The mechanisms by which Candida glabrata resists host defense peptides and caspofungin are incompletely understood. To identify transcriptional regulators that enable C. glabrata to withstand these classes of stressors, a library of 215 C. glabrata transcriptional regulatory deletion mutants was screened for susceptibility to both protamine and caspofungin.
KEYWORDS: Candida glabrata, antifungal agents, histone modification, host defense peptide, virulence regulation
ABSTRACT
The mechanisms by which Candida glabrata resists host defense peptides and caspofungin are incompletely understood. To identify transcriptional regulators that enable C. glabrata to withstand these classes of stressors, a library of 215 C. glabrata transcriptional regulatory deletion mutants was screened for susceptibility to both protamine and caspofungin. We identified eight mutants that had increased susceptibility to both host defense peptides and caspofungin. Of these mutants, six were deleted for genes that were predicted to specify proteins involved in histone modification. These genes were ADA2, GCN5, SPT8, HOS2, RPD3, and SPP1. Deletion of ADA2, GCN5, and RPD3 also increased susceptibility to mammalian host defense peptides. The Δada2 and Δgcn5 mutants had increased susceptibility to other stressors, such as H2O2 and SDS. In the Galleria mellonella model of disseminated infection, the Δada2 and Δgcn5 mutants had attenuated virulence, whereas in neutropenic mice, the virulence of the Δada2 and Δrpd3 mutants was decreased. Thus, histone modification plays a central role in enabling C. glabrata to survive host defense peptides and caspofungin, and Ada2 and Rpd3 are essential for the maximal virulence of this organism during disseminated infection.
INTRODUCTION
In the United States, Candida glabrata causes approximately one third of cases of hematogenously disseminated candidiasis (1, 2). The echinocandin antifungal agents are currently the primary treatment for disseminated candidal infections, especially those caused by C. glabrata (3). Of significant concern, strains of C. glabrata that are resistant to echinocandin antifungal drugs are being isolated with increasing frequency (4). In some medical centers, up to 25% of C. glabrata isolates are either intermediately susceptible or resistant to echinocandins (5). Many strains of C. glabrata are resistant to echinocandins because of mutations in FKS1 or FKS2, which encode 1,3 β-glucan synthases, the target enzymes of these drugs (6). However, other strains are resistant to echinocandins by other, unknown mechanisms (2, 7).
Host defense peptides play key roles in the host defense against C. glabrata infections. These peptides, such as α-defensins, β-defensins, cathelicidins, and LL-37, are produced by leukocytes and the epithelial cells that line cutaneous and mucosal barriers (8, 9). For C. glabrata to colonize the host and persist within the deep tissues, it must be able to resist inhibition or killing by these host defense peptides. While some strains of C. glabrata are readily killed by β-defensins, other strains are relatively resistant (10). The mechanisms by which C. glabrata resists host defense peptides are not fully understood.
To gain a deeper understanding of the mechanisms by which C. glabrata resists both echinocandins and host defense peptides, we screened a library of 215 C. glabrata transcriptional regulator mutants for strains that were susceptible to echinocandin, caspofungin, and the host defense peptide protamine. Remarkably, of the eight strains that were found to be hypersusceptible to both protamine and caspofungin, 6 contained mutations in genes whose products are predicted to function in histone modification, including acetylation, deacetylation, and methylation. Among these mutants, the Δada2 and Δgcn5 mutants were also found to have increased sensitivity to hydrogen peroxide and SDS. Both the Δada2 and the Δrpd3 mutants had attenuated virulence during disseminated infection in neutropenic mice. These results indicate that in C. glabrata, histone modification is required for the organism to resist caspofungin and protamine, and that Ada2 and Rbd3 govern virulence during disseminated infection.
RESULTS
Screening the transcription factor deletion mutant library identified eight genes that governed susceptibility to protamine and caspofungin.
In total, 215 C. glabrata deletion mutants were screened for increased susceptibility to protamine and caspofungin. This screen identified 17 mutants that had increased susceptibility to protamine alone and eight that had increased susceptibility to caspofungin alone, relative to the wild-type strain (Table S1 in the supplemental material). Twenty-three mutants had reduced susceptibility to protamine and four had reduced susceptibility to caspofungin; no strains had decreased susceptibility to both stressors (Table S1).
We identified eight mutants that had increased susceptibility to both protamine and caspofungin (Table 1; Fig. 1). Of the genes that were mutated in these eight strains, six specified proteins involved in histone modification. These proteins included Ada2, Gcn5, and Spt8, which are components of the SAGA histone acetyltransferase complex (11), Hos2 and Rpd3, which are members of same family of histone deacetylases (12), and Spp1, a component of the COMPASS histone methyltransferase complex (13). To verify that Ada2, Gcn5, Hos2, Rpd3, and Spp1 governed susceptibility to protamine and caspofungin, we complemented each of the C. glabrata mutants with an intact allele of the gene that was disrupted. Complementation restored the wild-type phenotype in all strains (Fig. 1). These results indicate that histone modification is a key mechanism by which C. glabrata resists host defense peptides and caspofungin.
TABLE 1.
List of genes required for resistance to both protamine and caspofungin within the mutant screen
| Systematic name | Gene name | Putative function |
|---|---|---|
| CAGL0K06193g | ADA2 | Transcriptional coactivator; component of the Spt-Ada-Gcn5 acetyltransferase (SAGA) complex |
| CAGL0F08283g | GCN5 | Histone acetyltransferase; component of the Spt-Ada-Gcn5 acetyltransferase (SAGA) complex |
| CAGL0F01837g | SPT8 | Component of the Spt-Ada-Gcn5 acetyltransferase (SAGA) complex |
| CAGL0B01441g | RPD3 | Histone deacetylase; regulates transcription and silencing; subunit of the Rpd3L complex |
| CAGL0A03322g | HOS2 | NAD-dependent histone deacetylase; subunit of the Set3 and Rpd3L complex |
| CAGL0M02585g | SPP1 | Transcriptional activator; histone methyltransferase; part of Set1/COMPASS complex |
| CAGL0M06831g | CRZ1 | Transcription factor; downstream component of the calcineurin signaling pathway |
| CAGL0J10274g | MKS1 | Pleiotropic negative transcriptional regulator |
FIG 1.
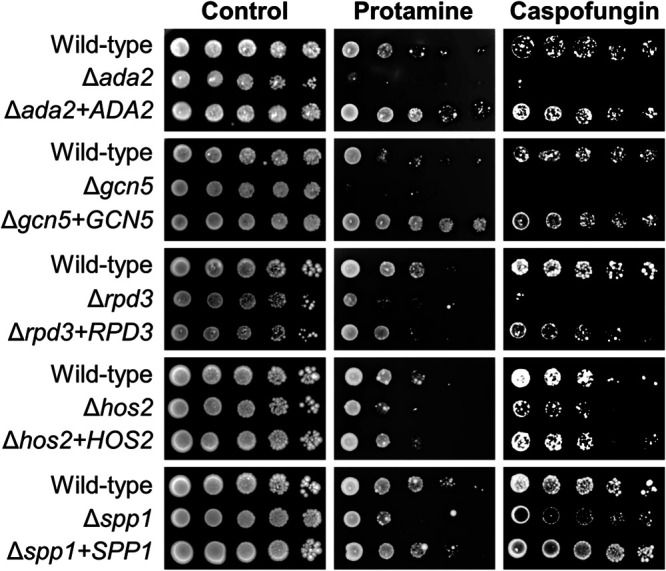
Susceptibility of the various C. glabrata mutants to protamine and caspofungin. Serial 10-fold dilutions of the indicated strains were grown on YPD agar alone or on agar containing either protamine or caspofungin at 30°C and then imaged after 48 h.
Two additional mutants, Δcrz1 and Δmks1, were identified that had increased susceptibility to both protamine and caspofungin. Neither Crz1 nor Mks1 are involved in histone modification (Table 1). Crz1 is a transcription factor in the calcineurin signaling pathway (14), and Mks1 is a negative transcriptional regulator that inhibits multiple signaling pathways, such as the target of rapamycin (TOR) and Ras-cyclic AMP pathways (15, 16). These data demonstrate that diverse signaling pathways govern susceptibility to protamine and caspofungin.
To assess the magnitude of the caspofungin resistance, we used the CLSI broth microdilution method to determine the MIC of caspofungin for the eight mutants that were hypersusceptible to both this drug and protamine. The caspofungin MICs of all of these mutants were consistently 1 to 2 dilutions less than the wild-type strain, except for that of the Δspt8 mutant, which was equivalent to the wild-type strain (Table 2). Thus, these mutants had a modest increase in their susceptibility to caspofungin in the broth microdilution assay.
TABLE 2.
MICs of caspofungin for the indicated strains
| Strain | Caspofungin MIC (mg/liter)a |
|---|---|
| Wild-type | 0.5 |
| Δada2 | 0.25 |
| Δgcn5 | 0.125 |
| Δspt8 | 0.5 |
| Δhos2 | 0.25 |
| Δrpd3 | 0.25 |
| Δspp1 | 0.25 |
| Δcrz1 | 0.25 |
| Δmks1 | 0.25 |
Data are the combined results from three independent experiments.
Multiple transcription factors govern susceptibility to host defense peptides.
Protamine, which is obtained from salmon, was used as a representative host defense peptide in the screening assays. To investigate the response of the various mutants to mammalian host defense peptides, we used a radial diffusion assay to assess susceptibility to protamine as well as to human neutrophil peptide-1 (hNP-1), human β-defensin-2 (hBD-2), LL-37, and γ-RP-1. The strains were tested at pH 5.5 and 7.5. Virtually all strains were more susceptible to the antimicrobial peptides at pH 5.5 relative to pH 7.5, as indicated by larger zones of inhibition (Fig. 2). At pH 5.5, the Δada2, Δspt8, Δhos2, and Δspp1 mutants had significantly increased susceptibility to protamine, whereas at pH 7.5, all mutants except for the Δspp1 strain had increased susceptibility to protamine. Susceptibility to the mammalian host defense peptides was dependent on the peptide, the pH, and the gene that was deleted. Overall, the Δada2, Δgcn5, and Δrpd3 mutants showed the greatest susceptibility to the largest number of host defense peptides. At pH 5.5, but not pH 7.5, the Δada2 mutant had increased susceptibility to hNP-1 and γ-RP-1. The Δgcn5 mutant had increased susceptibility to LL-37 at pH 5.5 and 7.5, and increased susceptibility to γ-RP-1 at pH 5.5. The Δrpd3 mutant had increased susceptibility to hNP-1 at pH 5.5 and to hBD-2 at pH 7.5. Collectively, these results indicate that while multiple transcriptional regulators are required for C. glabrata to withstand protamine, Ada2, Gcn5, and Rpd3 are the most important for resistance to mammalian host defense peptides.
FIG 2.
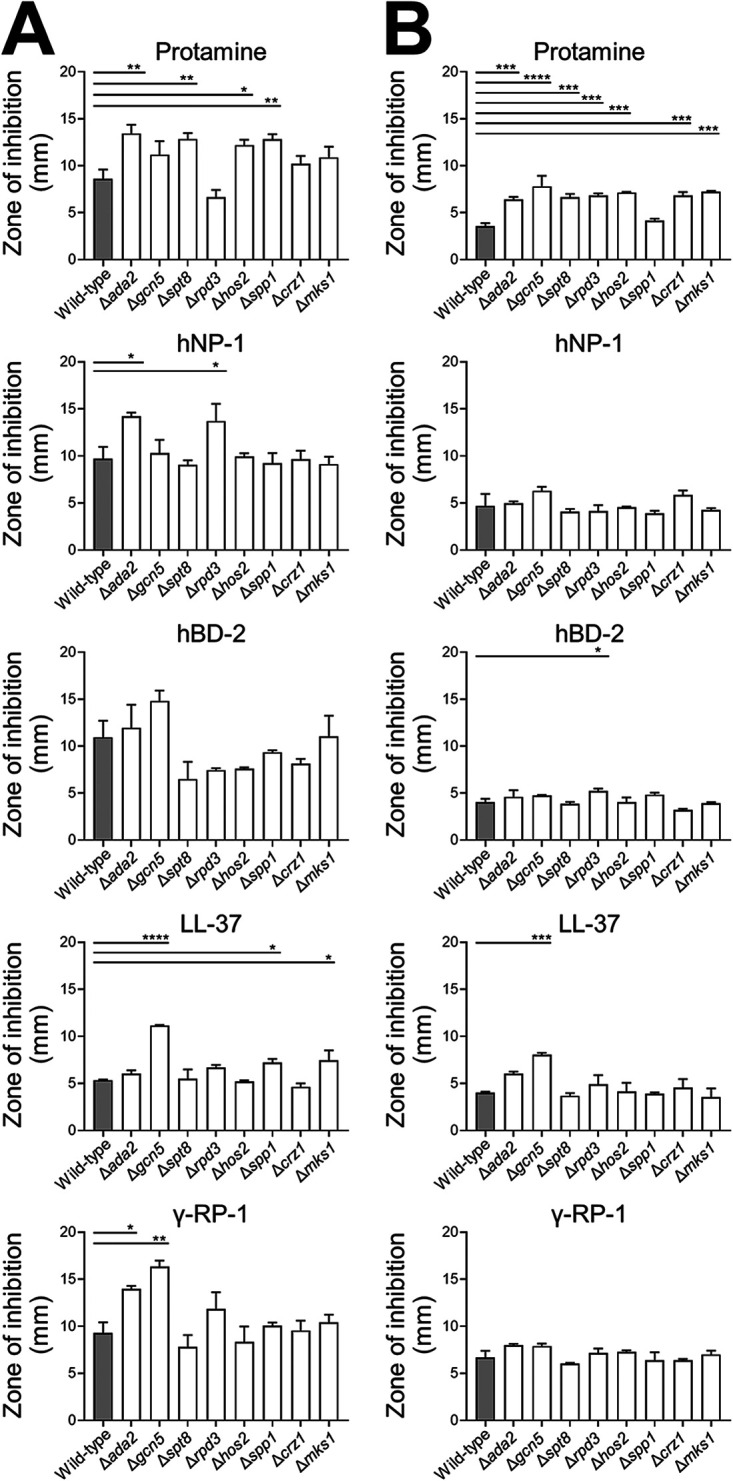
Susceptibility of C. glabrata mutants to host defense peptides. (A and B) The indicated strains of C. glabrata were tested for susceptibility to protamine, human neutrophil peptide-1 (hNP-1), human β-defensin-2 (hBD-2), LL-37, and γ-RP-1 using a radial diffusion assay at pH 5.5 (column A) and pH 7.5 (column B). Results are the means ± standard deviation (SD) of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 compared to the wild-type strain by analysis of variance with the Dunnett test for multiple comparisons.
Ada2 and Gcn5 govern resistance to multiple stressors.
Next, we evaluated whether histone modification is required for C. glabrata to withstand stressors other than protamine and caspofungin. We determined the Δada2 mutant had increased susceptibility to calcofluor white, H2O2, SDS, and NaCl relative to the wild-type and Δada2+ADA2 complemented strains (Fig. 3). The Δgcn5 mutant had increased susceptibility to H2O2 and SDS compared to the wild-type and Δgcn5+GCN5 complemented strains. In contrast, the Δrpd3, Δhos2, and Δspp1 mutants had wild-type susceptibility to all stressors tested. Thus, the members of the SAGA complex, particularly Ada2, are required for C. glabrata to withstand multiple different types of stress.
FIG 3.
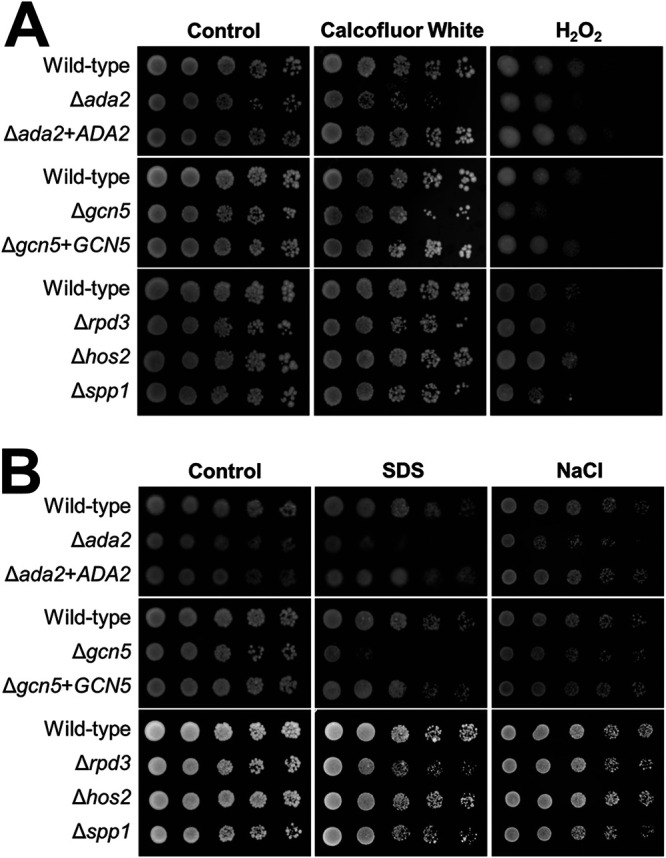
Susceptibility of C. glabrata mutants to additional stressors. Serial 10-fold dilutions of the indicated strains were grown on YPD agar alone or on agar containing the indicated stressors at 30°C and then imaged after 48 h.
Ada2 is required for virulence.
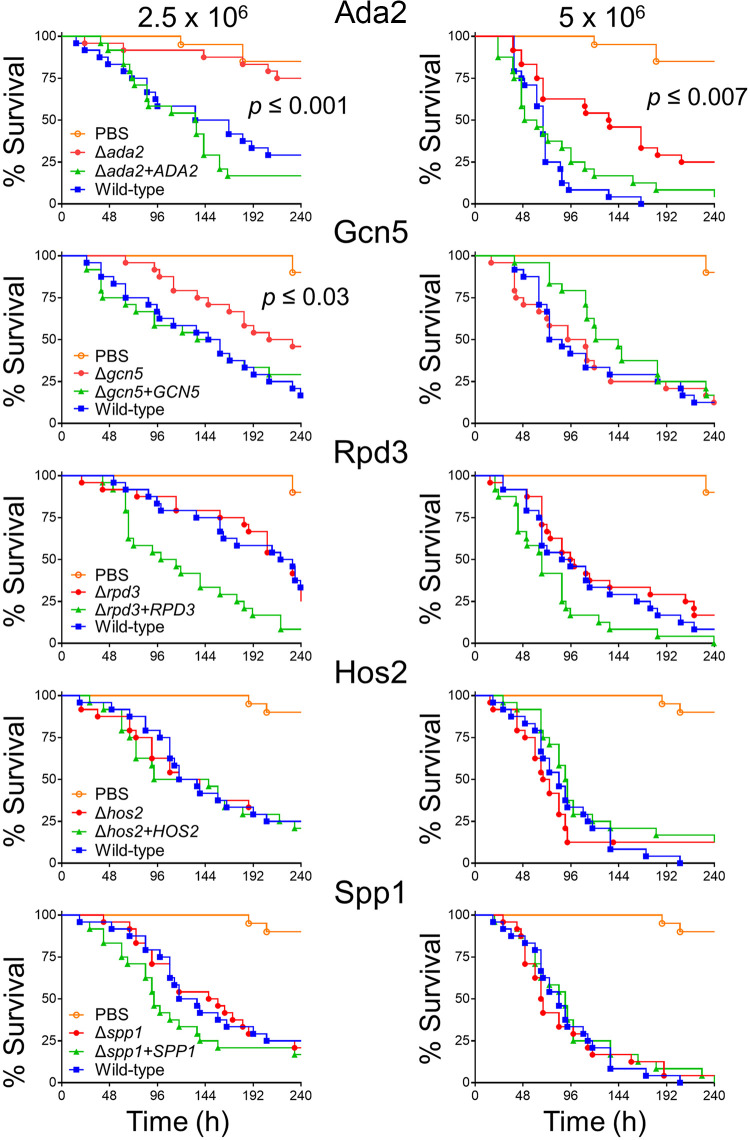
We hypothesized that mutants susceptible to both protamine and caspofungin would also be attenuated in virulence because they would have increased susceptibility to host defense peptides and impaired cell wall integrity. To test this hypothesis, we tested the virulence of the Δada2, Δgcn5, Δrpd3, Δhos2, and Δspp1 mutants in two models of disseminated candidiasis, Galleria mellonella larvae and immunosuppressed mice. In the G. mellonella model, each mutant was tested at inocula of 2.5 × 106 and 5 × 106 organisms per larvae. Of the five mutants tested, only the Δada2 and Δgcn5 mutants had virulence defects. Larvae infected with either the high or low inoculum of the Δada2 mutant had significantly longer survival than those infected with the wild-type strain and the Δada2+ADA2 complemented strain (Fig. 4). Larvae infected with the low inoculum of the Δgcn5 mutant also survived significantly longer than larvae infected with the control strains. However, when the higher inoculum was used, the virulence defect of the Δgcn5 mutant was no longer apparent. Collectively, these results demonstrate that Ada2, and to a lesser extent Gcn5, are required for the full virulence of C. glabrata in G. mellonella.
FIG 4.
Virulence testing in G. mellonella. G. mellonella larvae were infected with either 2.5 × 106 cells (left panels) or 5 × 106 (right panels) cells of the indicated strains of C. glabrata and then monitored twice daily for survival. Survival curves are the combined results of two independent experiments for a total of 24 larvae per strain. The indicated P values were determined using the Gehan-Breslow-Wilcoxon test, comparing the larvae infected with the deletion mutant with those infected with the wild-type and complemented strains.
The virulence of the five transcriptional regulatory mutants was also tested in the neutropenic mouse model of disseminated candidiasis using kidney fungal burden as the primary endpoint. Both the Δada2 and Δrpd3 mutants had significant virulence defects (Fig. 5). The median kidney fungal burdens of mice infected with these mutants was 50- to 80-fold lower than that of mice infected with the wild-type strain. In contrast to its attenuated virulence in G. mellonella, the Δgcn5 mutant had wild-type virulence in mice.
FIG 5.
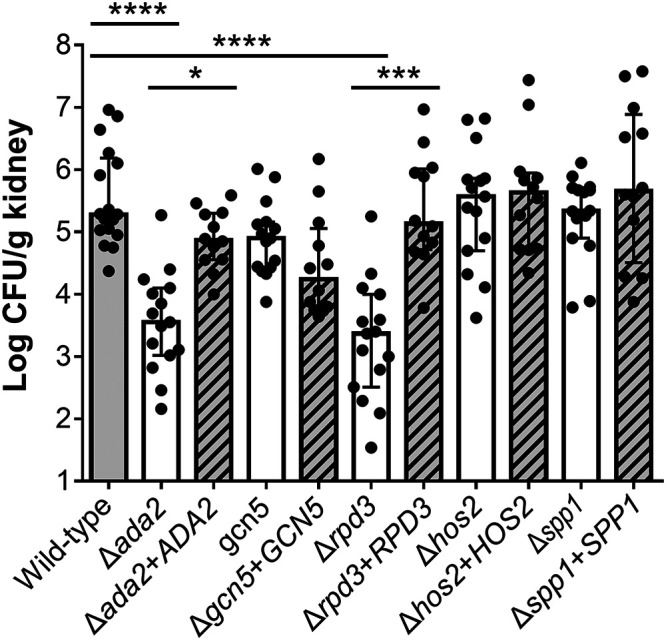
Virulence testing in mice. Neutropenic mice were inoculated intravenously with the indicated strains of C. glabrata. After 8 days, the mice were sacrificed and the kidney fungal burden was determined. Results are the median ± interquartile range of the combined results of two independent experiments for a total of 12 to 17 mice per strain. *, P < 0.05, ***, P < 0.001, ****, P < 0.0001 by the Kruskal-Wallis test for multiple comparisons.
DISCUSSION
The results obtained from screening a library of C. glabrata deletion mutants revealed that histone modification plays a central role in governing the susceptibility of the organism to protamine and caspofungin. Of the eight transcriptional regulators identified in this screen, six were predicted to be components of major regulatory complexes that modify histones. Furthermore, Ada2, an adapter protein that is a critical component of multiple histone acetyltransferase complexes, and Rpd3, a histone deacetylase, are required for the maximal virulence of C. glabrata in neutropenic mice.
Ada2, Gcn5, and Spt8 were found to be necessary for C. glabrata to withstand both protamine and caspofungin. These proteins are members of the SAGA histone acetylase complex (11, 17). Ada2 functions as an adapter protein, Gcn5 is an acetyltransferase, and Spt8 is a regulatory protein that is required for the function of the SAGA complex (11). Ada2 and Gcn5 are also members of multiple other histone acetyltransferase complexes (18, 19). Although the phenotypes of the Δada2 and Δgcn5 mutants were similar, the Δada2 mutant had increased susceptibility to a broader range of stressors than the Δgcn5 mutant. For example, the Δada2 mutant had increased susceptibility to calcofluor white and NaCl, whereas the Δgcn5 mutant did not. Also, while the Δada2 mutant had a virulence defect in both G. mellonella and mice, the Δgcn5 mutant had only a modest virulence defect in G. mellonella and had wild-type virulence in mice. In Candida albicans, deletion of both ADA2 and GCN5 results in attenuated virulence (20, 21). Our results indicate that C. glabrata differs from C. albicans in that Ada2 plays a greater role than Gcn5 in stress resistance and virulence.
The finding that Ada2 is particularly important for the C. glabrata to tolerate a wide variety of stressors provides a compelling explanation as to why the Δada2 mutant had reduced virulence in both G. mellonella and mice. Two other groups have also reported that C. glabrata Δada2 mutants have increased susceptibility to a broad range of stressors (22, 23). In agreement with our results, Kounatidis et al. found that a Δada2 mutant had attenuated virulence in a Drosophila model of gastrointestinal infection (22). In contrast, Yu et al. found that a Δada2 mutant actually had increased virulence in mice, as manifested by enhanced lethality and increased organ fungal burden (23). The reason for why the virulence data of Yue et al. differed from ours is unclear, but may be due to differences in the background strain of C. glabrata and the immunosuppression regimen that was used.
We found that both the Δrpd3 and Δhos2 mutants had increased susceptibility to protamine and caspofungin, and that the Δrpd3 mutant had attenuated virulence in mice. In agreement with our results, Schwarzmuller et al. also determined that a C. glabrata Δrpd3 mutant had increased susceptibility to caspofungin (24). Rpd3 and Hos2 are members of the same class of histone deacetylases, and in Saccharomyces cerevisiae, Δrpd3 and Δhos2 mutants have overlapping phenotypes (12). Other members of this class of histone deacetylases include Hda1, Hos1, and Hos3. We found that the Δhda1 mutant had increased resistance to protamine and wild-type susceptibility to caspofungin, and that the Δhos1 and Δhos3 mutants had wild-type susceptibility to both protamine and caspofungin. Thus, although these histone deacetylases are members of the same family, they have different functions in C. glabrata.
It was found the Δspp1 mutant had increased susceptibility to protamine and caspofungin. Spp1 is predicted to be a member of the COMPASS complex. In S. cerevisiae, this complex functions as a histone methyltransferase, adding three methyl groups to lysine 4 of histone H3 by the Set1 methyltransferase. Spp1 stabilizes Set1 and retards its degradation (25). Our data suggest the COMPASS complex is required for C. glabrata to withstand protamine and caspofungin. The phenotype of a C. glabrata Δspp1 mutant has not been reported previously and a Δset1 mutant was not present in the library. However, it is known that a C. albicans set1Δ/Δ mutant is hyperfilamentous and has attenuated virulence in mice (26). Our experimental results predict the Δset1 mutant would be likely susceptible to both protamine and caspofungin, and perhaps have attenuated virulence.
Testing the virulence of the various C. glabrata mutants in both G. mellonella and mice enabled us to determine the extent of concordance between these two models of disseminated candidiasis. The Δada2 mutant had attenuated virulence in both models, and the Δhos2 and Δspp1 mutants had wild-type virulence in both models. In contrast, the Δgcn5 mutant had reduced virulence only in G. mellonella, and the Δrpd3 mutant had reduced virulence only in mice. These results suggest that although the G. mellonella model has modest predictive value, any C. glabrata mutant that has reduced virulence in this model should be tested in mice to verify the results.
Our data on the susceptibility of the various mutants to host defense peptides provides a possible explanation for why the Δgcn5 mutant had attenuated virulence in G. mellonella, but wild-type virulence in mice. In G. mellonella, the host defense against fungal pathogens is mediated by phenol oxidase, which is involved in the synthesis of melanin and reactive quinones, and host defense peptides (27, 28). G. mellonella makes multiple host defense peptides, including gallerimycin, galiomicin, gloverin, cecropins, and moricins (28–30). Gloverin, the cecropins, and the moricins all form extended or helical structures similar to the human host defense peptide LL-37 (31–34). We found the Δgcn5 mutant was the only strain tested that was hypersusceptible to LL-37 at pH 5.5 and 7.5. The structural similarity between LL-37 and the host defense peptides of G. mellonella suggests that attenuated virulence of the Δgcn5 mutant in G. mellonella may have been due to enhanced susceptibility to gloverin, cecropins A or B, and the moricins. It is highly plausible that the Δgcn5 mutant had wild-type virulence in mice because it was able to resist other types of host defense peptides, such as the β-1 and β-2 defensins.
Although our data indicate that histone modification plays a major role in governing the virulence of C. glabrata and its response to host defense peptides and caspofungin, we do not yet know the precise mechanisms involved. Resistance to host defense peptides can be mediated by multiple mechanisms, including the production of proteases, expression of efflux pumps, membrane remodeling, and changes in membrane energetics (8, 35). Tolerance to echinocandins such as caspofungin can be mediated by activation of stress response pathways that govern cell wall integrity, maintenance, and architecture (36). How epigenetic changes mediated by histone modification activate these resistance mechanisms in C. glabrata is a fruitful area of study because this information holds promise to inform the development of new approaches to combat the growing problem of antifungal drug resistance.
MATERIALS AND METHODS
Deletion mutant library screening.
To identify transcriptional regulators that govern resistance to caspofungin and protamine, a representative host defense peptide, a library of 215 C. glabrata transcriptional regulator deletion mutants constructed in the ATCC 2001 strain background was generously provided by Brendan Cormack at the Johns Hopkins School of Medicine (24). The susceptibility of all the mutants to protamine and caspofungin was determined using an agar-dilution assay (37). Prior to screening, preliminary experiments were performed to determine the concentrations of protamine and caspofungin that partially inhibited the growth of the wild-type strain. These concentrations were found to be 3 mg/ml for protamine and 325 ng/ml for caspofungin. The various mutants and the wild-type control strain were grown in yeast extract peptone dextrose (YPD) broth at 30°C in a shaking incubator overnight. The next day, each culture was adjusted to an optical density at 600 nm (OD600) of 2.7 in YPD broth. Four serial 10-fold dilutions were made in phosphate-buffered saline (PBS), after which 3 µl of each dilution was spotted onto YPD agar containing either 325 ng/ml of caspofungin or 3 mg/ml of protamine. After incubation at 30°C for 48 h, the plates were imaged and the growth of each mutant was compared to that of the wild-type parent strain. Any mutant that differed from the wild-type strain in its susceptibility to caspofungin or protamine was independently tested at least twice to verify the results.
To assess whether selected mutants had increased susceptibility to additional stressors, the above agar dilution method was used to assess growth on calcofluor white (400 µg/ml), hydrogen peroxide (15 mM), SDS (0.2%), and NaCl (0.55 M).
Complementation.
To complement the Δada2, Δgcn5, Δrpd3, Δhos2, and Δspp1 mutants, the protein-coding regions plus 1,000 bp of upstream and 500 bp of downstream flanking sequences of each gene were PCR amplified from C. glabrata genomic DNA. Each resulting PCR product was cut with BamHI and XhoI and ligated into plasmid pGRB2.1+His3 (38), which had been linearized with BamHI and XhoI. The resulting plasmids were transformed into C. glabrata using the lithium acetate method.
Susceptibility testing.
The susceptibility of the various isolates to caspofungin was determined using the CLSI M27 A4 broth microdilution method (39).
Radial diffusion assay.
To assess the susceptibility of the various strains to mammalian host defense peptides, we employed an ultrasensitive radial diffusion assay at 37°C (40). The host defense peptides used were nNP-1 (classical defensin structure, predominantly found in neutrophils), hBD-2 (classical defensin structure, predominantly found in human epithelial tissue), LL-37 (prototypic extended helix structure, predominantly found in human immune cells), and γ-RP-1 (congener of prototypic CS-αβ kinocidn, predominantly released from platelets). Protamine was included as a control. Susceptibility to these host defense peptides was evaluated at pH 5.5 and pH 7.5.
Virulence testing.
The animal studies performed for this research were approved by the Lundquist Institute Animal Care and Use Committee. The virulence of the various C. glabrata mutants was assessed in G. mellonella larvae (41) and neutropenic mouse models of disseminated infection (42). G. mellonella larvae in the final instar stage were inoculated with 2.5 × 106 or 5 × 106 C. glabrata cells in a total volume of 5 μl using a Hamilton syringe. Twelve larvae were inoculated with each strain of C. glabrata and an additional 10 larvae were injected with 5 μl of PBS as a negative control. The larvae were kept in a humidified incubator at 37°C and monitored twice daily for survival. Each strain was tested twice and the results of the two experiments were combined.
The virulence of the mutants was also tested in the neutropenic mouse model of disseminated candidiasis (42). To induce neutropenia, male BALB/c mice were administered 5-fluorouracil via intraperitoneal injection at 150 mg/kg of body weight at day −1 relative to infection. The next day, they were inoculated with 107 C. glabrata cells via the lateral tail vein. After 8 days of infection, the mice were euthanized, after which their kidneys were harvested, weighed, homogenized, and quantitatively cultured. Each strain was tested twice in 6 to 8 mice per experiment and the results were combined.
Statistical analysis.
Using the GraphPad Prism software package, the survival of larvae infected with each mutant was compared with larvae infected with the wild-type strain and the complemented strain using the Gehan-Breslow-Wilcoxon test. The mouse organ fungal burden data were analyzed using the Kruskal-Wallis test. P values of ≤0.05 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Brendan Cormack for generously supplying the transcription factor deletion library.
This work was supported in part by NIH grant R01AI124566.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Cleveland AA, Farley MM, Harrison LH, Stein B, Hollick R, Lockhart SR, Magill SS, Derado G, Park BJ, Chiller TM. 2012. Changes in incidence and antifungal drug resistance in candidemia: results from population-based laboratory surveillance in Atlanta and Baltimore, 2008–2011. Clin Infect Dis 55:1352–1361. doi: 10.1093/cid/cis697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfaller M, Neofytos D, Diekema D, Azie N, Meier-Kriesche HU, Quan SP, Horn D. 2012. Epidemiology and outcomes of candidemia in 3648 patients: data from the Prospective Antifungal Therapy (PATH Alliance(R)) registry, 2004–2008. Diagn Microbiol Infect Dis 74:323–331. doi: 10.1016/j.diagmicrobio.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD. 2016. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 62:e1–e50. doi: 10.1093/cid/civ933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cleveland AA, Harrison LH, Farley MM, Hollick R, Stein B, Chiller TM, Lockhart SR, Park BJ. 2015. Declining incidence of candidemia and the shifting epidemiology of Candida resistance in two US metropolitan areas, 2008–2013: results from population-based surveillance. PLoS One 10:e0120452. doi: 10.1371/journal.pone.0120452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vallabhaneni S, Cleveland AA, Farley MM, Harrison LH, Schaffner W, Beldavs ZG, Derado G, Pham CD, Lockhart SR, Smith RM. 2015. Epidemiology and risk factors for echinocandin nonsusceptible Candida glabrata bloodstream infections: data from a large multisite population-based candidemia surveillance program, 2008–2014. Open Forum Infect Dis 2:ofv163. doi: 10.1093/ofid/ofv163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexander BD, Johnson MD, Pfeiffer CD, Jimenez-Ortigosa C, Catania J, Booker R, Castanheira M, Messer SA, Perlin DS, Pfaller MA. 2013. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin Infect Dis 56:1724–1732. doi: 10.1093/cid/cit136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shields RK, Kline EG, Healey KR, Kordalewska M, Perlin DS, Nguyen MH, Clancy CJ. 2018. Spontaneous mutational frequency and FKS mutation rates vary by echinocandin agent against Candida glabrata. Antimicrob Agents Chemother 63:e01692-18. doi: 10.1128/AAC.01692-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeaman MR, Yount NY. 2003. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev 55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 9.Yount NY, Weaver DC, Lee EY, Lee MW, Wang H, Chan LC, Wong GCL, Yeaman MR. 2019. Unifying structural signature of eukaryotic alpha-helical host defense peptides. Proc Natl Acad Sci U S A 116:6944–6953. doi: 10.1073/pnas.1819250116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vylkova S, Nayyar N, Li W, Edgerton M. 2007. Human beta-defensins kill Candida albicans in an energy-dependent and salt-sensitive manner without causing membrane disruption. Antimicrob Agents Chemother 51:154–161. doi: 10.1128/AAC.00478-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol 19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma VM, Tomar RS, Dempsey AE, Reese JC. 2007. Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol Cell Biol 27:3199–3210. doi: 10.1128/MCB.02311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acquaviva L, Szekvolgyi L, Dichtl B, Dichtl BS, de La Roche Saint Andre C, Nicolas A, Geli V. 2013. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science 339:215–218. doi: 10.1126/science.1225739. [DOI] [PubMed] [Google Scholar]
- 14.Miyazaki T, Yamauchi S, Inamine T, Nagayoshi Y, Saijo T, Izumikawa K, Seki M, Kakeya H, Yamamoto Y, Yanagihara K, Miyazaki Y, Kohno S. 2010. Roles of calcineurin and Crz1 in antifungal susceptibility and virulence of Candida glabrata. Antimicrob Agents Chemother 54:1639–1643. doi: 10.1128/AAC.01364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dilova I, Chen CY, Powers T. 2002. Mks1 in concert with TOR signaling negatively regulates RTG target gene expression in S. cerevisiae. Curr Biol 12:389–395. doi: 10.1016/S0960-9822(02)00677-2. [DOI] [PubMed] [Google Scholar]
- 16.Matsuura A, Anraku Y. 1993. Characterization of the MKS1 gene, a new negative regulator of the Ras-cyclic AMP pathway in Saccharomyces cerevisiae. Mol Gen Genet 238:6–16. doi: 10.1007/BF00279524. [DOI] [PubMed] [Google Scholar]
- 17.Daniel JA, Grant PA. 2007. Multi-tasking on chromatin with the SAGA coactivator complexes. Mutat Res 618:135–148. doi: 10.1016/j.mrfmmm.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eberharter A, Sterner DE, Schieltz D, Hassan A, Yates JR, 3rd, Berger SL, Workman JL. 1999. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol Cell Biol 19:6621–6631. doi: 10.1128/mcb.19.10.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sendra R, Tse C, Hansen JC. 2000. The yeast histone acetyltransferase A2 complex, but not free Gcn5p, binds stably to nucleosomal arrays. J Biol Chem 275:24928–24934. doi: 10.1074/jbc.M003783200. [DOI] [PubMed] [Google Scholar]
- 20.Sellam A, Askew C, Epp E, Lavoie H, Whiteway M, Nantel A. 2009. Genome-wide mapping of the coactivator Ada2p yields insight into the functional roles of SAGA/ADA complex in Candida albicans. Mol Biol Cell 20:2389–2400. doi: 10.1091/mbc.e08-11-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shivarathri R, Tscherner M, Zwolanek F, Singh NK, Chauhan N, Kuchler K. 2019. The fungal histone acetyl transferase Gcn5 controls virulence of the human pathogen Candida albicans through multiple pathways. Sci Rep 9:9445. doi: 10.1038/s41598-019-45817-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kounatidis I, Ames L, Mistry R, Ho HL, Haynes K, Ligoxygakis P. 2018. A host-pathogen interaction screen identifies ada2 as a mediator of Candida glabrata defenses against reactive oxygen species. G3 (Bethesda) 8:1637–1647. doi: 10.1534/g3.118.200182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu SJ, Chang YL, Chen YL. 2018. Deletion of ADA2 increases antifungal drug susceptibility and virulence in Candida glabrata. Antimicrob Agents Chemother 62:e01924-17. doi: 10.1128/AAC.01924-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarzmuller T, Ma B, Hiller E, Istel F, Tscherner M, Brunke S, Ames L, Firon A, Green B, Cabral V, Marcet-Houben M, Jacobsen ID, Quintin J, Seider K, Frohner I, Glaser W, Jungwirth H, Bachellier-Bassi S, Chauvel M, Zeidler U, Ferrandon D, Gabaldon T, Hube B, d'Enfert C, Rupp S, Cormack B, Haynes K, Kuchler K. 2014. Systematic phenotyping of a large-scale Candida glabrata deletion collection reveals novel antifungal tolerance genes. PLoS Pathog 10:e1004211. doi: 10.1371/journal.ppat.1004211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thornton JL, Westfield GH, Takahashi YH, Cook M, Gao X, Woodfin AR, Lee JS, Morgan MA, Jackson J, Smith ER, Couture JF, Skiniotis G, Shilatifard A. 2014. Context dependency of Set1/COMPASS-mediated histone H3 Lys4 trimethylation. Genes Dev 28:115–120. doi: 10.1101/gad.232215.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raman SB, Nguyen MH, Zhang Z, Cheng S, Jia HY, Weisner N, Iczkowski K, Clancy CJ. 2006. Candida albicans SET1 encodes a histone 3 lysine 4 methyltransferase that contributes to the pathogenesis of invasive candidiasis. Mol Microbiol 60:697–709. doi: 10.1111/j.1365-2958.2006.05121.x. [DOI] [PubMed] [Google Scholar]
- 27.García-Carnero LC, Clavijo-Giraldo DM, Gómez-Gaviria M, Lozoya-Pérez NE, Tamez-Castrellón AK, López-Ramírez LA, Mora-Montes HM. 2020. Early virulence predictors during the Candida species-Galleria mellonella interaction. J Fungi (Basel) 6:152. doi: 10.3390/jof6030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vertyporokh L, Wojda I. 2020. Immune response of Galleria mellonella after injection with non-lethal and lethal dosages of Candida albicans. J Invertebr Pathol 170:107327. doi: 10.1016/j.jip.2020.107327. [DOI] [PubMed] [Google Scholar]
- 29.Barros PP, Rossoni RD, Ribeiro FC, Silva MP, Souza CM, Jorge AOC, Junqueira JC. 2019. Two sporulated Bacillus enhance immunity in Galleria mellonella protecting against Candida albicans. Microb Pathog 132:335–342. doi: 10.1016/j.micpath.2019.05.023. [DOI] [PubMed] [Google Scholar]
- 30.Brown SE, Howard A, Kasprzak AB, Gordon KH, East PD. 2009. A peptidomics study reveals the impressive antimicrobial peptide arsenal of the wax moth Galleria mellonella. Insect Biochem Mol Biol 39:792–800. doi: 10.1016/j.ibmb.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Axén A, Carlsson A, Engström A, Bennich H. 1997. Gloverin, an antibacterial protein from the immune hemolymph of Hyalophora pupae. Eur J Biochem 247:614–619. doi: 10.1111/j.1432-1033.1997.00614.x. [DOI] [PubMed] [Google Scholar]
- 32.Hemmi H, Ishibashi J, Hara S, Yamakawa M. 2002. Solution structure of moricin, an antibacterial peptide, isolated from the silkworm Bombyx mori. FEBS Lett 518:33–38. doi: 10.1016/s0014-5793(02)02637-6. [DOI] [PubMed] [Google Scholar]
- 33.Gong Z, Ikonomova SP, Karlsson AJ. 2018. Secondary structure of cell-penetrating peptides during interaction with fungal cells. Protein Sci 27:702–713. doi: 10.1002/pro.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sancho-Vaello E, Gil-Carton D, François P, Bonetti EJ, Kreir M, Pothula KR, Kleinekathöfer U, Zeth K. 2020. The structure of the antimicrobial human cathelicidin LL-37 shows oligomerization and channel formation in the presence of membrane mimics. Sci Rep 10:17356. doi: 10.1038/s41598-020-74401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bechinger B, Gorr SU. 2017. Antimicrobial peptides: mechanisms of action and resistance. J Dent Res 96:254–260. doi: 10.1177/0022034516679973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perlin DS. 2015. Mechanisms of echinocandin antifungal drug resistance. Ann N Y Acad Sci 1354:1–11. doi: 10.1111/nyas.12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruno VM, Kalachikov S, Subaran R, Nobile CJ, Kyratsous C, Mitchell AP. 2006. Control of the C. albicans cell wall damage response by transcriptional regulator Cas5. PLoS Pathog 2:e21. doi: 10.1371/journal.ppat.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zordan RE, Ren Y, Pan SJ, Rotondo G, De Las Penas A, Iluore J, Cormack BP. 2013. Expression plasmids for use in Candida glabrata. G3 (Bethesda) 3:1675–1686. doi: 10.1534/g3.113.006908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clinical and Laboratory Standards Institute. 2017. Reference method for broth dilution antifungal susceptibility testing of yeasts M27-A4, 4th ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 40.Yeaman MR, Yount NY, Waring AJ, Gank KD, Kupferwasser D, Wiese R, Bayer AS, Welch WH. 2007. Modular determinants of antimicrobial activity in platelet factor-4 family kinocidins. Biochim Biophys Acta 1768:609–619. doi: 10.1016/j.bbamem.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brennan M, Thomas DY, Whiteway M, Kavanagh K. 2002. Correlation between virulence of Candida albicans mutants in mice and Galleria mellonella larvae. FEMS Immunol Med Microbiol 34:153–157. doi: 10.1111/j.1574-695X.2002.tb00617.x. [DOI] [PubMed] [Google Scholar]
- 42.Wiederhold NP, Najvar LK, Bocanegra R, Molina D, Olivo M, Graybill JR. 2007. In vivo efficacy of anidulafungin and caspofungin against Candida glabrata and association with in vitro potency in the presence of sera. Antimicrob Agents Chemother 51:1616–1620. doi: 10.1128/AAC.00105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.