Abstract
Rhodopsin, the light-sensitive visual pigment expressed in rod photoreceptors, is specialized for vision in dim-light environments. Aquatic environments are particularly challenging for vision due to the spectrally dependent attenuation of light, which can differ greatly in marine and freshwater systems. Among fish lineages that have successfully colonized freshwater habitats from ancestrally marine environments, croakers are known as highly visual benthic predators. In this study, we isolate rhodopsins from a diversity of freshwater and marine croakers and find that strong positive selection in rhodopsin is associated with a marine to freshwater transition in South American croakers. In order to determine if this is accompanied by significant shifts in visual abilities, we resurrected ancestral rhodopsin sequences and tested the experimental properties of ancestral pigments bracketing this transition using in vitro spectroscopic assays. We found the ancestral freshwater croaker rhodopsin is redshifted relative to its marine ancestor, with mutations that recapitulate ancestral amino acid changes along this transitional branch resulting in faster kinetics that are likely to be associated with more rapid dark adaptation. This could be advantageous in freshwater due to the redshifted spectrum and relatively narrow interface and frequent transitions between bright and dim-light environments. This study is the first to experimentally demonstrate that positively selected substitutions in ancestral visual pigments alter protein function to freshwater visual environments following a transition from an ancestrally marine state and provides insight into the molecular mechanisms underlying some of the physiological changes associated with this major habitat transition.
Keywords: rhodopsin spectral tuning, likelihood-based codon models, clade models of molecular evolution, underwater visual ecology, evolution of fish vision
Introduction
Resurrecting ancestral protein sequences has made it possible to test in vitro the functional effects of evolutionary change along an ancestral lineage (Chang and Donoghue 2000; Storz 2016). This approach combines the reconstruction of ancestral sequences with experimental assays of protein function. It is especially useful for investigating the molecular underpinnings of major evolutionary transitions and the adaptive molecular evolution of protein function in the context of gene family or organismal evolution (Chang et al. 2002; Gaucher et al. 2003; Ugalde et al. 2004; Projecto-Garcia et al. 2013; Kratzer et al. 2014; Dungan and Chang 2017; Isogai et al. 2018; Kaltenbach et al. 2018). Adaptive radiations associated with biogeographic events, such as the rise of mountain ranges, formation of volcanic islands, and glacial retreat, and the corresponding appearance of novel niches, have long served as model systems in evolutionary biology (Schluter 2000). However, although genomic signatures of adaptation have been observed in lineages that have experienced major habitat changes (Lamichhaney et al. 2015; Marques et al. 2018; Lim et al. 2019), the underlying molecular mechanisms associated with major biogeographical transitions are seldom studied by resurrecting ancestral sequences (Projecto-Garcia et al. 2013; Dungan and Chang 2017; Isogai et al. 2018). This may stem from the prohibitive cost associated with resurrecting multiple genes and lineages involved in these evolutionary events. Codon-based analyses of selection (Yang and Bielawski 2000; Anisimova and Liberles 2007), and in particular branch site and clade models (Yang and Nielsen 2002; Bielawski and Yang 2004; Kosakovsky Pond et al. 2011; Weadick and Chang 2012) can identify protein-coding genes where specific sites or lineages are likely to be the targets of positive selection. By focusing on positively selected substitutions, functional differences in ancestral sequences can be studied with respect to the changes in patterns of natural selection associated with ecology and biogeography (Zakon et al. 2006; Van Nynatten et al. 2015; Dungan and Chang 2017; Schott et al. 2018; Gower et al. 2019; Pisciottano et al. 2019).
One of the most striking biogeographical events involving aquatic ecosystems includes the massive marine incursions of South America during the Miocene (Lovejoy et al. 1998; Hoorn et al. 2010). During this time, vast regions of Amazonia were inundated with seawater, creating environments of intermediate salinity that facilitated evolutionary transitions from marine to freshwater. As a result, marine lineages, including fishes such as stingrays, anchovies, needlefishes, and croakers, as well as invertebrates including shrimps, crabs, and mollusks, adapted to freshwater and became components of South American freshwater aquatic communities (Lovejoy et al. 1998; Bloom and Lovejoy 2017). Marine to freshwater transitions require a suite of adaptations in physiological and life history traits including osmotic regulation (Tsai and Lin 2007; Lee et al. 2012), diet (Sanchez et al. 2019), metabolism (Kitano et al. 2010; Ishikawa et al. 2019), defense from predators (Jones et al. 2012), reproductive strategies (Reznick et al. 2007), and vision (Van Nynatten et al. 2015). Therefore, marine to freshwater habitat transitions offer an excellent opportunity to investigate the molecular and functional basis of a suite of interesting adaptations.
In terms of spectra of available light, marine and freshwater habitats differ considerably. Light penetrating the open ocean is known to become increasingly blueshifted with depth and can penetrate up to 1,000 m depending on water clarity (Jerlov 1968). Decades of research have shown that many deep-sea marine fishes, as well as those inhabiting deep and clear lakes, such as the Lake Baikal sculpins and African Rift Lake cichlids, have visual systems with spectral sensitivities optimized for the detection of blue–green light (Wald et al. 1957; Hunt et al. 1996, 2001; Marques et al. 2018). In contrast, the spectrum of light illuminating riverine environments attenuates much more rapidly, is more spectrally heterogeneous, but has been less studied from the perspective of the visual systems of riverine animals (Levine and MacNichol 1979; Costa et al. 2013). In general, higher concentrations of suspended particulate matter and organic compounds, which scatter and absorb short-wavelength light, respectively, result in a dramatically redshifted underwater visual environment, where overall light levels are dramatically reduced and attenuate rapidly, in some cases within a few meters of the surface (Levine and MacNichol 1979; Costa et al. 2013) (fig. 1). These differences in aquatic light environment are thought to affect the visual systems of fishes, with freshwater fishes generally expressing more redshifted visual pigments (Rennison et al. 2016; Musilova et al. 2019) and with higher proportions of the redshifted A2 chromophore (Toyama et al. 2008) than marine fishes. In addition, there is evidence of genetic signatures of positive selection and selective sweeps in freshwater fishes that may be associated with redshifts in visual sensitivity in visual pigments, but this has yet to be investigated experimentally (Van Nynatten et al. 2015; Hill et al. 2019).
Fig. 1.
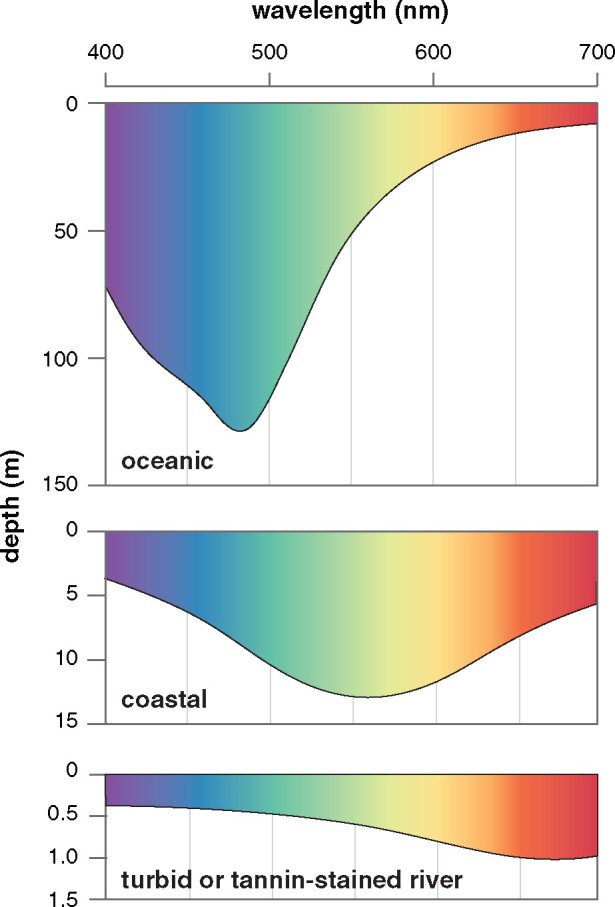
Schematic of underwater light environments. Data adapted from Jerlov (1968) and Costa et al. (2013).
Rhodopsin, contained in rod photoreceptors, is the visual pigment that initiates the first step in the visual transduction cascade and is essential for the perception of light in dim-light environments (Baylor 1996). It is often the most highly expressed visual pigment in fishes (Musilova et al. 2019) and thus the biochemical and biophysical characteristics of rhodopsin directly affect how an organism perceives its environment, particularly under dim-light conditions (Hauser and Chang 2017). Rhodopsin is comprised of an opsin G protein-coupled receptor and a light-sensitive chromophore bound within a pocket formed by the integral transmembrane helices of the opsin protein (Smith 2010). Upon the absorption of light, the chromophore undergoes an 11-cis to all-trans isomerization that initiates a series of conformational changes in the opsin protein, eventually forming the biologically active Meta II state (Smith 2010). These helical rearrangements facilitate the activation of the heterotrimeric G protein transducin, initiating the visual transduction cascade within the photoreceptors of the eye (Arshavsky et al. 2002). Rhodopsin serves as an excellent model system for comparative studies of molecular adaptation because several key aspects of its function can be isolated and studied experimentally (Bowmaker and Hunt 2006; Hofmann et al. 2009; Morrow and Chang 2010). These include spectral tuning (Chang et al. 2002; Musilova et al. 2019), and kinetics of both light activation (Sugawara et al. 2010), and recovery phase of the light-activated receptor (Hauser et al. 2017; Castiglione and Chang 2018; Gutierrez et al. 2018; Liu et al. 2019).
We focus on rhodopsin of fishes in the family Sciaenidae as a model system. Known as croakers or drum because many species use vibrations of their swim bladder to produce acoustic signals (Ramcharitar et al. 2006), the Sciaenids are a family of 286 species of fishes, distributed worldwide, mainly in tropical and subtropical regions (Sasaki 1989). Croakers inhabit nearshore marine habitats and estuaries, and freshwater species occur in rivers of southeast Asia, North America, and South America. Most croaker species are highly visual benthic predators, relying on vision despite often inhabiting waters where light is highly attenuated (Deary et al. 2016). They also have relatively large eyes and rod-dominated retinas, and like many other species adapted to dim-light environments, possess a tapetum lucidum to improve photon detection (Arnott et al. 1972). Croakers, like several other lineages of fishes, invaded South American freshwaters during Miocene marine incursions and then diversified into 21 species that occupy river habitats across the continent (Lo et al. 2015; Bloom and Lovejoy 2017). Croakers independently invaded freshwaters of southeast Asia and North America, but these freshwater lineages did not diversify as they did in South America (Lo et al. 2015).
Here, we investigate the effect of a major biogeographical event, a marine to freshwater transition, on molecular adaptations in the visual system of the croakers. Clade models of codon evolution are useful in detecting changes in selective constraint in different lineages (Yang and Bielawski 2000; Anisimova and Liberles 2007; Chang et al. 2012). We use these codon-based analyses of selection to identify if positive selection, and by implication adaptive evolution, took place in the dim-light visual pigment rhodopsin of croakers. Specifically, we used clade models to test if transitional lineages from marine environments to a much dimmer and redshifted freshwater environment are under elevated selection pressures associated with the change in habitat. We next mapped positively selected sites from different ecological partitions onto rhodopsin crystal structures with the expectation that positively selected substitutions along the transitional lineage would be in critical domains and as a result would have more significant impacts on protein function. To empirically characterize the impacts of positive selection associated with the transitions from marine to freshwater in South America, we reconstructed the ancestral sequences at the nodes bounding this major transitional event. Using spectroscopic assays, we tested the expectation that the transition to a more redshifted freshwater environment resulted in a more redshifted visual pigment. We further investigated the effects of four positively selected substitutions in close proximity to the chromophore in a model genetic background to test their contribution to the spectral shift as well as the kinetic properties of rhodopsin. This is the first study in which the nature and type of molecular adaptations associated with a major biogeographical transition to freshwater have been demonstrated by pairing ancestral reconstructions with experimental analyses.
Results
Positive Selection in Croaker Rhodopsin
We investigated if the diverse array of visual environments inhabited by croakers has resulted in higher levels of selection on rhodopsin by estimating among-site variation in dN/dS (Yang et al. 2000; Yang 2007). We found high average dN/dS estimates for the rhodopsin gene in croakers (m0: dN/dS = 0.27). This average dN/dS is greater than values observed in nonvision-related nuclear genes, EGR1, EGR2, and RAG1 used in this study and others for phylogenetic reconstructions. It is also greater than rates typical for rhodopsin in teleost fishes and protein-coding genes in general (Rennison et al. 2012). For croaker rhodopsin, random sites models in PAML that included a positively selected site class were found to fit the data significantly better than nested null models without (supplementary table S1–S3, Supplementary Material online), with several sites detected to be under pervasive positive selection across the croaker phylogeny (supplementary table S4, Supplementary Material online).
Increased Positive Selection in Rhodopsin Associated with a Major Transition to Freshwater in South America
To investigate whether there is evidence for positive selection concurrent with transitions to freshwater environments, we utilized branch site and clade models in PAML with transitional branches set as the foreground (fig. 2A) (Yang and Nielsen 2002; Bielawski and Yang 2004). The marine to freshwater transition in South America was found to be the target of positive selection under both branch site and clade models (branch site null vs. branch site: P = 0.0019 and M3 vs. Clade model D [CmD]: P = 0.0100), but this was not found for any other freshwater invasions (fig. 2B and supplementary tables S5–S7, Supplementary Material online). Branch-site REL, a complementary model implemented in the HYPHY package (Kosakovsky Pond et al. 2011), which does not require a priori partitioning of the phylogeny also identifies the South American transitional branch as being under positive selection.
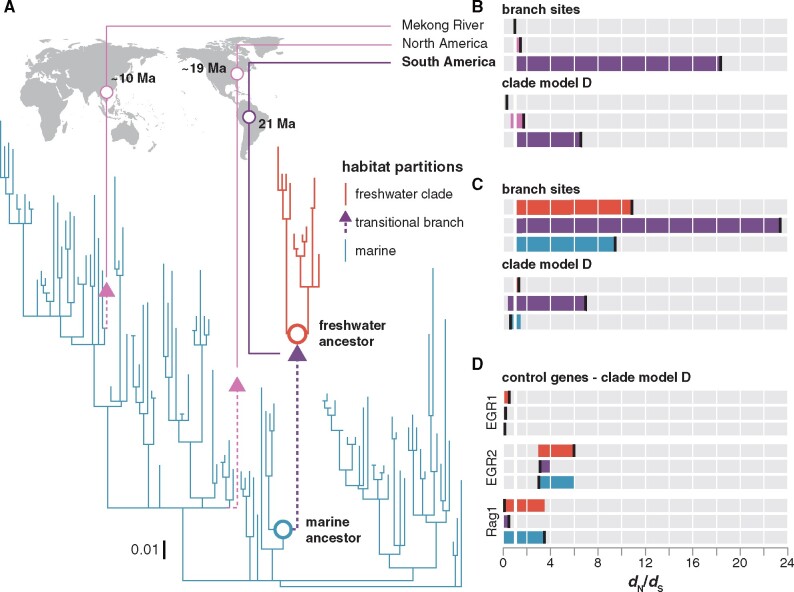
Fig. 2.
Tests for positive selection on rhodopsin associated with habitat transitions. (A) Bayesian phylogeny used for PAML analyses. Branch lengths scaled by the number of substitutions per codon. Divergence time estimates for invasions into freshwater from Lo et al. (2015). (B) Branch sites and CmD dN/dS estimates for the divergent site class with the marine to freshwater transitional branches in the world-wide croaker rhodopsin data set as the foreground. Each bar is colored to match foreground which is indicated by a vertical black bar. (C) Branch sites and CmD dN/dS estimates for the divergent site class for models with the marine lineages, the freshwater clade, and the transitional branch set as the foreground in the New World clade croaker rhodopsin data set. (D) CmD dN/dS estimates for nonvisual control genes using the same ecological partitioning scheme.
The dN/dS estimate for the branch, representing the marine to freshwater transition in South America, was found to be significantly higher than the dN/dS parameter estimated from all of the freshwater South American descendants as well as a large clade of North and South American marine species that the South American freshwater clade is nested within (fig. 2A and C). A model where only the marine to freshwater South American transitional branch was allowed an additional dN/dS parameter was not improved by a more parameter-rich partitioning scheme with all three partitions (CmD:transitional branch vs. CmD:transitional branch/freshwater clade: P = 0.1042, supplementary table S8, Supplementary Material online), supporting the idea that the marine and freshwater partitions have comparable dN/dS estimates and that these estimates are lower than that of the transitional branch.
Positive selection was not observed along the transitional branch in nonvisual nuclear genes sequenced in this study. Branch-site tests of the nonvisual control genes typically converged on the null model (dN/dS ≤ 1) (fig. 2D). Positive selection was only detected when marine lineages were set as the foreground in the Rag1 data set (branch-site null vs. branch site: P = 0.0121, supplementary tables S9 and S10, Supplementary Material online).
Different Sets of Positively Selected Sites in Marine and Freshwater Lineages
We investigated if the sites that were the targets of positive selection differed in the marine lineages, the freshwater clade and on the transitional branch by inferring positively selected sites in each foreground partition of branch-site tests using Bayes empirical Bayes (BEB) analysis in PAML (Yang 2005). Sites with posterior probability above 0.7 for inclusion in the positively selected site class are mostly unique to each phylogenetic partition and are distributed throughout the rhodopsin crystal structure (fig. 3A and C and supplementary table S4, Supplementary Material online). An increased proportion of the positively selected sites on the transitional branch falls within 10 Å of the chromophore, where spectral tuning sites have been previously observed (Bowmaker and Hunt 2006) (fig. 3B). Positively selected sites on the transitional lineage also undergo larger movements upon rhodopsin photoactivation, as measured using the root mean square deviation of dark (1U19) and active-state crystal structures of rhodopsin (3PQR) (fig. 3B) (Okada et al. 2004; Choe et al. 2011). Positively selected sites in the marine lineages and the freshwater clade primarily occur at external sites facing the plasma membrane (fig. 3C).
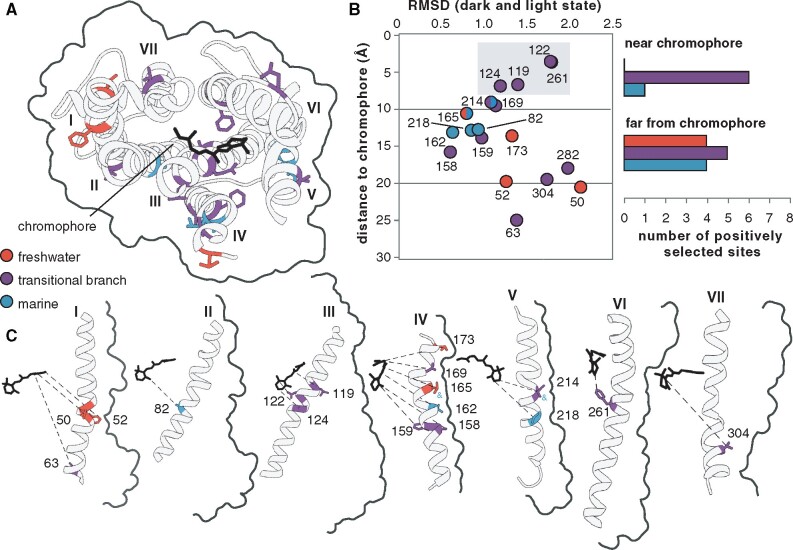
Fig. 3.
Habitat specificity of positively selected sites in croaker rhodopsin. (A) Positively selected sites on transmembrane helices shown on the rhodopsin dark-state crystal structure (1U19) looking down from the intradiscal face. (B) Distance from the chromophore and root mean square deviation between the dark (1U19) and light (3PQR) state rhodopsin crystal structures for each positively selected site. Histograms show the number of positively selected sites in each partition within ten angstroms of the chromophore where spectral tuning sites have been observed (Bowmaker and Hunt 2006). (C) Positively selected sites on each transmembrane helix shown arranged in order with the chromophore to the left. Sites under positive selection in more than one partition are indicated by an ampersand of the same color as the additional partition.
The multiple independent sets of positively selected sites within the data set interferes with the assumptions of the frequently employed Clade model C (CmC) (Bielawski and Yang 2004). CmC did not identify any difference in dN/dS on the transitional branch, at odds with inferences using branch sites, branch-site REL (HYPHY), CmD, and even the overly conservative two-ratio model. Comparisons of model parameters in CmC and CmD suggest that this is because positively selected sites, pervasive across the croaker phylogeny, are forced into the divergent site class in CmC, whereas in CmD, pervasive positively selected sites are better accommodated by the freely estimated second site class, resulting in a significantly better fit (CmC vs. CmD: P < 0.0001, supplementary table S11, Supplementary Material online).
Spectral Tuning in Marine and Freshwater Ancestral Croaker Rhodopsins
To investigate the substitutions and functional shifts associated with the transition from marine to freshwater in South America, we reconstructed the amino acid substitutions in rhodopsin along the transitional branch using maximum likelihood codon and amino acid methods (Yang et al. 1995). Ancestral rhodopsin sequences, representing the nodes bracketing the transitional branch into South American freshwater habitats, were supported by high posterior probabilities. In total, 20 substitutions were reconstructed along the transitional branch, the most of any branch in the phylogeny (supplementary table S12 and fig. S6, Supplementary Material online). Eight of these substitutions occur only along this branch, and the majority are subsequently conserved throughout the freshwater clade.
We used heterologous protein expression and spectroscopic assays to functionally characterize the spectral sensitivity of resurrected ancestral croaker rhodopsin sequences at the marine and freshwater transitional nodes (fig. 2A). The ancestral freshwater pigment was redshifted relative to the marine ancestor with peak spectral sensitivities of 504 and 498 nm, respectively (fig. 4A). The peak spectral sensitivity of the ancestral marine croaker rhodopsin is consistent with microspectrophotometry data for marine croaker rods (supplementary table S13, Supplementary Material online), and the magnitude of the shift is similar to the blueshifting amino acid substitutions observed in deep-water Lake Baikal cottoids (Hunt et al. 1996).
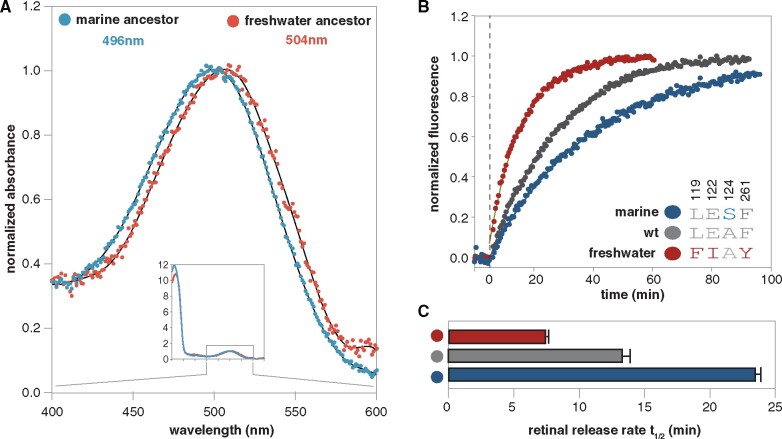
Fig. 4.
Functional characterization of resurrected croaker rhodopsin sequences bracketing the transitional branch. (A) Spectral absorbance curves of dark state rhodopsin shown for marine (blue) and freshwater (red) ancestral croaker rhodopsin pigments expressed in vitro (Govardovskii et al. 2000). (B) Retinal release rate for bovine rhodopsin and bovine rhodopsin with substitutions matching the marine and freshwater croaker sequences at site 119, 122, 124, and 261. Marine substitutions previously published in Castiglione and Chang (2018). (C) Bar chart comparing retinal release rate half-life for each pigment.
Positively Selected Sites on the Transitional Branch Increase Rate of Retinal Release in Rhodopsin
Because expression levels for the marine and freshwater ancestral pigments were not high enough to test the kinetic differences in marine and freshwater ancestral rhodopsin, we used site-directed mutagenesis (SDM) to investigate the effects of the four positively selected substitutions nearest the chromophore, F119L, E122I, S124A, and F261Y (fig. 3B). These four substitutions are observed in many other freshwater fishes, suggesting a more general basis for adaptation to freshwater (Van Nynatten et al. 2015; Castiglione and Chang 2018; Hill et al. 2019; Van Nynatten et al. 2019). We mutated bovine rhodopsin to match the identities observed at these sites in the marine and freshwater ancestrally reconstructed sequences. The amino acids at these sites are mostly conserved between bovine rhodopsin and the marine croaker ancestor and differ only at site 124. The ancestral freshwater sequence is identical to bovine rhodopsin at site 124 but differs at sites 119, 122, and 261 (fig. 4B). Compared with the ancestral croaker sequences, the spectral sensitivities of the freshwater-like and marine-like bovine pigments were slightly red- and blueshifted, respectively (506 and 496 nm, supplementary table S13 and fig. S7, Supplementary Material online). Retinal release rates also differed in freshwater- and marine-like bovine rhodopsin. The substitutions at sites 119, 122, and 124 in the freshwater-like bovine rhodopsin halved the half-life of the retinal release rate compared with wild-type bovine rhodopsin (7.44 vs. 16.08 min, fig. 4B and C) and is one third that estimated for marine-like bovine rhodopsin (7.44 vs. 24.3 min, fig. 4B and C) (Castiglione and Chang 2018). Our results indicate that these substitutions mediate an increase toward faster, slightly more cone-like retinal release rates in freshwater croaker rhodopsins. This is not likely to be due to issues with protein folding/stability in the ancestral pigments, as assays of the susceptibility to hydroxylamine attack for both ancestral croaker pigments were consistent with an inaccessible binding pocket typical of properly folded rhodopsins (supplementary fig. S8, Supplementary Material online).
Discussion
In this study, we investigate rates of molecular evolution in the dim-light rhodopsin gene of croakers during evolutionary transitions from marine to freshwater habitats, using the largest phylogenetic reconstruction of this family of fishes to date. We found significant evidence for a burst of positive selection on the branch representing the marine to freshwater transition in South America. Sites that were positively selected on this transitional branch differ from those positively selected in other croaker lineages and occur in domains likely to have substantial effect on rhodopsin function. In vitro spectroscopic assays of resurrected ancestral proteins bracketing this major transitional event result in an 8-nm redshift in the freshwater ancestral rhodopsin, consistent with the prevailing longer wavelengths of light in the freshwater visual environment. Four ancestral substitutions along this branch that also match the identities of other freshwater fishes (L119Y, E122I, S124A, and F261Y) were found to recapitulate the redshift. Unexpectedly, these substitutions also result in much faster retinal release rates that are likely to greatly improve dark adaptation, which may be important for fishes that experience sharp changes in light levels. However, these faster rates, in addition to the redshift, would have detrimental effects on photosensitivity in dim-light environments. This would appear counterintuitive for benthic fishes inhabiting dim-light environments but may represent a necessary trade-off in very redshifted freshwater ecosystems. Here, we discuss the implications of these results with respect to the evolution of rhodopsin structure and function, and the aquatic visual environments in which these fish lineages are thought to have occurred.
Mechanistic Studies of Major Evolutionary Transitions
The results of our study indicate that the significant functional changes observed in ancestral croaker rhodopsins during the evolutionary transition from marine to freshwater are associated with a strong burst of positive selection. Major changes in environment are known to be drivers of speciation and adaptive evolution (Schluter 2000; Stroud and Losos 2016), and genomewide scans of selection have identified evidence for selection in numerous species adapting to freshwater, such as the threespine stickleback (Jones et al. 2012), the Yangtze finless porpoise (Zhou et al. 2018), Lake Baikal sponges (Kenny et al. 2019), and Macrobrachium prawns (Rahi et al. 2019). However, few genomewide studies of selection attempt to identify the mechanisms underlying adaptation. Even when studies do identify specific genes putatively under selection, hypothetical links between the function and ecological niche are rarely tested in vitro (Storz 2005). Adaptive inferences made without functional characterization are rather risky and there are many factors that need to be considered. For example, epistatic interactions can constrain protein function (Worth et al. 2009) and alter the outcome of specific substitutions (Dungan and Chang 2017; Castiglione and Chang 2018). The optimal peak for protein function can also move because of the compartmentalization of expression (Zakon et al. 2006), the expression of other proteins with similar function (Schott et al. 2018), evolution of other novel physiological processes (Gower et al. 2019; Van Nynatten et al. 2019), or trade-offs with other functional properties of the protein (Tokuriki and Tawfik 2009; Castiglione and Chang 2018).
Spectral Tuning in Aquatic Environments
Our functional investigations of experimentally resurrected ancestral croaker rhodopsins bracketing the marine to freshwater transition in South America showed a clear redshift in visual sensitivity. It had long been hypothesized that riverine environments may have been associated with redshifts in visual sensitivity in freshwater fishes (Levine and MacNichol 1979; Schweikert et al. 2018), but prior to this study, this had never been demonstrated experimentally for an ancestral marine to freshwater transition. In general, freshwater environments are enriched in longer wavelength light because of increased concentrations of organic matter and suspended particulates that attenuate short-wavelength light rapidly (Lythgoe 1979; Costa et al. 2013). They also tend to be much dimmer compared with marine waters (Lythgoe 1979). The evolution of a redshifted spectrally tuned rhodopsin in freshwater fishes would likely have been accompanied by other adaptations to freshwater visual environments, including the utilization of an alternative redshifted chromophore (Toyama et al. 2008) and increased expression of rhodopsin and long-wavelength sensitive opsins (Rennison et al. 2016; Musilova et al. 2019).
Interestingly, some of the mechanisms of spectral tuning in riverine environments are similar to those observed in deep-sea species, but the direction of the spectral shift is the opposite. Amino acid substitutions observed in the freshwater croakers were found at some of the same sites associated with deep-sea spectral tuning adaptations in fishes invading deeper marine environments, where rhodopsin is tuned toward the blueshifted spectrum of available light typical of depths >200 m (Bowmaker and Hunt 2006). For example, A124S and Y261F mediate the blueshift observed in deep water sculpins of Lake Baikal (Hunt et al. 1996; Luk et al. 2016) as well as many deep-sea fishes (Hunt et al. 2001; Lin et al. 2017) and sharks (Hart et al. 2020). In croakers, the reverse substitutions are observed in association with redshifts in spectral sensitivity. Interestingly, the F261Y substitution in rhodopsin has been recently found to be associated with a selective sweep in the colonization of herring into the Baltic Sea (Hill et al. 2019). Other spectral tuning substitutions near the chromophore, L119F and E122I, observed in the riverine croakers, are rarely observed in deep-sea species but also appear to have arisen independently in many divergent freshwater lineages (Castiglione and Chang 2018; Musilova et al. 2019; Van Nynatten et al. 2019). Our study also demonstrates the dangers of attempting to predict functional shifts from sequence-based methods alone. In croakers, we observed a substitution (E122I) that initially appeared puzzling in the transition to freshwater as it results in a slight blueshift. However, our experimental assays show that this substitution shows a much larger effect on retinal release, an important nonspectral component of function in rhodopsin (Imai et al. 2007; Sugawara et al. 2010; Castiglione and Chang 2018), as discussed below.
Nonspectral Adaptations for Turbid Environments
In our study, ancestral rhodopsin amino acid substitutions that were targets of positive selection in the transition from marine to freshwater were found to increase the rate of light-activated retinal release when experimentally assayed in the laboratory. This rate is thought to be associated with physiological differences between rod and cone photoreceptors, affecting the rate of dark adaptation (Imai et al. 2007). Faster release of the retinal chromophore after light activation might facilitate more rapid recovery after a light bleach in cone photoreceptors (Imai et al. 2007), and may be one factor among several underlying the nearly 10-fold increase in the rate of dark adaptation in cones (Ingram et al. 2016). Indeed, these nonspectral changes in rhodopsin might help explain why some counterintuitive substitutions such as E122I are found to be the targets of selection, as well as why some positively selected sites in rhodopsin have been reported to have little impact on rhodopsin spectral sensitivity (Yokoyama et al. 2008). For freshwater fishes, faster recovery of dim-light vision might provide an ecological advantage during quick ascents and descents in the water column where light is attenuated very rapidly creating a very narrow interface between bright and dark-light environments. Interestingly, rapid rates of retinal release have been observed in other well-studied freshwater fishes, such as the zebrafish (Morrow and Chang 2015). Therefore, as with mesopelagic species possessing retinas tuned to intermediate light intensities (de Busserolles et al. 2017), highly turbid and tannin stained freshwater environments might pose important trade-offs between rod and cone functional properties.
Photosensitivity Trade-offs with Thermal Noise and Signal Amplification?
Although redshifted spectral sensitivity and increased rate of retinal release might be adaptive for some aspects of freshwater croaker vision, other studies suggest that these properties are likely to negatively impact other aspects of rhodopsin function such as photosensitivity. Computational modeling as well as experimental studies indicate that redshifts in visual pigment absorption maxima correlate with decreased barriers to thermal isomerization and therefore decreased photosensitivity (Ala-Laurila et al. 2003; Luo et al. 2011; Luk et al. 2016). The need to maintain a high barrier to thermal activation is considered a critical factor constraining the evolution of rhodopsin and is a major aspect that differentiates rhodopsin from cone opsins (Ingram et al. 2016; Yue et al. 2017). An endemic flock of cottoid fishes radiating into dim-light environments in the depths of Lake Baikal are thought to have acquired adaptations to rhodopsin that would increase photosensitivity by decreasing thermal noise, which ultimately sets the limit of light detection (Luk et al. 2016). However, many of the advantages of high photosensitivity in blueshifted visual pigments have focused on marine or deep lake species inhabiting blueshifted waters or terrestrial habitats (Luo et al. 2011; Luk et al. 2016). In contrast, the redshifted pigments observed in the benthic freshwater croakers, as well as most other freshwater fishes (Schweikert et al. 2018), especially benthic species (Levine and MacNichol 1979), are likely to result in decreased barriers to thermal activation and therefore decreased photosensitivity. This is also likely compounded by the fact that many fast-flowing turbid rivers are much more homogeneous in temperature with depth than marine waters. However, this trade-off with photosensitivity is likely offset by very rapid attenuation of short-wavelength light in most freshwater systems, necessitating a redshifted visual pigment. This highlights the importance of environmental context when considering the optimal functional properties of a protein, especially those involved in sensory systems.
Model Flexibility Is Important in Detecting Positive Selection
For this data set, the more flexible model, CmD was found to result in a better fit than CmC. CmC is by far the most frequently used of the clade models in PAML (Chang et al. 2012), but its failure to adequately accommodate the multiple classes of positive selected sites may become increasingly prevalent as data sets increase in size and sequence diversity. Issues associated with models designed to detect branch- and clade-specific positive selection in PAML have been previously identified and remedied. Modifications to the branch-site model (Zhang 2005) and the null model used in tandem with CmC have improved their reliability (Weadick and Chang 2012). Understanding the limitations of these methods and approaches is critical for interpreting results. For example, in data sets where pervasive positive selection is prevalent, such as in the rapidly evolving cichlid fishes, the assumptions of the branch-site tests may be violated, requiring the more flexible clade models (Schott et al. 2014). Few previous studies have utilized CmD, and fewer yet have compared its goodness of fit with CmC (Chang et al. 2012), but exploring comparisons between CmC and D may be important, particularly in cases where the dN/dS may be elevated across the phylogeny.
Conclusion
In this study, we link positive selection along a transitional branch, representing the invasion of South American rivers by croakers, to a shift in rhodopsin spectral and kinetic properties consistent with the intensity and wavelengths of light illuminating the ancestral marine and freshwater environments. We have observed a similar shift in rate of nonsynonymous substitutions in anchovies, another marine to freshwater transition in South America (Van Nynatten et al. 2015). Comparatively, the number and nature of substitutions observed along the transitional branch in the South American freshwater anchovies suggests a less substantial shift in protein function. This may reflect the highly visual nature in the primarily piscivorous croakers in contrast to the most planktivorous and less-active feeding anchovies (Sasaki 1989; Van Nynatten et al. 2015). It may also reflect the benthic habitats of croakers, where more light is attenuated and the need for spectral tuning is greater, resulting in more rapid molecular adaptation (Sasaki 1989; Horodysky et al. 2008). This additional level of complexity may be even more critical for understanding important but often overlooked aspects of functional adaptation, such as the kinetic rate of rhodopsin, potentially more closely associated with behavior than shifts in spectral sensitivity. Because major habitat transitions can result in rapid shifts in protein function, as we have observed in the croakers, using evolutionary transitions to investigate the mechanistic basis of physiological adaptations promises to be a productive avenue of future research in this field.
Materials and Methods
Sequencing and Sequence Alignment
DNA was extracted from 43 tissue samples preserved in ethanol, representing 36 species using a QIAGEN DNeasy kit (Qiagen Inc., Santa Clara CA, USA). Four nuclear loci, the recombination activating gene 1 (RAG1), early growth response protein 1 and 2 (EGR1 and EGR2), and rhodopsin, as well as two mitochondrial loci, Cytochrome c oxidase subunit I (coi) and cytochrome b (cytb) were amplified using polymerase chain reaction (PCR) using primers and protocols previously published in (Van Nynatten et al. 2015) and (Lo et al. 2015). PCR products were purified using ExoSAP-IT PCR Product Cleanup Reagent (Applied Biosystems, Foster City, CA, USA) and sequenced by Sanger sequencing at the Hospital for Sick Children TCAG Sequencing Facility. These sequences were combined with sequences for the same six molecular markers generated in a previous phylogenetic reconstruction of croakers (Lo et al. 2015) to form a data set of 139 taxa (supplementary tables S14 and S15, Supplementary Material online). Sequences for each gene in the final data set were aligned using MUSCLE (Edgar 2004). Terminal gaps were removed when present in more than half of the samples. We concatenated the alignments of all six molecular markers for a total data set length of 6,460 bp.
Phylogenetic Reconstructions
Partition Finder (Lanfear et al. 2017) was used to estimate the best-fitting model of molecular evolution for each gene and the optimal partitioning scheme for the concatenated data set (supplementary table S16, Supplementary Material online). A Bayesian phylogeny was generated with Mr. Bayes 3.2 (supplementary fig. S1, Supplementary Material online) (Ronquist et al. 2012), sampling from an Markov chain Monte Carlo chain every 1,000 steps from two independent runs of 5,000,000 generations, discarding the first 25% of samples as burn-in. RAXML (Stamatakis 2014) was used to reconstruct maximum-likelihood phylogenies of the concatenated data set and a rhodopsin-only data set with 1,000 bootstrap replicates to obtain support for nodes (supplementary figs. S2 and S3, Supplementary Material online).
Molecular Evolutionary Analyses
To model the selection pressures acting on croaker rhodopsin, we employed codon models of molecular evolution implemented in PAML (Yang 2007) and HYPHY (Kosakovsky Pond et al. 2005) estimating rates of nonsynonymous to synonymous substitutions (dN/dS). We tested for pervasive positive selection by comparing random sites models in PAML including a positively selected site class (M2a and M8) with nested null models without (m1a, M7, and M8a) using likelihood ratio tests (Yang et al. 2000). We also used FUBAR (HYPHY) to estimate positive selection on rhodopsin. FUBAR independently estimates the rate of synonymous substitutions and allows for greater flexibility in dN/dS site class parameter estimates (Murrell et al. 2013).
We used branch-site and clade models in PAML to test for episodic bouts of positive and divergent selection on a priori selected branches herein referred to as “foreground” branches. We tested for positive selection on branches representing the three freshwater invasion events and each ecological partition (fig. 2A) by comparing the branch-site models with the branch-site null models in PAML (Yang and Nielsen 2002). CmC and CmD (CmC and CmD) were also used to test for episodic shifts in selection pressures on these branches with comparison to nested null models M2aREL and M3, respectively (Bielawski and Yang 2004; Weadick and Chang 2012). As a compliment to models in PAML that allows for different rates across a priori selected branches, we employed the aBSREL model in HYPHY that estimates dN/dS on each branch of the phylogeny without any a priori input (Kosakovsky Pond et al. 2011). These tests were replicated on control gene data sets pruned to contain only species with sequence data for each of the nuclear genes included in this study. All tests of selection were replicated on each of the different tree topologies (supplementary figs. S1–S3, Supplementary Material online).
PAML was also used to reconstruct the ancestral sequences using the best-fitting codon models and the Dayhoff, JTT, and WAG empirical amino acid models of molecular evolution. For all PAML analyses sites under positive selection were inferred by BEB analysis when available or Naïve Empirical Bayes when not (Yang 2005). We visualized the location of positively selected sites and substitutions observed along transitional lineages on the crystal structure of rhodopsin in its dark-state (IU19) and active-state (3PQR) crystal structures using UCSF Chimera (Okada et al. 2004; Pettersen et al. 2004; Choe et al. 2011).
Protein Expression and Functional Characterization
Ancestral marine and freshwater rhodopsin sequences at the nodes bounding the marine to freshwater transition in South America were synthesized using GeneArt (Invitrogen) with N and C termini, and, where possibly, codons converted to match human rhodopsin for better expression in HEK cells. Restriction sites for insertion into the p1D4-hrGFP II expression vector were also included (Morrow and Chang 2010). Expression vectors containing the ancestral sequences and SDM bovine rhodopsin sequences were transiently transfected into HEK cells using Lipofectamine 2000 (Invitrogen) and harvested 48 h posttransfection. Rhodopsins were regenerated for 2 h in the dark with 5 µM 11-cis-retinal and immunoaffinity purified in the dark using the 1D4 monoclonal antibody. Because expression levels of these ancestral pigments were not sufficient for our fluorescence retinal release assays, we were unable to differentiate between the two ancestral pigments. However, because four positively selected substitutions nearest the chromophore have also been observed in many other freshwater taxa, we opted to investigate the effects of these substitutions in a model system genetic background using SDM to convert the amino acid identities in bovine rhodopsin at sites 119, 122, 124, and 261 to match the marine and freshwater croaker sequences via PCR (QuickChange II, Agilent). SDM sequences were verified using a 3730 DNA Analyzer (Applied Biosystems) at the Centre for Analysis of Genome Evolution and Function and expressed as described above.
The UV-visible absorption spectra of purified rhodopsin samples were recorded in the dark at 20 °C using a Cary 4000 double-beam absorbance spectrophotometer (Agilent). Peak spectral sensitivities were determined by fitting dark spectra to a standard template curve for A1 visual pigments (Govardovskii et al. 2000). Rhodopsin samples were bleached for 30 s with a fiber optic lamp (Dolan-Jenner), and fluorescence measurements of retinal release were recorded at 20 °C over 30-s intervals using a Cary Eclipse spectrophotometer (Varian). Data were fit to a three variable, first-order exponential equation (y = y0 + a(1 − e−bx)), and half-life values were calculated using the rate constant b (t1/2 = ln2/b). The accessibility of the rhodopsin-binding pocket was assayed with a hydroxylamine treatment (NH2OH; 50 mM).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Acknowledgements
This work was supported by Discovery grants to B.S.W.C. and N.R.L. from the Natural Sciences and Engineering Research Council of Canada, and a VSRP fellowship to A.V.N. We thank Hernan Lopez-Fernandez (Royal Ontario Museum), Devin Bloom (University of Western Michigan), and Mark Sabaj (Academy of Natural Sciences of Drexel University) for providing tissue samples, and Frances Hauser and two anonymous reviewers for their helpful comments and suggestions.
Data Availability
All sequence data and accession numbers, are provided in the Supplementary Materials online.
Supplementary Material
References
- Ala-Laurila P, Albert RJ, Saarinen P, Koskelainen A, Donner K.. 2003. The thermal contribution to photoactivation in A2 visual pigments studied by temperature effects on spectral properties. Vis Neurosci. 20(4):411–419. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Liberles DA.. 2007. The quest for natural selection in the age of comparative genomics. Heredity 99(6):567–579. [DOI] [PubMed] [Google Scholar]
- Arnott HJ, Nicol JAC, Querfeld CW.. 1972. Tapeta lucida in the eyes of the seatrout (Sciaenidae). Proc R Soc Lond B. 180:247–271. [Google Scholar]
- Arshavsky VY, Lamb TD, Pugh EN.. 2002. G proteins and phototransduction. Annu Rev Physiol. 64(1):153–187. [DOI] [PubMed] [Google Scholar]
- Baylor DA. 1996. How photons start vision. Proc Natl Acad Sci U S A. 93(2):560–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski JP, Yang Z.. 2004. A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J Mol Evol. 59(1):121–132. [DOI] [PubMed] [Google Scholar]
- Bloom DD, Lovejoy NR.. 2017. On the origins of marine-derived freshwater fishes in South America. J Biogeogr. 44(9):1927–1938. [Google Scholar]
- Bowmaker JK, Hunt DM.. 2006. Evolution of vertebrate visual pigments. Curr Biol. 16(13):R484–R489. [DOI] [PubMed] [Google Scholar]
- Castiglione GM, Chang BSW.. 2018. Functional trade-offs and environmental variation shaped ancient trajectories in the evolution of dim-light vision. eLife 7:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang BSW, Donoghue MJ.. 2000. Recreating ancestral proteins. Trends Ecol Evol. 15(3):109–114. [DOI] [PubMed] [Google Scholar]
- Chang BSW, Du J, Weadick CJ, Müller J, Bickelmann C, David Yu D, Morrow JM.. 2012. The future of codon models in studies of molecular function: ancestral reconstruction, and clade models of functional divergence. In: Cannarozzi GM, Schneider A, editors. Codon evolution: mechanisms and models. Oxford: Oxford University Press. p. 145–163. [Google Scholar]
- Chang BSW, Jönsson K, Kazmi MA, Donoghue MJ, Sakmar TP.. 2002. Recreating a functional ancestral archosaur visual pigment. Mol Biol Evol. 19(9):1483–1489. [DOI] [PubMed] [Google Scholar]
- Choe H-W, Kim YJ, Park JH, Morizumi T, Pai EF, Krauß N, Hofmann KP, Scheerer P, Ernst OP.. 2011. Crystal structure of metarhodopsin II. Nature 471(7340):651–655. [DOI] [PubMed] [Google Scholar]
- Costa MPF, Novo EMLM, Telmer KH.. 2013. Spatial and temporal variability of light attenuation in large rivers of the Amazon. Hydrobiologia 702(1):171–190. [Google Scholar]
- de Busserolles F, Cortesi F, Helvik JV, Davies WIL, Templin RM, Sullivan RKP, Michell CT, Mountford JK, Collin SP, Irigoien X, et al. 2017. Pushing the limits of photoreception in twilight conditions: the rod-like cone retina of the deep-sea pearlsides. Sci Adv. 3(11):eaao4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deary AL, Metscher B, Brill RW, Hilton EJ.. 2016. Shifts of sensory modalities in early life history stage estuarine fishes (Sciaenidae) from the Chesapeake Bay using X-ray micro computed tomography. Environ Biol Fish. 99(4):361–375. [Google Scholar]
- Dungan SZ, Chang BSW.. 2017. Epistatic interactions influence terrestrial–marine functional shifts in cetacean rhodopsin. Proc R Soc B. 284(1850):20162743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher EA, Thomson JM, Burgan MF, Benner SA.. 2003. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature 425(6955):285–288. [DOI] [PubMed] [Google Scholar]
- Govardovskii VI, Fyhrquist N, Reuter T, Kuzmin DG, Donner K.. 2000. In search of the visual pigment template. Vis Neurosci. 17(4):509–528. [DOI] [PubMed] [Google Scholar]
- Gower DJ, Sampaio FL, Peichl L, Wagner H-J, Loew ER, Mclamb W, Douglas RH, Orlov N, Grace MS, Hart NS, et al. 2019. Evolution of the eyes of vipers with and without infrared-sensing pit organs. Biol J Linn Soc Lond. 126(4):796–823. [Google Scholar]
- Gutierrez EA, Castiglione GM, Morrow JM, Schott RK, Loureiro LO, Lim BK, Chang BSW.. 2018. Functional shifts in bat dim-light visual pigment are associated with differing echolocation abilities and reveal molecular adaptation to photic-limited environments. Mol Biol Evol. 35(10):2422–2434. [DOI] [PubMed] [Google Scholar]
- Hart NS, Lamb TD, Patel HR, Chuah A, Natoli RC, Hudson NJ, Cutmore SC, Davies WIL, Collin SP, Hunt DM.. 2020. Visual opsin diversity in sharks and rays. Mol Biol Evol. 37(3):811–827. [DOI] [PubMed] [Google Scholar]
- Hauser FE, Chang BSW.. 2017. Insights into visual pigment adaptation and diversity from model ecological and evolutionary systems. Curr Opin Genet Dev. 47:110–120. [DOI] [PubMed] [Google Scholar]
- Hauser FE, Ilves KL, Schott RK, Castiglione GM, López-Fernández H, Chang BSW.. 2017. Accelerated evolution and functional divergence of the dim light visual pigment accompanies cichlid colonization of Central America. Mol Biol Evol. 34(10):2650–2664. [DOI] [PubMed] [Google Scholar]
- Hill J, Enbody ED, Pettersson ME, Sprehn CG, Bekkevold D, Folkvord A, Laikre L, Kleinau G, Scheerer P, Andersson L, et al. 2019. Recurrent convergent evolution at amino acid residue 261 in fish rhodopsin. Proc Natl Acad Sci U S A. 116(37):18473–18478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann KP, Scheerer P, Hildebrand PW, Choe H-W, Park JH, Heck M, Ernst OP.. 2009. A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem Sci. 34(11):540–552. [DOI] [PubMed] [Google Scholar]
- Hoorn C, Wesselingh FP, Steegeter H, Bermudez MA, Mora A, Sevink J, Sanmartín I, Sanchez-Meseguer A, Anderson CL, Figueiredo JP, et al. 2010. Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science 330(6006):927–931. [DOI] [PubMed] [Google Scholar]
- Horodysky AZ, Brill RW, Warrant EJ, Musick JA, Latour RJ.. 2008. Comparative visual function in five sciaenid fishes inhabiting Chesapeake Bay. J Exp Biol. 211(22):3601–3612. [DOI] [PubMed] [Google Scholar]
- Hunt DM, Dulai KS, Partridge JC, Cottrill P, Bowmaker JK.. 2001. The molecular basis for spectral tuning of rod visual pigments in deep-sea fish. J Exp Biol. 204:3333. [DOI] [PubMed] [Google Scholar]
- Hunt DM, Fitzgibbon J, Slobodyanyuk SJ, Bowmaker JK.. 1996. Spectral tuning and molecular evolution of rod visual pigments in the species flock of cottoid fish in Lake Baikal. Vision Res. 36(9):1217–1224. [DOI] [PubMed] [Google Scholar]
- Imai H, Kefalov V, Sakurai K, Chisaka O, Ueda Y, Onishi A, Morizumi T, Fu Y, Ichikawa K, Nakatani K, et al. 2007. Molecular properties of rhodopsin and rod function. J Biol Chem. 282(9):6677–6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram NT, Sampath AP, Fain GL.. 2016. Why are rods more sensitive than cones? J Physiol. 594(19):5415–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa A, Kabeya N, Ikeya K, Kakioka R, Cech JN, Osada N, Leal MC, Inoue J, Kume M, Toyoda A, et al. 2019. A key metabolic gene for recurrent freshwater colonization and radiation in fishes. Science 364(6443):886–889. [DOI] [PubMed] [Google Scholar]
- Isogai Y, Imamura H, Nakae S, Sumi T, Takahashi K-I, Nakagawa T, Tsuneshige A, Shirai T.. 2018. Tracing whale myoglobin evolution by resurrecting ancient proteins. Sci Rep. 8(1):16883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerlov NG, ed. 1968.Optical oceanography. Vol. 5. New York: Elsevier. p. 115–132. [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, et al. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484(7392):55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach M, Burke JR, Dindo M, Pabis A, Munsberg FS, Rabin A, Kamerlin SCL, Noel JP, Tawfik DS.. 2018. Evolution of chalcone isomerase from a noncatalytic ancestor. Nat Chem Biol. 14(6):548–555. [DOI] [PubMed] [Google Scholar]
- Kenny NJ, Plese B, Riesgo A, Itskovich VB.. 2019. Symbiosis, selection, and novelty: freshwater adaptation in the unique sponges of Lake Baikal. Mol Biol Evol. 36(11):2462–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano J, Lema SC, Luckenbach JA, Mori S, Kawagishi Y, Kusakabe M, Swanson P, Peichel CL.. 2010. Adaptive divergence in the thyroid hormone signaling pathway in the stickleback radiation. Curr Biol. 20(23):2124–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SLK, Frost SDW, Muse SV.. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21(5):676–679. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Murrell B, Fourment M, Frost SDW, Delport W, Scheffler K.. 2011. A Random effects branch-site model for detecting episodic diversifying selection. Mol Biol Evol. 28(11):3033–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratzer JT, Lanaspa MA, Murphy MN, Cicerchi C, Graves CL, Tipton PA, Ortlund EA, Johnson RJ, Gaucher EA.. 2014. Evolutionary history and metabolic insights of ancient mammalian uricases. Proc Natl Acad Sci U S A. 111(10):3763–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S, Berglund J, Almén MS, Maqbool K, Grabherr M, Martinez-Barrio A, Promerová M, Rubin C-J, Wang C, Zamani N, et al. 2015. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature 518(7539):371–375. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B.. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773. [DOI] [PubMed] [Google Scholar]
- Lee CE, Posavi M, Charmantier G.. 2012. Rapid evolution of body fluid regulation following independent invasions into freshwater habitats. J Evol Biol. 25(4):625–633. [DOI] [PubMed] [Google Scholar]
- Levine JS, MacNichol EF.. 1979. Visual pigments in teleost fishes: effects of habitat, microhabitat, and behavior on visual system evolution. Sens Processes. 3(2):95–131. [PubMed] [Google Scholar]
- Lim MCW, Witt CC, Graham CH, Dávalos LM.. 2019. Parallel molecular evolution in pathways, genes, and sites in high-elevation hummingbirds revealed by comparative transcriptomics. Genome Biol Evol. 11(6):1573–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J-J, Wang F-Y, Li W-H, Wang T-Y.. 2017. The rises and falls of opsin genes in 59 ray-finned fish genomes and their implications for environmental adaptation. Sci Rep. 7(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Cui Y, Chi H, Xia Y, Liu H, Rossiter SJ, Zhang S.. 2019. Scotopic rod vision in tetrapods arose from multiple early adaptive shifts in the rate of retinal release. Proc Natl Acad Sci U S A. 16:201900481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo P-C, Liu S-H, Chao NL, Nunoo FKE, Mok H-K, Chen WJ.. 2015. A multi-gene dataset reveals a tropical New World origin and Early Miocene diversification of croakers (Perciformes: Sciaenidae). Mol Phylogenet Evol. 88:132–143. [DOI] [PubMed] [Google Scholar]
- Lovejoy NR, Bermingham E, Martin AP.. 1998. Marine incursion into South America. Nature 396(6710):421–422. [Google Scholar]
- Luk HL, Bhattacharyya N, Montisci F, Morrow JM, Melaccio F, Wada A, Sheves M, Fanelli F, Chang BSW, Olivucci M.. 2016. Modulation of thermal noise and spectral sensitivity in Lake Baikal cottoid fish rhodopsins. Sci Rep. 6(1):W116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo DG, Yue WWS, Ala-Laurila P, Yau KW.. 2011. Activation of visual pigments by light and heat. Science 332(6035):1307–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lythgoe JN. 1979. The ecology of vision. Oxford; New York: Clarendon Press; Oxford University Press [Google Scholar]
- Marques DA, Jones FC, Di Palma F, Kingsley DM, Reimchen TE.. 2018. Experimental evidence for rapid genomic adaptation to a new niche in an adaptive radiation. Nat Ecol Evol. 2(7):1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow JM, Chang BSW.. 2010. The p1D4-hrGFP II expression vector: a tool for expressing and purifying visual pigments and other G protein-coupled receptors. Plasmid 64(3):162–169. [DOI] [PubMed] [Google Scholar]
- Morrow JM, Chang BSW.. 2015. Comparative mutagenesis studies of retinal release in light-activated zebrafish rhodopsin using fluorescence spectroscopy. Biochemistry 54(29):4507–4518. [DOI] [PubMed] [Google Scholar]
- Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K.. 2013. FUBAR: a fast, unconstrained Bayesian approximation for inferring selection. Mol Biol Evol. 30(5):1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musilova Z, Cortesi F, Matschiner M, Davies WIL, Patel JS, Stieb SM, de Busserolles F, Malmstrøm M, Tørresen OK, Brown CJ, et al. 2019. Vision using multiple distinct rod opsins in deep-sea fishes. Science 364(6440):588–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Sugihara M, Bondar A-N, Elstner M, Entel P, Buss V.. 2004. The retinal conformation and its environment in rhodopsin in light of a new 2.2Å crystal structure. J Mol Biol. 342(2):571–583. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE.. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem. 25(13):1605–1612. [DOI] [PubMed] [Google Scholar]
- Pisciottano F, Cinalli AR, Stopiello JM, Castagna VC, Elgoyhen AB, Rubinstein M, Gómez-Casati ME, Franchini LF.. 2019. Inner ear genes underwent positive selection and adaptation in the mammalian lineage. Mol Biol Evol. 36(8):1653–1670. [DOI] [PubMed] [Google Scholar]
- Projecto-Garcia J, Natarajan C, Moriyama H, Weber RE, Fago A, Cheviron ZA, Dudley R, McGuire JA, Witt CC, Storz JF.. 2013. Repeated elevational transitions in hemoglobin function during the evolution of Andean hummingbirds. Proc Natl Acad Sci U S A. 110(51):20669–20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahi ML, Mather PB, Ezaz T, Hurwood DA.. 2019. The molecular basis of freshwater adaptation in prawns: insights from comparative transcriptomics of three Macrobrachium species. Genome Biol Evol. 11(4):1002–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramcharitar J, Gannon DP, Popper AN.. 2006. Bioacoustics of fishes of the family Sciaenidae (croakers and drums). Trans Am Fish Soc. 135(5):1409–1431. [Google Scholar]
- Rennison DJ, Owens GL, Heckman N, Schluter D, Veen T.. 2016. Rapid adaptive evolution of colour vision in the threespine stickleback radiation. Proc R Soc B . 283(1830):20160242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennison DJ, Owens GL, Taylor JS.. 2012. Opsin gene duplication and divergence in ray-finned fish. Mol Phylogenet Evol. 62(3):986–1008. [DOI] [PubMed] [Google Scholar]
- Reznick D, Meredith R, Collette BB.. 2007. Independent evolution of complex life history adaptations in two families of fishes, live-bearing halfbeaks (Zenarchopteridae, Beloniformes) and poeciliidae (Cyprinodontiformes). Evolution 61(11):2570–2583. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP.. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JL, Bracken-Grissom HD, Trexler JC.. 2019. Freshwater-to-marine transitions may explain the evolution of herbivory in the subgenus Mollienesia (genus Poecilia, mollies and guppies). Biol J Linn Soc Lond. 127(4):742–761. [Google Scholar]
- Sasaki K. 1989. Phylogeny of the family Sciaenidae, with notes on its zoogeography (Teleostei, Percifomes). Memoirs of the Faculty of Fisheries, Hokkaido University. p. 36.
- Schluter D. 2000. The ecology of adaptive radiation. Oxford: Oxford University Press. [Google Scholar]
- Schott RK, Refvik SP, Hauser FE, López-Fernández H, Chang BSW.. 2014. Divergent positive selection in rhodopsin from lake and riverine cichlid fishes. Mol Biol Evol. 31(5):1149–1165. [DOI] [PubMed] [Google Scholar]
- Schott RK, Van Nynatten A, Card DC, Castoe TA, Chang BSW.. 2018. Shifts in selective pressures on snake phototransduction genes associated with photoreceptor transmutation and dim-light ancestry. Mol Biol Evol. 35(6):1376–1389. [DOI] [PubMed] [Google Scholar]
- Schweikert LE, Caves EM, Solie SE, Sutton TT, Johnsen S.. 2018. Variation in rod spectral sensitivity of fishes is best predicted by habitat and depth. J Fish Biol. 95(1):179–185. [DOI] [PubMed] [Google Scholar]
- Smith SO. 2010. Structure and activation of the visual pigment rhodopsin. Annu Rev Biophys. 39(1):309–328. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF. 2005. Invited Review: using genome scans of DNA polymorphism to infer adaptive population divergence. Mol Ecol. 14(3):671–688. [DOI] [PubMed] [Google Scholar]
- Storz JF. 2016. Causes of molecular convergence and parallelism in protein evolution. Nat Rev Genet. 17(4):239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud JT, Losos JB.. 2016. Ecological opportunity and adaptive radiation. Annu Rev Ecol Evol Syst. 47(1):507–532. [Google Scholar]
- Sugawara T, Imai H, Nikaido M, Imamoto Y, Okada N.. 2010. Vertebrate rhodopsin adaptation to dim light via rapid meta-II intermediate formation. Mol Biol Evol. 27(3):506–519. [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS.. 2009. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol. 19(5):596–604. [DOI] [PubMed] [Google Scholar]
- Toyama M, Hironaka M, Yamahama Y, Horiguchi H, Tsukada O, Uto N, Ueno Y, Tokunaga F, Seno K, Hariyama T.. 2008. Presence of rhodopsin and porphyropsin in the eyes of 164 fishes, representing marine, diadromous, coastal and freshwater species. A qualitative and comparative study. Photochem Photobiol. 84(4):996–1002. [DOI] [PubMed] [Google Scholar]
- Tsai J-R, Lin H-C.. 2007. V-type H+-ATPase and Na+,K+-ATPase in the gills of 13 euryhaline crabs during salinity acclimation. J Exp Biol. 210(Pt 4):620–627. [DOI] [PubMed] [Google Scholar]
- Ugalde JA, Chang BSW, Matz MV.. 2004. Evolution of coral pigments recreated. Science 305(5689):1433. [DOI] [PubMed] [Google Scholar]
- Van Nynatten A, Bloom DD, Chang BSW, Lovejoy NR.. 2015. Out of the blue: adaptive visual pigment evolution accompanies Amazon invasion. Biol Lett. 11(7):20150349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nynatten A, Janzen FH, Brochu K, Maldonado-Ocampo JA, Crampton WGR, Chang BSW, Lovejoy NR.. 2019. To see or not to see: molecular evolution of the rhodopsin visual pigment in neotropical electric fishes. Proc R Soc B. 286(1906):20191182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald G, Brown PK, Brown PS.. 1957. Visual pigments and depths of habitat of marine fishes. Nature 180(4593):969–971. [Google Scholar]
- Weadick CJ, Chang BSW.. 2012. An improved likelihood ratio test for detecting site-specific functional divergence among clades of protein-coding genes. Mol Biol Evol. 29(5):1297–1300. [DOI] [PubMed] [Google Scholar]
- Worth CL, Gong S, Blundell TL.. 2009. Structural and functional constraints in the evolution of protein families. Nat Rev Mol Cell Biol. 10(10):709–720. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 22(4):1107–1118. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Bielawski JP.. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15(12):496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Kumar S, Nei M.. 1995. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141(4):1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R.. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 19(6):908–917. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen A-MK.. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Tada T, Zhang H, Britt L.. 2008. Elucidation of phenotypic adaptations: molecular analyses of dim-light vision proteins in vertebrates. Proc Natl Acad Sci U S A. 105(36):13480–13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue WWS, Frederiksen R, Ren X, Luo DG, Yamashita T, Shichida Y, Cornwall CM, Yau KW.. 2017. Spontaneous activation of visual pigments in relation to openness/closedness of chromophore-binding pocket. eLife 6:e18492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakon HH, Lu Y, Zwickl DJ, Hillis DM.. 2006. Sodium channel genes and the evolution of diversity in communication signals of electric fishes: convergent molecular evolution. Proc Natl Acad Sci U S A. 103(10):3675–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22(12):2472–2479. [DOI] [PubMed] [Google Scholar]
- Zhou X, Guang X, Sun D, Xu S, Li M, Seim I, Jie W, Yang L, Zhu Q, Xu J, et al. 2018. Population genomics of finless porpoises reveal an incipient cetacean species adapted to freshwater. Nat Commun. 9(1):1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data and accession numbers, are provided in the Supplementary Materials online.