Abstract
Although centromeres have conserved function, centromere-specific histone H3 (CenH3) and centromeric DNA evolve rapidly. The centromere drive model explains this phenomenon as a consequence of the conflict between fast-evolving DNA and CenH3, suggesting asymmetry in female meiosis as a crucial factor. We characterized evolution of the CenH3 protein in three closely related, polyploid mitotic parthenogenetic species of the Meloidogyne incognita group, and in the distantly related meiotic parthenogen Meloidogyne hapla. We identified duplication of the CenH3 gene in a putative sexual ancestral Meloidogyne. We found that one CenH3 (αCenH3) remained conserved in all extant species, including in distant Meloidogyne hapla, whereas the other evolved rapidly and under positive selection into four different CenH3 variants. This pattern of CenH3 evolution in Meloidogyne species suggests the subspecialization of CenH3s in ancestral sexual species. Immunofluorescence performed on mitotic Meloidogyne incognita revealed a dominant role of αCenH3 on its centromere, whereas the other CenH3s have lost their function in mitosis. The observed αCenH3 chromosome distribution disclosed cluster-like centromeric organization. The ChIP-Seq analysis revealed that in M. incognita αCenH3-associated DNA dominantly comprises tandem repeats, composed of divergent monomers which share a completely conserved 19-bp long box. Conserved αCenH3-associated DNA is also confirmed in the related mitotic Meloidogyne incognita group species suggesting preservation of both centromere protein and DNA constituents. We hypothesize that the absence of centromere drive in mitosis might allow for CenH3 and its associated DNA to achieve an equilibrium in which they can persist for long periods of time.
Keywords: holocentromere, evolution, nematode, mitotic parthenogenesis, CenH3, gene duplication, centromeric DNA
Introduction
Centromeres are specific chromosomal regions that recruit components of the kinetochore complex to enable accurate chromosome segregation during mitosis and meiosis. High-fidelity segregation is vital for all eukaryotic organisms and centromeric defects lead to chromosome breakage and aneuploidy. Regarding centromere architecture, the majority of animal and plant species have monocentric chromosomes characterized by primary constriction with a single regional centromere. In contrast, holocentric or polycentric chromosomes, with the centromere function distributed at multiple sites along the chromosome length, were observed in some nematode, insect, and plant species (Dernburg 2001; Guerra et al. 2010; Melters et al. 2012). In total, approximately 800 species have been reported to possess holocentromeres (Cuacos et al. 2015).
In general, centromere identity is defined by epigenetic determinants. An epigenetic mark of almost all functional centromeres is the specialized histone H3 variant, CenH3, which replaces the canonical H3 in centromeric nucleosomes (Allshire and Karpen 2008). CenH3 is associated with the centromeric DNA (cenDNA) and its incorporation into centromeric nucleosomes is considered a prerequisite for the proper assembly and function of the kinetochore (Blower and Karpen 2001; Talbert et al. 2002; Steiner and Henikoff 2015). Despite the conserved role of CenH3 in maintaining centromere integrity, CenH3 demonstrates accelerated evolution which is especially pronounced at its N-terminal tail and loop 1 of the histone-fold domain (HFD) (Malik and Henikoff 2001; Talbert et al. 2004). In most diploid genomes CenH3 is encoded by a single gene, whereas multiple copies of CenH3 have been common in polyploid plants. Multiple copies usually show a different expression pattern and the efficiency of their incorporation at centromeres can vary among different tissues (Yuan et al. 2015). In addition, recent studies revealed duplication events of CenH3 genes in some diploid plants and in Drosophila species (Sanei et al. 2011; Kursel and Malik 2017). These genes encode for functional CenH3 paralogous that colocalize at centromere during cell division.
Centromere regions of monocentric chromosomes are often enriched in repetitive DNA families, mainly megabase-sized satellite DNAs (satDNAs). Centromeric repeats usually evolve rapidly, and significantly differ between closely related species (Plohl et al. 2014). Although many organisms possess a single satDNA which dominates in all centromeres (Hartley and O’Neill 2019), recent studies disclosed multiple satDNAs in centromeres, as it has been shown in the plant Pisum and related Fabeae species (Neumann et al. 2012; Ávila Robledillo et al. 2020). Extensive phylogenetic study of cenDNA candidates from 282 animal and plant species revealed astonishing diversity in their sequences which is difficult to associate with their conserved function (Melters et al. 2013). In the context of cenDNA role, evidences from studies of neocentromeres and dicentric chromosomes indicate that cenDNAs are neither necessary nor sufficient for centromere assembly (reviewed in Barra and Fachinetti 2018). On the other hand, studies on human alpha-satDNA show that, in contrast to other sequences, alpha-satDNA has property to facilitate assembly of CENPA (human CenH3) (Dumont and Fachinetti 2017). It has also been shown that existence of alpha-satDNA is necessary for de novo formation of human artificial chromosomes (HACs) (reviewed in McNulty and Sullivan 2018). In support, during the process of maturation, evolutionarily new centromeres rapidly accumulate satDNAs, and their recruitment increases segregation fidelity through binding with specific kinetochore proteins (Piras et al. 2010; Yang et al. 2018). In contrast to studies in monocentric species, the characterization of cenDNA in holocentric organisms is rare. For example, in the nematode Caenorhabditis elegans, the most studied holocentric species, centromere-specific sequences were not identified (Gassmann et al. 2012; Steiner and Henikoff 2014). Similarly, none of the identified high-copy repeats characterized in the holocentric plant Luzula showed colocalization with the centromere (Heckmann et al. 2013). On the other hand, a detailed CenH3-ChIP analysis in the holocentric plant Rhynchospora confirmed one satDNA as the underlying centromere sequence (Marques et al. 2015). However, the dilemma between exclusively epigenetic centromere definition and the role of cenDNAs in mediating centromere identity and function still remains unresolved (Talbert and Henikoff 2020).
The centromere drive model explains diversity of eukaryotic centromeres as a consequence of the conflict between rapidly evolving centromeric repeats and CenH3, suggesting asymmetry in female meiosis as the main factor responsible for rapid evolution of cenDNA and concomitant adaptive evolution of CenH3 (Malik 2009). Multiple reports in various animal and plant species with asymmetric meiosis have suggested that CenH3 evolves under positive selection to suppress the deleterious effect of rapid changes in cenDNA (Henikoff et al. 2001; Malik and Henikoff 2001; Cooper and Henikoff 2004; Malik and Bayes 2006; Hirsch et al. 2009; Talbert et al. 2009; Schueler et al. 2010; Zedek and Bureš 2012). It was also shown that centromeres with expanded cenDNA and higher amount of CenH3 (“stronger centromeres”) are more likely to segregate to the egg and thus be transmitted to offspring (Chmátal et al. 2014; Iwata-Otsubo et al. 2017). In support to this hypothesis, species with symmetric meiosis display a lower frequency of adaptive evolution of CenH3 compared with those with asymmetric meiosis (Zedek and Bureš 2016a). However, information about the possible role of centromere drive in species with holocentric chromosomes is scarce and controversial. The absence of positive selection on CenH3 in holocentric Luzula species suggests that holocentric chromosomes may suppress centromere drive (Zedek and Bureš 2016b). On the other hand, HCP-3 (CenH3) from C. elegans is rapidly evolving, even though HCP-3 was not required for oocyte meiotic divisions in this holocentric nematode (Monen et al. 2005).
In this work, we address the evolution and organization of centromere components, CenH3 and cenDNA, using the plant-parasitic nematode Meloidogyne incognita and its congeners as a model system. Among them, M. incognita, M. javanica, and M. arenaria (M. incognita group—MIG), are closely related species which have been determined as obligatory mitotic parthenogens which do not undergo meiosis and reproduce asexually (Castagnone-Sereno and Danchin 2014). On the contrary, the phylogenetically distant species M. hapla, reproduces by both meiotic parthenogenesis and cross fertilization (Castagnone-Sereno et al. 2013). In recent years, whole‐genome sequencing of MIG species and M. hapla, enabled comparative genome analyses which revealed substantial differences in their genome structure (Abad et al. 2008; Opperman et al. 2008; Blanc-Mathieu et al. 2017). Genome studies indicated that MIG species are polyploids with whole-genome duplications, whereas M. hapla is a diploid species with a small and compact genome. Interspecific hybridization has been highlighted as a critical force in the processes of polyploidization in MIG species (Lunt 2008; Blanc-Mathieu et al. 2017). Regarding the organization of the centromere, the absence of primary constriction and holocentric-like mitosis observed by classical cytological approach proclaimed Meloidogyne to be holocentric species (Triantaphyllou 1981). This classification is also supported by karyotype variability between M. incognita populations, fluctuating from 40 to 46 chromosomes (Triantaphyllou 1981). In general, the studies of centromere have been performed mostly in diploid sexual species with monocentric centromere organization. Therefore, Meloidogyne species offer the unique platform to explore the evolutionary dynamics of CenH3 and cenDNA components in exclusively asexual animal species which also possess holocentromere, the poorly investigated centromere organization. In addition, the fact that the CenH3 evolutionary trends in polyploidization have been explored very limitedly in animal species makes Meloidogyne a valuable model to address those studies.
In the present work, we characterized CenH3 proteins in the selected parthenogenetic Meloidogyne species and analyzed their evolution considering the complex species history. Our results suggested the duplication of a CenH3 gene in a common sexual ancestor of both mitotic and meiotic Meloidogyne species. We found that one CenH3 gene is preserved as nearly identical in all analyzed species, including in the distantly related M. hapla, whereas the other evolved rapidly and formed four different CenH3 variants. We further investigated the centromere DNA composition in M. incognita using chromatin immunoprecipitation (ChIP) and immunofluoreoscence (IF) techniques and unveiled the unique characteristics of the holocentromeres in an exclusively mitotic species.
Results
Meloidogyne Species Have Multiple and Divergent CenH3 Genes/Proteins
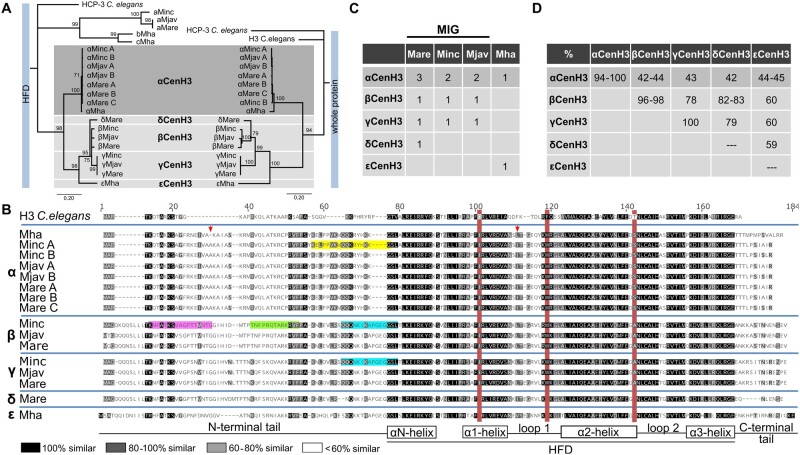
To identify CenH3 candidates in selected Meloidogyne species (three obligatory mitotic species; M. incognita, M. arenaria, and M. javanica [MIG] and facultative meiotic M. hapla) C. elegans CenH3 (HCP-3) was used as query for BLAST analysis against a protein database for each species. After elimination of truncated protein sequences (see Materials and Methods), the 21 CenH3 protein candidates with specific CenH3 features (Vermaak et al. 2002) were detected (supplementary fig. 1, Supplementary Material online). The sources of all CenH3 protein candidates together with abbreviated names are listed in supplementary table 1, Supplementary Material online. Given that N-terminal tails among CenH3 proteins were hypervariable (supplementary fig. 1, Supplementary Material online) an alignment of the more conserved histone-fold domains (HFD) was selected to estimate mutual sequence identities (supplementary table 2, Supplementary Material online) and phylogenetic relationships (fig. 1A, left). Phylogenetic tree showed the branch topology with two distant well-supported clades. Interestingly, although both groups show CenH3-specific features tree branching support their polyphyletic origin (fig. 1A, left). Among the CenH3 candidates, abcCenH3 group of sequences shows low HFD sequence identity to H3 (33–42%), whereas αβγδεCenH3 group shares considerably higher sequence identity to H3 histone (from 50% to 63%) (supplementary table 2, Supplementary Material online). The abcCenH3 group includes highly divergent CenH3 candidates divided in two subgroups (aCenH3s and bcCenH3s) which show the low mutual sequence identity in HFD (34–37%) (supplementary table 2, Supplementary Material online). One subgroup consists of three similar sequences (aCenH3; identity 85–100%) which belong to closely related MIG species whereas the other clade includes two rather divergent CenH3 candidates (bCenH3 and cCenH3, showing mutual HFD identity of 79%) from more distant M. hapla.
Fig. 1.
Identification of CenH3 proteins in Meloidogyne species (Minc, M. incognita; Mare, M. arenaria; Mjav, M. javanica; and Mha, M. hapla). (A) Phylogenetic analyses using NJ with a protein alignment of the (HFD) (left) and full length of CenH3 sequences (right) of all detected Meloidogyne CenH3s. Bootstrap values above 50 are displayed. CenH3 sequences are separated in the subgroups (a, b, c, α, β, γ, δ, and ε). (B) Amino acid alignment of CenH3 proteins separated into α, β, γ, δ, and ε subgroups. The red boxes indicate diagnostic amino acid changes in comparison to H3 from Caenorhabditis elegans. Secondary structure of histone-fold domain (HFD) is depicted below the alignment. The arrows indicate amino acids changes in αCenH3 from M. hapla in comparison to other αCenH3s. Highlighted are peptide sequence regions which were used as antigens to produce antibodies to αCenH3 (yellow), βCenH3-1 (green), βCenH3-2 (magenta), and βγCenH3 (cyan). (C) Copy number of CenH3 genes in Meloidogyne genomes. (D) Sequence identity matrix of CenH3 protein sequences.
In contrast, αβγδε CenH3 group represents a far more homogeneous CenH3 sequences with mutual HFD identity from 65% to 100%. As expected for CenH3 proteins, most of the sequence divergence was concentrated in the N-terminal tail (fig. 1B). The phylogenetic trees based on a multiple alignment of HFD domains exclusively (fig. 1A, left) and on alignment of complete α, β, γ, δ, and εCenH3 protein sequences (fig. 1A, right) showed the same branching topology with two monophyletic subclades. The α subclade consists of eight αCenH3 proteins. The multiple αCenH3s were detected in MIG species: two in M. incognita and in M. javanica, and three in M. arenaria (fig. 1C). On the other hand, only one αCenH3 was detected in M. hapla. All members of αCenH3 subclade are almost completely conserved within MIG species with only one change in the C-terminal tail (fig. 1B). Interestingly, in distant M. hapla, except at the very end of C-terminal tail which is substantial different, only two AA changes were found in comparison to MIG species. These changes include deletion of one AA within the N-tail and one AA change within Loop 1 in HFD region (fig. 1B). The other subclade comprises β, γ, and δCenH3s detected in MIG species. The β and γCenH3s were found in M. incognita and in M. javanica, whereas β, γ, and δCenH3s were found in M. arenaria. The εCenH3 was exclusively found in distant M. hapla (fig. 1C). Detected intragroup identity ranges from 96% to 98% for βCenH3s to 100% for γCenH3s (fig. 1D and supplementary table 3, Supplementary Material online). Concerning intergroup identity, αCenH3s share ∼43% identity with βγδεCenH3 sequence group, whereas identities between the members of βγδε CenH3 group were considerably higher, ranging from 59% for δCenH3 and εCenH3 to ∼83% for βCenH3 and δCenH3 (fig. 1D and supplementary table 3, Supplementary Material online).
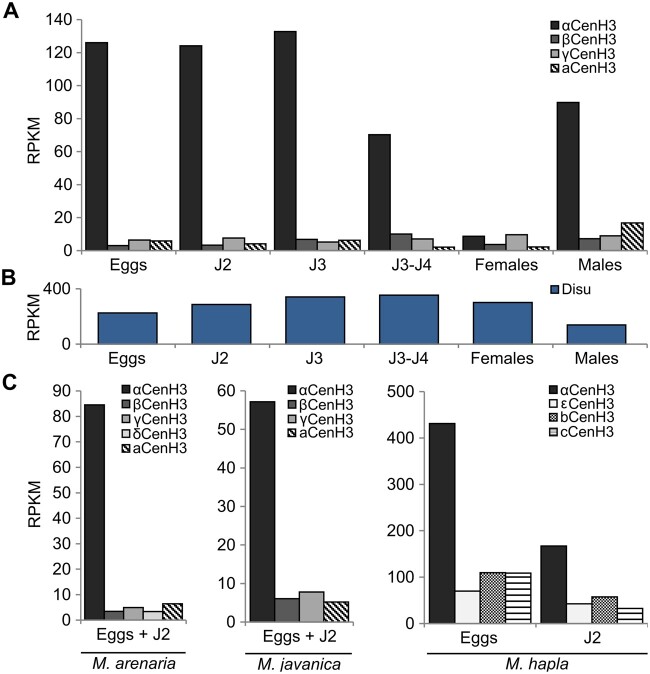
To test expression profile of all CenH3 candidates, publicly available raw Illumina transcriptome data from five clearly defined developmental stages including egg, juvenile J2, J3, J4, and female/male in M. incognita. Results showed that all copies of the CenH3 genes are actively transcribed (fig. 2A). However, through the development of M. incognita expression of αCenH3 was 20- to 50-fold higher in contrast to other CenH3 genes whose transcription proved to be extremely low. The exception was female stage where transcription of αCenH3 is decreased to the level of other CenH3. To test the quality of analyzed M. incognita transcriptomes expression of the reference gene disulfide-isomerase (Disu) (Hu and DiGennaro 2019) was performed. The results showed a comparable amount of Disu transcripts in all M. incognita developmental stages (fig. 2B) proving the reliability of the used RNA-seq data sets. In the closely related M. arenaria and M. javanica analyses on available mixed eggs and J2 stages also revealed significantly higher expression of αCenH3 and very low level of transcription of other CenH3 genes. A similar phenomenon was observed in diploid meiotic M. hapla where αCenH3 transcripts prevailed in analyzed samples (eggs and J2) in comparison to other CenH3s. In conclusion, expression profile of different CenH3 genes in all developmental stages as well as in different Meloidogyne species shows similar pattern characterized by dominant expression of αCenH3. It should be noted that RNA-seq from reproductive stages (females and males) were not available for M. arenaria, M. javanica, and M. hapla.
Fig. 2.
Expression profile of CenH3 genes in different species and developmental stages. (A) CenH3 expression through life cycle of Meloidogyne incognita. (B) Expression of Disu reference gene in M. incognita. (C) Expression of CenH3 in three closely related species; M. arenaria, M. javanica, and M. hapla. The relative expressions of CenH3 genes in different samples of RNA-seq data were analyzed using Bowtie2 v.2.3.0 mapper (Langmead and Salzberg 2012). Hits were normalized with RPKM (reads per kilobase of transcript per million mapped reads) method. Expression profile was shown as logarithmic transformation of RPKM values. The developmental stages include eggs, different juvenile stages (J2, J3, J4), females, and males. The RNA-seq data with accession numbers are listed in Materials and Methods section.
Different Evolutionary Dynamics of αβγδε CenH3 Group of Genes
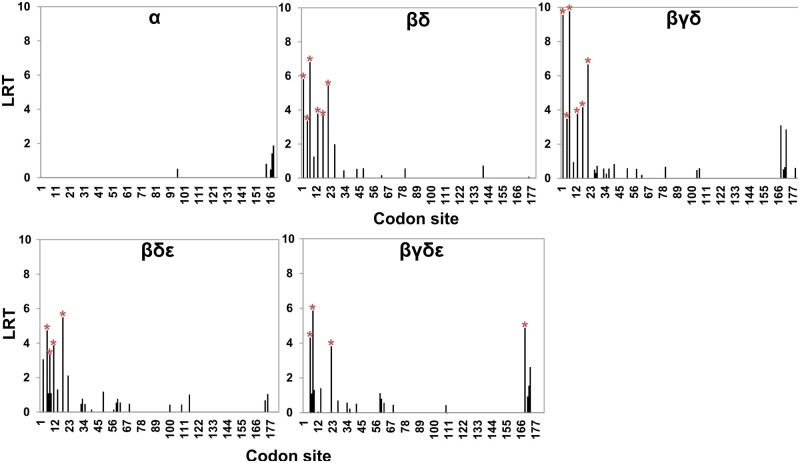
Dominant expression and conservation of αCenH3 gene in all analyzed species, including M. hapla provoked us to focus the study on αCenH3 and its closely related CenH3s grouped in the monophyletic αβγδε CenH3 clade. To evaluate how different CenH3 genes evolve, we examined the overall ω value for different gene comparisons (supplementary fig. 2, Supplementary Material online) using the substitution model implemented in MEGA version X (Kumar et al. 2018). First, intragroup comparisons were done on the full-length sequence alignment among α, β, and γ CenH3 variants originating from different species. All intragroup analyses revealed extremely purifying selective pressure where ω ratio varied from 0 to 0.5 (supplementary table 4, Supplementary Material online). Intergroup analyses of CenH3 genes were carried out on HFD and N-termini alignments separately. The results indicated purifying selective pressure on HFD domain in all pair-wise comparisons with ω values ranging from 0.01 to 0.1 (supplementary table 4, Supplementary Material online). In N-termini intergroup analyses, αCenH3 was excluded due to its high divergence to β, γ, δ, and εCenH3s, which resulted in incorrect alignments. N-termini selection analyses in β, γ, δ, and εCenH3 comparisons showed that ω value was higher in comparison with HFD analyses but still <1, which appears to be a signature of stabilizing selection (supplementary table 4, Supplementary Material online). Given that positive selection could act only on a few codons, to identify potential sites under positive selection, we carried out MEME model of codon substitution that allows ω value to vary across both codons and branches. The results are shown in figure 3 and supplementary table 5, Supplementary Material online. These analyses detected the same pattern of codon evolution in all gene comparisons among β,γ,δ, and εCenH3 group (βδ, βγδ, βγε, and βγδε) identifying few statistically significant positively selected sites in the first part of N-terminal tail (fig. 3). No codon under positive selection was found in comparison of αCenH3s from MIG species and distant M. hapla. Our results therefore confirm that αCenH3 sequences have evolved under purifying selection, whereas βγδεCenH3 sequences have undergone positive selection in the part of N-terminal domain.
Fig. 3.
Codon-specific tests for positive selection of CenH3 genes. Tests were inferred by mixed effects model of evolution (MEME) using the likelihood ratio test (LRT). LRT values with P values <0.1 were considered as codons under positive selection (red stars) (source data are supplied in the supplementary table 5, Supplementary Material online). Multiple alignments of CenH3 genes used for analyses are shown in supplementary figure 2, Supplementary Material online.
Distribution of αCenH3 Centromeres in M. incognita
Chromosomal localization of CenH3 proteins were assayed in female gonads of M. incognita using polyclonal antibodies against peptide corresponding to the N-terminus of CenH3s (fig. 1B). First analyses were done with αCenH3 because it is dominantly expressed compared with the rest of CenH3s. Western blot with rabbit-raised anti-αCenH3 demonstrated that the antibody recognizes proteins of the predicted molecular weight of 18 kDa in protein isolate of eggs, J2 stage and also in females (supplementary fig. 3A, Supplementary Material online). Although low transcription of αCenH3 gene has been shown in females, somewhat lower but comparable amount of αCenH3 proteins exist in females in comparison to J2 and eggs (supplementary fig. 3A, Supplementary Material online). This unusual phenomenon of low transcription in females could be explained by stability of αCenH3 protein in the process of transition from J4 stage (high αCenH3 transcription) to females (low αCenH3 transcription). In support to this, recent data on Mus musculus oocytes showed that centromere function of oocyte does not depend on the loading of newly transcribed CenH3 implying the stability of CenH3 protein (Smoak et al. 2016).
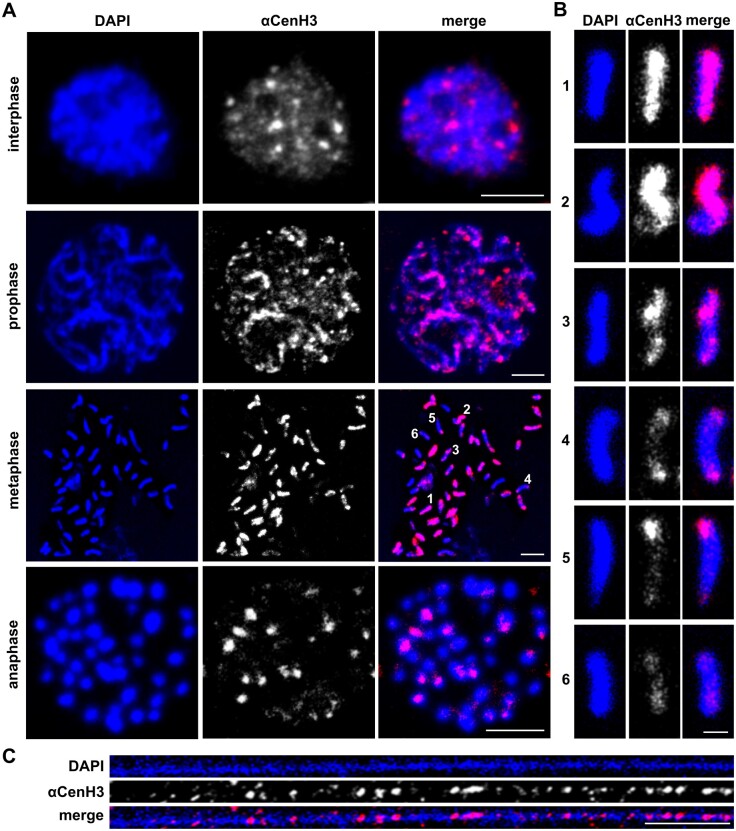
Given that M. incognita is a mitotic parthenogenetic species, cytosmear preparations from reproductive female tissue (ovaries and uterus) represent exclusively mitotic divisions. Chromosomal distribution of αCenH3 through different mitotic phases evaluated by anti-αCenH3 immunofluorescence (IF) is presented in figure 4A. In interphase nuclei, many αCenH3 signals differing in intensities were found. With the progression of the mitotic cycle when the chromosomes’ contours became visible, αCenH3 clusters that differ in intensity and representation become more apparent (fig. 4A, prophase). In addition to high number, M. incognita chromosomes are characterized by remarkable diminutives. Meloidogyne incognita population analyzed in this work has 46 chromosomes ranging in size from 0.4 to 1.5 µm in metaphase, whereas, for comparison C. elegans possess only five pairs of significantly bigger chromosomes (∼5 µm in length in metaphase). Immunofluorescence on M. incognita metaphase chromosomes revealed unexpected patterns of αCenH3 distribution according to which the chromosomes can be classified roughly into six types according to αCenH3 distribution (fig. 4A metaphase and fig. 4B). The chromosomes with strong αCenH3 signal which seems to occupy the entire chromosome length in the condensed metaphase (fig. 4B, chromosome type 1) are predominant (∼20 out of 46). The other group includes the chromosomes with uneven distribution of αCenH3 signal characterized by combining discrete and abundant αCenH3 regions in different chromosome areas (fig. 4B, chromosome types 2–5). Abundant αCenH3 regions may occupy even more than half of the chromosome length (4B, chromosome type 2) or appear as bicentric (fig. 4B, chromosome types 3 and 4) and telocentric clusters (fig. 4B, chromosome type 5). Finally, a few chromosomes show discrete αCenH3 clusters dispersed along the entire chromosome (fig. 4A and B, chromosome type 6). To examine the αCenH3 centromeric chromatin at higher resolution, immunostaining experiments were performed on chromatin fibers. The results show organizational pattern in which interspersed αCenH3 domains are interrupted by αCenH3-free subdomains (fig. 4C).
Fig. 4.
The organization of αCenH3 centromeres in Meloidogyne incognita. Slides were prepared from isolated reproductive tissue of females (ovaries and uterus). (A) Immunofluorescence of αCenH3-containing domains (red) during the mitosis cycle in M. incognita using anti-αCenH3 antibodies raised in rabbit 2 (supplementary fig. 3A, Supplementary Material online). Scale bar = 5 µm. (B) Distribution pattern of αCenH3-containing domains along metaphase chromosomes in six different chromosome types. Selected chromosome types were indicated in metaphase spread with numbers. Scale bar = 1 µm. (C) Immunofluorescence of αCenH3-containing domains (red) on chromatin fiber. Scale bar = 5 µm. All chromosomes and fibers were counterstained with DAPI (blue). Images were acquired with confocal microscopy and shown as z-stack projection.
To disclose chromosomal deposition of β and γCenH3s in M. incognita, polyclonal antibodies against two epitopes specific for βCenH3 (βCenH3-1 and βCenH3-2) as well as the epitope shared by β and γCenH3 (βγCenH3 epitope) (fig. 1B) were generated in parallel in rabbits and guinea pigs. The peptides selected for immunization encompassed even 55% of βCenH3 N-terminal sequence (41AA of 75AA) and represented all potential N-tail βCenH3-specific epitopes (fig. 1B). Western-blot analysis using anti-βCenH3-1 and anti-βCenH3-2 as well as anti-βγCenH3 did not detect any specific band of the expected molecular weight of 18 kDa (supplementary fig. 3A). Although, Western-blot results were in accordance with very low transcription of βCenH3 and γCenH3 genes in RNA-seq data (fig. 2) it remained doubtful whether antibodies recognized the epitopes and consequently βCenH3 and γCenH3 proteins. Peptide dot blot validation revealed that anti-βCenH3-1, anti-βCenH3-2, and anti-βγCenH3 generated in rabbits were sensitive for a peptide of interest without cross-reactivity with nonspecific peptides, including peptide specific for αCenH3. Among antibodies raised in guinea pig only anti-βCenH3-2 recognized specific peptide (supplementary fig. 3B).
Taking into account that β and γCenH3s could be present at a level below the detection limit of Western-blot analyses, we performed IF experiments in an additional attempt to verify if these CenH3s still participate in female centromeres. The double IF combining anti-αCenH3 (produced in rabbit) and anti-βCenH3-2 (produced in guinea pig) as well as immunostaining with anti-βCenH3-1/-2 or anti-βγCenH3 antibodies raised in rabbits were performed. Although approximately 100 cytological specimens were analyzed, neither of these experiments could confirm the presence of βCenH3 and/or γCenH3 on M incognita chromosomes (data not shown). Although, the participation of β and γCenH3 in M. incognita centromere cannot be completely ruled out due to limited methodological approaches in nonmodel organism their low transcription in all developmental stages together with efficient validation of antibodies by indirect assay of peptide dot blot, suggest that unsuccessfully detection of βCenH3 and γCenH3 by Western blot and IF most likely reflects the absence of βCenH3 and γCenH3 protein synthesis and consequently their lack in M. incognita centromere. Thus, it is the most probable that αCenH3 is, if not exclusively, then certainly predominantly incorporated into M. incognita centromeres.
In addition, to check if microtubules attach to the αCenH3 units, coimmunostaining with antibodies against α-tubulin and anti-αCenH3 was analyzed. The specificity of anti-α-tubulin was confirmed by Western blot on protein crude extract from M. incognita eggs (supplementary fig. 4A). Considering that the most of the oocytes present in the ovaries and uteri of M. incognita females appear to be in prophase/prometaphase (Triantaphyllou 1981), we were able to visualize α-tubulin and αCenH3 on chromosomes only in prometaphase (supplementary fig. 4B). Although, the results showed that the αCenH3 clusters mainly colocalized with the α-tubulin these images could not offer high-resolution view on the microtubule attachment at sites where αCenH3 was not detected nor detailed examination of α-tubulin distribution in comparison to αCenH3 along the chromosome length.
Composition of αCenH3-Associated DNA in M. incognita
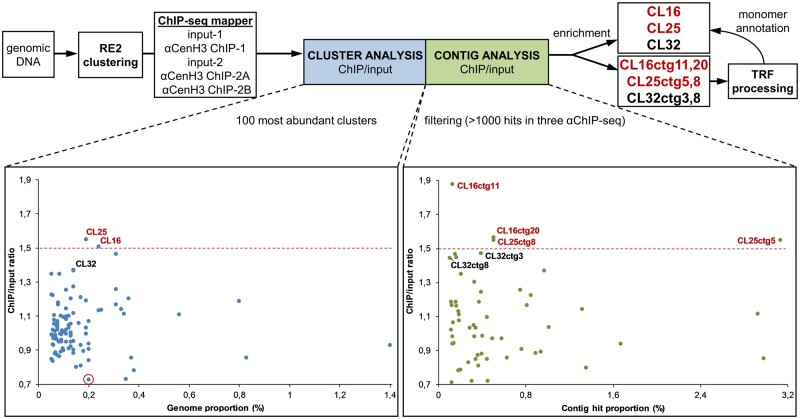
To map DNA sequences in αCenH3 centromere of M. incognita, native ChIP followed by next-generation sequencing (ChIP-seq) was performed. Three ChIPped DNA (supplementary fig. 5A) and input DNA libraries were generated and sequenced. To evaluate the enrichment of αCenH3-associated DNA in ChIP samples, ChIPped DNA (supplementary fig. 5B) was labeled and combined IF/fluorescence in situ hybridization (FISH) with anti-αCenH3 and labeled ChIPped DNA probe was conducted. The results showed highly abundant overlapping signals of ChIPped DNA and αCenH3 clusters thus confirming the specificity of ChIP experiments (supplementary fig. 5C). These strong FISH signals could likely have derived from repetitive DNA associated with αCenH3, because repeats enriched in ChIP DNA would result in strong hybridization. Mapping of ChIP reads on the reference genome using BWA (Li and Durbin 2009) produced high and unreliable peaks at very end of scaffolds or around placeholders (N stretches) which probable represent loci with repetitive sequences are only partially included in the genome assembly The fact that M. incognita genome is assembled into 12,091 scaffolds indicates high genome fragmentation and speak in favor of poor representation of repetitive regions in the assembled genome (Blanc-Mathieu et al. 2017). Following the above observations, we decided to focus our ChIP analysis on the repetitive genome fraction.
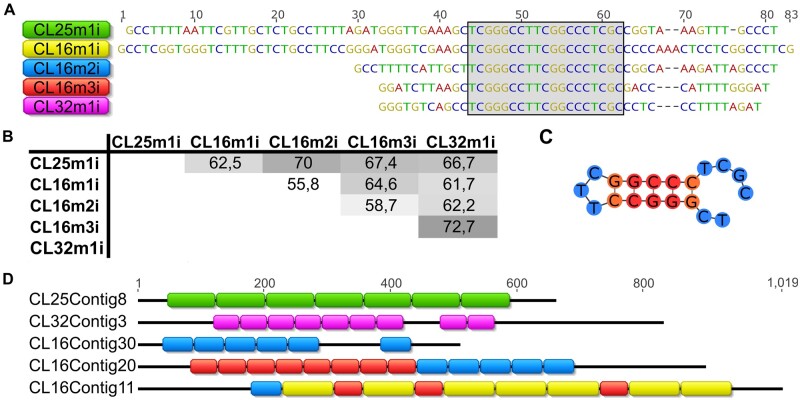
Therefore, an alternative approach of graph-based repeat clustering (Novák et al. 2013), which is independent of the assembled reference genome, was used for estimating read enrichment associated with repeat sequence types (fig. 5). The results of ChIP/input ratio for each ChIPped DNA together with values of SD are presented in supplementary table 6, Supplementary Material online. The ChIP/input ratio >1.5 was chosen as a threshold for considering the clusters enriched in the ChIP samples (fig. 5). Detailed analyses of enriched clusters, CL25 and CL16, revealed that they represent complex groups composed of different contigs, which made impossible to define the cluster consensus sequence. For that reason, more in-depth but complementary analyses of ChIP/input ratio on individual contig sequences (at least 0.002% genome abundance) were performed. The results disclosed that two contigs (ctg11 and ctg20) from the enriched cluster CL16 and two contigs (ctg8 and ctg5) from the enriched cluster CL25 show enrichment >1.5 suggesting that these sequences were most likely the αCenH3-associated DNAs (fig. 5 and supplementary table 6, Supplementary Material online). The analyses of enriched contigs by Tandem Repeat Finder (TRF) pipeline revealed that CL25ctg8 and CL25ctg5 represent arrays of tandem repeated 70-bp-long monomers (Cl25m1i) (supplementary fig. 6A, Supplementary Material online). In addition, the same analysis of CL16ctg11 and CL16ctg20 showed that these contigs are composed of three different tandem repeats (TRs) with monomer units of 83, 55, and 45 bp (CL16m1i, CL16m2i, and CL16m3i, respectively) (supplementary fig. 6B, Supplementary Material online). Aligned consensus sequences of monomers extracted from the enriched contigs (CL25m1i, CL16m1i, CL16m2i, and CL16m3i) are presented in figure 6A. Moreover, annotation analyses of all contigs from the cluster CL25 revealed that CL25 mostly comprised TR arrays with CL25m1i monomer (supplementary fig. 6A, Supplementary Material online). In addition, mapping of CL16-specific contigs with CL16m1i-m3i monomers showed that only 1/3 of contigs represent monomeric (monomers belonging to the same family) or mosaic TR arrays composed of monomers from different families (fig. 6D and supplementary fig. 6B, Supplementary Material online). The reason that only two contigs from each cluster (CL25 and CL16) proved to be enriched in contig analysis lies in the fact that these sequences are the longest TR arrays composed of αCenH3-associated monomers in enriched clusters. Interestingly, despite different monomer length (45–83 bp) and relatively low-sequence similarity (55.8–72.7%, fig. 6B), αCenH3-associated monomers show a completely conserved 19-bp sequence box (TCGGGCCTTCGGCCCTCGC, fig. 6C).
Fig. 5.
Identification of αCenH3 ChIP-enriched sequences. Strategies for identifying the most abundant repeat clusters and contigs associated with αCenH3 chromatin in Meloidogyne incognita (on the top). Relative enrichments of repeat DNA families in the ChIP-seq data are presented for clusters (left graph) and contigs (right graph) analysis. Clusters/contigs are represented by dots. The y axis is the ratio of the ChIP-seq reads to input-seq reads, representing the enrichment of each corresponding cluster/contig from the ChIP-seq data. The x axis is the genome proportion for each cluster (left graph) or hit proportion for each contig (right graph). A cluster rounded in red was used as a negative control in the IF-FISH experiment. Data with ChIP enrichment analyses of clusters and contigs with SDs are presented in supplementary table 6, Supplementary Material online.
Fig. 6.
Candidates for αCenH3-associated sequences in Meloidogyne incognita. (A) Alignment of consensus monomer sequences extracted from tandem repeated arrays (TRs) enriched in ChIP-seq analyses. The names of the consensus monomers were derived according to the cluster from which they originated. The conserved 19-bp box is indicated within the gray shaded area. (B) The percentage of identity among consensus monomer sequences. (C) Secondary structure of the conserved 19-bp box sequence. Folding free energy of the 19-bp fragment is −5.40 kcal/mol. (D) The most prominent examples of array organization of αCenH3-associated sequences (source data are supplied in supplementary fig. 6, Supplementary Material online). The color code of monomer sequences in arrays corresponds to monomer labels at the panel (A).
In order to detect possible additional αCenH3 centromeric candidates in ChIP data, all other clusters/contigs were searched against the conserved 19-bp box. The results revealed that the cluster CL32 together with the contigs CL32ctg8 and CL32ctg3 have tandem repeat organization of monomers which contain the conserved 19-bp box (fig. 6D and supplementary fig. 6C, Supplementary Material online). In support, the results of ChIP/input ratio for these cluster/contigs showed enrichment slightly below the defined threshold (fig. 5). Extraction of monomers from CL32ctg8 and CL32ctg3 contigs disclosed a new TR family, CL32m1i, which shares sequence similarity of 72.7% with CL16m3i monomer sequence (fig. 6A and B). Annotation of CL32m1i monomer to contigs of CL32 cluster showed that only 14 of 60 contigs contain short TR arrays composed of CL32m1i monomers (supplementary fig. 6C, Supplementary Material online). The other contigs from clusters CL25, CL16, and CL32 which are not involved in αCenH3-associated monomers, probably represent different TR surrounding regions (supplementary fig. 6, Supplementary Material online). This assertion is supported by the fact that different non-αCenH3-associated contigs assemble the flanking regions of αCenH3-containing TRs (supplementary fig. 6D, Supplementary Material online). The specific feature of all αCenH3-associated monomers is relatively high GC content (>50%), especially pronounced in the conserved 19-bp box (∼80%), in contrast to 70% AT-richness that characterizes the assembled part of the genome (Abad et al. 2008). In addition, sequence analyses of the conserved 19-bp box revealed two sequence segments repeated in an inverted orientation which has a high potential to form an energetically stable dyad structure (fig. 6C).
Validation of the ChIP-Identified αCenH3-Associated Sequences in M. incognita
The localization of αCenH3 on the chromosome level showed pattern with highly abundant and discrete αCenH3 clusters. It can be assumed that TRs associated with αCenH3-highly abundant cluster are not represented in the reference genome, although the existence of a genome-wide distribution of αCenH3 discrete clusters implies their presence (at least to some extent) in the genome assembly. Therefore, to further validate our results of ChIP analyzes obtained on reference repeat database, we performed an additional survey on the reference genome. The ChIP background was removed by subtracting input signal from the ChIP data. The reference genome was simultaneously mapped with ChIP sequences and the 19-bp box to assess the enrichment of TRs-containing the 19-bp box in the genome assembly. We identified 1,117 19-bp boxes on the reference genome and as expected from previous ChIP mapping data the most of the detected 19-bp box-containing TRs were located at the ends of the scaffolds thus confirming the underrepresentation of these sequences in the assembly. However, TRs distributed along the scaffolds could be associated with discrete αCenH3 clusters were also found. Some representative examples provided as genome browser views in supplementary figure 7A, Supplementary Material online, clearly showed that distribution of enrichment correlates with TRs containing the 19-bp box in all three ChIP samples, whereas the 19-bp motif-free regions of the scaffolds did not show enrichment for αCenH3-ChIPped sequences.
In addition, we mapped WGS, input, and ChIP data with the 19-bp box to compare abundance of αCenH3-associated TRs among those genome resources and comparing it also to genome assembly (supplementary fig. 7B, Supplementary Material online). WGS and input with ∼0.22% of TRs containing 19-bp box sequences showed about 5× higher abundance related to the assembled genome (0.04%). All three ChIP data showed the same ratio of TRs containing the 19-bp box enrichment (1.55×) in comparison to input data. This is in accordance to average enrichment obtained in ChIP analyses on repetitive database conducted by Repeat Explorer (fig. 5). Based on Repeat Explorer analysis, 6.3% of WGS reads were classified as tandem repeats. TRs containing the 19-bp box with 0.22% abundance of WGS thus make only 3.5% of the detected tandem repetitive fraction in M. incognita genome. Considering that the several methodological issues in ChIP and Illumina sequencing workflow (e.g. fragility or resistance to DNA fragmentation, A/T homopolymers, high %GC) may cause underrepresentation of different types of repetitive sequences in WGS and ChIP data, to estimate experimentally the genome abundance of αCenH3-associated TRs, we performed the dot blot of M. incognita genomic DNA with TRs probes (CL25m1, CL16m1i, and CL16m3i/CL32m1i; probe preparation is explained in the section below). Interestingly, the results of dot blots revealed that αCenH3-associated TRs comprise cumulatively about 2.3% of the genome (supplementary fig. 7C, Supplementary Material online), which is almost 10× higher than was estimated in WGS/input data and more than 50× higher than in the assembled genome (supplementary fig. 7B, Supplementary Material online), Since there is no significant discrepancy in abundance between WGS and inputs data related to αCenH3-associated TRs it can be assumed that the Illumina sequencing rather than DNA fragmentation step cause underrepresentation of these repetitive sequences in WGS/input data. It is known that high GC content interfere with PCR-based library amplification, causing a depletion of the GC-rich templates (Aird et al. 2011). Regarding to relatively high GC content (53–68%) of αCenH3-associated TRs, it is most likely that this factor causes a depletion of the corresponding sequences in WGS/input data.
Cytological Confirmation of the ChIP-Identified αCenH3-Centromeric Repeats in M. incognita
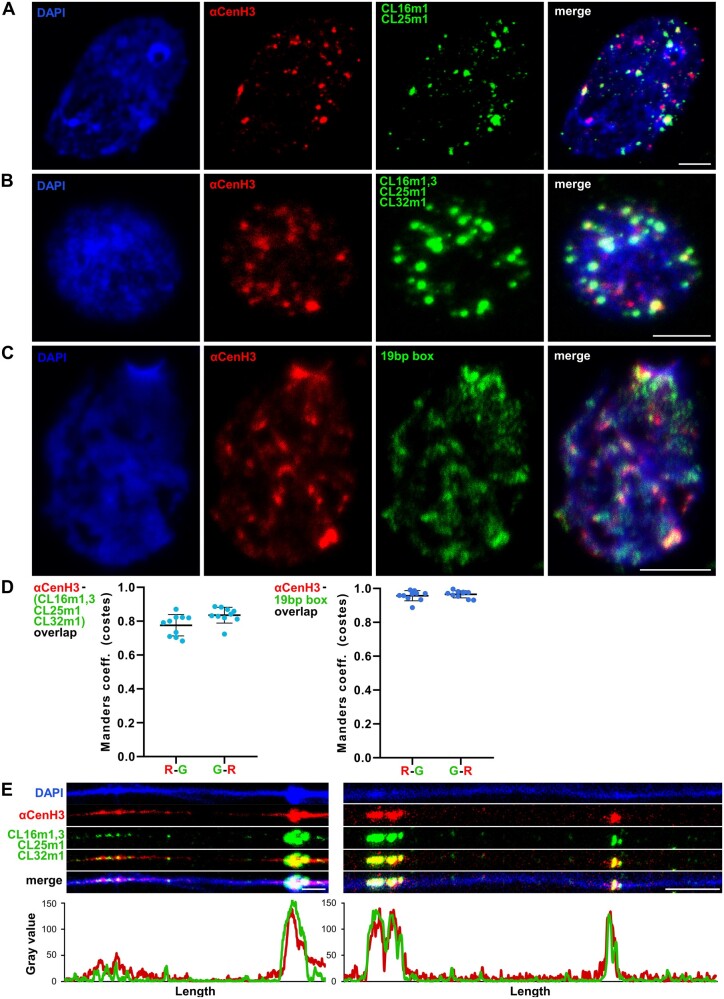
To test the association of ChIP-enriched TRs with functional αCenH3 centromere domains, IF-FISH using αCenH3-specific antibody and centromere associated monomers as hybridization probes was performed. The labeled DNA probes were generated for CL25m1i, CL16m1i, and CL16m3i (supplementary fig. 8A, Supplementary Material online). Primers specific for CL32m1i were difficult to design due to short sequence and high similarity with CL16m3i. Therefore, FISH was conducted under moderate stringency conditions which allow a single probe to hybridize to both variants, CL32m1i and CL16m3i. Since primers specific for CL16m2i did not produce relevant PCR profile, this probe was excluded from analyses. Given that metaphases are extremely rare in chromosomal preparation of M. incognita IF-FISH analyses were presented on interphases. To confirm αCenH3-specific localization and disclose distribution pattern of putative cenDNA sequences on αCenH3 domains, we first performed IF-FISH using CL25m1 and CL16m1i as hybridization probes. The results of combined IF-FISH analysis with CL25m1-i and CL16m1i probes showed that approximately half of the αCenH3 centromeres contain a considerable amount of these TRs (fig. 7A). In addition, IF-FISH with all TRs probes (CL25m1, CL16m1i, and CL16m3i/CL32m1i) showed that these sequences cover the majority of αCenH3 domains resulting in overlapped regions (yellow fluorescence signals at fig. 7B).
Fig. 7.
Simultaneous detection of αCenH3 centromere and αCenH3-associated DNA in Meloidogyne incognita. Slides were prepared from isolated reproductive tissue of females (ovaries and uterus). (A) Combined immunofluorescence with anti-αCenH3 raised in rabbit 2 (red) and FISH with CL16m1i and CL25m1i αCenH3-associated monomers as probes (green). (B) IF-FISH with anti-αCenH3 (red) and mixed probe for αCenH3-associated monomers, CL16m1i, CL16m3i, CL25m1i, and CL32m1i (green). The overlapped IF-FISH signals are yellow. (C) IF-PRINS with anti-αCenH3 raised in rabbit 2 (red) and centromeric 19-bp box sequence (green). (D) Quantification of signal colocalization for ten representative images with calculated Manders coefficients (costes thresholding) seen as high overlapping ratios for both channel pairs (R-G represents overlapping ratio of red vs. green signals; G-R is overlapping ratio of green vs. red signals); αCenH3 with αCenH3-associated monomers, CL16m1i, CL16m3i, CL25m1i, and CL32m1i (left panel) and αCenH3 with 19-bp box regions (right panel). Data are presented as mean ± SD (source data are listed in supplementary table 7, Supplementary Material online). (E) Dual-color fiber-IF/FISH using anti-αCenH3 (red) and αCenH3-associated monomers CL16m1i, CL16m3i, CL25m1i, and CL32m1i (green) as probes. The plots below the images represent intensities of the IF (red) and FISH (green) signals. DNA was counterstained with DAPI (blue). Images were acquired with confocal microscopy and shown as z-stack projection. Scale bar = 5 µm.
To provide a more accurate estimation of αCenH3/TRs colocalization, the quantification of IF (anti-αCenH3) and FISH signals (all TRs probes) on the original confocal images of ten interphases was performed. Based on the quantification, the IF signals overlapped average 78% of FISH signals, whereas FISH signals coincide with 83% of IF signals (fig. 7D, left and supplementary table 7, Supplementary Material online). We suppose that a minor fraction of αCenH3-specific domains, which are not overlapped with FISH signals, originate from αCenH3 domains enriched in monomeric TR type of CL16m2i which was absent from analyses or some other sequence(s)-containing 19-bp box. Thus, in order to confirm our assumption that αCenH3 associated sequences contain the 19-bp box, combined IF with anti-αCenH3 and primed in situ labeling (PRINS) assay using the 19-bp box as primer sequence was conducted. Although PRINS showed lower brightness of signals in contrast to FISH signals, which is probably due to a different methodology, it can be observed that the incidence of signal overlap is higher in comparison to IF/FISH experiment (fig. 7C vs. fig. 7B). This is especially evident in quantification graph of IF/PRINS overlapping, where the αCenH3 IF signals match >95% of the 19-bp box PRINS signals (fig. 7D right panel and supplementary table 7, Supplementary Material online). To examine organization of αCenH3-specific domains and selected TR repeats at higher resolution, sequential detection of αCenH3 and Cl25m1, Cl16m1i and Cl16m3i/Cl32m1i sequences on stretched chromatin fibers was performed. These results proved that αCenH3-associated DNA coincides with αCenH3 domains, following interspersed organization of αCenH3 centromere interrupted by αCenH3-free domains (fig. 7E).
To exclude the possibility that αCenH3-associated TRs, detected as sequences with relatively low enrichments, could be the result of a centromere inclination to accumulate any repetitive sequences a control test was performed. Satellite DNA of similar genome abundance that was not found to be enriched in the ChIP-seq analysis (marked in fig. 5) was selected for IF/FISH. Simultaneous detection of αCenH3 and nonenriched satDNA revealed that the regions of satellite DNAs did not coincide with αCenH3 clusters (supplementary fig. 9, Supplementary Material online). In addition, it can be observed that in contrast to αCenH3-associated TRs, noncentromeric satellite DNA showed a relatively comparable genome abundance that was estimated in WGS analysis (fig. 5), which further confirms the previous conclusions on abundance discrepancy of αCenH3-associated TRs in WGS/input data.
Prediction of αCenH3 Centromeric Repeats in M. incognita-Related Species
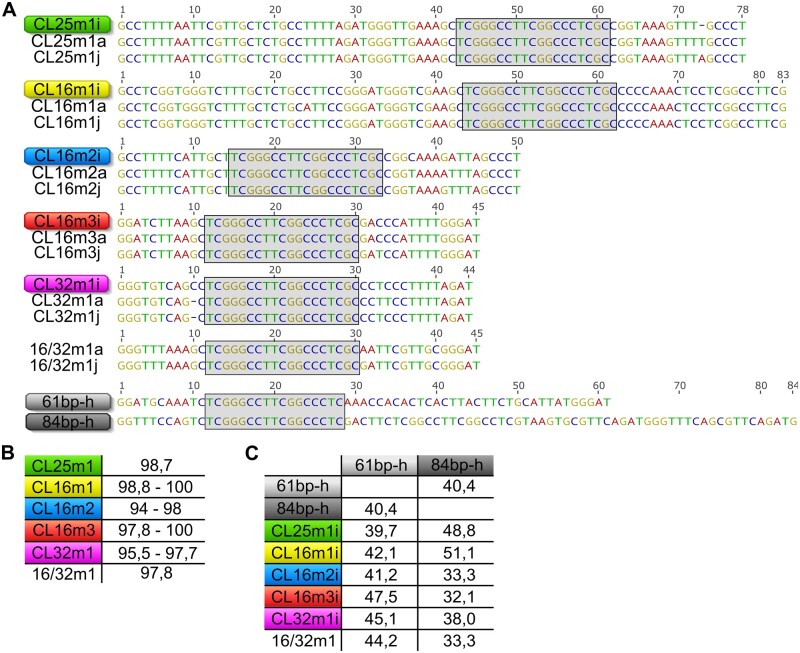
Since αCenH3 turned out to be conserved in all analyzed Meloidogyne species, we wondered whether the 19-bp box-containing sequences, as αCenH3-associated DNA in M. incognita remained conserved in other Meloidogyne genomes. We hypothesize that short TRs with conserved 19-bp box-containing monomers in other Meloidogyne species could disclose putative cenDNA regions of the corresponding species. For this analysis, we used publicly available Illumina WGS databases for two closely related species, M. arenaria and M. javanica (Blanc-Mathieu et al. 2017). Using as a criterion short TRs with the conserved 19-bp box, clustering of M. arenaria and M. javanica WGS reads followed by cluster annotation with conserved the 19-bp box was performed. The results revealed the appearance of the 19-bp box in repetitive form in the majority of contigs in clusters CL8 and CL7 in M. arenaria, and M. javanica, respectively (supplementary fig. 10A and B, Supplementary Material online). Using TRF pipeline, contigs with repeated organization of the 19-bp box were subjected to monomer unit extraction. Consensus sequences of extracted monomers were compared with αCenH3-associated monomers from M. incognita. The alignments show that the 19-bp box-associated monomers from M. arenaria and M. javanica grouped with all TR families of M. incognita (fig. 8A). Moreover, sequence comparison revealed that monomers remained almost completely conserved (94–100%) among these closely related species (fig. 8B). Monomer variants with the conserved 19-bp box and 45-bp monomer length specific for M. arenaria and M. javanica only (16/32m1-a and 16/32m1-j) were also identified (fig. 8A). In addition to the conserved sequence features of putative αCenH3-associated monomers, mapping of these monomers to contigs from M. arenaria and M. javanica revealed organization previously detected in M. incognita, with monomeric and mosaic TR arrays embedded in unrelated sequence environment (supplementary fig. 10A and B, Supplementary Material online). To confirm presumption that 19-bp box-containing TRs are associated with αCenH3 in M. arenaria and in M. javanica, combined IF using anti-αCenH3 and PRINS using the 19-bp box sequences as primer was performed on cytosmears. Although it was even more difficult than in M. incognita to obtain cytological preparation in these species due to their tetraploidy, similarly as in M. incognita the results showed high coincidence of overlapped αCenH3/19-bp box signals indicating αCenH3 deposition on their chromosomes as well as association of αCenH3 with 19-bp box-containing TRs (supplementary fig. 11).
Fig. 8.
Candidates for αCenH3 centromeric sequences in Meloidogyne incognita-related species. (A) Alignments of consensus monomers sequences (CL25m1, CL16m1, CL16m2, CL32m1, and 16/32m1) extracted from clusters with tandem repeated arrays containing the conserved 19-bp box from M. arenaria (a) and M. javanica (j) in comparison to M. incognita (i). 61 bp-h and 84 bp-h represent monomers from tandem repeats containing the 19-bp box found in M. hapla assembled genome. The conserved 19-bp box is indicated within the gray shaded areas. (B) The percentage of sequence identity among consensus monomers in MIG species. (C) The percentage of sequence identity of monomers from M. hapla in comparison to monomers from MIG species.
Considering that our analyses of CenH3s showed almost completely conserved αCenH3 protein sequence and highly transcribed αCenH3 gene in nonreproductive stages (eggs and juveniles) of distant meiotic M. hapla, we asked whether its genome also comprises putative αCenH3-associated sequences. We did not apply the same clustering strategy done for M. arenaria and M. javanica because Illumina WGS data for M. hapla were not public available. Instead, the 19-bp box was mapped to the assembled M. hapla genome, and monomers were extracted from detected TR arrays. Two types of short TRs, composed of 61- and 84-bp long monomers, were associated with the 19-bp box (fig. 8A and supplementary fig. 10C, Supplementary Material online). In contrast to putative αCenH3-associated monomers in M. arenaria and M. javanica which share high sequence identity (95–100%) with M. incognita, M. hapla putative αCenH3-associated monomers except conserved 19-bp box exhibit low-sequence identity in comparison to MIG species (32–51%) (fig. 8C).
Discussion
The availability of sequenced genomes and complex species evolution makes Meloidogyne an ideal system to study evolution of CenH3 proteins. We identified 21 CenH3 proteins in the three closely related mitotic M. incognita, M. arenaria, M. javanica (MIG) species and in the distantly related meiotic parthenogenetic species M hapla. Interestingly, phylogenetic analysis suggests the presence of two polyphyletic groups of CenH3s, and multiple copies of CenH3 genes in all analyzed species. The abcCenH3 group comprises rather divergent sequences with features of H3 variants but with relatively low HFD sequence identity related to the canonical H3. In the contrast, αβγδεCenH3s clade represents more homogenous group of sequences with identity to H3 considered as a common between H3 and CenH3s in many organisms analyzed so far (Malik and Henikoff 2003). These observations suggest independent evolution of two CenH3 groups from H3, indicating complex pattern of CenH3s in Meloidogyne species. The existence of multiple CenH3 candidates with polyphyletic origin in Meloidogyne species raises the question of their classification and function, The presence of two CenH3 genes is not uncommon in plant genomes (Kawabe et al. 2006; Moraes et al. 2011; Sanei et al. 2011; Finseth et al. 2015; Ishii et al. 2015; Neumann et al. 2015). In contrast, occurrence of paralogs in animals is considered to be a rare event. However, the recent comprehensive study of many high-quality sequenced genomes of Drosophila species revealed multiple copies of Cid histone (Drosophila CenH3) (Kursel and Malik 2017). The similar phenomenon was found in genomes of mosquitoes, where CenH3 paralogs evolve under different selective constraints, and have been coretained for over 150 My (Kursel et al. 2020). Interestingly, among detected CenH3s in Meloidogyne, only αCenH3 showed abundant expression in all analyzed species regardless of mode of reproduction (mitotic or meiotic), whereas the other CenH3s were dropped to a relatively low level of transcription. Anticipating that αCenH3 represents centromeric protein in Meloidogyne species, we focused our analyses on αCenH3 and on monophyletic group of βγδε CenH3s closely related to αCenH3s.
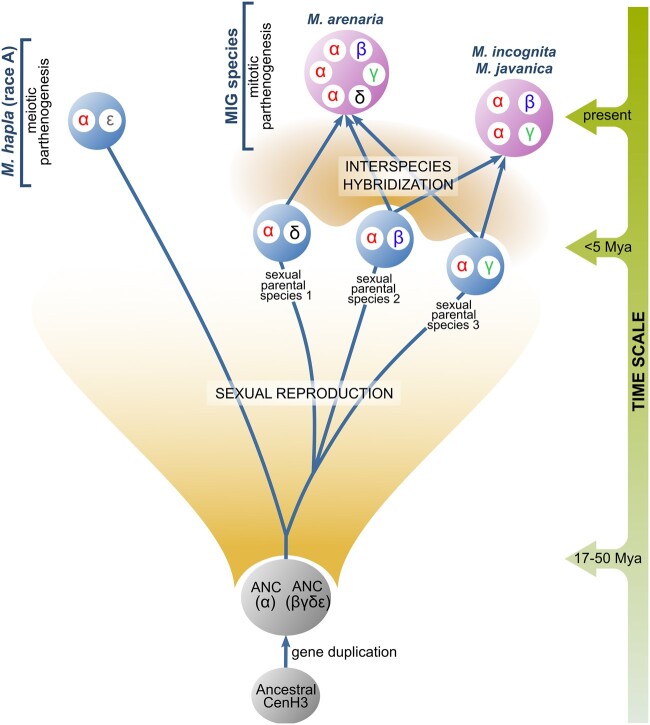
To understand evolution of αβγδε CenH3s in the selected Meloidogyne species, their complex species evolution history should be considered. MIG species have been determined as polyploids formed by recent and multiple interspecific hybridization events (Castagnone-Sereno et al. 2013). They reproduce exclusively asexually by mitotic parthenogenesis. On the contrary, M. hapla is a diploid species, and reproduces asexually by meiotic parthenogenesis, although alternatively can also be sexual (Castagnone-Sereno and Danchin 2014). The most parsimonious scenario of CenH3 evolution in the analyzed species is therefore based on the integration of results obtained in this work together with previous data on species evolution (fig. 9). The phylogenetic analysis of CenH3 sequences strongly suggests that CenH3 gene has undergone one duplication in an ancestral sexual species, a progenitor of MIG species and of M. hapla, resulting in an appearance of αCenH3 and (βγδε)CenH3 ancestral genes, (ANC [α] and ANC [βγδε] in fig. 9). Earlier phylogenetic studies based on different mitochondrial and nuclear markers revealed significant distance between MIG species and M. hapla, estimating their separation for 17–50 Ma (reviewed in Castagnone-Sereno et al. 2013). Unexpectedly, during this time the αCenH3 gene evolved under strong purifying selection in all analyzed species, resulting in almost completely conserved αCenH3 proteins in mitotic MIG species as well as in the distant meiotic M. hapla. So far, the only example of nearly identical protein sequences of CenH3s in related species were found in plant genus Secale (Evtushenko et al. 2017). The other copy of the gene, the ancestral βγδε CenH3, evolved rapidly into four different but related proteins: β, γ, δ, and εCenH3 (fig. 9). β, γ, and δCenH3s are specific for MIG species, whereas εCenH3 was found exclusively in M. hapla. In mitotic M. incognita, the αCenH3 showed high expression and chromosomal deposition on all chromosomes of the complement, whereas β and γCenH3s exhibit low expression and absence of chromosomal deposition. In support, two other MIG species, M. arenaria and M. javanica also showed the chromosome deposition of αCenH3 and silencing of β, γ, and δCenH3s. This indicates centromere competence of αCenH3 and most probably the loss of centromere-associated function for β, γ, and δCenH3s in exclusively mitotic MIG species. In support, detected dominant expression of αCenH3 in contrast to εCenH3 in the nonreproductive stages of M. hapla where mitosis is expected to occur, speaks in favor of dominant role of αCenH3 in mitotic cells of the meiotic M. hapla. The comparative analyses of MIG genomes and ITS markers suggest that polyploid genomes of MIG species result from additive interspecies hybridization between related parental sexual taxa (Hugall et al. 1999; Blanc-Mathieu et al. 2017; Szitenberg et al. 2017). Moreover, studies of mitochondrial DNA and genetic test of allelic sequence divergence suggest that hybridization events included in formation of MIG species have a recent origin (Giorgi et al. 2002; Lunt 2008; García and Sánchez-Puerta 2015; Blanc-Mathieu et al. 2017). In line with this scenario, MIG species possess multiple copies of αCenH3 and one copy of β, γ, and δCenH3s, whereas diploid M. hapla has only one copy of αCenH3 and of εCenH3 (fig. 9). Moreover, the copy number of αCenH3s and the presence of β, γ, and δ CenH3s in MIG species are consistent with the estimated ploidy levels based on protein-coding sequences (CDSs) data mapping in MIG genomes (Blanc-Mathieu et al. 2017). In addition, one copy of both αCenH3 and εCenH3 in M. hapla are corroborated by CDS mapping where one single locus was detected for the particular gene in M. hapla genome (Blanc-Mathieu et al. 2017). The observed pattern of CenH3s evolution in MIG species is in accordance with proposed species evolution characterized by independent evolution of α, β, γ, and δCenH3s in parental sexual taxa followed by polyploidization as result of recent species hybridization (fig. 9). In the light of the rapid evolution of β, γ, and δCenH3s, the evidence that these CenH3s remain almost completely conserved among MIG species suggests that MIG interspecific hybridization is a relatively recent event. Consequently, it is the most probable that β, γ, and δCenH3s diverged in sexual progenitors and that their redundancy in the nascent MIG coincided with recent species hybridization followed by mitotic mode of reproduction. Otherwise, if they were subjected to a long period of nonfunctionality, accumulation of random mutations and pseudogenization would be expected. The putative role of βγδ and εCenH3s in sexual progenitors has been additionally supported by the fact that they have been retained in both, the extant MIG and M. hapla species since their divergence for about 17–50 Ma (Castagnone-Sereno et al. 2013). In contrast to αCenH3 conservation, β, γ, δ, and εCenH3s evolved under the purified selection with the positive evolution trend on the first several amino acids of the N-terminal tail. The similar selection pattern was found in CenH3 proteins of holocentric chromosomes among the related species of the nematode genus Caenorhabditis which reproduces sexually, by asymmetric meiosis (Zedek and Bureš 2012). Rapid evolution of CenH3s with positive selection has been detected so far exclusively in sexual lineages with asymmetric meiosis where centromere drive occurs (Henikoff et al. 2001; Talbert et al. 2004; Hirsch et al. 2009; Schueler et al. 2010; Zedek and Bureš 2012; Finseth et al. 2015).
Fig. 9.
The most parsimonious evolution of α, β, γ, δ, and εCenH3 variants in Meloidogyne species (M. hapla [race A], M. incognita, M. arenaria, M. javanica). Presented species evolution with evolutionary time scale was based on different previous studies. Separation between MIG species and M. hapla was estimated between 17 and 50 Ma (reviewed in Castagnone-Sereno et al. 2013). Interspecies hybridization between related sexual parental species was proposed in formation of MIG species (Blanc-Mathieu et al. 2017; Szitenberg et al. 2017). Interspecific hybridization is estimated as relatively recent event (<5 Ma) (Giorgi et al. 2002; García and Sánchez-Puerta 2015; Blanc-Mathieu et al. 2017). ANC(α) represents the ancestral αCenH3, ANC(βγδε) represents the ancestral CenH3 of βγδε variants.
The major question raised by our observation is why the two subgroups of CenH3s, αCenH3, and βγδεCenH3s, have completely different evolutionary rate. We propose that different evolutionary dynamics of analyzed Meloidogyne CenH3s might be due to distinct requirements posed on αCenH3 in contrast to β, γ, δ, and εCenH3s in Meloidogyne centromere. Based on recent comprehensive evolutionary studies of CenH3 duplication in Drosophila and mosquito species the authors suggested that gene duplications of CenH3 could be required for multiple centromeric functions, for example, in mitosis versus meiosis (Kursel and Malik 2017; Kursel and Malik 2019). The observed evolution pattern characterized by purified selection with positive trends of β, γ, δ, and ε CenH3 could be predicted by the centromeric drive model which implies asymmetric meiosis and sexual reproduction. In that case, β, γ, and δCenH3 could have evolved in parental sexual lineages as a consequence of centromere drive during meiosis and become redundant in MIG species due to transition from sexual to mitotic parthenogenesis. It has been considered that transition from sexual reproduction to mitotic parthenogenesis in MIG species correlates with recent species hybridization (Lunt 2008; Blanc-Mathieu et al. 2017).in MIG species. In support, the major sperm protein which is a meiotic-specific gene, shows no increase in evolutionary rate nor change in substitution pattern in the mitotic Meloidogyne taxa, indicating that the locus has been maintained by selection (Lunt 2008). In MIG species there are also no morphological abnormalities in the sperm development, and insemination still occurs sporadically but without fertilization (Triantaphyllou 1981). Given that β, γ, and δCenH3s show no signs of pseudogenization and sequence degeneration which could be caused by random mutations, it is likely that these CenH3s, similarly as in the case of the sperm protein represent a “meiotic relict” in exclusively mitotic MIG species. Analogously, the αCenH3 conservation through the same period of the time could be due to the possible subspecialization. The main prediction of centromere drive is that CenH3 coevolves with cenDNA in order to suppress the deleterious effect of rapidly evolving cenDNAs in meiosis. In that case, the possible subspecialization of αCenH3 for mitosis could release αCenH3 from the adaptive conflict imposed in meiosis. This could result in strong amino acid conservation of αCenH3 among distant species. In support to different CenH3 functions in cell divisions, holocentric nematode C. elegans harbors two CenH3-related proteins, HCP-3 and CPAR-1, which indicates centromere functional specialization. HCP-3 has been proven to be essential for mitosis but is not required for meiotic kinetochore formation or chromosome segregation (Monen et al. 2005). Although the functional importance of Cpar-1 is not completely understood, its enrichment on meiotic chromosomes was documented (Monen et al. 2015). In addition, recent data on Arabidopsis suggest that in species with a single-copy CenH3 gene, one protein probably must be customized for different centromere functions (Ravi et al. 2011). The experiments showed that impaired CenH3 lost its function in meiotic centromeres of Arabidopsis, whereas the C-terminal region and HFD were sufficient for centromere function during mitosis (Lermontova et al. 2006; Lermontova et al. 2011; Ravi et al. 2011).
Predicted rapid evolution of CenH3s as a response to the deleterious effect of extremely divergent cenDNAs motivated the investigation of the genetic landscape features of M. incognita centromere determined by long-term conserved αCenH3. The αCenH3-associated cenDNA found in M. incognita is organized in a form of short arrays of tandem repeats (TRs), up to 1 kb in length, composed of five different families based on 50- to 80-bp monomers. Although presence of tandem repeats, in the form of long arrays of satDNAs is a common characteristic found in many monocentric species (Plohl et al. 2014), holocentric organisms investigated so far exhibit different patterns of cenDNAs. The robust ChIP CenH3-based studies on the whole-genome scale showed that C. elegans holocentromeres do not coincide with satDNAs but do coincide with nonspecific binding sites for multiple transcription factors (Steiner and Henikoff 2014). The native ChIP-seq of the parasitic nematode Ascaris also suggested absence of centromere-specific DNA sequence (Kang et al. 2016). So far, the only example of holocentric chromosomes which possess satDNAs as the centromere-specific sequence has been found in the plant Rhynchospora (Marques et al. 2015; Ribeiro et al. 2018). Comparative analyses of αCenH3-associated centromeric repeats revealed an exceptional feature in form of the 19-bp long-conserved box shared by extremely divergent monomers, suggesting selective pressure imposed on this sequence part regardless of the fast-evolving nature of repetitive DNAs. In addition to high GC content, the 19-bp conserved box exhibits a specific potential to form a stable dyad structure. In spite of enormous diversity in cenDNA detected in many species, the recent study of structural features of centromeric satDNAs from diverse eukaryotes pointed out two major characteristics to be crucial for putative cenDNA: a specific DNA sequence as a binding site for proteins and/or a specific feature of the sequence itself such as DNA secondary structure (Kasinathan and Henikoff 2018). Several studies have shown that centromeric satDNAs may form various types of non-B-form including single-stranded DNA, hairpins, R-loops, and i-motifs (Garavís et al. 2015; Kabeche et al. 2018; Kasinathan and Henikoff 2018). Consistent with this, if the conserved 19-bp box is a binding site for αCenH3 in M. incognita, primary as well as secondary structure of the 19-bp box could be crucial for its binding capacity. In contrast to AT-rich DNA which is common feature in centromeres (Talbert and Henikoff 2020) centromeric TRs in M. incognita show the extremely high GC content compared with the high AT composition of the genome (Abad et al. 2008). Therefore, we proposed that these unique sequence features, such as primary and secondary structures of αCenH3-associated DNA in the form of the 19-bp box incorporated into GC-rich short TRs, act in concert to ensure the faithful formation of an αCenH3 centromere in M. incognita. Another important finding that arose from this work is the existence of completely preserved αCenH3 centromere associated TRs in terms of sequence and organization in closely related M. incognita, M. javanica, and M. arenaria. The colocalization of αCenH3 and the 19-bp box in M. arenaria and M. javanica, similar to M. incognita, suggested the preservation of the αCenH3 centromere in protein and DNA aspects. Moreover, divergent TR short arrays with monomers containing almost completely conserved 19-bp box were shown in the distant M. hapla, implying the functional constraints imposed on this sequence part even in distantly related species. Concerning cenDNA with conserved sequence features, the recent study of early-diverging fungi showed the presence a 41-bp unique DNA motif in all nine core centromeres which has been proposed as a binding site for some kinetochore proteins (Navarro-Mendoza et al. 2019). The most prominent example of cenDNA sequence conservation is the CENP-B box, the conserved 17-bp long-sequence motif specific for alpha-satDNA in humans (Ohzeki et al. 2002) as well as in alphoid repeats in mammalian species (Alkan et al. 2011). This motif proved to be a binding site for centromeric protein CENP-B that is involved in kinetochore formation (Masumoto et al. 2004). Interestingly, interspecifically preserved motifs that probably evolve under functional constraints whose potential role(s) remain elusive were observed in many satDNAs, including in satDNAs of Meloidogyne species (Meštrović et al. 2006a, 2006b, 2013).
Regarding to chromosome organization of αCenH3 centromere in M. incognita an unusual pattern characterized by uneven distribution of αCenH3 among and along the chromosomes has been shown. Immunofluorescence on prophase chromosomes and on extended chromatin fibers, revealed discontinuous pattern of αCenH3 domains separated by αCenH3-lacking chromatin. The observed αCenH3 distribution pattern can be defined as cluster-like centromeric organization. In more condensed metaphase chromosomes, αCenH3 encompasses the entire chromosome length in the form of abundant or discrete signals or exhibits extremely uneven distribution with highly abundant domains in different chromosome regions. The α-tubulin was observed to be mostly colocalized with αCenH3 domains thus indicating functional potential of αCenH3 centromere in mitosis. In contrast to the point centromere subunits in C. elegans (Steiner and Henikoff 2014) the observed cluster-like organization of M. incognita is similar to the nematode Ascaris where CenH3 is organized into 1–15 kb domains distributed across the chromosomes (Kang et al. 2016), Recent data on the nematode C. elegans (Buchwitz et al. 1999; Moore et al. 1999), and plant Rhynchospora (Marques et al. 2015) suggest different organization of CenH3 domains in mitotic and meiotic holocentromeres In addition, holocentromere of the plant Cuscuta showed CenH3 restricted only to one to three regions per chromosome, whereas the rest of the chromatin appeared to be devoid of CenH3 (Oliveira et al. 2020). Even more extreme situation has been revealed in holocentric insects characterized by complete loss of CenH3s in at least four lineages (Drinnenberg et al. 2014). Observations based on different holocentric species analyzed so far, including Meloidogyne, lead to the conclusion that in contrast to monocentromere, holocentromeres show greater flexibility in the organization of CenH3 domains at the chromosome level.
In conclusion, our study represents the first insight into the centromere evolution and composition in an exclusively mitotic species belonging to higher eukaryotes. By generating and analyzing CenH3s from different Meloidogyne species, we have for the first time demonstrated almost complete conservation of one CenH3 protein among distant animal species and hypothesized its subspecialization, presumably associated with mitosis. We confirmed that the TRs arrays with the conserved 19-bp box span almost the entire αCenH3 centromere and represent the underlying DNA sequence of M. incognita centromere. Moreover, conserved αCenH3 and αCenH3-associated DNA in the form of 19-bp box was found in related MIG species suggesting preservation of αCenH3 centromere across mitotic Meloidogyne species. Our study disclosed for the first time a long-term conservation of CenH3 and its association with a conserved box regardless to highly evolved centromeric tandem repeats, thus suggesting the state where CenH3 and cenDNA achieved an equilibrium in which they can coexist for a long period of time. An exciting line of future investigation concerning specialization of CenH3s in Meloidogyne species would be to address the potential of the αCenH3 and εCenH3 in mitosis in comparison to meiosis in meiotic parthenogenetic M. hapla.
Materials and Methods
Nematodes
Meloidogyne incognita was cultivated on tomato (Solanum lycopersicum cultivar Saint Pierre) in greenhouse at 20 °C in laboratories from INRAE (Sophia Antipolis, France) and Agricultural institute of Slovenia (Ljubljana, Slovenia). Plants were inoculated with one to three second-stage juveniles per ml silver sand. Females or egg masses were harvested from roots under stereo microscope (SteREO Discovery.V20, Zeiss) and collected into an isotonic salt solution (M9 buffer). Egg masses were shaken in 15% bleach for 5 min to release eggs and eggs were isolated by successively passing through the sieves.
DNA and Protein Isolation
DNA was isolated from eggs using DNeasy Blood and Tissue Kit (Qiagen) in accordance to manufacturer’s protocol and quantification was done by Qubit fluorimeter (Invitrogen). For protein isolation, eggs, J2, or females were transferred in cold RIPA buffer supplemented with 10 mM PMSF and cOmplete (Roche) protease inhibitors and homogenized in Dounce homogenizer with 10–15 strokes. The homogenate was incubated with rotation on 7 rpm for 2 h at 4 °C and centrifuged for 20 min at 12,850 g at 4 °C. Supernatant containing whole cell proteins was collected and stored at −80 °C. Protein concentration was estimated using Bradford assay.
Identification and Sequence Analyses of CenH3 Proteins
To identify CenH3 sequences in four Meloidogyne species (M. incognita, M. arenaria, M. javanica, and M. hapla) a nonredundant database of protein sequences generated from the automatic annotation of sequenced genomes available at INRA website (http://meloidogyne.inra.fr/) and WormBase ParaSite (http://parasite.wormbase.org/, Howe et al. 2017) were used. BLAST search for CenH3 proteins was done using C. elegans H3 protein sequence (NCBI accession number P08898) as query. Among 23 detected CenH3 candidates two of them were truncated in N-terminal tail (supplementary fig. 1B, Supplementary Material online). Assuming that these truncated copies are the result of assembly/annotation error or represent CenH3 pseudogenes, we omitted them from the further analysis. The WormBase proteins and genes IDs with the list of CenH3s and corresponding species are available in supplementary table 1, Supplementary Material online. Multiple alignments of CenH3 candidates were generated using MUSCLE with default parameters implemented in Geneious v9.1. The structural features of CenH3 candidates such as histone-fold domain (HFD), N-terminal tail, αhelix, loops, and C-terminus were defined in accordance with Malik and Henikoff (2003). CenH3 candidates were tested for diagnostic features in HFD which include longer loop1 region and absence of glutamine, phenylalanine, and threonine at positions 69, 85, and 118, respectively, in comparison to canonical H3 (Malik and Henikoff 2003). Neighbor-joining trees of CenH3 proteins and pairwise percent identity calculations were generated using Geneious v9.1. Bootstrap values were calculated from at 1,000 replicates. Phylogenetic trees were drawn and edited using the FigTree 1.4.4 software (Rambaut 2018).
Tests for Selective Pressure
CenH3 gene sequences (supplementary fig. 2, Supplementary Material online) related to detected CenH3 protein candidates (supplementary table 1, Supplementary Material online) were generated from full-length transcripts databases from WormBase ParaSite (http://parasite.wormbase.org/, Howe et al. 2017). Multiple alignments of CenH3 nucleotide sequences were done using MUSCLE algorithm with default parameters. Alignments were further refined manually and used for downstream analyses. To determine the selective pressures acting on a CenH3 genes using the nonsynonymous/synonymous substitution rate ratio (dN/dS = ω), distance computation using Nei–Gojobori (Jukes–Cantor) substitution model implemented in MEGA version X was done (Kumar et al. 2018). All obtained comparison values (dN/dS = ω) are shown in supplementary table 4, Supplementary Material online. Generally, ω = 1 indicates neutral selection, ω < 1 purifying selection and ω > 1 positive selection. If purifying selection is relaxed, ω tends to be elevated toward 1. To assess the positive selection at the level of individual codons, Mixed Effects Model of Evolution (MEME) model was used (Murrell et al. 2012). MEME allows ω to vary across both codons and branches and infers selective regimes independently for each codon of a given alignment pooling information over branches. MEME analyses with a significance level cutoff of 0.1, correspondingly were performed through the Datamonkey server (http://datamonkey.org/). Analysis of the positive selection at individual codons was carried out on αCenH3 variants and also among different CenH3s using likelihood ratio test (LRT) values plotted against each codon site for visualization (supplementary table 5, Supplementary Material online).
Expression Profile of CenH3 Candidates
To compare gene expression among CenH3s in analyzed species or developmental stages RNA-seq data from M. incognita (PRJEB8846, Danchin et al. 2013), M. arenaria (PRJEB8845, Blanc-Mathieu et al. 2017), M. javanica (PRJEB8843; Blanc-Mathieu et al. 2017), and M. hapla (PRJEB14142) were used. The relative expressions of CenH3 genes and reference gene Disu (Hu and DiGennaro 2019) were analyzed using Bowtie2 v.2.3.0 mapper (Langmead and Salzberg 2012). Single-end reads were mapped with parameters -a and –very-sensitive for each transcriptome separately to CenH3 genes. Hits were normalized with RPKM (reads per kilobase of transcript per million mapped reads) method. This approach takes in account different CenH3 variant length and size of RNA-seq libraries dividing CenH3 hits by number of mapped reads per million reads and gene length in kilobase.
Production of CenH3 Antibodies
Polyclonal IgG antibodies against αCenH3 were raised in rabbits using peptide KELPPVKMQQKRYHKKGC. Two another antibodies were raised in rabbits and in guinea pigs using two peptides specific exclusively for βCenH3 (the peptide CTNFPRQTARKRVF specific for βCenH3-1; and the peptide KNFATKSVAGPTTMNTG specific for βCenH3-2) and peptide specific for both, β and γ CenH3 (the peptide QQQNKIKAPGEGGSL specific for βγ CenH3). Selected peptides correspond to region of divergent N-terminal tails of CenH3s (fig. 1B) and meet the parameters (amino acid phosphorylation, glycosylation profile, and secondary structure; Parker et al. 1986) that were prerequisite for suitable antibodies production and specificity. Peptide synthesis, immunization, and peptide affinity purification were performed by Pineda Service (Berlin, Germany). The preimmune sera as well as the sera samples were tested during the immunization process by Western blot monthly, to monitor the immune response. Immunizations were stopped after 90–120 days and affinity purification of the monospecific IgG fraction of CenH3 antisera was performed. Purified monospecific IgG fraction was concentrated 25× using Amicon Ultra-0.5 centrifugal filter device (Merck), and used in all downstream applications.
Western Blot
For Western blot 20 μg/reaction of whole protein extract from eggs, J2 or females were denatured in 1xLaemmli buffer (50 mM Tris–HCl pH 6.8, 10% glycerol, 2% SDS, 0.005% bromophenol blue) with 0.1 M dithiothreitol (DTT) at 65 °C for 15 min. The protein samples were separated on 4–20% Mini-PROTEAN TGX (Bio-Rad) SDS–PAGE gels at 200 V for 30 min followed with protein transfer for 40 min at 200 mA onto Amersham Protran 0.2-μm nitrocellulose membrane (GE Healthcare Life Sciences). Membranes were simultaneously incubated for 1 h in blocking solution of 5% BSA in TBST buffer (20 mM Tris, 150 mM NaCl, pH 7.6, 0.1% Tween 20) followed by overnight incubation at 4 °C with CenH3 rabbit or guinea pig polyclonal primary antibodies (dilution 1:500). HRP-linked goat antirabbit (Cell Signaling Technology 7074) or antiguinea pig (Invitrogen, A18769) antibodies diluted 1:2,000 were used as secondary antibodies. Dilution of primary and secondary antibodies was performed in TBST buffer with 5% BSA. Signals were detected using the Pierce ECL Western Blotting substrate (Thermo Scientific) and Amersham Hyperfilm ECL X-ray films (GE Healthcare Life Sciences). The α-tubulin mouse monoclonal antibody (Sigma Aldrich, T6199) used for combined α-tubulin and αCenH3 immunofluorescence assay was tested by Western blot as described above. H3K9 polyclonal antibody (Abcam, ab8898) was used as a positive control in Western-blot experiments.
Peptide Dot Blot
The assay was performed using specific antibodies and dilutions of several peptides which were plotted onto nitrocellulose membrane. The specificity of antibodies we produced against CenH3s was tested on specific and nonspecific peptides. Synthetized 14–18mer peptides were dissolved in PBS to 1 mg/ml and then further diluted to concentration of 1, 0.1, and 0.01 µg/µl. The series of peptide dilutions in form of 2 µl spots were applied to the nitrocellulose membrane; the membrane was dried for 30 min and blocked in 5% BSA in TBST buffer for 1 h with mild shaking at RT. Affinity purified monospecific IgG fractions of anti-CenH3s were used as primary antibodies, diluted 1:500 in blocking buffer. For βCenH3-1, βCenH3-2, βγCenH3 antibody testing, two rabbits or three guinea pigs antibodies were pooled together when testing rabbit of guinea pig antibodies, respectively. After 1 h incubation at RT, membranes were washed 3 × 5 min with TBST and incubated with secondary antibody (HRP-conjugated antirabbit; Cell Signaling Technology, 7074) or antiguinea pig (Invitrogen, A18769) antibody diluted 1:1,000 in blocking buffer for 1 h at RT followed by 3 × 5 min washes with TBST. The final detection was carried out as described for the Western blot.
Microscope Slide Preparation
Slides were prepared from isolated reproductive tissue (ovaries and uterus) of M. incognita, M. arenaria, and M. javanica females using cytospin technique. Samples were collected in 10 μg/ml colcemid (Roche) and pierced using needle. The ovaries and uteri were isolated and incubated for 1 h to overnight at 4 °C. Samples were washed with PBS and then mixed for 30 s with microtube homogenizer. Suspension was transferred to Dounce homogenizer and tissue parts were broken with 30 strokes of pestle A followed by straining through 100- and 40-μm cell strainers. Volume of suspension is adjusted with PBS up to 400 μl for loading into one Cytospin funnel that corresponds to five to ten females per one coated Cytoslide (Shandon, ThermoFisher Scientific). Slides were spun for 10 min at 1,200 rpm by Cytospin 4 cytocentrifuge (Shandon, ThermoFischer Scientific), dried, and fixed by immersing into ice-cold fixative for 20 min, completely dried and stored. Several fixatives were tested and methanol: acetone (1:1) incubation for 20 min at −20 °C showed the best results in IF and IF-FISH analyses. For chromatin fiber preparation the best results were obtained following the protocol described in Frum et al. (2013) with some modifications. Briefly, slides were dried after cytospinning and incubated with 15 μl of freshly prepared mild SDS lysis buffer (0.2% SDS, 200 mM Tris–HCl pH 7.5, 50 mM EDTA, 1 mM PMSF) by covering it with 18 mm square coverslips. Lysis reaction was performed at room temperature for 30 min and coverslip was carefully removed with a blade. Slides were then fixed in methanol:acetone (1:1) for 20 min at −20 °C, completely dried, and stored at −80 °C until use.
DNA Probe Preparation
FISH probes for centromere candidates were obtained by PCR labeling with biotin-16-dUTP (Jena BioScience) using genomic DNA. Primers for centromeric candidates were designed based on monomer sequences enriched in ChIP analyses (Cl25m1i, Cl16m1i, Cl16m2i, and Cl16m3i) using Primer 3 (Rozen and Skaletsky 2000); the primer sequences are listed in supplementary figure 8A, Supplementary Material online. PCR reactions were performed in 25 μl reaction volume containing 0.02 ng of gDNA, 0.1 μM primers, 2.5 mM MgCl2, 1× Green GoTaq Reaction Buffer, 0.1 mM dNTPs, and 0.5 U of GoTaq G2 DNA Polymerase (Promega). Thirty-five amplification cycles (20 s at 95 °C, 20 at 58 °C annealing temperature and 40 s at 72 °C) were run. ChIPped-DNA was labeled using random priming approach with Klenow fragment in accordance to manufacturer’s protocol (New England BioLabs). Characteristic ladder-like profile with expected fragment sizes was taken as proof for specificity of hybridization probes (supplementary fig. 8B, Supplementary Material online).
DNA Dot Blot
For genomic DNA, 50, 100, and 200 ng were spotted onto positively charged nylon membrane (Roche). PCR products corresponding to CL25m1, CL16m1, and CL16m3/32m1 fragments were spotted in the amounts of 0.5, 1, 2, 4, and 8 ng. Hybridization was done in 250 mM phosphate buffer pH 7.2, 1 mM EDTA pH 8, 20% SDS, and 0.5% Blocking Reagent (Roche) with 50 ng of biotin-labeled probes at 65 °C with agitation overnight. Posthybridization washes were done in 20 mM Na2HPO4, 1 mM EDTA, and 1% SDS 3 × 20 min at 62 °C. Detection was carried out using streptavidin-AP-conjugate (1:5,000, Roche) followed by chemiluminescence with AP substrate CDP-Star (1:50, Roche). Dot blot intensities were compared using ImageJ with measurement of mean gray values that were inverted and normalized for background.
Immunofluorescence
Affinity purified monospecific IgG fractions of anti-CenH3 were concentrated using Amicon Ultra-0.5 Centrifugal Filter Unit (Merck) with 30 kDa cutoff. Slides with cytosmear or chromatin fibers were blocked with 2.5% BSA in PBST (PBS, 0.2% Tween 20) and incubated with anti-CenH3 (1:400 dilution) at 37 °C overnight. After 3 × 5 min washes in PBST, slides were incubated with secondary Alexa594 antirabbit (Abcam, ab150080) or Alexa488 antiguinea pig (Abcam, ab150158) antibodies, diluted 1:1,000 dilution in blocking solution for 1 h at 37 °C. After two washes in PBST for 5 min and one wash in PBS for 5 min, the slides were counterstained with DAPI or continued with the FISH protocol. In double immunostaining with two anti-CenH3, the primary as well as secondary antibodies were incubated together.
For combined α-tubulin and αCenH3 immunofluorescence, slides were after O/N incubation with αCenH3 antibody the slides were washed 3 × 5 min with PBST and then incubated with α-tubulin mouse monoclonal antibody (Sigma–Aldrich, T6199) diluted 1:1,000 in PBST with 2.5% BSA for 3 h at 37 °C. After 3 × 5 min washes in PBST and additional blocking for 1 h at 37 °C, slides were consecutively incubated with secondary Alexa594 antirabbit (Abcam, ab150080) and CF488 antimouse (Sigma–Aldrich, SAB4600035) antibodies followed by washes and DAPI staining as described above.
Fluorescence In Situ Hybridization
For combined detection of the αCenH3 and centromeric candidates, immunodetection procedure was followed by FISH. After IF detection and washing, slides were immediately pretreated for FISH washing in 45% acetic acid for 10 min and in 2xSSC for 5 min. After RNase A treatment for 30 min at 37 °C slides were washed 3 × 5 min with PBS and fixed with 1% formaldehyde in PBS with 50 mM MgCl2. After washing 2 × 5 min with PBS, slides were dehydrated in a series of cold ethanol. Denaturation was carried out in 70% formamide in 2xSSC at 70 °C for 2 min and slides were dehydrated and air dried. Lyophilized-specific probe (100 ng/slide) was denatured at 75 °C for 5 min in 15 μl of hybridization buffer (60% formamide in dextran sulfate buffer [20% DeSO4, 4xSSC, 50 mM Na-phosphate pH 7.0]) and chilled on ice. Hybridization was performed at 37 °C overnight. Posthybridization washes were carried out with 50% formamide in 2xSSC for four times during 5 min at 37 °C. Slides were blocked with 5% Blocking Reagent (Roche) in 4xSSC. Immunodetection was performed with fluorescein avidin D and biotinylated antiavidin D system (Vector Laboratories). Slides were counterstained with DAPI, dried, and embedded in Mowiol 4-88 mounting medium (Sigma–Aldrich). To minimize nonspecific staining slides with chromatin fibers were incubated in Image-iT FX Signal Enhancer (Invitrogen) for 30 min.
Primed In Situ Labeling
For combined detection of the αCenH3 and 19-bp box sequence immunodetection procedure was followed by primed in situ labeling (PRINS). Slides were pretreated and denatured as for FISH experiments. The reaction mixture was prepared in 50 µl containing 2 µM primer (19-bp box; TCGGGCCTTCGGCCCTCGC), 2.5 µM MgCl2, 150 µM each of dATP, dCTP, dGTP, 96 µM dTTP and 54 µM biotin-16-dUTP, 1 U of GoTaq G2 DNA Polymerase (Promega), and 1× Colorless GoTaq Reaction Buffer (Promega). On each prewarmed slide, 25 µl of prepared mixture was applied, covered with coverslip, sealed, and continued to heat at appropriate annealing and elongation temperature of 65 °C for 30 min. Reaction was stopped by washing in 50 mM NaCl, 50 mM EDTA, pH 8 buffer for 5 min at 65 °C followed by 3 × 5 min washes in 4× SSC with 0.05% Tween 20. Immunodetection and DAPI staining were afterward performed in the same manner as for FISH.
Images Processing and Quantification
Microscopic images were recorded using confocal laser scanning microscope Leica TCS SP8 X (Leica Microsystems) equipped with an HC PL APO CS2 63×/1.40 oil objective, 405 nm diode laser, and a supercontinuum excitation laser (Leica Microsystems). Images were acquired as z-stacks with five slices and average step size of 0.5 µm per cytosmear. Each fluorochrome was capture separately and images were merged and analyzed using Image J and Adobe Photoshop software. Images quantification was done using CellProfiler (https://cellprofiler.org) with align and measure colocalization modules. Ten nuclei were selected as separate regions of interests on original images acquired with confocal microscopy where grayscale separated red and green channels were analyzed. Manders coefficients with Costes automated thresholding were calculated for channel interrelationship and all values were shown on graphs using GraphPad Prism version 8.
Native Chromatin Immunoprecipitation Followed by Sequencing (ChIP-Seq)
Native ChIP was performed, with some modifications, according to protocol previously described for nematode C. elegans (Steiner and Henikoff 2014). Briefly, approximately 500 mg of frozen M. incognita eggs were ground using liquid nitrogen. Suspension was homogenized for 2 min with pestle A and 4 min with pestle B in 3 ml of ice-cold buffer (15 mM Tris–HCl pH 7.5, 2 mM MgCl2, 340 mM sucrose, 0.2 mM spermine, 0.5 mM spermidine, 0.1% Triton X-100, and 0.5 mM PMSF) using Dounce homogenizer. Prolonged homogenization step has previously shown to be crucial for higher chromatin yield in M. incognita (Perfus-Barbeoch et al. 2014). Cellular debris was removed by spinning for 2 min at 100 g. Supernatant was centrifuged at 1,000 × g for 10 min and nuclei were gently resuspended in 250 μl of buffer (10 mM Tris–HCl pH 7.5, 2 mM MgCl2, 0.5 mM PMSF) and incubated for 5 min at 37 °C with addition of 2 mM CaCl2. Different MNase (Thermo Scientific, Cat No. 88216) concentrations were tested and optimal digestion with prevalent fraction of mononucleosome ∼150 bp was obtained with 0.2 U/µl eggs and 0.4 U/µl eggs for 2 min. The reactions were stopped by adding EDTA (final concentration of 20 mM). Chromatin was solubilized by cavitation using needle extraction (ten times with 26 gauge) and suspension was centrifuged at 1,000 × g for 5 min. The supernatant containing well-digested chromatin was used for ChIP (supplementary fig. 5A, Supplementary Material online).
ChIP was done using Dynabeads Protein A Immunoprecipitation Kit (Invitrogen) in accordance to manufacturer’s protocol, with some modifications. Three ChIP experiments with chromatin from two different MNase digestions using two different αCenH3 antibody concentration (10 and 30 μg) were performed (supplementary fig. 5B, Supplementary Material online). Beads were first washed with PBS followed by binding of αCenH3 antibody in 200 μl of Ab Binding and Washing Buffer for 2 h at 4 °C. Beads–antibody complex were washed and separated using a magnetic rack. For input control sample, 10% of starting chromatin was stored at 4 °C until the last step of elution. ChIP reactions were done with 50 μl of beads–antibody complex and 200 μl of isolated chromatin fraction (∼6 μg). Chromatin was diluted 1:2 with ChIP dilution buffer (50 mM NaCl, 20 mM Tris–HCl, pH 7.5, 5 mM EDTA, 0.2 mM PMSF, 1× cOmplete protease inhibitor; Roche) as used in Neumann et al. (2012). Antibody bound beads were incubated with diluted chromatin O/N on rotation at 4 °C. Precipitated immunocomplexes were washed three times for 5 min with 200 μl of Washing buffer (Invitrogen) and ChIPped chromatin was eluted two times for 15 min at 65 °C with 100 μl of elution buffer (1% SDS, 0.1 M NaHCO3). After washing, beads were resuspended in TE buffer followed by RNase and Proteinase K treatment to release DNA from the immunoprecipitated nucleosomes. Finally, ChIPped DNA and input were isolated with DNA purification kit (Qiagen) and eluted in 50 μl of 10 mM Tris–Cl, pH 8.5 buffer. About 5 μg of normal rabbit IgG (Cell Signaling Technology, No. 2729) was included as a negative control in each ChIP experiment in order to optimize the experimental conditions. The relatively high ratio of antialpha CenH3 ChIPed DNA versus rabbit IgG ChIPed DNA was used as an indicator of a successful ChIP experiment. Genomic DNA for WGS sequencing was isolated from eggs using DNeasy Blood and Tissue Kit (Qiagen). Library construction (KAPA Hyper Prep kit) and sequencing were done via multiplexing using Illumina HiSeq technology which produced 151-bp paired-end reads (AdmeraHealth). Raw Illumina Input and ChIP-sequencing reads have been deposited to NCBI BioProject database under the study accession number (PRJNA639449).
ChIP-Seq Data Analysis
Sequencing data were first tested with FastQC (Andrews 2010) and preprocessed to get high-quality reads. About 2 million of 151-bp pair-end sequence reads were mapped to the M. incognita genome reference genome GCA_900182535.1 (Blanc-Mathieu et al. 2017) using the Burrows–Wheeler Aligner (BWA) with default parameters (Li and Durbin 2009).
Repetitive part of the M. incognita genome was analyzed using graph-based clustering which utilizes grouping of reads based on their shared similarity. A total of 1 million paired reads (0.8 genome coverage) was used for RepeatExplorer (Novák et al. 2013) clustering. The output of analysis was clusters, composed of contigs with overlapping sequences, where each cluster represents a repetitive element. One million of single-end reads of ChIP and input were mapped to the clusters with ChIP-seq Mapper (Neumann et al. 2012). Repeats enriched in ChIPped DNA were identified by elevated proportions of reads from ChIP versus input data. The threshold for ChIP versus input ratio >1.5 was chosen. This relatively low threshold for ChIP enrichment was also selected previously in studies where CenH3 was associated with many different sequences (e.g., in the plant Beta; Kowar et al. 2016). Recently, it has been proposed that both the abundance and enrichment have to be taken into account in ChIP analyses to estimate of sequences associated with the centromeres (Talbert et al. 2018). Therefore, 100 most abundant repeat clusters, with at least 0.02% M. incognita genome content were presented. In addition to cluster analysis, mapping of ChIP and input on 10,000 contigs (with at least 0.002% genome proportion) using criteria that each read can only be assigned to one contig was done. The analysis was performed for all three ChIP replicates and enriched contigs were determined based on calculated ChIP/input hit ratio (supplementary table 6, Supplementary Material online). The centromeric candidates (from clusters and contigs analyses) were further analyzed for tandem repeats using the Tandem Repeats Finder server (https://tandem.bu.edu/trf/trf.html) with default parameters (Benson 1999). Sequence monomer analyses such as multiple sequence comparison, GC content, pairwise identity, and motif search were done using Genious v9.1, together with implemented DNA-fold Vienna package for secondary structure analysis. Surrounding TR regions were investigated by assembling contigs using de novo assembler. In order to find putative centromere candidates in related M. incognita species, the first step was clustering of raw Illumina WGS data of M. arenaria (SRR4242477) and M. javanica (SRR4242459) raw Illumina WGS data (Szitenberg et al. 2017) using RepeatExplorer. The second step included detection of clusters/contigs which contain TRs-associated with M. incognita-specific centromere sequence motif. The selected putative centromeric TR arrays in closely related species were further analyzed as described for M. incognita.
For validation of ChIP enrichment obtained on repetitive part of the M. incognita genome preprocessed and subsampled ChIP and input reads for each replicate were mapped to M. incognita genome assembly (Blanc-Mathieu et al. 2017) using bowtie2 (Langmead and Salzberg 2012) with –very-sensitive-local and -a option on the Galaxy platform. For normalizing read counts bamCompare (Ramírez et al. 2016) was used with default parameters, where difference was computed by subtracting read number of input from corresponding ChIP sample. Results were visualized using Integrative Genomics Viewer (Robinson et al. 2011).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank M. Pavlek for performing the preliminary Western blot experiments and L. Horvat for assistance in high-resolution microscopy. This work has been supported by grant IP-2014-09-3183 from Croatian Science Foundation (https://www.hrzz.hr/default.aspx?id=47) to M.P. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
References
- Abad P, Gouzy J, Aury J-M, Castagnone-Sereno P, Danchin EGJ, Deleury E, Perfus-Barbeoch L, Anthouard V, Artiguenave F, Blok VC, et al. 2008. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol. 26(8):909–915. [DOI] [PubMed] [Google Scholar]
- Aird D, Ross MG, Chen W-S, Danielsson M, Fennell T, Russ C, Jaffe DB, Nusbaum C, Gnirke A.. 2011. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 12(2):R18–R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C, Cardone MF, Catacchio CR, Antonacci F, O’Brien SJ, Ryder OA, Purgato S, Zoli M, Della Valle G, Eichler EE, et al. 2011. Genome-wide characterization of centromeric satellites from multiple mammalian genomes. Genome Res. 21(1):137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Karpen GH.. 2008. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 9(12):923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Ávila Robledillo L, Neumann P, Koblížková A, Novák P, Vrbová I, Macas J.. 2020. Extraordinary sequence diversity and promiscuity of centromeric satellites in the legume tribe Fabeae. Mol Biol Evol. 37(8):2341–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barra V, Fachinetti D.. 2018. The dark side of centromeres: types, causes and consequences of structural abnormalities implicating centromeric DNA. Nat Commun. 9(1):4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc-Mathieu R, Perfus-Barbeoch L, Aury JM, Da Rocha M, Gouzy J, Sallet E, Martin-Jimenez C, Bailly-Bechet M, Castagnone-Sereno P, Flot JF, et al. 2017. Hybridization and polyploidy enable genomic plasticity without sex in the most devastating plant-parasitic nematodes. PLoS Genet. 13(6):e1006777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Karpen GH.. 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 3(8):730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S.. 1999. A histone-H3-like protein in C. elegans. Nature 401(6753):547–548. [DOI] [PubMed] [Google Scholar]
- Castagnone-Sereno P, Danchin EGJ.. 2014. Parasitic success without sex – the nematode experience. J Evol Biol. 27(7):1323–1333. [DOI] [PubMed] [Google Scholar]
- Castagnone-Sereno P, Danchin EGJ, Perfus-Barbeoch L, Abad P.. 2013. Diversity and evolution of root-knot nematodes, genus Meloidogyne: new insights from the genomic era. Annu Rev Phytopathol. 51(1):203–220.23682915 [Google Scholar]
- Chmátal L, Gabriel SI, Mitsainas GP, Martínez-Vargas J, Ventura J, Searle JB, Schultz RM, Lampson MA.. 2014. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr Biol. 24(19):2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JL, Henikoff S.. 2004. Adaptive evolution of the histone fold domain in centromeric histones. Mol Biol Evol. 21(9):1712–1718. [DOI] [PubMed] [Google Scholar]
- Cuacos M, Franklin FCH, Heckmann S.. 2015. Atypical centromeres in plants – what they can tell us. Front Plant Sci. 6:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchin EGJ, Arguel MJ, Campan-Fournier A, Perfus-Barbeoch L, Magliano M, Rosso MN, Da Rocha M, Da Silva C, Nottet N, Labadie K, et al. 2013. Identification of novel target genes for safer and more specific control of root-knot nematodes from a Pan-genome mining. PLoS Pathog. 9(10):e1003745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF. 2001. Here, there, and everywhere: kinetochore function on holocentric chromosomes. J Cell Biol. 153:33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg IA, deYoung D, Henikoff S, Malik HI.. 2014. Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. Elife 3:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont M, Fachinetti D.. 2017. DNA sequences in centromere formation and function. Prog Mol Subcell Biol. 56:305–336. [DOI] [PubMed] [Google Scholar]
- Evtushenko EV, Elisafenko EA, Gatzkaya SS, Lipikhina YA, Houben A, Vershinin AV.. 2017. Conserved molecular structure of the centromeric histone CENH3 in SecaleS and its phylogenetic relationships. Sci Rep. 7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finseth FR, Dong Y, Saunders A, Fishman L.. 2015. Duplication and adaptive evolution of a key centromeric protein in mimulus, a genus with female meiotic drive. Mol Biol Evol. 32(10):2694–2706. [DOI] [PubMed] [Google Scholar]
- Frum RA, Deb S, Deb SP.. 2013. Use of the DNA fiber spreading technique to detect the effects of mutant p53 on DNA replication. Methods Mol Biol. 962:147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavís M, Méndez-Lago M, Gabelica V, Whitehead SL, González C, Villasante A.. 2015. The structure of an endogenous Drosophila centromere reveals the prevalence of tandemly repeated sequences able to form i-motifs. Sci Rep. 5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García LE, Sánchez-Puerta MV.. 2015. Comparative and evolutionary analyses of Meloidogyne spp. based on mitochondrial genome sequences. PLoS One 10(3):e0121142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R, Rechtsteiner A, Yuen KW, Muroyama A, Egelhofer T, Gaydos L, Barron F, Maddox P, Essex A, Monen J, et al. 2012. An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature 484(7395):534–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi C, De VP, De LF, Natilla A, Lanave C, Pesole G.. 2002. Structural and evolutionary analysis of the ribosomal genes of the parasitic nematode Meloidogyne artiellia suggests its ancient origin. Mol Biochem Parasitol. 124(1–2):91–94. [DOI] [PubMed] [Google Scholar]
- Guerra M, Cabral G, Cuacos M, González-García M, González-Sánchez M, Vega J, Puertas M.. 2010. Neocentrics and holokinetics (Holocentrics): chromosomes out of the centromeric rules. Cytogenet Genome Res. 129(1–3):82–96. [DOI] [PubMed] [Google Scholar]
- Hartley G, O’Neill RJ.. 2019. Centromere repeats: hidden gems of the genome. Genes (Basel). 10(3):223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann S, Macas J, Kumke K, Fuchs J, Schubert V, Ma L, Novák P, Neumann P, Taudien S, Platzer M, et al. 2013. The holocentric species Luzula elegans shows interplay between centromere and large-scale genome organization. Plant J. 73(4):555–565. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS.. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293(5532):1098–1103. [DOI] [PubMed] [Google Scholar]
- Hirsch CD, Wu Y, Yan H, Jiang J.. 2009. Lineage-specific adaptive evolution of the centromeric protein CENH3 in diploid and allotetraploid Oryza species. Mol Biol Evol. 26(12):2877–2885. [DOI] [PubMed] [Google Scholar]
- Howe KL, Bolt BJ, Shafie M, Kersey P, Berriman M.. 2017. WormBase ParaSite − a comprehensive resource for helminth genomics. Mol Biochem Parasitol. 215:2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, DiGennaro PM.. 2019. Identification of suitable Meloidogyne spp. housekeeping genes. J Nematol. 51:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugall A, Stanton J, Moritz C.. 1999. Reticulate evolution and the origins of ribosomal internal transcribed spacer diversity in apomictic Meloidogyne. Mol Biol Evol. 16(2):157–164. [DOI] [PubMed] [Google Scholar]
- Ishii T, Karimi-Ashtiyani R, Banaei-Moghaddam AM, Schubert V, Fuchs J, Houben A.. 2015. The differential loading of two barley CENH3 variants into distinct centromeric substructures is cell type- and development-specific. Chromosome Res. 23(2):277–284. [DOI] [PubMed] [Google Scholar]
- Iwata-Otsubo A, Dawicki-Mckenna JM, Akera T, Falk SJ, Chmátal L, Yang K, Sullivan BA, Schultz RM, Lampson MA, Black BE.. 2017. Expanded satellite repeats amplify a discrete CENP-A nucleosome assembly site on chromosomes that drive in female meiosis. Curr Biol. 27(15):2365–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeche L, Nguyen HD, Buisson R, Zou L.. 2018. A mitosis-specific and R loop–driven ATR pathway promotes faithful chromosome segregation. Science 359(6371):108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Wang J, Neff A, Kratzer S, Kimura H, Davis RE.. 2016. Differential chromosomal localization of centromeric histone CENP-A contributes to nematode programmed DNA elimination. Cell Rep. 16(9):2308–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinathan S, Henikoff S.. 2018. Non-B-form DNA is enriched at centromeres. Mol Biol Evol. 35(4):949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe A, Nasuda S, Charlesworth D.. 2006. Duplication of centromeric histone H3 (HTR12) gene in Arabidopsis halleri and A. lyrata, plant species with multiple centromeric satellite sequences. Genetics 174(4):2021–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowar T, Zakrzewski F, Macas J, Kobližková A, Viehoever P, Weisshaar B, Schmidt T.. 2016. Repeat composition of CenH3-chromatin and H3K9me2-marked heterochromatin in sugar beet (Beta vulgaris). BMC Plant Biol. 16(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursel LE, Malik HS.. 2017. Recurrent gene duplication leads to diverse repertoires of centromeric histones in Drosophila species. Mol Biol Evol. 34(6):1445–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursel LE, Malik HS.. 2019. Gametic specialization of centromeric histone paralogs in Drosophila virilis. bioRxiv doi:10.1101/530295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursel LE, Welsh FC, Malik HS.. 2020. Ancient coretention of paralogs of Cid centromeric histones and Cal1 chaperones in mosquito species. Mol Biol Evol. 37(7):1949–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I, Koroleva O, Rutten T, Fuchs J, Schubert V, Moraes I, Koszegi D, Schubert I.. 2011. Knockdown of CENH3 in Arabidopsis reduces mitotic divisions and causes sterility by disturbed meiotic chromosome segregation. Plant J. 68(1):40–50. [DOI] [PubMed] [Google Scholar]
- Lermontova I, Schubert V, Fuchs J, Klatte S, Macas J, Schubert I.. 2006. Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell 18(10):2443–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt DH. 2008. Genetic tests of ancient asexuality in Root Knot Nematodes reveal recent hybrid origins. BMC Evol Biol. 16:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS. 2009. The centromere-drive hypothesis: a simple basis for centromere complexity. Prog Mol Subcell Biol. 48:33–52. [DOI] [PubMed] [Google Scholar]
- Malik HS, Bayes JJ.. 2006. Genetic conflicts during meiosis and the evolutionary origins of centromere complexity. Biochem Soc Trans. 34(4):569–573. [DOI] [PubMed] [Google Scholar]
- Malik HS, Henikoff S.. 2001. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157(3):1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Henikoff S.. 2003. Phylogenomics of the nucleosome. Nat Struct Mol Biol. 10(11):882–891. [DOI] [PubMed] [Google Scholar]
- Marques A, Ribeiro T, Neumann P, Macas J, Novák P, Schubert V, Pellino M, Fuchs J, Ma W, Kuhlmann M, et al. 2015. Holocentromeres in Rhynchospora are associated with genome-wide centromere-specific repeat arrays interspersed among euchromatin. Proc Natl Acad Sci U S A. 112(44):13633–13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Nakano M, Ohzeki J.. 2004. The role of CENP-B and α-satellite DNA: de novo assembly and epigenetic maintenance of human centromeres. Chromosome Res. 12(6):543–556. [DOI] [PubMed] [Google Scholar]
- McNulty SM, Sullivan BA.. 2018. Alpha satellite DNA biology: finding function in the recesses of the genome. Chromosome Res. 26(3):115–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters DP, Bradnam KR, Young HA, Telis N, May MR, Ruby JG, Sebra R, Peluso P, Eid J, Rank D, et al. 2013. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 14(1):R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters DP, Paliulis LV, Korf IF, Chan SWL.. 2012. Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 20(5):579–593. [DOI] [PubMed] [Google Scholar]
- Meštrović N, Castagnone-Sereno P, Plohl M.. 2006a. High conservation of the differentially amplified MPA2 satellite DNA family in parthenogenetic root-knot nematodes. Gene 376(2):260–267. [DOI] [PubMed] [Google Scholar]
- Meštrović N, Castagnone-Sereno P, Plohl M.. 2006b. Interplay of selective pressure and stochastic events directs evolution of the MEL172 satellite DNA library in root-knot nematodes. Mol Biol Evol. 23(12):2316–2325. [DOI] [PubMed] [Google Scholar]
- Meštrović N, Pavlek M, Car A, Castagnone-Sereno P, Abad P, Plohl M.. 2013. Conserved DNA motifs, including the CENP-B Box-like, are possible promoters of satellite DNA array rearrangements in nematodes. PLoS One 8(6):e67328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monen J, Hattersley N, Muroyama A, Stevens D, Oegema K, Desai A.. 2015. Separase cleaves the N-tail of the CENP-A related protein CPAR-1 at the meiosis I metaphase-anaphase transition in C. elegans. PLoS One 10(4):e0125382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monen J, Maddox PS, Hyndman F, Oegema K, Desai A.. 2005. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat Cell Biol. 7(12):1248–1255. [DOI] [PubMed] [Google Scholar]
- Moore LL, Morrison M, Roth MB.. 1999. HCP-1, a protein involved in chromosome segregation, is localized to the centromere of mitotic chromosomes in Caenorhabditis elegans. J Cell Biol. 147(3):471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes ICR, Lermontova I, Schubert I.. 2011. Recognition of A. thaliana centromeres by heterologous CENH3 requires high similarity to the endogenous protein. Plant Mol Biol. 75(3):253–261. [DOI] [PubMed] [Google Scholar]
- Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL.. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8(7):e1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Mendoza MI, Pérez-Arques C, Panchal S, E.Nicolás F, Mondo SJ, Ganguly P, Pangilinan J, Grigoriev IV, Heitman J, Sanyal K, et al. 2019. Early diverging fungus Mucor circinelloides lacks centromeric histone CENP-A and displays a mosaic of point and regional centromeres. Curr Biol. 29(22):3791–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, Navrátilová A, Schroeder-Reiter E, Koblížková A, Steinbauerová V, Chocholová E, Novák P, Wanner G, Macas J.. 2012. Stretching the rules: monocentric chromosomes with multiple centromere domains. PLoS Genet. 8(6):e1002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, Pavlíková Z, Koblížková A, Fuková I, Jedličková V, Novák P, Macas J.. 2015. Centromeres off the hook: massive changes in centromere size and structure following duplication of CenH3 gene in Fabeae species. Mol Biol Evol. 32(7):1862–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P, Neumann P, Pech J, Steinhaisl J, Macas J.. 2013. RepeatExplorer: a galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29(6):792–793. [DOI] [PubMed] [Google Scholar]
- Ohzeki J, Nakano M, Okada T, Masumoto H.. 2002. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol. 159(5):765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira L, Neumann P, Jang TS, Klemme S, Schubert V, Koblížková A, Houben A, Macas J.. 2020. Mitotic spindle attachment to the holocentric chromosomes of Cuscuta europaea does not correlate with the distribution of CENH3 chromatin. Front Plant Sci. 10:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman CH, Bird DM, Williamson VM, Rokhsar DS, Burke M, Cohn J, Cromer J, Diener S, Gajan J, Graham S, et al. 2008. Sequence and genetic map of Meloidogyne hapla: A compact nematode genome for plant parasitism. Proc Natl Acad Sci U S A. 105(39):14802–14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JMR, Guo D, Hodges RS.. 1986. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry 25(19):5425–5432. [DOI] [PubMed] [Google Scholar]
- Perfus-Barbeoch L, Castagnone-Sereno P, Reichelt M, Fneich S, Roquis D, Pratx L, Cosseau C, Grunau C, Abad P.. 2014. Elucidating the molecular bases of epigenetic inheritance in non-model invertebrates: the case of the root-knot nematode Meloidogyne incognita. Front Physiol. 5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras FM, Nergadze SG, Magnani E, Bertoni L, Attolini C, Khoriauli L, Raimondi E, Giulotto E.. 2010. Uncoupling of satellite DNA and centromeric function in the genus Equus. PLoS Genet. 6(2):e1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plohl M, Meštrović N, Mravinac B.. 2014. Centromere identity from the DNA point of view. Chromosoma 123(4):313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. 2018. FigTree v1.4.4, a graphical viewer of phylogenetic trees. Available from: http://tree.bio.ed.ac.uk/software/figtree/.
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T.. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44(W1):W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi M, Shibata F, Ramahi JS, Nagaki K, Chen C, Murata M, Chan SWL.. 2011. Meiosis-specific loading of the centromere-specific histone CENH3 in Arabidopsis thaliana. PLoS Genet. 7(6):e1002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro T, Buddenhagen CE, Thomas WW, Souza G, Pedrosa-Harand A.. 2018. Are holocentrics doomed to change? Limited chromosome number variation in Rhynchospora Vahl (Cyperaceae). Protoplasma 255(1):263–272. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP.. 2011. Integrative genomics viewer. Nat Biotechnol. 29(1):24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H.. 2000. WWW for general users and for biologist programmers. In: Misener S, Krawetz SA, editors. Bioinformatics methods and protocols. Methods in molecular biology. Vol. 132. Totowa (NJ: ): Humana Press. p. 365–386. [DOI] [PubMed] [Google Scholar]
- Sanei M, Pickering R, Kumke K, Nasuda S, Houben A.. 2011. Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc Natl Acad Sci U S A. 108(33):E498–E505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler MG, Swanson W, Thomas PJ, Program NCS, Green ED.. 2010. Adaptive evolution of foundation kinetochore proteins in primates. Mol Biol Evol. 27(7):1585–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoak EM, Stein P, Schultz RM, Lampson MA, Black BE.. 2016. Long-term retention of CENP-A nucleosomes in mammalian oocytes underpins transgenerational inheritance of centromere identity. Curr Biol. 26(8):1110–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner FA, Henikoff S.. 2014. Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. Elife 3:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner FA, Henikoff S.. 2015. Diversity in the organization of centromeric chromatin. Curr Opin Genet Dev. 31:28–35. [DOI] [PubMed] [Google Scholar]
- Szitenberg A, Salazar-Jaramillo L, Blok VC, Laetsch DR, Joseph S, Williamson VM, Blaxter ML, Lunt DH.. 2017. Comparative genomics of apomictic root-knot nematodes: hybridization, ploidy, and dynamic genome change. Genome Biol Evol. 9(10):2844–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Bayes JJ, Henikoff S.. 2009. Evolution of centromeres and kinetochores: a two-part fugue. In: De Wulf P, Earnshaw WC, editors. The kinetochore: from molecular discoveries to cancer therapy. New York: Springer New York. p. 1–37. [Google Scholar]
- Talbert PB, Bryson TD, Henikoff S.. 2004. Adaptive evolution of centromere proteins in plants and animals. J Biol. 3(4):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S.. 2020. What makes a centromere? Exp Cell Res. 389(2):111895. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S.. 2002. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14(5):1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Sivakanthan K, Henikoff S.. 2018. Simple and complex centromeric satellites in Drosophila sibling species. Genetics 208(3):977–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantaphyllou AC. 1981. Oogenesis and the chromosomes of the parthenogenetic root-knot nematode Meloidogyne incognita. J Nematol. 13:95–104. [PMC free article] [PubMed] [Google Scholar]
- Vermaak D, Hayden HS, Henikoff S.. 2002. Centromere targeting element within the histone fold domain of Cid. Mol Cell Biol. 22(21):7553–7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zhao H, Zhang T, Zeng Z, Zhang P, Zhu B, Han Y, Braz GT, Casler MD, Schmutz J, et al. 2018. Amplification and adaptation of centromeric repeats in polyploid switchgrass species. New Phytol. 218(4):1645–1657. [DOI] [PubMed] [Google Scholar]
- Yuan J, Guo X, Hu J, Lv Z, Han F.. 2015. Characterization of two CENH3 genes and their roles in wheat evolution. New Phytol. 206(2):839–851. [DOI] [PubMed] [Google Scholar]
- Zedek F, Bureš P.. 2012. Evidence for centromere drive in the holocentric chromosomes of Caenorhabditis. PLoS One 7(1):e30496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zedek F, Bureš P.. 2016a. CenH3 evolution reflects meiotic symmetry as predicted by the centromere drive model. Sci Rep. 6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zedek F, Bureš P.. 2016b. Absence of positive selection on CenH3 in Luzula suggests that holokinetic chromosomes may suppress centromere drive. Ann Bot. 118(7):1347–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.