Abstract
Zebrafish are an ideal cell transplantation model. They are highly fecund, optically clear and an excellent platform for preclinical drug discovery studies. Traditionally, xenotransplantation has been carried out using larval zebrafish that have not yet developed adaptive immunity. Larval engraftment is a powerful short-term transplant platform amenable to high-throughput drug screening studies, yet animals eventually reject tumors and cannot be raised at 37 °C. To address these limitations, we have recently developed adult casper-strain prkdc−/−, il2rgα−/− immune compromised zebrafish that robustly engraft human cancer cells for in excess of 28 days. Because the adult zebrafish can be administered drugs by oral gavage or intraperitoneal injection, our model is suitable for achieving accurate, preclinical drug dosing. Our platform also allows facile visualization of drug effects in vivo at single cell resolution over days. Here, we describe the procedures for xenograft cell transplantation into the prkdc−/−, il2rgα−/− model, including refined husbandry protocols for optimal growth and rearing of immune suppressed zebrafish at 37 °C; optimized intraperitoneal and periocular muscle cell transplantation; and epifluorescence and confocal imaging approaches to visualize the effects of administering clinically relevant drug dosing at single cell resolution in vivo. Following identification of adult homozygous animals, this procedure takes 35 days to complete. 7 days are required to acclimate adult fish to 37°C and 28 days are required for engraftment studies. Our protocol provides a comprehensive guide for using immune compromised zebrafish for xenograft cell transplantation and credentials the model as a new preclinical drug discovery platform.
EDITORIAL SUMMARY
This protocol describes how to engraft human cancer cells in immune compromised adult zebrafish. The fish are first adapted to 37°C, followed by intraperitoneal or periocular muscle transplantation of xenograft cells and fluorescence imaging.
Introduction
Xenograft cell transplantation of human malignant cells into immune compromised mice is an invaluable method for assessing cancer cell growth, clonal evolution, intratumoral heterogeneity, as well as cancer stem cell self-renewal, proliferation, and differentiation [1–5]. In translational clinical research, xenograft engraftment studies are indispensable for drug discovery and are required for investigational new drug filings that lead to clinical trials. Specifically, demonstration of potent therapeutic responses, low toxicity, as well as safe and tolerable pharmacokinetics using mouse xenografts is an essential prerequisite for FDA approval of new drugs [6]. Currently, these xenotransplantation studies are carried out almost exclusively using NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) immune deficient mice [7].
Despite important and wide-ranging application of engraftment studies using immune compromised mice, these models have inherent disadvantages. Caliper measurement of tumor volume and luciferase bioluminescence imaging are the two most common methods for quantifying engraftment and tumor growth, yet neither are able to provide an accurate measure of cell growth at single cell resolution [8–10]. Although intravital imaging of engrafted cells is possible using immunocompromised mice, these procedures require surgical implantation of an optical imaging window and multi-photon imaging of immobilized mice. These approaches are expensive, technically challenging, and require a priori knowledge by the investigator where to image tumor growth following imaging window creation [11].
Comparison with other methods
The zebrafish is an excellent cell transplantation model with many inherent advantages, including high fecundity, low cost, and facile high-resolution single cell imaging. Xenotransplantation of human cancer cells has traditionally been carried out using 2 to 7 day-old larvae that have yet to develop a fully functional acquired immune system [12–14]. Aside from being tolerant to xenotransplanted cells, the larval model has important added advantages such as being fully transparent, readily available in large quantities, as well as easy dosing of drugs by immersion therapy - making them suitable for high-throughput and large scale drug screening approaches. Despite many important findings coming from larval xenograft experiments, there are also inherent limitations. First, experiments must be performed in 2–7 day-old larvae at a stage prior to the development of the adaptive immune system in order to prevent graft rejection[15]. Second, the small body size of zebrafish larvae restricts the number of transplanted cells to 100–200 per animal. Thus, these studies are ill-suited for analysis of rare cell populations and would necessarily limit the study of cancer stem cells that drive continued tumor growth. Third, most larval xenotransplantation experiments are conducted at ≤35 °C [16]; however, a subset of human cancers likely do not thrive at these non-physiological temperatures and many do not form tumor masses akin to what is found in xenografted mice or human primary tumor. Lastly, drug delivery in larval animals is carried out by immersion therapy; failing to accurately achieve clinically relevant dosing and pharmacokinetic studies are technically challenging.
Development of protocol
To address the fundamental gap in using larval xenograft experiments, we have recently created optically clear, adult immunocompromised prkdc−/−, il2rgα−/− zebrafish that lack T, B and Natural Killer cells (Figure 1) [17]. These mutant animals allow robust, long-term engraftment of human cancer cells and are an excellent model for preclinical drug discovery studies. One can directly visualize therapy responses in vivo by administering clinically relevant drug doses using either oral gavage or intraperitoneal injection. Traditional pharmacokinetic analysis can also be used to assess drug stability and retention in blood, akin to those carried out in mouse and human. Unlike the larval transplantation model that is inherently suitable for high-throughput, short term drug screening, the adult immunocompromised zebrafish model is best suited for intermediate throughput assessment of combination therapies and studies limited to evaluation of 10–30 drugs. Adult transplant approaches would also be good for secondary screening approaches, helping to prioritize in vitro drug candidates for best assessing in mice xenograft studies.
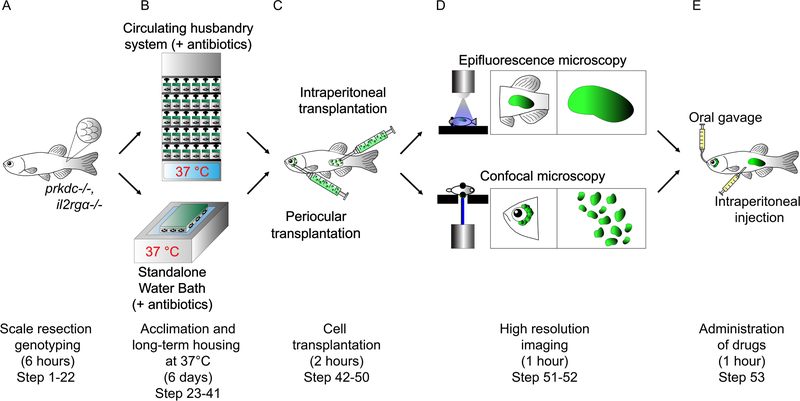
Figure 1. Schematic of a xenotransplantation experiment using prkdc−/−, il2rgα−/− adult zebrafish.
(A) Incrossed progeny from prkdc−/−, il2rgα+/− matings are genotyped at 2.5–3 months of age using a minimally invasive scale resection approach (step 1–22). (B) Following DNA extraction, PCR, and digestion analysis, verified prkdc−/−, il2rgα−/− mutant animals are acclimated to 37 °C for long-term housing using either a stand-alone heated fish system with automatic antibiotic dosing (step 25 option A) or 8L tanks heated to 37 °C using a general purpose water bath (step 25 option B) (step 23–41). (C) Acclimated prkdc−/−, il2rgα−/− zebrafish are then transplanted with fluorescent labeled cells into the intraperitoneal cavity (step 49 option A) or the periocular musculature (step 49 option B) (step 42–50). (D) Animals are then imaged by epifluorescence (step 52 option A) or confocal microscopy (step 52 option B) to assess relative tumor growth over time, respectively (step 51–52). (E) Engrafted prkdc−/−, il2rgα−/− zebrafish can also be subjected to clinically relevant drug dosing by either oral gavage (step 53 option A) or intraperitoneal injection (step 53 option B) (step 53).
Despite the wide utility of engrafting adult immune compromised fish with human cancers and assessing drug responses in vivo, the immunocompromised zebrafish are prone to infection following wounding and rearing fish at 37 °C is uncommon in the field. We have therefore developed optimized husbandry approaches for their genotyping, feeding, and growth at 37 °C. For example, adult prkdc−/−, il2rgα−/− zebrafish must be genotyped following a minimally invasive scale-resection approach and then acclimatized to human physiological temperature of 37 °C using serial increases in temperature cline, antibiotic treatment, and specialized food that includes supplemental amino acids, vitamins and other essential nutrients.
One of the main advantages afforded the optically-clear immunocompromised zebrafish model is the facile imaging of fluorescent-labeled tumor cells. For example, we have optimized two different engraftment approaches by injecting tumor cells into the intraperitoneal cavity or periocular musculature. The former allows facile imaging of tumor growth over time using conventional stereo epifluorescence microscopy while single cell imaging of tumor growth, migration, and therapy responses can best be assessed by confocal microscopy of cells engrafted into the periocular muscle at a location depth of less than 300 μm.
Finally, we have developed/optimized two methods for drug administration. We have achieved clinically relevant and accurate dosing by using daily oral gavage and traditional pharmacokinetic approaches to show drug stability and retention in the blood at similar levels when compared with mouse and human. Moreover, intraperitoneal injection drug delivery can be achieved using ultra fine 30-gauge syringe and needles.
Overview of procedure
This protocol provides a comprehensive blueprint for using adult immunocompromised zebrafish for xenograft cell transplantation. Most commonly used protocols in our field cannot be readily applied to immunocompromised zebrafish because animals are susceptible to opportunistic pathogen infections and animals are housed at elevated temperature. To circumvent these problems, we have tailored many well established protocols (see Experimental Design for details) and provide tips on how best to utilize the model based on our experience. Specifically, we employ a minimally invasive scale resection genotyping approach (Step 1 to 23) and define approaches to acclimatize zebrafish to be grown in sterile conditions at 37 °C in either on a standalone fish system or static water bath (Step 24 to 29). Next, we outline methods to engraft and detect fluorescent labeled human cancer cells using stereo-microscopy and confocal imaging following injection into the peritoneal cavity or the periocular musculature (Step 30 to 52). Lastly, our protocol provides methods to administer clinically relevant drugs by either intraperitoneal injection or oral gavage (Step 53 to 58). Notably, in this protocol, we describe an improved acclimating procedure as well as type and antibiotic dosing, varying slightly from procedures outlined in our initial publication, which further improves general health and survival of immunocompromised zebrafish. In sum, these procedures ensure successful engraftment of human cancer cells into immunocompromised zebrafish for a preclinical drug discovery study.
Application of the method
Our initial published studies have used the prkdcD6312fs/D6312fs, il2rgαY91fs/Y91fs immune deficient zebrafish model for engraftment of human cancers to visualize therapy responses in vivo [17]. In the oncology context, the adult immunocompromised zebrafish model is an ideal tool for preclinical drug discovery studies, where the model can be put through clinically relevant treatment to assess therapy effects. These studies are largely similar to experiments carried out using NSG mice, but performed at lower cost and increased sample size. Notably, the model is also amenable to in vivo single cell imaging, thus allowing real time visualization of cellular responses to drugs. Yet, we envision widespread utility of our model that is not limited to cancer biology. For example, in the future, pluripotency of transplanted embryonic stem cells or induced pluripotent stem cells could be carried out at large scale using immune compromised zebrafish. In addition, future studies will likely involve engraftment of human blood cells including CD34+ cord blood cells and peripheral blood mononuclear cells, allowing direct visualization of human blood cell development and identifying drugs that alter stem cell self-renewal divisions in vivo. Lastly, the zebrafish may provide new models for assessing regenerative potential for a wide array of tissue-restricted stem cells including muscle, liver, intestine, and kidney, identifying new drugs and pathways that modulate normal stem cell growth and regeneration.
Expertise needed and limitations
Utilizing adult immune compromised zebrafish for xenograft cell transplantation studies requires basic skills in zebrafish handling. Zebrafish mating and husbandry are largely similar to conventional husbandry approaches. We estimate about 20 h of hands on time with the animal model will be required before the user is familiar with optimized genotyping, transplantation and imaging techniques.
The largest barrier to using the xenograft cell transplantation approaches is the dedicated husbandry and housing requirements for completing experiments. Adult immunocompromised zebrafish engrafted with human cancer cells need to be maintained at 37 °C and supplemented with antibiotics to curb bacterial infection. Specifically, Aquarius Fish Systems (Aquatic Enterprises) has developed a custom heated, re-circulating zebrafish system that rears fish at 37 °C and doses antibiotics every 4 h. Despite our labs use of this stand-alone system, we have also successfully reared animals in 4 L tanks off line and grew fish in 37 °C using a 23 L general-purpose water bath. It is important to rear only 8 fish/4 L tank, keeping animals in the dark until feeding times (30 minutes/feed/three times daily), and performing daily water changes with antibiotic containing media daily.
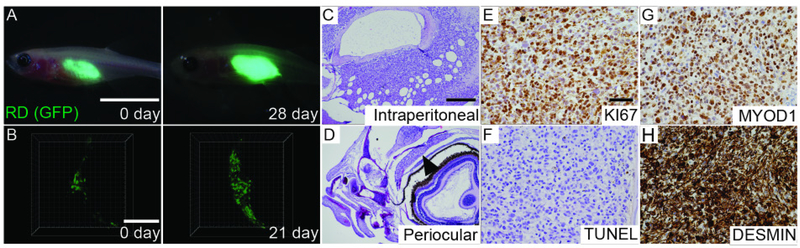
In addition, the prkdc−/−, il2rga−/− zebrafish model has several important limitations to consider. First, engraftment efficiency of most human cancer types ranges from 50% to 90%, with robust engraftment being observed for melanoma, brain cancers, chronic myeloid leukemia, mantle cell lymphoma, breast cancer, renal carcinoma, rhabdomyosarcoma and Ewing’s Sarcoma. However, some tumor types, such as gastrointestinal cancers and reproductive cancers, fail to engraft into the model. Rejection in a subset of recipient animals is likely due to retention of small numbers of phagocytic B cells, NK-lysin+ NK cells and macrophages in prkdc−/−, il2rgα−/− animals [18–20]. Ongoing work is focused on developing new genetic models with enhanced immune deficiencies that fully ablate T, B, and NK cell function, with the hope that these more severely immune-deficient mutants will further enhance engraftment rates. Second, quantification of cell growth, or single cell resolution imaging of engrafted cells, necessarily requires cells to express fluorescence protein, which can be difficult or impractical, especially when dealing with primary patient samples. Circumventing this problem will likely require use of non-invasive imaging modalities such as secondary-harmonic differences in engrafted tissues compared to zebrafish, X-ray histotomography and/or ultrasound biomicroscopy.
EXPERIMENTAL DESIGN
Distinction from current protocols.
Parts of this protocol are based on commonly used protocols in the field, including tail-resection genotyping [21, 22], drug administration by either intraperitoneal or oral-gavage [23–25], and imaging of transplanted cells in casper-zebrafish [21, 23–27]. While these published protocols are widely used in the zebrafish community, we have adapted many steps for use in rearing and imaging of engrafted cells grown in adult immunocompromised zebrafish. Specifically, we have developed a genotyping assay that uses minimally-invasive scale resection rather than genotyping by fin clip. This procedure is critical, as tail genotyping creates a large wound and causes elevated risk of infection. Furthermore, we have developed procedures to serially increase temperature to 37 °C, optimized antibiotics recipes to prevent opportunistic infections, and developed specific food supplements to support the high metabolic rates of fish reared at 37 °C and that support overall tumor growth. Our protocol provides refined methods for oral gavage and intraperitoneal injection that have modified specific technical details so as to inflict minimal injury to the animal and ensure high survival rates following repeated drug dosing. Lastly, imaging tumor growth has been accomplished using both stereo epifluorescence microscopy [28] and confocal imaging [29]. Our refined approach now extends these protocols to imaging engrafted human tumor cells grown in the periocular space and at 37 °C.
Genotyping by minimally invasive scale resection.
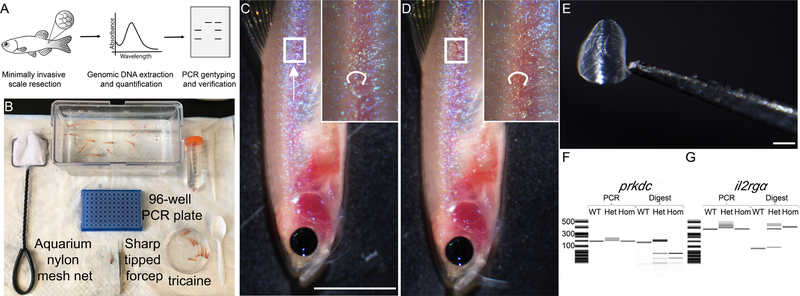
prkdc−/−, il2rgα+/− mutant zebrafish are maintained as stock lines and bred from 3–9 months of age. Following crossing, progeny is raised under conventional husbandry approaches and genotyped at 2.5–3 months of age. We highly recommend genotyping adult progeny by 3 months of age to identify prkdc−/−, il2rgα−/− mutant zebrafish to be used in cell transplantation experiments. Caudal fin resection is the most commonly used approach to harvest tissues for genomic DNA (gDNA) extraction in zebrafish [21]. Yet, we have found that this approach leads to high morbidity in these immunodeficient lines due to large wound size and subsequent infection. Instead, we utilize minimally invasive scale resection genotyping (Figure 1A) [26, 30, 31]. Scales, unlike fin cartilage, are easily detached from the animal’s body using a pair of sharp tipped dissection forceps, and does not cause overt injuries to the animal [32]. Single or small clusters of scales can be removed from the dorsal posterior region of the animal without causing bleeding or infection, with one investigator completing up to 200 scale resections per hour (Steps 3–19). Following scale resection, animals are housed in 24 mg/L penicillin, 36mg/L streptomycin, 100 μg/L amphotericin B, 7 mg/L cephalexin and 67 μl/L praziquantel antibiotic containing fish water at 28.5 °C for 2–4 days allowing ample time to identify mutant animals and to recover from the resection procedure with reduced risk of infection. Meanwhile, scale tissues are lysed, extracted for genomic DNA (gDNA), and PCR amplification performed akin to procedures optimized for traditional tail-fin resection (Steps 20–22). Mutated genes are identified by diagnostic restriction digest analysis of amplified PCR fragments. Genomic mutation at prkdcD6312fs introduces a de novo HinfI site while il2rgαY91fs mutation removes the existing RsaI site found in the wild-type allele.
Acclimation of mutant zebrafish to 37 °C.
Engraftment, growth and proliferation of human cancer cells is best assessed at 37 °C, yet zebrafish are conventionally reared at 28 °C. In the wild, zebrafish experience temperature fluctuations and wide seasonal variation in temperature, ranging from 18 °C to 38 °C, suggesting these animals can tolerate and survive at human body temperature [33]. Several published studies have demonstrated reduced proliferation of human cancer cells engrafted into larvae at colder temperatures [16, 34]. In addition, it is well known that lower temperature other than 37 °C decreases human cancer cell growth in vitro [35–37]. Our own experimental approaches showed in vitro human cell lines become shocked following moving from 37 °C to 35 °C and must be “cold adapted” to regain proliferative capacity, typically taking 7–10 days based on the cell lines tested [38]. We anticipate similar results would hold whether human cancer cells were grown in an incubator in culture, or engrafted into larval or adult zebrafish. Moreover, optimal growth of human cancers requires essential amino acids, vitamins and salts that are not commonly found in zebrafish food. To address these hurdles, we have developed approaches to acclimatize adult immunocompromised zebrafish to be grown at 37 °C and support the efficient engraftment and proliferation of human cancer cells by providing an optimized, enriched diet (Figure 1B). Specifically, zebrafish are acclimatized to 37 °C by gradual changes in temperature cline. Animals are first moved from 28 °C to 32 °C in zebrafish water on a dedicated recirculating aquaculture system supplemented with penicillin, streptomycin, amphotericin B, cephalexin and praziquantel antibiotics (Step 23). Antibiotics are included starting at this stage in order to reduce microbial load in the recirculating system and to reduce risk of infection and animal loss during subsequent steps. Temperature is then increased daily by 1 °C for six days (Steps 24–25). During this acclimation period, fish are fed a special diet composed of zebrafish food infused with cell culture medium three times daily. This can be prepared by mixing powdered cell culture media with conventional commercially available zebrafish dietary fed. The amount of food proportioned is the amount that can be consumed in 10 minutes of active feeding in each tank. Artemia is also fed once daily, to satiety. We have found that this optimized feeding regimen and schedule are critical to the successful engraftment and growth of human cancer cells.
Transplantation and imaging human cancers cells grown in immune-deficient zebrafish.
Transplantation and imaging of human cancer cells in immune deficient zebrafish is largely similar to that conducted in immunocompromised mice (Figure 1C) [8, 10]. In our studies, we have successfully engrafted a wide array of human cancer cell lines and patient derived xenografts (PDXs) from rhabdomyosarcoma, glioma, melanoma and breast cancer. Importantly, cell line and PDXs engraft with overall comparable efficiency. The transplantation procedure is largely similar when transplanting human cancer cell lines or PDXs, including the use of similar numbers of cells and overall injection techniques. However, the major difference is that most PDXs do not lenti-virally express fluorescence proteins. Rather, PDXs should be stained with a long-term lineage tracking dye, such as DiL or CSFE, for engraftment visualization (Step 33). A critical advantage of using the fish model when compared to mouse xenograft studies is that less elaborate transplantation and imaging procedures can be employed. For example, in vivo imaging of engrafted cells does not require surgical implantation of optical imaging windows or special immobilization chambers and multi-photon imaging set ups. Rather, imaging can be accomplished using traditional confocal microscopy of live zebrafish positioned onto a glass-bottomed petri dish containing anesthetics. In our experience, a sample size of n ≥10 animals per treatment arm and up to 6 treatment groups is very manageable for one investigator to complete. This includes day of transplantation and subsequent drug administration and imaging subsequently.
To best visualize and track growth of the engrafted cells, cancer cells should be genetically engineered to express a fluorescence reporter following lentiviral delivery. In the event that such genetic manipulation is impractical or undesired, such as when one wishes to engraft PDXs, transplanted cells can be stained with long-term tracking dyes such as CSFE or Dil A major drawback to these staining approaches is that dyes are diluted following cell division and thus, cell growth cannot be quantified by overall increases in fluorescent intensity. Moreover, fluorescence is usually too low to detect following 10 days of tumor growth.
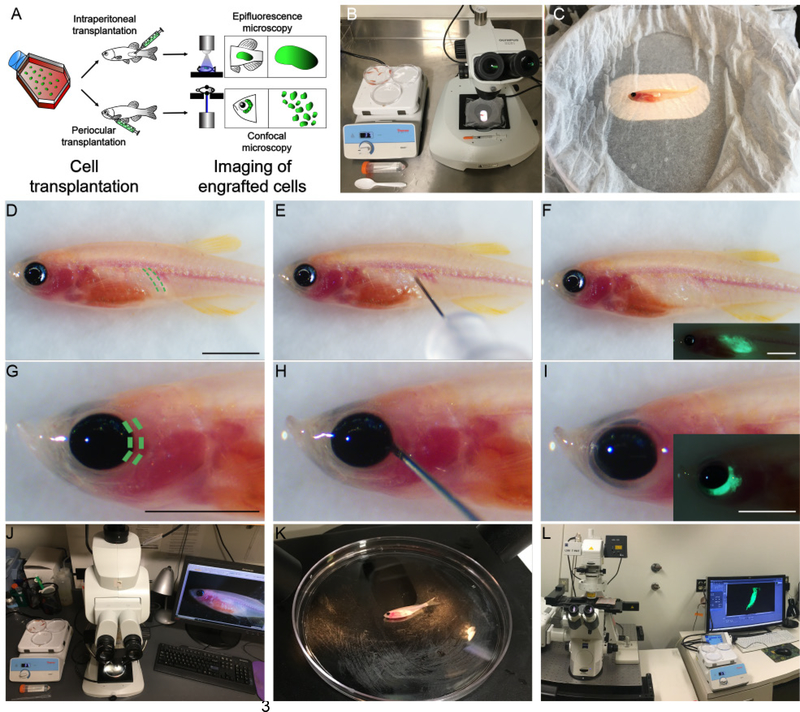
Pending labeling of cancer cells using one of these approaches, cells can next be engrafted into recipient zebrafish. In general, adult 3-month-old immune compromised zebrafish are ~2 cm in length and weigh between 0.5 g to 1 g, considerably smaller than a typical 12-week-old NSG mouse commonly used for xenotransplantation experiments (approximately 20 g). Despite this size difference, up to 10 μl of volume can be injected into the peritoneal cavity of zebrafish. In general, our studies have used 5 × 105 tumor cells engrafted into each adult zebrafish (10 μl of volume) (Step 49 Option A). However, up to 2 × 106 tumor cells can be injected per animal (10 μl of volume). Importantly, cells are resuspended in 2x cell culture media and then supplemented with an equal volume of matrigel, as well as 1% total volume of clodronate liposome that is added to deplete infiltrating macrophages [39]. Optical clarity of the casper-strain immunocompromised zebrafish allows for easy visualization of the transplanted cells following engraftment into the peritoneal cavity using a traditional stereo-fluorescent upright microscope (Step 52 Option A). Growth is quantified by multiplying area by fluorescence intensity, similar to bioluminescence luciferase imaging commonly used in mice (Figure 1D). Tumor cells can also be injected into the periocular space between sclera and eye musculature (Step 49 Option B). In total, up to 5 μl of matrigel embedded cells can be injected into this location. We recommend engrafting 5 × 104 tumor cells into the periocular space, but up to 5 × 105 tumor cells can readily be injected into this site. This site permits growth of human tumor cells at a shallow enough depth for direct confocal microscopy imaging and facilitates single cell visualization of engrafted cells for live cell tracking (Step 52 Option B), assessing cell cycle kinetics, and quantifying responses to therapy (Figure 1D) [29].
Clinically relevant drug administration into adult zebrafish.
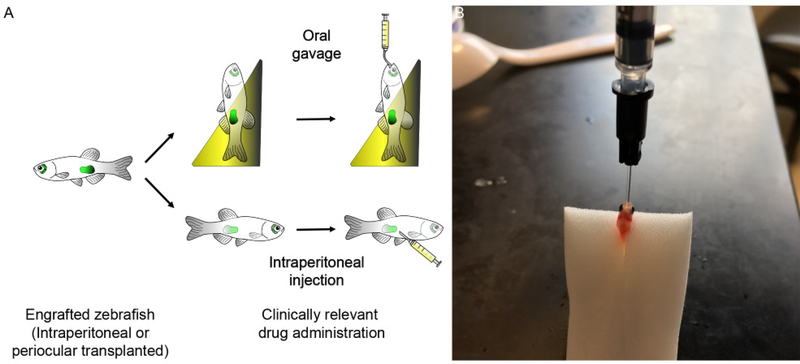
Drug studies using larval zebrafish are completed using immersion therapy where drugs are dissolved in the water and taken up by the fish through a combination of skin permeability or water ingestion. As a result, uptake and absorption likely differ greatly between animals and, since pharmacokinetics (PK) studies are challenging in small larvae, accurate drug uptake is usually unknown. By contrast, we have optimized two delivery approaches to mimic drug administering procedures most often carried out in patients and in preclinical mouse models. Specifically, oral gavage of drugs can be completed using 22-gauge catheters commonly used for mice pups (Step 53 Option A). Up to 10 μl of solution can be fed into the animal at each gavage session (1x daily, 5 times/week). In addition, drugs can be microinjected into the abdominal cavity of the animal daily using a 30 gauge ultra-fine syringe and needle (1x daily, every 3 days, for 2 weeks) (Step 53 Option B). We recommend injecting a maximum of 5 μl of solution daily. Both approaches are minimally invasive, are well tolerated by animals dosed over several days, and do not cause overt injuries to the animal (Figure 1E).
Material
Biological Materials
-
PrkdcD3612fs/il2rgαY91fs casper-strain mutant zebrafish (Available upon request from https://www.langenaulab.com/fish-orders)
CAUTION All animal experiments must be performed in accordance with national and institutional regulations. Institutional approval for the experiments described in this protocol was obtained from the Massachusetts General Hospital (protocol #2011-N-00027).
-
Cells of interest for transplantation. In the example described in this study, we use and embryonal rhabdomyosarcoma cell line (RRID: CVCL_1649) that was labeled with EGFP by lentiviral infection (Available upon request from David Langenau, dlangenau@mgh.harvard.edu) [40].
CAUTION: The cell lines used in your research should be regularly checked to ensure they are authentic and are not infected with mycoplasma.
Antibodies used in these study are rabbit monoclonal anti-Ki67 (Abcam, RRID: AB-302459), rabbit polyclonal Desmin (Abcam, RRID: AB_306653) and Mouse monoclonal MYOD1 (Dako, RRID: 2721191).
Reagents
Paramecium multimicronucleatum (ParameciaVAp cat. no. Par)
Gemma micro 300 (Skretting Zebrafish)
Tricaine-S (MS222) (Western Chemicals, cat. no. ANADA 200–226)
1M Tris buffer pH 9.0 (G-Bioscience, cat. no. 786–476)
Sodium Bicarbonate ACS grade (MedSupply Partners, cat. no. EM-SX0320–1)
Instant Ocean Sea salt for Aquariums (Instant Ocean, cat. no. SS15–10)
Sodium Hydroxide (Sigma Aldrich, cat. no. 221465)
Ethanol, 200 proof (Fisher Scientific, cat. no. 04355222)
Penicillin G sodium salt (Sigma Aldrich, cat. no. P3032)
Streptomycin Sulfate salt (Sigma Aldrich, cat. no. S9137)
Amphotericin B solubilized powder (Sigma Aldrich, cat. no. A9528)
Pimenta racemoa (API, cat. no. 10J)
Fish Flex Forte - Cephalexin (Thomas Labs Cat# 00–12140-C03)
Hikari PraziPro Freshwater & Marine Aquarium Treatment (<5 % w/v praziquantel) (Hikari, cat. no. 142240)
Stress coat water conditioner (1 % v/v aloe vera) (API, cat. no. 165876)
PimaFix (1 % v/v Pimenta racemose) (API, cat. no. 10J)
10x NEB standard Taq Pol buffer (NEB, cat. no. B9014S)
100 mM dNTP set (Life Technologies, cat. no. 10297018)
Taq DNA Polymerase (NEB, cat. no. M0273S)
Nuclease-free H2O (Life Technologies, cat. no. AM9939)
HinfI (NEB, cat. no. R0155S)
RsaI (NEB, cat. no. R0167S)
Cutsmart buffer (NEB, cat. no. B7204S)
Ultrapure Agarose (Life Technologies, cat. no. 165005001)
Tris-acetate-EDTA buffer (Life Technologies, cat. no. 15558026)
RPMI 1640 Medium, powder (Thermo Fisher, cat. No. 31800022)
Fetal Bovine Serum, heat inactivated (Altanta Biologicals, cat. No. S11550H)
PSG (Life Technologies cat. no. 10378–016)
Phenol red-free matrigel (VWR, cat. no. 47743–715)
Human Serum (Lyophilized) (VWR, cat. no. 75928–834)
Standard macrophage depletion kit (Clodrosome, cat. no. 8901)
Trypsin/EDTA Solution (Life Technologies, cat. no. R001100)
PBS (Life Technologies, cat. no. 10010049)
DMEM (Life Technologies, cat. no. 11995073)
0.4% (wt/vol) Tryphan blue solution (Thermo Fisher, cat. no. 115250061)
Viafluor Cell proliferation assay (Biotium, cat. no. 30050)
Primers
prkdc forward: 5’-CAGGACTGGTGGGATGAGGT-3’
prkdc reverse: 5’-CATAGCATATCAGAATTTTGGGCTT-3’
il2rgα forward: 5’-TTTGACATCGAAGACTGTCCTG-3’
il2rgα reverse: 5’-GTCCTGTAACGAACTTCGCTCT-3’
Equipment
Zebrafish husbandry rack (Aquarius Enterprise, custom aquarium systems)
4 Liter husbandry tank (Aquarius Enterprise)
8 Liter husbandry tank (Aquarius Enterprise)
40 mm disposable Petri dish (VWR, cat. no. 25384–342)
PCR plate, 96 well, semi-skirted (Thermo Fisher, cat. no. AB2400)
Marina Fine Blue Nylon Net with Handle (Marina, cat. no. 103989)
Disposable sterile plastic spoon (Fisher Scientific, cat. no. 14–375-254)
Sharp tipped Durmont forceps (World Precision Instruments, cat. no. 500235)
Kimwipes 4.4in × 8.4in (Thermo Fisher, cat. no. 06666A)
PCR adhesive seals (Thermo Fisher, cat. no. AB0558)
T100 Thermo cycler (Biorad, cat. no. 1861096)
Nanodrop microvolume spectrophotometer (Thermo Scientific, cat. no. ND2000)
Scientific Industries Vortex Genie2 (Scientific industries, cat. no. SI-0236)
Falcon 15mL Conical Centrifuge Tubes (Fisher Scientific, cat. no. 1495970C)
Falcon 50mL Conical Centrifuge Tubes (Fisher Scientific, cat. no. 05526B)
1.5ml Eppendorf Microcentrifuge tube (Thermo Fisher, cat. no. 13698791)
QiAxcel Advanced system (Qiagen, cat. no. 9001941)
Qiaxcel DNA screening kit (Qiagen, cat. no. 929004)
Qiaxcel DNA alignment marker 15bp-600bp (Qiagen cat. no. 929530)
Qiaxcel DNA alignment marker 15bp-1kb (Qiagen cat. no. 929526)
23 L General purpose water bath (Chemglass, cat. no. CLS-4953–022)
Multi-Channel Aquarium Auto Dosing Pump (Jebao, cat. no. 131704)
Tissue culture T75 flask (Thermo Fisher, cat. no. 1368065)
1cc insulin syringe only (BD, cat. no. 329650)
Disposable 30 gauge needle (Owens & Minor, cat. no. BD305106)
Glass Pasteur Disposable Aspirating Pipets (Thermo Fisher, cat. no. 136751)
Cellometer Auto 1000 Bright Field Cell Counter (Nexcelom)
Hemacytometer Neubauer counting chamber (Medicus Health, cat. no. 4085M1)
Stereo dissection microscope (Olympus America Inc, cat. no. SZ61)
Stereo macrozoom microscope (Olympus America Inc, cat. no. MVX10)
Stereo macrozoom microscope camera (Olympus America Inc, cat. no. DP74)
Microscope Temperature Control Stage Slide Warmer (AMScope, cat. no. TCS-100)
Fluorescent lamp light source X-CITE 120Q (Thermo Fisher, cat. no.14395231)
150W Fiber Optic Dual Gooseneck Microscope Illuminator (AMscope, cat. no. HL250-AY)
Zeiss LSM710 inverted microscope (Zeiss, cat. no. LSM710)
Glass bottom 35-mm petri dish (Thermo Fisher, cat. no. 50823005)
22 gauge 38mm feeding tube (Thermo Fisher, cat. no. 5081047)
Cosmetic sponge (CVS)
Image analysis software, such as ImageJ (https://fiji.sc) or Imaris (https://imaris.oxinst.com)
Reagents setup
0.4mg/ml Tricaine
Prepare the stock solution (4 mg/ml) by mixing 400 mg tricaine powder with 97.9ml deionized water, adjust to pH 7 with 2.1 ml of 1 M Tris (pH 9). Aliquots can be stored up to 6 months in the dark at −20 °C. Prepare the working solution (0.4 mg/ml) by mixing 1 ml of stock solution with 9 ml of fish system water. Make fresh working solution aliquots per use.
25 g/l Sodium Bicarbonate
Dissolve 25g of sodium bicarbonate powder in 1 litre of deionized water. Make 10 litre volume and store at room temperature (25–28°C) for up to 1 month.
25 g/l Sea salt
Dissolve 25g of sea salt in 1 litre of deionized water. Make 10 litre volume and store at room temperature for up to 1 month.
Cell culture media
The embryonal rhabdomyosarcoma cell line is grown in DMEM supplemented with 10 % (v/v) FBS and 1 % (v/v) PSG, stored at 4 °C for up to 2 weeks.
Fish water
Fish water is prepared using 1 liter deionized water, buffering with 25g/l sodium bicarbonate and 25g/l sea salt, to reach pH of 7.4 and conductivity of 650 V. Fish water can be stored at room temperature for 2 weeks.
Antibiotics-supplemented fish water
Prepare 1 liter fish water containing 24 mg/L penicillin, 36 mg/L streptomycin, 100 μg/L amphotericin B 7mg/L Cephalexin, 67 μg/L PraziPro, 387 μg/L Stress coat water conditioner, 53 μg/L Pimafix. Prepare the antibiotics-supplemented fish water fresh before use.
Cell culture infused food
Cell culture infused food are prepared by first mixing 81 g powdered RPMI 1640 medium with 10 g GEMMA Micro 300 (SKRETTING) fish food in a 500 ml glass beaker. Using 10 ml heat inactivated FBS and 5 ml 1% v/v PSG antibiotics solution (cell culture grade) as solvent, homogenise all components thoroughly with a stirring rod. Aliquot mixture as 5 g pellets into 50 ml Falcon tubes, freeze down at −20°C and store for up to 1 month. Take frozen down food aliquots out of the freezer 5 min prior to feeding, feed ample cell culture infused food to the animals.
Brine shrimp
Hatched brine shrimp were harvested 24 h after seeding 60 g of cysts into 17 L of fish water and grown at 28 °C over night with aeration. Collect hatched brine shrimp into 1 L squeeze bottle and fill with fish water. Keep brine shrimp in squeeze bottle aerated. 5 ml of Brine shrimps are fed three times a day to an 8 L tank with adult zebrafish. Prepare fresh batch of brine shrimp daily.
Lysis buffer
Lysis buffer for resected scales is prepared by mixing 100 mg sodium hydroxide in 50 ml nuclease-free water. Make 50 ml volume and store at room temperature for up to 1 month.
Tris buffer
1M, pH 8.0 Tris buffer used to neutralise lysis buffer is prepared by mixing 4.44 g of Tris-HCL salt with 2.65 g of Tris-base salt, dissolved in 50 ml of nuclease-free water. Make 50 ml volume and store at room temperature for up to 1 month.
Cell culture media used in transplantation mix
Cell line specific media supplemented with 10 % (v/v) FBS and 1 % (v/v) PSG, stored at 4 °C for up to 2 weeks.
Procedure
CAUTION All animal experiments must be performed in accordance with national and institutional regulations. Institutional approval for the experiments described in this protocol was obtained from Massachusetts General Hospital.
Stock maintenance and breeding of prkdc−/−, il2rgα+/− adult zebrafish. (●Timing 3 months/generation of animal)
-
1
2–3 months prior to planned transplantation experiments, set up incrosses between prkdc−/−, il2rgα+/− adult zebrafish.
-
2
Transfer 100 five-day-old larvae into 8 L tanks supplemented with 1 × 105 paramecium/ml. Raise animals using conventional husbandry protocols, including splitting fish into additional tanks after 4 weeks of age to facilitate growth. In total, 25 % of progeny are expected to be homozygous mutant for both prkdc and il2rgα gene (prkdc−/−, il2rgα−/−). prkdc−/−, il2rgα+/− progeny are expected at 50% frequency and should be retained for line maintenance.
▲CRITICAL STEP: When raising larval animals to adulthood, general husbandry practices can be applied. Animals are raised in circulating 28 °C, pH 7.4, 550 V fish water, fed with brine shrimp three times a day.
Scale resection and genomic DNA extraction (●Timing 2 h/96 animals)
-
3
When the prkdc−/−, il2rgα+/− incrossed progeny are 2–3 months old, remove the animals from the fish facility to a dedicated laboratory space assigned for husbandry of immune deficient fish.
▲CRITICAL STEP: We highly recommend that quarantine procedures be maintained following removal and handling fish outside the main fish facility. This is important as these fish lines will be subject to antibiotic treatment and prone to possible infection.
-
4
Place 1 ml of 4 mg/ml Tris-buffered pH 7.0 tricaine into a 40 mm petri dish containing 9 ml of zebrafish facility water.
-
5
Set up a 96-well PCR plate by pipetting 25 μl of 50 mM NaOH (lysis buffer) into every well (if sampling 96 animals). The PCR plate should be held on ice throughout the resection procedure.
-
6
Using a nylon mesh net, transfer the first animal into tricaine infused zebrafish water. Wait 10–30 sec for the animal to become anesthetized.
▲CRITICAL STEP Prolonged exposure to tricaine will lead to respiratory acidosis, cardiac arrest and death. Move on to step 7 immediately when animal shows minimal visible movement.
-
7
Using a plastic spoon, gently transfer the anesthetized animal onto an absorbent pad. Remove any residual moisture on the skin using kimwipes.
-
8
With a sharp tipped forceps, gently remove 1–5 scales along the trunk region posterior to the dorsal fin (Figure 2C–E).
▲CRITICAL STEP Applying excessive pressure when removing scales from animal might lead to bleeding and injury to the animal.
-
9
Ensure scales are resected by visual confirmation.
-
10
Place scales into a single well in the 96-well PCR plate from Step 5.
-
11
Agitate forceps vigorously within the well to ensure scales are detached from the forceps and tissue becomes fully submerged in the NaOH solution.
-
12
Using a plastic spoon, move the scale-resected animal into a dedicated isolation recovery tank with antibiotics-supplemented zebrafish water.
-
13
Ensure to clean forceps between each scale resection with 95 % ethanol and wiping down with a kimwipe.
▲CRITICAL STEP Failure to clean forceps can lead to contamination of DNA and failure to identify homozygous mutant fish for future studies.
-
14
Repeat steps 4 to 13 as needed, with scales from each animal being submerged into different wells within the 96-well PCR plate.
-
15
After sampling all animals, seal the 96-well PCR plate with microplate cover. Vortex briefly, followed by spin down at 400 g for 15 sec at room temperature. Visually inspect the plate to ensure that all scales are now fully submerged in NaOH lysis buffer.
-
16
Using a thermocycler, incubate the 96-well PCR plate at 95 °C for 20 min, and let chill to 4°C.
-
17
Add 2.5 μl of pH 8.0 1 M Tris buffer to each well to neutralize the lysis reaction. Pipette 10 to 20 times to mix thoroughly.
-
18
Seal the 96-well PCR plate with a microplate cover. Vortex briefly. Centrifuge at 300 g for 15 sec at room temperature to expel bubbles at the bottom of each well. Genomic DNA (gDNA) is now extracted.
-
19
Measure the DNA concentration in each well using Nanodrop microvolume spectrophotometer to ensure enough gDNA is extracted (extracted gDNA concentration should be in the range of 20 to 200 ng/μl).
■PAUSE POINT Extracted gDNA can be stored at 4 °C for an extended period. Sampled mutant zebrafish can be kept in isolation tanks containing antibiotics supplemented water (see Reagent Setup) at 28 °C for a few days if fed daily and water changes are performed.
? TROUBLESHOOTING
Figure 2. Genotyping for prkdc and il2rgα mutations.
(A) Schematic of the genotyping experiment using the scale resection approach described in Steps 3–22. (B) Experimental setup for a 96-animal genotyping experiment. (C) Animal is imaged under the dissecting microscope prior to scale resection. Scales are resected from the trunk region by a scrapping downward using a sharp tipped forceps (noted by arrow direction). (D) Same animal following scale resection. Note no visible bleeding was caused by the procedure. (E) Microscopic image of a single resected scale held by the sharp tipped forceps. (F) Qiaxcel DNA screening analysis of PCR amplicons and those digested with Hinfl enzyme (prkdc) and RsaI enzyme (il2rgα). Scale bar = 0.5 cm (C, D), 1 mm (E). Institutional approval from Massachusetts General Hospital was obtained for the experiments shown in this figure.
Genotyping by PCR and restriction enzyme digestion (●Timing 4 h/96 animals)
CRITICAL All genotyping procedures in this section are based on genotyping 96 animals using a 96-well microtiter plate.
-
20
Thaw dNTP, primers, Taq polymerase and standard Taq polymerase buffer on ice 30 min prior to procedure.
-
21
For amplifying the prkdc gene, follow option A. For amplifying the il2rgα gene, follow option B. We recommend validating mutation in both genes to confirm the genotypes of all experimental animals.
-
Genotyping the prkdc mutation
-
Pipette all components of the PCR master mix into a 15 ml Falcon tube.
Reagent 1 Reaction (μl) 1 plate (100 reactions, μl) 10x NEB standard Taq Pol buffer 2.5 250 100 mM dNTP 0.2 20 10 μM Forward prkdc primer 0.5 50 10 μM Reverse prkdc primer 0.5 50 Taq DNA Polymerase 0.1 10 Nuclease-free H2O 18.7 1870 Total 22.5 2250 ▲CRITICAL STEP Always prepare 100 reaction worth of PCR or digestion reaction mix to account for pipetting error.
Pipette 22.5 μl of master PCR mix into each well of a 96 well PCR plate. The plate should be held on ice at 4°C.
Pipette 2.5 μl of gDNA solution from each well from Step 18 into the corresponding well in the 96-well PCR plate containing PCR master mix.
Pipette 10–20 times to mix solutions thoroughly. Seal the 96-well PCR plate with microplate sealer. Vortex briefly. Spin down at 300 g at 15 sec at room temperature.
- Perform PCR using the following cycling parameters:
Denature Anneal Extend Hold Denature 94 °C, 2 min Cycle 1–35 94 °C, 30 s 53 °C, 30 s 68 °C, 25 sec Final extension 68 °C, 5 min 4 °C, ∞ After the PCR reaction is completed, analyze 5 μl of PCR product per well by gel electrophoresis (2.5 % w/v agarose in 1x TAE) or by Qiaxcel. A single band of 207 bp or 199 bp should be amplified for wild type or mutant prkdc, respectively.
- Prepare a master mix for DNA enzyme digestion by combining the following into a 1.5 ml Eppendorf tube.
Reagent 1 Reaction (μl) 1 plate (100 reactions, μl) HinfI 0.3 30 NEB Buffer 2.1 or 3.1 or CutSmart 2 200 Nuclease-free water 2.7 270 Total 5 500 Pipette 5 μl of enzyme digestion mix into each well of a 96 well PCR plate. The plate should be held on ice at 4 °C.
Pipette 15 μl of PCR product from Step V per well into the corresponding well in the 96-well PCR plate containing digestion mix.
Pipette 10–20 times to mix solutions thoroughly. Seal the 96-well plate with microplate sealer. Vortex briefly. Spin down at 300 g at 15 sec at room temperature.
-
Incubate the 96-well plate at 37 °C for 1 h.
▲CRITICAL STEP Incubation of amplified prkdc fragment with Hinfl enzyme should not be less than 1 h, and no more than 3 h.
Restriction enzyme digestion with Hinfl cuts the wild type PCR fragment once and the mutant PCR fragment twice. Resolve the digested DNA using a 2.5 % w/v TAE agarose gel or with Qiaxcel DNA using the 15 bp to 1 kb alignment marker. Homozygous wild-type (WT) animals will produce two bands at 183 bp and 24 bp; prkdc+/− animals will have four bands at 183 bp, 107 bp, 68 bp and 24 bp; prkdc−/− mutant will have three bands at 107 bp, 68 bp, and 24 bp (Figure 2F).
-
-
Genotyping il2rgα mutation
-
Pipette all components of the PCR master mix into a 15 ml Falcon tube.
Reagent 1 Reaction (μl) 1 plate (100 reactions, μl) 10x NEB standard Taq Pol buffer 2.5 250 100 mM dNTP 0.2 20 10 μM Forward il2rgα primer 0.5 50 10 μM Reverse il2rgα primer 0.5 50 Taq DNA Polymerase 0.1 10 Nuclease-free H2O 18.7 1870 Total 22.5 2250 ▲CRITICAL STEP Always prepare 100 reaction worth of PCR or digestion reaction mix to account for pipetting error.
Pipette 22.5 μl of master PCR mix into each well of a 96 well PCR plate. The plate should be held on ice at 4 °C.
Pipette 2.5 μl of gDNA solution from each well into the corresponding well in the 96-well PCR plate containing PCR master mix.
Pipette 10–20 times to mix solutions thoroughly. Seal 96-well PCR plate with microplate sealer. Vortex briefly. Spin down at 300 g at 15 sec at room temperature.
- Using a thermocycler, set up PCR cycling parameter as such:
Denature Anneal Extend Hold Denature 94 °C, 2 min Cycle 1–36 94 °C, 30 s 60 °C, 30 s 68 °C, 30 sec Final extension 68 °C, 5 min 4 °C, ∞ After PCR reaction is completed, pipette 5 μl of PCR product per well for a test run on either a 2.5% w/vTAE agarose gel or with a Qiaxcel DNA screening cartridge. There should be a single band of 373 bp or 360 bp for amplified wild type and mutant il2rgα genomic DNA fragment respectively.
- Prepare a master mix for DNA enzyme digestion by combining the following into a 1.5 ml Eppendorf tube.
Reagent 1 Reaction (μl) 1 plate (100 reactions, μl) RsaI 0.3 30 CutSmart buffer 2 200 Nuclease-free water 2.7 270 Total 5 500 Pipette 5 μl of enzyme digestion mix into each well of a 96-well PCR plate. The plate should be held on ice at 4 °C.
Pipette 15 μl of PCR product from Step V per well into the corresponding well in the 96-well PCR plate containing digestion mix.
Pipette 10–20 times to mix solutions thoroughly. Seal 96-well PCR plate with microplate sealer. Vortex briefly. Spin down at 300 g at 15 sec at room temperature.
-
Incubate 96-well PCR plate containing amplified PCR fragment and RsaI enzyme at 37 °C for 1 h.
▲CRITICAL STEP Incubation of amplified prkdc fragment with RsaI enzyme should not be less than 1 h and no more than 3 h.
Restriction enzyme RsaI cuts wild type PCR fragment once, but not mutant PCR fragment. Analyze digested reactions on a 2.5% w/v TAE agarose gel or Qiaxcel using the 15bp to 1kb alignment marker. WT animals for il2rgα gene should have 2 bands at 261 bp and 112 bp; il2rgα+/− animals should have 3 bands at 360 bp, 261 bp, 112 bp; il2rgα−/− mutant should have 1 band at 360 bp (Figure 2G).
-
-
-
22
Following assignment of genotypes as outlined in Steps 20–21, combine animals based on their genotypes. We recommend keeping prkdc−/−, il2rgα+/− animals as future breeder pairs, while the prkdc−/−, il2rgα−/− animals will be used as transplant recipients. There should be approximately 24 prkdc−/−, il2rgα−/− animals identified per 96-well plate.
▲CRITICAL STEP From this point onwards, prkdc−/−, il2rgα−/− animals should be housed in antibiotics-supplemented water (see Reagent Setup), until sacrificed or euthanized.
? TROUBLESHOOTING
Acclimation and long-term housing of mutant zebrafish (●Timing 6 d)
-
23
6 d prior to transplantation experiment, move verified prkdc−/−, il2rgα−/− zebrafish into an 8 L isolation tank that is placed within a temperature controlled 23 L general-purpose water bath (Figure 3A and B).
-
24
Set the bath temperature to 32 °C and gradually increase the bath temperature by 1 °C every 24 h, starting at 32 °C on the first day. During the acclimation procedure, feed animals three times a day with 5 ml of brine shrimp and perform daily water change with antibiotics supplemented fish water.
▲CRITICAL STEP Do not increase temperature by more than 1 °C/d, doing so will elicit the heatshock response and kill animals.
-
25
After 6 d of incremental temperature increases, animals are now acclimated to surviving long-term at 37 °C. If available, transfer mutant animals onto a heated standalone circulating husbandry system that supplies antibiotics and circulates water (option A). Alternatively, animals can be housed in a general-purpose water bath at 37 °C (option B).
-
Housing fish in a standalone 37 °C heated fish system with automatic antibiotic dosing.
Add a water heater to the reservoir of husbandry rack of an Aquarius™ 5 shelve system (12 valves/row), and set at 37.0°C ± 0.5°C. Set water purge of 8 L occurs three times a day at 9am, 1pm and 5pm (total 24 L water purged/d).
House prkdc−/−, il2rgα−/− zebrafish in the Aquarius™ 5 shelve system circulating fish water supplemented with antibiotics. Use 25 mg/L Sodium Bicarbonate and 25 mg/L Sea salt to adjust and maintain pH of circulating water between pH 7.0 to 7.4 and conductivity at 650 V-850 V (Figure 3C, D). ▲CRITICAL STEP Minimise pH, temperature and conductivity fluctuations. Abrupt change in these physiological parameters can cause death to animals.
To compensate for the loss of antibiotics during water purge, use a programmable doser to dispense 1 L of 4X antibiotics into fish system reservoir every 4 h (8 am, 12 noon, 4 pm, 8 pm, 12 midnight, 4 am). Feed the animals 3 times a day at 9 am, 1 pm and 5 pm, with cell culture infused GEMMA Micro 300 (SKRETTING) fish food (see Reagent Setup). Supplement diet with 5 ml of brine shrimp 10 min after feeding with cell culture infused fish food.■PAUSE POINT prkdc−/−, il2rgα+/− breeders can be housed indefinitely in conventional zebrafish husbandry facilities. prkdc−/−, il2rgα−/− should be housed under the aforementioned husbandry procedures indefinitely until the end of each experiment.
-
Housing fish in a 37°C heated water bath
House prkdc−/−, il2rgα−/− zebrafish in tanks containing antibiotics and submersed into a general purpose water bath set a 37 °C. Use 25 mg/L Sodium Bicarbonate and 25 mg/L Sea salt to adjust and maintain pH of circulating water between pH 7.0 to 7.4 and conductivity at 650 V-850 V (Figure 3C, D). ▲CRITICAL STEP Minimise pH, temperature and conductivity fluctuations. Abrupt change in these physiological parameters can cause death to animals.
Feed the animals 3 times a day at 9 am, 1 pm and 5 pm, with cell culture infused GEMMA Micro 300 (SKRETTING) fish food (see Reagent Setup). Supplement diet with 5 ml of brine shrimp 10 min after feeding with cell culture infused fish food.
-
Perform daily water change. Pre-heat 8 L of zebrafish water to 37 °C supplemented with 1X antibiotics mix. Use an aquarium nylon mesh net to transfer animals into the new 8 L tank at the end of the day, after all 3 feedings have been carried out.
■PAUSE POINT prkdc−/−, il2rgα+/− breeders can be housed indefinitely in conventional zebrafish husbandry facilities. prkdc−/−, il2rgα−/− should be housed under the aforementioned husbandry procedures indefinitely until the end of each experiment.
-
Figure 3. Acclimating prkdc−/−, il2rgα−/− zebrafish for long term housing at 37 °C.
(A) Schematic of acclimation process described in Steps 23–25. (B) prkdc−/−, il2rgα−/− zebrafish are moved to a general-purpose water bath initially set at 32 °C. The temperature is then gradually increased to 37 °C at 1 °C/d. The animals can be housed long-term in the 37°C water bath or in a dedicated standalone husbandry unit. In both cases, water needs to be supplemented with antibiotics. (C-E) Representative images of the antibiotics supplemented, heated stand-alone fish system produced by Aquarius Fish Systems. Institutional approval from Massachusetts General Hospital was obtained for the experiments shown in this figure.
Cell Preparation (●Timing 1 h)
-
26
Five days prior to the transplantation experiment, plate approximately 5 × 105 cells into a T75 cell culture flask. In the example described in this Protocol we use an embryonal rhabdomyosarcoma cell line that was EGFP labeled by lentiviral infection (EGFP+ RD cells). Incubate the flasks at 37 °C, 5 % CO2, and 95 % humidity. Perform cell media changes every 2 d. There will be approximately 1 × 107 EGFP+ RD cells on the day of transplantation.
▲CRITICAL STEP If another cancer cell line is used, precise plating density should be determined by proliferation rate of the cell line of interest. In general, we recommend an initial transplantation cell number of 5 × 105 cells/animal for intraperitoneal transplantation and 5 × 104 cells/animal for periocular transplantation.
-
27On the day of experiment, pre-chill all the following, either in an ice bucket or in a cold room at 4°C, for at least 2 h:
- 1 cc syringe attached with 30 Gauge needle
- 1 box of 1000 μl pipette tip
- 1 vial of 100 μl phenol red-free matrigel
- 1 vial of 20 μl human serum dissolved in FBS
- 1 vial of 2 μl clondronate liposome
- 1 vial of 2 μl 100X Penicillin/Streptomycin/Glutamine
- 1 vial of 100 μl cell culture media
-
28
Harvest adherent EGFP+ RD cells from T75 flask in a tissue culture hood (option A). If suspension cells are used for transplantation go to option B.
-
Harvesting adherent cells
Remove the T75 flask containing EGFP+ RD from the tissue culture incubator. Aspirate off the cell culture medium. Wash once with 5 ml pre-warmed PBS.
Aspirate off the PBS and add 2 ml of cold 0.05 % wt/v trypsin. Incubate cells at 37 °C, 5 % CO2, and 95 % humidity for 2 min to dissociate attached cells.
Observe cell detachment using an inverted tissue culture microscope and then neutralize by addting 4 ml of cell culture media.
Transfer detached cells from the T75 flask into a 15 ml Falcon tube using a Pasteur pipette. Spin cells at 300 g for 3 min, at room temperature. There should be a sizable cell pellet following centrifugation.
Aspirate supernatant from the tube and re-suspend in 1 ml cell culture media.
-
Harvesting suspension cells
-
I.
Transfer cell culture medium containing the cells of interest from the T75 flask into a 15 ml Falcon tube using a Pasteur pipette. Rinse the T75 with another 5ml cell culture media and transfer into the same falcon tube.
-
II.
Spin cells at 300 g for 3 min at room temperature. There should be a sizable cell pellet following centrifugation.
-
VI.
Aspirate supernatant from the tube and re-suspend in 1 ml cell culture media.
-
I.
-
-
29
Assess cell viability, for example using the tryphan blue exclusion approach [41]. Continue with the transplantation experiment only if cell viability is more than 90 %.
-
30
Count the total number of cells/ml using either an automated cell counter or a hemacytometer.
-
31
Pellet the cells at 300 g for 3 min at room temperature. Aspirate supernatant.
▲CRITICAL STEP If the cell of interest is not engineered to express any fluorescence protein, they should be stained with long term tracking fluorescent cell cytoplasmic dye to facilitate visualization of engraftment (Step 36–39). Otherwise go straight to step 40.
-
32
In a 15 ml falcon tube, suspend cells at 1 × 106 cells/ml in pre-warmed PBS containing
-
33
Optional; In order to stain the cells, add 1 μM CSFE or Dil dye and incubate cells at 37 °C, 5 % CO2, and 95 % air humidity for 15 min. Protect stained cells from light.
-
34
Add an equal volume of cell culture medium into the 15ml falcon tube, and incubate at 37 °C, 5 % CO2, and 95 % air humidity for 5 min.
-
35
Pellet the cells at 300 g for 3 min at room temperature. The stained cell pellet should appear yellow-green.
-
36
Pre-chill the cell pellet on ice for 3 min.
-
37
Prepare the following transplantation master mix and store on ice. Using a pre-chilled p1000 pipette and tip, gently pipette the solution 5–10 times to mix. Ensure the p1000 is set to a volume of 200 μl to avoid incorporation of air bubbles at the time of mixing.
▲CRITICAL STEP Take care not to introduce air bubbles. Injection air bubbles into zebrafish recipient can affect buoyancy of animal and is often fatal.
- ? TROUBLESHOOTING
Reagent Volume Phenol-red matrigel 100 μl Cell culture medium 100 μl FBS 20 μl PSG antibiotics 2 μl Clondronate liposome 2 μl
-
38
Resuspend the cell pellet from Step 36 in the transplantation master mix, ensuring a final volume of 5×105 cells/10 μl for intraperitoneal injections or 5 × 104 cells/5 μl for intra-ocular injections.
-
39
Remove the plunger from a 1cc insulin syringe (fitted with 30 gauge needle) and transfer the cell mixture from Step 38 directly into the syringe cylinder. Load at least 100 μl into the syringe.
▲CRITICAL STEP Transplantation mixture containing matrigel can be difficult to pipette when volume is small. We recommend loading at least 100 μl of transplantation mixture into the syringe.
-
40
Insert the plunger back into the syringe and gently push mixture to the top of the syringe.
-
41
Incubate the syringe at 37 °C, 5 % CO2, and 95 % air humidity for 30 min. After incubation, the mixture inside the syringe should appear to be partially transparent and semi-viscous. EGFP+ RD cells are now ready to be injected into mutant animals.
▲CRITICAL STEP Cold transplantation mixture has a liquid like consistency and can be difficult to inject into animals. Ensure the injection mixture is preheated to 37 °C prior to injection
? TROUBLESHOOTING
Preparing fish for transplantation procedure (●Timing 5 min)
-
42
On the day of transplantation, mutant prkdc−/−, il2rgα−/− animals (from Step 25) should not be fed 4 h prior to the transplantation procedure.
Cell transplantation (●Timing 1 h/10 animals)
-
43
Prior to the transplantation procedure, set up the dissection microscope and temperature-controlled stage warmer to 37 °C. Use a 40 mm petri dish cover as the injection platform for the procedure. Lay a piece of Kimwipe on the dish cover and wet it with the 0.4 mg/ml tricaine anesthetizing solution in zebrafish water. Place the petri dish cover on top of heated stage warmer (Figure 4A–D).
-
44
30 min before the transplantation procedure, set up a tabletop hot plate adjacent to dissection microscope. Place three 40 mm petri dishes onto the 37 °C hot plate. Two petri dishes should contain 10 ml of antibiotics supplemented water and one with 10 ml fish water supplemented with 0.4 mg/ml tricaine and antibiotics.
-
45
Using an aquarium nylon mesh net, transfer 4–8 prkdc−/−, il2rgα−/− fish from Step 42 into one of the 40mm petri dishes that contain antibiotics supplemented zebrafish system water. Ensure to cover the dish so fish do not jump and escape.
-
46
Remove the syringe containing EGFP+ RD cells (from Step 41) from the cell culture incubator and place at room temperature. Remove the needle cap and expel any air bubbles introduced within the needle.
-
47
Using a plastic spoon, transfer a single prkdc−/−, il2rgα−/− fish into the petridish supplemented with and 0.4 mg/ml tricaine. Wait for the animal to become anesthetized before moving on to the next step.
-
48
Use a plastic spoon to transfer one fish onto the dissecting microscope platform. Place the left side of the fish facing up (Figure 4C).
-
49
Perform transplantation of the cells, either using intraperitoneal (option A) or periocular (option B) transplantation. We recommend intraperitoneal transplantation when evaluating the overall efficiency of engraftment or for assessing therapy responses. By contrast, periocular transplantation is best used for single cell visualization of engrafted cells to assess migration and cellular mechanisms governing responses to treatment.
-
Intraperitoneal transplantation
-
Using the dissection microscope, identify the last 2 pleural ribs that run perpendicular along the peritoneal cavity of the animal, closest to the anus of animal. Insert the 30-gauge needle in between the 2 highlighted pleural ribs (Figure 4D). Stop inserting once the entire bevel is inserted into the cavity.
▲CRITICAL STEP Ensure that the needle is not inserted too deep into the peritoneal cavity. Stop inserting the needle once the bevel has fully inserted into the cavity. Inserting the needle deeply will cause internal organ damage, bleeding and possible death of the animal.
? TROUBLESHOOTING
Slowly inject the cell mixture from Step 46 into intraperitoneal cavity (10 μl). 10 μl of cell mixture corresponds to 1 step interval on the syringe (Figure 4E).
The injected mixture should appear to be semi-solid and take up a significant volume behind the swimming bladder. If correctly injected, the intraperitoneal cavity will visibly distend. Slowly retract needle from cavity ensuring no injected mixture leaks out from the injection site.
Using a plastic spoon, move the injected animal into the remaining 40mm petri dish that contains antibiotics supplemented water. Allow animal to recover (Figure 4F). Ensure to cover the petridish to stop jumping fish from escaping once they have recovered from the anesthesia.
Repeat step I to IV until all animals are transplanted with cell of interest.
-
-
Periocular transplantation
-
I.
Under the dissection microscope, identify the periocular muscle space (Figure 4G).
-
II.
Gently insert the needle at the 3 O’Clock position, with bevel of needle pointing toward the eye at a 45-degree angle (Figure 4H).
▲CRITICAL STEP Ensure the tip of needle is not inserted into the cornea of the animal. Insert the needle until the bevel has completely pierced the muscle space. Inserting the needle to deeply will cause serious damage and bleeding in the animal.
-
III.
Slowly inject 5 μl of cell mixture from Step 46. 5 μl corresponds to half a step interval on the syringe. The eye of animal should rise slowly following the injection. Slowly retract the needle ensuring no leakage of cells. After injection, the cell mixture should coalesce into a semi-solid opaque plug (Figure 4I).
▲CRITICAL STEP Do not inject more than 5 μl or inject too fast, both of which will lead to rupture of tissue around the eye.
-
VI.
Repeat step I to III until all animals are transplant experiments are completed.
-
I.
-
-
50
Return transplanted animal into a 8 liter fish tank, housed either on the standalone heated system or in the static water bath set at 37 °C. Wait 4 h after the transplantation procedure to feed the animals again. Allow animals to recover for at least 24 hours before imaging for engrafted cells.
Figure 4. Transplantation of EGFP-expressing human RD cells into prkdc−/−, il2rgα−/− zebrafish by either intraperitoneal or periocular transplantation.
(A) schematic of transplantation experiment described in Steps 43–50. (B) Experimental set up of a transplantation experiment. (C) Anesthetized prkdc−/−, il2rgα−/− zebrafish are oriented with the left side facing up in a 40 mm petri dish covered with a tricaine soaked kimwipe. The Petri dish is placed onto a 37C heated stage affixed to the stereomicroscope. (D) Site of intraperitoneal injection denoted by green dotted lines. (E) A 30-gauge needle syringe inserted between the 2 pleural ribs of prkdc−/−, il2rgα−/− zebrafish. (F) Transplanted animal imaged for EGFP fluorescence immediately following transplantation. (G) Site of periocular muscle injection denoted by green dotted lines. (H) Transplant syringe should be inserted at the 3 O’clock direction outside the eye of animal. (I) Transplanted animal imaged for EGFP fluorescence immediately following transplantation. (J) Experimental setup for imaging intraperitoneal transplantations using epifluorescence stereomicroscopy. (K) During epifluorescence imaging, animal should be positioned with its left side facing up in the 40 mm petri dish. (L) Experimental setup for imaging of periocular engrafted cells using confocal microscopy. Scale bar equals 1 cm in (D-I, including insets). Institutional approval from Massachusetts General Hospital was obtained for the experiments shown in this figure.
Imaging of engrafted cells (●Timing 1 h/10animals)
-
51
30 min before the imaging procedure, set up a 37 °C hot plate adjacent to the microscope. Place three 40mm petri dishes onto the hot plate. Two dishes contain 10ml of antibiotics supplemented zebrafish water and one contains 10 ml of antibiotics supplemented water containing 0.4 mg/ml tricaine.
CRITICAL STEP: Do not feed animals 4 h prior to imaging. Cell culture infused fish food has high autofluorescence;
-
52
Image the fluorescent-labeled cells by epifluorescence microscopy (option A). Periocular injected cells can also be imaged by confocal microscopy (option B).
? TROUBLESHOOTING
-
Epifluorescence imaging of intraperitoneal transplanted cells
Position one of the 40 mm petri dishes containing 10 ml of antibiotics supplemented water onto the microscope stage.
Transfer all animals to be imaged into the other pre-warmed 40 mm petri dish containing 10 ml of antibiotics supplemented water.
Using a plastic spoon, transfer one prkdc−/−, il2rgα−/− into the 40 mm petri dish containing antibiotics supplemented water containing 0.4 mg/ml tricaine.
Transfer the anesthetized animal into the 40 mm petri dish that is positioned on the 37 °C microscope stage. Adjust the animal so it is centered and focused for imaging (Figure 4K). Use a low magnification that allows visualization of most of the animal, particularly the intraperitoneal region (Figure 4J).
-
Take a brightfield image of the animal, followed by a low and a high exposure fluorescence image. GFP images are captured using a wide band blue fluorescence filter cube at an excitation of 450–480nm and a 510 long pass filter.
▲CRITICAL STEP To avoid acquiring images that are saturated and result in photobleaching and phototoxicity to the animal, we recommend imaging at low excitation intensity and capturing images at 3 exposure times.
Repeat step II to V until all engrafted animals have been imaged.
Quantify tumor cell growth by multiplying pixel intensity by total 2D surface area of engrafted cells. Normalization should be made to animals imaged at 1 day post-transplantation using the same magnification and exposure conditions.
-
Image and quantify tumor cell growth every 7 days until proceeding to drug injection in Step 53.
▲CRITICAL STEP A subset of animals will reject tumors by 7 days post-injection. Engraftment efficiencies typically range between 50% to 90% of injected animals and differs based on tumor cell line and type. Animals that are rejecting human tumor cells will display a gradual reduction in fluorescence intensity and/or cell number by 7 days post-injection when imaged by either stereo-epifluroscence microscope or confocal microscope. At the end of experiment, animals are sacrificed by tricaine overdose or methods compliant with institutional and government regulations.
-
Confocal imaging of periocular transplanted cells
-
I.
On the day of imaging, place two heat packs on the confocal microscope imaging stage 30 min prior to mounting animal.
-
II.
Transfer all animals to be imaged into one of the pre-warmed 40 mm petri dishes containing 10 ml of antibiotics supplemented water.
-
III.
Using a plastic spoon, transfer a single prkdc−/−, il2rgα−/− into the 40 mm petri dish containing 0.4 mg/ml tricaine and wait for animal to become anesthetized
-
IV.
Using a plastic spoon, transfer the immobilized animal onto a 23 mm glass bottom Petri dish. Drip three drops of 37 °C 0.4 mg/ml tricaine onto animal and transfer to the confocal microscope stage.
-
V.
Perform confocal imaging using the desired objectives and fluorophore detectors. For imaging GFP images, 488 nm laser is used. A Z-stack image should be captured to reconstitute the 3-dimensional architecture of engrafted cells. Use no more than 5 microns per stack layer to ensure adequate speed for acquiring images. The total imaging time should not exceed 5 minutes per animal.
▲CRITICAL STEP To avoid acquiring images that are saturated and the risk of photobleaching and phototoxicity, laser power and gain should be set using the automated set exposure function within the imaging software.
? TROUBLESHOOTING
-
VI.
Repeat step III to V until all engrafted animals are imaged.
-
IX.
Quantify tumor cell growth by counting absolute cell number. This can be completed using image analysis software such as ImageJ or Imaris.
-
X.
Image and quantify tumor cell growth every day until proceeding to drug injection in Step 53. CRITICAL STEP A subset of animals will reject tumors by 7 days post-injection. Engraftment efficiencies typically range between 50% to 90% of injected animals and differs based on tumor cell line and type. Animals that are rejecting human tumor cells will display a gradual reduction in fluorescence intensity and/or cell number by 7 days post-injection when imaged by either stereo-epifluroscence microscope or confocal microscope. At the end of experiment, animals are sacrificed by tricaine overdose or methods compliant with institutional and government regulations.
-
I.
Administering drugs to engrafted zebrafish (●Timing 30 min/10animals)
-
53
After 7 days of engraftment, clinically relevant doses of drugs can be administered to transplanted prkdc−/−, il2rgα−/− mutant animals in two different ways: For using oral gavage, follow option A. For intraperitoneal injection (which is similar to intravenous injections completed in mice), follow option B.
CRITICAL: Animals that fail to engraft tumor cells generally show rejection within the first five days after transplantation. Therefore, we recommend that drug administration studies be initiated after 7 d of engraftment.
-
Oral gavage of drugs
Determine an estimate of the average wet weight for the cohort of transplanted zebrafish. Average weight will be used to calculate the amount of drugs that should be given. ▲CRITICAL STEP A maximum of 10 μl can be orally administered.
Using a hot plate set to 37 °C, heat a glass beaker containing 500 ml of antibiotics-supplemented water, two 40 mm petri dishes with 10 ml of antibiotics-supplemented water, and a 40 mm petri dish containing 10 ml of antibiotics-supplemented fish water containing 0.4 mg/ml tricaine. Solutions should be heated 30 min before procedure.
Mount a 22-gauge soft tip oral tube onto a 1 ml syringe. Pipette 10 μl of drug onto a clean 40mm petri dish, forming a droplet. Load the solution by suctioning the entire droplet into syringe. Ensure no air bubbles are introduced.
Cut a single 3 cm slit into a piece of triangular cosmetic sponge. The slit should be large enough to accommodate the engrafted zebrafish that will be gavaged. Soak the cosmetic sponge in the pre-heated fish water from the beaker.
Transfer 4–8 animals to be gavaged into one of the 40 mm petri dishes containing antibiotic-supplemented fish water. Ensure to cover the Petri dish to stop jumping fish from escaping.
Transfer a single animal, into the antibiotics-supplemented fish water containing 0.4 mg/ml tricaine using a plastic spoon. Wait for animal to become anethesized.
Once the animal shows minimal movement, place the animal mouth-up into the slit. The animal’s head should be exposed while the body is embedded within the sponge.
-
Using the drug-loaded syringe from Step iii, gently open mouth of zebrafish and insert the soft tip of feeding catheter into mouth of the animal. Stop inserting the catheter once obstruction is felt, where the pharyngeal sphincter leading into intestine of the animal is located. Slowly administer the drugs, avoiding regurgitation through gills. When correctly administered, there should be not leakage of solution from the gills and the peritoneum should visibly distend.
CRITICAL STEP To avoid regurgitation through gills of the animal, ensure the tube is introduced into the pharyngeal sphincter and ensure slow delivery of the solution (Figure 5B).
? TROUBLESHOOTING
Move injected animal into the second plate containing antibiotics-supplemented fish water on the hot plate.
Repeat step vi to ix for all animals that require oral dose of drug.
Image and quantify tumor cell growth for the duration of the experiment as described in Step 52 Option A.VII (epifluorescence) or Option B.V (confocal).
-
Intraperitoneal injection of drugs
Determine an estimate of the average wet weight for the cohort of transplanted zebrafish. Average weight will be used to calculate the amount of drugs that should be given. ▲CRITICAL STEP A maximum of 5 μl can be intraperitoneally injected.
Using a hot plate set to 37 °C, heat two 40 mm petri dishes with 10 ml of antibiotics-supplemented water, and a 40 mm petri dish containing 10 ml of antibiotics-supplemented fish water containing 0.4 mg/ml tricaine. Solutions should be heated 30 min before procedure.
With a 30-gauge needle mount on a 1cc syringe. Pipette 5 μl of drug onto a clean 40mm petri dish, forming a droplet. Load the solution by suctioning the entire droplet into syringe. Ensure no air bubbles are introduced.
Transfer 4–8 animals into the 40 mm petri dish containing antibiotics-supplemented fish water on the hot plate set to 37 °C.
Transfer a single animal into the plate with antibiotics-supplemented fish water containing 0.4 mg/ml tricaine using a plastic spoon. Wait for animal to become antethesized.
When the animal shows minimal movement, place animal onto stage of dissection microscope with the right side facing up (opposite side to the injection site).
Under the dissection microscope, gently insert needle at a location in the cavity so that it does not perturb the engrafted intraperitoneal tumor cells. Like intraperitoneal transplantation of cancer cells, the needle should be inserted between the last 2 pleural ribs to avoid injuring the bone structure of animal (Figure 5).
Slowly inject drug solution into the animal.
Move injected animal into the second plate containing antibiotics-supplemented fish water on the hot plate.
Repeat step iii to vii for all animals that require intraperitoneal injection.
Image and quantify tumor cell growth for the duration of the experiment as described in Step 52 Option A.VII (epifluorescence) or Option B.V (confocal).
-
Figure 5. Clinically relevant dosing of drugs in prkdc−/−, il2rgα−/− zebrafish.
(A) schematic of dosing of drugs by either oral gavage or IP injection as described in Step 53. (B) Representative image of animal being oral gavaged using a 22-gauage oral feeding catheter. Institutional approval from Massachusetts General Hospital was obtained for the experiments shown in this figure.
Troubleshooting
Troubleshooting guidance can be found in Table 1.
Table 1.
Troubleshooting Table
| Step | Problem | Reason | Solution |
|---|---|---|---|
| 19 | Efficiency of gDNA extraction is poor. | Not enough tissue was sampled. | Resect more scales from individual animals. Submerge forceps with scales in NaOH lysis solution for 10 second before agitation. |
| 22 | Verified prkdc−/−, il2rgα−/− are sick. | Verified prkdc−/−, il2rgα−/− shows signs of infection, such as clamped up fins, lethargy, and/or disorientation. | Animals might be infected from genotyping procedure. Either euthanize animals or move them into an isolation tank and supplement animals with 2X recommended dose of antibiotics for a week. Verify the correct dosing and efficacy of antibiotics is being used. |
| 37 | An air bubble was trapped in syringe containing transplant mixture. | Air bubble was introduced during mixing. | Be more cautious and pipette slowly when mixing different components of the transplantation mix. |
| 41 | Low engraftment rate | Some cell lines do not engraft well in our prkdc−/−, il2rgα−/− animals | Increase cell number transplanted from 5 × 105 to 2 × 106 cells per animal. |
| 49 | Excessive bleeding post IP injection | Needle broke pleural ribs of animal, or was inserted too deep into the intraperitoneal cavity causing injurty to internal organs. | Make sure to insert the needle between ribs and stop inserting needle once the bevel has fully entered the cavity. |
| 52 | High mortality post imaging of engrafted cells | Imaging time was too long or tricaine dosing was incorrect. | Limit procedure time to a maximum of 5 min per animal when imaging. Use 0.1 mg/ml tricaine to anesthetize animal. |
| 53 | Drugs are expelled from the gills during oral gavage | The oral gavage catheter was not inserted deep enough into the animal or drugs were delivered to fast. | Make sure the rubber tip of feeding catheter is inserted completely into the animal, with the animal vertically oriented, with mouth and syringe aligned in a straight line. Inject slowly. |
Timing
From our experience, 20 h of hands on time are required to master the protocol outlined here, assuming the investigator has rudimentary experience in working with zebrafish. The procedure can be largely divided into two sections. The first involves sampling, genotyping and acclimating prkdc−/−, il2rgα−/− animals to long-term husbandry at 37 °C (Steps 3–25), which takes 7 d. About 6 h of the first day is spend sampling and genotyping for mutations in prkdc and il2rgα genes. These genotyped animals are then acclimated to 37 °C in a general-purpose water bath, at 1 °C/d. Workload during these six days of acclimation is only about 30 min a day, which includes feeding with brine shrimp three times daily and water change. Experimental transplant animals can be housed either on a 37 °C antibiotics supplemented zebrafish rack or in tanks submerged within a 37 °C water bath indefinitely. The second part of these experiments are the transplantation and follow up imaging studies (Steps 26–53). The transplantation experiments take about 2 h in total, with half of the time spend on prepping cells, and the rest used for the transplant procedure (n=24 animals). Once animals are transplanted with human cancer cells, follow up imaging studies can range from 7 to 28 d, depending on the context of experiment. Procedures, such as imaging of engrafted cells or drug administration can be routinely completed over a duration of 30 min for 10 experimental animals. See below for detailed timing information for each section of the procedure:
Steps 3–19, Scale resection and genomic DNA extraction: 2 h/96 animals
Steps 20–22, Genotyping PCR and restriction enzyme digestion: 4 h/96 animals
Steps 23–25, Acclimation and long-term housing of mutant zebrafish: 6 d/10animals
Steps 26–41, Cell Preparation: 1 h
Step 42, Fish Preparation: 5 min/10animals
Steps 43–50, Cell transplantation: 1 h/10animals
Steps 51–52, Imaging of engrafted cells: 1 h/10animals
Step 53, Clinically relevant dosing of drugs in animal: 30 min/10animals
Anticipated results
Using the described protocol, we expect ~95% engraftment of EGFP-expressing RD cells into prkdc−/−, il2rgα−/− zebrafish. Growth of intraperitoneal transplanted tumor cells can be assessed over time by comparison of fluorescent tumor volume from day 1 and quantified using ImageJ or Imaris. In contrast, growth rates of periocular transplanted cells can be quantified by counting absolute cell number, also using the aforementioned image analysis software (Figure 6). Using our protocol, we have engrafted 15 different cancer types to date, including 20 cell lines and 6 patient derived xenografts. For example, >90% of animals successfully engraft GFP-expressing human RD cells and show significantly increased fluorescence intensity over the course of the 28 day experiment (Figure 6A). Similarly, single cell resolution imaging of engrafted cells was achieved following periocular transplantation of cells and subsequent confocal microscopy (Figure 6B). While engraftment efficiencies vary between cancer types, engraftment efficiency often exceeds 50% for a given tumor type, with exception to gastro-intestinal and reproductive cancers that do not engraft well into prkdc−/−, il2rgα−/− zebrafish [17]. In addition, mutant zebrafish with engrafted cells can be fixed, sectioned and stained for H&E, proliferation (KI67) and apoptosis (TUNEL) markers, as well as clinically relevant IHC diagnostic pathology markers, providing more accurate depiction of tumor proliferation and apoptosis, as well as disease pathology (Figure 6C–H) [38, 42]. Together, both live animal imaging of engrafted cells and subsequent histopathological analysis of tumors on section confirm high proliferation capacity and low overall rates of apoptosis in RD cancer cells grown in prkdc−/−, il2rgα−/− zebrafish.
Figure 6. Engraftment of EGFP-expressing RD cells into prkdc−/−, il2rgα−/− zebrafish.
(A) Representative epifluorescence images of prkdc−/−, il2rgα−/− zebrafish engrafted intraperitoneally with EGFP-expressing RD cells at day 0 (left) and day 28 (right). (B) Representative confocal images of prkdc−/−, il2rgα−/− zebrafish engrafted periocularly with EGFP-expressing RD cells at day 0 (left) and day 21 (right). (C, D) Hematoxylin and eosin staining of intraperitoneally (C) and periocularly (D) engrafted EGFP-expressing RD cells. (E-H) Immunohistochemistry staining of intraperitoneally engrafted EGFP-expressing RD cells stained with proliferation marker KI67 (E), apoptosis marker TUNEL (F), as well as clinically relevant histopathology marker MYOD1 (G) and Desmin (H). Scale bar equals 0.5cm (A), 200 μm (B), 5mm (C, D), and 1mm (E-H). Institutional approval from Massachusetts General Hospital was obtained for the experiments shown in this figure.
Acknowledgements
We thank the MGH histopathology core, as well as the MGH Cancer Center Molecular Pathology Confocal Core for their technical help. We thank Drs. Qin Tang, John Moore, and Jessica McCann for their advice and intellectual support for the initial development of the protocol. This work is supported by NIH grants R24OD016761 (D.M.L.), R01CA154923 (D.M.L.), R01CA215118, (D.M.L.), R01CA211734 (D.M.L.), R01CA226926 (D.M.L.); the Liddy Shriver Sarcoma Initiative (D.M.L.); the MGH Research Scholars Program (D.M.L.); the Tosteson & Fund for Medical Discovery Fellowship from MGH (C.Y.); and the Alex’s Lemonade Stand Foundation Young Investigator Award (C.Y.).
Footnotes
Declaration of interest
The General Hospital Corporation has a patent pending on the creation and use of immune-compromised zebrafish for engraftment of human cancers. D.M.L. is also a co-inventor on patent application number USSN 14/903,940.
Data Availability
All data generated in this study are included in this published article. The original data files are available upon request from the corresponding author.
Related links
Key reference using this protocol:
1. Yan C, et al., Cell. 2019 Jun 13;177(7):1903-1914.e14. https://www.sciencedirect.com/science/article/pii/S0092867419303903
References
- 1.Bakhoum SF, et al. , Chromosomal instability drives metastasis through a cytosolic DNA response. Nature, 2018. 553(7689): p. 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stewart E, et al. , Orthotopic patient-derived xenografts of paediatric solid tumours. Nature, 2017. 549(7670): p. 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian L, et al. , Mutual regulation of tumour vessel normalization and immunostimulatory reprogramming. Nature, 2017. 544(7649): p. 250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das R, et al. , Microenvironment-dependent growth of preneoplastic and malignant plasma cells in humanized mice. Nat Med, 2016. 22(11): p. 1351–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goyama S, Wunderlich M, and Mulloy JC, Xenograft models for normal and malignant stem cells. Blood, 2015. 125(17): p. 2630–40. [DOI] [PubMed] [Google Scholar]
- 6.Day CP, Merlino G, and Van Dyke T, Preclinical mouse cancer models: a maze of opportunities and challenges. Cell, 2015. 163(1): p. 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito M, et al. , NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood, 2002. 100(9): p. 3175–82. [DOI] [PubMed] [Google Scholar]
- 8.Yu M, et al. , Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science, 2014. 345(6193): p. 216–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drapkin BJ, et al. , Genomic and Functional Fidelity of Small Cell Lung Cancer Patient-Derived Xenografts. Cancer Discov, 2018. 8(5): p. 600–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes MN, et al. , Vangl2/RhoA Signaling Pathway Regulates Stem Cell Self-Renewal Programs and Growth in Rhabdomyosarcoma. Cell Stem Cell, 2018. 22(3): p. 414–427 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellenbroek SI and van Rheenen J, Imaging hallmarks of cancer in living mice. Nat Rev Cancer, 2014. 14(6): p. 406–18. [DOI] [PubMed] [Google Scholar]
- 12.Fior R, et al. , Single-cell functional and chemosensitive profiling of combinatorial colorectal therapy in zebrafish xenografts. Proc Natl Acad Sci U S A, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman A, et al. , Heterogeneous tumor subpopulations cooperate to drive invasion. Cell Rep, 2014. 8(3): p. 688–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welker AM, et al. , Changes in tumor cell heterogeneity after chemotherapy treatment in a xenograft model of glioblastoma. Neuroscience, 2017. 356: p. 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novoa B and Figueras A, Zebrafish: model for the study of inflammation and the innate immune response to infectious diseases. Adv Exp Med Biol, 2012. 946: p. 253–75. [DOI] [PubMed] [Google Scholar]
- 16.Cabezas-Sainz P, et al. , Improving zebrafish embryo xenotransplantation conditions by increasing incubation temperature and establishing a proliferation index with ZFtool. BMC Cancer, 2018. 18(1): p. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan C, et al. , Visualizing Engrafted Human Cancer and Therapy Responses in Immunodeficient Zebrafish. Cell, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pereiro P, et al. , Zebrafish Nk-lysins: First insights about their cellular and functional diversification. Dev Comp Immunol, 2015. 51(1): p. 148–59. [DOI] [PubMed] [Google Scholar]
- 19.Page DM, et al. , An evolutionarily conserved program of B-cell development and activation in zebrafish. Blood, 2013. 122(8): p. e1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida N, Frickel EM, and Mostowy S, Macrophage-Microbe Interactions: Lessons from the Zebrafish Model. Front Immunol, 2017. 8: p. 1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meeker ND, et al. , Method for isolation of PCR-ready genomic DNA from zebrafish tissues. Biotechniques, 2007. 43(5): p. 610, 612, 614. [DOI] [PubMed] [Google Scholar]
- 22.Botstein D, et al. , Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet, 1980. 32(3): p. 314–31. [PMC free article] [PubMed] [Google Scholar]
- 23.Samaee SM, Seyedin S, and Varga ZM, An Affordable Intraperitoneal Injection Setup for Juvenile and Adult Zebrafish. Zebrafish, 2017. 14(1): p. 77–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dang M, et al. , Long-term drug administration in the adult zebrafish using oral gavage for cancer preclinical studies. Dis Model Mech, 2016. 9(7): p. 811–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collymore C, Rasmussen S, and Tolwani RJ, Gavaging adult zebrafish. J Vis Exp, 2013(78). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White RM, et al. , Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell, 2008. 2(2): p. 183–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westerfield M, The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). 5th Edition. ed. 2007: University of Oregon Press, Eugene.. [Google Scholar]
- 28.Ignatius MS and Langenau DM, Fluorescent imaging of cancer in zebrafish. Methods Cell Biol, 2011. 105: p. 437–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Q, et al. , Imaging tumour cell heterogeneity following cell transplantation into optically clear immune-deficient zebrafish. Nat Commun, 2016. 7: p. 10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore JC, et al. , T cell immune deficiency in zap70 mutant zebrafish. Mol Cell Biol, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang Q, et al. , Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. J Exp Med, 2017. 214(10): p. 2875–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sire JY and Akimenko MA, Scale development in fish: a review, with description of sonic hedgehog (shh) expression in the zebrafish (Danio rerio). Int J Dev Biol, 2004. 48(2–3): p. 233–47. [PubMed] [Google Scholar]
- 33.Scott GR and Johnston IA, Temperature during embryonic development has persistent effects on thermal acclimation capacity in zebrafish. Proc Natl Acad Sci U S A, 2012. 109(35): p. 14247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu W, et al. , Characterization of prostate cancer cell progression in zebrafish xenograft model. Int J Oncol, 2018. 52(1): p. 252–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalamida D, et al. , Fever-range hyperthermia vs. hypothermia effect on cancer cell viability, proliferation and HSP90 expression. PLoS One, 2015. 10(1): p. e0116021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu S, et al. , Culture at a Higher Temperature Mildly Inhibits Cancer Cell Growth but Enhances Chemotherapeutic Effects by Inhibiting Cell-Cell Collaboration. PLoS One, 2015. 10(10): p. e0137042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mantso T, et al. , Hyperthermia induces therapeutic effectiveness and potentiates adjuvant therapy with non-targeted and targeted drugs in an in vitro model of human malignant melanoma. Sci Rep, 2018. 8(1): p. 10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore JC, et al. , Single-cell imaging of normal and malignant cell engraftment into optically clear prkdc-null SCID zebrafish. J Exp Med, 2016. 213(12): p. 2575–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Rooijen N and Hendrikx E, Liposomes for specific depletion of macrophages from organs and tissues. Methods Mol Biol, 2010. 605: p. 189–203. [DOI] [PubMed] [Google Scholar]
- 40.McAllister RM, et al. , Cultivation in vitro of cells derived from a human rhabdomyosarcoma. Cancer, 1969. 24(3): p. 520–6. [DOI] [PubMed] [Google Scholar]
- 41.Strober W, Trypan blue exclusion test of cell viability. Curr Protoc Immunol, 2001. Appendix 3: p. Appendix 3B. [DOI] [PubMed] [Google Scholar]
- 42.Tang Q, et al. , Optimized cell transplantation using adult rag2 mutant zebrafish. Nat Methods, 2014. 11(8): p. 821–4. [DOI] [PMC free article] [PubMed] [Google Scholar]