Abstract
Revealing the mechanisms underlying the breathtaking morphological diversity observed in nature is a major challenge in Biology. It has been established that recurrent mutations in hotspot genes cause the repeated evolution of morphological traits, such as body pigmentation or the gain and loss of structures. To date, however, it remains elusive whether hotspot genes contribute to natural variation in the size and shape of organs. As natural variation in head morphology is pervasive in Drosophila, we studied the molecular and developmental basis of differences in compound eye size and head shape in two closely related Drosophila species. We show differences in the progression of retinal differentiation between species and we applied comparative transcriptomics and chromatin accessibility data to identify the GATA transcription factor Pannier (Pnr) as central factor associated with these differences. Although the genetic manipulation of Pnr affected multiple aspects of dorsal head development, the effect of natural variation is restricted to a subset of the phenotypic space. We present data suggesting that this developmental constraint is caused by the coevolution of expression of pnr and its cofactor u-shaped (ush). We propose that natural variation in expression or function of highly connected developmental regulators with pleiotropic functions is a major driver for morphological evolution and we discuss implications on gene regulatory network evolution. In comparison to previous findings, our data strongly suggest that evolutionary hotspots are not the only contributors to the repeated evolution of eye size and head shape in Drosophila.
Keywords: evolutionary hotspots, repeated evolution, gene regulatory networks, evolution of development, Drosophila, eye–antennal disc
Introduction
The morphological diversity present in nature is a key prerequisite for organisms to adapt to an ever-changing environment. As the genome of an organism contains instructive information about its morphology, one of the major goals in biological research is to establish genotype–phenotype correlations for a given morphological trait (Lewontin 1974; Alberch 1991). The genetic architecture of some traits has been successfully determined at high resolution. For instance, natural variation in body pigmentation in the vinegar fly Drosophila melanogaster and the beach mouse Peromyscus polionotus has been mapped to individual nucleotides affecting gene expression (Jeong et al. 2006; Rebeiz et al. 2009) or protein function (Hoekstra et al. 2006), respectively. Also, the genetic basis of gain or loss of structures like trichomes in Drosophila (McGregor et al. 2007), armor plates in three spine stickleback fish (Shapiro et al. 2004), or the repeated loss of eyes in cave fish (Menuet et al. 2007; Stemmer et al. 2015) has been successfully revealed. Intriguingly, the analysis of similar morphological differences in various lineages showed that the same genes were repeatedly affected. For instance, differences in abdominal pigmentation and the occurrence of wing spots across the phylogeny of Drosophilidae have been mapped to the bric-à-brac (bab) (Gompel and Carroll 2003) and yellow (y) (Prud’homme et al. 2006) loci, respectively. Also the evolution of larval trichome patterns in various Drosophila species is caused by variation in the regulation of the shavenbaby (svb) gene (Sucena et al. 2003) and recurrent natural variation in the Pitx1 locus has been causally linked to pelvic spine reduction in different stickleback lineages (Shapiro et al. 2006; Chan et al. 2010). Although recurrent natural variation in such hotspot genes seems to be common for the evolution of those traits, it remains elusive, whether evolutionary hotspots play a major role in the evolution of the size and shape of organs.
The identification of the molecular changes underlying variation in the size and shape of morphological traits is difficult because many genomic loci with small effect sizes that are spread throughout the genome contribute to trait differences (Mackay 2001; Mackay et al. 2009; Boyle et al. 2017). For instance, natural variation in mandible and craniofacial shape in mouse is influenced by various loci located on most of the chromosomes (Burgio et al. 2009; Pallares et al. 2014, 2015; Maga et al. 2015), and studies in Drosophila revealed loci on several chromosomes associated with differences in eye size and head shape (Arif et al. 2013; Norry and Gomez 2017; Gaspar et al. 2020; Reis et al. 2020). The omnigenic model suggests that most genomic loci can influence the phenotypic outcome even though they are not functionally linked to the trait (“peripheral genes”) because they are connected to loci with a direct effect (“core genes”) in highly wired gene regulatory networks (GRNs) (Boyle et al. 2017). As many other biological networks (Jeong et al. 2000; Uetz et al. 2000; Ito et al. 2001; Featherstone and Broadie 2002), GRNs are thought to be scale-free, meaning that most nodes within the network have only very few connections and these low-degree nodes are connected by a few highly connected nodes, so-called hubs (Barabási and Albert 1999; Barabási and Oltvai 2004). Genes that act as hubs relay complex incoming information to a high number of target genes and they often code for evolutionary conserved proteins with crucial functions within the network (Fraser et al. 2002; Krylov et al. 2003). Therefore, it is conceivable that core genes proposed by the omnigenic model have features of hub genes within GRNs. It has been suggested that the recurrent evolution in hotspot genes is facilitated by their hub position within GRNs (Sucena et al. 2003; Stern and Orgogozo 2008, 2009), as well as the observation that only few such hubs are necessary to orchestrate the formation of rather simple traits, such as trichomes which develop from a single cell (Stern 2000). In contrast, the development of complex organs relies on the interplay of many hub genes (Peter and Davidson 2011; Potier et al. 2014), suggesting that evolutionary hotspots are less likely to be relevant for more complex trait evolution.
An excellent model to test, whether natural variation in organ size and shape is caused by recurrent changes in hotspot genes, is the Drosophila head that harbors major sensory organs, such as the compound eyes (Snodgrass 1935). Natural intra- and interspecific variation in eye size is pervasive in various Drosophila lineages, and increased eye size is often associated with a reduction of the interstitial cuticle between the eyes (Norry et al. 1997, 2000; Posnien et al. 2012; Keesey et al. 2019; Gaspar et al. 2020). Although the genetic architecture of this developmental trade-off is starting to be revealed in various species (Hammerle and Ferrus 2003; Arif et al. 2013; Norry and Gomez 2017; Gaspar et al. 2020), the most comprehensive genetic and developmental analysis established that intra- and interspecific differences in the developmental trade-off are caused by variation in the early subdivision of the eye–antennal imaginal disc (Ramaekers et al. 2019), the larval tissue from which most of the adult head develops (Haynie and Bryant 1986). Genetic analyses between different D. melanogaster strains revealed a single nucleotide polymorphism in the regulatory region of the eyeless/pax6 (ey) gene affecting its temporal expression and thus eye–antennal disc subdivision (Ramaekers et al. 2019). Ey regulates various target genes and signaling pathways (Ostrin et al. 2006; Yeung et al. 2018) to initiate retinal development, suggesting that it acts as a hub gene during eye development. Additionally, the recurrent variation in ey expression suggests that ey might also be a hotspot gene regulating the developmental trade-off between eye and head cuticle development (Ramaekers et al. 2019). We sought to test whether variation in the early subdivision of the eye–antennal disc is a common mechanism underlying the development of a head trade-off in Drosophila. As model we studied head development in the two closely related Drosophila species, D. melanogaster and D. mauritiana, which vary in adult eye size and head shape (Posnien et al. 2012; Hilbrant et al. 2014; Gaspar et al. 2020). Specifically, D. mauritiana develops larger compound eyes with more ommatidia and the bigger eyes of D. mauritiana are accompanied by a narrower interstitial head cuticle (Posnien et al. 2012).
We could show that differences in retinal differentiation occur only late during eye–antennal disc development, suggesting that the early subdivision of the disc may not be affected. To reveal candidate genes responsible for eye size differences, we developed a new method to identify evolutionary relevant hub genes with crucial positions within the developmental GRN. Applying comparative transcriptomics and functional genomics, we revealed the GATA transcription factor (TF) Pannier (Pnr) and its transcriptional cofactor U-shaped (Ush) as candidate hub genes. We functionally confirmed that the overexpression of pnr in D. melanogaster indeed phenocopies aspects of the D. mauritiana head shape as well as eye size. In summary, we argue that similar complex morphological differences can be caused by different developmental processes in different lineages. Therefore, our data suggest that evolutionary hotspots may play a less prominent role during the evolution of organ size and shape.
Results
Progression of Retinal Differentiation Varies during Third Larval Instars between D. melanogaster and D. mauritiana
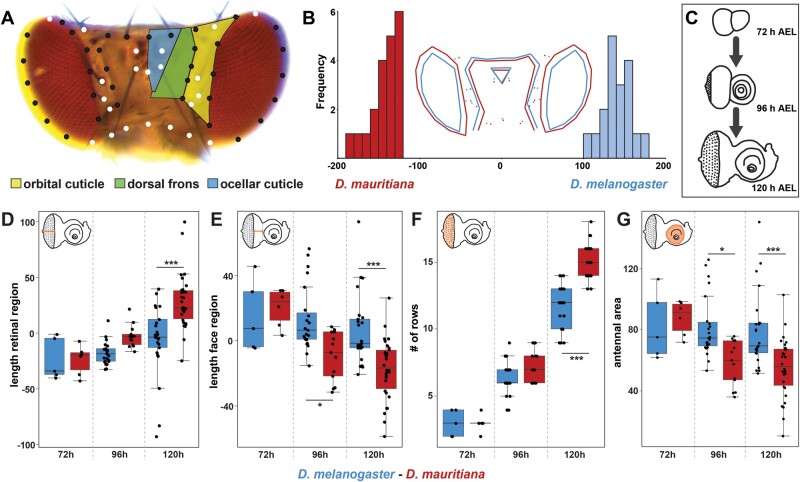
Drosophila eye size and head shape vary extensively between D. melanogaster and D. mauritiana with the latter having bigger eyes due to more ommatidia at the expense of interstitial head cuticle (Posnien et al. 2012; Arif et al. 2013; Hilbrant et al. 2014). As eye size differences are most pronounced in the dorsal part (Posnien et al. 2012), we first quantified differences in the dorsal head morphology to test whether dorsal head shape varies as well. We placed 57 landmarks on pictures of dorsal heads and applied a geometric morphometrics analysis (fig. 1A). A discriminate function analysis clearly distinguished the head shapes of D. melanogaster and D. mauritiana (fig. 1B) (P = 0.0001). In accordance with previous data (Posnien et al. 2012), we found main differences in dorsal eye size with the eye area protruding more toward the back of the head in D. mauritiana (fig. 1B). The expansion of the eye area in D. mauritiana was accompanied by a narrower dorsal head region, which affected both the orbital cuticle and the dorsal frons region. The ocellar complex was slightly shifted ventrally in D. mauritiana (fig. 1B). Therefore, D. melanogaster and D. mauritiana do not only differ in the size of dorsal eye, but they also exhibit variation in the relative contribution of different head regions to the dorsal head.
Fig. 1.
Quantitative differences in dorsal head shape are defined late during eye–antennal disc development. (A) Dorsal view of a Drosophila melanogaster head. The head cuticle consists of three morphologically distinguishable regions, namely the orbital cuticle (yellow) next to the compound eye, the dorsal frons (green), and the ocellary cuticle (blue). The 57 landmarks that were used to analyze head shape are shown as fixed (white) and sliding landmarks (black). (B) Discriminant function analysis distinguishes mean dorsal head shapes of D. melanogaster (blue) and D. mauritiana (red). Difference between means: Procrustes distance: 0.094, P value = 0.0001. (C) The development was characterized, and transcriptomic data sets were generated for developing eye–antennal discs for both species at three developmental stages: 72 h AEL (early L3), 96 h AEL (mid L3), and 120 h AEL (late L3). (D) Distance from the optic stalk to the morphogenetic furrow was measured along the equator region. Significant differences were observed at 120 h AEL (F5,96 = 15.61, P = 3.2e−11). (E) Distance from the morphogenetic furrow to the antennal anlagen was measured. From 96 h AEL on, we observed significant differences (F5,96 = 10.23, P = 7e−8). (F) Number of ommatidial precursor rows was counted along the equator region of the eye–antennal disc. Significant differences were observed at 120 h AEL (F5,96 = 210.8, P < 2e−16). (G) Area of the antennal region of the eye–antennal disc. Significant differences were observed at 96 and 120 h AEL (F5,96 = 7.86, P = 3.08e−6). One-way ANOVA followed by Tukey multiple comparisons: ***<0.001, *<0.05.
In Drosophila, most adult head structures develop from larval eye–antennal imaginal discs (Haynie and Bryant 1986; supplementary fig. S1A, Supplementary Material online). A crucial process during Drosophila eye development is retinal differentiation during the third larval instar stages. Retinal differentiation in the disc is characterized by the progression of a morphologically visible indentation, the morphogenetic furrow (supplementary fig. S1, Supplementary Material online). Cells in front of the furrow are mitotically active, whereas cells in the wake of the furrow eventually stop dividing and differentiate to give rise to evenly space ommatidia precursors (Treisman 2013). We therefore compared the progression of differentiation during eye–antennal disc development between species throughout third larval instar stages (fig. 1C). The progression of differentiation was estimated by measuring the width of the retinal region posterior (fig. 1D;supplementary fig. S1B–D, Supplementary Material online) and anterior (fig. 1E;supplementary fig. S1B–D, Supplementary Material online) to the morphogenetic furrow, as well as by counting the number of ommatidia rows (fig. 1F;supplementary fig. S1B–D, Supplementary Material online). In the light of the trade-off between eye size and interstitial head cuticle size (Posnien et al. 2012), we also measured the area of the antennal region to compare our retinal measurements to the antennal region (fig. 1G;supplementary fig. S1B–D, Supplementary Material online). None of these measurements was different between species at the earliest third instar stage (72 h after egg laying [AEL]), suggesting that eye–antennal discs are comparable in size and state of differentiation at 72 h AEL. In contrast, all measured traits were significantly different at the latest third instar stage (120 h AEL; fig. 1D–G) and the antennal region and the width of the retinal region anterior to the morphogenetic furrow were significantly different at an intermediate stage (96 h AEL; fig. 1D and F). The length of the retinal region including cells anterior and posterior of the morphogenetic furrow (i.e., the sum of the distances shown in fig. 1E and F) was the same at all stages (one-way ANOVA with multiple comparisons of means; F5,96 = 1.87, P = 0.107), suggesting that overall growth of the retinal region was not different between species. Those differences suggest that retinal differentiation progresses faster in D. mauritiana compared with D. melanogaster, implying that this process diverges species, specifically throughout third larval instar development.
Developmental Differences between D. melanogaster and D. mauritiana Are Associated with Variation in the Developmental Transcriptomic Landscape
Next, we tested whether differences in adult head morphology and retinal differentiation during eye–antennal disc development were associated with variation in the developmental transcriptomic landscape. As we observed differences in eye–antennal disc development starting at 96 h AEL, we obtained comparative transcriptomes covering 72 h AEL to capture any differences that may induce the observed developmental differences, as well as 96 and 120 h AEL (fig. 1C and supplementary fig. S2, supplementary Materials and Methods, Supplementary Material online, for details) (Torres-Oliva et al. 2018). In line with the overall functional conservation of head morphology between species, we first confirmed that genes with stable interspecific expression represent central processes involved in eye–antennal disc development (supplementary fig. S3, Supplementary Material online). A principal component analysis (PCA) of those genes that were differentially expressed between species revealed that 72% of variation in the data set was due to differences between 72 h and the other two stages (supplementary fig. S4, Supplementary Material online). This observation was confirmed by a stage-specific pairwise differential expression analysis where we found the highest number of differentially expressed genes (DEGs) between species at 72 h AEL (6,683 genes), whereas 3,260 and 2,380 genes were differentially expressed at 96 and 120 h AEL, respectively. The excess of DEGs at 72 h AEL may hint toward extensive variation in gene regulation that predetermines the interspecific differences in differentiation progression observed from 96 h AEL onward (fig. 1D–G). Differential expression was not biased toward one species as we observed an equal number of DEGs with higher expression in D. melanogaster and D. mauritiana, respectively (supplementary table S1, Supplementary Material online).
Variation in the Transcriptomics Landscape Is Associated with Variation in Expression of Hub Genes
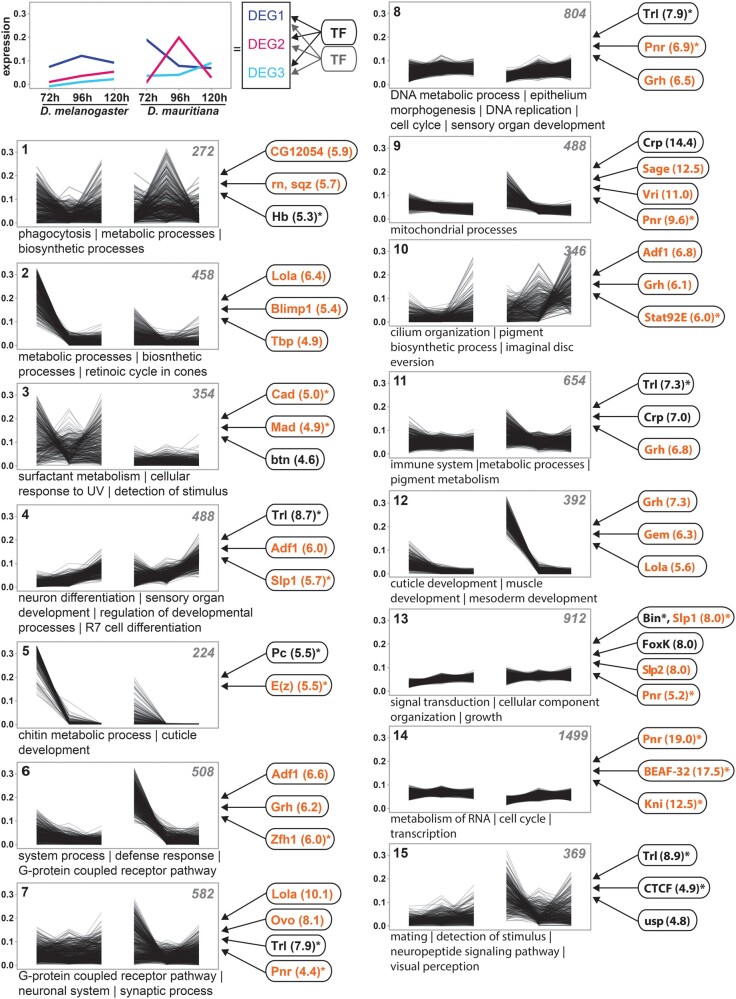
The interspecific differences in gene expression dynamics must be the result of variation in the underlying regulatory interactions. To identify putative TFs that regulate the expression of DEGs, we assume that DEGs with similar expression profiles are regulated by similar TFs; that is, we treated coexpression clusters (clusters in fig. 2) as “gene modules” (Bar-Joseph et al. 2003; Segal et al. 2003; Arda and Walhout 2010). We established stage-specific chromatin accessibility data (ATACseq) to identify enriched TF-binding motifs for each cluster (see Extended Materials and Methods and supplementary fig. S2, Supplementary Material online, for details). Each cluster showed enrichment of motifs for a unique combination of TFs (fig. 2 and supplementary table S2, Supplementary Material online), suggesting that variation in different gene modules is influenced by different regulatory interactions. Overall, we identified 136 unique transcriptional regulators (normalized enrichment score, NES > 4.0) that were expressed in at least one developmental stage. At 72 h AEL, 55.9% of the expressed transcriptional regulators were differentially expressed at false discovery rate (FDR) 0.05 (compared with 44.1% of a random set of transcriptional regulators; X2 (1, N = 279) = 3.9, P = 0.0483; fig. 2 and supplementary table S2, Supplementary Material online), suggesting that the high number of DEGs at this stage may partially be the result of differential expression of the transcriptional regulators identified in each cluster.
Fig. 2.
Extensive remodeling of the transcriptome underlying eye–antennal disc development. The scheme in the upper left corner illustrates the rational of the analysis. DEGs with similar expression profiles were clustered. Assuming that coexpressed genes are regulated by similar TFs, enriched TF-binding motifs were identified in accessible chromatin regions of genes within each cluster (see Materials and Methods for details). Cluster analysis of all differentially expressed genes between Drosophila melanogaster and D. mauritiana. The number of genes in each cluster is given in the upper right corner. Significantly enriched GO terms are provided for each cluster. TFs refer to potential upstream regulators and the respective NES values (nonredundant occurrence and excluding non-Drosophila motifs). Factors written in orange are significantly differentially expressed in at least one stage between the two species. Factors marked with an asterisk are among the top 10% of highly connected transcriptional regulators (i.e., hub genes). A complete list of enriched GO terms and potential upstream factors is available in supplementary table S2, Supplementary Material online.
As hub genes occupy central positions in developmental GRNs affecting many target genes (Barabási and Albert 1999; Barabási and Oltvai 2004) and the clusters of DEGs contained many genes (fig. 2), we asked whether highly connected hub genes are among the identified transcriptional regulators. We defined hubs by being the top 10% genes with highest number of target genes in the DroID reference network (see Extended Materials and Methods for details; supplementary fig. S5, Supplementary Material online) (Seo et al. 2009; Murali et al. 2011; Liu et al. 2019). Forty of the 136 transcriptional regulators were present in the DroID reference network and 28 of them were hubs (70%, fig. 2 and supplementary table S2, Supplementary Material online). Seventeen of the hub genes (60.7%) were differentially expressed in at least one stage (fig. 2 and supplementary table S2, Supplementary Material online). Therefore, we identified differentially expressed hub genes among the transcriptional regulators that drive expression divergence between species. However, it is unlikely that the observed expression differences are specifically driven by hub genes because the number of hub genes and the number DEGs among those hubs was not significantly different from a randomly sampled set of hub genes expressed during eye–antennal disc development (supplementary table S2, Supplementary Material online).
Many hub genes play central roles during development (Fraser et al. 2002; Krylov et al. 2003), which we confirmed by an extensive enrichment of Gene Ontology (GO) terms related to developmental processes for the hub genes in the DroID network (supplementary table S2, Supplementary Material online). Accordingly, some of the identified hub genes have previously been described to be involved in eye–antennal disc development. For instance, Slp1 (fig. 2; clusters 4 and 13) controls dorsal–ventral axis formation during retinal development (Sato and Tomlinson 2007) and Stat92E (fig. 2; cluster 10), a member of the JAK/STAT pathway, regulates eye growth and ommatidial polarity by regulating many target genes, and signaling pathways (Zeidler et al. 1999; Ekas et al. 2006; Flaherty et al. 2009). Similarly, Mad (fig. 2; clusters 3, 4, and 15), the TF that mediates Decapentaplegic (Dpp)—signaling (Sekelsky et al. 1995), is involved in various processes in the eye–antennal disc (Wiersdorff et al. 1996; Cordero et al. 2007; Wells et al. 2017). Additionally, in 5 of 15 clusters (NES > 4) (fig. 2; e.g., clusters 7–9, 13, and 14) (in 9 of 15 clusters, NES > 3), we found a strong enrichment of motifs predicted to be bound by the GATA TF Pnr, which is involved in establishing the early dorsal–ventral axis of the eye (Maurel-Zaffran and Treisman 2000; Singh and Choi 2003; Pereira et al. 2006). In summary, we identified many transcriptional regulators, some of which are highly connected hubs, which regulate DEGs between D. melanogaster and D. maurtiana.
Pnr Is a Phenotypically Relevant Hub Gene in the GRN Underlying Dorsal Head Development
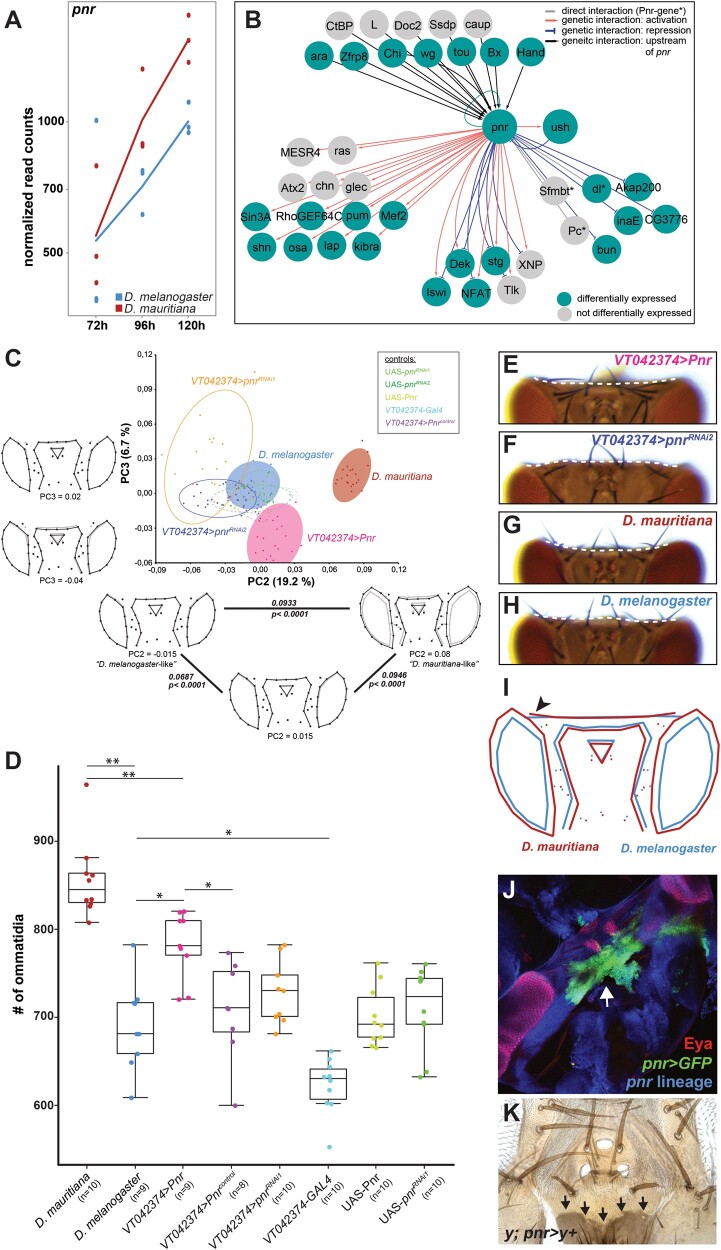
The observation that many of the identified hub genes were differentially expressed between species suggests that these changes may be causal in defining the observed differences in eye size and head shape. However, developmental processes (Waddington 1942; Kitano 2004) tend to be robust against perturbations because variation in the expression of developmental genes often does not result in phenotypic differences due to extensive buffering within GRNs (Gavin-Smyth et al. 2013; Fear et al. 2016; Cannavo et al. 2017). Therefore, it remains questionable, whether the differential expression of the hub genes and thus the variation in expression of their target genes are indeed relevant for the differences in head morphology between D. melanogaster and D. mauritiana. To test this, we further investigated the role of Pnr, which is an interesting hub gene for the following reasons: 1) Our global clustering and motif enrichment analyses suggest that Pnr regulates many DEGs between both species and it is among the top 10% of highly connected transcriptional regulators (fig. 2); 2) pnr itself is differentially expressed between D. melanogaster and D. mauritiana, with higher expression in the latter species (fig. 3A); and 3) Pnr is known to be expressed in the dorsal eye–antennal disc (Maurel-Zaffran and Treisman 2000) and it is involved in defining the dorsal–ventral axis of the eye (Maurel-Zaffran and Treisman 2000; Singh and Choi 2003; Singh et al. 2005). Later during eye–antennal disc development, Pnr influences the ratio of retinal and head cuticle fate in the dorsal disc by repressing retinal determination genes (García-García et al. 1999; Maurel-Zaffran and Treisman 2000; Oros et al. 2010).
Fig. 3.
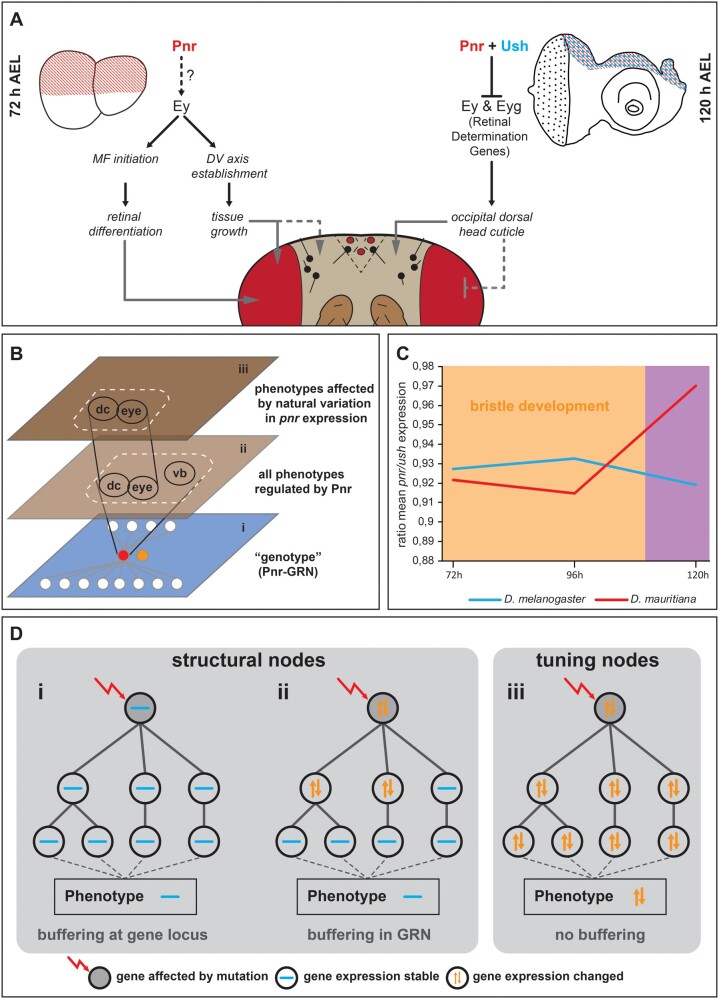
Pnr is a phenotypically relevant hub gene during eye–antennal disc development. (A) Expression dynamics of the pnr transcript at the three developmental stages in Drosophila melanogaster (blue) and D. mauritiana (red) based on rlog transformed read counts. (B) Network reconstruction of known interactions upstream and downstream of Pnr that overlaps with our Pnr target gene list. Fourteen of the 29 target genes are activated (red edges) and 8 genes are repressed (blue edges) by Pnr. Six target genes showed both types of interactions. Twenty-one of the 30 putative Pnr target genes (68%) were differentially expressed (green nodes). (C) Principal component analysis of dorsal head shapes. Shape outlines for principal component (PC) values are given along the PC2 and PC3 axes, respectively, as indicated below each outline. In the triangle along PC2, the Procrustes distance between each shown shape outline is provided, as well as the P value after permutation test with 1,000 cycles. Overexpression of pnr (VT042374 > Pnr) shifts the dorsal head shape toward a D. mauritiana head shape, whereas reduction of pnr expression (VT042374 > pnrRNAi1 and VT042374 > pnrRNAi2) resulted in an opposite shift. (D) Number of ommatidia counted in one compound eye per individual. The significance was calculated by ANOVA and each pairwise comparison was calculated using Tukey’s Honestly Significant Difference technique test; **≤0.01; *≤0.05. The color code in (C) and (D) is the same, and control flies were either the Gal4 driver line (VT042374-Gal4) or the UAS effector lines (UAS-Pnr, UAS-pnrRNAi1, UAS-pnrRNAi2) without crossing them. VT042374 > Pnrcontrol are CyO carrying offspring of the VT042374 > Pnr cross that do not contain the UAS-Pnr transgene and thus no overexpression. (E) Representative dorsal view of an adult head after overexpression of pnr and (F) after knockdown of pnr. See supplementary figure S9, Supplementary Material online, for quantification and controls for E and F. (G) Representative dorsal view of an adult head D. mauritiana. (H) Representative dorsal view of an adult head of D. melanogaster. The dotted lines in E–H mark the border of the occipital region. (I) Mean shapes of D. melanogaster (blue) and D. mauritiana (red) heads after Discriminant Function Analysis including seven additional landmarks in the occipital region. The black arrowhead marks the lateral bending of the occipital region in D. mauritiana (Procrustes distance: 0.094; P < 0.0001 based on permutation test with 1,000 runs; nD. mauritiana = 20; nD. melanogaster = 18). (J) pnr expression in developing pupal head structures. Cells marked with pnr > GFP accumulated in the future occipital region (green) behind the developing ocelli (red), and the head region where the two discs fuse (white arrow). (K) pnr lineage in adult dorsal heads shown by cells in which “yellow” expression is restored (black arrows).
To validate the hub gene status of pnr, we first refined the list of its potential target genes. We searched for the Pnr-specific GATA motif in genomic regions that were accessible during eye–antennal imaginal disc development (see supplementary Materials and Methods and supplementary fig. S2, Supplementary Material online, for details). In 14,511 open chromatin regions (across all timepoints), we found 1,335 Pnr-specific GATA motifs associated with 1,060 genes expressed in our RNAseq data set (supplementary table S3, Supplementary Material online), suggesting that those genes were active during eye and head development. In the regulatory regions of these 1,060 Pnr target genes, we found a strong enrichment of Pnr-binding sites (supplementary fig. S6A, Supplementary Material online), and many of the genes were also predicted to be regulated by Pnr in our cluster analysis (see fig. 2 and supplementary fig. S6B, Supplementary Material online). In line with the known functions of Pnr during eye–antennal disc development, the Pnr target genes were highly enriched in processes like compound eye development and growth as well as cell cycle progression (supplementary fig. S6C, Supplementary Material online). Additionally, a crossvalidation of our Pnr target genes with the DroID interaction database (Yu et al. 2008; Murali et al. 2011) showed that our list contained 3 known direct target genes (i.e., Pnr-regulatory sequence interaction; dl, Pc, and Sfmbt) and 23 genes with known genetic interactions (fig. 3B), suggesting that these genes are also direct targets of Pnr. Seventeen out of these 26 genes (65.38%) (fig. 3B) and 714 of the full set of 1,060 target genes (67.4%) showed expression differences between D. melanogaster and D. mauritiana. Note that the number of DEGs among Pnr target genes was not significantly different from those of other TFs involved in retinal development or from transcriptional regulators with similar connectivity (Extended Materials and Methods and supplementary table S3, Supplementary Material online), suggesting that also other highly connected transcriptional regulators contribute to the differences in the transcriptomic landscape. Taken together, we confirmed that Pnr regulates many DEGs during eye–antennal disc development.
To test if variation in pnr expression can explain naturally occurring differences in eye size and dorsal head shape (fig. 1), we overexpressed and knocked down pnr in D. melanogaster in the dorsal eye–antennal disc in a domain that is reminiscent of endogenous pnr expression (supplementary fig. S7A–E, Supplementary Material online). Successful manipulation of the expression was confirmed by reduced and enhanced protein expression in knockdown and overexpression discs, respectively (supplementary fig. S7F–H, Supplementary Material online). Subsequently, we quantitatively compared the adult head morphology applying geometric morphometrics and ommatidia counting (see Extended Materials and Methods, Supplementary Material online, for details). A PCA of head shape variation showed that principal component 2 (PC2) explained 19.2% of the observed variation in head shape and PC3 explained 6.7% (fig. 3C; note that PC1 captured a technical artifact, see supplementary fig. S8, Supplementary Material online). Variation along PC2 mainly captured differences in the proportion of eye versus interstitial cuticle in the dorsal head as well as the location of the ocellar region. Intriguingly, the overexpression of pnr in the dorsal head region resulted in a shift from a “D. melanogaster”-like shape toward a more “D. mauritiana”-like shape along PC2, with an enlargement of the eyes at the expense of a slight reduction of the interstitial cuticle (fig. 3C). Ommatidia counting in entire eyes confirmed that the increase in eye area upon pnr overexpression was indeed due to an increase in number of ommatidia (fig. 3D). Reduction of pnr expression resulted in a more extreme D. melanogaster-like head shape (fig. 3C), whereas no effect on ommatidia number was observed (fig. 3D).
PC3 mostly explained differences in the dorsal–posterior head cuticle and the location of the ocellar region (fig. 3C). We analyzed the dorsal–posterior head cuticle in more detail and observed that the occipital region was more convex upon pnr overexpression (fig. 3E; for quantification, see supplementary fig. S9A, B, E, Supplementary Material online), whereas downregulation consistently led to an enlargement of these regions (fig. 3F; for quantification, see supplementary fig. S9A–D, Supplementary Material online). Intriguingly, in accordance with a higher expression of pnr in D. mauritiana, we found a more convex occipital region in D. mauritiana (fig. 3G) compared with a more concave shape in D. melanogaster (fig. 3H). The quantification of this qualitative difference using seven extra landmarks in the occipital head region confirmed the interspecific shape differences (fig. 3I). Lineage tracing experiments showed consistent expression of pnr during eye–antennal disc development (supplementary fig. S10, Supplementary Material online) and detection of pnr expression in pupae (fig. 3J) as well as the analysis of pnr-expressing clones in adult heads (fig. 3K) confirmed that pnr was indeed expressed in the future occipital region. In summary, we showed that Pnr is a phenotypically relevant hub gene during eye–antennal disc development because we were able to phenocopy aspects of the D. mauritiana-like head shape and eye size by upregulating pnr expression in the developing eye–antennal disc of D. melanogaster.
U-Shaped Modulates Pnr Function during Eye–Antennal Disc Development
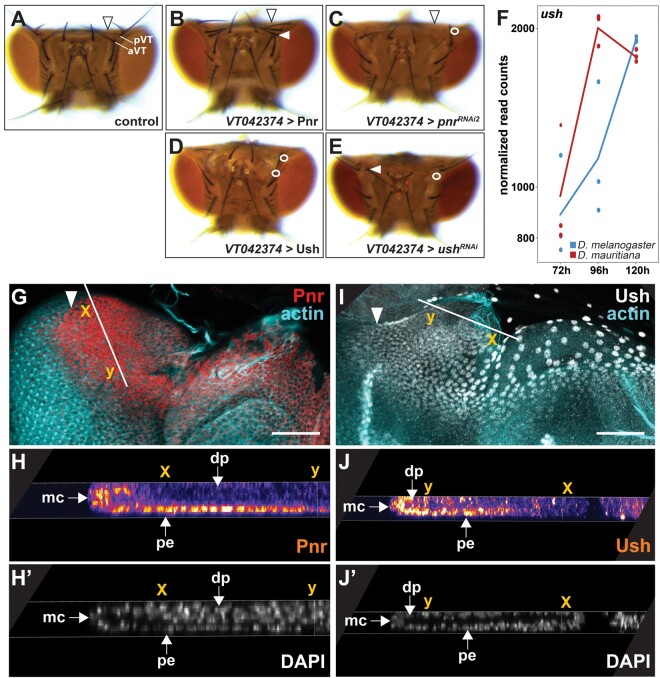
The detailed analysis of the heads of pnr gain and loss of function flies revealed a consistent gain and loss of vertical bristles, respectively (fig. 4A–C). A similar role in sensory bristle development has been shown in the wing imaginal disc, where Pnr controls the positioning sensory bristles in the thorax (Heitzler et al. 1996; Cubadda et al. 1997; Gomez-Skarmeta et al. 2003). The role of Pnr in thoracic sensory bristle development is antagonized by the presence of its cofactor Ush (Cubadda et al. 1997; Haenlin et al. 1997). Upon heterodimerization with Ush, Pnr that normally acts as a transcriptional activator, acquires a repressive function (Haenlin et al. 1997; Fossett et al. 2001; Sorrentino et al. 2007). During eye–antennal disc development, Pnr activates target genes during dorsal–ventral axis formation (Maurel-Zaffran and Treisman 2000; Singh and Choi 2003), whereas it represses retinal genes later during development (Oros et al. 2010), suggesting that it plays a dual (i.e., activating or repressing) role in this process as well. In line with a dual role of Pnr as transcriptional activator and repressor, we found genes that were activated as well as repressed by Pnr among its differentially expressed high confidence target genes during eye–antennal disc development (supplementary fig. S11, Supplementary Material online and fig. 3B). Additionally, the knockdown and overexpression of ush during eye–antennal disc development resulted in loss or misplacement of vertical bristles (fig. 4D and E), suggesting that Pnr and Ush may coordinate similar processes. Therefore, we asked whether Ush may also modulate the function of Pnr during eye–antennal disc development. To answer this question, we tested whether 1) Pnr and Ush are colocalized in the same cells in the eye–antennal disc and 2) both genes interact genetically.
Fig. 4.
Pnr and Ush interact during eye–antennal disc development. (A) Dorsal view of an adult control head of D rosophila melanogaster (CyO carrying offspring of the VT042374 > Pnr cross that do not contain the UAS-Pnr transgene and thus no overexpression). Posterior (pVT) and anterior vertical bristles (aVT) are labeled. The arrowhead marks the occipital region. (B) Overexpression of pnr did not lead to major irregularities in the dorsal head cuticle, but to duplication of the pVT bristle next to the compound eye (white arrowhead) (9/9 females and 10/10 males showed the phenotype). (C) Knockdown of pnr resulted in visible enlargement of the occipital region (black arrowhead) and to the loss of the pVT bristle next to the compound eye (white circle) (10/10 males and females, respectively, showed the phenotype). (D) Overexpression of ush resulted in irregularities in the dorsal head cuticle and to the loss of the pVT andaVT bristles (white circles) next to the compound eye (10/10 females showed the phenotype, males were not tested). (E) Knock-down of ush resulted in slight irregularities of the dorsal head cuticle (11/11 females showed the phenotype, males were not tested), to loss (white circle) (10/10 females showed the phenotype, males were not tested) or misplacement (white arrowhead) of the aVT bristle (2/10 females showed the phenotype, males were not tested). (F) Expression dynamics of the ush transcript at the three developmental stages in D. melanogaster (blue) and D. mauritiana (red) based on rlog transformed read counts. (G, H) Pnr protein location in third instar eye–antennal discs in D. melanogaster. The Pnr protein is present in the dorsal peripodial epithelium (pe) of the developing disc (G), including a few cells of the margin cells (mc) and the disc proper (dp) (H′). The white arrow in G marks the morphogenetic furrow, the solid white line marks region of the cross-section shown in H and H′, and the x and y coordinates indicate the same location in G, H, and H′. (I, J) Ush protein location in third instar eye–antennal discs in D. melanogaster. The Ush protein is, similar to Pnr (compare with G), expressed in the dorsal peripodial epithelium (pe) of the developing disc (I), including a few cells of the margin cells (mc) and the disc proper (dp) (J′). The white arrow in I marks the morphogenetic furrow, the solid white line marks region of the cross-section shown in J and J′, and the x and y coordinates indicate the same location in I, J, and J′. G and I are maximum intensity projections of confocal sections throughout the eye–antennal disc. The scale bars represent 50 µm.
Although, it was previously stated that ush is not expressed in the eye–antennal disc (Maurel-Zaffran and Treisman 2000; Fossett et al. 2001), we observed expression in all three studied stages in our RNAseq data (fig. 4F). As shown for pnr (fig. 3A), ush expression was mostly higher in D. mauritiana (fig. 4F). We confirmed the transcriptomics data using newly generated antibodies to show that both proteins were expressed in largely overlapping domains (fig. 4G–J and supplementary fig. S12, Supplementary Material online) in the large nuclei of the dorsal peripodial epithelium (compare fig. 4G to 4I) and in a subset of the cuboidal margin cells in the disc proper (compare fig. 4H to 4J).
As both proteins were colocalized in the eye–antennal disc, we tested for genetic interactions. We asked whether manipulation of pnr expression affected ush expression and vice versa. To this end, we overexpressed and knocked down both genes in the dorsal eye–antennal disc in a domain that is reminiscent of pnr expression (supplementary fig. S7A–E, Supplementary Material online). We confirmed the successful overexpression and knockdown of both genes by enhanced and loss of protein expression, respectively (supplementary fig. S7F–K, Supplementary Material online). Due to the qualitative nature of our experimental setup, we could not obtain conclusive results for some of the tested combinations. However, we observed a reduced Ush signal upon knockdown of pnr (supplementary fig. S7L, Supplementary Material online), suggesting a positive interaction between pnr and ush. Additionally, upregulation of ush resulted in slightly reduced Pnr protein levels (supplementary fig. S7M, Supplementary Material online), suggesting a negative impact of Ush on pnr expression. Further support for this idea comes from the induction of a double-antenna phenotype when ush was overexpressed using a stronger pnr driver line (supplementary fig. S7N, Supplementary Material online) (Fossett et al. 2001), a phenotype that we also observed upon pnr knockdown (supplementary fig. S7O, Supplementary Material online) (Maurel-Zaffran and Treisman 2000). In summary, we could show that Pnr and its cofactor Ush are coexpressed, interact genetically (supplementary fig. S7P, Supplementary Material online) and are involved in dorsal sensory head bristle development. We suggest that a dual role of Pnr during eye–antennal disc development is most likely mediated by its cofactor Ush.
Discussion
The different investment in compound eye and interstitial cuticle (referred to as trade-off here) in Drosophila heads is an excellent model to study the developmental and molecular mechanisms underlying complex trait evolution (Posnien et al. 2012; Arif et al. 2013; Norry and Gomez 2017; Keesey et al. 2019; Ramaekers et al. 2019; Gaspar et al. 2020). We established that this trade-off is present in the dorsal head of D. melanogaster and D. mauritiana, with the latter having larger compound eyes with significantly more ommatidia (this work and Posnien et al. 2012). The comparison of larval eye–antennal disc development in both species revealed differences in the progression of differentiation starting at 96 h AEL. To identify candidate genes driving these developmental differences, we combined comparative transcriptomics with chromatin accessibility data obtained prior to and after interspecific differences were observable (i.e., 72, 96, and 120 h AEL). We show that the GATA TF Pnr is a prime candidate hub gene: It showed higher expression in D. mauritiana and up to 67% of its more than 1,000 predicted direct target genes were differentially expressed between species. In the following, we propose a developmental model explaining how variation in pnr expression can simultaneously affect ommatidia number and interstitial cuticle size and we discuss our findings with respect to evolutionary hotspots and GRN evolution.
Developmental Mechanism Underlying Eye Size and Head Shape Differences between D. melanogaster and D. mauritiana
The spatially restricted overexpression of pnr in its endogenous domain resulted in a significant increase in ommatidia number, implying a direct effect on retinal development and thus final eye size. Our lineage tracing experiment showed that pnr positive cells contribute to the entire dorsal peripodial epithelium and to cells of the dorsal posterior margin where the morphogenetic furrow, and thus retinal differentiation, is initiated at late second/early third instar stages (Casares and Almudi 2016). It has recently been shown that Eyeless/Pax6 (Ey) activity in the peripodial epithelium and in these posterior margin cells is necessary for morphogenetic furrow initiation and placement of the dorsal–ventral boundary (Baker et al. 2018). The latter is tightly linked to reported functions of Pnr and tissue growth in the retinal region of the eye–antennal disc (Maurel-Zaffran and Treisman 2000; Singh and Choi 2003; Singh et al. 2005). We found ey among the putative direct Pnr target genes, suggesting that Pnr may activate ey expression in the peripodial epithelium and in posterior margin cells. As ey was not differentially expressed in our data set, it remains to be tested if Pnr may regulate the final number of ommatidia clusters and thus final eye size through Ey-mediated initiation of differentiation and regulation of overall retinal growth prior to the stages studied here (fig. 5A).
Fig. 5.
Developmental model and hypothesis about GRN evolution. (A) Summary of expression and functional data obtained in this work. Black connections indicate genetic interactions among genes. Gray connections represent observed phenotypic effects upon manipulation of gene expression. (B) Schematic representation of functional constraints on vertical bristle development. The genotype level (i) is depicted as GRN centered on Pnr (red node in i). Pnr regulates the development of the dorsal occipital cuticle (dc), the compound eye (eye), and vertical bristles (vb) (ii). Due to a functional constraint on vertical bristle development, only the effect of differences of pnr expression on dc and eye are phenotypically relevant during the evolution of head morphology (iii). We propose that the constraint effect of pnr expression differences between D. melanogaster and D. mauritiana is mediated by coevolution of ush expression (orange node in i). (C) The ratio of pnr and ush expression is stable at 72 and 96 h AEL when vertical bristles are defined. Later expression of both genes is not constraint anymore by a function in bristle development. (D) Schematic representation of structural (i, ii) and tuning nodes (iii) in developmental GRNs.
Besides a putative role in morphogenetic furrow initiation, Pnr may also regulate the progress of differentiation. Faster progression of differentiation in D. mauritiana is correlated with larger adult eye size, whereas previous experiments manipulating the speed of the morphogenetic furrow suggest that faster progression results in smaller adult eye sizes (Brown et al. 1995; Singh and Choi 2003). However, many genes that affect the progression of the morphogenetic furrow, such as extramacrochaetae (Brown et al. 1995; Bhattacharya and Baker 2009) or daughterless (Brown et al. 1996), are also tightly linked to cell cycle control, making it difficult to clearly disentangle these two functions in the disc proper (Kumar 2011). Intriguingly, manipulation of morphogenetic furrow progression through abolished signaling from the peripodial epithelium resulted in highly reduced adult eyes (Gibson and Schubiger 2000, 2001; Atkins and Mardon 2009). It will thus be interesting to further study the role of Pnr in the peripodial epithelium and to test whether variation in pnr expression may directly affect differential progression of the morphogenetic furrow as observed between D. melanogaster and D. mauritiana. Additionally, it remains to be tested why pnr overexpression affected adult ommatidia number, whereas its knockdown did not. This may be due to a tight interplay of Pnr and its cofactor Ush as our preliminary data suggest that both genes interact genetically in the peripodial epithelium. This interaction may also explain another interesting observation. Although we showed an increase in eye size upon overexpression of pnr, previous work reported dorsal retinal overgrowth and ectopic eye fields after the induction of loss of function clones using an ey-Gal4 driver (Maurel-Zaffran and Treisman 2000; Oros et al. 2010). As the ey-Gal4 driver is active from early L1 stages on in the disc proper (Baker et al. 2018), the retinal overgrowth very likely results from the induction of an ectopic dorsal–ventral equator region (Oros et al. 2010). In contrast, the spatially defined overexpression of pnr in its endogenous peripodial epithelium domain that also expresses ush (this work) allows studying variation in gene expression in the natural trans-regulatory background. Interestingly, in line with our results, cuticle overgrowth has been observed for some ey-Gal4 induced loss of function clones (Oros et al. 2010), suggesting that Pnr plays spatially and temporally restricted roles during eye antennal disc development.
We found that pnr-expressing cells contribute to the dorsal interstitial head region in pupae, suggesting that the phenotypic effects in the occipital head region most likely highlight late functions of Pnr. Indeed, Pnr represses members of the retinal determination network throughout the third instar stage to regulate the ratio of retinal cells versus head cuticle cells (Oros et al. 2010). In addition to ey, we found eyegone (eyg) as second member of the retinal determination network among putative direct Pnr targets. Both genes must be repressed to ensure proper dorsal interstitial cuticle development (Wang et al. 2010; Magri et al. 2018), suggesting that Pnr may act through transcriptional repression during late dorsal head development. Such a repressive role could be mediated upon heterodimerization with its cofactor Ush as shown for the wing disc (Haenlin et al. 1997; García-García et al. 1999; Fromental-Ramain et al. 2008; Fromental-Ramain et al. 2010). Indeed, we observed that despite previous reports (Maurel-Zaffran and Treisman 2000), the Ush protein is colocalized with Pnr in dorsal peripodial epithelium and in the cuboidal cells of the disc margin. In accordance with their coexpression, both genes interact genetically and they are both necessary for proper dorsal head development and the formation of the same head bristles (this work and see Cubadda et al. [1997] for bristle phenotype). Therefore, we propose that Ush may mediate a repressive late function of Pnr in the dorsal peripodial epithelium to regulate retinal versus dorsal head cuticle development during the third larval instar stage (fig. 5A).
Taken together, our functional data suggest that variation in pnr expression during third instar larval development can simultaneously affect the compound eyes (representing an early function) and the size of the dorsal interstitial head cuticle (representing a late function) (fig. 5A). According to the two proposed developmental roles of Pnr (fig. 5A), increasing pnr expression levels in D. melanogaster mimics higher pnr expression as observed in D. mauritiana and phenocopied major aspects of the dorsal D. mauritiana head shape and eye size. It is important to note that the Gal4 driver line used for the functional assays is active throughout third instar stages and we cannot exclude Gal4 activity prior to these stages. Therefore, our experiments do not allow distinguishing between the two temporal functions and the exact time points when Pnr affects each process remain to be established. Future comparative studies on the temporal regulation of pnr expression and stage- and species-specific regulatory interactions of Pnr will allow revealing the exact mechanisms responsible for the two distinct roles of Pnr during eye–antennal disc development and the evolution of eye size and head shape.
Consistent with the fact that variation in complex traits is caused by multiple loci (Mackay et al. 2009; Boyle et al. 2017), pnr overexpression was not sufficient to fully phenocopy D. mauritiana head shape and eye size. We also identified other transcriptional regulators that regulated many DEGs, suggesting their involvement in remodeling the transcriptomic landscape and thus regulating variation in adult head morphology. Furthermore, it remains to be established whether the pnr and/or ush loci contain genetic variants associated with eye size and head shape differences or if causative genetic changes affect the expression or function of regulators of both genes. Although quantitative genetics approaches are not applicable due to the infertility of F1 females in interspecific crosses between D. melanogaster and D. mauritiana (Lachaise et al. 1986), reciprocal hemizygosity tests (Stern 2014) for Pnr, Ush, and putative regulators of these two factors represent a powerful tool to further dissect the causative genetic variants in the future.
In summary, based on our developmental model, the increase in eye size in D. mauritiana that goes along with a reduction of dorsal interstitial cuticle and a convex bending of the occipital head region can partially be explained by natural variation in pnr expression. As Pnr is a highly connected pleiotropic hub gene that regulates many differentially expressed genes between D. melanogaster and D. mauritiana, we argue that its differential expression is phenotypically relevant and may underly natural variation in head morphology.
Functional Constraints Drive Coevolution of Pnr and Ush Expression
In line with the pleiotropic role of Pnr during eye–antennal disc development, classical gain and loss of function experiments resulted in extensive phenotypic consequences ranging from retinal overgrowth to the entire loss of the compound eyes (Maurel-Zaffran and Treisman 2000; Oros et al. 2010). These rather coarse experiments based on highly artificial systems, such as ectopic expression of pnr induced by an eyeless/Pax6-Gal4 driver line that is active from early stages on in most of the eye–antennal disc (Baker et al. 2018), are suitable to unravel the combined functions of a pleiotropic gene. However, it is a major challenge to disentangle the distinct tissue- and stage-specific functions of pleiotropic factors and find out which of these are phenotypically relevant in an evolutionary context (Stern 2000; Wagner and Zhang 2011). This is particularly relevant because developmental processes (Waddington 1942; Kitano 2004) and the expression of the genes regulating them (Batada and Hurst 2007; Landry et al. 2007; Lehner 2008) tend to be robust against genetic or environmental perturbations. Such an intrinsic robustness of developmental systems may be the result of constraints imposed by the crucial function of the developing organ. Therefore, functional constraints may lead to robustness that limits the evolutionarily relevant phenotypic effects of pleiotropic genes (fig. 5B).
We observed an excellent example for such a constraint. Although individual gain and loss of function of pnr and ush in D. melanogaster affected the formation of anterior and posterior vertical sensory bristles in the dorsal head, we never observed gain or loss of these bristles between species. Pnr and Ush have been shown to be involved in thoracic sensory bristle development by controlling the positioning of proneural cell clusters in the wing disc (Heitzler et al. 1996; Cubadda et al. 1997; Gomez-Skarmeta et al. 2003). Thoracic sensory bristles develop in regions with high pnr and low ush expression (Cubadda et al. 1997; Haenlin et al. 1997) and their balanced expression is facilitated by genetic interactions between both genes (Fromental-Ramain et al. 2008). We observed that pnr overexpression in the eye–antennal disc consistently resulted in duplication of the anterior vertical bristles, suggesting that extra bristles arise where pnr overexpression cannot be compensated by Ush (see also Heitzler et al. 1996). Similarly, overexpression of ush in most of the dorsal peripodial epithelium resulted in loss of the anterior and posterior vertical bristles, showing that extra Ush above a certain threshold completely antagonizes Pnr function and subsequent sensory bristle formation. Hence, the stoichiometry between Pnr and its cofactor Ush is crucial for proper sensory bristle development in the dorsal head as well.
How can natural variation in pnr and ush expression cause differences in head morphology without affecting the sensory bristle pattern? As most of the proneural cell clusters are specified 18–6 h prior to puparium formation (Huang et al. 1991), the ratio between Pnr and Ush must remain stable at 72 and 96 h AEL to ensure proper sensory bristle formation. However, once bristles are specified, variation in the ratio of both factors at later stages (e.g., 120 h AEL) can cause natural variation in the shape of the dorsal occipital head region without affecting bristle formation (fig. 5C). Accordingly, both genes showed higher expression in D. mauritiana at 72 and 96 h AEL resulting in a similar ratio of both factors (fig. 5C). In contrast, a different ratio was observed at 120 h AEL, with pnr expression being higher in D. mauritiana, whereas ush expression was more similar in both species at this later stage (fig. 5C). We propose that the functional constraint on stereotypic sensory bristle development drove the coevolution of pnr and ush expression during early third instar disc development in D. melanogaster and D. mauritiana (fig. 5C). Indeed, according to their crucial function in environmental perception (Kernan 2007), vertical bristles have been shown to be robust against temperature fluctuations during development (Matamoro-Vidal et al. 2018). Therefore, the context-dependent presence and action of a transcriptional cofactor provides a versatile mechanism to restrict the phenotypic effects of variation in expression of a pleiotropic developmental factor in an evolutionary context (fig. 5B).
Repeated Evolution of Hotspot Genes May Play a Minor Role for Complex Trait Evolution
Natural intra- and interspecific variation in eye size and a developmental trade-off between retinal tissue and head cuticle is a common feature of Drosophila (Norry et al. 2000; Posnien et al. 2012; Arif et al. 2013; Hilbrant et al. 2014; Norry and Gomez 2017; Keesey et al. 2019; Ramaekers et al. 2019). The most comprehensive genetic and developmental characterization of this trait suggested that differences in the early subdivision of the eye–antennal disc driven by variation in the temporal regulation of eyeless/Pax6 expression is a common mechanism driving the developmental trade-off (Ramaekers et al. 2019). In line with the identification of evolutionary hotspot genes underlying variation in pigmentation patterns (Gompel and Carroll 2003; Prud’homme et al. 2006) and the gain and loss of structures (Sucena et al. 2003; Shapiro et al. 2006; Chan et al. 2010), this finding suggests that repeated natural variation in hotspot genes may be a major driver for the evolution of size and shape of complex organs as well (Ramaekers et al. 2019). In contrast to these results, our comparison of eye–antennal disc development between D. melanogaster and D. mauritiana suggest that differences in the progression of differentiation during later third instar stages contribute to the observed differences in eye size and head shape. Accordingly, our developmental model involving natural variation in pnr and ush expression (fig. 5A) provides an alternative developmental mechanism that can explain the simultaneous effects on ommatidia number variation and differences in the interstitial head cuticle.
Recent advances in studying the developmental and genetic basis of the different investment in eye and interstitial cuticle tissue in Drosophila revealed that differences in developmental processes affecting both traits simultaneously seem to be common in Drosophila (Norry and Gomez 2017; Gaspar et al. 2020). However, eye and interstitial cuticle size can also be genetically and developmentally uncoupled as shown by a detailed characterization of the head trade-off in D. mauritiana and D. simulans (Arif et al. 2013). Quantitative genetics data for intraspecific variation in the head trade-off in D. melanogaster and D. simulans show that its genetic basis is highly diverse (Gaspar et al. 2020), supporting our observation that repeated evolution through hotspot genes may not be prevalent in the evolution of head morphology.
The insect compound eye is composed of hundreds of functional subunits, the ommatidia, and they themselves are built by 14 unique cells (Cagan and Ready 1989). Therefore, the size of an insect compound eye can vary due to the number of ommatidia or due to their size. Indeed, a detailed morphological analysis showed that differences in the number of ommatidia explains intra- and interspecific variation in various Drosophila species (this work and Posnien et al. 2012; Ramaekers et al. 2019; Gaspar et al. 2020), whereas eye size differences between D. simulans and D. mauritiana are caused by variation in ommatidia size (Posnien et al. 2012; Arif et al. 2013). As these two features are regulated at different timepoints by different developmental processes, it is expected that the molecular and developmental basis of eye size differences varies in different groups. Therefore, depending on the cellular basis of observed morphological differences, different developmental and molecular mechanisms can be affected.
A potential explanation for the convergent evolution of the head trade-off may be the complex modular nature of head development. The eye–antennal disc comprises compartments for all future head structures, including the eye, the interstitial cuticle, and the antennae (supplementary fig. S1A, Supplementary Material online). Besides the already established mechanisms (this work and Ramaekers et al. 2019), it is conceivable that differences in adult head morphology can be the result of variation in different developmental processes in each compartment of the disc at different time points. Additionally, the development of a complex organ, such as the insect compound eye, is controlled by several hub genes. This is best exemplified by the observation that the experimental mis-expression of different members of the retinal determination network can induce an ectopic retinal developmental program (Halder et al. 1995; Bonini et al. 1997; Shen and Mardon 1997; Pan and Rubin 1998; Seimiya and Gehring 2000), suggesting that they all act as input–output devices in the retinal GRN. In contrast, the development of rather simple morphological traits is regulated by very few hub genes that act as input–output devices integrating positional information and activating most target genes required to initiate the entire developmental program to form the structure (Stern and Orgogozo 2008, 2009). In summary, our current knowledge based on quantitative genetics, developmental as well as morphological data suggest that different nodes within the GRN underlying head and eye development evolve to give rise to variation in head morphology in Drosophila. We argue that the complexity and modularity of complex organ development facilitate the convergent evolution of complex morphological traits.
Context-Dependent Regulatory Interactions Facilitate the Evolution of Pleiotropic Hub Genes
Our data and the work of others consistently revealed variation in the expression and function of crucial pleiotropic genes contributing to morphological evolution (e.g., this work and Shapiro et al. 2004; McGregor et al. 2007; Ramaekers et al. 2019). On the first glance, this observation is counter-intuitive for two reasons: 1) the mutation rates of central developmental regulators, such as TFs and signaling molecules, are low (Wagner and Lynch 2008; Alvarez-Ponce et al. 2009; Davila-Velderrain et al. 2014) and such factors tend to be functionally conserved across the animal kingdom (Halder et al. 1995) and 2) systematic analyses of GRN connectivity showed that developmental regulators are often highly connected (Borneman et al. 2006; Vermeirssen et al. 2007). As GRNs are thought to be scale-free with few highly connected hub genes, mutations are more likely to occur in the many weakly connected peripheral genes (Barabási and Oltvai 2004; Boyle et al. 2017). Therefore, the question arises, why morphological evolution is often driven by natural variation in expression of highly connected pleiotropic genes?
First, although a high level of network connectivity ensures network stability, data obtained in yeast showed that genes that are regulated by many other factors (trans-mutational target size) and genes that contain many TF-binding motifs (cis-mutational target size) are more prone to accumulate mutations (Promislow 2005; Landry et al. 2007). A study of Mendelian and complex disease genes revealed that they are often highly connected and that they act as brokers, connecting genes that are themselves poorly connected. Such broker positions could represent fragile nodes and variation in such genes may more likely result in phenotypic differences (Cai et al. 2010). Although it remains to be tested if highly connected developmental regulators occupy similar fragile positions within GRNs, the position of pleiotropic genes within GRNs may influence the evolvability of their expression or function.
Second, highly connected genes often contain a complex regulatory landscape with many cis-regulatory elements allowing for their incorporation in different developmental GRNs (Promislow 2005; Borneman et al. 2006; Vermeirssen et al. 2007). Consequently, a complex gene regulation facilitates the remodeling of regulatory interactions in a temporally and spatially defined manner (reviewed in Macneil and Walhout 2011). Eye–antennal disc development is highly complex and the regulatory interactions within the underlying GRN are variable both throughout time (Torres-Oliva et al. 2018) and in different parts of the disc (Potier et al. 2014), requiring the use of the same developmental gene products in different contexts. For instance, genes of the retinal determination network are essential for the initial proliferation and growth of the entire eye–antennal disc (Bessa et al. 2002; Lopes and Casares 2010; Neto et al. 2017) and later they play a pivotal role in retinal specification (Treisman 2013; Casares and Almudi 2016). These distinct roles have been suggested to be achieved by considerable rewiring of the respective GRNs, which allows them to fulfill temporally and even spatially restricted tasks (Palliyil et al. 2018). The various described roles for Pnr during eye development (Maurel-Zaffran and Treisman 2000; Oros et al. 2010), its continuous expression in the dorsal eye–antennal disc, and the observation that variation in pnr expression affects overall head shape and eye size simultaneously (this work), strongly suggest that Pnr as well is involved in several subnetworks during eye and head development. The interaction with cofactors, such as Ush provides a mechanism of modulating the role of Pnr from an activating to a repressing TF and its usage in spatially defined GRNs. Hence, the context-specific modulation of expression of a highly pleiotropic developmental factor, such as pnr and its target genes, is facilitated by complex and modular gene regulation mechanisms and a highly dynamic underlying GRN.
A Versatile Method to Identify Tuning Nodes within Developmental GRNs Underlying Morphological Evolution
As natural variation in gene expression is a major driver of morphological divergence (Stern and Orgogozo 2008; Buchberger et al. 2019), many studies aiming at revealing candidate genes underlying the evolution of morphological traits employ comparative transcriptomics methods to study differential gene expression. However, changes in gene function and expression are often buffered in GRNs to maintain a stable phenotypic outcome (Giaever et al. 2002; Kamath et al. 2003; Hollenhorst et al. 2007; Costanzo et al. 2010; Frankel et al. 2010; Gavin-Smyth et al. 2013; Tsai et al. 2019). This is best exemplified by the observation that the individual loss of function of many genes in yeast (Giaever et al. 2002) and the nematode Caenorhabditis elegans (Kamath et al. 2003) rarely has deleterious effects for the organism. In accordance with a generally conserved adult head morphology in D. melanogaster and D. mauritiana, we found that the expression of many genes representing key developmental and metabolic processes was conserved between the two species. However, the observed quantitative interspecific differences in eye size and head shape must be due to variation in some parts of the GRN which can be selected for if it translates into phenotypic diversity that is advantageous for the organism (Tsuda and Kawata 2010; Thompson et al. 2015; Chen et al. 2019).
In the light of the observed discrepancy between a generally high network stability and the flexibility in some parts of the network, we propose that developmental GRNs must contain at least two types of nodes (i.e., genes): structural nodes and tuning nodes (fig. 5D). Structural nodes confer phenotypic stability because variation in such nodes (e.g., variation in expression of the gene) is either prevented by tight regulation at the locus (fig. 5Di) or buffered downstream in the GRN (fig. 5Dii). Such a robustness has been demonstrated to be often the result of redundancies (Costanzo et al. 2010), either on the level of cis-regulatory elements (Frankel et al. 2010; Tsai et al. 2019) or on TFs and signaling molecules (Hollenhorst et al. 2007; Gavin-Smyth et al. 2013). In contrast, tuning nodes represent genes for which variation is tolerated within the GRN and results in quantitative phenotypic variation (fig. 5Diii). Natural variation in ey (Ramaekers et al. 2019) and pnr expression (this work), which translates into head shape and eye size differences, as well as the previously identified hotspot genes underlying variation in trichome formation (Sucena et al. 2003) and body pigmentation (Gompel and Carroll 2003; Prud’homme et al. 2006) are excellent examples of such tuning nodes. It is important to note here that developmental genes may represent tuning nodes in some contexts, whereas they are structural nodes in most other contexts. Our observation that interspecific differences in pnr expression affected the dorsal head shape and eye size, but not sensory bristle formation is an excellent example for the context dependence of tuning and structural nodes. Revealing structural and tuning nodes in different developmental GRNs allows gaining new insights into constrained and variable developmental processes, respectively.
As many highly connected hub genes with pleiotropic functions have been shown to regulate morphological differences, they can be identified based on their effect on their target genes (Peter and Davidson 2011). A systematic analysis of GRN properties suggests that TF-to-gene interactions are scale-free, meaning that some TFs regulate a high number of genes (Barabási and Oltvai 2004). Therefore, we hypothesized that putative tuning nodes (e.g., genes coding for TFs) can be elucidated by their impact on the expression of their target genes (i.e., gene modules; Bar-Joseph et al. 2003; Segal et al. 2003; Arda and Walhout 2010). Accordingly, our methodological framework combines the identification of DEGs, clustering of DEGs based on their developmental expression dynamics and the identification of putative shared upstream regulators integrating genomewide chromatin accessibility data (fig. 2). As fluctuations in gene expression are often buffered within GRNs, not all expression differences of putative tuning nodes will result in phenotypic differences. Therefore, a functional validation is crucial to reveal phenotypically relevant tuning nodes. Such a validation can be achieved by applying classical developmental genetics methods as shown in this work. If such tools are not established, a tuning node could be validated using a hemizygosity test based on widely applicable CRISPR/Cas9 aided genome editing (Stern 2014).
Our approach unfolds its full potential if complemented with quantitative genetics data to identify exact genetic variants constituting the tuning node. This is relevant because mutations affecting the expression or function of tuning nodes could be in its locus itself or it could be the result of genetic changes in an upstream regulator. In the latter case, it is expected that the number of potential candidates upstream of the tuning node is low because systematic analyses of GRN architecture revealed that the in-degree within networks follows a random distribution, suggesting that most TFs tend to be regulated by only a few upstream factors (Barabási and Oltvai 2004). Therefore, once putative tuning nodes are identified by comparative transcriptomics, the number of candidate genes that regulate the tuning node is expected to be low facilitating further molecular characterization.
In summary, our comparative transcriptomics approach can be used as entry point to study the evolution of complex morphological traits or it can be applied to link already identified genetic variation to nodes within developmental GRNs and to developmental processes. As the individual steps are applicable in many organisms, including those for which quantitative genetics approaches are not applicable, we are convinced that the approach represents a versatile framework to study the molecular and developmental basis of morphological evolution.
Materials and Methods
A detailed description of all methods is available as Extended Materials and Methods in the Supplementary Material, Supplementary Material online.
Comparative Transcriptomics and ATACseq
Eye–antennal imaginal discs were dissected 72, 96, and 120 h AEL from larvae of D. melanogaster (Oregon R) and D. mauritiana (TAM16), respectively. Total RNA was extracted from the discs and libraries for Illumina HiSeq2000 sequencing were generated using the TruSeq RNA Sample Preparation Kit (Illumina, catalog ID RS-122-2002). Reads were mapped using Bowtie2 v. 2.3.4.1 (Langmead and Salzberg 2012), mapping data were processed using samtools version 1.9 (Li et al. 2009; Li 2011), and differential expression analysis was performed with the DESeq2 package (DESeq2_1.22.2, Love et al. 2014; R version 3.5.2). Metascape was used to analyze differential enrichment of GO terms for pairwise comparisons. Clustering of genes according to their developmental expression dynamics was performed with the coseq package (version 1.6.1) (Rau and Maugis-Rabusseau 2018; Godichon-Baggioni et al. 2019) and we searched for potential upstream factors using the i-cisTarget tool (Herrmann et al. 2012; Imrichová et al. 2015). ATACseq to assess chromatin accessibility was perfromed as described before (Buenrostro et al. 2013). The pipeline used to define a high confidence target gene list of Pnr is described in detail in the Extended Materials and Methods, Supplementary Material online.
Genetic Crosses, Immunohistology, and Phenotyping
All fly crosses to knock down and overexpress pnr and ush as well as crosses for lineage tracing experiments were performed at 25 °C and at a constant 12:12 h light:dark cycle. We generated polyclonal antibodies against Pnr (Junion et al. 2012) and Ush (Fossett et al. 2001) based on previous knowledge (Proteintech, Rosemont, IL), and immunohistology was performed as described in the Extended Materials and Methods, Supplementary Material online. Adult heads were either mounted in Hoyer’s medium:Lactic Acid (50:50) or directly imaged for ommatidia counting or shape analysis using Geometric Morphometrics.
Supplementary Material
Acknowledgments
Many thanks to Marita Büscher and Kolja Eckermann for sharing antibodies and to Alistair P. McGregor, Marc Haenlin, and Gerd Vorbrüggen for sharing Drosophila stocks. We are grateful for financial support for the ATACseq data as well as for critical comments on an earlier version of this manuscript to Alistair P. McGregor. Stocks obtained from the Bloomington Drosophila Stock Center (National Institute of Health [NIH] P40OD018537) were used in this study. Many thanks to the Deep-Sequencing Core Facility of the Universitätsmedizin Göttingen (UMG) for next generation sequencing. We are grateful for many fruitful discussions and feedback from members of the Department of Developmental Biology and Posnien Lab members. Also, many thanks to the three anonymous reviewers whose suggestions helped to improve the manuscript considerably. N.P. and E.B. are funded by the Deutsche Forschungsgesellschaft (DFG, Grant No. PO 1648/3-1 to N.P.). F.C. acknowledges funding through grants BFU2015-66040-P, PGC2018-093704-B-I00, and MDM-2016-0687 from Ministerio de Ciencia, Innovación y Universidades of Spain. We acknowledge support by the Open Access Publication Funds of the Göttingen University.
Author Contributions
E.B.: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing—original draft, Writing—review and editing; A.B.: Investigation, Writing—review and editing; S.A.: Investigation, Writing—review and editing; D.S.: Investigation, Writing—review and editing; C.M.: Investigation, Writing—review and editing; A.N.: Investigation, Writing—review and editing; I.A.: Resources, Writing—review and editing; M.T.-O.: Investigation, Resources, Writing—review and editing; F.C.: Investigation, Resources, Writing—review and editing; N.P.: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Visualization, Writing—original draft, Writing—review and editing.
Data Availability
All RNAseq and ATACseq reads are accessible in the Short Read Archive through umbrella BioProject PRJNA666691 (containing PRJNA374838 and PRJNA666524). All additional files containing measurements and statistics for morphological comparisons as well as intermediate data for the transcriptomic and ATACseq analyses are available at the Göttingen Research Online Repository (https://doi.org/10.25625/VUJOJS and https://doi.org/10.25625/HICPAZ).
References
- Alberch P. 1991. From genes to phenotype: dynamical systems and evolvability. Genetica 84(1):5–11. [DOI] [PubMed] [Google Scholar]
- Alvarez-Ponce D, Aguadé M, Rozas J.. 2009. Network-level molecular evolutionary analysis of the insulin/TOR signal transduction pathway across 12 Drosophila genomes. Genome Res. 19(2):234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda HE, Walhout AJM.. 2010. Gene-centered regulatory networks. Brief Funct Genomics. 9(1):4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif S, Hilbrant M, Hopfen C, Almudi I, Nunes MDS, Posnien N, Kuncheria L, Tanaka K, Mitteroecker P, Schlötterer C, et al. 2013. Genetic and developmental analysis of differences in eye and face morphology between Drosophila simulans and Drosophila mauritiana. Evol Dev. 15(4):257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins M, Mardon G.. 2009. Signaling in the third dimension: the peripodial epithelium in eye disc development. Dev Dyn. 238(9):2139–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker LR, Weasner BM, Nagel A, Neuman SD, Bashirullah A, Kumar JP.. 2018. Eyeless/Pax6 initiates eye formation non-autonomously from the peripodial epithelium. Development (Cambridge, England) 145(15):dev163329.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Joseph Z, Gerber GK, Lee TI, Rinaldi NJ, Yoo JY, Robert F, Gordon DB, Fraenkel E, Jaakkola TS, Young RA, et al. 2003. Computational discovery of gene modules and regulatory networks. Nat Biotechnol. 21(11):1337–1342. [DOI] [PubMed] [Google Scholar]
- Barabási A-L, Albert R.. 1999. Emergence of scaling in random networks. Science 286(5439):509–512. [DOI] [PubMed] [Google Scholar]
- Barabási AL, Oltvai ZN.. 2004. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 5(2):101–113. [DOI] [PubMed] [Google Scholar]
- Batada NN, Hurst LD.. 2007. Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat Genet. 39(8):945–949. [DOI] [PubMed] [Google Scholar]
- Bessa J, Gebelein B, Pichaud F, Casares F, Mann RS.. 2002. Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev. 16(18):2415–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Baker NE.. 2009. The HLH protein Extramacrochaetae is required for R7 cell and cone cell fates in the Drosophila eye. Dev Biol. 327(2):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonini NM, Bui QT, Gray-Board GL, Warrick JM.. 1997. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development 124(23):4819–4826. [DOI] [PubMed] [Google Scholar]
- Borneman AR, Leigh-Bell JA, Yu H, Bertone P, Gerstein M, Snyder M.. 2006. Target hub proteins serve as master regulators of development in yeast. Genes Dev. 20(4):435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EA, Li YI, Pritchard JK.. 2017. An expanded view of complex traits: from polygenic to omnigenic. Cell 169(7):1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Paddock SW, Sattler C, Cronmiller C, Thomas BJ, Carroll SB.. 1996. daughterless is required for Drosophila photoreceptor cell determination, eye morphogenesis, and cell cycle progression. Dev Biol. 179(1):65–78. [DOI] [PubMed] [Google Scholar]
- Brown NL, Sattler CA, Paddock SW, Carroll SB.. 1995. Hairy and EMC negatively regulate morphogenetic furrow progression in the Drosophila eye. Cell 80(6):879–887. [DOI] [PubMed] [Google Scholar]
- Buchberger E, Reis M, Lu T-H, Posnien N.. 2019. Cloudy with a chance of insights: context dependent gene regulation and implications for evolutionary studies. Genes 10(7):492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ.. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 10(12):1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgio G, Baylac M, Heyer E, Montagutelli X.. 2009. Genetic analysis of skull shape variation and morphological integration in the mouse using interspecific recombinant congenic strains between C57BL/6 and mice of the Mus spretus species. Evolution 63(10):2668–2686. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Ready DF.. 1989. The emergence of order in the Drosophila pupal retina. Dev Biol. 136(2):346–362. [DOI] [PubMed] [Google Scholar]
- Cai JJ, Borenstein E, Petrov DA.. 2010. Broker genes in human disease. Genome Biol Evol. 2:815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, Koelling N, Harnett D, Garfield D, Casale FP, Ciglar L, Gustafson HE, Viales RR, Marco-Ferreres R, Degner JF, et al. 2017. Genetic variants regulating expression levels and isoform diversity during embryogenesis. Nature 541(7637):402–406. [DOI] [PubMed] [Google Scholar]
- Casares F, Almudi I.. 2016. Fast and Furious 800. The retinal determination gene network in Drosophila. In: Castelli-Gair Hombría J, Bovolenta P, editors. Organogenetic gene networks. Genetic control of organ formation. Cham (Switzerland: ): Springer International Publishing. p. 95–124. [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, et al. 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327(5963):302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhang D, Hazbun TR, Zhang M.. 2019. Inferring gene regulatory networks from a population of yeast segregants. Sci Rep. 9(1):1197.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero JB, Larson DE, Craig CR, Hays R, Cagan R.. 2007. Dynamic decapentaplegic signaling regulates patterning and adhesion in the Drosophila pupal retina. Development 134(10):1861–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JLY, Toufighi K, Mostafavi S, et al. 2010. The genetic landscape of a cell. Science (New York, N.Y.) 327(5964):425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubadda Y, Heitzler P, Ray RP, Bourouis M, Ramain P, Gelbart W, Simpson P, Haenlin M.. 1997. u-shaped encodes a zinc finger protein that regulates the proneural genes achaete and scute during the formation of bristles in Drosophila. Genes Dev. 11(22):3083–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila-Velderrain J, Servin-Marquez A, Alvarez-Buylla ER.. 2014. Molecular evolution constraints in the floral organ specification gene regulatory network module across 18 angiosperm genomes. Mol Biol Evol. 31(3):560–573. [DOI] [PubMed] [Google Scholar]
- Ekas LA, Baeg G-H, Flaherty MS, Ayala-Camargo A, Bach EA.. 2006. JAK/STAT signaling promotes regional specification by negatively regulating wingless expression in Drosophila. Development 133(23):4721–4729. [DOI] [PubMed] [Google Scholar]
- Fear JM, León-Novelo LG, Morse AM, Gerken AR, van Lehmann K, Tower J, Nuzhdin SV, McIntyre LM.. 2016. Buffering of genetic regulatory networks in Drosophila melanogaster. Genetics 203(3):1177–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone DE, Broadie K.. 2002. Wrestling with pleiotropy: genomic and topological analysis of the yeast gene expression network. Bioessays 24(3):267–274. [DOI] [PubMed] [Google Scholar]
- Flaherty MS, Zavadil J, Ekas LA, Bach EA.. 2009. Genome-wide expression profiling in the Drosophila eye reveals unexpected repression of notch signaling by the JAK/STAT pathway. Dev Dyn. 238(9):2235–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossett N, Tevosian SG, Gajewski K, Zhang Q, Orkin SH, Schulz RA.. 2001. The Friend of GATA proteins u-shaped, FOG-1, and FOG-2 function as negative regulators of blood, heart, and eye development in Drosophila. Proc Natl Acad Sci U S A. 98(13):7342–7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL.. 2010. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466(7305):490–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW.. 2002. Evolutionary rate in the protein interaction network. Science 296(5568):750–752. [DOI] [PubMed] [Google Scholar]
- Fromental-Ramain C, Taquet N, Ramain P.. 2010. Transcriptional interactions between the pannier isoforms and the cofactor u-shaped during neural development in Drosophila. Mech Dev. 127(9–12):442–457. [DOI] [PubMed] [Google Scholar]
- Fromental-Ramain C, Vanolst L, Delaporte C, Ramain P.. 2008. pannier encodes two structurally related isoforms that are differentially expressed during Drosophila development and display distinct functions during thorax patterning. Mech Dev. 125(1–2):43–57. [DOI] [PubMed] [Google Scholar]
- García-García MJ, Ramain P, Simpson P, Modolell J.. 1999. Different contributions of pannier and wingless to the patterning of the dorsal mesothorax of Drosophila. Development 126(16):3523–3532. [DOI] [PubMed] [Google Scholar]
- Gaspar P, Arif S, Sumner-Rooney L, Kittelmann M, Bodey AJ, Stern DL, Nunes MDS, McGregor AP.. 2020. Characterization of the genetic architecture underlying eye size variation within Drosophila melanogaster and Drosophila simulans. G3 (Bethesda, MD) 10(3):1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin-Smyth J, Wang YC, Butler I, Ferguson EL.. 2013. A genetic network conferring canalization to a bistable patterning system in Drosophila. Curr Biolo. 23(22):2296–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418(6896):387–391. [DOI] [PubMed] [Google Scholar]
- Gibson MC, Schubiger G.. 2000. Peripodial cells regulate proliferation and patterning of Drosophila imaginal discs. Cell 103(2):343–350. [DOI] [PubMed] [Google Scholar]
- Gibson MC, Schubiger G.. 2001. Drosophila peripodial cells, more than meets the eye? Bioessays 23(8):691–697. [DOI] [PubMed] [Google Scholar]
- Godichon-Baggioni A, Maugis-Rabusseau C, Rau A.. 2019. Clustering transformed compositional data using K-means, with applications in gene expression and bicycle sharing system data. J Appl Stat. 46(1):47–65. [Google Scholar]
- Gomez-Skarmeta JL, Campuzano S, Modolell J.. 2003. Half a century of neural prepatterning: the story of a few bristles and many genes. The story of a few bristles and many genes. Nat Rev Neurosci. 4(7):587–598. [DOI] [PubMed] [Google Scholar]
- Gompel N, Carroll SB.. 2003. Genetic mechanisms and constraints governing the evolution of correlated traits in drosophilid flies. Nature 424(6951):931–935. [DOI] [PubMed] [Google Scholar]
- Haenlin M, Cubadda Y, Blondeau F, Heitzler P, Lutz Y, Simpson P, Ramain P.. 1997. Transcriptional activity of Pannier is regulated negatively by heterodimerization of the GATA DNA-binding domain with a cofactor encoded by the u-shaped gene of Drosophila. Genes Dev. 11(22):3096–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ.. 1995. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science (New York, N.Y.) 267(5205):1788–1792. [DOI] [PubMed] [Google Scholar]
- Hammerle B, Ferrus A.. 2003. Expression of enhancers is altered in Drosophila melanogaster hybrids. Evol Dev. 5(3):221–230. [DOI] [PubMed] [Google Scholar]
- Haynie JL, Bryant PJ.. 1986. Development of the eye-antenna imaginal disc and morphogenesis of the adult head in Drosophila melanogaster. J Exp Zool. 237(3):293–308. [DOI] [PubMed] [Google Scholar]
- Heitzler P, Haenlin M, Ramain P, Calleja M, Simpson P.. 1996. A genetic analysis of pannier, a gene necessary for viability of dorsal tissues and bristle positioning in Drosophila. Genetics 143(3):1271–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann C, van de Sande B, Potier D, Aerts S.. 2012. i-cisTarget. An integrative genomics method for the prediction of regulatory features and cis-regulatory modules. Nucleic Acids Res. 40(15):e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbrant M, Almudi I, Leite DJ, Kuncheria L, Posnien N, Nunes MD, McGregor AP.. 2014. Sexual dimorphism and natural variation within and among species in the Drosophila retinal mosaic. BMC Evol Biol. 14(1):240.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP.. 2006. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science 313(5783):101–104. [DOI] [PubMed] [Google Scholar]
- Hollenhorst PC, Shah AA, Hopkins C, Graves BJ.. 2007. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 21(15):1882–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Dambly-Chaudière C, Ghysen A.. 1991. The emergence of sense organs in the wing disc of Drosophila. Development 111(4):1087–1095. [DOI] [PubMed] [Google Scholar]
- Imrichová H, Hulselmans G, Atak ZK, Potier D, Aerts S.. 2015. i-cisTarget 2015 update. Generalized cis-regulatory enrichment analysis in human, mouse and fly. Nucleic Acids Res. 43(W1):W57–W64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y.. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 98(8):4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL.. 2000. The large-scale organization of metabolic networks. Nature 407(6804):651–654. [DOI] [PubMed] [Google Scholar]
- Jeong S, Rokas A, Carroll SB.. 2006. Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell 125(7):1387–1399. [DOI] [PubMed] [Google Scholar]
- Junion G, Spivakov M, Girardot C, Braun M, Gustafson EH, Birney E, Furlong EEM.. 2012. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell 148(3):473–486. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421(6920):231–237. [DOI] [PubMed] [Google Scholar]
- Keesey IW, Grabe V, Gruber L, Koerte S, Obiero GF, Bolton G, Khallaf MA, Kunert G, Lavista-Llanos S, Valenzano DR, et al. 2019. Inverse resource allocation between vision and olfaction across the genus Drosophila. Nat Commun. 10(1):1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernan MJ. 2007. Mechanotransduction and auditory transduction in Drosophila. Pflugers Arch. 454(5):703–720. [DOI] [PubMed] [Google Scholar]
- Kitano H. 2004. Biological robustness. Nat Rev Genet. 5(11):826–837. [DOI] [PubMed] [Google Scholar]
- Krylov DM, Wolf YI, Rogozin IB, Koonin EV.. 2003. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 13(10):2229–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP. 2011. My what big eyes you have. How the Drosophila retina grows. Dev Neurobiol. 71(12):1133–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise D, David JR, Lemeunier F, Tsacas L.. 1986. The reproductive relationships of Drosophila sechellia with D. mauritiana, D. simulans, and D. melanogaster from the Afrotropical region. Evolution 40(2):262–271. [DOI] [PubMed] [Google Scholar]
- Landry CR, Lemos B, Rifkin S, Dickinson WJ, Hartl DL.. 2007. Genetic properties influencing the evolvability of gene expression. Science 317(5834):118–121. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B. 2008. Selection to minimise noise in living systems and its implications for the evolution of gene expression. Mol Syst Biol. 4(1):170.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC. 1974. The genetic basis of evolutionary change. New York: Columbia University Press. [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27(21):2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gu H-Y, Zhu J, Niu Y-M, Zhang C, Guo G-L.. 2019. Identification of hub genes and key pathways associated with bipolar disorder based on weighted gene co-expression network analysis. Front Physiol. 10:1081.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes CS, Casares F.. 2010. Hth maintains the pool of eye progenitors and its downregulation by Dpp and Hh couples retinal fate acquisition with cell cycle exit. Dev Biol. 339(1):78–88. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF. 2001. Quantitative trait loci in Drosophila. Nat Rev Genet. 2(1):11–20. [DOI] [PubMed] [Google Scholar]
- Mackay TFC, Stone E, Ayroles JF.. 2009. The genetics of quantitative traits. Challenges and prospects. Nat Rev Genet. 10(8):565–577. [DOI] [PubMed] [Google Scholar]
- Macneil LT, Walhout AJM.. 2011. Gene regulatory networks and the role of robustness and stochasticity in the control of gene expression. Genome Res. 21(5):645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga AM, Navarro N, Cunningham ML, Cox TC.. 2015. Quantitative trait loci affecting the 3D skull shape and size in mouse and prioritization of candidate genes in-silico. Front Physiol. 6:92.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri MS, Domínguez-Cejudo MA, Casares F.. 2018. Wnt controls the medial-lateral subdivision of the Drosophila head. Biol Lett. 14(7):20180258.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamoro-Vidal A, Tully T, Courtier-Orgogozo V.. 2018. Robustness of bristle number to temperature and genetic background varies with bristle type and is regulated by miR-9a. bioRxiv. 295485.
- Maurel-Zaffran C, Treisman JE.. 2000. pannier acts upstream of wingless to direct dorsal eye disc development in Drosophila. Development 127(5):1007–1016. [DOI] [PubMed] [Google Scholar]
- McGregor AP, Orgogozo V, Delon I, Zanet J, Srinivasan DG, Payre F, Stern DL.. 2007. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature 448(7153):587–590. [DOI] [PubMed] [Google Scholar]
- Menuet A, Alunni A, Joly J-S, Jeffery WR, Rétaux S.. 2007. Expanded expression of Sonic Hedgehog in Astyanax cavefish. Multiple consequences on forebrain development and evolution. Development 134(5):845–855. [DOI] [PubMed] [Google Scholar]
- Murali T, Pacifico S, Yu J, Guest S, Roberts GG, Finley RL.. 2011. DroID 2011: a comprehensive, integrated resource for protein, transcription factor, RNA and gene interactions for Drosophila. Nucleic Acids Res. 39(Database issue):D736–D743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto M, Naval-Sánchez M, Potier D, Pereira PS, Geerts D, Aerts S, Casares F.. 2017. Nuclear receptors connect progenitor transcription factors to cell cycle control. Sci Rep. 7(1):4845.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norry FM, Gomez FH.. 2017. Quantitative trait loci and antagonistic associations for two developmentally related traits in the Drosophila head. J Insect Sci. 17(1):19.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norry FM, Vilardi JC, Hasson E.. 1997. Genetic and phenotypic correlations among size-related traits, and heritability variation between body parts in Drosophila buzzatii. Genetica 101(2):131–139. [DOI] [PubMed] [Google Scholar]
- Norry FM, Vilardi JC, Hasson E.. 2000. Negative genetic correlation between traits of the Drosophila head, and interspecific divergence in head shape. Heredity 85(2):177–183. [DOI] [PubMed] [Google Scholar]
- Oros SM, Tare M, Kango-Singh M, Singh A.. 2010. Dorsal eye selector pannier (pnr) suppresses the eye fate to define dorsal margin of the Drosophila eye. Dev Biol. 346(2):258–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrin EJ, Li Y, Hoffman K, Liu J, Wang K, Zhang L, Mardon G, Chen R.. 2006. Genome-wide identification of direct targets of the Drosophila retinal determination protein eyeless. Genome Res. 16(4):466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallares LF, Carbonetto P, Gopalakrishnan S, Parker CC, Ackert-Bicknell CL, Palmer AA, Tautz D.. 2015. Mapping of craniofacial traits in outbred mice identifies major developmental genes involved in shape determination. PLoS Genet. 11(11):e1005607.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallares LF, Harr B, Turner LM, Tautz D.. 2014. Use of a natural hybrid zone for genomewide association mapping of craniofacial traits in the house mouse. Mol Ecol. 23(23):5756–5770. [DOI] [PubMed] [Google Scholar]
- Palliyil S, Zhu J, Baker LR, Neuman SD, Bashirullah A, Kumar JP.. 2018. Allocation of distinct organ fates from a precursor field requires a shift in expression and function of gene regulatory networks. PLoS Genet. 14(1):e1007185.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Rubin GM.. 1998. Targeted expression of teashirt induces ectopic eyes in Drosophila. Proc Natl Acad Sci U S A. 95(26):15508–15512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira PS, Pinho S, Johnson K, Couso JP, Casares F.. 2006. A 3' cis-regulatory region controls wingless expression in the Drosophila eye and leg primordia. Dev Dyn. 235(1):225–234. [DOI] [PubMed] [Google Scholar]
- Peter IS, Davidson EH.. 2011. Evolution of gene regulatory networks controlling body plan development. Cell 144(6):970–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posnien N, Hopfen C, Hilbrant M, Ramos-Womack M, Murat S, Schönauer A, Herbert SL, Nunes MDS, Arif S, Breuker CJ, et al. 2012. Evolution of eye morphology and rhodopsin expression in the Drosophila melanogaster species subgroup. PLoS One 7(5):e37346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potier D, Davie K, Hulselmans G, Naval Sanchez M, Haagen L, Huynh-Thu VA, Koldere D, Celik A, Geurts P, Christiaens V, et al. 2014. Mapping gene regulatory networks in Drosophila eye development by large-scale transcriptome perturbations and motif inference. Cell Rep. 9(6):2290–2303. [DOI] [PubMed] [Google Scholar]
- Promislow D. 2005. A regulatory network analysis of phenotypic plasticity in yeast. Am Nat. 165(5):515–523. [DOI] [PubMed] [Google Scholar]
- Prud’homme B, Gompel N, Rokas A, Va K, Williams TM, Yeh S-D, True JR, Carroll SB.. 2006. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440(7087):1050–1053. [DOI] [PubMed] [Google Scholar]
- Ramaekers A, Claeys A, Kapun M, Mouchel-Vielh E, Potier D, Weinberger S, Grillenzoni N, Dardalhon-Cuménal D, Yan J, Wolf R, et al. 2019. Altering the temporal regulation of one transcription factor drives evolutionary trade-offs between head sensory organs. Dev Cell. 50(6):780–792.e7. [DOI] [PubMed] [Google Scholar]
- Rau A, Maugis-Rabusseau C.. 2018. Transformation and model choice for RNA-seq co-expression analysis. Brief Bioinform. 19(3):425–436. [DOI] [PubMed] [Google Scholar]
- Rebeiz M, Pool JE, Kassner V, Aquadro CF, Carroll SB.. 2009. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 326(5960):1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis M, Wiegleb G, Claude J, Lata R, Horchler B, Ha N-T, Reimer C, Vieira CP, Vieira J, Posnien N, et al. 2020. Multiple loci linked to inversions are associated with eye size variation in species of the Drosophila virilis phylad. Sci Rep. 10(1):12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Tomlinson A.. 2007. Dorsal-ventral midline signaling in the developing Drosophila eye. Development 134(4):659–667. [DOI] [PubMed] [Google Scholar]
- Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N.. 2003. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 34(2):166–176. [DOI] [PubMed] [Google Scholar]
- Seimiya M, Gehring WJ.. 2000. The Drosophila homeobox gene optix is capable of inducing ectopic eyes by an eyeless-independent mechanism. Development 127(9):1879–1886. [DOI] [PubMed] [Google Scholar]
- Sekelsky JJ, Newfeld SJ, Raftery LA, Chartoff EH, Gelbart WM.. 1995. Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics 139(3):1347–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo CH, Kim J-R, Kim M-S, Cho K-H.. 2009. Hub genes with positive feedbacks function as master switches in developmental gene regulatory networks. Bioinformatics 25(15):1898–1904. [DOI] [PubMed] [Google Scholar]
- Shapiro MD, Bell MA, Kingsley DM.. 2006. Parallel genetic origins of pelvic reduction in vertebrates. Proc Natl Acad Sci U S A. 103(37):13753–13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jónsson B, Schluter D, Kingsley DM.. 2004. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 428(6984):717–723. [DOI] [PubMed] [Google Scholar]
- Shen W, Mardon G.. 1997. Ectopic eye development in Drosophila induced by directed dachshund expression. Development 124(1):45–52. [DOI] [PubMed] [Google Scholar]
- Singh A, Chan J, Chern JJ, Choi K-W.. 2005. Genetic interaction of Lobe with its modifiers in dorsoventral patterning and growth of the Drosophila eye. Genetics 171(1):169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Choi K-W.. 2003. Initial state of the Drosophila eye before dorsoventral specification is equivalent to ventral. Development 130(25):6351–6360. [DOI] [PubMed] [Google Scholar]
- Snodgrass RE. 1935. Principles of insect morphology. Ithaca (NY: ): McGraw-Hill Book Co. [Google Scholar]
- Sorrentino RP, Tokusumi T, Schulz RA.. 2007. The friend of GATA protein u-shaped functions as a hematopoietic tumor suppressor in Drosophila. Dev Biol. 311(2):311–323. [DOI] [PubMed] [Google Scholar]
- Stemmer M, Schuhmacher L-N, Foulkes NS, Bertolucci C, Wittbrodt J.. 2015. Cavefish eye loss in response to an early block in retinal differentiation progression. Development 142(4):743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL. 2000. Perspective. evolutionary developmental biology and the problem of variation. Evolution 54(4):1079–1091. [DOI] [PubMed] [Google Scholar]
- Stern DL. 2014. Identification of loci that cause phenotypic variation in diverse species with the reciprocal hemizygosity test. Trends Genet. 30(12):547–554. [DOI] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V.. 2008. The loci of evolution: how predictable is genetic evolution? How predictable is genetic evolution? Evolution 62(9):2155–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V.. 2009. Is genetic evolution predictable? Science 323(5915):746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucena E, Delon I, Jones I, Payre F, Stern DL.. 2003. Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature 424(6951):935–938. [DOI] [PubMed] [Google Scholar]
- Thompson D, Regev A, Roy S.. 2015. Comparative analysis of gene regulatory networks: from network reconstruction to evolution. Annu Rev Cell Dev Biol. 31(1):399–428. [DOI] [PubMed] [Google Scholar]
- Torres-Oliva M, Schneider J, Wiegleb G, Kaufholz F, Posnien N.. 2018. Dynamic genome wide expression profiling of Drosophila head development reveals a novel role of Hunchback in retinal glia cell development and blood-brain barrier integrity. PLoS Genet. 14(1):e1007180.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman JE. 2013. Retinal differentiation in Drosophila. Wires Dev Biol. 2(4):545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai A, Alves MR, Crocker J.. 2019. Multi-enhancer transcriptional hubs confer phenotypic robustness. eLife 8:e45325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda ME, Kawata M.. 2010. Evolution of gene regulatory networks by fluctuating selection and intrinsic constraints. PLoS Comput Biol. 6(8):e1000873.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403(6770):623–627. [DOI] [PubMed] [Google Scholar]
- Vermeirssen V, Barrasa MI, Hidalgo CA, Babon JAB, Sequerra R, Doucette-Stamm L, Barabási A-L, Walhout AJM.. 2007. Transcription factor modularity in a gene-centered C. elegans core neuronal protein-DNA interaction network. Genome Res. 17(7):1061–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. 1942. Canalization of development and the inheritance of acquired characters. Nature 150(3811):563–565. [Google Scholar]
- Wagner GP, Lynch VJ.. 2008. The gene regulatory logic of transcription factor evolution. Trends Ecol Evol. 23(7):377–385. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Zhang J.. 2011. The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat Rev Genet. 12(3):204–213. [DOI] [PubMed] [Google Scholar]
- Wang LH, Huang YT, Tsai YC, Sun YH.. 2010. The role of eyg Pax gene in the development of the head vertex in Drosophila. Dev Biol. 337(2):246–258. [DOI] [PubMed] [Google Scholar]
- Wells BS, Pistillo D, Barnhart E, Desplan C.. 2017. Parallel activin and BMP signaling coordinates R7/R8 photoreceptor subtype pairing in the stochastic Drosophila retina. eLife 6:e25301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersdorff V, Lecuit T, Cohen SM, Mlodzik M.. 1996. Mad acts downstream of Dpp receptors, revealing a differential requirement for dpp signaling in initiation and propagation of morphogenesis in the Drosophila eye. Development 122(7):2153–2162. [DOI] [PubMed] [Google Scholar]
- Yeung K, Wang F, Li Y, Wang K, Mardon G, Chen R.. 2018. Integrative genomic analysis reveals novel regulatory mechanisms of eyeless during Drosophila eye development. Nucleic Acids Res. 46(22):11743–11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Pacifico S, Liu G, Finley RL.. 2008. DroID: the Drosophila Interactions Database, a comprehensive resource for annotated gene and protein interactions. BMC Genomics. 9:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler MP, Perrimon N, Strutt DI.. 1999. Polarity determination in the Drosophila eye. A novel role for unpaired and JAK/STAT signaling. Genes Dev. 13(10):1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNAseq and ATACseq reads are accessible in the Short Read Archive through umbrella BioProject PRJNA666691 (containing PRJNA374838 and PRJNA666524). All additional files containing measurements and statistics for morphological comparisons as well as intermediate data for the transcriptomic and ATACseq analyses are available at the Göttingen Research Online Repository (https://doi.org/10.25625/VUJOJS and https://doi.org/10.25625/HICPAZ).