Abstract
Eutherian dentition has been the focus of a great deal of studies in the areas of evolution, development, and genomics. The development of molar teeth is regulated by an antero-to-posterior cascade mechanism of activators and inhibitors molecules, where the relative sizes of the second (M2) and third (M3) molars are dependent of the inhibitory influence of the first molar (M1). Higher activator/inhibitor ratios will result in higher M2/M1 or M3/M1. Pax9 has been shown to play a key role in tooth development. We have previously shown that a G-quadruplex in the first intron of Pax9 can modulate the splicing efficiency. Using a sliding window approach with we analyzed the association of the folding energy (Mfe) of the Pax9 first intron with the relative molar sizes in 42 mammalian species, representing 9 orders. The Mfe of two regions located in the first intron of Pax9 were shown to be significantly associated with the M2/M1 and M3/M1 areas and mesiodistal lengths. The first region is located at the intron beginning and can fold into a stable G4 structure, whereas the second is downstream the G4 and 265 bp from intron start. Across species, the first intron of Pax9 varied in G-quadruplex structural stability. The correlations were further increased when the Mfe of the two sequences were added. Our results indicate that this region has a role in the evolution of the mammalian dental pattern by influencing the relative size of the molars.
Keywords: Pax9, mammalian dentition, G-quadruplex
Introduction
The identification of regulatory domains in DNA and RNA sequences responsible for morphological evolution is one of the major challenges in the areas of evolution, development, and genomics. Evolutionary changes in body shape are thought to be mainly the result of distinct spatiotemporal expression of genes, which can be provoked by variations in conserved cis-acting regulatory sequences (Carroll 2008; Kvon et al. 2016; Roscito et al. 2018). Eutherian dentition has been the focus of a great deal of studies in this subject matter. The major gene networks and mechanisms governing tooth development have been described (Thesleff 2018). Morphological characteristics of dentition observed in mutant phenotypes and variation of dental pattern among species have been associated with trait inheritance, evolutionary aspects, and gene function (Line 2003; Kavanagh et al. 2007; Hlusko 2016). Therefore, the quest for domains responsible for the evolution of dental pattern may be eased by the facts that: 1) Dentition can be morphologically and developmentally divided in incisor, canine, premolar, and molar fields. These fields develop and evolve as relatively independent units (Line 2003; Hlusko et al. 2011); 2) The relative size and number of molar teeth in eutherian mammals can be predicted by an antero-to-posterior cascade mechanism of activators and inhibitors molecules. The activators are produced by the mesenchyme whereas inhibitors are produced by the predecessor teeth (Kavanagh et al. 2007). The activator/inhibitor ratio will influence initiation time, earlier initiation results in faster growth and larger size; 3) Gene mutations causing tooth agenesis with low pleiotropic effects have been described (Fauzi et al. 2018).
Despite of these insights, the search for genomic variations responsible for the diversification of eutherian molar teeth may be hindered by the fact that teeth can vary in size and number, and tooth loss seems to be an extreme case of reduction in tooth size. In mice third molar agenesis, the absent teeth start to develop, but in most cases epithelial cell proliferation arrests at the late bud‐cap stages (Grewall 1962). In Sciurids and Lagomorphs small maxillary and mandibular vestigial incisors are formed but fail to develop beyond early bell stage (Luckett 1984; Brook et al. 2009). The association between tooth size and number suggests that variations in dental phenotype are likely not to be caused by an all or nothing mechanism (i.e. creation/inactivation of cis-regulatory elements). Consequently, the key regulatory elements may not be conserved among eutherian species, making their identification a complex and challenging task.
Among the genes associated with tooth development Pax9 deserves special attention. Over 50 different mutations in this gene have been reported in human families (Bonczek et al. 2017), making this the most frequent gene associated with tooth agenesis. The relatively high frequency of Pax9 mutations in humans is likely to be due to the fact that, in most cases, Pax9 haploinsufficiency only affects dental phenotype, inducing mainly molar agenesis and size reduction of the remaining teeth (Brook et al. 2009). These aspects make Pax9 a strong candidate gene responsible for the evolution of mammalian molar dental pattern. We have previously shown that G-quadruplexes formed at the 5ʹ-end of the first intron of human and mice Pax9 gene can modulate mRNA splicing efficiency (SE) (Ribeiro et al. 2015), where higher stability was associated with higher SE. G-quadruplexes (G4) are noncanonical nucleic acids secondary structures formed from single-stranded G-rich sequences (Davis 2004). G4 motifs contain guanine blocks (G-runs) spaced by one or more nucleotides (loop). A G-run is usually formed by three or more guanines and a typical G4-motif contains four G-runs. The self-stacking of G4 motifs is stabilized by monovalent cations located in the inner core of the structure (Huppert and Balasubramanian 2005; Burge et al. 2006). G4 structural stability increases linearly for G-runs lengths of 4–7 nt (Rachwal et al. 2007), and is also influenced by loop sequence and length (Smirnov and Shafer 2000). G4 motifs are enriched at gene promoters in humans (Huppert and Balasubramanian 2007; Verma et al. 2008), yeast (Capra et al. 2010), and bacteria (Rawal 2006), suggesting that these structures act as transcriptional regulatory elements. G4 motifs are overrepresented in conserved transcription factor binding sites, such as SP1 (Eddy and Maizels 2008). Their functional role is supported by experimental evidences showing that binding of transcription factors to G4 motifs can modulate gene transcription (Thakur et al. 2009; Raiber et al. 2012).
Altogether, the above-mentioned evidences prompted us to study a possible association between the stability of the sequences present in the first intron of Pax9 and the relative size of molar teeth in eutherian mammals.
Results
Relative Molar Ratios Are Associated with Folding Stability of Pax9 Intronic Sequences
Using the sliding window approach with original and phylogenetically independent contrasted data, two regions were identified in association with both M2md/M1md (fig. 2) and M2area/M1area (supplementary fig. S1, Supplementary Material online). These regions were selected as they presented the highest absolute correlation coefficients and smallest P-values (≤0.01) for both area and mesiodistal length proportions. The first region (Seq1, supplementary table S2, Supplementary Material online) located 60 bp downstream the 5′-end of human PAX9 intron 1 exhibited significant negative correlations with M2area/M1area (Spearman rho = −0.55, P = 0.0001, sf = 0, and rho = −0.38, P = 0.01, sf = 0.012 with phylogenetically independent contrasted data [pic], supplementary fig. S1, Supplementary Material online), and with M2md/M1md (rho = −0.57, P = 9e−05, sf = 0.001; and rho = −0.47, P = 0.002, sf = 0.001 with pic) (fig. 2). This region contained 16 bases forming a predicted G-quadruplex sequence (supplementary table S2, Supplementary Material online). Seq1 exhibited low level of identity conservation among the orders included in our study, which could be translated into different G-quadruplex structural potentials. No significant correlations between M2M1 and minimum free energy (Mfe) were observed when -g option of RNAfold was not used (not shown).
Fig. 2.
Spearman correlations between M2/M1 mesiodistal lengths (M2md/M1md) and minimum folding energy of 16 bp sliding window. (A) Analysis of data without phylogenetically independent contrasted data. (B) Analysis of data with phylogenetic independent contrast (pic).
The second region (Seq2, supplementary table S2, Supplementary Material online), located 265 bp downstream the 5′-end of human PAX9 intron 1, also exhibited significant negative correlations with M2area/M1area (rho = −0.48, P = 0.001, sf = 0; and rho = −0.57, P = 0.0001, sf = 0 with pic, supplementary fig. S1, Supplementary Material online) and with M2md/M1md (rho = −0.49, P = 0.0009, sf = 0; and rho = −0.56, P = 0.0001, sf = 0 with pic) (fig. 2). The stability of both sequences varied among species, more stable sequences tended to have higher M2md/M1md and M2area/M1area ratios.
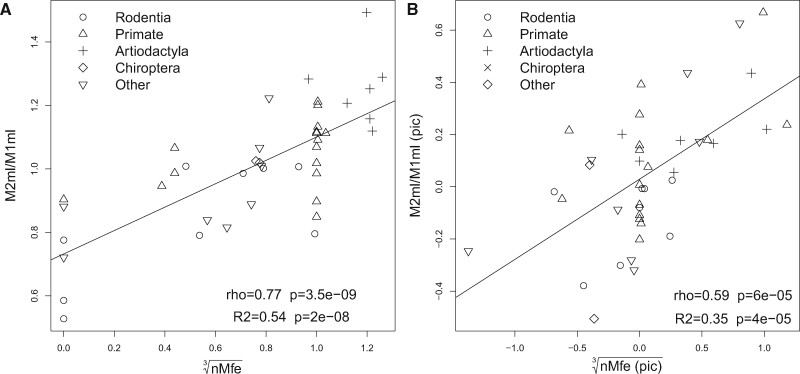
In all further analysis, Seq1 and Seq2 Mfe were normalized by dividing Mfe by the minimum value of Mfe (nMfe = Mfe/min[Mfe]). Therefore, normalized Mfe values (nMfe) ranged from 0 to 1 (Mfe are negative values), where score 1 represented RNA structures with the highest structural stability. Normalization did not affect the absolute values of correlations between M2/M1 area or mesiodistal length with Seq1 and Seq2 nMfe (fig. 3). Correlations could be further increased by summing the nMfe values of Seq1 and Seq2 (Seq1Seq2 nMfe), which produced significant correlations with M2area/M1area (rho = 0.74, P = 1.7e−08, sf = 0 and, rho = 0.49, P = 0.002, sf = 0.005 with pic) and with M2md/M1md (Spearman rho = 0.77, P = 3e−09, sf = 0; and rho = 0.59, P = 5e−05, sf = 0 with pic), (fig. 3). Seq1Seq2 nMfe and M2/M1 area and mesiodistal length were used as independent and dependent variables in linear regression analysis, respectively. The highest linear associations were found with cubic roots of nMfe with both area (R2 = 0.45, P = 1e−06; and R2 = 0.26, P = 6e−03 with pic, supplementary fig. S2, Supplementary Material online) and mesiodistal length (R2 = 0.54, P = 2e−08; and R2 = 0.35, P = 4e−05 with pic, fig. 4). Smaller and mostly nonsignificant correlations and linear associations were obtained with buccolingual width. In summary, correlations and linear associations between nMfe and M2/M1 ratios were more significant for mesiodistal length followed by area. Poor correlations were observed with buccolingual width. The sum of Seq1 and Seq2 nMfe produced higher and more significant correlations and linear associations. Maximum correlations were obtained when folding energies were obtained for RNA sequences (i.e. allowing G–U interactions) rather than DNA.
Fig. 3.
Folding energies of Pax9 intronic sequences correlate with molar proportions. Barplots showing scores of Spearman correlations (rho) between M2/M1 mesiodistal (M2M1md), buccolingual (M2M1bl) and area (M2M1area) with Seq1, Seq2, and Seq1Seq2 nMfe (upper panels). Seq1pic, Seq2pic, and Seq1Seq2pic are correlations performed with phylogenetically independent contrasted data. Lower panels show correlations with M3/M1. Note that highest and most significant correlation scores were obtained with M2/M1 mesiodistal length and area.
Fig. 4.
Folding energies of Pax9 intronic sequences correlate with M2md/M1md. Dot plots, Spearman correlations scores (rho), and linear associations obtained by least square regression (R2) of M2md/M1md ratio and cubic roots of Seq1Seq2 * nMfe. (A) Original values. (B) Phylogenetically independent contrasted values. nMfe represents the normalized minimum free energy values.
We have also performed correlations between Seq1 and Seq2 nMfe with M3/M1. Seq1 nMfe exhibited significant correlations with M3area/M1area (rho = 0.53, P = 0.0003, sf = 0; and rho = 0.32, P = 0.04, sf = 0.04 with pic) and with M3md/M1md (rho = 0.48, P = 0.001, sf = 0; and rho = 0.32, P = 0.04, sf = 0.03 with pic). Seq2 nMfe exhibited significant correlations with M3area/M1area (rho = 0.48, P = 0.001, sf = 0.001; and rho = 0.51, P = 0.0006, sf = 0 with pic) and with M3md/M1md (rho = 0.44, P = 0.003, sf = 0.002; and rho = 0.42, P = 0.006, sf = 0.007 with pic). The correlations between Seq1Seq2 Mfe with M3area/M1area (rho = 0.66, P = 1.6e−06, sf = 0; and rho = 0.42, P = 0.006, sf = 0 with pic) and M3md/M1md (rho = 0.57, P = 7.5e−05, sf = 0; and rho = 0.35, P = 0.02, sf = 0.02 with pic) were higher with the original data than with pic (fig. 3).
Different from the other orders included the Order Rodentia is monophyodont (only a single set of teeth throughout life), and most included rodents lack all premolars. Due to these particularities, we also performed correlation analysis without rodents. The exclusion of rodents did not significantly alter the pattern of correlations (supplementary fig. S3, Supplementary Material online). More significant correlations scores were observed between Seq1Seq2 nMfe and M2mdM1md (rho = 0.74, P = 1.6e−06, sf = 0; and rho = 0.64, P = 8e−05, sf = 0 with pic), followed by M2area/M1area (rho = 0.72, P = 3e−06, sf = 0; and rho = 0.52, P = 0.002, sf = 0.002 with pic).
Interestingly, when comparing M2md/M1md correlations obtained with and without rodents, it is possible to observe that scores obtained with Seq1Seq2 nMfe were of the same magnitude (compare fig. 3 [rodents included] and supplementary fig. S3, Supplementary Material online [rodents removed]). Seq1 nMfe produced higher scores than Seq2 nMfe in correlations with all species; whereas Seq2 nMfe produced higher scores than Seq1 nMfe when rodents were removed. This may indicate that these regions may have variable influence across taxa.
Folding Stability of Pax9 Intronic Sequences Can Increase Predictability of the Molar Proportions Model
Since correlations between M3/M1 and nMfe were of smaller magnitude than M2/M1, we hypothesized that this could be due to a smaller influence of Pax9 on the development of M3 than in M2. We have tested the influence of nMfe on the molar proportion model (i.e. correlations between M2area/M1area and M3area/M1area, Kavanagh et al. 2007). These correlation scores (rho = 0.81, P = 1e−10, sf = 0; and rho = 0.74, P = 2e−07, sf = 0 with pic, supplementary fig. S4, Supplementary Material online) were used as parameters (controls) to estimate the effect of Mfe. Our assumption is that if Pax9 intronic Seq1 and Seq2 nMfe are associated with molar proportions, the product M3/M1 * nMfe will produce higher and more significant correlations and linear associations than controls. The correlations decreased when M3area/M1area ratios were multiplied by Seq1 nMfe values (rho = 0.67, P = 1.4e−06, sf = 0; and rho = 0.59, P = 4.2e−05, sf = 0 with pic), Seq2 nMfe values (rho = 0.54, P = 2e−07, sf = 0; and rho = 0.56, P = 1e−04, sf = 0 with pic), and when M3area/M1area ratios were multiplied by Seq1Seq2 nMfe values (rho = 0.82, P = 3.6e−11, sf = 0; and rho = 0.62, P = 2.5e−05, sf = 0 with pic, supplementary fig. S4, Supplementary Material online). However, correlations higher than controls were obtained with the cubic root of Seq1Seq2 nMfe (rho = 0.86, P = 3e−13, sf = 0; and rho = 0.77, P = 8e−08, sf = 0 with pic, supplementary fig. S5, Supplementary Material online).
M2md/M1md versus M3md/M1md (controls, without nMfe) exhibited a significant correlation (rho = 0.7, P = 3e−07, sf = 0; and rho = 0.62, P = 3.5e−05, sf = 0 with pic, supplementary fig. S4, Supplementary Material online). The correlations were of the same magnitude when the M3md/M1md * Seq1 nMfe was used in the correlation analysis (rho = 0.67, P = 1e−06, sf = 0; and rho = 0.62, P = 1.6e−05, sf = 0 with pic, supplementary fig. S4, Supplementary Material online), and smaller when M3md/M1md * Seq2 nMfe was used in the correlation analysis (rho = 0.54, P = 3e−04, sf = 0; and rho = 0.57, P = 2e−05, sf = 0 with pic, supplementary fig. S4, Supplementary Material online). Correlation scores were higher than controls (rho = 0.77, P = 3e-09, sf = 0; and rho = 0.66, P = 4.6e-06, sf = 0 with pic) when M3md/M1md ratios were multiplied by Seq1Seq2 nMfe values (supplementary fig. S4, Supplementary Material online). Correlation coefficients and linear associations were further increased when cubic root of Seq1Seq2 nMfe were used (fig. 5).
Fig. 5.
Folding energies of Pax9 intronic sequences increase correlation between M3md/M1md and M2md/M1md. Dot plots, Spearman correlations, and linear associations of molar proportions. (A) M2md/M1md and M3md/M1md. (B) M2md/M1md and M3md/M1md multiplied by the cubic root of Seq1Seq2 nMfe. (C) M2md/M1md and M3md/M1md with phylogenetically independent contrasted values (pic). (D) M2md/M1md and M3md/M1md multiplied by the cubic root of Seq1Seq2 nMfe (pic). Best fit line was obtained with least square method. nMfe represents the normalized minimum free energy values. Note that the product of M3md/M1md and the cubic root of Seq1Seq2 nMfe (B, D) resulted in improved correlations and linear associations when compared with their respective controls (A, C).
It is worth mentioning that the correlations scores and R2 obtained between M2bl/M1bl and M3bl/M1bl * nMfe (bl = buccolingual width) were smaller than the correlations obtained with area, mesiodistal width and controls (M2bl/M1bl with M3bl/M1bl, not shown).
When comparing correlations between M2md/M1md and M3md/M1md * nMfe obtained with and without rodents it is possible to observe that scores obtained with Seq1Seq2 nMfe were of the same magnitude (supplementary fig. S6, Supplementary Material online). Seq1 nMfe produced higher scores than Seq2 nMfe in correlations with all species; whereas Seq2 nMfe produced higher scores than Seq1 nMfe when rodents were removed (compare fig. S6 [rodents removed] and supplementary fig. S4, Supplementary Material online [rodents included]).
Folding Stability of Pax9 Intronic Sequences as a Proxi of Activators in the Inhibitory Cascade Model of Molar Development
In order to further associate the stability of Seq1Seq2 nMfe with molar proportions, we tested the nMfe as activators in the inhibitory cascade model proposed by Kavanagh et al. (2007). In this model, molar proportions are obtained by the formula Mx/M1 = 1 + [(a—i)/i] (x-1), where for each tooth position (x), (a) represents the relative strength of activators, and (i) the inhibitors. This model allows (a/i) to be estimated from the dental phenotypes and to be compared with data obtained in experimental analysis. The ratio between mesiodistal length of M1 and of secondary palate (M1md/Pal2) was used as an estimate of inhibitory effect (i) of first molar on the growth of subsequent molars. This parameter was chosen because the length of Pal2 represents the anatomical region where posterior teeth are formed (Ribeiro et al. 2013). Therefore, relatively larger developing M1 would secrete higher amounts of diffusible inhibitory factors. M1md/Pal2 proportions were normalized by dividing the values by the maximum value among species. As a parameter to estimate the relevance of nMfe on the formula output, the nMfe of all taxa were replaced by the number 1 (could be any number different from 0) in the formula. In this case, the correlation scores between molar proportions and the formula estimates were only dependent on the ratio between M1md/Pal2 (inhibitor estimate, controls). Therefore, meaningful results should present coefficients higher than controls.
The control coefficients, obtained in the correlations between M2area/M1area and the formula output without nMfe, were rho = 0.51, P = 5e−04, sf = 0; and rho = 0.34, P = 0.03, sf = 0.033 with pic (supplementary fig. S7, Supplementary Material online). These correlations were used as thresholds on further correlation analyses. Higher correlation coefficients were found between M2area/M1area and the formula output using Seq1 nMfe (rho = 0.62, P = 1.2e−05, sf = 0; and rho = 0.41, P = 0.009, sf = 0.012 with pic), and Seq2 nMfe (rho = 0.52, P = 4e−04, sf = 0.001; and rho = 0.45, P = 0.003, sf = 0.001 with pic) (supplementary fig. S7, Supplementary Material online). The use of Seq1Seq2 nMfe as activator resulted in improved correlations (rho = 0.76, P = 4.7e−09, sf = 0; and rho = 0.47, P = 0.002, sf = 0.001 with pic, supplementary fig. S7, Supplementary Material online) and linear associations (supplementary fig. S8, Supplementary Material online). Highest linear associations were obtained with cubic root of Seq1Seq2 nMfe (R2 = 0.52, P = 7e−08, and R2 = 0.31, P = 1e−04 with pic). The results show that the use of nMfe as activator can increase predictability of M2area/M1area in the inhibitory cascade model proposed by Kavanagh et al. (2007).
The control coefficients, obtained in the correlations and linear regressions between M2md/M1md and the formula output without nMfe, were rho = 0.62, P = 5e−04, sf = 0, and rho = 0.44, P = 0.004, sf = 0.003, with pic (supplementary fig. S7, Supplementary Material online). Higher coefficients were found in the correlation analyses between M2md/M1md and the formula output using Seq1 nMfe (rho = 0.65, P = 2.8e−06, sf = 0; and rho = 0.52, P = 0.009, sf = 0.001 with pic), Seq2 nMfe (rho = 0.54, P = 2e−04, sf = 0; and rho = 0.49, P = 1e−03, sf = 0.004 with pic), and Seq1Seq2 nMfe (rho = 0.8, P = 1.5e−10, sf = 0; and rho = 0.58, P = 1e−04, sf = 0 with pic) (supplementary fig. S7, Supplementary Material online, fig. 6). The highest linear associations were also obtained with cubic roots of Seq1Seq2 nMfe (R2 = 0.63, P = 3e−10, and R2 = 0.42, P = 5e−06 with pic). Smaller (rho < 0.3) and predominantly nonsignificant (P > 0.01, sf > 0.005) coefficients were found when buccolingual width was used in the correlation analyses. Since the area was obtained by the product of mesiodistal and buccolingual distances, the results indicate that the predictability of M2area/M1area is imparted by the influence of Pax9 nMfe on mesiodistal length.
Fig. 6.
Folding stability of Pax9 intronic sequences as a proxi of activators in the inhibitory cascade model. Dot plots, Spearman correlations (rho), and linear association (R2) of M2md/M1md ratio and the A/I model given by the formula Mx/M1 = 1 + [(a—i)/i] (x-1), where for each tooth position (x), (a) represents the relative strength of activators, and (i) the inhibitors. The ratio between mesiodistal length of M1 (M1md) and of secondary palate (M1md/Pal2) was used as an estimate of inhibitory effect (i) and nMfe the activator (a). (A) Associations between M2md/M1md and control (nMfe replaced by 1). (B) Associations obtained between M2md/M1md and the formula output using Seq1Seq2 nMfe as activator. (C) and (D) represent the associations shown on (A) and (B) using phylogenetically independent contrasted data, respectively. Note that the use of Seq1Seq2 nMfe as activator (B, D) resulted in improved correlations and linear associations when compared with their respective controls (A, C).
The control coefficients, obtained in the correlations between M3area/M1area and the formula output without nMfe, were rho = 0.68, P = 7e−07, sf = 0, R2 = 0.45; and rho = 0.60, P = 4.8e−05, sf = 0.001, R2 = 0.36 with pic (supplementary fig. S7, Supplementary Material online). Smaller coefficients were found in the correlation analyses between M3area/M1area and the formula output using Seq1 nMfe (rho = 0.65, P = 3.3e−06, sf = 0; and rho = 0.34, P = 0.03, sf = 0.025 with pic), Seq2 nMfe (rho = 0.54, P = 2e−04, sf = 0.001; and rho = 0.56, P = 1e−04, sf = 0.001 with pic), and Seq1Seq2 nMfe (rho = 0.75, P = 1.2e−08, sf = 0; and rho = 0.49, P = 0.001, sf = 0.009 with pic) (supplementary fig. S7, Supplementary Material online). Highest linear associations were obtained with cubic roots of Seq1Seq2 nMfe in the formula (R2 = 0.58, P = 5e−09; R2 = 0.44, P = 2.5e−06 with pic). Correlation scores between M3md/M1md and the formula output without nMfe (control) were higher than when nMfe were included in the formula (supplementary fig. S7, Supplementary Material online).
The control coefficients, obtained in the correlations between M3bl/M1bl and the formula output without nMfe, were rho = 0.28, P = 0.07, sf = 0.06; and rho = 0.33, P = 0.04, sf = 0.04 with pic. Higher coefficients, but with nonsignificant pic sf values, were only obtained between the correlations between M3bl/M1bl and the formula when Seq1Seq2 nMfe was used (rho = 0.55, P = 1e−04, sf = 0.001; and rho = 0.35, P = 0.03, sf = 0.026 with pic). These results show that nMfe has a small nonsignificant effect on the predictability of M3/M1 area, buccolingual width or mesiodistal length in the inhibitory cascade model (Kavanagh et al. 2007).
When comparing correlations between M2md/M1md and nMfe obtained with and without rodents it is possible to note that scores obtained with Seq1Seq2 nMfe were of the same magnitude. Seq1 nMfe produced higher scores than Seq2 nMfe in correlations with all species; whereas Seq2 nMfe increased to similar values of Seq1 nMfe and were higher than Seq1 nMfe when rodents were removed, in nonpic and pic, respectively (compare supplementary fig. S9, Supplementary Material online [rodents removed] with supplementary fig. S7, Supplementary Material online [rodents included]).
Correlation scores smaller than the ones observed when M1md/Pal2 was used as the inhibitor estimate was obtained by its replacement by M1bl/Pal2 or M1area/Pal2.
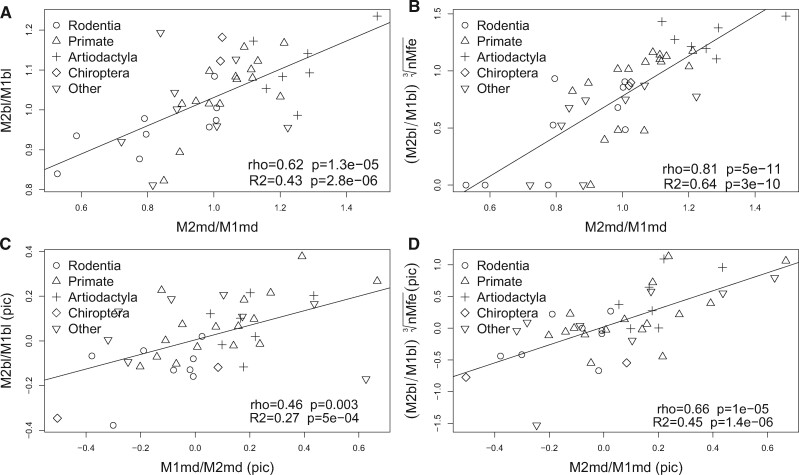
Differences in Relative Mesiodistal and Buccolingual Molar Proportions Are Associated with Folding Stability of Pax9 Intronic Sequences
As shown above the structural stability of the intronic sequences of Pax9 gene exhibited strongest correlations when molar proportions were based on mesiodistal distances. Molar proportions estimated on buccolingual measures yielded small nonsignificant correlations. In order to understand this phenomenon, we analyzed the effect of nMfe in the correlation of molar proportions obtained with mesiodistal and buccolingual measures. The coefficients of the correlations between M2md/M1md with M2bl/M1bl (rho = 0.62, P = 1e−05, sf = 0; and rho = 0.46, P = 0.002, sf = 0.004 with pic, supplementary fig. S10, Supplementary Material online) were set as a threshold, above which nMfe was considered to have a significant effect. The coefficients were higher than controls in the correlations between M2md/M1md and M2bl/M1bl * Seq1 nMfe (rho = 0.64, P = 5.9e−06, sf = 0; and rho = 0.56, P = 1.5e−04, sf = 0 with pic), and between M2md/M1md and M2bl/M1bl * Seq1Seq2 nMfe (rho = 0.79 P = 4e−10; sf = 0; and rho = 0.64, P = 1e−05, sf = 0 with pic) (supplementary fig. S10, Supplementary Material online). The correlations between M2bl/M1bl * Seq2 nMfe and M2md/M1md (rho = 0.51, P = 2e−04, sf = 0; and rho = 0.55, P = 5e−04, sf = 0 with pic), produced nonpic scores bellow threshold (supplementary fig. S10, Supplementary Material online). It is worth mentioning that the correlations obtained between M2bl/M1bl * nMfe and M2md/M1md were higher and more significant than the ones obtained with these single variables alone (M2bl/M1bl or nMfe). Linear associations higher than controls (M2md/M1md ∼ M2bl/M1bl) were only obtained with Seq1Seq2 nMfe. Maximum associations were obtained with cubic roots of Seq1Seq2 nMfe (fig. 7).
Fig. 7.
Differences in relative mesiodistal and buccolingual molar proportions are associated with folding stability of Pax9 intronic sequences. Dot plots, Spearman correlations (rho) and linear associations (R2) of M2md/M1md and M2bl/M1bl. (A) M2md/M1md and M2bl/M1bl (control). (B). M2md/M1md and M2bl/M1bl multiplied by the cubic root of Seq1Seq2 nMfe. (C) and (D) represent the same experiments of (A) and (B), respectively, using phylogenetically independent contrasted data. Note that the product of M3bl/M1bl and the cubic root of Seq1Seq2 nMfe (B, D) resulted in improved correlations and linear associations when compared with their respective controls (A, C).
The correlations between M3md/M1md with M3bl/M1bl * Seq1 nMfe (rho = 0.56, P = 1e−04, sf = 0; and rho = 0.38, P = 0.01, sf = 0.02 with pic), M3bl/M1bl * Seq2 nMfe (rho = 0.45, P = 0.003, sf = 0.003; and rho = 0.48, P = 0.002, sf = 0.002 with pic), and Seq1Seq2 nMfe (rho = 0.61 P = 1e−05, sf = 0; and rho = 0.40, P = 0.01, sf = 0.011 with pic) presented and nonpic coefficients higher than control (M3md/M1md and M3bl/M1bl, rho = 0.44, P = 1.4e−04, sf = 0.005; and rho = 0.43, P = 9e−04, sf = 0.06 with pic, supplementary fig. S10, Supplementary Material online). Linear associations higher than control (M3md/M1md ∼ M3bl/M1bl) were only obtained with Seq1Seq2 nMfe. Maximum associations were obtained with cubic roots of Seq1Seq2 nMfe (supplementary fig. S11, Supplementary Material online). In summary, the effect of nMfe on the correlations is more evident on M2/M1 proportions, where the product of Seq1 and Seq1Seq2 nMfe with M2bl/M1bl can significantly increase the correlations with M2md/M1md.
Similar to the observed in previous analyses, when comparing correlations between M2md/M1md with M2bl/M1bl * nMfe obtained with and without rodents it was possible to observe that scores obtained with Seq1Seq2 nMfe were of the same magnitude (supplementary fig. S12, Supplementary Material online). Seq1 nMfe produced higher scores than Seq2 nMfe in correlations with all species; whereas Seq2 nMfe produced higher scores than Seq1 nMfe when rodents were removed (compare supplementary fig. S12, Supplementary Material online [rodents removed] and supplementary fig. S10, Supplementary Material online [rodents included]).
Folding Stability of Pax9 Intronic Sequences Improves Associations of the Molar-Module-Component and Premolar-Molar-Module
The Molar-Module-Component (MMC) and Premolar-Molar-Module (PMM) represent the ratios between mesiodistal lengths of M3/M1 and P4/M2 (Hlusko et al. 2016). These modules were shown to be heritable, independent of body size, and to vary significantly across primates (Monson, Coleman, et al. 2019). Since results presented here show that the highest associations were obtained with mesiodistal length of molar proportions we tested if nMfe can improve correlation and linear associations between MMC and PMM. Rodents were removed from the analyses as most species lack P4. Significant correlations and linear associations between MMC and PMM (controls) were only observed in nonpic data (fig. 8A), as P-values of pic analysis were higher than 0.1 (fig. 8C). Significant correlations and linear associations were found between MMC and PMM * Seq1 nMfe (rho = 0.57, P = 6e−06, sf = 0, R2 = 0.29; and rho = 0.39, P = 6e−06, sf = 0.002, R2 = 0.15. with pic), and between MMC and PMM * Seq1Seq2 nMfe (rho = 0.65, P = 4e−05, sf = 0, R2 = 0.31; and rho = 0.44, P = 0.01, sf = 0.001, R2 = 0.2. with pic, fig. 8B and D). The use of buccolingual width in the analysis produced nonsignificant associations smaller than controls (not shown).
Fig. 8.
Folding stability of Pax9 intronic sequences improves associations of the MMC and PMM. Dot plots, Spearman correlations, and linear associations of PMM and MMC. (A) M3md/M1md and P4md/M2md. (B) M3md/M1md with P4md/M2md * Seq1Seq2 nMfe. (C) and (D) represent the same experiments of (A) and (B) using phylogenetically independent contrasted data. Best fit line was obtained with least square method. nMfe represents the normalized minimum free energy values. Note that P4md/M2md * Seq1Seq2 nMfe (B, D) resulted in improved correlations and linear associations when compared with their respective controls (A, C).
Discussion
Our results indicate that variations within Pax9 intronic locus contributed to dental phenotype diversification in mammalian species. In summary, correlations and linear associations between nMfe and M2/M1 ratios were more significant for mesiodistal length followed by area. Poor correlations were observed with buccolingual width. The sum of Seq1 and Seq2 nMfe (Seq1Seq2 nMfe) produced higher and more significant correlations and linear associations. Correlations were more significant with M2/M1 ratios than with M3/M1. These results suggest an “activator” effect driven by the Pax9 gene, promoting the relative growth of M2, and to a lesser extent M3. Since tooth area was obtained by the product of mesiodistal and buccolingual distances, the positive correlations obtained with area are likely to be imparted by influence of Pax9 on variations in mesiodistal length.
Besides the direct correlations between M2md/M1md, the associations of nMfe were tested in other four distinct experiments (predictability of the molar proportions, activators in the AI cascade model, mesiodistal/buccolingual proportions, and MMC/PMM). Seq1 nMfe improved associations, producing scores higher than controls in three of these tests for both original and pic values (failed in predictability of the molar proportions), Seq2 nMfe passed only in the last test, whereas Seq1Seq2 nMfe passed in all tests, producing the highest correlation scores and best linear associations. The performance of Seq2 nMfe improved when rodents were removed from the analyses (passed the last three tests). It is also worth mention correlations scores decreased in that phylogenetic independent contrasts (pic) transformed data. This seems to indicate that the Pax9 Mfe effects underlie the phylogenetic signal that is observed in the relative sizes of the molars.
The exact role of Seq2 nMfe in Pax9 expression remains to be elucidated. Recently, Jolma et al. (2020) have identified 86 RNA-binding proteins, from which 17 only bind to structured motifs. These motifs are formed by stem-loop structure with stems containing from 3 to 15 pairs of bases and loops with median of five bases. The most stable Seq2 structures are found in Order Artiodactyla with stems with 3–4 pairs of bases and loops ranging from 3 to 6 bases, whereas primates have the least stable structures (supplementary fig. S13, Supplementary Material online).
Variations in noncoding DNA regulatory sequences play an important role in morphological evolution (Britten and Davidson 1969; King and Wilson 1975; Wray 2007; Carroll 2008; Kvon et al. 2016; Xie et al. 2019). Haploinsufficiency of PAX9 in human families affects mainly the dental phenotype (Line 2003), in agreement with its key role during tooth development. Using genetically engineered mutant mice with Pax9 mutations, Kist et al. (2005) showed that the severity of changes in dental phenotype followed a dose–response relationship. Mice (lacz/neo) expressing 7% of wild-type Pax9 had severe oligodontia, with absence of all third molars, lower second molars and incisors, and frequent agenesis of lower first molars. Mice (neo/neo) expressing 20% of wild type Pax9 molars presented hypodontia with frequent agenesis of lower third molars and incisors. Moreover, the role of Pax9 intronic quadruplex (Seq1) acting as regulatory element during mRNA splicing has been demonstrated (Ribeiro et al. 2015). Synthetic oligonucleotides containing the human Seq1 can form highly stable G-quadruplexes and replacement of the human Pax9 first intron quadruplex for the less stable rat sequences resulted in a 20% decrease in SE. The expression of spliced Pax9 intron 1 decreased (88%) when G-quadruplex formation was abolished by replacing guanines by adenines. The SE, for a given intron, can be defined and measured as the ratio between the number of spliced RNA (sRNA) and the total number of mRNA molecules that is the sum of spliced and unspliced (uRNA, Xia 2020) (SE=sRNA/(sRNA+uRNA)). Therefore, decrease in SE will decrease the amount of mRNA molecules that are translated. The control of SE is an important mechanism to regulate gene expression and cell function (Gotic et al. 2016; Pecce et al. 2018; Ding and Elowitz 2019). Delayed unspliced mRNA molecules are rapidly degraded by an active surveillance mechanism (Sayani and Chanfreau 2012; Bresson and Tollervey 2018). Therefore, retention of Pax9 intron 1 would likely result in degradation of its pre-mRNA.
In all tests performed here the correlations with M2/M1 area and mesiodistal length ratios were more significant than M3/M1. These results may be explained by the fact that like other quantitative body characters such as body weight and body height, tooth size is determined by genetic and environmental factors. Third molar is the most frequent tooth missing in humans and also the most affected molar tooth in nonsyndromic haploinsufficiency of PAX9, MSX1, and AXIN2 genes in human families (Line 2003; Lammi et al. 2004; Yue et al. 2016). Pax9+/− Msx1+/− mice have agenesis of third molar (Nakatomi et al. 2010). Quantitative genetic analysis in rats showed that the size of the first and the second molars was more significantly controlled by genetic effect than maternal effect, whereas maternal effect could not be ignored for the size of the third molar in addition to genetic effect (Otani et al. 1990). Third molars of CBA and CH3 mice strains exposed to dioxin during development were frequently missing, whereas M1 and M2 were always present (Keller et al. 2007). Maxillary third molar development is retarded in CBA mice exposed to ethanol during maternal pregnancy (Stuckey and Berry 1984). In CBA mice strain the agenesis of the third molars is strikingly more frequent in large litters than in small litters, indicating that third molar development may be more susceptible to changes of maternal physiology than the other molar teeth (Gruneberg 1963). It seems that during the sequential initiation and development of molar tooth row the strength of genetic network of activating cascade progressively decreases, leading to a consequent increase in the influence of environmental and epigenetic factors. This assumption is supported by the fact that the loss of M3 in callitrichids has been attributed to slower prenatal growth rates, which would result in delayed initiation of M3 (Monson, Coleman, et al. 2019).
The AI model (Kavanagh et al. 2007) predicts that reduced a/i ratios will delay initiation of subsequent teeth resulting in smaller M2 and M3. Therefore, correlation between genetic signatures of activator molecules and third molar morphology is expected to be less significant than in the other molar teeth. Accordingly, the correlation scores between M2md/M1md and M3md/M1md were maximized when M3md/M1md was multiplied by cubic root of nMfe (fig. 5). A possible and plausible explanation for this phenomenon is that nMfe represents a virtual effect of Pax9 gene compensating for the overall decrease of genetic influence on M3md/M1md ratio. Cubic roots will produce a smaller range of values than plain nMfe. Therefore, it is plausible to infer that small variations in Pax9 gene expression can result large changes in the relative size of M2 and M3.
Significant associations between Pax9 intronic sequences nMfe and molar proportions were obtained when molar area and mesiodistal lengths were used. Buccolingual width produced the smallest, mostly nonsignificant associations. This fact seems to be contradictory as Pax9 is expressed at early stages of tooth development, before differences between buccolingual and mesiodistal shape appear (Lin et al. 2007; Nakatomi et al. 2010). Besides, haploinsufficiency of PAX9 in humans causes a decrease in both mesiodistal and buccolingual dimensions in the remaining teeth (Brook et al. 2009).
Although molar mesiodistal/buccolingual ratios are variable among species, the mechanisms that modulate this variability have been largely overlooked. Evidence of a differential mechanisms on the growth of mesiodistal and buccolingual dimensions come from genetic correlation studies in baboon molar teeth, showing a small but significant overlapping additive genetic effect between buccolingual width and trunk length, which was not observed in mesiodistal length (Hlusko et al. 2006). These observations were expanded by Hlusko et al. (2016) and Monson, Boisserie, et al. (2019) showing that primate mandibular mesiodistal posterior length is primarily influenced by genetic factor determining relative tooth size, whereas buccolingual width has a higher influence of systemic effects (i.e. body size and sex dimorphism). Our results add to these previously published data confirming the influence of genetic factors, as Seq1 and Seq1Seq2 nMfe improved MMC and PMM correlations. nMfe values were not effective when buccolingual proportions were used.
More recently, the mechanisms driving the independence between mesiodistal and buccolingual growth of molar teeth have been demonstrated (Renvoisé et al. 2017). By limiting the buccolingual growth of cultured mouse molars with braces, the authors showed that mechanical constrains were able to alter the first molar cusp formation to a pattern similar to those observed in voles. Moreover, cultured molars grow wider (bucolingual width) than in vivo teeth. However, in contrast to the lateral cusps, longitudinal patterning is largely unaffected. Finally, it was suggested that space limitation imposed by jawbone is a relevant factor to determine tooth crown morphology. These works indicate that mesiodistal and buccolingual edges seem to be distinctly influenced by local constraints during molar development, where small influences can be translated into significant phenotypic variations. These external influences interfere with the manifestation of intrinsic genetic network on phenotype.
Our results add to these observations, as the interaction Seq1Seq2 nMfe with the relative buccolingual molar proportions can significantly improve the correlation with the relative molar area and mesiodistal proportions. As previously discussed, a plausible reason for the increase in the correlation scores is that nMfe can recapitulate the potential effect of Pax9 gene on buccolingual width, which has been partially restrained by the external/environmental influences.
The activator/inhibitor model (Kavanagh et al. 2007) was based on in vivo experiments performed in mice and then successfully expanded to other mammals (Poly 2007). The relative length of M1, given by the M1md/Pal2 ratio, in rodents without P4 (last premolar) seems to be larger in rodents with P4 and also higher than in most taxa. Five out of six rodents without P4 had M1md/Pal2 ratio larger than 0.2, whereas all three rodent taxa with P4 had M1md/Pal2 smaller than 0.16. The mean M1md/Pal 2 in the analysis without rodents is 0.16. In fact, mice had the largest M1md/Pal2 (0.29) among the 42 taxa included in the study. The increase in mesiodistal length in mice has been attributed to the incorporation of a rudimentary signaling center, possibly from P4 (Prochazka et al. 2010; Sadier et al. 2019). When comparing correlations obtained between M2md/M1md and nMfe, or the product of nMfe, with or without rodents, it is possible to observe that scores obtained with Seq1Seq2 nMfe were of the same magnitude. Seq1 produced higher scores than Seq2 nMfe in correlations with all species; whereas Seq2 nMfe produced higher scores than Seq1 nMfe when rodents were removed (compare fig. 3 and supplementary fig. S3, Supplementary Material online, supplementary figs S4 and S6, figs. S7 and S9, figs. S10 and S12, Supplementary Material online). This is an interesting aspect indicating that Seq1 and Seq2 may have distinct influence in Pax9 expression in different taxa, and in rodents Seq1 seems to be more relevant than Seq2.
The slide window approach used in the present study has not, to our knowledge, been used elsewhere. A possible reason is because it was designed specifically to correlate variations in RNA structural stability with molar proportions. Prior to automated analysis, the process requires that nucleotide sequences to be individually downloaded from different sources and aligned. Furthermore, statistical analysis requires customized and complex coding. Despite these difficulties, this approach allowed us to provide evidences that changes in the structural stability of a G4 at the intron 1 of Pax9 gene are associated with variations in mammalian dental phenotype. It is striking that the structural stability of a small region in the Pax9 first intron bears a significant correlation with a complex trait. Besides changes in Pax9 intronic sequences, the changes in molar proportions in the species analyzed are likely to be also influenced by variations in other regulatory regions in Pax9 or other genes associated with tooth development. The rates of gene expression are mainly determined by DNA sequences that modulate transcription and translation (Schwanhäusser et al. 2011) and the selection for efficient usage of expression machinery seems to be a major force that shapes the evolution of gene sequences (Shah and Gilchrist 2011; Line et al. 2013). Optimum gene expression requires a balanced participation of these processes, as a gene that is transcribed at high rates should also be efficiently translated (Coghlan and Wolfe 2000). Therefore, regulatory sequences that participate in a common developmental network are expected to work in an integrated manner and co-evolve in order to cope with demands for efficient gene expression.
The relevance of G4 sequences in cell function is highlighted by the fact that the transcription factors binding sites of over 850 human, chimp, mouse, and rat genes are located near (less than 100 bp) a G4 motif (Kumar et al. 2011). Besides, the presence of G4 motifs in loci with elevated recombination and rearrangements, such as telomeres, immunoglobulin switch regions, ribosomal DNA loci, and breakpoints associated with translocations and somatic copy number variations in malignant neoplastic processes (Katapadi et al. 2012; Bacolla et al. 2016; Wang et al. 2019) indicates that G4 motifs can drive genome instability. Stable G4 sequences require unwinding by DNA helicases during replication. Inactivation of DNA helicases induces deletions upstream G4 sequences (Cheung 2002). This evidence, allied with the functional role of G4 motifs in gene expression, make these sequences potential evolutionary hot spots. Since evolutionary variations in molar patterning frequently result in loss or decrease in the size of the last molars, it is likely that decreased functional requirement for these teeth result in a relaxation of selective pressure on Pax9 SE, favoring mutations that decrease the structural stability of Pax9 intronic G-quadruplex (Seq1). In this sense, it is possible that besides dental patterning, changes in G-quadruplex structural stability are associated with the evolution of other mammalian phenotypes.
Materials and Methods
Phenotypic Data
In this study, 42 placental mammalian species, representing nine orders, were included. Measurements of 231 specimens of 40 taxa were made on skulls of specimens held in the collections of the National Museum of Natural History (USNM) in Washington, DC. Measurements were performed with a pair of digital Neiko 6″ calipers, or Fowler High Precision 24″ for the larger specimens. Mesiodistal length and buccolingual width of upper molars were obtained drawing a straight line between distal and mesial contact points on the occlusal surface. The length of secondary palate (Pal2) was obtained by the distance between the distal of M3 and the point where incisal suture cross alveolar ridge (fig. 1). Measurements used in the analyses were from the left size of skull, right side was used when teeth were missing on the left. Definitive dimensions were obtained by averaging the measurements in each taxa analyzed. Dental dimensions from the other 2 taxa, Homo sapiens (n = 133) and Pan paniscus (n = 46) were obtained from Zorba et al. (2011) and Pilbrow (2003), respectively. For these two taxa Pal2 was obtained by adding the mesiodistal length of posterior teeth (canine, premolars, and molars). Morphological measurements are shown on supplementary table S1, Supplementary Material online. The sample size was limited by genomic data availability, since G4 sequences were not available for all species. Occlusal area was obtained by the product of mesiodistal and buccolingual dimensions. Only animals with three molars were used in the analysis. Species with residual teeth or anomalous dental patterns (ex: Loxodonta africana, Odobenus rosmarus, Dasypus novemcinctus) were excluded from the analysis.
Fig. 1.
Dental and cranial measurements. Pal2 is the distance between the distal surface of M3 to the point where incisal suture (IS) cross alveolar ridge. M1md and M1bl are the mesiodistal length and buccolingual width of the first molar, respectively. Similar measurements were made for the second (M2md, M2bl) and third (M3md, M3bl) molars.
Genomic Data
Human PAX9 was downloaded from the Ensembl 85 database (http://www.ensembl.org/index.html), and used in the Multiz Alignments of 100 Vertebrates Genome browser tool, from Genome Browser Gateway (http://www.ucsc.edu/), in order to obtain the corresponding placental mammalian alignments. Some primate sequences were obtained from GenBank (Perry et al. 2006).
Pax9 sequences ranging from 100 nt upstream to 400 nt downstream from the exon 1-intron 1 boundary were aligned using Mafft alignment tool (Katoh and Standley 2013), and scanned from 5 to 3 end using sliding windows with sizes ranging between 15 and 30 bp with a shift of 1 bp. Sequences in each window were analyzed using the RNAfold-g tool (http://www.tbi.univie.ac.at/RNA/, Lorenz et al. 2011) in order to calculate their Mfe. The -g option allows the RNAfold software to identify and calculate the Mfe for G4 motifs. The Mfe values were used as a genetic variable in correlation tests.
Phylogeny Analysis
Mitochondrial RNA sequences of all placental mammals analyzed were obtained from Nucleotide database (https://www.ncbi.nlm.nih.gov/nuccore/) and aligned using the MAFFT alignment algorithm (Katoh and Standley 2013). Finally, we obtained a phyla class object which was used for computing phylogenetically independent contrasts (pic) using the method described by Felsenstein (1985). For that, we used the pic() function from the “phytools” R package (Revell 2012). The R package “msa” was used in order to assay the Pax9 intron 1 and G4FS region nucleotide conservation (Bodenhofer et al. 2015).
Statistical Analysis
Statistical analyzes were performed using R software version 3.4.4 (https://www.r-project.org/, R Core Team 2019). Spearman correlations and linear regressions were performed using both the original variables and data obtained using phylogenetically independent contrasts (pic; Felsenstein 1985), which take the nonindependence of observations due to phylogeny into account (Bininda-Emonds et al. 2007). The consistency of correlations was tested by randomly reshuffling the y-axis values 1000 times, for both original and phylogeny independent contrasted data. The frequency of reshuffled correlations presenting higher correlation scores than the nonreshuffled variables was denominated sf.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
S.R.P.L. and M.J.E. are supported by a fellowship from the Brazilian National Council for Scientific and Technological Development (CNPq, grants 305783/2018-1 and 141004/2018-5). G.B.F. is supported by a fellowship from Sao Paulo Research Foundation (FAPESP, grant 2018/23038-7).
References
- Bacolla A, Tainer JA, Vasquez KM, Cooper DN.. 2016. Translocation and deletion breakpoints in cancer genomes are associated with potential non-B DNA-forming sequences. Nucleic Acids Res. 44(12):5673–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bininda-Emonds ORP, Cardillo M, Jones KE, MacPhee RDE, Beck RMD, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A.. 2007. The delayed rise of present-day mammals. Nature. 446(7135):507–512. [DOI] [PubMed] [Google Scholar]
- Bresson S, Tollervey D.. 2018. Surveillance-ready transcription: nuclear RNA decay as a default fate. Open Biol. 8(3):170270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenhofer U, Bonatesta E, Horejš-Kainrath C, Hochreiter S.. 2015. Msa: an R package for multiple sequence alignment. Bioinformatics. 31(24):3997–3999. [DOI] [PubMed] [Google Scholar]
- Bonczek O, Balcar VJ, Šerý O.. 2017. PAX9 gene mutations and tooth agenesis: a review. Clin Genet. 92(5):467–476. [DOI] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH.. 1969. Gene regulation for higher cells: a theory. Science. 165(3891):349–357. [DOI] [PubMed] [Google Scholar]
- Brook AH, Elcock C, Aggarwal M, Lath DL, Russell JM, Patel PI, Smith RN.. 2009. Tooth dimensions in hypodontia with a known PAX9 mutation. Arch Oral Biol. 54:S57–S62. [DOI] [PubMed] [Google Scholar]
- Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S.. 2006. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 34(19):5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra JA, Paeschke K, Singh M, Zakian VA.. 2010. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput Biol. 6(7):e1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 134(1):25–36. [DOI] [PubMed] [Google Scholar]
- Cheung I, Schertzer M, Rose A, Lansdorp PM.. 2002. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat Genet. 31(4):405–409. [DOI] [PubMed] [Google Scholar]
- Coghlan A, Wolfe H.. 2000. Relationship of codon bias to mRNA concentration and protein length in Saccharomyces cerevisiae. Yeast. 16(12):1131–1145. [DOI] [PubMed] [Google Scholar]
- Davis JT. 2004. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew Chem Int Ed Engl. 43(6):668–698. [DOI] [PubMed] [Google Scholar]
- Ding F, Elowitz MB.. 2019. Constitutive splicing and economies of scale in gene expression. Nat Struct Mol Biol. 26(6):424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy J, Maizels N.. 2008. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 36(4):1321–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauzi NH, Ardini YD, Zainuddin Z, Lestari W.. 2018. A review on non-syndromic tooth agenesis associated with PAX9 mutations. Jpn Dent Sci Rev. 54(1):30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Phylogenies and the comparative method. Am Nat. 125(1):1–15. [Google Scholar]
- Gotic I, Omidi S, Fleury-Olela F, Molina N, Naef F, Schibler U.. 2016. Temperature regulates splicing efficiency of the cold-inducible RNA- binding protein gene Cirbp. Genes Dev. 30(17):2005–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewall MS. 1962. The development of an inherited tooth defect in the mouse. J Embryol Exp Morphol. 10:202–211. [Google Scholar]
- Gruneberg H. 1963. The pathology of development. In: Minor skeletal variations. New York: John Willey and Sons. p. 239. [Google Scholar]
- Hlusko LJ, Lease LR, Mahaney MC.. 2006. Evolution of genetically correlated traits: tooth size and body size in baboons. Am J Phys Anthropol. 131(3):420–427. [DOI] [PubMed] [Google Scholar]
- Hlusko LJ, Sage RD, Mahaney MC.. 2011. Modularity in the mammalian dentition: mice and monkeys share a common dental genetic architecture. J Exp Zool Part B Mol Dev Evol. 316:21–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlusko LJ, Schmitt CA, Monson TA, Brasil MF, Mahaney MC.. 2016. The integration of quantitative genetics, paleontology, and neontology reveals genetic underpinnings of primate dental evolution. Proc Natl Acad Sci U S A. 113(33):9262–7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert JL, Balasubramanian S.. 2005. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 33(9):2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert JL, Balasubramanian S.. 2007. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 35(2):406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Zhang J, Mondragón E, Morgunova E, Kivioja T, Laverty KU, Yin Y, Zhu F, Bourenkov G, Morris Q, et al. 2020. Binding specificities of human RNA-binding proteins toward structured and linear RNA sequences. Genome Res. 30(7):962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katapadi VK, Nambiar M, Raghavan SC.. 2012. Potential G-quadruplex formation at breakpoint regions of chromosomal translocations in cancer may explain their fragility. Genomics. 100(2):72–80. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh KD, Evans AR, Jernvall J.. 2007. Predicting evolutionary patterns of mammalian teeth from development. Nature. 449(7161):427–432. [DOI] [PubMed] [Google Scholar]
- Keller JM, Huet-Hudson YM, Leamy LJ.. 2007. Qualitative effects of dioxin on molars vary among inbred mouse strains. Arch Oral Biol. 52(5):450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M, Wilson A.. 1975. Evolution at two levels in humans and chimpanzees. Science. 188(4184):107–116. [DOI] [PubMed] [Google Scholar]
- Kist R, Watson M, Wang X, Cairns P, Miles C, Reid DJ, Peters H.. 2005. Reduction of Pax9 gene dosage in an allelic series of mouse mutants causes hypodontia and oligodontia. Hum Mol Genet. 14(23):3605–3617. [DOI] [PubMed] [Google Scholar]
- Kumar P, Yadav VK, Baral A, Kumar P, Saha D, Chowdhury S.. 2011. Zinc- finger transcription factors are associated with guanine quadruplex motifs in human, chimpanzee, mouse and rat promoters genome-wide. Nucleic Acids Res. 39(18):8005–8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Kamneva OK, Melo US, Barozzi I, Osterwalder M, Mannion BJ, Tissières V, Pickle CS, Plajzer-Frick I, Lee EA, et al. 2016. Progressive loss of function in a limb enhancer during snake evolution. Cell. 167(3):633–642.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammi L, Arte S, Somer M, Jarvinen H, Lahermo P, Thesleff I, Pirinen S, Nieminen P.. 2004. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet. 74(5):1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Huang Y, He F, Gu S, Zhang G, Chen Y, Zhang Y.. 2007. Expression survey of genes critical for tooth development in the human embryonic tooth germ. Dev Dyn. 236(5):1307–1312. [DOI] [PubMed] [Google Scholar]
- Line SRP. 2003. Variation of tooth number in mammalian dentition: connecting genetics, development, and evolution. Evol Dev. 5(3):295–304. [DOI] [PubMed] [Google Scholar]
- Line SRP, Liu X, de Souza AP, Yu F.. 2013. Translational signatures and mRNA levels are highly correlated in human stably expressed genes. BMC Genomics. 14(1):268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL.. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol. 6(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckett WP. 1984. Superordinal and intraordinal affinities of rodents: developmental evidence from the dentition and placentation. In Luckett WP, Hartenberger JL, editors. Evolutionary relationships among rodents: a multidisciplinary analysis. NATO ASI series. Vol. 92. New York: Plenum. p. 227–276. [Google Scholar]
- Monson TA, Boisserie JR, Brasil MF, Clay SM, Dvoretzky R, Ravindramurthy S, Schmitt CA, Souron A, Takenaka R, Ungar PS, et al. 2019. Evidence of strong stabilizing effects on the evolution of boreoeutherian (Mammalia) dental proportions. Ecol Evol. 9(13):7597–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monson TA, Coleman JL, Hlusko LJ.. 2019. Craniodental allometry, prenatal growth rates, and the evolutionary loss of the third molars in new world monkeys. Anat Rec. 302(8):1419–1433. [DOI] [PubMed] [Google Scholar]
- Nakatomi M, Wang XP, Key D, Lund JJ, Turbe-Doan A, Kist R, Aw A, Chen Y, Maas RL, Peters H.. 2010. Genetic interactions between Pax9 and Msx1 regulate lip development and several stages of tooth morphogenesis. Dev Biol. 340(2):438–349. [DOI] [PubMed] [Google Scholar]
- Otani H, Nonaka K, Nakata M.. 1990. Genetic study on the mandibular molar size of rats. J Craniofac Genet Dev Biol. 10(3):311–327. [PubMed] [Google Scholar]
- Pecce V, Sponziello M, Damante G, Rosignolo F, Durante C, Lamartina L, Grani G, Russo D, di Gioia CR, Filetti S, et al. 2018. A synonymous RET substitution enhances the oncogenic effect of an in-cis missense mutation by increasing constitutive splicing efficiency. PLoS Genet. 14(10):e1007678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Verrelli BC, Stone AC.. 2006. Molecular evolution of the primate developmental genes MSX1 and PAX9. Mol Biol Evol. 23(3):644–654. [DOI] [PubMed] [Google Scholar]
- Pilbrow VC. 2003. Dental variation in African apes with implications for understanding patterns of variation in species of fossil apes [PhD thesis]. New York University. p. 117–118.
- Poly D. 2007. Development with a bite. Nature. 449:413–414. [DOI] [PubMed] [Google Scholar]
- Prochazka J, Pantalacci S, Churava S, Rothova M, Lambert A, Lesot H, Klein O, Peterka M, Laudet V, Peterkova R.. 2010. Patterning by heritage in mouse molar row development. Proc Natl Acad Sci U S A. 107(35):15497–15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2019. R: A language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing. Available from: https://www.R- project.org.
- Rachwal PA, Brown T, Fox KR.. 2007. Effect of G-tract length on the topology and stability of intramolecular DNA quadruplexes. Biochemistry. 46(11):3036–3044. [DOI] [PubMed] [Google Scholar]
- Raiber EA, Kranaster R, Lam E, Nikan M, Balasubramanian S.. 2012. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucleic Acids Res. 40(4):1499–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawal P. 2006. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 16(5):644–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renvoisé E, Kavanagh KD, Lazzari V, Häkkinen TJ, Rice R, Pantalacci S, Salazar-Ciudad I, Jernvall J.. 2017. Mechanical constraint from growing jaw facilitates mammalian dental diversity. Proc Natl Acad Sci U S A. 114(35):9403–9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ. 2012. Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 3(2):217–223. [Google Scholar]
- Ribeiro MM, de Andrade SC, de Souza AP, Line SRP.. 2013. The role of modularity in the evolution of primate postcanine dental formula: integrating jaw space with patterns of dentition. Anat Rec. 296(4):622–629. [DOI] [PubMed] [Google Scholar]
- Ribeiro MM, Teixeira GS, Martins L, Marques MR, de Souza AP, Line SRP.. 2015. G-quadruplex formation enhances splicing efficiency of PAX9 intron 1. Hum Genet. 134(1):37–44. [DOI] [PubMed] [Google Scholar]
- Roscito JG, Sameith K, Parra G, Langer BE, Petzold A, Moebius C, Bickle M, Rodrigues MT, Hiller M.. 2018. Phenotype loss is associated with widespread divergence of the gene regulatory landscape in evolution. Nat Commun. 9(1):4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadier A, Twarogowska M, Steklikova K, Hayden L, Lambert A, Schneider P, Laudet V, Hovorakova M, Calvez V, Pantalacci S.. 2019. Modeling Edar expression reveals the hidden dynamics of tooth signaling center patterning. PLoS Biol. 17(2):e3000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayani S, Chanfreau GF.. 2012. Sequential RNA degradation pathways provide a fail-safe mechanism to limit the accumulation of unspliced transcripts in Saccharomyces cerevisiae. RNA. 18(8):1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M.. 2011. Global quantification of mammalian gene expression control. Nature. 473(7347):337–342. [DOI] [PubMed] [Google Scholar]
- Shah P, Gilchrist MA.. 2011. Explaining complex codon usage patterns with selection for translational efficiency, mutation bias, and genetic drift. Proc Natl Acad Sci U S A. 108(25):10231–10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov I, Shafer RH.. 2000. Effect of loop sequence and size on DNA aptamer stability. Biochemistry. 39(6):1462–1468. [DOI] [PubMed] [Google Scholar]
- Stuckey E, Berry CL.. 1984. The effects of high dose sporadic (binge) alcohol intake in mice. J Pathol. 142(3):175–180. [DOI] [PubMed] [Google Scholar]
- Thakur RK, Kumar P, Halder K, Verma A, Kar A, Parent J-L, Basundra R, Kumar A, Chowdhury S.. 2009. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 37(1):172–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thesleff I. 2018. From understanding tooth development to bioengineering of teeth. Eur J Oral Sci. 126(Suppl 1):67–71. [DOI] [PubMed] [Google Scholar]
- Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S.. 2008. Genome-wide computational and expression analyzes reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J Med Chem. 51(18):5641–5649. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yang J, Wild AT, Wu WH, Shah R, Danussi C, Riggins GJ, Kannan K, Sulman EP, Chan TA, et al. 2019. G-quadruplex DNA drives genomic instability and represents a targetable molecular abnormality in ATRX-deficient malignant glioma. Nat Commun. 10(1):943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 8(3):206–216. [DOI] [PubMed] [Google Scholar]
- Xia X. 2020. RNA-Seq approach for accurate characterization of splicing efficiency of yeast introns. Methods. 176:25–33. [DOI] [PubMed] [Google Scholar]
- Xie KT, Wang G, Thompson AC, Wucherpfennig JI, Reimchen TE, MacColl ADC, Schluter D, Bell MA, Vasquez KM, Kingsley DM.. 2019. DNA fragility in the parallel evolution of pelvic reduction in stickleback fish. Science. 363(6422):81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue H, Liang J, Yang K, Hua B, Bian Z.. 2016. Functional analysis of a novel missense mutation in AXIN2 associated with non-syndromic tooth agenesis. Eur J Oral Sci. 124(3):228–233. [DOI] [PubMed] [Google Scholar]
- Zorba E, Moraitis K, Manolis SK.. 2011. Sexual dimorphism in permanent teeth of modern Greeks. Forensic Sci Int. 210(1–3):74–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.