Abstract
Negative genetic regulators of phenotypic heterogeneity, or phenotypic capacitors/stabilizers, elevate population average fitness by limiting deviation from the optimal phenotype and increase the efficacy of natural selection by enhancing the phenotypic differences among genotypes. Stabilizers can presumably be switched off to release phenotypic heterogeneity in the face of extreme or fluctuating environments to ensure population survival. This task could, however, also be achieved by positive genetic regulators of phenotypic heterogeneity, or “phenotypic diversifiers,” as shown by recently reported evidence that a bacterial divisome factor enhances antibiotic resistance. We hypothesized that such active creation of phenotypic heterogeneity by diversifiers, which is functionally independent of stabilizers, is more common than previously recognized. Using morphological phenotypic data from 4,718 single-gene knockout strains of Saccharomyces cerevisiae, we systematically identified 324 stabilizers and 160 diversifiers and constructed a bipartite network between these genes and the morphological traits they control. Further analyses showed that, compared with stabilizers, diversifiers tended to be weaker and more promiscuous (regulating more traits) regulators targeting traits unrelated to fitness. Moreover, there is a general division of labor between stabilizers and diversifiers. Finally, by incorporating NCI-60 human cancer cell line anticancer drug screening data, we found that human one-to-one orthologs of yeast diversifiers/stabilizers likely regulate the anticancer drug resistance of human cancer cell lines, suggesting that these orthologs are potential targets for auxiliary treatments. Our study therefore highlights stabilizers and diversifiers as the genetic regulators for the bidirectional control of phenotypic heterogeneity as well as their distinct evolutionary roles and functional independence.
Keywords: phenotypic heterogeneity, morphological trait, evolutionary capacitance, drug resistance, evolutionary genomics
Introduction
Most phenotypic traits vary among individuals within a genetically identical population. The level of such phenotypic heterogeneity is crucial to biological evolution. On the one hand, low heterogeneity may increase the efficacy of natural selection via better phenotypic separation between populations of different genotypes (Wu et al. 2009). On the other hand, high heterogeneity may be deleterious, as frequent deviation from an optimal phenotype should lower the average fitness of the population (Wang and Zhang 2011), but it may also facilitate evolution by granting a fitness advantage to subpopulations with adaptive phenotypes in changing environments (Sharma et al. 2010). These adaptive phenotypes can eventually be (epi-)genetically fixed or can simply keep the population alive to allow adaptation (Sharma et al. 2010; Schmutzer and Wagner 2020). Moreover, phenotypic heterogeneity has been shown to have broad medical relevance, such as implications for enhancement of antibiotic resistance in bacteria (Rego et al. 2017) and drug resistance in cancer (Sharma et al. 2010; Shaffer et al. 2017).
Given the critical role of phenotypic heterogeneity, it is unsurprising that genetic regulatory mechanisms for phenotypic heterogeneity exist. These mechanisms include, for example, chromatin state modifiers that regulate gene expression heterogeneity (Sharma et al. 2010), chaperones that suppress protein misfolding (Rutherford and Lindquist 1998), and cyclin-dependent kinases that maintain cell size homeostasis (Patterson et al. 2019). One of the best known regulators of phenotypic heterogeneity is the molecular chaperone Hsp90, which suppresses the phenotypic consequences of protein misfolding by facilitating correct folding and thereby reduces phenotypic heterogeneity (Rutherford and Lindquist 1998; Queitsch et al. 2002). Because genes such as Hsp90 “store” phenotypic variation and “release” it upon mutation or pharmacologically induced functional impairment (Rutherford and Lindquist 1998), they have been termed “phenotypic capacitors,” as they are reminiscent of electrical capacitors that store and release electric energy (Rutherford and Lindquist 1998). To date, multiple efforts have been made to identify and characterize the properties of phenotypic capacitors (Levy and Siegal 2008; Wang et al. 2011).
Nevertheless, much less is known about “phenotypic diversifiers,” which function to actively increase phenotypic heterogeneity, whereas phenotypic capacitors can increase phenotypic heterogeneity only when they are functionally impaired. To better differentiate them, we, respectively, refer to the positive and negative regulators of phenotypic heterogeneity as phenotypic diversifiers and stabilizers (instead of capacitors). The current underascertainment of phenotypic diversifiers may be due to the common presumption of a narrow optimal phenotype, such that the phenotypic variation created by phenotypic diversifiers (beyond the intrinsic and environmental variation) will most likely be detrimental for the average fitness of the population (Wagner 2013).
In this context, a recent study demonstrated that a mycobacterial divisome factor, LamA, is a positive regulator of asymmetric polar growth and, therefore, of phenotypic heterogeneity in Mycobacterium smegmatis (Rego et al. 2017). In that study, deletion of lamA sensitized a mycobacterial population to rifampicin, linking drug resistance to phenotypic heterogeneity and phenotypic diversifiers (Rego et al. 2017). Following this demonstration of the existence of phenotypic diversifiers, several important questions have arisen. For example, do other phenotypic diversifiers exist? What are their relationships with phenotypes and phenotypic stabilizers? How did they evolve? Are their links with drug resistance generally applicable across species? These questions may be answered by systematic screening of phenotypic diversifiers and investigation of their roles in cellular drug resistance.
In this study, we took advantage of existing morphological phenotypic data from 4,718 single-gene knockout strains of Saccharomyces cerevisiae (Ohya et al. 2005). Using this data set, we systematically identified phenotypic stabilizers and diversifiers by, respectively, searching for significant increases and decreases in phenotypic heterogeneity upon gene deletion. We found that diversifiers resemble stabilizers with respect to evolutionary conservation and protein indispensability but differ from stabilizers in that they regulate more traits with weaker effects. We further confirmed the distinct roles of stabilizers and diversifiers by revealing a general “division of labor” between the two types of regulators. Finally, we confirmed the relevance of stabilizers and diversifiers in cancer drug resistance by analyzing their one-to-one orthologs in humans using the NCI-60 data set. Our results revealed important properties of phenotypic diversifiers/stabilizers, highlighted their functional independence and distinct evolutionary roles, and suggested the general relevance of these diversifiers/stabilizers in drug resistance.
Results
Systematic Identification of Phenotypic Diversifiers of Morphological Traits in Yeast
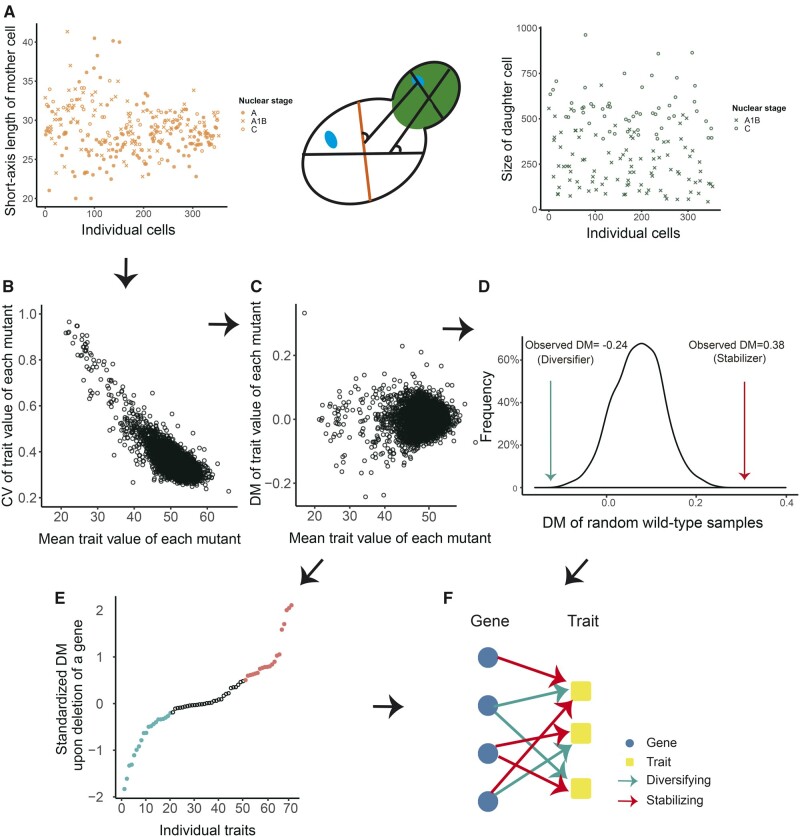
A data set on quantitative morphological phenotypes in 4,718 mutant haploid strains of S. cerevisiae was previously used to identify putative phenotypic stabilizers (Ohya et al. 2005; Levy and Siegal 2008). In each strain, one nonessential gene was deleted, and various morphological traits were measured for an average of >100 cells in each of three cell cycle stages (fig. 1A). Using this data set and previously described methods, we identified phenotypic diversifiers by searching for genes whose deletion significantly suppressed phenotypic heterogeneity. Briefly, for each trait and each strain, we calculated the coefficient of variation (CV) (fig. 1B) and its deviation from the running median CV of strains with similar mean trait values (DM) (fig. 1C; see Materials and Methods), thereby controlling for confounding effects of the mean trait value on the CV (supplementary fig. S1 and supplementary table S1, Supplementary Material online, for the 4,718 × 220 raw DMs). The DMs of each trait from different strains were then standardized to allow comparison across phenotypes.
Fig. 1.
Systematic identification of phenotypic diversifiers and stabilizers of morphological traits in yeast. (A) Morphological traits measured for hundreds of cells were used here to quantify phenotypic heterogeneity. Two traits, the short axis length of mother cell (red) and size of daughter cell (green), are shown as examples. (B) The CVs of individual traits in mutants (with gene deletion), which were clearly dependent on the mean trait values, are shown. (C) The deviation from the running median of CV (DM) was used to control for the confounding effects of the mean. (D) Randomly sampled wild-type cells were used to estimate a null distribution of the DM of a trait (black curve). The observed DM for a gene was compared with this null distribution. If it was significantly increased, the gene was deemed a stabilizer of the trait. Conversely, if it was significantly decreased, the gene was deemed a diversifier of the trait. (E) The standardized DMs are shown for all traits for a gene. The top/bottom 20 standardized DMs (red/cyan dots) were summed to estimate the phenotypic potential of the candidate stabilizer/diversifier, which was used to control the FDR of the putative stabilizer/diversifier (see Materials and Methods). (F) Schematic diagram of the resulting bipartite network between genes (blue circles) and traits (yellow squares), with green/red arrows indicating diversification/stabilization relationship between genes and traits.
Based on a matrix of the standardized DMs, we extracted a nonredundant list of phenotypes consisting of 70 representative morphological traits, minimizing interdependence between phenotypes (see Materials and Methods). To identify putative regulatory relationships between these genes and morphological traits, we compared the observed DM of a yeast knockout (YKO) strain for a particular trait with 1,000 mock DMs calculated from random samples of wild-type cells (fig. 1D; see Materials and Methods). If the observed DM was significantly lower (higher) than the mock DMs (P < 0.001), the gene deleted in the YKO strain was deemed to have a diversifying (stabilizing) effect over the trait. To identify putative diversifiers and stabilizers, we averaged the bottom and top 20 (of 70) standardized DMs for each gene (fig. 1E). The resulting score, which has previously been termed “phenotypic potential” was compared with its random expectation (see Materials and Methods) to assess the false discovery rate (FDR). For an FDR of 12%, we identified 160 diversifiers and 324 stabilizers with significantly lower and higher phenotypic potentials than expected, respectively, among which 27 genes had dual roles (supplementary table S2, Supplementary Material online). Finally, we constructed a bipartite network including 457 genes and 70 morphological traits, with the links in the network representing the diversification or stabilization relationships between the genes and traits (fig. 1F, see supplementary table S3, Supplementary Material online, for the complete network). In support of the reliability of our computational pipeline, we found that the stabilizers were likely to be hubs in protein–protein interaction networks (supplementary fig. S2, Supplementary Material online), which recapitulates a major finding in a previous report (Levy and Siegal 2008).
Significant Functional Genomic Differences between Diversifiers and Stabilizers
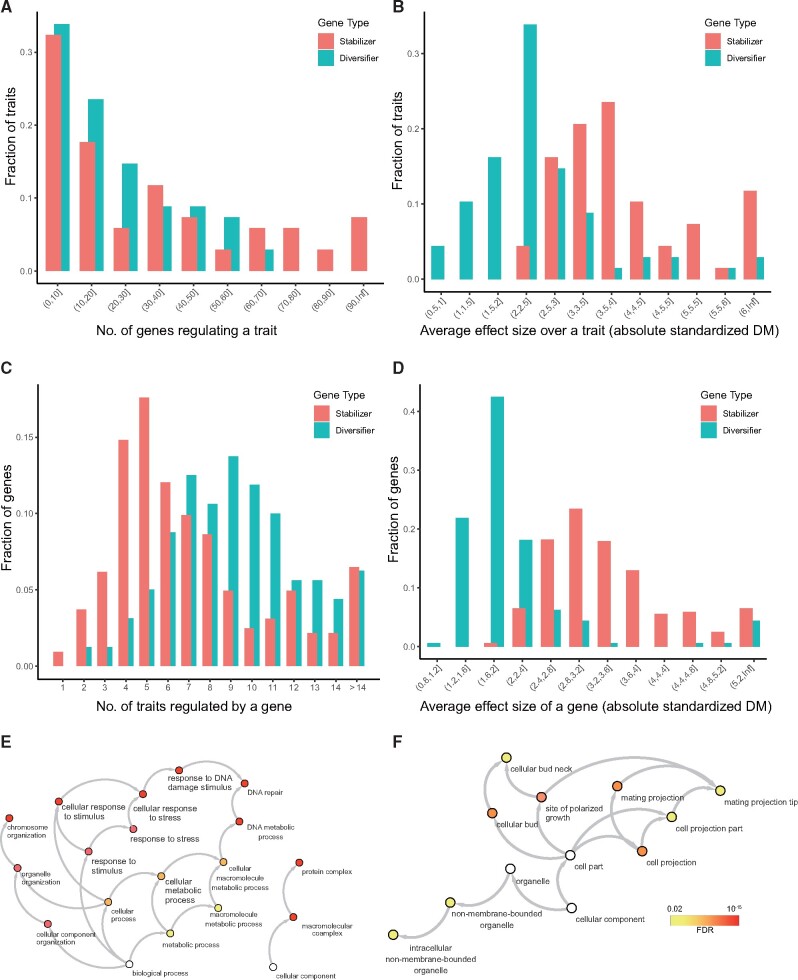
We next investigated the structure of the aforementioned bipartite network of relationships between phenotypic diversifiers/stabilizers and morphological traits. We found that each trait was diversified by an average of 22 diversifiers and stabilized by an average of 17 stabilizers, which were not significantly different from each other (P = 0.131, Mann–Whitney U test, fig. 2A). However, in terms of the effect size, the absolute average DM of the diversification effect mediated by diversifiers equaled 2.32, which was significantly lower than the average absolute DM of 3.67 found for the stabilization effect mediated by stabilizers (P < 10−12, Mann–Whitney U test, fig. 2B). From a gene-centric perspective, we found that the median number of traits stabilized by a stabilizer was 6, whereas the median number of traits diversified by a diversifier was 9 (P < 10−12, Mann–Whitney U test, fig. 2C). However, the magnitude of the average diversifier-mediated increase in phenotypic heterogeneity of a trait was only ∼58% of that of the stabilizer-mediated decrease in phenotypic heterogeneity (average absolute DM of 1.87 vs. 3.24, P < 10−49 by Mann–Whitney U test, fig. 2D). These findings suggest that phenotypic diversifiers tend to be weaker but more promiscuous regulators of phenotypic heterogeneity than stabilizers. Note that the weaker effect of diversifiers may alternatively be explained by a biologically or technically bounded minimum of phenotypic heterogeneity.
Fig. 2.
Functional properties of diversifier and stabilizer genes. Histogram showing the distribution of the (A) number of genes regulating a trait, (B) average effect size (absolute standardized DM) of a stabilizer/diversifier for a trait, (C) number of traits regulated by a stabilizer/diversifier, and (D) average effect size (absolute standardized DM) of a stabilizer/diversifier. (E) GO functional enrichment results for the 324 stabilizers and 160 diversifiers (F).
We also examined the functional enrichment of 160 diversifiers for Gene Ontology terms. The same type of analysis was performed using 324 stabilizers for comparison. Consistent with previous reports (Levy and Siegal 2008), we found that the stabilizers were enriched for several terms broadly related to DNA stability (fig. 2E). The 160 diversifiers were enriched for bud growth and cell projection (fig. 2F), which was also largely consistent with the aforementioned LamA, a diversifier identified in bacteria (Rego et al. 2017). The biological relevance of the identified diversifiers was further supported by some specific genes with known functions. For example, one diversifier identified by our pipeline was RGA1, which encodes an activating protein for CDC42 (a GTPase required for polarity establishment and bud emergence). It has been found that RGA1 can prevent rebudding at old division sites, especially before G1 phase (Miller et al. 2017). Consistent with this known function, our pipeline identified RGA1 as a diversifier for trait “C106-A1B: bud direction.” Another example was KEL1 that controls actin cable assembly. Loss of KEL1 resulted in long, bent, and hyper-stable actin cables (Gould et al. 2014), as well as an elongated cell shape (Philips and Herskowitz 1998), which should effectively homogenize the ratio of the long and short axis of the cell. Consistent with this known function of KEL1, we found it to be a diversifier for traits such as “C114-C: bud axis ratio” and “C115-A1B: mother axis ratio.”
Differential Evolutionary Profiles of Diversifiers and Stabilizers
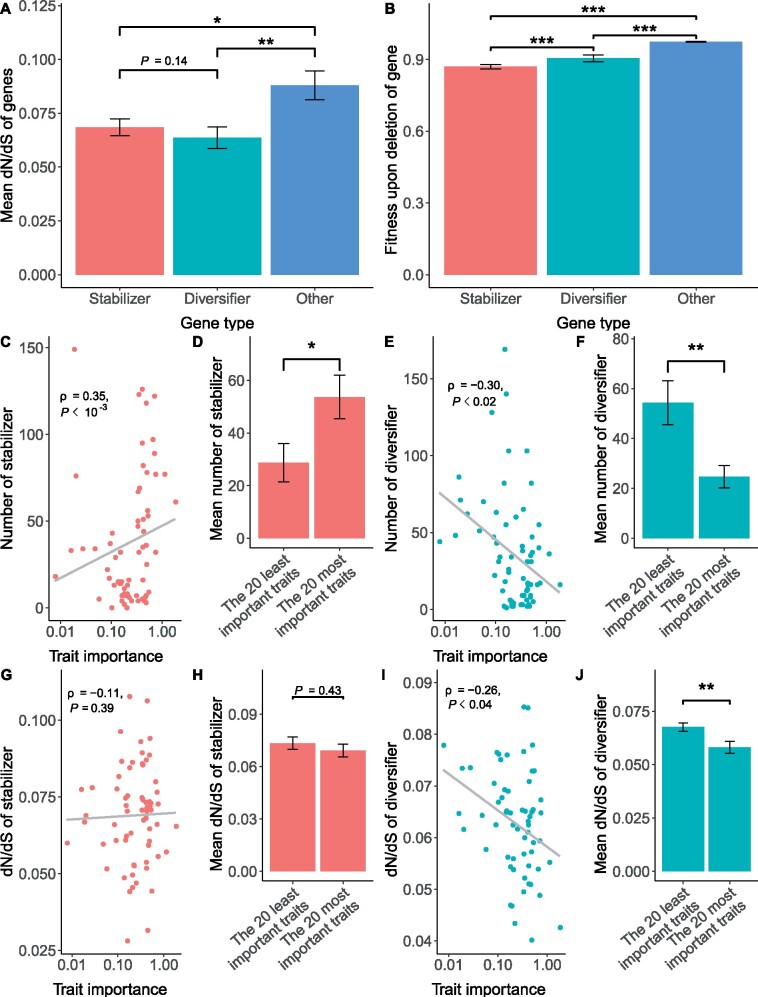
Given the critical importance of phenotypic heterogeneity, the significant differences in functional genomic properties between diversifiers and stabilizers prompted a closer assessment of the evolutionary forces underlying them. Accordingly, we assessed whether diversifiers and stabilizers are evolutionarily more conserved than other nonessential genes. Here, evolutionary conservation was estimated inversely by the ratio between the number of nonsynonymous substitutions per nonsynonymous site (dN) and the number of synonymous substitutions per synonymous site (dS) calculated from one-to-one orthologs between S. cerevisiae and five different yeast species (see Materials and Methods). We found that both diversifiers and stabilizers were more conserved than other nonessential genes (P < 0.05, one-tailed Wilcoxon rank-sum test, fig. 3A), suggesting that the ability to generate phenotypic heterogeneity is maintained by purifying selection. We further examined the effects of diversifier/stabilizer deletion on fitness and found that deletion of both diversifiers and stabilizers tended to be more harmful to cells than deletion of other nonessential genes (P < 10−12 and 10−40 for diversifiers and stabilizers, respectively, fig. 3B). Interestingly, deletion of diversifiers seemed to be less deleterious than deletion of stabilizers (P < 10−4, fig. 3B). Note that the disparity between fitness effect (fig. 3B) and sequence conservation (dN/dS, fig. 3A) could also be because both fitness and morphology were measured in the constant lab environment, whereas sequence conservation was the result of evolution in the fluctuating (e.g., temperature, humidity, etc.) natural environment. The natural environment is different from the lab, and the frequent environmental change should further intensity the purifying selection for diversifiers, thereby lowering their dN/dS. Nevertheless, these results suggest that although diversifiers are more dispensable than stabilizers in the constant laboratory environments, their functions are at least equally (if not more strongly) maintained by purifying selection.
Fig. 3.
Differential evolutionary profiles of diversifiers and stabilizers. (A) Evolutionary conservation of a gene was estimated inversely by the ratio between the number of nonsynonymous substitutions per nonsynonymous site (dN) and the number of synonymous substitutions per synonymous site (dS) calculated from one-to-one orthologs between Saccharomyces cerevisiae and five other yeast species (see Materials and Methods). The average dN/dS values are shown for stabilizers, diversifiers, and other yeast genes. (B) The fitness upon deletion of the gene is shown for stabilizers, diversifiers, and other yeast genes. In both (A) and (B), error bars indicate standard errors, and one-tailed Wilcoxon rank-sum tests were carried out to assess the statistical significance of differences, with the P values indicated. *P < 0.05, **P < 0.01, ***P < 0.001. (C–J) The importance of each trait (x axis) was estimated as the decrease in fitness per unit of change in the trait value relative to the fitness of the wild-type strains (see Materials and Methods). The important traits tended to be regulated by more stabilizers (C and D) with slightly lower average dN/dS values (G and H). In contrast, the important traits tended to be regulated by fewer diversifiers (E and F) with lower average dN/dS values (I and J). In (C/E/G/I), the gray line indicates the fitted linear model of each panel. In (D/F/H/J), the error bars indicate standard errors, and two-tailed Wilcoxon rank-sum tests were carried out to assess the statistical significance of differences, with the P values indicated. *P < 0.05, **P < 0.01.
To further contrast the evolutionary forces underlying diversifiers and stabilizers, we separately analyzed the diversifiers/stabilizers for each of the 70 traits. In particular, we estimated the importance of each trait by the decrease in fitness per unit of change in trait value relative to the fitness of the wild-type strains (see Materials and Methods). We found that important traits tended to have more stabilizers (fig. 3C and D) but fewer diversifiers (fig. 3E and F) than unimportant traits, although the regulatory effects per trait did not change with trait importance (data not shown). In other words, suppression of phenotypic heterogeneity was stronger for important traits than for unimportant traits. This is not unexpected, as phenotypic heterogeneity of important traits should be suppressed more strongly than that of unimportant traits due to it deleterious effect on population average fitness (Wang and Zhang 2011; Metzger et al. 2015). However, we also found that the evolutionary conservation of stabilizers for important traits was not significantly higher than that for unimportant traits (fig. 3G and H), whereas diversifiers of important traits tended to be more conserved than those of unimportant traits (fig. 3I and J). This result suggests that diversifiers are under stronger purifying selection than stabilizers, especially when they regulate traits that are tightly coupled with organismal fitness.
Collectively, the above results suggest different evolutionary scenarios for stabilizers versus diversifiers. With respect to stabilizers, more stabilizers are recruited to maintain homeostasis of important traits than to maintain homeostasis of unimportant traits, but the strength of purifying selection exerted on each stabilizer is not correlated with trait importance. With respect to diversifiers, although fewer diversifiers act on traits that are important (in laboratory environments) than on traits that are unimportant, the diversifiers are individually highly constrained by purifying selection, especially for diversifiers of important traits. These differential evolutionary profiles between stabilizers and diversifiers, along with the findings that diversifiers are generally weaker but more promiscuous regulators than stabilizers, suggest the existence of fundamental differences between these two types of regulators and highlight the evolutionary significance of diversifiers.
Functional “Division of Labor” between Diversifiers and Stabilizers
As diversifiers and stabilizers are similar to two faces of the same coin, we sought to investigate the relationship between their regulatory capacities. In a simple mechanistic model in which stabilizers or diversifiers regulate the allocation of core resources to buffer or diversify various biological processes, respectively, such as chaperone activity in protein folding, one would expect that the resulting reallocation of the core resources should cause reciprocal changes in opposite directions in heterogeneity of some other phenotypes (the left half of fig. 4A). For example, recruitment of chaperones to support the folding of one protein should lead to increased misfolding of another protein. In contrary to such “resource allocation” model, it is also possible that both diversifiers and stabilizers are independent regulators of phenotypic heterogeneity, a scenario we called the division of labor model (the right half of fig. 4A). The applicability of these two models could be tested by comparing the number of traits stabilized by a gene with that diversified by the same gene (fig. 4A), which gave rise to a significant negative correlation (Spearman’s ρ = −0.42, P < 10−20, fig. 4B). This result suggests a lack of reciprocal changes in phenotypic heterogeneity, at least for the stabilizers and diversifiers examined here.
Fig. 4.
Division of labor between diversifiers and stabilizers. (A) Schematic diagram showing the different predictions for the resource allocation model versus the division of labor model. (B) The number of traits diversified by a gene (x axis) was compared with the number of traits stabilized by the same gene (y axis). Spearman’s rank correlation coefficient is indicated. The red line indicates the fitted linear model. (C) The anticorrelation observed in (B), which is indicated by the red arrow, was compared with the expectations (black and orange histograms) estimated in two control experiments, each involving 1,000 correlations derived from randomly shuffled data sets (see Materials and Methods). (D) GO functional enrichment results for the 27 dual-role genes (i.e., the genes identified as being both putative stabilizers and putative diversifiers).
To exclude the possibility that this correlation was a computational artifact, we performed two control experiments. In the first experiment, we shuffled the links (representing stabilizing or diversifying regulation) in the bipartite network between diversifiers/stabilizers and morphological traits while maintaining the number of links attached to each gene and each trait. In the second experiment, we shuffled all the raw DMs in the original matrix (4,718 strains × 220 morphological traits) and recalculated the whole bipartite network. Both control experiments were repeated 1,000 times. For each control experiment, we assessed the anticorrelation between the number of traits stabilized by a gene and that diversified by the same gene. We found that the observed anticorrelation (Spearman’s ρ = −0.42) was always stronger than those observed in the control experiments (fig. 4C), thereby confirming the validity of our findings.
Notwithstanding, there was a minor group of 27 genes with dual roles (identified as both putative diversifiers and stabilizers), which appeared consistent with the resource allocation model. We examined the functional annotations of the proteins encoded by these genes and found that they were enriched for proteins within macromolecular complexes and the molecular function of binding (fig. 4D). We therefore speculated that these dual-role regulators exert their functions on phenotypic heterogeneity indirectly by recruiting other direct regulators to specific pathways, thereby enhancing the phenotypic heterogeneity of some traits while suppressing that of others. Indeed, the known functions of some of the dual-role genes are compatible with our speculation base regarding the resource allocation model. For example, several dual-role genes, such as BFR1, POP2, and SCP160, seemed to be diversifying for bud/daughter cell morphology (e.g., trait “C113_C: distance from bud tip to mother cell’s long axis along bud direction on nucleus C”), but stabilizing for mother morphology (e.g., trait “D108_C: distance from neck to mother cell’s nucleus in nucleus C” or “D103_C: distance from nuclear center to mother tip in nucleus C”). These genes are known to be associated with the recruitment of mRNA into or formation of P-bodies (Teixeira and Parker 2007; Simpson et al. 2014; Weidner et al. 2014). On the one hand, the formation of P-bodies enhances cellular viability and suppresses morphological abnormalities (Luo et al. 2018), thereby stabilizing morphological phenotypes of mother cells. On the other hand, as P-body is associated with enlarged bud/daughter cells, the unidirectional P-body transportation to daughter cells (Garmendia-Torres et al. 2014) may have contributed to the heterogeneity of bud/daughter cell morphology. Regardless of their specific functions, the observation that dual-role regulators were a minority among genes that regulate phenotypic heterogeneity again suggested that there was a division of labor between diversifiers and stabilizers.
Roles of Diversifiers and Stabilizers in Cancer Drug Resistance
A recent study has connected lamA, a positive regulator of phenotypic heterogeneity in bacteria, to antibiotic resistance (Rego et al. 2017). We, therefore, investigated whether a similar association with drug resistance is generally applicable to other regulators of phenotypic heterogeneity, such as the diversifiers and stabilizers identified in this study. Theoretically, like the case of lamA, a diversifier should increase drug resistance because enhanced phenotypic heterogeneity increases the likelihood of survival of a small population of cells via biological bet hedging (Slatkin 1974). In contrast, a stabilizer should homogenize a population of cells, thereby increasing the likelihood of the elimination of the whole population by a single drug.
Assuming functional conservation between human and yeast one-to-one orthologs (Glover et al. 2019; Stamboulian et al. 2020), we aimed to assess the roles of diversifiers/stabilizers in drug resistance of cancer cells. The relevance of cancer in this context is supported by a previous discovery that carcinogenesis exhibits strong signals of reverse evolution from multicellularity to unicellularity (Chen et al. 2015), whereas yeast is the best studied unicellular eukaryote. In addition, recent studies have suggested the certain morphological phenotypes of breast (Sirois et al. 2019) and colon (Pasqualato et al. 2012) cancer cells are associated with chemoresistance. We used the NCI-60 data set produced via assay of the responses of 60 typical cancer cell lines with known transcriptional profiles to 21,121 types of natural chemicals that are anticancer drug candidates. In this data set, the response of a cancer cell line to a chemical is represented by the concentration that was lethal to 50% of the cells (LC50). We searched for the one-to-one human orthologs of the 160 diversifiers and 324 stabilizers identified in yeast and identified 29 and 46 genes (among which 28 and 42 genes have NCI-60 data), respectively; these genes are hereinafter referred to as human diversifiers and stabilizers, respectively, for simplicity. Under the assumption of functional conservation, the hypothesized association between phenotypic diversifiers and drug resistance should predict that highly expressed human stabilizers will reduce cellular resistance to (or strengthen the cytotoxicity of) drug candidates by decreasing the phenotypic heterogeneity of a cancer cell population. Conversely, highly expressed human diversifiers should enhance cellular resistance to (or undermine the cytotoxicity of) drug candidates by increasing phenotypic heterogeneity of a cancer cell population.
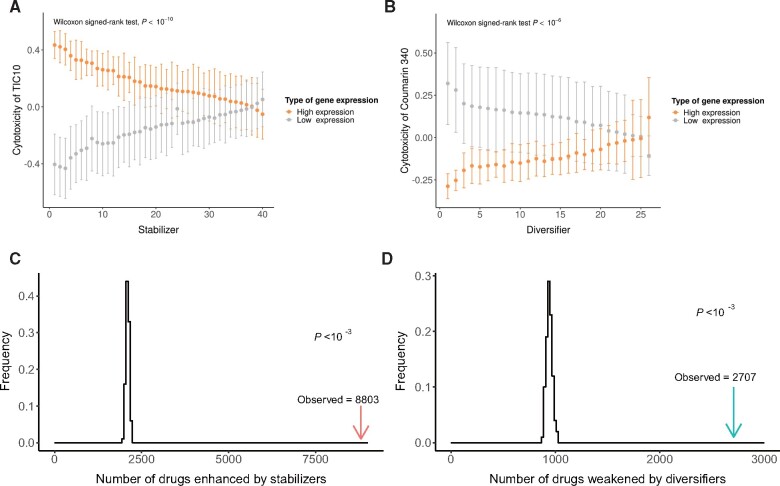
We first tested the function of human diversifiers (stabilizers) as a group rather than individually (fig. 5A and B, see Materials and Methods), as the latter analysis has been deemed less powerful than the former for the NCI-60 data (Covell 2012). Consistent with our predictions, we found that the human stabilizers strengthened the cytotoxicity of 8,803 drug candidates (fig. 5C), whereas the human diversifiers undermined the cytotoxicity of 2,707 drug candidates (fig. 5D). Both numbers were significantly higher than the random expectations for groups of the same numbers of human genes (P < 0.001, permutation test), suggesting a general role for regulators of phenotypic heterogeneity in cancer cell drug resistance.
Fig. 5.
Roles of diversifiers and stabilizers in cancer drug resistance. (A) The average LC50 of a drug candidate (TIC10) in the 30 cancer cell lines in which a specific human stabilizer had the HE-LC50 level (orange dots) was compared with that in the 30 cell lines in which the stabilizer had the LE-LC50 level (gray dots) by the Wilcoxon signed-rank test. The error bars represent the standard errors, and the one-tailed P value (high expression of the focal gene will increase the cytotoxicity of the drug candidate) is indicated. (B) Same as (A), except that diversifiers were tested for another drug candidate (Coumarin 340), and the opposite one-tailed P value (high expression of the focal gene will decrease the cytotoxicity of the drug candidate) is indicated. (C) A total of 1,000 random sets of human genes were constructed. Each set contained the same number of genes as the number of putative human stabilizers and was tested for functional association with all drug candidates by the method shown in (A) (see also Materials and Methods). The distribution of the number of drugs whose cytotoxicity was enhanced by these gene sets is shown in a histogram (black line), which was significantly different from that of the putative human stabilizers (red arrow). (D) A total of 1,000 random sets of human genes were constructed. Each set contained the same number of genes as the number of putative human diversifiers and was tested for functional association with all drug candidates by the method shown in (B) (see also Materials and Methods). The distribution of the number of drugs whose cytotoxicity was undermined by these gene sets is shown in a histogram (black line), which was significantly different from that of the putative human diversifiers (green arrow).
To better understand the regulators of phenotypic heterogeneity and their roles in anticancer drug resistance, we also sought to analyze human diversifiers and stabilizers individually. On the basis of the above results, we constructed a bipartite network between the drug candidates in NCI-60 and the human genes identified as diversifiers/stabilizers (by orthology with yeast diversifiers/stabilizers). In this network, a link between a gene and a drug candidate indicated that an increase in the expression of the gene would significantly enhance or weaken the cellular resistance to the drug candidate (see Materials and Methods). The biological relevance of the constructed network was supported by some human stabilizers/diversifiers known to be associated with cancer drug sensitivity/resistance. For example, the human diversifier identified as weakening the largest number of drug candidates was HDLBP (high-density lipoprotein binding protein), which has been found as overexpressed in vincristine resistant versus nonresistant gastric cancer cells (Hu et al. 2010). On the other hand, the human stabilizer identified as an enhancer for the largest number of drug candidates was MSH2 (MutS Homolog 2), a major component of the DNA mismatch repair system. It has been found that the loss of DNA mismatch repair would lead to cellular resistance of doxorubicin, taxanes, and topoisomerase poisons, which are commonly used to treat breast cancer (Fedier et al. 2001). Interestingly, we found that human stabilizers undermining the resistance of more drug candidates tended to have yeast orthologs stabilizing more morphological traits (Spearman’s ρ = 0.335, P < 0.03, supplementary fig. S3A, Supplementary Material online). A similar trend was found for human/yeast diversifiers, although the correlation was not significant (Spearman’s ρ = 0.051, P = 0.8, supplementary fig. S3B, Supplementary Material online).
For yeast diversifiers/stabilizers, we investigated whether gene deletion causes reciprocal changes in the heterogeneity of other traits and found that the answer is a resounding “no” (fig. 4A and B). We therefore asked a similar question with respect to human diversifiers/stabilizers: do expression changes in human diversifiers/stabilizers tend to decrease cellular resistance to some drug candidates while simultaneously increasing cellular resistance to some other drug candidates? If the answer is yes, targeting human diversifiers/stabilizers to enhance drug efficacy may lead to increased resistance of some other drugs, thereby undermining the value of human diversifiers/stabilizers as therapeutic targets. However, using the bipartite network, we found a depletion of such collateral resistance between the NCI-60 drug candidates and the human diversifiers/stabilizers (Spearman’s ρ = −0.61, P < 10−7, supplementary fig. S3C, Supplementary Material online). This result also suggests that the ancient evolutionary history of unicellular organisms has shaped fundamental characteristics of cancer cells in multicellular organisms. Collectively, our results highlight the involvement of genetic regulation of phenotypic heterogeneity in cancer cell resistance to anticancer drugs.
Discussion
In the current study, we analyzed the morphological phenotype data of single-gene deletion yeast strains and identified positive and negative genetic regulators of phenotypic heterogeneity (phenotypic diversifiers and stabilizers, respectively). We found that diversifiers tended to be weaker but more promiscuous regulators than stabilizers. Evolutionary analyses revealed that both diversifiers and stabilizers were likely maintained by purifying selection, but stabilizers tended to target traits that were tightly coupled with organismal fitness, whereas diversifiers tended to target other traits. Furthermore, there was a general division of labor between stabilizers and diversifiers such that genetic enhancement of phenotypic heterogeneity in one trait tended not to occur at the cost of phenotypic heterogeneity of other traits. Finally, we discovered a role for these genetic regulators of phenotypic heterogeneity in cancer cell drug resistance, in which overexpression of human one-to-one orthologs of yeast stabilizers (diversifiers) tended to reduce (increase) cellular resistance to candidate cancer drugs in the NCI-60 data set. More importantly, the division of labor between stabilizers and diversifiers recapitulated by their human orthologs resulted in a depletion of collateral resistance, making some stabilizers/diversifiers ideal targets for potential auxiliary treatments sensitizing cancer cells to anticancer drugs.
A few caveats of our analyses are worth discussion. First, the numbers of phenotypic stabilizers and diversifiers were likely underestimated because we focused our study on only morphological traits. The use of morphological data rather than other types of phenotypic heterogeneity data, such as single-cell transcriptomes (molecular phenotypes) or flow cytometry data, was, to the best of our knowledge, the optimal way to strike an ideal balance between data throughput (in terms of number of traits and genes) and measurement accuracy. For example, the state-of-the-art methods of single-cell RNA sequencing (scRNA-seq) scalable to >1,000 cells (e.g., droplet-based methods such as inDrop and 10x Genomics Chromium) have a CV of at least 0.5 for the counts per million of the majority of genes (Zhang et al. 2019), suggesting strong technical noise compared with our targeted biological noise. Consistent with this notion, it was recently estimated that the signal of gene expression cofluctuation among individual cells was at least an order of magnitude underestimated via scRNA-seq data (Sun and Zhang 2019). Nevertheless, it will be desirable to search for regulators of phenotypic heterogeneity using other types of phenotype data in the future. Second, associating human orthologs of diversifiers and stabilizers with the drug resistance of cancer cells assumes not only functional conservation of these genes between humans and yeast, which is commonly accepted (Chen and Zhang 2012), but also an association between phenotypic heterogeneity and diversity of drug responses of cancer cells, which is less well established. However, it was previously found that nongenetic variability in phenotypes may increase the drug resistance of tumors (Slack et al. 2008; Brock et al. 2009; Snijder and Pelkmans 2011). Furthermore, we found a significant functional resemblance between yeast diversifiers/stabilizers and their human orthologs, such as a division of labor between diversifiers and stabilizers, as well as a positive correlation between the number of traits stabilized by a yeast stabilizer and the number of drugs strengthened by a human stabilizer. This evidence strongly suggests that the above assumptions are at least partially valid.
One unexpected finding by our analysis was that the evolutionary conservation of diversifiers is comparable with that of stabilizers and were more conserved than other genes (fig. 3A). As assumed by most theoretical models of biological evolution, the optimal fitness could only be achieved by a narrow range of phenotypes. In these models, phenotypic variations and, therefore, the phenotypic diversifiers will likely be detrimental to the population average fitness. Our unexpected observation of the generally strong conservation of diversifiers, which is comparable with stabilizers, seemed to be incompatible with this (assumed) detrimental role of diversifiers. We think that there are three potential nonexclusive explanations for such incompatibility. First, it is possible that the optimal fitness could actually be achieved by a wide array of phenotype(s), such that phenotypic variation is mostly not detrimental. This is unlikely as numerous studies have shown the deleterious effect of phenotypic heterogeneity (Metzger et al. 2015). Second, it could be caused by the functional constraints imposed on diversifiers to avoid the diversification of important traits. This was supported by our observation that diversifiers regulating unimportant traits were generally less conserved (fig. 3I and J). Third, it could be due to the fluctuation of optimal phenotype(s) in natural environments, which warrants further assessment by an experimental evolution within fluctuating environments.
Our entire analysis was based on morphological data of single-gene deletion yeast strains, and we interpreted the data by the functional effect of gaining the gene, but not losing the gene. Nevertheless, from a molecular function point of view, diversifiers and stabilizers are similar to two faces of the same coin. Thus, switching on a diversifier and switching off a stabilizer should have very similar phenotypic heterogeneity-enhancing effects; likewise, switching off a diversifier and switching on a stabilizer should have very similar phenotypic heterogeneity-suppressing effects. However, from an evolutionary point of view, stabilizers and diversifiers have subtle yet critical differences. On the one hand, stabilizers enhance organismal fitness by suppressing harmful deviation of some traits from their optimal phenotypes and grant phenotypic robustness to evolving populations, thereby making “exploration” of new genotypes possible without detrimental consequences (Payne and Wagner 2019). Therefore, stabilizers should preferentially act on traits that are tightly coupled to fitness. On the other hand, as diversifiers enhance the chance of survival by elevating phenotypic heterogeneity, the number of traits a diversifier regulates should not be too limited, as environmental changes (and therefore traits that should enhance survivability once heterogenized) are unpredictable. Our observations that diversifiers are weaker and more promiscuous regulators than stabilizers and that diversifiers exhibit a preference for unimportant traits therefore highlight the evolutionary and functional importance of genetic control for phenotypic heterogeneity and support the hypothesis that genetic regulation of phenotypic heterogeneity is favored by natural selection.
With our result demonstrating the existence of stabilizers and diversifiers, it is straightforward to ask about their evolutionary origin. Specifically, do stabilizers/diversifiers evolve because regulating the heterogeneity of a specific phenotype improves the efficacy of phenotypic adaptation or do phenotypes with stabilizers/diversifiers intrinsically tend to be more/less fitness-coupled? Although we cannot guess how the stabilizers/diversifiers first arose, we did speculate that their evolutionary maintenance appears controlled by purifying selection and is therefore adaptive. This is because if stabilizers/diversifiers are intrinsically associated with more/less fitness-coupled traits, we expected neither the stronger sequence conservation of stabilizers/diversifiers compared with other genes nor the correlation between trait importance and the sequence conservation of stabilizers/diversifiers.
In addition to the aforementioned theoretical value, our results regarding the roles of human diversifiers/stabilizers in the drug resistance of cancer cells also point to the possibility of auxiliary treatments involving diversifiers and stabilizers in improving the efficacy of anticancer drugs. Specifically, according to the NCI-60 data, disruption of diversifiers or functional enhancement of stabilizers could sensitize cancer cells to a considerable number of drug candidates presumably via phenotypic homogenization of cancer cell populations. Additional features of human diversifiers/stabilizers, such as depletion of dual-role genes, which enhance some anticancer drugs while weakening others, increase the medical value of this potential strategy. We also want to emphasize that these results were obtained based on the assumption of functional conservation between yeast and human one-to-one orthologs. Although it is a widely held null hypothesis about sequence conservation and functional conservation, the result should be interpreted with caution, such that specific functional experiments should be conducted to determine the actual role of individual human diversifier/stabilizers on cancer drug resistance.
Materials and Methods
General Statistical Analysis
All statistical analyses were conducted with custom R (R Core Team 2013) scripts, which are all available on GitHub (https://github.com/moningsysu/Phenotypic-heterogeneity).
Phenotypic, Genomic, and Comparative Genomic Data
Morphological data for S. cerevisiae deletion strains were downloaded from a previous study (Ohya et al. 2005). The importance of each trait, that is, the decrease in fitness per unit of change in the trait value relative to the fitness of the wild-type strains, was calculated according to a previously described procedure (Ho and Zhang 2014). Data on the indispensability of 4,718 yeast genes, that is, the magnitude of the decrease in fitness upon deletion of each gene, were obtained from a previous study (Steinmetz et al. 2002). The genome sequence and annotations of S. cerevisiae (R64-1-1) were downloaded from the Saccharomyces Genome Database (Cherry et al. 2012). To quantify the evolutionary conservation of yeast genes, we estimated the dN and dS for one-to-one orthologs between S. cerevisiae and five different yeast species (S. paradoxus, S. mikatae, S. bayanus, Candida glabrata, and S. castellii) following previously described pipelines (Zhang and Yang 2015). Taking S. bayanus as an example, all S. bayanus coding sequences were retrieved from the Fungal Orthogroups Repository (Wapinski et al. 2007). Orthologos proteins were identified by reciprocal best hits of BlastP (Camacho et al. 2009) searches between the proteomes of the S. bayanus and S. cerevisiae, with the criteria of E value <10−20, alignment covering at least 80% of both orthologos sequences, and a length of at least 30 amino acids. To avoid the influence of gene duplication, we used only one-to-one orthologos proteins; that is, we excluded any protein from a species that was the best hit for more than one protein in the other species. The orthologos gene pairs were realigned by MUSCLE (Edgar 2004), filtered for gaps in alignment, and processed by PAML (Yang 2007) to calculate dN and dS. The dN/dS ratios from all five pairwise comparisons between S. cerevisiae and other species were then used to calculate the average dN/dS ratio of each gene. A list of human genes and their orthology to yeast genes was collected from Ensembl v93 (Zerbino et al. 2018). The degrees of connectivity in a protein–protein interaction network of the yeast genes were collected from a previous report (Levy and Siegal 2008).
Identification of Putative Phenotypic Stabilizers and Diversifiers
Using the morphological data of yeast deletion strains, we followed a previously proposed pipeline (Levy and Siegal 2008) for identification of phenotypic stabilizers with minor modifications to identify putative phenotypic stabilizers and diversifiers in S. cerevisiae. The pipeline involved four major steps: 1) estimation of phenotypic heterogeneity, 2) removal of redundancy in morphological traits, 3) identification of genes with significant impacts on phenotypic heterogeneity, and 4) screening for high-confidence diversifiers and stabilizers according to their phenotypic potential. Additional details are given below.
First, we collected the means and variances from quantitative morphological phenotype data collected from 4,718 yeast deletion strains in a previous study (Ohya et al. 2005). For each of the 220 traits, we used a running median strategy over the 4,718 strains to estimate the expected CV based on the mean, as the CVs were typically confounded by the mean (supplementary fig. S1, Supplementary Material online). Specifically, we calculated the expected CV of a strain as the median CV of 100 strains with similar means. We then used the deviation of the observed CV from the running median, or DM, as a measurement of the level of phenotypic heterogeneity controlled by the mean phenotypic value. If the observed CV was higher than the expected CV (DM > 0), the phenotypic heterogeneity was considered to be increased. Conversely, if the observed CV was lower than the expected CV (DM < 0), the phenotypic heterogeneity was considered to be decreased. The 4,718 × 220 raw DMs are listed in supplementary table S1, Supplementary Material online. Finally, the DM of each trait was standardized, that is, transformed into a Z score, to give every trait the same weight.
Second, using the 4,718 × 220 matrix of standardized DMs, we attempted to eliminate redundancy among the 220 traits, as some of the traits may be highly correlated with each other for biological or physical reasons. We used the Partitioning Around Medoids (PAM) algorithm (Reynolds et al. 2006) to cluster the traits and find representative traits from the clusters. We chose 70 clusters to strike a balance between having a large average silhouette width and a smaller chance of having noninformative clusters with a silhouette width of 0 (Levy and Siegal 2008). Ultimately, we obtained a redundancy-removed matrix of the DMs of 4,718 strains and 70 traits.
Third, to determine whether deletion of an individual gene could significantly impact the heterogeneity of a certain trait, we compared the observed DM for the trait and the corresponding deletion strain with mock DMs calculated through the two aforementioned steps using randomly sampled cells from the wild-type strain. Here, the number of cells sampled from the wild-type strain was the same as the number of cells from the deletion strain used to calculate the observed DM. For each gene and each trait, we repeated this process 1,000 times and obtained 1,000 mock DMs. An observed DM was said to be significant if it ranked first or last among the mock DMs, which corresponded to a P value of 0.001. A significant DM suggests a gene with a significant impact on the phenotypic heterogeneity of a trait.
Fourth, to identify high-confidence stabilizers, we averaged the top 20 (of 70) DMs for a gene to obtain a score that was similar to the previously proposed “phenotypic potential” (Levy and Siegal 2008). The logic behind the usage of phenotypic potential was that a gene with a physiological function of regulating phenotypic heterogeneity should, theoretically, have a relatively large effect on relatively more traits compared with that of other genes that only slightly affect the heterogeneity of a small number of traits as a by-product of its physiological function. In other words, the actual diversifier/stabilizer should have a larger phenotypic potential relative to that of other genes. We estimated the null distribution of phenotypic potential scores by averaging 100 mock distributions of phenotypic potential scores that were generated by shuffling of all DMs within each trait. We chose a phenotypic potential score >1.743 as the cutoff for stabilizers, as this cutoff yields a false positive rate of 1%, which corresponds to an FDR of 12% (284 true positives among 324 discoveries). Similarly, high-confidence diversifiers were identified with a phenotypic potential score (average of the bottom 20 DMs) cutoff of <−1.437, as this cutoff yields a false-positive rate of 0.5%, which corresponds to an FDR of 12% (141 true positives among 160 discoveries). Note that the number of top/bottom DMs chosen (20) was arbitrary, but the vast majority of the identified stabilizers/diversifiers were insensitive to the choice of this parameter within a reasonable range. For example, if we changed this number to 15 and looked for the same number of genes with the most extreme phenotypic potential (324 genes with the highest phenotypic potential for stabilizers, 160 genes with the lowest phenotypic potential for diversifiers), 96.0% stabilizers and 88.1% diversifiers retained their classification. If we changed the parameter to 30, these numbers became 93.5% and 84.4%, respectively. We therefore used 20 traits across our study.
Altogether, the final bipartite network was composed of 70 traits (selected at the second step), 457 genes (324 stabilizers and 160 diversifiers identified at the fourth step, among which 27 were dual-role genes), and 5,013 links between genes and traits (identified at the third step), which represented 2,528 capacitance and 2,485 potentiation relationships between genes and traits. The network is presented in supplementary table S3, Supplementary Material online.
There is one potential caveat worth discussing in our method of identifying diversifiers. Basically, it is possible that some or many morphological traits have a physical limit (e.g., the smallest possible nucleic size that all yeast chromosomes could fit within). If the deletion of a gene pushes certain traits toward their physical limits, the CV and DM may appear lowered because the value of the trait cannot extend beyond the physical limit, rather than because the deleted gene is a diversifier. If this alternative possibility could explain the majority of the DM decrease observed for diversifier-deleted strains, we should expect that, for a specific trait, mutant strains whose mean trait values are closer to the physical limit of the trait should have smaller DMs. To exclude this alternative possibility, we estimated the physical limit by the range of mean trait values of all mutant/wild-type strains. Then for each trait, we calculated the Spearman’s rank correlation between DM and the distance to the physical limit (the smaller one between the distance to the upper limit and the distance to the lower limit). When we used all 4,718 strains to calculate this correlation, none of the traits (supplementary fig. S4A, Supplementary Material online) gave rise to statistically significant correlations after multiple testing correction (Benjamini and Hochberg 1995). When we used only the 160 strains whose diversifiers were deleted, only 3 out of the 70 traits (supplementary fig. S4B, Supplementary Material online) showed significantly positive correlation. In other words, few (if any) traits showed lower DM for deletion strains closer to the physical limit of the trait. Note that it is possible that our estimations of the physical limits were conservative, such that the actual physical limits were wider than our estimation. In this case, the distance of the observed mean trait value to its physical limit is larger than our estimation. Therefore, the variation of the trait should become even less likely to reach the physical limit, such that the probability of false identification of diversifier due to the constraints of physical limit should be even lower. To further guard against nonlinear relationship between DM and distance to physical limits (e.g., only traits really close to their physical limits were affected), we also extracted for each trait 5% of strains that were closest to its physical limit and compared their average DM with that of the other strains. Consistent with the results from Spearman’s correlation, we found no evidence for smaller DM in strains closer to the traits’ physical limit (supplementary fig. S4C and D, Supplementary Material online). Collectively, these analyses suggested that the false identification of diversifier due to the constraints of physical limit was a minor problem in our analyses.
Functional Annotation Based on GO
Functional annotation and gene ontology (GO) term enrichment analyses of yeast diversifiers, stabilizers, and dual-role genes were performed by the BiNGO (Maere et al. 2005) plugin of Cytoscape (Shannon et al. 2003) with the default parameters.
NCI-60 Data
We downloaded the NCI-60 data set from the CellMiner (Reinhold et al. 2012) website. The data set includes the transcriptome profiles of 60 typical cancer cell lines from 9 different types of cancers and the responses of these cell lines to 21,121 types of natural chemicals that are anticancer drug candidates. In this data set, the response of a cancer cell line to a drug candidate is represented by the LC50. To test for functional associations between a group of genes and cancer cell resistance to a specific drug, for each gene within the group, we calculated the average LC50 of the 30 cancer cell lines in which the focal gene had the highest expression level (HE-LC50) and that of the 30 cell lines in which the focal gene had the lowest expression level (LE-LC50). If the HE-LC50 of the group of genes was significantly higher/lower than the LE-LC50 of that group of genes (P < 0.05 by the Wilcoxon signed-rank test, fig. 5A and B), the group was considered capable of decreasing/increasing the cellular resistance to the specific drug. Note that the results presented in the Results section were not corrected for multiple testing, as we considered testing for different combinations of drugs and genes was associated with different hypotheses. Nevertheless, we also tried to perform multiple testing correction (Benjamini and Hochberg 1995), and the number of drugs strengthened (weakened) by stabilizers (diversifiers) became 6,238 (823), which is still significantly higher than expected from random groups of human genes.
To investigate the association between a specific gene and a specific drug candidate, the HE-LC50s of the focal gene were compared by the Wilcoxon rank-sum test with the LE-LC50s of the focal gene. If the HE-LC50s were significantly higher/lower than the LE-LC50s, the gene was said to decrease/increase the cellular resistance to the specific drug, thereby constituting a link between the gene and the drug. A bipartite network was constructed after testing all 67 human diversifiers/stabilizers with all 21,121 drug candidates.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the National Key R&D Program of China (Grant No. 2017YFA0103504 to X.C. and 2018ZX10301402 to J.-R.Y.), the National Natural Science Foundation of China (Grant Nos. 31871320, 81830103, and 31671320 to J.-R.Y. and 31771406 to X.C.), the National Special Research Program of China for Important Infectious Diseases (Grant No. 2018ZX10302103 to X C.), and a start-up grant from “100 Top Talents Program” of Sun Yat-sen University (Grant No. 50000-18821112 to X.C. and 50000-18821117 to J.-R.Y.).
Author Contributions
X.C. and J.-R.Y. conceptualized and supervised the study; N.M., X.Z., W.S., G.Y., X.C., and J.-R.Y. collected various data sets and analyzed the data; N.M., X.C., and J.-R.Y. prepared the original draft; and N.M., X.C., and J.-R.Y. revised the manuscript, with input from all authors.
Data Availability
No new data were created in this study. The sources of all analyzed data were explicitly mentioned in the text.
References
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological). 57(1):289–300. [Google Scholar]
- Brock A, Chang H, Huang S.. 2009. Non-genetic heterogeneity: a mutation-independent driving force for the somatic evolution of tumours. Nat Rev Genet. 10(5):336–342. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10(1):421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lin F, Xing K, He X.. 2015. The reverse evolution from multicellularity to unicellularity during carcinogenesis. Nat Commun. 6(1):6367. [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang J.. 2012. The ortholog conjecture is untestable by the current gene ontology but is supported by RNA sequencing data. PLoS Comput Biol. 8(11):e1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. 2012. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 40(D1):D700–D705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covell DG. 2012. Integrating constitutive gene expression and chemoactivity: mining the NCI60 anticancer screen. PLoS One 7(10):e44631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedier A, Schwarz VA, Walt H, Carpini RD, Haller U, Fink D.. 2001. Resistance to topoisomerase poisons due to loss of DNA mismatch repair. Int J Cancer. 93(4):571–576. [DOI] [PubMed] [Google Scholar]
- Garmendia-Torres C, Skupin A, Michael SA, Ruusuvuori P, Kuwada NJ, Falconnet D, Cary GA, Hansen C, Wiggins PA, Dudley AM.. 2014. Unidirectional P-body transport during the yeast cell cycle. PLoS One 9(6):e99428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover N, Dessimoz C, Ebersberger I, Forslund SK, Gabaldón T, Huerta-Cepas J, Martin M-J, Muffato M, Patricio M, Pereira C, et al. 2019. Advances and applications in the quest for orthologs. Mol Biol Evol. 36(10):2157–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould CJ, Chesarone-Cataldo M, Alioto SL, Salin B, Sagot I, Goode BL.. 2014. Saccharomyces cerevisiae Kelch proteins and Bud14 protein form a stable 520-kDa Formin regulatory complex that controls actin cable assembly and cell morphogenesis. J Biol Chem. 289(26):18290–18301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho WC, Zhang J.. 2014. The genotype-phenotype map of yeast complex traits: basic parameters and the role of natural selection. Mol Biol Evol. 31(6):1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HD, Ye F, Zhang DZ, Hu P, Ren H, Li SL.. 2010. iTRAQ quantitative analysis of multidrug resistance mechanisms in human gastric cancer cells. J Biomed Biotechnol. 2010:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Siegal ML.. 2008. Network hubs buffer environmental variation in Saccharomyces cerevisiae. PLoS Biol. 6(11):e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Na Z, Slavoff SA.. 2018. Bodies: composition, properties, and functions. Biochemistry 57(17):2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M.. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21(16):3448–3449. [DOI] [PubMed] [Google Scholar]
- Metzger BP, Yuan DC, Gruber JD, Duveau F, Wittkopp PJ.. 2015. Selection on noise constrains variation in a eukaryotic promoter. Nature 521(7552):344–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KE, Lo WC, Lee ME, Kang PJ, Park HO.. 2017. Fine-tuning the orientation of the polarity axis by Rga1, a Cdc42 GTPase-activating protein. Mol Biol Cell. 28(26):3773–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya Y, Sese J, Yukawa M, Sano F, Nakatani Y, Saito TL, Saka A, Fukuda T, Ishihara S, Oka S, et al. 2005. High-dimensional and large-scale phenotyping of yeast mutants. Proc Natl Acad Sci U S A. 102(52):19015–19020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualato A, Palombo A, Cucina A, Mariggiò MA, Galli L, Passaro D, Dinicola S, Proietti S, D’Anselmi F, Coluccia P, et al. 2012. Quantitative shape analysis of chemoresistant colon cancer cells: correlation between morphotype and phenotype. Exp Cell Res. 318(7):835–846. [DOI] [PubMed] [Google Scholar]
- Patterson JO, Rees P, Nurse P.. 2019. Noisy cell-size-correlated expression of cyclin B drives probabilistic cell-size homeostasis in fission yeast. Curr Biol. 29(8):1379–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne JL, Wagner A.. 2019. The causes of evolvability and their evolution. Nat Rev Genet. 20(1):24–38. [DOI] [PubMed] [Google Scholar]
- Philips J, Herskowitz I.. 1998. Identification of Kel1p, a kelch domain-containing protein involved in cell fusion and morphology in Saccharomyces cerevisiae. J Cell Biol. 143(2):375–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S.. 2002. Hsp90 as a capacitor of phenotypic variation. Nature 417(6889):618–624. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
- Rego EH, Audette RE, Rubin EJ.. 2017. Deletion of a mycobacterial divisome factor collapses single-cell phenotypic heterogeneity. Nature 546(7656):153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold WC, Sunshine M, Liu H, Varma S, Kohn KW, Morris J, Doroshow J, Pommier Y.. 2012. CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 72(14):3499–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AP, Richards G, de la Iglesia B, Rayward-Smith VJ.. 2006. Clustering rules: a comparison of partitioning and hierarchical clustering algorithms. J Math Model Algor. 5(4):475–504. [Google Scholar]
- Rutherford SL, Lindquist S.. 1998. Hsp90 as a capacitor for morphological evolution. Nature 396(6709):336–342. [DOI] [PubMed] [Google Scholar]
- Schmutzer M, Wagner A.. 2020. Gene expression noise can promote the fixation of beneficial mutations in fluctuating environments. PLoS Comput Biol. 16(10):e1007727. [DOI] [PMC free article] [PubMed]
- Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C, Beqiri M, Sproesser K, Brafford PA, Xiao M, et al. 2017. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546(7658):431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T.. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. 2010. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141(1):69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CE, Lui J, Kershaw CJ, Sims PF, Ashe MP.. 2014. mRNA localization to P-bodies in yeast is bi-phasic with many mRNAs captured in a late Bfr1p-dependent wave. J Cell Sci. 127(6):1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirois I, Aguilar-Mahecha A, Lafleur J, Fowler E, Vu V, Scriver M, Buchanan M, Chabot C, Ramanathan A, Balachandran B, et al. 2019. A unique morphological phenotype in chemoresistant triple-negative breast cancer reveals metabolic reprogramming and PLIN4 expression as a molecular vulnerability. Mol Cancer Res. 17(12):2492–2507. [DOI] [PubMed] [Google Scholar]
- Slack MD, Martinez ED, Wu LF, Altschuler SJ.. 2008. Characterizing heterogeneous cellular responses to perturbations. Proc Natl Acad Sci U S A. 105(49):19306–19311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. 1974. Hedging one’s evolutionary bets. Nature 250(5469):704–705. [Google Scholar]
- Snijder B, Pelkmans L.. 2011. Origins of regulated cell-to-cell variability. Nat Rev Mol Cell Biol. 12(2):119–125. [DOI] [PubMed] [Google Scholar]
- Stamboulian M, Guerrero RF, Hahn MW, Radivojac P.. 2020. The ortholog conjecture revisited: the value of orthologs and paralogs in function prediction. Bioinformatics 36(Supplement_1):i219–i226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz LM, Scharfe C, Deutschbauer AM, Mokranjac D, Herman ZS, Jones T, Chu AM, Giaever G, Prokisch H, Oefner PJ, et al. 2002. Systematic screen for human disease genes in yeast. Nat Genet. 31(4):400–404. [DOI] [PubMed] [Google Scholar]
- Sun M, Zhang J.. 2019. Chromosome-wide co-fluctuation of stochastic gene expression in mammalian cells. PLoS Genet. 15(9):e1008389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Parker R.. 2007. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell. 18(6):2274–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. 2013. Robustness and evolvability in living systems. Princeton (NJ: ): Princeton University Press. [Google Scholar]
- Wang GZ, Liu J, Wang W, Zhang HY, Lercher MJ.. 2011. A gene’s ability to buffer variation is predicted by its fitness contribution and genetic interactions. PLoS One 6(3):e17650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhang J.. 2011. Impact of gene expression noise on organismal fitness and the efficacy of natural selection. Proc Natl Acad Sci U S A. 108(16):E67–E76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A.. 2007. Automatic genome-wide reconstruction of phylogenetic gene trees. Bioinformatics 23(13):i549–i558. [DOI] [PubMed] [Google Scholar]
- Weidner J, Wang C, Prescianotto-Baschong C, Estrada AF, Spang A.. 2014. The polysome-associated proteins Scp160 and Bfr1 prevent P body formation under normal growth conditions. J Cell Sci. 127(9):1992–2004. [DOI] [PubMed] [Google Scholar]
- Wu CI, Shen Y, Tang T.. 2009. Evolution under canalization and the dual roles of microRNAs: a hypothesis. Genome Res. 19(5):734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, et al. 2018. Ensembl 2018. Nucleic Acids Res. 46(D1):D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yang JR.. 2015. Determinants of the rate of protein sequence evolution. Nat Rev Genet. 16(7):409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Li T, Liu F, Chen Y, Yao J, Li Z, Huang Y, Wang J.. 2019. Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-Seq systems. Mol Cell. 73(1):130–142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data were created in this study. The sources of all analyzed data were explicitly mentioned in the text.