Protective immunity against the obligate intracellular bacterium Chlamydia has long been thought to rely on CD4 T cell-dependent gamma interferon (IFN-γ) production. Nevertheless, whether IFN-γ is produced by other cellular sources during Chlamydia infection and how CD4 T cell-dependent and -independent IFN-γ contribute differently to host resistance have not been carefully evaluated.
KEYWORDS: IFN-γ, innate, CD4 T cells, infection, Chlamydia
ABSTRACT
Protective immunity against the obligate intracellular bacterium Chlamydia has long been thought to rely on CD4 T cell-dependent gamma interferon (IFN-γ) production. Nevertheless, whether IFN-γ is produced by other cellular sources during Chlamydia infection and how CD4 T cell-dependent and -independent IFN-γ contribute differently to host resistance have not been carefully evaluated. In this study, we dissected the requirements of IFN-γ produced by innate immune cells and CD4 T cells for resolution of Chlamydia muridarum female reproductive tract (FRT) infection. After C. muridarum intravaginal infection, IFN-γ-deficient and T cell-deficient mice exhibited opposite phenotypes for survival and bacterial shedding at the FRT mucosa, demonstrating the distinct requirements for IFN-γ and CD4 T cells in host defense against Chlamydia. In Rag1-deficient mice, IFN-γ produced by innate lymphocytes (ILCs) accounted for early bacterial control and prolonged survival in the absence of adaptive immunity. Although type I ILCs are potent IFN-γ producers, we found that mature NK cells and ILC1s were not the sole sources of innate IFN-γ in response to Chlamydia. By conducting T cell adoptive transfer, we showed definitively that IFN-γ-deficient CD4 T cells were sufficient for effective bacterial killing in the FRT during the first 21 days of infection and reduced bacterial burden more than 1,000-fold, although mice receiving IFN-γ-deficient CD4 T cells failed to completely eradicate the bacteria from the FRT like their counterparts receiving wild-type (WT) CD4 T cells. Together, our results revealed that innate IFN-γ is essential for preventing systemic Chlamydia dissemination, whereas IFN-γ produced by CD4 T cells is largely redundant at the FRT mucosa.
INTRODUCTION
Chlamydia trachomatis is the obligate intracellular bacterium that causes the most prevalent sexually transmitted infection worldwide. The prevalence of Chlamydia infection is partially attributed to its nature as a “silent infection,” as most women infected with C. trachomatis are asymptomatic and can resolve the infection spontaneously (1). Unfortunately, undiagnosed infections not only facilitate silent disease transmissions but can also lead to severe adverse effects, such as pelvic inflammatory disease, ectopic pregnancy, and infertility (2, 3). There is no licensed human Chlamydia vaccine available at present, partially owing to the lack of complete understanding of protective immune mechanisms (4–6).
CD4 T helper 1 (Th1) cell-dependent gamma interferon (IFN-γ) production promotes macrophage activation to eliminate intracellular pathogens within these professional phagocytes. This defense mechanism operates efficiently against bacterial pathogens that exhibit marked infection tropism for macrophages (7, 8). For Chlamydia, infection is largely restricted to epithelial cells at barrier tissues such as the eyes, lungs, and reproductive tract. Recent studies have shed light on the mechanisms of cell autonomous immunity against both human pathogen C. trachomatis and the mouse-adapted pathogen Chlamydia muridarum in epithelial cells in response to IFN-γ (9–11). Nevertheless, the cellular source for IFN-γ at different anatomic sites and their roles at different stages of infection remain to be characterized. Moreover, with increased knowledge of CD4 T cell biology, it is speculated that protective T cell responses to intracellular bacterium such as Chlamydia are likely to be more complex than Th1-dependent IFN-γ production (12).
The mouse models of C. muridarum infection of the female reproductive tract (FRT) provide an invaluable tool for studying host immunity to Chlamydia infection. Research in the field has established a predominant role for CD4 T cells and antibody in host resistance and vaccine-afforded protection against Chlamydia (4, 13–15). After C. muridarum intravaginal infection, WT B6 mice exhibit a self-limiting infection that resolves within 4 to 5 weeks. Similar courses of infection have also been observed in mice lacking B cells, CD8 T cells, and γδT cells, demonstrating the redundant roles of these cells in C. muridarum clearance from the female reproductive tract (16–18). In contrast, nude mice and mice lacking major histocompatibility complex class II (MHC-II)-restricted CD4 T cells shed persistent high levels of bacteria from the FRT without showing obvious signs of wasting (18–20). While a large body of evidence points to a role for Th1 but not other Th lineage cells in protective immunity against Chlamydia (17, 21–26), it remains perplexing that mice lacking the major Th1 effector cytokine IFN-γ are capable of eliminating >99% of C. muridarum from the FRT, although they suffer from multiorgan disseminated infections (17, 27).
The distinct phenotypes of CD4 T cell-deficient and IFN-γ-deficient mice after C. muridarum infection prompted us to investigate the definitive contributions of CD4 T cell-dependent and -independent IFN-γ production in host defense against Chlamydia. We hypothesized that IFN-γ produced during innate immune response prevents lethal disseminated infection, while CD4-dependent IFN-γ production is largely dispensable at the FRT mucosa. We tested these hypotheses using loss- and gain-of-function approaches in gene-deficient mouse models in which contributions of innate and adaptive IFN-γ production can be dissected separately.
RESULTS
IFN-γ-deficient and T cell-deficient mice exhibit opposite phenotypes after Chlamydia muridarum intravaginal infection.
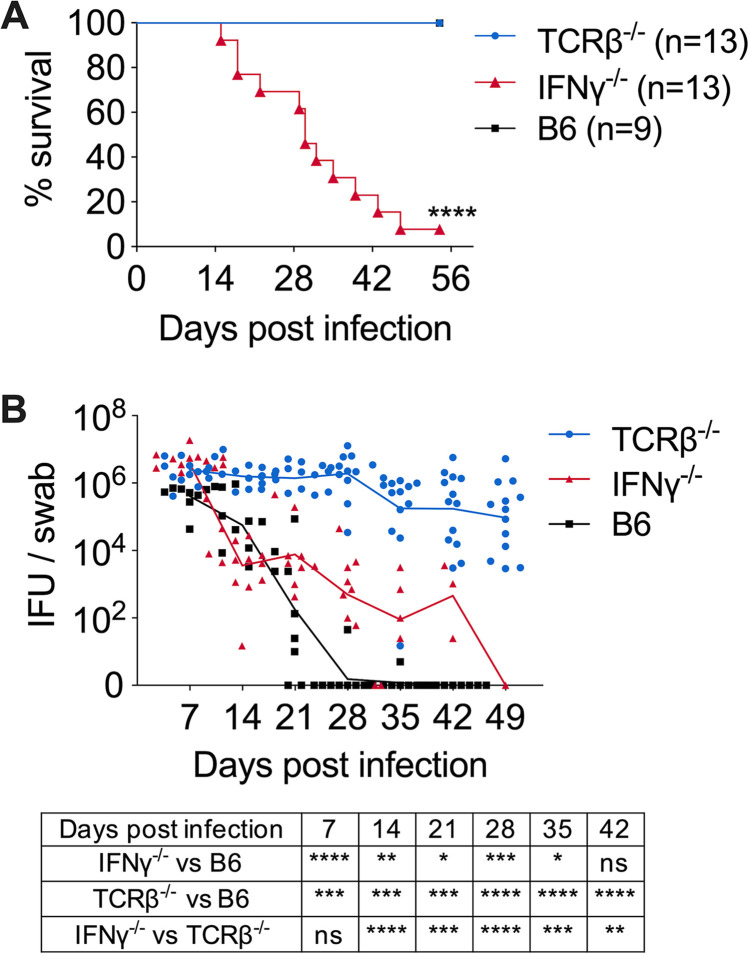
In order to directly compare the phenotypes of IFN-γ-deficient and T cell-deficient mice, we infected IFN-γ−/− and TCRβ−/− mice intravaginally with C. muridarum strain Nigg II and compared survival and bacterial shedding during primary infections. Consistent with previous findings (17), 100% of TCRβ−/− mice survived the infection with no obvious sign of wasting disease (Fig. 1A). Meanwhile, these mice manifested high-grade, persistent bacterial shedding with an average of >105 bacteria recovered from the FRT for at least 60 days (Fig. 1A and B). In contrast, over 90% of IFN-γ−/− mice succumbed to infection between days 15 and 47 after infection (Fig. 1A). Notably, IFN-γ−/− mice that survived by day 35 postinfection exhibited an ∼10,000-fold reduction in FRT bacterial burden compared to day 7 (Fig. 1B). These opposite phenotypes of T cell-deficient and IFN-γ-deficient mice suggest that distinct host defense mechanisms are involved in IFN-γ- and T cell-dependent Chlamydia containment in systemic versus mucosal tissues. Importantly, a non-αβ T cell source of IFN-γ in TCRβ−/− mice must have contributed to systemic Chlamydia control for their long-term survival.
FIG 1.
IFN-γ-deficient mice and αβ T cell-deficient mice exhibit opposite phenotypes after Chlamydia muridarum intravaginal infection. B6, IFN-γ−/−, and TCRβ−/− mice were infected intravaginally with 1 × 105 C. muridarum. Survival (A) and bacterial shedding (B) from the lower female reproductive tract (FRT) were monitored by vaginal swabs. Data are combined results of three independent experiments with 9 to 13 mice per group. Each data point represents an individual mouse. Lines represent mean log10-transformed values. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
ILCs are essential for systemic bacterial control and prolonged survival of Rag1−/− mice.
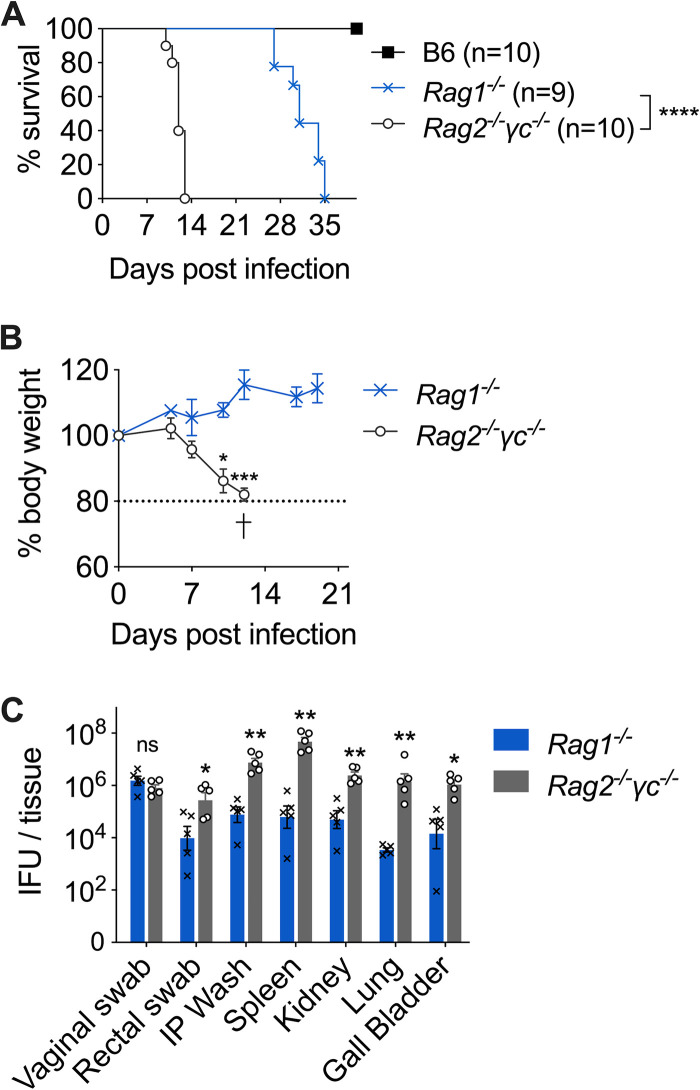
The nonlethal phenotype of TCRβ−/− mice led us to hypothesize that innate lymphocytes (ILCs) are essential for preventing lethal Chlamydia dissemination. To test this hypothesis, we infected Rag1−/− and Rag2−/− γc−/− mice intravaginally with C. muridarum and monitored survival. Rag2−/− γc−/− mice quickly succumbed to infection, with a median survival time of 12 days. This is significantly shorter than the average 30 days of survival in Rag1−/− mice (Fig. 2A). Fast weight loss was observed in Rag2−/− γc−/− mice starting from day 5 postinfection (dpi), but not in Rag1−/− mice (Fig. 2B). By the time Rag2−/− γc−/− mice were moribund (10 to 13 dpi), we detected widespread bacterial dissemination in these mice, with high bacterial burdens in systemic tissues, including spleen, kidney, lung, ball bladder, and peritoneal cavity, as well as vaginal and rectal mucosa (Fig. 2C). Notably, while all systemic tissues of Rag2−/− γc−/− mice exhibit 1- to 4-log-higher bacteremia than Rag1−/− mice, Chlamydia burdens at the FRT mucosa were not significantly different between the two strains (Fig. 2C). These results demonstrate that ILCs are essential for systemic Chlamydia control but are redundant for host resistance to Chlamydia at the FRT mucosa in the absence of adaptive immunity.
FIG 2.
Innate lymphocytes (ILCs) are essential for systemic bacterial control and prolonged survival of Rag1-deficient mice. B6, Rag1−/−, and Rag2−/− γc−/− mice were infected intravaginally with 1 × 105 C. muridarum organisms. (A) Survival. (B) Body weight. (C) Bacterial burdens in vaginal swabs, rectal swabs, and systemic organs determined at 12 days postinfection. Data shown are combined results of two independent experiments with 9 or 10 mice per group (A) or representative results of two independent experiments with 3 to 5 mice per group in each experiment (B and C). Each data point represents an individual mouse. Bars and error bars represent means and standard errors of the means (SEM). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
Innate IFN-γ production, partially by NK cells and ILC1s, is essential for early control of Chlamydia dissemination.
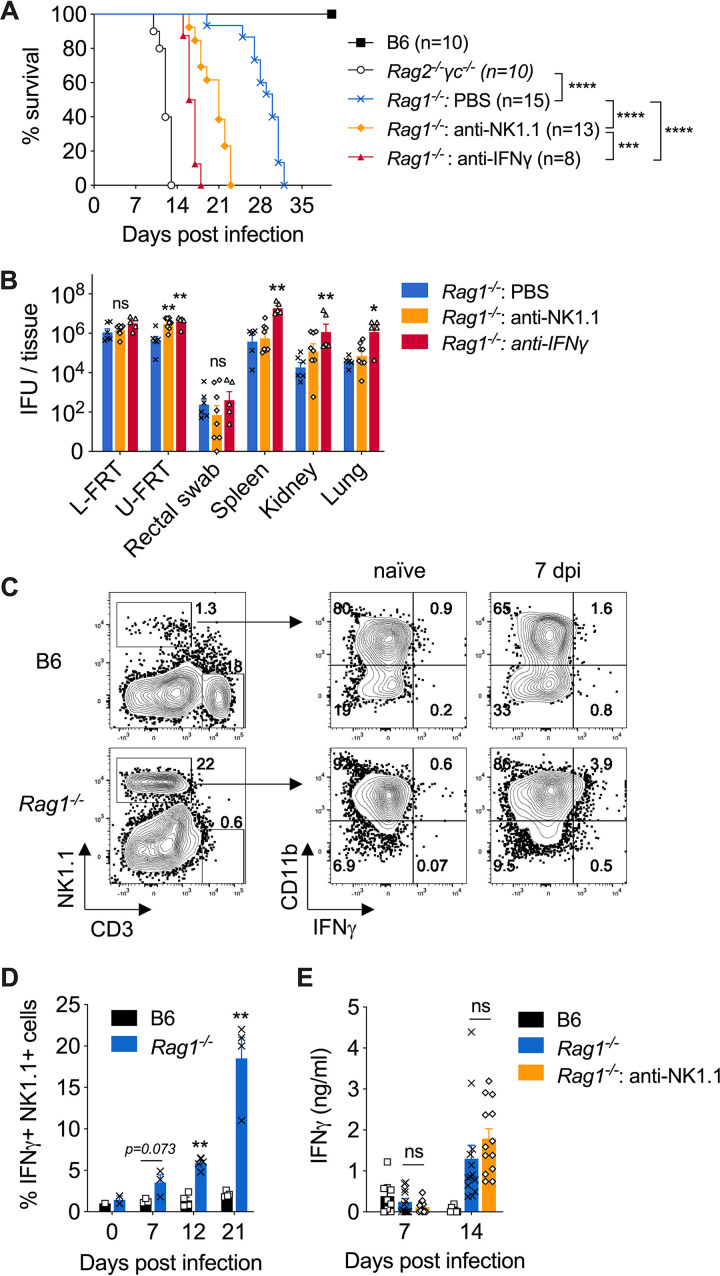
We next investigated whether IFN-γ production by ILCs, in particular the group 1 ILCs, including NK cells and ILC1s, accounts for the key innate effector mechanism for early control of C. muridarum systemic dissemination. To do this, we depleted either IFN-γ or NK1.1+ cells from Rag1−/− mice and monitored survival. As shown in Fig. 3A, both IFN-γ- and NK1.1-depleted groups displayed accelerated death compared to the phosphate-buffered-saline (PBS)-treated group, with average survival times of 16.5 and 21 days, respectively. At day 14 postinfection, bacterial burdens in the lower FRT and on rectal swabs were not affected by either depletion, while significantly more C. muridarum organisms were detected in both IFN-γ- and NK1.1-depleted groups in the upper FRT (Fig. 3B). Anti-IFN-γ treatment resulted in more than a 1.5-log increase in C. muridarum burdens in all systemic tissues, including spleen, kidney, and lung. The anti-NK1.1-treated group showed trends of higher systemic bacteria burdens than PBS-treated group, but such differences did not reach statistical significance at day 14, which is 1 week earlier than their average survival time (Fig. 3B). IFN-γ secretion by mature group 1 ILCs (CD11b+ NK1.1+) were readily detectable in the spleens of Rag1−/− mice throughout the course of infection but were not evident in wild-type (WT) B6 controls (Fig. 3C and D). Unexpectedly, circulating IFN-γ levels in Rag1−/− mice were not significantly affected by NK1.1 depletion at both days 7 and 14 postinfection (Fig. 3E). Together, these findings led us to conclude that both innate IFN-γ and group 1 ILCs are essential for early containment of Chlamydia dissemination. While NK1.1+ group 1 ILCs are potent IFN-γ producers, they do not seem to be the only source for IFN-γ derived from innate immune responses following C. muridarum intravaginal infection.
FIG 3.
Innate IFN-γ and group 1 ILCs contribute to host resistance to lethal Chlamydia dissemination in Rag1−/− mice. B6, Rag1−/−, and Rag2−/− γc−/− mice were infected intravaginally with 1 × 105 C. muridarum organisms. Groups of Rag1−/− mice were treated with either anti-IFN-γ or anti-NK1.1 depleting Abs throughout the infection. (A) Survival. (B) Bacterial burdens in lower FRT (L-FRT), upper FRT (U-FRT), rectal swabs, and systemic organs determined at 14 days postinfection. (C) Representative flow cytometry plots showing IFN-γ secretion by CD11b+ NK1.1+ cells detected by IFN-γ secretion assay. (D) Percentages of IFN-γ-producing CD11b+ NK1.1+ cells quantified based on the flow cytometry analysis in panel C. (E) Serum IFN-γ level on days 7 and 14 after infection, as measured by IFN-γ cytokine enzyme-linked immunosorbent assay (ELISA). Data in panels A, B, D, and E are combined results of at least two independent experiments with 3 to 5 mice per group in each experiment. Each data point in panels B, D, and E represents an individual mouse. Bars and error bars represent means and SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
IFN-γ production by CD4 T cells is largely redundant at the FRT mucosa.
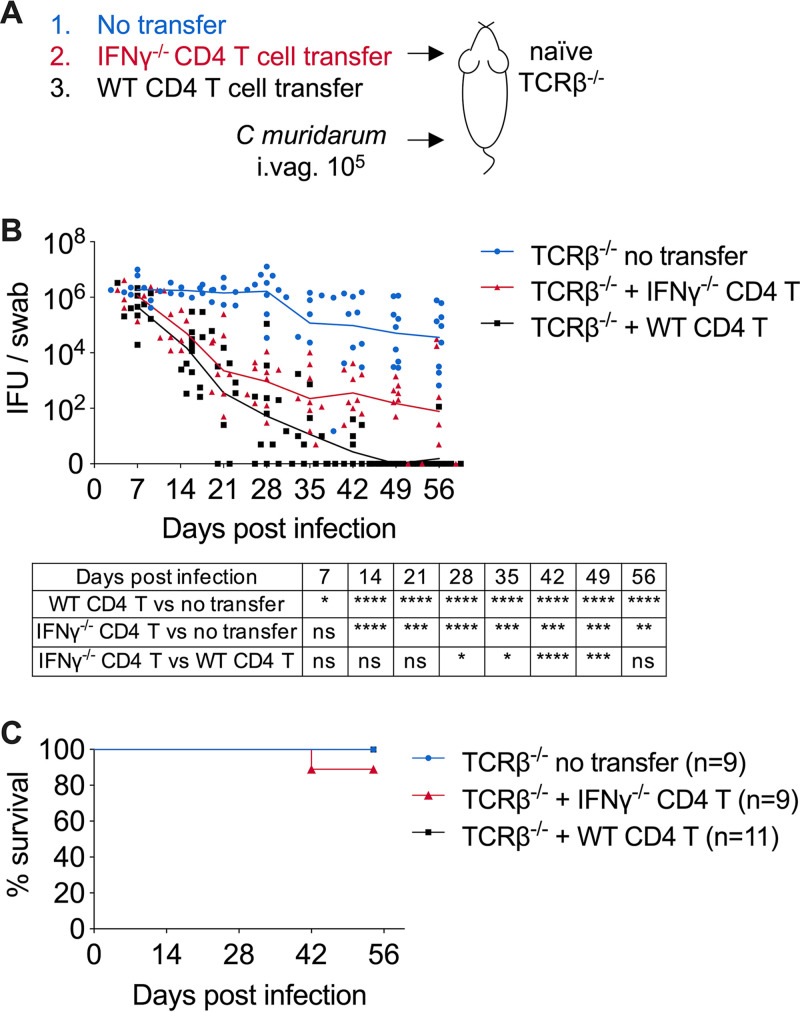
The complete lack of IFN-γ production in IFN-γ−/− mice allows the evaluation of only the global effect of this cytokine, which derives from numerous cellular sources. To specifically define the contribution of CD4 T cell-dependent IFN-γ production to host resistance to Chlamydia, we conducted T cell adoptive transfer experiments in which we transferred either IFN-γ-sufficient (WT) or IFN-γ-deficient (IFN-γ−/−) CD4 T cells into innate immunity-intact TCRβ−/− mice and challenged the recipients intravaginally with C. muridarum (Fig. 4A). Compared to the WT CD4 T cell transfer group, TCRβ−/− mice receiving IFN-γ-deficient CD4 T cells exhibited similar rates of bacterial containment for the first 21 days (fold change in log10, 3.1 ± 1.8 in WT CD4 transfer versus 2.7 ± 1.2 in IFN-γ−/− CD4 transfer; P = 0.21) (Fig. 4B). Chlamydia burden continued to decrease another 10-fold in the IFN-γ−/− CD4 T cell transfer group before these mice entered the chronic, low-grade shedding phase around day 35 (Fig. 4B). The cumulative ∼5,000-fold decrease in bacterial burden demonstrated that IFN-γ produced by CD4 T cells is dispensable for eliminating vast majority of the pathogen from the FRT. Last, as a result of the functional innate immunity in TCRβ−/− recipient mice, the lethal dissemination phenotype was completely rescued in this adoptive transfer model compared to IFN-γ−/− mice, reinforcing the idea that innate IFN-γ is sufficient for systemic Chlamydia containment (Fig. 4C).
FIG 4.
CD4 T cell-dependent IFN-γ production is largely redundant at the FRT mucosa. CD4 T cells isolated from WT B6 or IFN-γ−/− mice were adoptively transferred to TCRβ−/− mice. Recipient TCRβ−/− mice were infected intravaginally with 1 × 105 C. muridarum organisms. (A) Schematic depicting the TCRβ−/− adoptive transfer experimental setup. (B and C) Bacterial shedding from the FRT (B) and survival of TCRβ−/− recipient mice (C) after receiving naive CD4 T cells from either WT or IFN-γ−/− donors and intravaginal infection. Data are combined results of three independent experiments with 6 to 11 mice per group. Each data point in panels B and C represents an individual mouse. Lines represent mean log10-transformed values. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
DISCUSSION
The ability of T cells to produce IFN-γ in response to infection is an important readout for their antigen specificity and effector function. Although Th1 cells are essential for combating intracellular parasites, the reliance on IFN-γ to confer protection appears to be pathogen specific. Mice lacking the IFN-γ receptor or the Th1 transcription factor T-bet are highly susceptible to Salmonella infection (28, 29). In contrast, effective control of Mycobacterium tuberculosis infection depends on Th1 cells but has a minimal requirement of IFN-γ in the lung (30–32). The importance of CD4 T cells in host resistance to Chlamydia is highlighted by the absolute requirement of MHC-II and αβTCR for bacterial control in the FRT mucosa, while the most prominent phenotype of IFN-γ−/− mice after C. muridarum intravaginal infection is systemic bacterial dissemination (17, 18, 27). By conducting side-by-side comparison of TCRβ−/− and IFN-γ−/− mice in this study, we confirmed these previous findings and demonstrated unambiguously that hosts had distinct requirements for CD4 T cells and IFN-γ for Chlamydia resistance.
TCRβ−/− mice lack αβ T cells but retain relatively intact innate immunity and several adaptive immune components, such as γδ T cells and T-independent antibody, all of which may contribute to the long-term survival of the hosts. As part of the innate immune system, ILCs are known for functions that parallel their T cell counterparts but differ from T cells in their embryonic origin, tissue residency, lack of rearranged receptor for Ag recognition, and unique contribution to tissue integrity (33). Immune responses of the ILCs are critical for early control of pathogen replication before the host launches an effective adaptive immune response. This defense mechanism could be particularly important for containing mucosal pathogens like Chlamydia, since the target FRT tissue lacks defined lymphoid structure, and consequently, effective adaptive immune responses are significantly delayed (34, 35). Using the Rag1−/− and Rag2−/− γc−/− models, we showed in this study that removing ILCs from the innate immune system had a detrimental effect on host resistance to Chlamydia, as Rag2−/− γc−/− mice quickly succumbed to disseminated infection with high-grade bacteremia. In contrast, ILC-sufficient Rag1−/− mice exhibited significantly longer survival and stable body weights. These observations made it evident that ILCs are essential for preventing early systemic Chlamydia dissemination in the absence of adaptive immunity. Given the early responses of ILCs, it is unlikely that ILCs are completely redundant with adaptive responses to prevent early bacterial dissemination in immunocompetent hosts (36), although this notion needs to be confirmed experimentally.
Our IFN-γ and NK1.1 depletion experiment directly addressed the idea that both innate IFN-γ and NK1.1+ group 1 ILCs contribute to host resistance to lethal Chlamydia dissemination in Rag1−/− mice. Group I ILCs, including cytotoxic NK cells and noncytotoxic ILC1s, are the major source of IFN-γ during innate immune responses, along with neutrophils, macrophages, dendritic cells (DCs), and a subset of ILC3s (37–39). In immunocompetent hosts, IFN-γ production by NK cells can be detected as early as 4 h after intravaginal C. muridarum infection (40). The early response of NK cells dictates CD4 T cell differentiation and memory responses during C. muridarum lung infections (41, 42). Recently, Poston et al. showed that T cell-independent IFN-γ cooperates with B cells to prevent lethal dissemination caused by the highly virulent C. muridarum strain CM001 (20). Our data add to previous findings by showing ex vivo IFN-γ secretion on the surface of CD11b+ NK1.1+ cells, indicating that group 1 ILCs carry out a protective function, as least in part, by their production of IFN-γ in the absence of T cells. It should be noted that both IFN-γ-depleted and NK1.1-depleted Rag1−/− mice exhibited slightly longer survival times than Rag2−/− γc−/− mice, indicating that additional cellular sources, perhaps ILC3s, may also produce IFN-γ and confer protection against Chlamydia, as shown in mouse endometrium tissue in recent studies (43, 44). Additionally, other protective mechanisms independent of IFN-γ may be involved in ILC-mediated protection. It is intriguing to observe that serum IFN-γ was not significantly affected by anti-NK1.1 treatment in Rag1−/− mice and was minimally detected in WT animals. These results suggest two non-mutually exclusive probabilities. First, circulating IFN-γ is neither necessary nor sufficient for protection. Instead, close proximity between IFN-γ-secreting cells and responders are essential for bacterial control (45). Second, circulating IFN-γ is more efficiently removed by IFN-γ receptor-expressing cells in immune-sufficient mice (46). Future experiments are required to address these issues directly.
The sustained Chlamydia replication in the FRT of TCRβ−/− mice suggested that innate immune cells, γδ T cells, and T-independent antibodies (Abs) are incapable of restraining Chlamydia growth in the FRT mucosa. In contrast, adoptive transfer of CD4 T cells, regardless of their ability to produce IFN-γ, reduced bacterial burden to less than 0.1% of the peak burden within 3 weeks. These results are in agreement with previous studies using IFN-γ−/− mice, but this study differs from them by retaining intact innate IFN-γ in our system, thereby preventing lethal disseminated infection (17, 27). By solely manipulating the CD4 T cell compartment, we showed that CD4 T cells are both necessary and sufficient for C. muridarum control in the FRT. More importantly, we showed unequivocally that CD4 T cell-derived IFN-γ is largely redundant for protective immunity in the FRT, at least during the early phase of bacterial replication.
Numerous studies in the field have firmly established the importance of the pleiotropic cytokine IFN-γ in C. muridarum and C. trachomatis infection, in vitro and in vivo (10, 47–57). Our study, along with many others, has demonstrated that early IFN-γ production by ILCs is essential for preventing bacterial dissemination and achieving complete eradication of C. muridarum from the FRT at a late stage of infection relies on CD4 T cell-derived IFN-γ (17, 27). Non-lymphogranuloma venereum (LGV) strains of C. trachomatis rarely cause disseminated infection in humans. In contrast, low-grade C. trachomatis shedding is commonly observed in otherwise healthy individuals (58). Therefore, it is reasonable to speculate that IFN-γ effectively blocks Chlamydia dissemination in humans, whereas a further evolved adaptive immune response is likely required to overcome C. trachomatis evasion of IFN-γ-induced cell-autonomous immunity in FRT epithelium (10). In line with this argument, our CD4 T cell adoptive transfer studies in mice emphasized that a highly effective mechanism of T helper cell-mediated protection against mucosal Chlamydia infection independent of IFN-γ is yet to be discovered. Efforts to search for such a protective mechanism are urgently needed, as precise understanding of protective immunity is fundamental to the rational design of a much-needed Chlamydia vaccine.
Fortunately, recent studies have started to shed light on several important aspects of CD4 T cell biology related to host protective responses beyond IFN-γ production. Yu et al. showed that vaccination using live or dead C. muridarum EB elicits different degrees of protection that correlates with the frequency of multifunction Th1 cells (23). Likewise, a recently developed C. muridarum TCR-transgenic model has revealed that the ability of monoclonal Ag-specific CD4 T cells to coproduce IFN-γ, tumor necrosis factor alpha (TNF-α), and interleukin 2 (IL-2) is essential for their protective efficacy (59). Using MHC-II tetramers, we demonstrated that C. muridarum infection induces a highly heterogenous T helper response dominated by Th1 and accompanied by fractions of Chlamydia-specific Treg and Th17 cells in lymphoid and mucosal tissues (35). Other Th lineages, such as an IFN-γ- and IL-13-producing CD4 T cell clone, and a Th2-dominant response in human C. trachomatis infection have also been documented (60, 61). Finally, an elegant C. trachomatis vaccine study conducted by Stary et al. revealed that T cell activation, effector functions, and formation of tissue-resident memory should all be taken into consideration when protective efficacy afforded by a vaccine is evaluated (62). Our knowledge of CD4 T cell differentiation is evolving quickly as a result of groundbreaking technologies. It is proposed that a continuum of CD4 T cell states will likely replace our traditional understanding of defined CD4 T cell lineages (63). With the increased knowledge and higher resolution tool to understand CD4 T cell biology, a refreshed notion of protective immunity against Chlamydia will likely emerge.
MATERIALS AND METHODS
Mice.
C57BL/6 (B6), IFN-γ−/− (B6.129S7-Ifngtm1Ts/J), Rag1−/− (B6.129S7-Rag1tm1Mom/J), and TCRβ−/− (B6. 129P2-Tcrbtm1Mom/J) mice were purchased from The Jackson Laboratory. Rag2−/− γc−/− mice were purchased from Taconic Biosciences. All mice used for experiments were 6 to 16 weeks old, unless otherwise noted. Mice were maintained under specific-pathogen-free (SPF) conditions, and all mouse experiments were approved by the University of Arkansas for Medical Sciences (UAMS) Institutional Animal Care and Use Committee (IACUC).
Bacteria.
Chlamydia muridarum strain Nigg II was originally purchased from ATCC (VR-123; Manassas, VA). The organism was propagated in HeLa 229 cells. Elementary bodies (EBs) were purified by renografin discontinuous density gradient centrifugation, aliquoted, and stored at −80°C until use. EBs were titrated on HeLa 229 cells as previously described (35).
Infection and bacteria enumeration.
Mice were synchronized for estrus by subcutaneous injection of 2.5 mg Depo-Provera (Greenstone, NJ), 5 to 7 days prior to intravaginal infection. For intravaginal infection, 1 × 105 C. muridarum organisms in sucrose-phosphate-glutamic acid (SPG) buffer were deposited directly into the vaginal vault using a pipet tip. To enumerate bacterial shedding from the FRT, vaginal swabs were collected, suspended in SPG buffer, and disrupted with glass beads. Inclusion-forming units (IFUs) were determined by plating serial dilutions of swab samples on HeLa 229 cells, staining with anti-major outer membrane porin (MOMP) monoclonal antibody (MAb) and counting under microscope. To enumerate bacteria burden within tissues, intraperitoneal (i.p.) wash fluid was collected in SPG buffer, and spleens, kidneys, lungs, and gallbladders were homogenized in SPG buffer. Tissue homogenates were disrupted with glass beads and centrifuged at 500 × g for 10 min, supernatants were collected, and serial dilutions were plated on HeLa 229 cells for IFU counts.
Ab-mediated depletion.
IFN-γ in vivo depletion was performed by i.p. injection of 0.25 mg anti-IFN-γ (XMG1.2; BioXcell) on days −1 and 1 postinfection and every 3 days thereafter. NK cells were depleted by i.p. injection of 0.3 mg anti-NK1.1 (purified MAb from PK136 hybridoma; gift from Richard Morrison, UAMS) on days −3, −1, and 1 postinfection and every 3 days thereafter.
CD4 T cell adoptive transfer.
Total CD4 T cells from donor mice were purified from spleens using a STEMCELL EasySep CD4 T cell isolation kit according to the manufacturer’s instructions (STEMCELL Technologies). Depending on individual experiment, 5 to 20 million purified CD4 T cells were transferred intravenously into recipient mice via the tail vein.
IFN-γ secretion assay and flow cytometry.
An IFN-γ secretion assay was conducted according to the manufacturer’s instructions (mouse IFN-γ secretion assay detection kit; Miltenyi). Briefly, spleens were harvested, and single-cell suspensions were prepared in RPMI with 5% fetal calf serum (FCS). Cells were labeled with IFN-γ catch reagents for 5 min on ice and incubated for 45 min at 37°C. IFN-γ-producing cells were stained with IFN-γ detection antibody in conjunction with cell surface markers (listed below) and analyzed on an LSRFortessa flow cytometer (BD Biosciences). Antibodies used included CD3e (145-2C11), CD11b (M1/70), and NK1.1 (PK136) (BioLegend). Data were analyzed using FlowJo software (Tree Star).
Statistical analysis.
Statistical analysis was performed with GraphPad Prism 8. An unpaired t test was used for normally distributed continuous-variable comparisons; a Mann-Whitney U test was used for nonparametric comparisons. The log-rank Mantel-Cox test was used for survival curves.
ACKNOWLEDGMENTS
We thank Richard Morrison and Sandra Morrison for providing us the PK136 monoclonal antibody. We thank Andrea Harris at the UAMS Flow Cytometry Core for technical assistance.
This study was supported by grants from the National Institutes of Health to L.-X.L. (AI139124 and GM103625).
We declare no financial or commercial conflict of interest.
REFERENCES
- 1.Morré SA, Brule A, Rozendaal L, Boeke AJP, Voorhorst FJ, Blok SD, Meijer CJLM. 2002. The natural course of asymptomatic Chlamydia trachomatis infections: 45% clearance and no development of clinical PID after one-year follow-up. Int J STD AIDS 13:12–18. doi: 10.1258/095646202762226092. [DOI] [PubMed] [Google Scholar]
- 2.Darville T, Hiltke TJ. 2010. Pathogenesis of genital tract disease due to Chlamydia trachomatis. J Infect Dis 201:S114–S125. doi: 10.1086/652397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rekart ML, Gilbert M, Meza R, Kim PH, Chang M, Money DM, Brunham RC. 2013. Chlamydia public health programs and the epidemiology of pelvic inflammatory disease and ectopic pregnancy. J Infect Dis 207:30–38. doi: 10.1093/infdis/jis644. [DOI] [PubMed] [Google Scholar]
- 4.de la Maza LM, Zhong G, Brunham RC. 2017. Update on Chlamydia trachomatis vaccinology. Clin Vaccine Immunol 24:e00543-16. doi: 10.1128/CVI.00543-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poston TB, Gottlieb SL, Darville T. 2019. Status of vaccine research and development of vaccines for Chlamydia trachomatis infection. Vaccine 37:7289–7294. doi: 10.1016/j.vaccine.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 6.Phillips S, Quigley BL, Timms P. 2019. Seventy years of Chlamydia vaccine research—limitations of the past and directions for the future. Front Microbiol 10:70. doi: 10.3389/fmicb.2019.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaufmann SHE. 1993. Immunity to intracellular bacteria. Annu Rev Immunol 11:129–163. doi: 10.1146/annurev.iy.11.040193.001021. [DOI] [PubMed] [Google Scholar]
- 8.Tubo NJ, Jenkins MK. 2014. CD4+ T cells: guardians of the phagosome. Clin Microbiol Rev 27:200–213. doi: 10.1128/CMR.00097-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coers J, Gondek DC, Olive AJ, Rohlfing A, Taylor GA, Starnbach MN. 2011. Compensatory T cell responses in IRG-deficient mice prevent sustained Chlamydia trachomatis infections. PLoS Pathog 7:e1001346. doi: 10.1371/journal.ppat.1001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haldar AK, Piro AS, Finethy R, Espenschied ST, Brown HE, Giebel AM, Frickel E-M, Nelson DE, Coers J. 2016. Chlamydia trachomatis is resistant to inclusion ubiquitination and associated host defense in gamma interferon-primed human epithelial cells. mBio 7:e01417-16. doi: 10.1128/mBio.01417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finethy R, Coers J. 2016. Sensing the enemy, containing the threat: cell-autonomous immunity to Chlamydia trachomatis. FEMS Microbiol Rev 40:875–893. doi: 10.1093/femsre/fuw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labuda JC, McSorley SJ. 2018. Diversity in the T cell response to Chlamydia-sum are better than one. Immunol Lett 202:59–64. doi: 10.1016/j.imlet.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrison RP, Caldwell HD. 2002. Immunity to murine chlamydial genital infection. Infect Immun 70:2741–2751. doi: 10.1128/iai.70.6.2741-2751.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gondek DC, Olive AJ, Stary G, Starnbach MN. 2012. CD4+ T cells are necessary and sufficient to confer protection against Chlamydia trachomatis infection in the murine upper genital tract. J Immunol 189:2441–2449. doi: 10.4049/jimmunol.1103032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L-X, McSorley SJ. 2015. A re-evaluation of the role of B cells in protective immunity to Chlamydia infection. Immunol Lett 164:88–93. doi: 10.1016/j.imlet.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su H, Feilzer K, Caldwell HD, Morrison RP. 1997. Chlamydia trachomatis genital tract infection of antibody-deficient gene knockout mice. Infect Immun 65:1993–1999. doi: 10.1128/IAI.65.6.1993-1999.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perry LL, Feilzer K, Caldwell HD. 1997. Immunity to Chlamydia trachomatis is mediated by T helper 1 cells through IFN-gamma-dependent and -independent pathways. J Immunol 158:3344–3352. [PubMed] [Google Scholar]
- 18.Morrison RP, Feilzer K, Tumas DB. 1995. Gene knockout mice establish a primary protective role for major histocompatibility complex class II-restricted responses in Chlamydia trachomatis genital tract infection. Infect Immun 63:4661–4668. doi: 10.1128/IAI.63.12.4661-4668.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rank RG, Soderberg LS, Barron AL. 1985. Chronic chlamydial genital infection in congenitally athymic nude mice. Infect Immun 48:847–849. doi: 10.1128/IAI.48.3.847-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poston TB, O'Connell CM, Girardi J, Sullivan JE, Nagarajan UM, Marinov A, Scurlock AM, Darville T. 2018. T cell-independent gamma interferon and B cells cooperate to prevent mortality associated with disseminated Chlamydia muridarum genital tract infection. Infect Immun 86:e00143-18. doi: 10.1128/IAI.00143-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cain TK, Rank RG. 1995. Local Th1-like responses are induced by intravaginal infection of mice with the mouse pneumonitis biovar of Chlamydia trachomatis. Infect Immun 63:1784–1789. doi: 10.1128/IAI.63.5.1784-1789.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hawkins RA, Rank RG, Kelly KA. 2002. A Chlamydia trachomatis-specific Th2 clone does not provide protection against a genital infection and displays reduced trafficking to the infected genital mucosa. IAI 70:5132–5139. doi: 10.1128/IAI.70.9.5132-5139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu H, Karunakaran KP, Kelly I, Shen C, Jiang X, Foster LJ, Brunham RC. 2011. Immunization with live and dead Chlamydia muridarum induces different levels of protective immunity in a murine genital tract model: correlation with MHC class II peptide presentation and multifunctional Th1 cells. J Immunol 186:3615–3621. doi: 10.4049/jimmunol.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scurlock AM, Frazer LC, Andrews CW, O'Connell CM, Foote IP, Bailey SL, Chandra-Kuntal K, Kolls JK, Darville T. 2011. Interleukin-17 contributes to generation of Th1 immunity and neutrophil recruitment during Chlamydia muridarum genital tract infection but is not required for macrophage Influx or normal resolution of infection. Infect Immun 79:1349–1362. doi: 10.1128/IAI.00984-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrew DW, Cochrane M, Schripsema JH, Ramsey KH, Dando SJ, O'Meara CP, Timms P, Beagley KW. 2013. The duration of Chlamydia muridarum genital tract infection and associated chronic pathological changes are reduced in IL-17 knockout mice but protection is not increased further by immunization. PLoS One 8:e76664. doi: 10.1371/journal.pone.0076664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miguel RDV, Calla NEQ, Pavelko SD, Cherpes TL. 2016. Intravaginal Chlamydia trachomatiscChallenge infection elicits TH1 and TH17 immune responses in mice that promote pathogen clearance and genital tract damage. PLoS One 11:e0162445. doi: 10.1371/journal.pone.0162445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cotter TW, Ramsey KH, Miranpuri GS, Poulsen CE, Byrne GI. 1997. Dissemination of Chlamydia trachomatis chronic genital tract infection in gamma interferon gene knockout mice. Infect Immun 65:2145–2152. doi: 10.1128/IAI.65.6.2145-2152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hess J, Ladel C, Miko D, Kaufmann SH. 1996. Salmonella typhimurium aroA- infection in gene-targeted immunodeficient mice: major role of CD4+ TCR-alpha beta cells and IFN-gamma in bacterial clearance independent of intracellular location. J Immunol 156:3321–3326. [PubMed] [Google Scholar]
- 29.Ravindran R, Foley J, Stoklasek T, Glimcher LH, McSorley SJ. 2005. Expression of T-bet by CD4 T cells is essential for resistance to Salmonella infection. J Immunol 175:4603–4610. doi: 10.4049/jimmunol.175.7.4603. [DOI] [PubMed] [Google Scholar]
- 30.Cowley SC, Elkins KL. 2003. CD4+ T cells mediate IFN-γ-independent control of Mycobacterium tuberculosis infection both in vitro and in vivo. J Immunol 171:4689–4699. doi: 10.4049/jimmunol.171.9.4689. [DOI] [PubMed] [Google Scholar]
- 31.Gallegos AM, Heijst JWJ, van Samstein M, Su X, Pamer EG, Glickman MS. 2011. A gamma interferon independent mechanism of CD4 T cell mediated control of M. tuberculosis infection in vivo. PLoS Pathog 7:e1002052. doi: 10.1371/journal.ppat.1002052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakai S, Kauffman KD, Sallin MA, Sharpe AH, Young HA, Ganusov VV, Barber DL. 2016. CD4 T cell-derived IFN-γ plays a minimal role in control of pulmonary Mycobacterium tuberculosis infection and must be actively repressed by PD-1 to prevent lethal disease. PLoS Pathog 12:e1005667. doi: 10.1371/journal.ppat.1005667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie ANJ, Mebius RE, Powrie F, Spits H. 2018. Innate lymphoid cells: 10 years on. Cell 174:1054–1066. doi: 10.1016/j.cell.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 34.Brunham RC, Rey-Ladino J. 2005. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. 2. Nat Rev Immunol 5:149–161. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- 35.Li L-X, McSorley SJ. 2013. B cells enhance antigen-specific CD4 T cell priming and prevent bacteria dissemination following Chlamydia muridarum genital tract infection. PLoS Pathog 9:e1003707. doi: 10.1371/journal.ppat.1003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bando JK, Colonna M. 2016. Innate lymphoid cell function in the context of adaptive immunity. Nat Immunol 17:783–789. doi: 10.1038/ni.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sturge CR, Benson A, Raetz M, Wilhelm CL, Mirpuri J, Vitetta ES, Yarovinsky F. 2013. TLR-independent neutrophil-derived IFN-γ is important for host resistance to intracellular pathogens. Proc Natl Acad Sci U S A 110:10711–10716. doi: 10.1073/pnas.1307868110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frucht DM, Fukao T, Bogdan C, Schindler H, O'Shea JJ, Koyasu S. 2001. IFN-γ production by antigen-presenting cells: mechanisms emerge. Trends Immunol 22:556–560. doi: 10.1016/S1471-4906(01)02005-1. [DOI] [PubMed] [Google Scholar]
- 39.Artis D, Spits H. 2015. The biology of innate lymphoid cells 7534. Nature 517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 40.Tseng C-TK, Rank RG. 1998. Role of NK cells in early host response to chlamydial genital infection. Infect Immun 66:5867–5875. doi: 10.1128/IAI.66.12.5867-5875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Dong X, Zhao L, Wang X, Wang Y, Yang X, Wang H, Zhao W. 2016. Natural killer cells regulate Th1/Treg and Th17/Treg balance in chlamydial lung infection. J Cell Mol Med 20:1339–1351. doi: 10.1111/jcmm.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H, Li J, Dong X, Zhou X, Zhao L, Wang X, Rashu R, Zhao W, Yang X. 2020. NK cells contribute to protective memory T cell mediated immunity to Chlamydia muridarum infection. Front Cell Infect Microbiol 10:296. doi: 10.3389/fcimb.2020.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu H, Su X, Zhao Y, Tang L, Chen J, Zhong G. 2020. Innate lymphoid cells are required for endometrial resistance to Chlamydia trachomatis infection. Infect Immun 88:e00152-20. doi: 10.1128/IAI.00152-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He Y, Xu H, Song C, Koprivsek JJ, Arulanandam B, Yang H, Tao L, Zhong G. 2020. Adoptive transfer of group 3-like innate lymphoid cells restores mouse colon resistance to colonization of an IFN-γ-susceptible Chlamydia muridarum mutant. Infect Immun 88:e00533-20. doi: 10.1128/IAI.00533-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Müller AJ, Filipe-Santos O, Eberl G, Aebischer T, Späth GF, Bousso P. 2012. CD4+ T cells rely on a cytokine gradient to control intracellular pathogens beyond sites of antigen presentation. Immunity 37:147–157. doi: 10.1016/j.immuni.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 46.Farrar MA, Schreiber RD. 1993. The molecular cell biology of interferon-gamma and its receptor. Annu Rev Immunol 11:571–611. doi: 10.1146/annurev.iy.11.040193.003035. [DOI] [PubMed] [Google Scholar]
- 47.Kazar J, Gillmore JD, Gordon FB. 1971. Effect of interferon and interferon inducers on infections with a nonviral intracellular microorganism, Chlamydia trachomatis. Infect Immun 3:825–832. doi: 10.1128/IAI.3.6.825-832.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams DM, Byrne GI, Grubbs B, Marshal TJ, Schachter J. 1988. Role in vivo for gamma interferon in control of pneumonia caused by Chlamydia trachomatis in mice. Infect Immun 56:3004–3006. doi: 10.1128/IAI.56.11.3004-3006.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong GM, Peterson EM, Czarniecki CW, Schreiber RD, de la Maza LM. 1989. Role of endogenous gamma interferon in host defense against Chlamydia trachomatis infections. Infect Immun 57:152–157. doi: 10.1128/IAI.57.1.152-157.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rank RG, Ramsey KH, Pack EA, Williams DM. 1992. Effect of gamma interferon on resolution of murine chlamydial genital infection. Infect Immun 60:4427–4429. doi: 10.1128/IAI.60.10.4427-4429.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beatty WL, Byrne GI, Morrison RP. 1993. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc Natl Acad Sci U S A 90:3998–4002. doi: 10.1073/pnas.90.9.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johansson M, Schön K, Ward M, Lycke N. 1997. Genital tract infection with Chlamydia trachomatis fails to induce protective immunity in gamma interferon receptor-deficient mice despite a strong local immunoglobulin A response. Infect Immun 65:1032–1044. doi: 10.1128/IAI.65.3.1032-1044.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nelson DE, Virok DP, Wood H, Roshick C, Johnson RM, Whitmire WM, Crane DD, Steele-Mortimer O, Kari L, McClarty G, Caldwell HD. 2005. Chlamydial IFN-γ immune evasion is linked to host infection tropism. Proc Natl Acad Sci U S A 102:10658–10663. doi: 10.1073/pnas.0504198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li W, Murthy AK, Guentzel MN, Seshu J, Forsthuber TG, Zhong G, Arulanandam BP. 2008. Antigen-specific CD4+ T cells produce sufficient IFN-γ to mediate robust protective immunity against genital Chlamydia muridarum infection. J Immunol 180:3375–3382. doi: 10.4049/jimmunol.180.5.3375. [DOI] [PubMed] [Google Scholar]
- 55.Gondek DC, Roan NR, Starnbach MN. 2009. T cell responses in the absence of IFN-γ exacerbate uterine infection with Chlamydia trachomatis. J Immunol 183:1313–1319. doi: 10.4049/jimmunol.0900295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naglak EK, Morrison SG, Morrison RP. 2016. Gamma interferon is required for optimal antibody-mediated immunity against genital Chlamydia infection. Infect Immun 84:3232–3242. doi: 10.1128/IAI.00749-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Helble JD, Gonzalez RJ, von Andrian UH, Starnbach MN. 2020. Gamma interferon is required for Chlamydia clearance but is dispensable for T cell homing to the genital rract. mBio 11:e00191-20. doi: 10.1128/mBio.00191-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Price M, Ades A, Soldan K, Welton N, Macleod M, Simms I, DeAngelis D, Turner K, Horner P. 2016. The natural history of Chlamydia trachomatis infection in women: a multi parameter evidence synthesis. Health Technol Assess 20:1–250. doi: 10.3310/hta20220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poston TB, Qu Y, Girardi J, O'Connell CM, Frazer LC, Russell AN, Wall M, Nagarajan UM, Darville T. 2017. A Chlamydia-specific TCR-transgenic mouse demonstrates Th1 polyfunctionality with enhanced effector function. J Immunol 199:2845–2854. doi: 10.4049/jimmunol.1700914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson RM, Yu H, Strank NO, Karunakaran K, Zhu Y, Brunham RC. 2017. B cell presentation of Chlamydia antigen selects out protective CD4γ13 T cells: implications for genital tract tissue-resident memory lymphocyte clusters. Infect Immun 86:e00614-17. doi: 10.1128/IAI.00614-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miguel RDV, Harvey SAK, LaFramboise WA, Reighard SD, Matthews DB, Cherpes TL. 2013. Human female genital tract infection by the obligate intracellular bacterium Chlamydia trachomatis elicits robust type 2 immunity. PLoS One 8:e58565. doi: 10.1371/journal.pone.0058565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stary G, Olive A, Radovic-Moreno AF, Gondek D, Alvarez D, Basto PA, Perro M, Vrbanac VD, Tager AM, Shi J, Yethon JA, Farokhzad OC, Langer R, Starnbach MN, von Andrian UH. 2015. A mucosal vaccine against Chlamydia trachomatis generates two waves of protective memory T cells. Science 348:aaa8205. doi: 10.1126/science.aaa8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zemmour D, Kiner E, Benoist C. 2020. CD4+ teff cell heterogeneity: the perspective from single-cell transcriptomics. Curr Opin Immunol 63:61–67. doi: 10.1016/j.coi.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]