Abstract
Anthozoan corals are an ecologically important group of cnidarians, which power the productivity of reef ecosystems. They are sessile, inhabit shallow, tropical oceans and are highly dependent on sun- and moonlight to regulate sexual reproduction, phototaxis, and photosymbiosis. However, their exposure to high levels of sunlight also imposes an increased risk of UV-induced DNA damage. How have these challenging photic environments influenced photoreceptor evolution and function in these animals? To address this question, we initially screened the cnidarian photoreceptor repertoire for Anthozoa-specific signatures by a broad-scale evolutionary analysis. We compared transcriptomic data of more than 36 cnidarian species and revealed a more diverse photoreceptor repertoire in the anthozoan subphylum than in the subphylum Medusozoa. We classified the three principle opsin classes into distinct subtypes and showed that Anthozoa retained all three classes, which diversified into at least six subtypes. In contrast, in Medusozoa, only one class with a single subtype persists. Similarly, in Anthozoa, we documented three photolyase classes and two cryptochrome (CRY) classes, whereas CRYs are entirely absent in Medusozoa. Interestingly, we also identified one anthozoan CRY class, which exhibited unique tandem duplications of the core functional domains. We next explored the functionality of anthozoan photoreceptors in the model species Exaiptasia diaphana (Aiptasia), which recapitulates key photo-behaviors of corals. We show that the diverse opsin genes are differentially expressed in important life stages common to reef-building corals and Aiptasia and that CRY expression is light regulated. We thereby provide important clues linking coral evolution with photoreceptor diversification.
Keywords: Cnidaria, Aiptasia, cryptochromes, opsins, photolyases, phylogenetics
Introduction
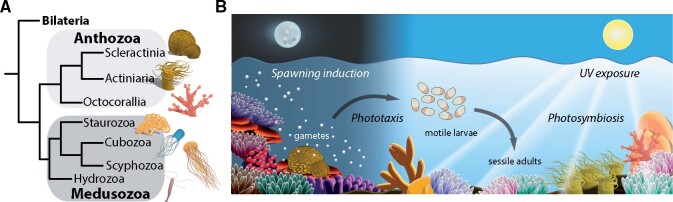
Light from both the sun and moon dominates the life of many organisms and has had a profound impact on their evolution. Notable examples are the highly light-dependent reef-building corals and anemones. These are members of the subphylum Anthozoa that together with the Medusozoa (which include jellyfish) constitute the phylum Cnidaria (fig. 1A)—a major group of nonbilaterian, eumetazoan animals (Zapata et al. 2015). The reef-building corals exploit a complexity of sunlight and moonlight-based cues to regulate many aspects of their physiology and behavior (fig. 1B). Many live in an evolutionary ancient symbiotic relationship with eukaryotic, photosynthetic dinoflagellates of the Symbiodiniaceae family (Janouskovec et al. 2017; LaJeunesse et al. 2018). The symbionts use sunlight to provide essential photosynthetically fixed nutrients to their hosts for supporting host survival in otherwise oligotrophic tropical oceans. Indeed, the nutrient transfer from dinoflagellate symbionts to the reef-building corals powers the productivity of reef ecosystems, which are home to more than 25% of all marine species (Fisher et al. 2015). The majority of these “photosynthetic Anthozoa” have a sessile lifestyle in shallow sunlit waters and are mobile only during early development at the larval stage (fig. 1B). Most corals harness light from both the sun and moon to orchestrate gamete release during sexual reproduction, including the synchronous mass-spawning events of reef-building corals worldwide (Sweeney et al. 2011; Kaniewska et al. 2015). Other important photo-induced behaviors include phototaxis of motile larvae and light-induced tentacle retraction in sessile adults (Bielecki et al. 2014; Oakley and Speiser 2015; Kanaya et al. 2019; Foo et al. 2020). Due to their almost “plant-like” lifestyle, sessile Anthozoa also face similar challenges as plants such as sustained exposure to intense sunlight, which also bears the risk of temperature stress and UV-induced DNA damage. Given the ecological niches that reef-building corals occupy, it is likely that their photoreceptors and light-responsive systems are essential elements in temporally and spatially coordinating behavior and physiology as well as combating the damaging effects of sunlight. Therefore, the question arises, has the adaptation of Anthozoa to their challenging photic environments influenced photoreceptor evolution and function?
Fig. 1.
The anthozoan lifestyle is dependent on sun- and moonlight. (A) Cladogram revealing phylogenetic relationships within the phylum Cnidaria showing the two main classes Anthozoa and Medusozoa and their major subclasses. (B) Schematic overview of how environmental light impacts reef-building corals. Light by the moon induces spawning and synchronizes gamete release. Motile larvae use environmental light for orientation and during settlement. Similar to larvae, adult corals are phototactic and exploit sunlight for photosymbiosis with photosynthetic dinoflagellates. Due to their sedentary lifestyle, corals face the challenge of DNA damage by exposure to sustained elevated levels of UV light.
Two main groups of light-sensing proteins exist in Metazoa. Opsins are eumetazoan-specific 7-transmembrane G-protein-coupled receptors, which typically incorporate a retinal chromophore and evolved from metazoan hormone-responsive receptors (Terakita 2005; Plachetzki et al. 2010; Feuda et al. 2012; Oakley and Speiser 2015). In most animal groups, they function as membrane-bound photoreceptors that mediate visual as well as nonvisual light sensing and trigger intracellular signaling events upon detection of specific wavelengths. However, recent findings have also implicated them in other nonphotic sensing functions (Leung and Montell 2017; Dalesio et al. 2018). To date, ten distinct opsin classes have been identified across all Metazoa, three of which, namely the cnidopsins (originally named “Cnidops” [Plachetzki et al. 2007]) as well as the Anthozoan-specific opsins I (ASO-I) and II (ASO-II) occur in cnidarians (Vöcking et al. 2017). The cnidopsins are relatively well studied and are often expressed in a distinct tissue- and stage-specific manner. For example, cnidopsin expression has been studied in the light-sensitive cilia of medusozoan eyes, in the hydrozoan battery complex, and more ubiquitously in sensory nerve cells (Suga et al. 2008; Plachetzki et al. 2012; Bielecki et al. 2014; Picciani et al. 2018). Moreover, light-induced gamete release has been shown to be opsin-dependent in the medusozoan Clytia hemisphaerica (Artigas et al. 2018). Anthozoa lack comparable eye-like sensory organs, yet they possess cnidopsins and Anthozoan-specific ASO-I and ASO-IIs both of which are thought to be generally restricted to Anthozoa and absent in the Medusozoa (Ramirez et al. 2016; Vöcking et al. 2017). A study of three out of nine Acropora opsins, comprising two cnidopsins and one ASO-I shows that the Acropora ASO-I can activate G-proteins in a light-dependent manner in vitro (Mason et al. 2012). However, due to a lack of extensive taxon sampling, in-depth assessment of function and diversity for the ASO-I and ASO-II opsins has been lacking.
A second major light-sensing group, comprising photolyases (PLs) and cryptochromes (CRYs), is a set of highly conserved flavoproteins involved in harvesting light energy to drive the repair of DNA damage as well as to regulate the circadian clock in response to light. Specifically, PLs enzymatically repair pyrimidine–pyrimidone (6-4) and cyclobutane pyrimidine dimer (CPD) DNA lesions generated by UV radiation. They were already present in the common ancestors of Bacteria, Archaea, and Eukarya and are classified according to the type of DNA damage that they repair ([6-4] PLs and CPD-PLs) (Cashmore 1999; Chaves et al. 2011; Benjdia 2012; Yokoyama and Mizutani 2014). CRYs, which generally lack PL enzyme activity, appear to have evolved independently several times from PLs at later stages of Eukarya evolution (Malhotra et al. 1995; Stanewsky et al. 1998; Thompson and Sancar 2002; Sancar 2003; Lucas-Lledo and Lynch 2009; Chaves et al. 2011). CRY1s are directly light sensitive and sometimes also referred to as Drosophila-type CRYs (Emery et al. 1998), whereas CRY-IIs (also called vertebrate-type CRYs) exhibit no light-dependent function. They instead regulate clock gene transcription in the negative limb of the circadian clock feedback loop (Kume et al. 1999; Shearman et al. 2000). CRY-DASHs ((Drosophila, Arabidopsis, Synechocystis, Human)-type CRYs) are functionally intermediate between PLs and CRYs and considered photoreceptors with residual DNA repair activity (Chaves et al. 2011; Gindt et al. 2015). However, all PLs and CRYs share an amino-terminal photolyase-related (PHR) region that contains a DNA-binding PL domain (also called alpha/beta domain which binds 5,10-methenyltetrahydrofolate pterin) and a flavin adenine dinucleotide (FAD) domain, which binds to a FAD chromophore (Chaves et al. 2011). To date, the phylogeny of CRYs and PL flavoproteins has not been assessed in great detail in cnidarians (Levy et al. 2007; Reitzel et al. 2010; Rivera et al. 2012) with only few studies on CRY function in relation to circadian rhythms in Acropora and Nematostella (Levy et al. 2011).
In order to gain some initial insights as to how evolution in challenging photic environments may have influenced photoreceptor evolution in corals, we first screened the cnidarian photoreceptor gene repertoire for anthozoan-specific signatures. Specifically, we classified extant anthozoan photoreceptors in comparison with other diverse cnidarian species using a thorough phylogenomics approach. Based on this phylogenetic analysis, we then investigated the expression and regulation of photoreceptors in Exaiptasia diaphana (formerly E. pallida; commonly Aiptasia) (Weis et al. 2008; ICZN 2017), an anthozoan model species which is widely used to investigate the molecular mechanisms underlying coral-dinoflagellate photosymbiosis (Bucher et al. 2016; Wolfowicz et al. 2016; Matthews et al. 2017; Neubauer et al. 2017; Hambleton et al. 2019) as well as its breakdown, a phenomenon known as “coral bleaching” (Tolleter et al. 2013). Moreover, analogous to most corals, Aiptasia also exhibits synchronous, blue (moon) light-induced gamete release (Grawunder et al. 2015) to produce motile larvae that must find a suitable niche for their light-dependent lifestyle (fig. 1B). Here, we reveal a large and highly diverse photoreceptor repertoire in Anthozoa. In turn, by scrutinizing the regulation of photoreceptor gene expression in Aiptasia, we shed new light on how the evolution of reef-building corals in their challenging shallow marine environments has shaped photoreceptor evolution and function.
Results
Opsin Complexity in Anthozoa
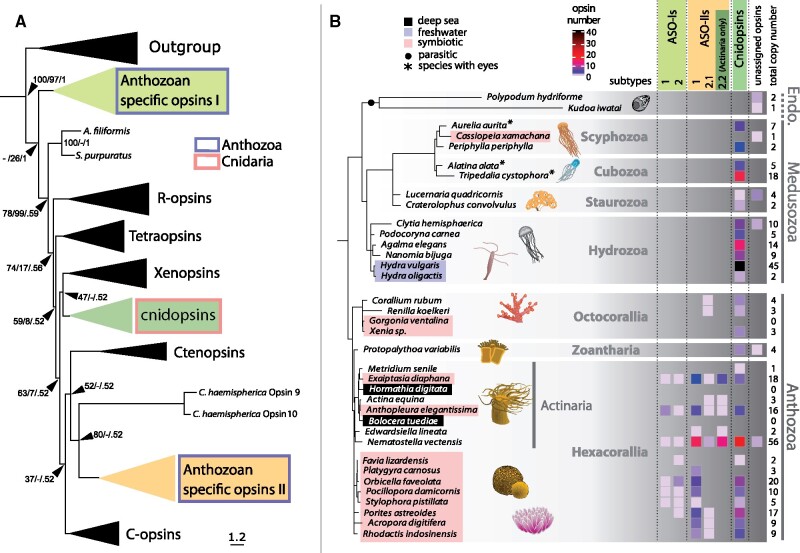
In order to screen for anthozoan-specific signatures within the cnidarian photoreceptor gene repertoire, we generated a detailed molecular phylogeny, based on RNA sequence data and gene structure analysis. We mined genomic data from 36 cnidarians (7 Anthozoa and 29 Medusozoa) and our large-scale phylogeny resolved the ten previously identified distinct opsin classes including three cnidarian types (ASO-Is, ASO-IIs, and cnidopsins; fig. 2A). All cnidarians (Anthozoa and Medusozoa) possess cnidopsins, which are monophyletic and sister to the animal xenopsins. The monophyletic ASO-I group appears as a sister group to all other opsins and like ASO-II opsins is only present in Anthozoa (fig. 2A). However, gene family trees (GFTs) sometimes fail to capture the complexity of the evolutionary processes that shape gene sequences. They are typically based on multiple sequence alignments of shortened and heavily curated genes and use statistical models of sequence evolution. Although this allows for comparisons and grouping of different sequence groups it does not necessarily account for the underlying taxonomic relationship of the included species. Therefore, to more directly relate the divergence of cnidarian light receptors to Anthozoa or Medusozoa and to distinguish between intraspecies gene duplication and gene loss versus gene diversification, we used a novel, automated species-tree-aware tool called GeneRax (Morel et al. 2020). This method exploits the relationship between a GFT and the species tree via appropriate statistical models to map the gene family members onto the species phylogeny with high accuracy as well as to provide insight into the evolutionary trajectory of gene families. GeneRax corrects the GFTs using the species tree and by incorporating and inferring gene duplication, gene loss, and, optionally, gene transfer events (not used here).
Fig. 2.
Anthozoa have a more diverse opsin repertoire than Medusozoa. (A) A majority-rule extended consensus tree topology generated with IQTree constructed from the combined bootstrap trees showing that cnidarians have at least three opsin paralogs. Anthozoans possess ASO-Is and ASO-IIs, which are monophyletic. ASO-I appears basal to all other animal opsins. All cnidarians (Anthozoa and Medusozoa) possess Cnidopsins. Cnidopsins are also monophyletic and sister to the animal xenopsins. ASO-IIs resolve as an opsin class that is distinct from ctenopsin and c-opsin despite their common ancestry lacking any conserved homologous intron positions (supplementary fig. 2, Supplementary Material online). Note: Clytia hemisphaerica opsin 9 and opsin 10 resolve at the base of the ASO-IIs, however elsewhere they are included within the Cnidopsins albeit with long branches (Artigas et al. 2018). Support values are SH-aLRT bootstrap percentages/UFBoots bootstrap percentages/MrBayes Bayesian posterior probabilities. Branch lengths are proportional to the mean number of expected substitutions per site. (B) Annotated phylogenetic tree (derived from Kayal et al. [2018]) including 36 cnidarian species. For each individual species, relevant characteristics including ecological distribution (freshwater [blue shading], deep sea [black shading]), symbiotic lifestyle (pink shading), and parasitism (black circle) are indicated. Cnidarian species with eyes are indicated by asterisks (*). Note that eyes are generally absent in Anthozoa, but the medusozoan classes Scyphozoa, Cubozoa, and Hydrozoa separately evolved eyes in certain taxa (Picciani et al. 2018). The exclusively parasitic Endocnidozoa (Endo.) are placed as sister to the Medusozoa. The total opsin gene copy numbers per species is indicated. The respective opsin copy numbers per opsin class/subtype are displayed in a heatmap with colors ranging from white to blue to red to black as shown in the key. The number of opsin classes and subtypes is generally higher in Anthozoa when compared with Medusozoa suggesting greater opsin diversity in this subphylum.
We therefore applied GeneRax to an additional data set of 36 cnidarian transcriptomes comprising 13 Medusozoa, 21 Anthozoa (including two deep-sea species), and two Endocnidozoa—an exclusively parasitic sister group to the Medusozoa. Using custom-developed python scripts for downstream analysis of the corrected gene tree and the position of the inferred gene duplication events, we revealed several novel opsin subtypes amongst the two anthozoan-specific opsin classes ASO-I and ASO-II. Specifically, we revealed that the ASO-Is are split into two distinct subtypes and named these ASO-I subtype 1 and ASO-I subtype 2. The ASO-IIs have been reported previously to be split into two groups (Vöcking et al. 2017; Picciani et al. 2018) and our analysis confirmed this. However, we now found that the ASO-IIs are actually split into three distinct subtypes, which we name, in accordance with their phylogenetic relationship, ASO-II subtype 1 and ASO-II subtype 2.1 and subtype 2.2 (fig. 2B). We also noticed that the ASO-II subtype 2.2 occurs exclusively in sea anemones (Actiniaria) and we confirmed this using reciprocal BLAST searches against and phylogenetic inference on a more extensive number of ASO-IIs using data from Picciani et al. (2018) (supplementary fig. 1, Supplementary Material online). Additionally, intron-phase analysis revealed that the ASO-II subtypes 1 and 2 could be distinguished from subtype 2.2 ASO-IIs by the different position of an intron (supplementary fig. 2A, red boxes, Supplementary Material online). Moreover, we observed that the ASO-II intron distribution is distinct from the ctenopsin and c-opsin intron distribution despite their common ancestry since there is a lack of any conserved homologous intron positions (supplementary fig. 2A, Supplementary Material online). This suggests that ancestrally, at least one intron-less ASO-II gene occurred which then duplicated. Subsequently, the resulting subtypes acquired distinct introns before the ASO-II subtypes further diversified. Interestingly, the exclusively parasitic Endocnidozoa only possess one or two opsins that cannot be assigned to any of the known cnidarian opsin classes, whereas Anthozoa occurring in the deep sea appear to entirely lack opsins (fig. 2B). The lack of opsins in deep-sea species was confirmed in 3 additional genera not show in figure 2B, namely: Dendrophyllia sp., Eguchipsammia fistula, and Rhizotrochus typus.
The monophyletic ASO-I group appears as a sister group to all other opsins and it has been proposed that both groups, ASO-I and ASO-II, have been secondarily lost in the Medusozoa (fig. 2A; Ramirez et al. 2016; Vöcking et al. 2017). Our GeneRax analysis indeed indicates that all three cnidarian opsin classes were present before the divergence of Medusozoa and Anthozoa. Specifically, we find that one ASO-Is, six ASO-IIs, and one cnidopsins were present in the last common ancestor of the Anthozoa and Medusozoa (supplementary file 7, Supplementary Material online). Thus, our results are consistent with the notion that the Anthozoa did not gain ASO-Is and ASO-IIs, but that they were lost secondarily in the Medusozoa instead (supplementary fig. 7, Supplementary Material online). Taken together, this demonstrates that the opsin repertoire in Anthozoa is more diverse than in the Medusozoa, with diversity being defined as the quantity of distinct classes of opsins and subtypes within those classes. Furthermore, it is tempting to speculate that the observed differences in the extant photoreceptor repertoire may be related to the various distinct lifestyles found in cnidarians (fig. 2B).
A second measure of opsin evolution and functional diversification are changes in key functional amino acid residues (Hope et al. 1997; Imai et al. 1997; Hunt et al. 2001; Terakita 2005; Musilova et al. 2019). To assess the overall sequence diversity of the opsins occurring in Cnidaria, we used pairwise identity mapping of more than 500 opsins (including 237 cnidarian opsins). We found that at the protein level most cnidarian opsins are indeed highly diverse (supplementary fig. 3, Supplementary Material online). For example, although the ASO-Is cluster tightly and form one distinct group with a highly similar amino acid composition, the cnidopsins and ASO-IIs are subdivided into various clusters which are interspersed with several classes of opsins associated with higher animals including vertebrate-specific c-opsins (supplementary fig. 3, Supplementary Material online). Opsin amino acid composition and photosensory function are correlated and specific amino acid residues interact with the chromophore to tune peak spectral sensitivities (Terakita and Nagata 2014; Musilova et al. 2019). Therefore, in line with our results from the species-tree-aware maximum-likelihood (ML)-based GFT inference, the high levels of diversity we observe in cnidarian opsins may reflect photosensory diversity rather than being the result of synonymous gene duplications that merely created functionally identical opsin paralogs.
Interestingly, we specifically noted that opsins within the ASO-II subtypes differ in one major functional amino acid. Typically, opsins contain a highly conserved glutamate residue at position 181 (E181), which stabilizes retinal, the light-sensitive chromophore forming a so-called protonated Schiff base when bound to opsin. Light absorption triggers retinal cis-to-trans isomerization, which, in turn, results in conformational changes that reveal a cytoplasmic G-protein binding site and thereby enables the activation of signaling cascades. Free retinal is maximally sensitive to UV light but its absorption maximum is shifted toward visible light when covalently bound to opsins (Terakita 2005). However, although most ASO-IIs subtype 2.1 opsins, similarly to the cnidopsins and ASO-Is, have a glutamate [E] at the equivalent position, all members of the other two anthozoan-wide ASO-IIs subtypes (subtypes 1 and 2) lack this conserved feature (supplementary fig. 2C, Supplementary Material online). To date, E181 has been found to be conserved in all opsins (Terakita et al. 2000; Terakita and Nagata 2014) with the exception of vertebrate visual opsins where it occurs together with, or is replaced by E113 (Sakmar et al. 1989; Zhukovsky and Oprian 1989; Imai et al. 1997; Terakita 2005; Tsutsui and Shichida 2010; Gerrard et al. 2018), a vertebrate-specific feature associated with even higher fidelity visual photoreception. Interestingly, one Aiptasia ASO-II (ASO-II.4—a subtype 2.1 ASO-II) contains both E113 and E181 (supplementary fig. 2C, Supplementary Material online), suggesting that this presumed vertebrate-specific feature may also have arisen independently in some cnidarians and therefore may represent an example of convergent evolution conferring higher-fidelity photoreception (Gerrard et al. 2018). Furthermore, these specific amino acid differences are consistent with functional diversification during evolution of the ASO-II opsin group.
Novelty in the Cnidarian PL and CRY Repertoire
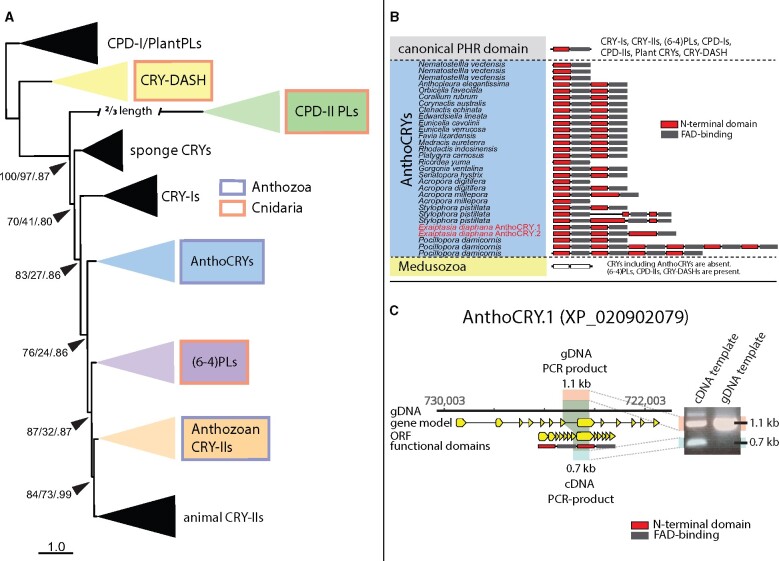
Next, we compared the repertoire of CRYs and PLs between Anthozoa and Medusozoa using phylogenomic analysis and considering the position of conserved introns. We revealed that although both possess CRY-DASH, CPD-II PLs, and (6-4) PLs, CPD-Is and CRY-Is are absent from all cnidarians (fig. 3A and supplementary fig. 5A, Supplementary Material online). CRY-IIs are present in the subphylum Anthozoa, however they are absent from the Medusozoa that are represented by 29 taxa in our analysis. We identified two distinct CRY groups in the Anthozoa. One is a sister group to “animal CRY-IIs” that we named Anthozoan CRY-IIs. Due to their phylogenetic position at the base of animal CRY-IIs we speculate that these are likely to be involved in circadian clock function (Emery et al. 1998; Lin and Todo 2005; Michael et al. 2017). However, a second, novel CRY group appears to diverge at the base of (6-4) PLs and “animal CRY-IIs” but is distinct from CRY-Is and sponge CRYs. We thus named this group Anthozoan-specific CRYs (AnthoCRYs) (fig. 3A and B). Previous, preliminary analysis of both CRY groups lead to them being classified within the animal CRY-IIs, presumably due to a lack of cnidarian taxa representation in associated phylogenies (Levy et al. 2007; Reitzel et al. 2010; Rivera et al. 2012). Our study now clarifies their phylogenetic position and proposes their name based on identity. Thus, similar to the situation for the opsin genes, the Anthozoa possess a more diverse repertoire of CRYs when compared with the Medusozoa.
Fig. 3.
A novel anthozoan-specific CRY class is characterized by tandemly duplicated PHR domains. (A) A majority rule extended consensus tree topology generated with IQTree constructed from the combined bootstrap trees showing that PLs (CPD-II, PLs, and (6-4) PLs) and CRY-DASHs are common amongst all Cnidaria. Other CRYs including CRY-II occur only in Anthozoa but are entirely absent from Medusozoa. The present phylogeny is supported by highly conserved and specific exon–intron patterns (supplementary fig. 5, Supplementary Material online) suggesting that CRY-IIs and (6-4) PLs share a common origin. A well supported but previously unidentified, anthozoan-specific CRY family resolves at the base of animal CRY-IIs and (6-4) PLs which we name Anthozoan-specific CRYs (AnthoCRYs). Support values are SH-aLRT bootstrap percentages/UFBoots bootstrap percentages/MrBayes Bayesian posterior probabilities. A full tree can be accessed through supplementary file 5, Supplementary Material online. (B) CRYs (except CRY-DASH) are absent in Medusozoa, whereas the Anthozoa possess a novel class of cryptochromes (AnthoCRYs), which contain unique tandem duplications including up to six copies of the PHR region comprising the N-terminal DNA-binding PL domain (also called alpha/beta domain, red) and the chromophore-binding FAD domain (gray). (C) A comparison between cDNA- and genomic DNA-derived exon-spanning amplicon sequencing confirms this PHR region duplication in Aiptasia AnthoCRY.1 (XP_020902079).
A hallmark of PLs and CRYs is the PL-homologous region (PHR region) that contains a N-terminal DNA-binding PL domain and a C-terminal FAD chromophore-binding domain (Cashmore 1999; Lin and Todo 2005; Benjdia 2012) that is involved in light harvesting. All PLs and CRYs described to date contain a single PHR region. Strikingly, however, here we reveal that AnthoCRYs contain up to six tandemly repeated PHR regions (fig. 3B). The loci encoding these duplicated regions exhibit a conserved intron-phase structure (supplementary fig. 5A, Supplementary Material online) and we confirmed this PHR region duplication independently in the sea anemone Aiptasia by PCR and sequencing (fig. 3C) of the Aiptasia AnthoCRY.1 cDNA. Such a PHR region expansion has not been described for any PL or CRY to date. This expansion occurs across all Anthozoa including nonsymbiotic and symbiotic members indicating that the tandem duplication of PHR domains in Anthozoan CRYs might serve a common purpose in this animal group. Interestingly, it appears to be absent from Nematostella.
Taken together, our broad-scale approach based on phylogenetics and computational structure–function analysis to compare the major differences of the light receptor repertoire between Anthozoa and Medusozoa, as well as key features of the anthozoan-specific light sensors support the hypothesis that the divergence of light-sensing molecules may be related to the highly distinct lifestyles found in cnidarians.
Life-Stage, Symbiotic State, and Tissue Type-Specific Opsin Expression in Aiptasia
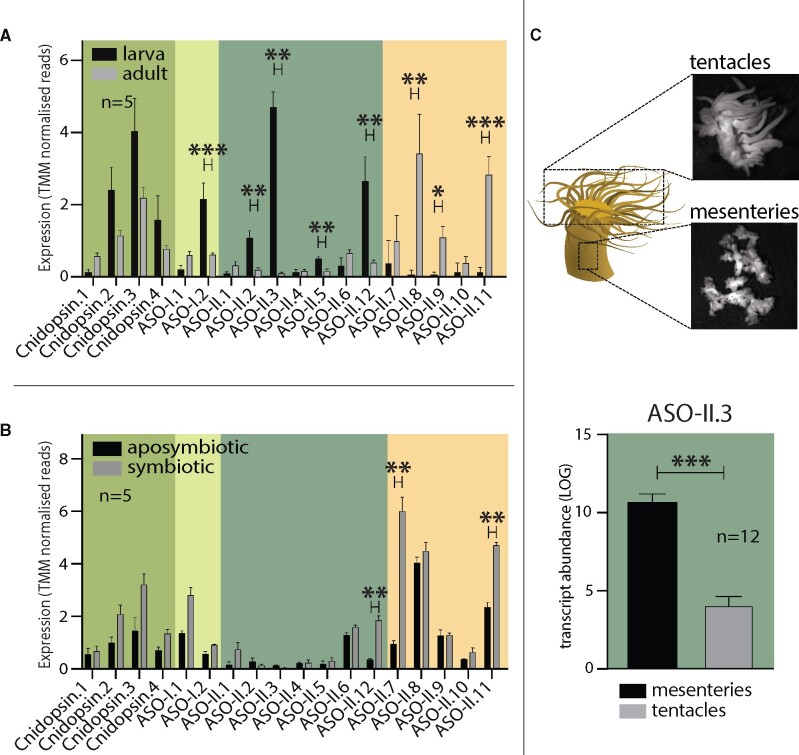
The lifestyle of reef-building corals is highly light dependent. Gamete release is synchronized by moonlight and both motile larvae as well as sessile adults are phototactic. Most strikingly though, corals satisfy most of their nutritional needs by photosymbiosis, yet this also requires them to withstand sustained high levels of solar radiation and thus UV-induced DNA damage. As a first step toward relating the coral photoreceptor repertoire to their unique lifestyle, we exploited Aiptasia, a model species for fundamental aspects of coral biology. In Aiptasia, we identified 18 distinct opsins: four cnidopsins (XP_020899757.1, XP_020913977.1, XP_020904301.1, and XP_020897723.2), two ASO-Is (XP_020902074.1 and XP_028515325.1), and 12 ASO-IIs (XP_020903100.1, AXN75743.1, XP_020897790.2, XP_028514120.1, XP_020909716.1, XP_020914799.1, XP_020906239.1, XP_020909580.1, XP_020907384.1, XP_020910007.1, XP_020893775.2, and XP_020914907.2) (see fig. 2B and supplementary file 1, Supplementary Material online). To assess whether this broad opsin repertoire is actively expressed and if so, whether certain opsins are expressed in distinct life stages, we compared opsin expression levels using publicly available Aiptasia RNA-Seq data (Baumgarten et al. 2015; Wolfowicz et al. 2016). We found that with one exception, all Aiptasia opsins are ubiquitously expressed in both larvae and adults with expression levels of three opsins elevated specifically in adults and four during larval stages suggesting the existence of opsins with larval- and adult-specific functions (fig. 4A). Likewise, opsin expression varies depending on the symbiotic state (fig. 4B). For example, one opsin from the actinarian-specific subtype 2.2 ASO-II group (ASO-II.12, dark green), and two from the subtype 1 and subtype 2.1 ASO-II group (ASO-II.7 and ASO-II.11, yellow) show significantly higher expression levels in symbiotic anemones when compared with their aposymbiotic (non-symbiotic) counterparts. This is consistent with previous findings that symbiotic association influences photo-movement in Aiptasia (Foo et al. 2020) and suggests that host perception of environmental light by opsin-mediated light sensing may change in response to symbiosis, for example to adjust the levels of sunlight exposure for optimal photosynthesis rates.
Fig. 4.
Opsin expression varies between life stages and symbiotic state in Aiptasia. (A) Bar chart comparing the expression (TMM normalized reads) of Aiptasia opsins in adults and larvae. ASO-I.2, ASO-II.2, ASO-II.3, ASO-II.5, and ASO-II.12 are significantly upregulated in larva. ASO-II.8, ASO-II.9, and ASO-II.11 are significantly upregulated in adults. (B) Bar chart comparing the expression (TMM normalized reads) of Aiptasia opsins in symbiotic and aposymbiotic adults. ASO-II.12, ASO-II.7, and ASO-II.11 are significantly upregulated in symbiotic adults. (C) Tissue-specific qPCR after Methacarn fixation of adult Aiptasia polyps reveals that ASO-II.3 is significantly upregulated in mesenteries, when compared with tentacle tissue. Data for all other opsins are shown in supplementary figure 3, Supplementary Material online. For all charts significant differences are *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001; and error bars are SEM.
Opsins have been implicated in “sensing” moonlight to synchronize gamete release in corals (Gorbunov and Falkowski 2002; Kaniewska et al. 2015). Specifically, it is predicted that physiologically relevant blue shifts in the irradiance spectrum measurable during twilight on several days before and after the full moon acts as a trigger for a potential opsin-mediated dichromatic visual system where readouts from a blue- and red-light-sensitive opsin are integrated to induce spawning (Sweeney et al. 2011). Accordingly, exposure for five nights to LED-based blue light has been shown to specifically induce synchronized spawning in Aiptasia (Grawunder et al. 2015). By analogy with the jellyfish C. hemisphaerica in which the opsin relevant for spawning is specifically expressed in the gonadal tissue (Artigas et al. 2018), we therefore asked whether any of the opsin genes present in Aiptasia showed a gonad-specific expression pattern (supplementary fig. 4, Supplementary Material online). By using quantitative reverse transcription PCR (RT-qPCR) analysis, we revealed that Aiptasia ASO-II.3 is indeed expressed in a tissue-specific manner and significantly upregulated in mesenteries of sexually mature adults, when compared with the tentacles (fig. 4C). The mesenteries contain muscle and digestive cells, but they are also highly enriched in gonadal tissue (Grawunder et al. 2015). Thus, Aiptasia ASO-II.3 likely represents a candidate opsin involved in spawning induction in this species.
Light-Regulated CRY Gene Expression in Cnidaria
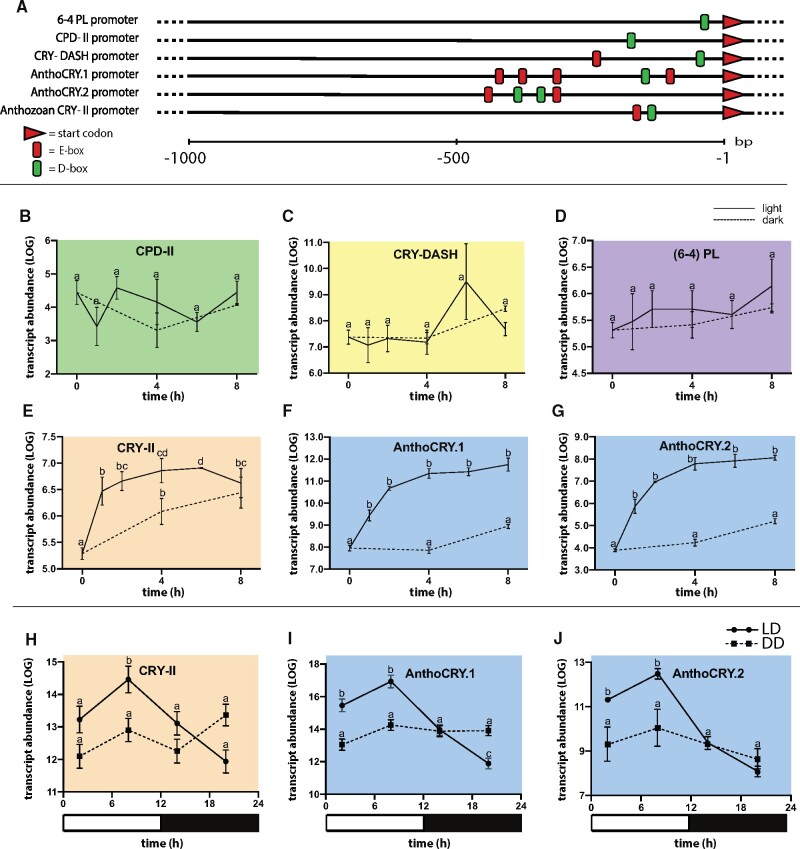
In contrast to opsins, which are typically constitutively expressed, CRY and PL expression is often regulated by light as a key mechanism to regulate the circadian clock or to upregulate DNA repair capacity in response to prolonged sunlight exposure. We have previously studied the mechanisms underlying light-driven gene expression of CRYs and PLs in vertebrates (Mracek et al. 2012; Zhao et al. 2018). We revealed a conserved role for light-induced transcription of these genes, mediated by the D-box element, an enhancer that binds to the PAR/bZIP family of transcription factors (TFs). The D-box, together with the E-box enhancer has also been associated with circadian clock regulation (Vatine et al. 2009). We therefore first tested whether D-box or E-box enhancer elements might be encountered within the promoter regions of the various Aiptasia CRY and PL genes. By scanning the genomic regions 1 kb upstream of the respective gene START codon (ATG), single D-box enhancers were identified in 6-4 PL and CPD-II. However, in the case of the AnthoCRY and CRY-II genes, we identified several D-boxes (fig. 5A), located proximally to E-box enhancers. Furthermore, consistent with the existence of functional D-box regulatory pathways in Anthozoa, we show that Aiptasia possesses four putative orthologs of the PAR/bZIP TF family (three TFs basal to hepatic leukemia factor-type PAR/bZIP TFs and one D-box binding PAR/bZIP) TF (supplementary fig. 6, Supplementary Material online) which have been shown to bind to and regulate transcription from the D-box enhancer in vertebrates (Mitsui 2001; Gachon et al. 2006; Pagano et al. 2018).
Fig. 5.
Light-induced expression profiles of Aiptasia CRYs and PLs. (A) Schematic representation of the D- and E-box distribution (green and red boxes, respectively) in Aiptasia PL and CRY promoter regions extending up to 1,000 bp upstream from the ATG start codons. (B–G) qPCR analysis of CRY and PL gene expression in response to light exposure in adult Aiptasia. Animals were adapted for 4 days to constant darkness and then exposed to light for a period of 8 h; sampling every 2 h. Control animals were kept in constant darkness. Aiptasia CPD-II, Aiptasia DASH-CRY, and Aiptasia (6-4) PL are unresponsive to light treatment. AnthoCRY.1, AnthoCRY.2, and CRY-II expression is rapidly induced by light. (H–J) qPCR analysis of CRY-II, AnthoCRY.1, and AnthoCRY.2 expression in LD-adapted Aiptasia polyps. Animals were exposed to LD (12 h:12 h) cycles and then transferred to constant darkness (DD), sampling at 2, 8, 14, and 20 h (LD) and 26, 32, 38, and 44 h (DD). Under LD conditions, we observed rhythmic expression with elevated expression, peaking at 8 h after lights on, and then decreasing during the dark period, with a trough at 8 h after lights off. In contrast, immediately upon transfer to DD conditions, rhythmic expression was absent. (B–J) Analysis of variance was performed to confirm statistically significant differences at each time point; P < 0.01 (J).
We next wished to directly investigate whether expression of these flavoprotein genes in Aiptasia is light-dependent. In Nematostella, three CRYs have been reported, two of which are upregulated in response to light (Reitzel et al. 2010). Similarly, at least two different CRYs (Cry1a [XP_001631029] and Cry1b [XP_001632849]) are expressed in a light-dependent manner in the coral Acropora millepora (Levy et al. 2007). In Aiptasia, two CRYs were identified (without accession numbers) and shown to be expressed rhythmically in the presence of a day–night cycle (Sorek et al. 2018). We therefore explored to which extent light regulates the eight different Aiptasia PL and CRY genes that we identified (two CPD_II isoforms [XP_020910442.1 and XP_020910516.1], one CRY-DASH [XP_020903321.1], two [6-4] PL isoforms [XP_020915076.1 and XP_020915067.1], one anthozoan CRY-II [XP_020904995.1], and two AnthoCRYs [XP_020902079.1 and XP_020917737.1]; see also: supplementary file 2, Supplementary Material online). With the exception of AnthoCRY.2, where mRNA levels are undetectable in Aiptasia larvae, we showed that all CRY and PL genes are generally ubiquitously expressed in both Aiptasia larvae and adults (supplementary fig. 5B, Supplementary Material online). We next adapted Aiptasia for four days to constant darkness and then exposed them for a period of 8 h to light, collecting whole animals at 2 h intervals for RT-qPCR. Our results revealed that the expression of Aiptasia PLs and CRYs was differentially affected by light exposure. Specifically, Aiptasia CPD-II, Aiptasia DASH-CRY, and Aiptasia (6-4) PL were largely unresponsive to light treatment (fig. 5A–C). In contrast, AnthoCRY and CRY-II expression was rapidly induced upon exposure to light (fig. 5B–G). Thus, light-induced gene expression correlates with the presence of multiple D-boxes in proximity to E-box enhancers in the AnthoCRYs and CRY-II gene promoter regions.
A light-inducible expression pattern may be the consequence of acutely light-driven gene expression or alternatively of regulation by the circadian clock that is also likely to be synchronized during the period of light exposure. Furthermore, E-box elements are located in the promoters of the light-induced AnthoCRY and CRY-II genes and are direct regulatory targets of the circadian clock. Therefore, to distinguish between these two possibilities, we tested AnthoCRY and CRY-II expression by exposing Aiptasia to light-dark (LD) cycles and then transferring animals to constant darkness (DD). Clock regulation would be revealed by the appearance of rhythmic expression under a LD cycle that would persist following transfer to constant darkness. Therefore, samples were prepared at 6-h intervals during either exposure to a LD cycle or immediately following transfer from LD to DD conditions and then CRY gene expression was assayed by qPCR. Under LD conditions, we observed rhythmic expression with elevated expression during the light period, peaking at 8 h after lights on, and then decreasing during the dark period, with a trough at 8 h after lights off (fig. 5H–J). This pattern is consistent with the induced expression of AnthoCRYs and CRY-II observed following acute exposure to light (fig. 5E–G). In contrast, immediately upon transfer to DD conditions, rhythmic expression was absent, showing that changes in CRY gene expression are indeed lightdriven, rather than clockdriven (fig. 5H–J).
Discussion
Reef-building corals and sea anemones represent critically important members of the ecosystems in shallow, oligotrophic, tropical oceans and their physiology is dominated by light. They exploit sunlight and moonlight to regulate their sexual reproduction, phototaxis and photosymbiosis. Furthermore, their exposure to sustained high levels of sunlight puts them at particular risk from elevated levels of DNA damage. In order to explore the molecular mechanisms linking light with anthozoan biology, we present an in-depth and detailed phylogenetic analysis of two major light-sensing protein groups: the opsins and the CRY/PL flavoproteins. Within the broader context of the ancient metazoan phylum Cnidaria, we reveal that the Anthozoa have a substantially expanded and diversified photoreceptor repertoire possessing more photoreceptor classes than their Medusozoa sister group. This striking observation raises several fundamental questions concerning how this expanded photoreceptor capacity may be linked with adaptation to their challenging, shallow water environments.
The Origins of Opsin Diversification in Cnidaria
The last common ancestor of the Cnidaria and Bilateria possessed at least three classes of distinct opsins giving rise to the cnidopsins, ASO-Is, and ASO-IIs, yet the Medusozoa only retain the cnidopsins. The extant Anthozoa on the other hand possess multiple ASO-Is and substantially expanded and diversified the ASO-IIs. The ASO-I is the most ancient opsin class in the metazoan lineage and represents phylogenetically and, based on protein sequence, a coherent group, but to date its function remains mostly unknown. However, a single ASO-I that was functionally studied to date was shown to activate G-protein signaling in response to light (Mason et al. 2012). Interestingly, the ASO-IIs are much more diverse at the sequence level and share a common ancestry with the tetraopsins and r-opsins as well as the cnidopsins, xenopsins, ctenopsins, and the well-studied c-opsins responsible for visual photoreception in vertebrates. This suggests that an urmetazoan animal possessed an ancestral but now extinct opsin that during its early stages of evolution gave rise to multiple opsin classes. Indeed, the GeneRax-based analysis of the GFTs inference analysis suggests that in Cnidaria, at least eight opsins genes were present before the divergence of Medusozoa and Anthozoa (supplementary file 7, Supplementary Material online). Frequent lineage-specific gains and losses then shaped the broad repertoire of light-sensing mechanisms and opsin classes that we see to date (Ramirez et al. 2016; Vöcking et al. 2017; Picciani et al. 2018; this study). This capacity for diversity is still reflected by the novel ASO-II subtype 2.2 that is restricted to anthozoan anemones. Interestingly, if cnidopsins are indeed early xenopsins as suggested by our study and also Ramirez et al. (2016) and if ASO-IIs are indeed sister to the c-opsins as suggested here, by Ramirez et al. (2016) and Vöcking et al. (2017) the cnidarians may be the only animals where xenopsins and c-opsins (or at least their direct cousins) co-occur.
In accordance with the notion that ASO-IIs play an important role in the adaptation of Anthozoa to their environments, we find that the highly conserved E181 amino acid residue is absent in all ASO-IIs subtypes except subtype 2.2. That suggests that this key position has been modified, possibly to shift the ASO-IIs wavelength specificity more toward blue light (shorter wavelengths) absorption, which may be a specific adaptation to aquatic marine environments where the penetration of longer wavelengths is reduced with increasing water depth (Cunningham et al. 2013; Renema 2018). Indeed, by using computational modeling based on a vertebrate opsin crystal structure, it was shown that the loss of E181 causes an absorption shift of more than 100 nm toward blue light (Sekharan et al. 2010). Nevertheless, studies in other animals show that opsins are surprisingly polymodal and can also act as non-light sensory receptors. In the fruit fly Drosophila melanogaster, for example, opsins can act light-independently in temperature sensation and hearing (Leung and Montell 2017). Furthermore, in mouse and humans, rhodopsins play a role in sperm thermotaxis modulating temperature-induced movement of spermatozoa. Moreover, this function can be chemically inhibited using opsin inhibitors and is also defective in rhodopsin-knockout mice (Pérez-Cerezales et al. 2015). In the future, a functional analysis of the wavelength specificities and potential light-independent functions of anthozoan opsins will provide fundamental new insight into the ability of non-bilaterian animals to exploit various combinations of lighting and sensory cues.
Adaptations to Light-Induced Sexual Reproduction
Coral sexual reproduction is a vital process for species viability. It is key for genetic diversity, dispersal by motile larvae and affects the abundance of juvenile corals to replenish aging coral communities. However, for sessile Anthozoa such as reef-building corals, achieving efficient fertilization rates is challenging because it occurs effectively only a few hours after gamete release by the parental colonies and gametes are easily diluted within the open space of the ocean. Accordingly, corals have evolved a precise spawning synchrony within populations integrating various environmental cues including temperature and solar irradiance to set the exact month, lunar cycles to set the exact night, and circadian light cues to set the exact hour (see Levy et al. 2007; Sweeney et al. 2011; Rosenberg et al. 2017; Shlesinger and Loya 2019, and references therein). In this context, it is worth noting that Aiptasia recapitulates key aspects of light-induced synchronized spawning observed in corals. Under laboratory conditions, delivery of LED-based blue light for five consecutive nights, simulating full moon, triggers gamete release peaking 9 -10 days after the last exposure to blue light. This effect is wavelength-specific as it only works effectively with light at 400- 460 nm, whereas white light is ineffective (Grawunder et al. 2015). Gamete release occurs ∼5.5 h after “sunset” and after release, fertilization efficiency drops from 100% to ∼25% within the first hour (Jones et al. 2018) underlining the need for precise timing of sexual reproduction within the Anthozoa.
Opsins have been implicated as light sensors to trigger spawning in corals (Sweeney et al. 2011; Reitzel et al. 2013; Kaniewska et al. 2015). Moreover, a functional relationship between a light-induced, opsin-dependent spawning mechanism has been shown in the medusozoan jellyfish C. hemisphaerica. Here, in response to blue light, neurosecretory cells of the gonadal tissue were found to release an oocyte maturation hormone triggering spawning upon dark-light transitions (Artigas et al. 2018). Thus, it is tempting to speculate that Aiptasia ASO-II.3, which is also expressed at elevated levels in gonads (fig. 3C) may serve a similar function for synchronous gamete release in this anthozoan species. Aiptasia represents a tractable model to experimentally dissect the mechanisms of light-induced sexual reproduction in Anthozoa including the integration of lunar cycles as well as circadian periodicities to trigger gamete maturation and synchronous release. A mechanistic understanding of how Anthozoa have adapted the timing of sexual reproduction to their environments, together with analyzing how environmental changes affect the spawning synchronicity within the ecosystem, is key to direct future research and conservation efforts (Sweeney et al. 2011; Shlesinger and Loya 2019).
Adaptations to Increased UV-Induced DNA Damage
Our results have also revealed that in the Anthozoa, more classes of CRY and PL genes occur when compared with the Medusozoa, suggesting expanded functional diversity. Although both Anthozoa and Medusozoa possess CRY-DASH, CPD-II PLs, and (6-4) PLs, Anthozoa have two additional CRY classes (AnthoCRYs and anthozoan CRY-IIs), which are absent from Medusozoa. Thus, strikingly, the Medusozoa completely lack CRY genes. The AnthoCRYs represent a phylogenetically well supported but previously unidentified, anthozoan-specific CRY family, which resolves at the base of animal CRY-IIs and (6-4) PLs. A hallmark of all CRY/PL proteins analyzed to date is the highly conserved structure of the PHR domain consisting of a N-terminal domain and a FAD domain. However, here we reveal that the AnthoCRYs exhibit an extensive and unprecedented PHR region expansion, which has never been observed before in any other eukaryotic or prokaryotic species. Although the structure and function of the PHR has been studied in great detail in the context of PLs, revealing its light-dependent DNA repair function, the role of the PHR region in the CRYs remains rather unclear (Benjdia 2012). Nevertheless, until this current report, only single PHR regions have been identified in PL or CRY proteins. The identification of the tandem duplication of the PHR region in Anthozoa provides some tantalizing insights as to the functional significance of this domain. It is tempting to speculate that AnthoCRYs, which are clearly distinct from vertebrate CRY-IIs and CRY-Is and sister also to (6-4) PLs, have not yet lost their light-dependent DNA repair activity and in fact have evolved in the Anthozoa to provide higher UV-damage repair capacities reflected by their domain duplications. Furthermore, the observation that both AnthoCRY.1 and AnthoCRY.2 exhibit light-inducible expression would also be consistent with these genes playing a key role in the response to intensive sunlight exposure, a threat that sessile corals likely face on a daily basis in their sunlit tropical habitats.
Adaptations of the Circadian Clock during Cnidarian Evolution
Transcriptional control of gene expression in response to light serves as a central regulatory element within the circadian clock core mechanism. This enables regular adjustment of the phase of the circadian clock to match that of the environmental day–night cycle. In non-mammalian vertebrates, D-boxes mediate light-inducible gene expression, alone or in combination with other enhancers such as the E-box (Vatine et al. 2009; Mracek et al. 2012; Zhao et al. 2018). Furthermore, in fish, D-boxes also activate transcription in response to oxidative stress and UV exposure (Pagano et al. 2018). Thus, in the majority of vertebrates D-boxes coordinate the transcription of a set of genes that includes certain clock genes as well as genes involved in the repair of UV-damaged DNA (Pagano et al. 2018; Zhao et al. 2018) to constitute a cellular response to sunlight exposure. This contrasts with the situation in mammals where D-boxes exclusively direct clock-controlled rhythms of gene expression (Bozek et al. 2009). Therefore, the discovery of an enrichment of proximally spaced E- and D-box enhancer elements in the promoters of the light-inducible AnthoCRY genes supports the view that ancestrally the D-box plays a sunlight-responsive role. Furthermore, this may also predict a function for AnthoCRYs in the complex cellular response to the damaging effects of sunlight which may involve responses to visible and UV light as well as oxidative stress.
CRYs are key regulators of the circadian clock in animals. Together with the Period proteins, they serve as negative regulators within the core transcription-translation feedback loop mechanism (Kume et al. 1999). Based on previous studies of plant and animal circadian clocks, it can be predicted that clock function is of fundamental importance for many anthozoan species. For example, the adaptation of the host cell physiology to the daily cycles of photosynthetic activity of the dinoflagellate symbionts as well as the sessile lifestyle of Anthozoa is likely to rely heavily on this endogenous timing mechanism that anticipates the course of the day–night cycle. This may well account for the conservation of CRY function in the Anthozoa. The absence of CRY in Medusozoa suggests that in these species, circadian clocks may be based upon fundamentally different mechanisms. Interestingly, canonical circadian clock genes were previously reported to be absent in Hydra and thus possibly from all Medusozoa (Chapman et al. 2010; Reitzel et al. 2013; Oren et al. 2015; Kanaya et al. 2019). As medusozoans have been shown to possess diurnal rhythmic behaviors they seem to have evolved alternative mechanisms to generate circadian rhythmicity (Garm et al. 2012; Nath et al. 2017). The mechanisms underlying these behaviors represent a fascinating topic for future investigation.
The Origins of Photoreceptor Diversification
One key difference between the Anthozoa and Medusozoa is that Medusozoa have a free-swimming medusa phase during their lifecycle, whereas Anthozoa do not. Instead, Anthozoa are typically sessile animals, which are only motile during larval stages (fig. 1A). Possibly connected with this fundamental difference is that the Anthozoa lack eyes. Thus, we speculate that the evolution of relatively sophisticated eye-like structures based on light-sensitive cilia expressing cnidopsins allows for the rapid and spatially well-defined perception of various lighting cues in certain Medusozoa. Instead, in the Anthozoa the repertoire of peripheral, nonvisual opsins expanded in order to acquire and integrate lighting information from different photic environments with less spatial precision but over longer time frames and during distinct life stages. This expansion could allow fine-tuning of animal physiology and behavior including gamete release and phototaxis as well as optimizing conditions for their photosynthetic symbionts even in the absence of eye-like structures. Another striking example of how the expansion and sequence diversification of opsins allows adaptation to specific environments has recently been elucidated in deep-sea fish. Although classically all vertebrates rely on only a single rod opsin rhodopsin 1 (RH1) for obtaining visual information in dim lighting conditions, some deep sea fish have independently expanded their single RH1 gene to generate multiple RH1-like opsins that are tuned to different wavelengths of light by modulating key functional residues (Musilova et al. 2019).
Our demonstration that 18 distinct opsins and eight distinct PLs and CRYs are expressed in Aiptasia, in either larval or adult stages, in symbiotic or aposymbiotic animals, in a tissue-specific manner and in some cases, in a light-inducible manner suggests that symbiotic anthozoans possess a remarkable, functional diversity in their photoreception mechanisms. The augmented complexity of photoreceptors in the Anthozoa is likely due to gene loss in the Medusozoa as well as to continued gene expansion and diversification within the Anthozoa and is indicative of distinct light-sensing mechanisms associated with different lifestyles. It may well be that the increased photoreceptor diversity of Anthozoa including corals and sea anemones represents an essential adaptation to their predominantly sessile lifestyle. Ultimately, complex light-sensing mechanisms may permit the integration of sun- and moonlight to regulate physiology and behavior, and to facilitate adaptation to their challenging environments: shallow, highly sunlit, tropical oceans where food is scarce and there is an enhanced risk of UV-induced DNA damage (fig. 1B). To date, only one anthozoan-specific photoreceptor has been functionally characterized. Acropora palmata Acropsin-3 that according to our analysis belongs to the ASO-Is was shown to be able to activate specific G-protein signaling upon light exposure (Mason et al. 2012) confirming that it acts, at least in part as a light receptor. Here, we have generated an essential framework to experimentally further analyze this diverse repertoire of anthozoan, non-visual photoreceptors using Aiptasia as a tractable model. A future, exhaustive functional characterization of photoreceptors to uncover the mechanisms of light sensing of cnidarians will provide profound new insight into the basic principles whereby metazoans adapt to challenging photic environments via evolution of their photoreceptor repertoires.
Materials and Methods
Aiptasia Culture and Spawning
Aiptasia stocks were cultured as described (Grawunder et al. 2015). Briefly, animals were reared from pedal lacerates for at least 6 months. For the spawning experiments, animals with a pedal disk diameter of 1 cm were separated into individual, small-sized, food-grade translucent polycarbonate tanks (GN 1/4– 100 cm height, #44 CW; Cambro, Huntington Beach, CA) filled with artificial seawater (ASW) (Coral Pro Salt; Red Sea Aquatics Ltd, Houston, TX or REEF PRO; Tropic Marin, Switzerland) at 31–34 ppt salinity at 26 °C. They were fed with Artemia salina nauplius larvae five times a week during the entire experimental period. ASW was exchanged twice per week and the tanks were cleaned using cotton tipped swabs as required.
Sampling Regimes
Circadian Rhythmicity of CRY/PL Expression
First, Aiptasia polyps were adapted for 4 days to constant darkness and then exposed to light from white fluorescent bulbs with an intensity of ∼20–25 μmol m−2 s−1 of photosynthetically active radiation (PAR), as measured with an Apogee PAR quantum meter (MQ-200; Apogee, Logan, UT) for a period of 8 h, sampling at 2-h intervals. Additionally, Aiptasia polyps were exposed to LD (12 h:12 h) cycles and then transferred to constant darkness (DD), sampling at 2, 8, 14, and 20 h (LD) and 26, 32, 38, and 44 h (DD).
Computational Methods
Identification of CRY, PL, and Opsin Photoreceptors and PAR/bZIP TF Homologs
Potential CRY, PL, and opsin sequences were recovered from Aiptasia genomic data (NCBI Bioproject PRJNA261862; Baumgarten et al. 2015) by searching for annotation keywords and BLAST search using specific query sets. For opsins we used bovine (P51490), Acropora palmata (L0ATA4), Carybdea rastonii (B6F0Y5), honeybee (B7X752), and C. hemisphaerica (A0A2I6SFS3) opsins. For PLs and CRYs queries, we used a set off previously published well-defined CRY/PL proteins, which we expanded to include known sponge and anthozoan CRYs (Brudler et al. 2003; Rivera et al. 2012). A set of known vertebrate PAR/bZIP TFs were used as query sets to identify Aiptasia PAR/bZIP homologues via BLAST searches. The longest open reading frames (ORFs) from recovered putative opsin genes were translated and aligned with the query sequences using ClustalW (GONNET, goc = 3, gec = 1.8). For opsins, we excluded sequences, which did not contain lysine K296, which is essential and indicative of chromophore binding, were excluded. For Aiptasia genes, we noticed that some of the associated gene models contained Ns (resulting in Xs in their amino acid sequences). Thus, we verify existing gene models by mapping reads from published short read RNA-seq libraries (Adult-apo: SRR1648359, SRR1648361, and SRR1648362; Adult-intermediate: SRR1648363, SRR1648365, SRR1648367, and SRR1648368; Adult-sym: SRR1648369, SRR1648370, SRR1648371, and SRR1648372; Larvae-apo: SRR1648373 and SRR1648374; Larvae-sym: SRR1648375 and SRR1648376) to these gene models using HiSAT2 (https://ccb.jhu.edu/software/hisat2/index.shtml; last accessed December 04, 2020) with default settings (Kim et al. 2015). Uniquely mapped reads were extracted using samtools 1.2 and sequences then manually curated prior to alignment and phylogenetic inference (supplementary files 1–3, Supplementary Material online).
Phylogenetic Analyses
A custom BLAST database comprising more than 70 eukaryotic genera including Nematostalla vectensis, Pocillopora, Stylophora, Orbicella, Acropora millepora, A. digitifera, Exaiptasia diaphana (Cnidaria, Anthozoa), Abylopsis tetragona, Aegina citrea, Agalma elegans, Alatina alata, Atolla vanhoeffeni, Aurelia aurita, Calvadosia cruxmelitensis, Cassiopea xamachana, Chironex fleckeri, Chrysaora fuscescens, C. hemisphaerica, Craseoa lathetica, Craspedacusta sowerbyi, Craterolophus convolvulus, Cyanea capillata, Ectopleura larynx, Haliclystus sanjuanensis, Hydractinia echinata, H. polyclina, Hydra oligactis, Hydra viridissima, Hydra vulgaris, Leucernaria quadricornis, Nanomia bijuga, Physalia physalis, Podocoryna carnea, Stomolophus meleagris, Tripedalia cystophora, Turritopsis sp. SK-2016 (Cnidaria, Medusozoa), Aplysia californica (Mollusca), Amphimedon queenslandica (Porifera), Caenorhabditis elegans (Nematoda), Drosophila melanogaster (Arthropoda), Homo sapiens, Mus musculus, Danio rerio, Xenopus laevis (all Vertebrata), Monosiga brevicollis (Choanoflagellate), Pleurobrachia bachei (Ctenophora), Saccoglossus kowalevskii (Hemichordata), Strongylocentrotus purpuratus (Echinodermata), Saccharomyces cerevisiae (Fungi), Toxoplasma gondii, Plasmodium falciparum, Perkinsus marinus, Tetrahymena thermophila (all Alveolata), and Trichoplax adherens (Placozoa) was used to assemble a CRY/PL data set for phylogenetic analysis. The majority of cnidarian sequences were obtained from published transcriptomes (Kayal et al. 2018) but manually curated and translated in KNIME using in-house workflows comprising EMBOSS getorf. All other sequences were obtained from NCBI. For opsin phylogenies existing data sets were modified by replacing Exaiptasia sequences with our newly verified Aiptasia opsin set (Vöcking et al. 2017; Artigas et al. 2018). Outgroups were defined according to Vöcking et al. (2017) comprising several GPCR receptor family members (melatonin, octopamine, serotonin and adrenergic receptors) and Trichoplax opsin-like sequences, which all belong to class α rhodopsin-like GPCRs. For the detailed analysis of the ASO-II subtypes, additional ASO-II candidates were identified from the same database used for inferring CRY/PL phylogenies using the previously identified Aiptasia ASO-IIs as query. Longest ORFs from recovered putative Aiptasia PAR/bZIP genes were translated and aligned to query sequences comprising multiple metazoan PAR/bZIPs and a CEBP (CCAAT-enhancer-binding proteins) outgroup. In all cases, sequences were aligned using ClustalW (GONNET, goc: 3, gec: 1.8). Automated trimming was performed using trimAI using default parameters (Capella-Gutierrez et al. 2009). Unstable leaves were identified and excluded using phyutilities with “-tt 100” settings (Smith and Dunn 2008). Best-fitting amino acid substitution models were determined using PROTTEST3 (-JTT -LG -DCMut -Dayhoff –WAG -G -I -F -AIC –BIC; https://github.com/ddarriba/prottest3; last accessed December 04, 2020; Darriba et al. 2011) and ModelFinder within IQTree (-m MF -msub nuclear -nt AUTO; Kalyaanamoorthy et al. 2017). ML trees were inferred using IQTree at default settings under the previously determined best-fitting amino acid substitution model (opsins: -m LG+F+R10 -bb 10000 -bnni -nt AUTO -alrt 10000 –abayes; CRY/PLs: -m LG±R6 -bb 10000 -bnni -nt AUTO -alrt 10000 –abayes; we measured branch support using the Ultrafast Bootstrap [UFBoot] algorithm with 1,000 replicates, the Shimodaira–Hasegawa approximate likelihood ratio test [SH-aLRT] and abayes [approximate transformation Bayes test] and made sure that the ultrafast bootstrap support values converged [Nguyen et al. 2015]). Bayesian inference was carried out using MrBayes 3.2.6 (lset rates = gamma ngammacat = 5; prset brlenspr = unconstrained: gammadir(1.0,0.1,1.0,1.0) aamodelpr = fixed(lg); mcmc ngen = 1100000 samplefreq = 200 printfreq = 1000 nchains = 4 temp = 0.2 savebrlens = yes; starttree = random; set seed = 518; sumt burnin = 500; sump burnin = 500; Ronquist et al. 2012). We made sure that the average standard deviation of split frequencies approached 0 and the potential scale reduction factor values approached 1 indicating that runs converged. For opsin and CRY/PL phylogenies, the support values of respectiveML and Baysian inferences were combined using Treegraph2.14.0-771 (Stöver and Müller 2010). Resulting trees were finalized using FigTree 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/; last accessed December 04, 2020) and Adobe Illustrator CC 2018. All alignments, tree files, and accession numbers are provided in nexus format (supplementary file 4–6, Supplementary Material online).
Subtype Assignment
We inferred an initial gene tree under standard statistical models of sequence evolution using RAxML-NG (v. 0.9.0; Kozlov et al. 2019; RAxML-NG command: -model LG+F+R10 - search). We conducted 20 ML searches on distinct starting trees under the previously determined best-fitting amino acid substitution model (see above). We then corrected the best-scoring initial gene tree (out of the 20 ML searches), estimated the numbers of ancestral gene copies per species and inferred the position of gene duplication events using GeneRax to account for duplication and loss events (–rec-model UndatedDL –max-spr-radius 5; subst_model = LG + F + R10; Morel et al. 2020). We provided the species tree (from Kayal et al. 2018) containing data from 36 Cnidaria and one outgroup (Drosophila) as reference species tree to GeneRax. Using custom-made python scripts, we extracted the subtree for each opsin class (ASO-I, ASO-II, and cnidopsins) under the lowest common ancestor node of all Aiptasia genes belonging to a given class. We then identified potentially relevant subtypes by listing all nodes in the gene tree corresponding to duplication events and sorting them by decreasing number of species covered by their two child subtrees. We then determined all opsin classes and subtypes that existed in the last common ancestor of Anthozoa and Medusozoa and reclassified the existing opsin classes accordingly revealing novel subtypes. Lastly, we counted the gene copy numbers per species (at the leaves of the gene tree) within each assigned subtype. The counts were then transformed into a heatmap using R (3.6.3). The species tree, scripts and results are provided in supplementary file 7, Supplementary Material online.
Domain Structure Analysis, Sequence Logos, and Duplicate Domain Confirmation
The domain structures of the different CRYs and opsins were predicted using InterProScan v5.44 in Geneious R10 (Biomatters). Sequence logos were generated using Weblogo (Crooks et al. 2004). Sequencing a region spanning exon 7 and 8, which encode the C-terminal end of the FAD-binding domain of the AnthoCRY.1 PHR region 1 and the start of the N-terminal sequence of the AnthoCRY.1 PHR region 2 confirmed the presence of the PHR region tandem duplication. Here, RNA (as cDNA) and gDNA were PCR amplified using exon-specific primers (supplementary table 1, Supplementary Material online), cloned into pJET2.1, and then Sanger sequenced. An alignment of the sequenced fragments confirmed that the genomic sequence contains an intron and that the genomic and transcript sequences of AnthoCRY.1 span two individual PHR regions and that both are expressed.
Conserved Intron Structure Analysis
Reference sequences were chosen at random to represent the canonical exon–intron structure of the respective Opsin/CRY/PL types. Reference and Aiptasia opsin gene models were generated using WebScipio (Odronitz et al. 2008) and conserved introns were identified using GenePainter 2.0 (Mühlhausen et al. 2015).
Expression Quantification of PLs, CRYs, and Opsins
We analyzed expression of the CRY, PL, and opsin genes using the same published short read RNA-seq libraries that we used for gene model verification comprising data for adult and larval life stages from aposymbiotic and symbiotic states including 2–4 biological replicates per sample treatment (Lehnert et al. 2014). The ultrafast, bias-aware short read mapper Salmon (Patro et al. 2017) and in-house R scripts were used to generate a TMM normalized expression quantitation matrix across all conditions and samples. Average expression data for adult and larval Aiptaisa was used irrespective of their symbiotic state to analyze the differential developmental expression of CRYs and opsins. To compare the effect of symbiosis, the average expression in symbiotic and aposymbiotic adults was compared. Graphs were drawn and significance levels were determined using a multiple t-test in Prism 8.1.1 (GraphPad).
D-Box and E-Box Searches
Potential PAR/bZIP binding sites in the genomic region 1 kb upstream of the CRY/PL TSS were identified using MATCH 1.0 Public (http://gene-regulation.com/pub/programs.html; last accessed December 04, 2020) (binding sites for Hlf [TransFac ID: T01071] and VBP [TransFac ID: T00881] were considered D-boxes). The identified potential sites were aligned to the canonical D-box elements identified in zebrafish using ClustalW (GONNET, goc: 3, gec: 1.8). Statistically overrepresented E-box motifs were identified in the same genomic regions using Clover (Frith 2004) and a library of 9 TF binding motifs including several know (MITF and USF TF binding motifs) and one manually generated E-box motif. An ∼6-Mb Aiptasia genomic scaffold (GenBank Accession NW_018384103.1) was used as a background sequence.
Gene Expression
RNA Extraction and qPCR
For circadian rhythmicity qPCR analysis, polyps were macerated in Trizol at a concentration of 500 mg per ml, snap frozen in liquid nitrogen, and stored at −80 °C. Total RNA was extracted as described, but replacing phenol–chloroform with Trizol (Hambleton et al. 2019). For determination of opsin expression levels in mesentery and tentacle tissues, a number of adult male Aiptasia were anesthetized in 7% MgCl2 (w/v) in ASW (1:1) for 1 h and then transferred into Methacarn fixative (6:3:1 methanol:chloroform:acetic acid). Following two Methacarn changes in the first hour, the samples were then incubated for 48 h at RT. Individual polyps were then transferred into PBS and dissected to separate tentacle and mesentery tissues. Total RNA was extracted from dissected tissue samples as described in Hambleton et al. (2019) replacing phenol–chloroform with Trizol. In all cases, cDNA was synthesized with 1 µg of total RNA per sample using a ReadyScript cDNA Synthesis Mix (Sigma-Aldrich). Primers for qPCR were determined using NCBI Primer BLAST (standard settings optimized for 100-bp exon-spanning amplicons) or designed manually using the same exon-spanning rules when NCBI gene models were not available (for qPCR primers see: supplementary table 1, Supplementary Material online). All qPCRs were run on a BioSystems StepOne Real-Time PCR System (ThermoFisher) using a Luna Universal qPCR Master Mix (NEB) at the fast setting following the manufacturer’s instructions to determine dCT levels in triplicate. Genes encoding 40S Ribosomal Proteins S7 and L11 (RPS7 and RPL11) and actin were chosen as comparison/baseline genes. Primers were validated in triplicate by amplicon sequencing of qPCR products. Melt curves were generated after each run confirming only a single product per reaction. Amplification efficiencies of each primer pair were determined through dilution series. Results were analyzed according to a standard protocol (https://matzlab.weebly.com/data--code.html; last accessed December 04, 2020) using in-house KNIME (https://www.knime.com/; last accessed December 04, 2020) workflows comprising an R integration of the Bayesian analysis pipeline MCMC.qPCR (Matz et al. 2013).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) (Emmy Noether Program Grant GU 1128/3–1 to A.G.), the H2020 European Research Council (ERC Consolidator Grant 724715 to A.G.), the Helmholtz Association (BioInterfaces in Technology and Medicine [BIFTM] program to N.S.F.), and the Klaus Tschira Foundation (to B.M and A.S.). We thank Steffen Lemke for critical feedback on our manuscript.
Data Availability
All data underlying this article are either accessible in the public domain, available in a repository and can be accessed using a unique identifier or available in the article and in its online supplementary material.
References
- Artigas GQ, Lapébie P, Leclère L, Takeda N, Deguchi R, Jékely G, Momose T, Houliston E.. 2018. A gonad-expressed opsin mediates light-induced spawning in the jellyfish Clytia. Elife 7:e29555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten S, Simakov O, Esherick LY, Liew YJ, Lehnert EM, Michell CT, Li Y, Hambleton EA, Guse A, Oates ME, et al. 2015. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc Natl Acad Sci U S A. 112(38):11893–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjdia A. 2012. DNA photolyases and SP lyase: structure and mechanism of light-dependent and independent DNA lyases. Curr Opin Struct Biol. 22(6):711–720. [DOI] [PubMed] [Google Scholar]
- Bielecki J, Zaharoff AK, Leung NY, Garm A, Oakley TH.. 2014. Ocular and extraocular expression of opsins in the Rhopalium of Tripedalia cystophora (Cnidaria: Cubozoa). PLoS One 9(6):e98870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozek K, Relógio A, Kielbasa SM, Heine M, Dame C, Kramer A, Herzel H.. 2009. Regulation of clock-controlled genes in mammals. PLoS One 4(3):e4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudler R, Hitomi K, Daiyasu H, Toh H, Kucho K-I, Ishiura M, Kanehisa M, Roberts VA, Todo T, Tainer JA, et al. 2003. Identification of a new cryptochrome class. Mol Cell 11(1):59–67. [DOI] [PubMed] [Google Scholar]
- Bucher M, Wolfowicz I, Voss PA, Hambleton EA, Guse A.. 2016. Development and symbiosis establishment in the cnidarian endosymbiosis model Aiptasia sp. Sci Rep. 6(1):19867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore AR. 1999. Cryptochromes: blue light receptors for plants and animals. Science 284(5415):760–765. [DOI] [PubMed] [Google Scholar]
- Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D, et al. 2010. The dynamic genome of Hydra. Nature 464(7288):592–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen L-O, van der Horst GTJ, Batschauer A, Ahmad M.. 2011. The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol. 62(1):335–364. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE.. 2004. WebLogo: a sequence logo generator. Genome Res. 14(6):1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham A, Ramage L, McKee D.. 2013. Relationships between inherent optical properties and the depth of penetration of solar radiation in optically complex coastal waters. J Geophys Res Oceans 118(5):2310–2317. [Google Scholar]
- Dalesio NM, Barreto Ortiz SF, Pluznick JL, Berkowitz DE.. 2018. Olfactory, taste, and photo sensory receptors in non-sensory organs: it just makes sense. Front Physiol. 9:1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27(8):1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P, So WV, Kaneko M, Hall JC, Rosbash M.. 1998. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95(5):669–679. [DOI] [PubMed] [Google Scholar]
- Feuda R, Hamilton SC, McInerney JO, Pisani D.. 2012. Metazoan opsin evolution reveals a simple route to animal vision. Proc Natl Acad Sci U S A. 109(46):18868–18872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R, O’Leary RA, Low-Choy S, Mengersen K, Knowlton N, Brainard RE, Caley MJ.. 2015. Species richness on coral reefs and the pursuit of convergent global estimates. Curr Biol. 25(4):500–505. [DOI] [PubMed] [Google Scholar]
- Foo SA, Liddell L, Grossman A, Caldeira K.. 2020. Photo-movement in the sea anemone Aiptasia influenced by light quality and symbiotic association. Coral Reefs 39(1):47–54. [Google Scholar]
- Frith MC. 2004. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 32(4):1372–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F, Olela FF, Schaad O, Descombes P, Schibler U.. 2006. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 4(1):25–36. [DOI] [PubMed] [Google Scholar]
- Garm A, Bielecki J, Petie R, Nilsson D-E.. 2012. Opposite patterns of diurnal activity in the box jellyfish Tripedalia cystophora and Copula sivickisi. Biol Bull. 222(1):35–45. [DOI] [PubMed] [Google Scholar]
- Gerrard E, Mutt E, Nagata T, Koyanagi M, Flock T, Lesca E, Schertler GFX, Terakita A, Deupi X, Lucas RJ.. 2018. Convergent evolution of tertiary structure in rhodopsin visual proteins from vertebrates and box jellyfish. Proc Natl Acad Sci U S A. 115(24):6201–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindt YM, Messyasz A, Jumbo PI.. 2015. Binding of substrate locks the electrochemistry of CRY-DASH into DNA repair. Biochemistry 54(18):2802–2805. [DOI] [PubMed] [Google Scholar]
- Gorbunov MY, Falkowski PG.. 2002. Photoreceptors in the cnidarian hosts allow symbiotic corals to sense blue moonlight. Limnol Oceanogr. 47(1):309–315. [Google Scholar]
- Grawunder D, Hambleton EA, Bucher M, Wolfowicz I, Bechtoldt N, Guse A.. 2015. Induction of gametogenesis in the cnidarian endosymbiosis model Aiptasia sp. Sci Rep. 5(1):15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambleton EA, Jones VAS, Maegele I, Kvaskoff D, Sachsenheimer T, Guse A.. 2019. Sterol transfer by atypical cholesterol-binding NPC2 proteins in coral-algal symbiosis. Elife 8:e43923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope AJ, Partridge JC, Dulai KS, Hunt DM.. 1997. Mechanisms of wavelength tuning in the rod opsins of deep-sea fishes. Proc R Soc Lond B 264(1379):155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt DM, Dulai KS, Partridge JC, Cottrill P, Bowmaker JK.. 2001. The molecular basis for spectral tuning of rod visual pigments in deep-sea fish. J Exp Biol. 204:3333–3344. [DOI] [PubMed] [Google Scholar]
- ICZN. 2017. Opinion 2404: Dysactis pallida Agassiz in Verrill, 1864 (currently Aiptasia pallida; Cnidaria, Anthozoa, Hexacorallia, Actiniaria): precedence over Aiptasia diaphana (Rapp, 1829), Aiptasia tagetes (Duchassaing de Fombressin & Michelotti, 1864), Aiptasia mimosa (Duchassaing de Fombressin & Michelotti, 1864) and Aiptasia inula (Duchassaing de Fombressin & Michelotti, 1864) not approved. Bull Zool Nomencl. 74. [Google Scholar]
- Imai H, Kojima D, Oura T, Tachibanaki S, Terakita A, Shichida Y.. 1997. Determinants of visual pigment absorbance: identification of the retinylidene Schiff’s base counterion in bovine rhodopsin. Proc Natl Acad Sci U S A. 94(6):2322–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouskovec J, Gavelis GS, Burki F, Dinh D, Bachvaroff TR, Gornik SG, Bright KJ, Imanian B, Strom SL, Delwiche CF, et al. 2017. Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proc Natl Acad Sci U S A. 114(2):E171–E180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones VAS, Bucher M, Hambleton EA, Guse A.. 2018. Microinjection to deliver protein, mRNA, and DNA into zygotes of the cnidarian endosymbiosis model Aiptasia sp. Sci Rep. 8(1):679–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Haeseler von A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya HJ, Kobayakawa Y, Itoh TQ.. 2019. Hydra vulgaris exhibits day–night variation in behavior and gene expression levels. Zool Lett. 5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniewska P, Alon S, Karako-Lampert S, Hoegh-Guldberg O, Levy O.. 2015. Signaling cascades and the importance of moonlight in coral broadcast mass spawning. Elife 4:e09991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayal E, Bentlage B, Pankey MS, Ohdera AH, Medina M, Plachetzki DC, Collins AG, Ryan JF.. 2018. Phylogenomics provides a robust topology of the major cnidarian lineages and insights on the origins of key organismal traits. BMC Evol Biol. 18:68. [Google Scholar]
- Kim D, Langmead B, Salzberg SL.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 12(4):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A.. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35(21):4453–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM.. 1999. Reppert SM. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98(2):193–205. [DOI] [PubMed] [Google Scholar]
- LaJeunesse TC, Parkinson JE, Gabrielson PW, Jeong HJ, Reimer JD, Voolstra CR, Santos SR.. 2018. Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr Biol. 28(16):2570–2580.e2576. [DOI] [PubMed] [Google Scholar]
- Lehnert EM, Mouchka ME, Burriesci MS, Gallo ND, Schwarz JA, Pringle JR.. 2014. Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. G3 (Bethesda) 4:277–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung NY, Montell C.. 2017. Unconventional roles of opsins. Annu Rev Cell Dev Biol. 33(1):241–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy O, Appelbaum L, Leggat W, Gothlif Y, Hayward DC, Miller DJ, Hoegh-Guldberg O.. 2007. Light-responsive cryptochromes from a simple multicellular animal, the coral Acropora millepora. Science 318(5849):467–470. [DOI] [PubMed] [Google Scholar]
- Levy O, Kaniewska P, Alon S, Eisenberg E, Karako-Lampert S, Bay LK, Reef R, Rodriguez-Lanetty M, Miller DJ, Hoegh-Guldberg O.. 2011. Complex diel cycles of gene expression in coral-algal symbiosis. Science 331(6014):175. [DOI] [PubMed] [Google Scholar]
- Lin C, Todo T.. 2005. The cryptochromes. Genome Biol. 6(5):220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas-Lledo JI, Lynch M.. 2009. Evolution of mutation rates: phylogenomic analysis of the photolyase/cryptochrome family. Mol Biol Evol. 26(5):1143–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra K, Kim ST, Batschauer A, Dawut L, Sancar A.. 1995. Putative blue-light photoreceptors from Arabidopsis thaliana and Sinapis alba with a high degree of sequence homology to DNA photolyase contain the two photolyase cofactors but lack DNA repair activity. Biochemistry 34(20):6892–6899. [DOI] [PubMed] [Google Scholar]
- Mason B, Schmale M, Gibbs P, Miller MW, Wang Q, Levay K, Shestopalov V, Slepak VZ.. 2012. Evidence for multiple phototransduction pathways in a reef-building coral. PLoS One 7(12):e50371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews JL, Crowder CM, Oakley CA, Lutz A, Roessner U, Meyer E, Grossman AR, Weis VM, Davy SK.. 2017. Optimal nutrient exchange and immune responses operate in partner specificity in the cnidarian-dinoflagellate symbiosis. Proc Natl Acad Sci U S A. 114(50):13194–13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz MV, Wright RM, Scott JG.. 2013. No control genes required: Bayesian analysis of qRT-PCR data. PLoS One 8(8):e71448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael AK, Fribourgh JL, Van Gelder RN, Partch CL.. 2017. Animal cryptochromes: divergent roles in light perception, circadian timekeeping and beyond. Photochem Photobiol. 93(1):128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui S. 2001. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 15(8):995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel B, Kozlov AM, Stamatakis A, Szöllősi GJ.. 2020. GeneRax: a tool for species-tree-aware maximum likelihood-based gene family tree inference under gene duplication, transfer, and loss. Mol Biol Evol. 37(9):2763–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mracek P, Santoriello C, Idda ML, Pagano C, Ben-Moshe Z, Gothilf Y, Vallone D, Foulkes NS.. 2012. Regulation of per and cry genes reveals a central role for the D-box enhancer in light-dependent gene expression. PLoS One 7(12):e51278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlhausen S, Hellkamp M, Kollmar M.. 2015. GenePainter v. 2.0 resolves the taxonomic distribution of intron positions. Bioinformatics 31(8):1302–1304. [DOI] [PubMed] [Google Scholar]
- Musilova Z, Cortesi F, Matschiner M, Davies WIL, Patel JS, Stieb SM, de Busserolles F, Malmstrøm M, Tørresen OK, Brown CJ, et al. 2019. Vision using multiple distinct rod opsins in deep-sea fishes. Science 364(6440):588–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath RD, Bedbrook CN, Abrams MJ, Basinger T, Bois JS, Prober DA, Sternberg PW, Gradinaru V, Goentoro L.. 2017. The jellyfish Cassiopea exhibits a sleep-like state. Curr Biol. 27(19):2984–2990.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer E-F, Poole AZ, Neubauer P, Detournay O, Tan K, Davy SK, Weis VM.. 2017. A diverse host thrombospondin-type-1 repeat protein repertoire promotes symbiont colonization during establishment of cnidarian-dinoflagellate symbiosis. Elife 6:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, Haeseler von A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley TH, Speiser DI.. 2015. How complexity originates: the evolution of animal eyes. Annu Rev Ecol Evol Syst. 46(1):237–260. [Google Scholar]
- Odronitz F, Pillmann H, Keller O, Waack S, Kollmar M.. 2008. WebScipio: an online tool for the determination of gene structures using protein sequences. BMC Genomics. 9(1):422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren M, Tarrant AM, Alon S, Simon-Blecher N, Elbaz I, Appelbaum L, Levy O.. 2015. Profiling molecular and behavioral circadian rhythms in the non-symbiotic sea anemone Nematostella vectensis. Sci Rep. 5(1):11418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano C, Siauciunaite R, Idda ML, Ruggiero G, Ceinos RM, Pagano M, Frigato E, Bertolucci C, Foulkes NS, Vallone D.. 2018. Evolution shapes the responsiveness of the D-box enhancer element to light and reactive oxygen species in vertebrates. Sci Rep. 8(1):13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C.. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 14(4):417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cerezales S, Boryshpolets S, Afanzar O, Brandis A, Nevo R, Kiss V, Eisenbach M.. 2015. Involvement of opsins in mammalian sperm thermotaxis. Sci Rep. 5(1):R304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picciani N, Kerlin JR, Sierra N, Swafford AJM, Ramirez MD, Roberts NG, Cannon JT, Daly M, Oakley TH.. 2018. Prolific origination of eyes in cnidaria with co-option of non-visual opsins. Curr Biol. 28(15):2413–2419.e4. [DOI] [PubMed] [Google Scholar]
- Plachetzki DC, Degnan BM, Oakley TH.. 2007. The origins of novel protein interactions during animal opsin evolution. PLoS One 2(10):e1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachetzki DC, Fong CR, Oakley TH.. 2010. The evolution of phototransduction from an ancestral cyclic nucleotide gated pathway. Proc R Soc B 277(1690):1963–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachetzki DC, Fong CR, Oakley TH.. 2012. Cnidocyte discharge is regulated by light and opsin-mediated phototransduction. BMC Biol. 10(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez MD, Pairett AN, Pankey MS, Serb JM, Speiser DI, Swafford AJ, Oakley TH.. 2016. The last common ancestor of most bilaterian animals possessed at least nine opsins. Genome Biol Evol. 8(12):3640–19693652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Behrendt L, Tarrant AM.. 2010. Light entrained rhythmic gene expression in the sea anemone Nematostella vectensis: the evolution of the animal circadian clock. PLoS One 5(9):e12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Tarrant AM, Levy O.. 2013. Circadian clocks in the cnidaria: environmental entrainment, molecular regulation, and organismal outputs. Integr Comp Biol. 53(1):118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renema W. 2018. Terrestrial influence as a key driver of spatial variability in large benthic foraminiferal assemblage composition in the Central Indo-Pacific. Earth-Sci Rev. 177:514–544. [Google Scholar]
- Rivera AS, Ozturk N, Fahey B, Plachetzki DC, Degnan BM, Sancar A, Oakley TH.. 2012. Blue-light-receptive cryptochrome is expressed in a sponge eye lacking neurons and opsin. J Exp Biol. 215(8):1278–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP.. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg Y, Doniger T, Harii S, Sinniger F, Levy O.. 2017. Canonical and cellular pathways timing gamete release in Acropora digitifera, Okinawa, Japan. Mol Ecol. 26(10):2698–2710. [DOI] [PubMed] [Google Scholar]
- Sakmar TP, Franke RR, Khorana HG.. 1989. Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc Natl Acad Sci U S A. 86(21):8309–8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A. 2003. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 103(6):2203–2237. [DOI] [PubMed] [Google Scholar]
- Sekharan S, Altun A, Morokuma K.. 2010. Photochemistry of visual pigment in a G q protein-coupled receptor (GPCR)-insights from structural and spectral tuning studies on squid rhodopsin. Chem Eur J. 16(6):1744–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, et al. 2000. Interacting molecular loops in the mammalian circadian clock. Science 288(5468):1013–1019. [DOI] [PubMed] [Google Scholar]
- Shlesinger T, Loya Y.. 2019. Breakdown in spawning synchrony: a silent threat to coral persistence. Science 365(6457):1002–1007. [DOI] [PubMed] [Google Scholar]
- Smith SA, Dunn CW.. 2008. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics 24(5):715–716. [DOI] [PubMed] [Google Scholar]
- Sorek M, Schnytzer Y, Ben-Asher HW, Caspi VC, Chen C-S, Miller DJ, Levy O.. 2018. Setting the pace: host rhythmic behavior and gene expression patterns in the facultatively symbiotic cnidarian Aiptasia are determined largely by Symbiodinium. Microbiome 6:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC.. 1998. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95(5):681–692. [DOI] [PubMed] [Google Scholar]
- Stöver BC, Müller KF.. 2010. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinf. 11(1):275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, Schmid V, Gehring WJ.. 2008. Evolution and functional diversity of jellyfish opsins. Curr Biol. 18(1):51–55. [DOI] [PubMed] [Google Scholar]
- Sweeney AM, Boch CA, Johnsen S, Morse DE.. 2011. Twilight spectral dynamics and the coral reef invertebrate spawning response. J Exp Biol. 214(5):770–777. [DOI] [PubMed] [Google Scholar]
- Terakita A. 2005. The opsins. Genome Biol. 6(3):213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakita A, Nagata T.. 2014. Functional properties of opsins and their contribution to light-sensing physiology. Zool Sci. 31(10):653–659. [DOI] [PubMed] [Google Scholar]
- Terakita A, Yamashita T, Shichida Y.. 2000. Highly conserved glutamic acid in the extracellular IV-V loop in rhodopsins acts as the counterion in retinochrome, a member of the rhodopsin family. Proc Natl Acad Sci U S A. 97(26):14263–14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CL, Sancar A.. 2002. Photolyase/cryptochrome blue-light photoreceptors use photon energy to repair DNA and reset the circadian clock. Oncogene 21(58):9043–9056. [DOI] [PubMed] [Google Scholar]
- Tolleter D, Seneca FO, DeNofrio JC, Krediet CJ, Palumbi SR, Pringle JR, Grossman AR.. 2013. Coral bleaching independent of photosynthetic activity. Curr Biol. 23(18):1782–1786. [DOI] [PubMed] [Google Scholar]
- Tsutsui K, Shichida Y.. 2010. Multiple functions of Schiff base counterion in rhodopsins. Photochem Photobiol Sci. 9(11):1426–1434. [DOI] [PubMed] [Google Scholar]
- Vatine G, Vallone D, Appelbaum L, Mracek P, Ben-Moshe Z, Lahiri K, Gothilf Y, Foulkes NS.. 2009. Light directs zebrafish period2 expression via conserved D and E boxes. PLoS Biol. 7(10):e1000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vöcking O, Kourtesis I, Tumu SC, Hausen H.. 2017. Co-expression of xenopsin and rhabdomeric opsin in photoreceptors bearing microvilli and cilia. Elife 6:e23435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis VM, Davy SK, Hoegh-Guldberg O, Rodriguez-Lanetty M, Pringle JR.. 2008. Cell biology in model systems as the key to understanding corals. Trends Ecol Evol. 23(7):369–376. [DOI] [PubMed] [Google Scholar]
- Wolfowicz I, Baumgarten S, Voss PA, Hambleton EA, Voolstra CR, Hatta M, Guse A.. 2016. Aiptasia sp. larvae as a model to reveal mechanisms of symbiont selection in cnidarians. Sci Rep. 6(1):32366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama H, Mizutani R.. 2014. Structural biology of DNA (6-4) photoproducts formed by ultraviolet radiation and interactions with their binding proteins. IJMS 15(11):20321–20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata F, Goetz FE, Smith SA, Howison M, Siebert S, Church SH, Sanders SM, Ames CL, McFadden CS, France SC, et al. 2015. Phylogenomic analyses support traditional relationships within cnidaria. PLoS One 10(10):e0139068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Di Mauro G, Lungu-Mitea S, Negrini P, Guarino AM, Frigato E, Braunbeck T, Ma H, Lamparter T, Vallone D, et al. 2018. Modulation of DNA repair systems in blind cavefish during evolution in constant darkness. Curr Biol. 28(20):3229–3243.e4. [DOI] [PubMed] [Google Scholar]
- Zhukovsky E, Oprian D.. 1989. Effect of carboxylic acid side chains on the absorption maximum of visual pigments. Science 246(4932):928–930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data underlying this article are either accessible in the public domain, available in a repository and can be accessed using a unique identifier or available in the article and in its online supplementary material.