131I is an important method used for the treatment of distant metastatic DTC. However, ~50% of distant metastatic DTCs are resistant to 131I therapy, which is currently the main cause of DTC-related mortality (2,4,26). Reversing the 131I resistance of distant metastatic lesions to 131I therapy has been associated with the upregulated expression of NIS, which increases radiosensitivity and may represent a valuable research focal point for the clinical treatment of DTC (26). miRNAs are a class of short-stranded RNAs that play important roles in regulating the biological functions of tumor cells, including the response to cancer therapy. However, research into the role of miRNAs in the resistance of thyroid cancer to 131I remains limited.
The expression levels of miR-221 and miR-222 were previously found to be upregulated in DTC (27,28). The RT-qPCR results obtained in the present study also revealed that the expression levels of miR-221-3p and miR-222-3p were upregulated in thyroid cancer cell lines compared with those in normal thyroid cells. Few previous studies have investigated the mechanism of miR-221-3p and miR-222-3p in the resistance to 131I therapy. In the present study, through bioinformatics analysis prediction, SOCS3 was identified as a potential target gene of miR-221-3p and miR-222-3p. Analysis of thyroid cancer tissues from the Oncomine database revealed that the expression levels of SOCS3 were downregulated in thyroid cancer. Several previous studies have also confirmed that the miR-221/miR-222 cluster can bind to the 3'-UTR of SOCS3 and negatively regulate the translation of SOCS3 (9,29-31). The results of the present study demonstrated that the overexpression of miR-221-3p or miR-222-3p inhibited the expression of SOCS3 in thyroid cancer cells, and inhibiting the expression of miR-221-3p and miR-222-3p upregulated the expression levels of SOCS3. Whether miR-221-3p and miR-222-3p directly target 3'-UTR of SOCS3 and suppress the expression of SOCS3 was not been further verified in the present study. SOCS3 has been previously observed to negatively regulate the JAK/STAT signaling pathway (9,29,32). The JAK/STAT signaling pathway participates in the regulation of EMT in tumor cells and was found to inhibit 131I uptake in thyroid cancer (16,33). Furthermore, the upregulated expression of SOCS3 was shown to inhibit metastasis by downregulating p-STAT3 and vimentin expression and upregulating E-cadherin expression (32,34).
In the present study, the expression levels of miR-221-3p and miR-222-3p were found to be higher in the BCPAP cell line compared with those in the FTC and TPC1 cell lines. Therefore, BCPAP cells were transfected with miR-221-3p and miR-222-3p inhibitors or NC to establish miR-221-3p and miR-222-3p knockdown cell models. The results revealed that the simultaneous inhibition of the expression of miR-221-3p and miR-222-3p downregulated the expression levels of p-STAT3 and vimentin, and upregulated E-cadherin expression levels. In addition, the simultaneous inhibition of miR-221-3p and miR-222-3p restored the expression of NIS, which is a key gene required for the uptake of 131I by thyroid cancer cells (26). In addition, FTC133 and TPC1 cells were transfected with lentiviral vectors to establish miR-221-3p and miR-222-3p overexpression cell models. Following overexpression of miR-221-3p or miR-222-3p, the expression levels of p-STAT3 and vimentin were upregulated, while the expression levels of E-cadherin and NIS were significantly downregulated. These findings further suggested that the upregulated expression of miR-221-3p and miR-222-3p may be associated with the mechanisms underlying iodine metabolism dysfunction in thyroid cancer cells. In the miR-221-3p and miR-222-3p overexpression cell models, the radiosensitivity of the cells was verified using a colony formation assay. The results demonstrated that the overexpression of miR-221-3p or miR-222-3p in FTC133 and TPC1 cells could significantly reduce their radiosensitivity.
A limitation of the present study is that rescue assay or dual-luciferase reporter gene assay would be necessary to further verify the role of the miR-221/222-3p-SOCS3 axis. Furthermore, although overexpression of miR-221/222-3p decreased NIS expression, this result must be verified by iodine uptake test.
In conclusion, the results of the present study indicated that the overexpression of miR-221-3p and miR-222-3p in thyroid cancer cells may downregulate NIS expression levels and thereby reduce radiosensitivity, which indicates that the overexpression of miR-221-3p and miR-222-3p in DTC may simultaneously inhibit iodine metabolism and reduce radiosensitivity. The potential underlying mechanism was hypothesized to be associated with the miR-221-3p- and miR-222-3p-targeted inhibition of SOCS3, which may subsequently lead to the activation of the STAT3 signaling pathway in DTC cells.
Acknowledgements
Not applicable.
Funding Statement
Funding: The present study was supported by the National Natural Science Foundation of China (grant no. 81502318) and the Zhejiang Province Natural Science Foundation of China (grant no. LY20H180007).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HY conceived and designed the study, contributed to the experiments and drafted the manuscript. TY and LZ performed all the experiments. XY and JL performed the data analysis. LL contributed to the conception and designed the study. TY and HY have seen and can confirm the authenticity of all the raw data. All the authors have read and approved the final manuscript.
Ethics approval and consent to participate
The experimental protocols were approved by the Ethics Committee of Zhejiang Cancer Hospital (approval no. IRB-2020-337). All patients provided consent for the publication of their data.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. Erratum in: CA Cancer J Clin 70, 313, 2020. [DOI] [PubMed] [Google Scholar]
- 2.Song HJ, Qiu ZL, Shen CT, Wei WJ, Luo QY. Pulmonary metastases in differentiated thyroid cancer: Efficacy of radioiodine therapy and prognostic factors. Eur J Endocrinol. 2015;173:399–408. doi: 10.1530/EJE-15-0296. [DOI] [PubMed] [Google Scholar]
- 3.Agrawal N, Akbani R, Aksoy BA, Ally A, Arachchi H, Asa SL, Auman JT, Balasundaram M, Balu S, Baylin SB, et al. Cancer Genome Atlas Research Network: Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Nostrand D. Radioiodine refractory differentiated thyroid cancer: Time to update the classifications. Thyroid. 2018;28:1083–1093. doi: 10.1089/thy.2018.0048. [DOI] [PubMed] [Google Scholar]
- 5.Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20:5–20. doi: 10.1038/s41580-018-0059-1. Erratum in: Nat Rev Mol Cell Biol 19, 808, 2018. [DOI] [PubMed] [Google Scholar]
- 6.Tsikrika FD, Avgeris M, Levis PK, Tokas T, Stravodimos K, Scorilas A. miR-221/222 cluster expression improves clinical stratification of non-muscle invasive bladder cancer (TaT1) patients' risk for short-term relapse and progression. Genes Chromosomes Cancer. 2018;57:150–161. doi: 10.1002/gcc.22516. [DOI] [PubMed] [Google Scholar]
- 7.Chen WX, Hu Q, Qiu MT, Zhong SL, Xu JJ, Tang JH, Zhao JH. miR-221/222: Promising biomarkers for breast cancer. Tumour Biol. 2013;34:1361–1370. doi: 10.1007/s13277-013-0750-y. [DOI] [PubMed] [Google Scholar]
- 8.Amini S, Abak A, Sakhinia E, Abhari A. MicroRNA-221 and microRNA-222 in common human cancers: Expression, function, and triggering of tumor progression as a key modulator. Lab Med. 2019;50:333–347. doi: 10.1093/labmed/lmz002. [DOI] [PubMed] [Google Scholar]
- 9.Xu CH, Liu Y, Xiao LM, Chen LK, Zheng SY, Zeng EM, Li DH, Li YP. Silencing microRNA-221/222 cluster suppresses glioblastoma angiogenesis by suppressor of cytokine signaling-3-dependent JAK/STAT pathway. J Cell Physiol. 2019;234:22272–22284. doi: 10.1002/jcp.28794. [DOI] [PubMed] [Google Scholar]
- 10.Jikuzono T, Kawamoto M, Yoshitake H, Kikuchi K, Akasu H, Ishikawa H, Hirokawa M, Miyauchi A, Tsuchiya S, Shimizu K, et al. The miR-221/222 cluster, miR-10b and miR-92a are highly upregulated in metastatic minimally invasive follicular thyroid carcinoma. Int J Oncol. 2013;42:1858–1868. doi: 10.3892/ijo.2013.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Mao H, Lv Z. MicroRNA role in thyroid cancer pathogenesis. Front Biosci. 2013;18:734–739. doi: 10.2741/4135. [DOI] [PubMed] [Google Scholar]
- 12.Braun J, Hoang-Vu C, Dralle H, Hüttelmaier S. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene. 2010;29:4237–4244. doi: 10.1038/onc.2010.169. [DOI] [PubMed] [Google Scholar]
- 13.Yu S, Liu Y, Wang J, Guo Z, Zhang Q, Yu F, Zhang Y, Huang K, Li Y, Song E, et al. Circulating microRNA profiles as potential biomarkers for diagnosis of papillary thyroid carcinoma. J Clin Endocrinol Metab. 2012;97:2084–2092. doi: 10.1210/jc.2011-3059. [DOI] [PubMed] [Google Scholar]
- 14.Gómez-Pérez AM, Cornejo Pareja IM, García Alemán J, Coín Aragüez L, Sebastián Ochoa A, Alcaide Torres J, Molina Vega M, Clu Fernández C, Mancha Doblas I, Tinahones FJ. New molecular biomarkers in differentiated thyroid carcinoma: Impact of miR-146, miR-221 and miR-222 levels in the evolution of the disease. Clin Endocrinol (Oxf) 2019;91:187–194. doi: 10.1111/cen.13972. [DOI] [PubMed] [Google Scholar]
- 15.Ravegnini G, Cargnin S, Sammarini G, Zanotti F, Bermezo JL, Hrelia P, Terrazzino S, Angelini S. Prognostic role of miR-221 and miR-222 expression in cancer patients: A systematic review and meta-analysis. Cancers (Basel) 2019;11(970) doi: 10.3390/cancers11070970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tseng LM, Huang PI, Chen YR, Chen YC, Chou YC, Chen YW, Chang YL, Hsu HS, Lan YT, Chen KH, et al. Targeting signal transducer and activator of transcription 3 pathway by cucurbitacin I diminishes self-renewing and radiochemoresistant abilities in thyroid cancer-derived CD133+ cells. J Pharmacol Exp Ther. 2012;341:410–423. doi: 10.1124/jpet.111.188730. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019;47 (D1):D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. TargetScanHuman 5.2 Custom. http://www.targetscan.org/vert_50/seedmatch.html. Accessed April 14, 2021. [Google Scholar]
- 20.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 22.Wu C, Guo E, Ming J, Sun W, Nie X, Sun L, Peng S, Luo M, Liu D, Zhang L, et al. Radiation-induced DNMT3B promotes radioresistance in nasopharyngeal carcinoma through methylation of p53 and p21. Mol Ther Oncolytics. 2020;17:306–319. doi: 10.1016/j.omto.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie C, Wu Y, Fei Z, Fang Y, Xiao S, Su H. MicroRNA-1275 induces radiosensitization in oesophageal cancer by regulating epithelial-to-mesenchymal transition via Wnt/β-catenin pathway. J Cell Mol Med. 2020;24:747–759. doi: 10.1111/jcmm.14784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou S, Zhang M, Zhou C, Wang W, Yang H, Ye W. The role of epithelial-mesenchymal transition in regulating radioresistance. Crit Rev Oncol Hematol. 2020;150(102961) doi: 10.1016/j.critrevonc.2020.102961. [DOI] [PubMed] [Google Scholar]
- 25.Gomez-Casal R, Bhattacharya C, Ganesh N, Bailey L, Basse P, Gibson M, Epperly M, Levina V. Non-small cell lung cancer cells survived ionizing radiation treatment display cancer stem cell and epithelial-mesenchymal transition phenotypes. Mol Cancer. 2013;12(94) doi: 10.1186/1476-4598-12-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y, Zhong L, Yi H. A review on the mechanism of iodide metabolic dysfunction in differentiated thyroid cancer. Mol Cell Endocrinol. 2019;479:71–77. doi: 10.1016/j.mce.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Marini F, Luzi E, Brandi ML. MicroRNA role in thyroid cancer development. J Thyroid Res. 2011;2011(407123) doi: 10.4061/2011/407123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li S, Li Q, Lü J, Zhao Q, Li D, Shen L, Wang Z, Liu J, Xie D, Cho WC, et al. Targeted inhibition of miR-221/222 promotes cell sensitivity to cisplatin in triple-negative breast cancer MDA-MB-231 cells. Front Genet. 2020;10(1278) doi: 10.3389/fgene.2019.01278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ying X, Wu Q, Wu X, Zhu Q, Wang X, Jiang L, Chen X, Wang X. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget. 2016;7:43076–43087. doi: 10.18632/oncotarget.9246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie J, Wen JT, Xue XJ, Zhang KP, Wang XZ, Cheng HH. miR-221 inhibits proliferation of pancreatic cancer cells via down regulation of SOCS3. Eur Rev Med Pharmacol Sci. 2018;22:1914–1921. doi: 10.26355/eurrev_201804_14714. [DOI] [PubMed] [Google Scholar]
- 31.Zhou QY, Peng PL, Xu YH. miR-221 affects proliferation and apoptosis of gastric cancer cells through targeting SOCS3. Eur Rev Med Pharmacol Sci. 2020;24(7565) doi: 10.26355/eurrev_202007_22226. [DOI] [PubMed] [Google Scholar]
- 32.Wang H, Zhan M, Liu Q, Wang J. Glycochenodeoxycholate promotes the metastasis of gallbladder cancer cells by inducing epithelial to mesenchymal transition via activation of SOCS3/JAK2/STAT3 signaling pathway. J Cell Physiol. 2020;235:1615–1623. doi: 10.1002/jcp.29080. [DOI] [PubMed] [Google Scholar]
- 33.Riesco-Eizaguirre G, Rodríguez I, De la Vieja A, Costamagna E, Carrasco N, Nistal M, Santisteban P. The BRAFV600E oncogene induces transforming growth factor beta secretion leading to sodium iodide symporter repression and increased malignancy in thyroid cancer. Cancer Res. 2009;69:8317–8325. doi: 10.1158/0008-5472.CAN-09-1248. [DOI] [PubMed] [Google Scholar]
- 34.Zhou QX, Jiang XM, Wang ZD, Li CL, Cui YF. Enhanced expression of suppresser of cytokine signaling 3 inhibits the IL-6-induced epithelial-to-mesenchymal transition and cholangiocarcinoma cell metastasis. Med Oncol. 2015;32(105) doi: 10.1007/s12032-015-0553-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.