Abstract
India represents an intricate tapestry of population substructure shaped by geography, language, culture, and social stratification. Although geography closely correlates with genetic structure in other parts of the world, the strict endogamy imposed by the Indian caste system and the large number of spoken languages add further levels of complexity to understand Indian population structure. To date, no study has attempted to model and evaluate how these factors have interacted to shape the patterns of genetic diversity within India. We merged all publicly available data from the Indian subcontinent into a data set of 891 individuals from 90 well-defined groups. Bringing together geography, genetics, and demographic factors, we developed Correlation Optimization of Genetics and Geodemographics to build a model that explains the observed population genetic substructure. We show that shared language along with social structure have been the most powerful forces in creating paths of gene flow in the subcontinent. Furthermore, we discover the ethnic groups that best capture the diverse genetic substructure using a ridge leverage score statistic. Integrating data from India with a data set of additional 1,323 individuals from 50 Eurasian populations, we find that Indo-European and Dravidian speakers of India show shared genetic drift with Europeans, whereas the Tibeto-Burman speaking tribal groups have maximum shared genetic drift with East Asians.
Keywords: India, population structure, South Asia, genomics, algorithms, data mining
Introduction
The genetic structure of human populations reflects gene flow around and through geographic, linguistic, cultural, and social barriers (Cavalli-Sforza et al. 1988; Sokal 1991). The intricate tapestry of population substructure and complexity in India undoubtedly showcases the interplay among them. The Indian subcontinent encompasses 3,200 km from North to South, complex topography with elements ranging from the Himalayas to the Thar desert, plateaux, and rain forests, almost 800 spoken languages, a long history of migrations and invasions and a strict caste system imposing endogamy.
The strata within India can be summarized into the so-called backward castes and forward castes (Desai and Dubey 2012), whereas 8.2% of the total population belongs to tribes (1991 census) representing minorities that are unassimilated into the caste system. The tribes in India continue to live in forest hills and naturally isolated regions with a largely hunting-gathering subsistence mode. They practice endogamy, a matrimonial rule governing mate-exchange within local groups (Vidyarthi and Rai 1977). On the other hand, the caste system is a rigorous social hierarchy of endogamous groups in which individuals are born (Olcott 1944; Wooding et al. 2004). Prior to the establishment of the caste system there was wide admixture among them, which came to an abrupt end 1,900 to 4,200 years before present (Moorjani et al. 2013). Historically, the so-called forward castes have been associated with socio-economic privileges, whereas the backward castes and tribal groups faced social segregation (Desai and Dubey 2012). Although discrimination on the basis of caste was abolished by the Indian constitution in 1950, this strict social structure has existed for thousands of years (Thapar 1990).
Numerous studies have attempted to dissect the genetic components and origins of Indian populations (Bamshad et al. 2001; Majumder 2001; Roychoudhury et al. 2001; Basu et al. 2003, 2016; Brahmachari et al. 2005; Reich et al. 2009; Metspalu et al. 2011; ArunKumar et al. 2012; Moorjani et al. 2013; Silva et al. 2017; Pathak et al. 2018) along with ancient individuals from Central and South Asia (Narasimhan et al. 2019). Studies of Indian populations based on groupings of tribal versus nontribal, geographic regions, or linguistic affiliation have shown that the observed genetic structure resulted from admixture of five ancestral populations. These are Ancestral North Indians, which loosely captures Indo-European (IE) speakers in Northern India; Ancestral South Indians, who are mostly Dravidian (DR) speakers of Southern India; Ancestral Austroasiatic with Austroasiatic (AA) speakers of Central and Eastern India; Ancestral Tibeto-Burman speakers constituted of Tibeto-Burman (TB) speakers in Northeast and the tribal populations, Jarawa and Onge, from Andaman (AND) archipelago (Basu et al. 2016). Great Andamanese is considered as the sixth language family of India, being a linguistic isolate, typologically and genealogically different from other AND languages (Abbi 2009). However, to date, no study has attempted to model how different spatiocultural features acted in concert in order to create the observed genetic structure across the Indian subcontinent and to evaluate the relative contribution of each factor.
Earlier attempts to investigate the covariance of allele frequencies and nongenetic factors on genetic structure either depended heavily on assumptions and a computationally expensive Bayesian framework (Bradburd et al. 2013) or did not provide any statistical significance or feature selection to identify the most relevant structure-related factors (Schlebusch et al. 2012). To dissect the population substructure in Indian populations, we designed a quantitative framework for the evaluation of the relative contribution of geodemographic features such as geography, spoken language, and social structure to the architecture of the genetic pool of human populations. Our work provides a general model that may be used to study the significance of each underlying factor on the genetic substructure of a given population.
New Approaches
In order to understand the genetic substructure of India, considering the strongly endogamous social structure as well as the presence of multiple language families and their geographical distribution, we developed Correlation Optimization of Genetics and Geodemographics (COGG). COGG is a deterministic algorithm that may be used to simultaneously correlate genome-wide genotypes, with multiple factors that may have acted to shape population genetic substructure. In the context of this study, we correlate genetic structure as depicted by the top two principal components (PCs) with geography (longitude and latitude) and sociolinguistic factors (social and language group information in this case) as shown in equation (1). We encoded four language groups AA, DR, IE, and TB as well as the social group information as indicator variables i. e., if a sample belongs to a social or language group, we use 1 and 0 otherwise. We refrain from using terms that could be considered socially stigmatizing and instead refer to Social Group A (SGA) for forward castes and Social Group B (SGB) for backward castes, respectively. For the seminomadic tribes in India, we assign Social Group C (SGC) (more details in supplementary note, Supplementary Material online).
Given information on m samples, the objective of COGG is to maximize the correlation between u, the genetic component as represented by either of the top two PCs of the genetic covariance matrix formed by the genotype data and a geodemographic matrix where k is the number of demographic features.
| (1) |
Therefore, COGG solves the following optimization problem,
| (2) |
where a be the k-dimensional vector whose elements are (k = 9 in this case). Recall that denotes the i-th column vector of G. Let for and let d be the vector of the di’s. Also, let for all and let M be the matrix of Mij. Then the optimizer for COGG is given by
We also check for statistical significance of the maximum squared Pearson correlation coefficient r2, returned by COGG, by conducting 1,000 permutation tests on the sociolinguistic variables in G. On top of COGG, we used a greedy feature selection algorithm to select the most significant factors which influence genetic variation in India.
To further study the interplay between these factors, we propose a simple analytic procedure using the so-called ridge leverage score (RLS) statistic that highlights the significant populations capturing genetic diversity in India. The RLS of the i-th row of any matrix is defined as:
| (3) |
where is the regularization parameter.
Starting from the mean-centered (subtracting each column by its respective mean) genotype matrix where n is the number of markers for each of m samples and G as described above, we compute population level RLS (median RLS of the samples in the population) for each matrix (details in Materials and Methods and supplementary note, Supplementary Material online). Thereafter, we compute an additive RLS statistic for each population highlighting the ethnic groups which represent and capture the greatest portion of observed genetic diversity across India. Our analysis aims to better understand the intricate details of admixture, substructure, and genetic variation across social and language groups in the Indian subcontinent. The need for methods such as COGG has been previously underlined by many studies (Bamshad et al. 2001; Roychoudhury et al. 2001; Basu et al. 2003, 2016; Majumder 2010). The ability to correlate genomic background with geographic, sociolinguistic, and cultural differences opens new avenues to study genomic structure of extant human populations.
Results and Discussion
Description of Compiled Data Sets
We begin by briefly introducing the different data sets that are presented throughout our analysis (supplementary table S1, Supplementary Material online). We initially compiled a pan-Indian data set of 891 individuals across 90 populations (supplementary table S1A and fig. S1A, Supplementary Material online) and 47,283 SNPs from various sources (Reich et al. 2009; Chaubey et al. 2011; Metspalu et al. 2011; Moorjani et al. 2013; Basu et al. 2016). This data set presented unequal representations of the five language families IE, DR, AA, TB, and AND as well as uneven distribution across social groups and geographical regions. To create a normalized subset across these spatiocultural features, we selected a subset of 33 populations spanning 368 individuals (supplementary table S1B, Supplementary Material online and fig. 1A) in which four language families AA, DR, IE, and TB are represented (supplementary note, Supplementary Material online) and used it for COGG and subsequent feature selection analyses. For other analyses such as the RLS statistic identifying representative ethnic groups contributing to the genetic diversity in India and relationship between sociolinguistic groups, we used the pan-Indian data set. Furthermore, in order to interrogate the shared ancestry between Indian sociolinguistic groups and Eurasia, we merged the normalized subset with 1,323 individuals from 50 populations and 42,975 SNPs across Eurasia (supplementary table S1C, Supplementary Material online). For the outgroup f3 analysis, we present later in this section, we used 124 samples of Yorubans in Nigeria (YRI) from the 1000 Genomes phase 3 data set (Auton et al. 2015) and merged it with the Eurasian data set.
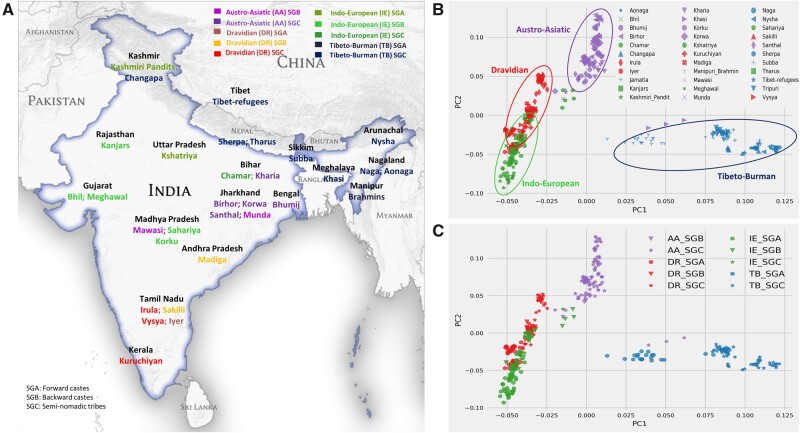
Fig. 1.
A map of locations of the 33 populations in the normalized set and the results of principal component analysis. (A) Map of India showing the locations of the 368 individuals in the normalized subset across 33 well-defined populations, 47,283 SNPs (see supplementary fig. S1A, Supplementary Material online, for the pan-Indian data set of 90 ethnic groups and supplementary fig. S2, Supplementary Material online, for the corresponding PCA plot). The populations are colored by their sociolinguistic group. (B) Top two PCs of the normalized data set show clustering by language groups. (C) PCA plot colored and marked by sociolinguistic groups shows the genetic structure stratified by sociolinguistic groups.
Geography versus Population Structure within India
Studies of populations in different parts of the world have shown that when top two PCs are extracted from genome-wide genotypes, individuals from the same geographic region cluster together with the PCs being well correlated with geographic coordinates, namely longitude and latitude (Lao et al. 2006; Rosenberg et al. 2006; Chen et al. 2009; Paschou et al. 2010).
For instance, Novembre and Stephens (2008) showed that within Europe, the Pearson correlation coefficient (r2) (hereafter r2) between PC1 versus latitude (north–south) is equal to 0.77 and 0.78 for PC2 versus longitude (east–west). In order to explore whether Indian genetic information mirrors geography, we computed principal component analysis (PCA) on the normalized data set of 33 Indian populations and plotted the top two PCs (fig. 1B and C and supplementary fig. S1B, Supplementary Material online, for language, sociolinguistic, and geographical groupings, respectively). The first three PCs explained 32%, 15%, and 10% of the total variance, respectively. Along PC1, we observed a separation of TB speakers from the rest of the Indian populations. On the other hand, the IE and DR speaking populations formed a cline separated from AA speakers on PC2 (fig. 1B). Next, we computed r2 between the top two PCs of the covariance matrix and the geographic coordinates (longitude and latitude) of the samples under study. We observed () for PC1 versus longitude and () for PC2 versus latitude. Thus, PC1 correlates well with longitude due to the East–West cline of language families with IE and TB speakers in Northwestern and Northeastern Frontiers, respectively and AA speakers dwelling in the forests of Central India between them. However, PC2 only minimally correlates with latitude, just barely picking up a previously reported North–South cline of IE and DR speakers (Reich et al. 2009). We note that IE and DR speakers also share significant ancestry among SGA and SGB groups as indicated by the result of ADMIXTURE analysis (Alexander et al. 2009) (supplementary fig. S3, Supplementary Material online). Interestingly, we observe clusters of sociolinguistic groups which become more prominent in the second and third PCs (supplementary fig. S4, Supplementary Material online) with the SGCs distinguished from SGA and SGB within their language group.
This weak correlation between geography and genetics in Indian context is confirmed by Mantel tests between genetic (FST) and geographic distances which returned a low (P = 0.0001, Z = 5.71) when run on the normalized data set with 33 groups. These findings are in sharp contrast with findings within the European continent (Novembre and Stephens 2008; Drineas et al. 2010) and highlight the need for social and linguistic factors to be accounted for, as noted in prior work (Bamshad et al. 2001; Roychoudhury et al. 2001; Brahmachari et al. 2005; Majumder 2010; Basu et al. 2016). We performed linear discriminant analysis (LDA) (supplementary fig. S5, Supplementary Material online) in order to gain further understanding of the relationship between genetics, geography, language, and social groups in shaping the structure of the data. We run LDA on the normalized data set with the language groups set as classes (supplementary fig. S5A, Supplementary Material online) followed by the geographic regions (supplementary fig. S5B, Supplementary Material online). In the LDA performed by language group, three separate clusters capturing IE social groups (SGA, SGB, and SGC) appear in one axis of variation. The second axis captures the rest of the language groups again stratified by social group. In the LDA performed by geography, we see an east–west cline with TB speakers in the left and IE speakers in the right along the first discriminant. However, the second discriminant does not pick up the north–south cline as was expected, further indicating confounding by sociolinguistic groups.
Correlation Optimization of Genetics and Geodemographics
Having shown that geography alone cannot explain the genetic structure within India, we applied COGG to explore whether integrating information on spoken language and social structure as shaped by endogamy can lead to an improved model. Indeed, solving the optimization problem that underlies COGG (see Materials and Methods and supplementary note, Supplementary Material online, for the exact formulation) and plugging in the solution, we observe almost perfect correlation with PC1 and PC2 representing the genetic structure of the Indian subcontinent using the geodemographic matrix G instead of just longitude and latitude: r2 increases from 0.6 to 0.93 () for PC1 versus G and from 0.06 to 0.85 () for PC2 versus G.
Our results clearly show that endogamy and language families are pivotal in studying the genetic stratification of Indian populations. This is in sharp contrast to what has been seen in other parts of the world where geography is a major contributor in shaping genetic structure of populations (Cann et al. 2002; Novembre and Stephens 2008; Auton et al. 2015). Our results are statistically significant (supplementary fig. S6, Supplementary Material online) over 1,000 iterations with permutation of the variables related to social factors and languages (see supplementary note, Supplementary Material online).
We further explored an extension of COGG in order to jointly analyze multiple PCs simultaneously and not just each component individually. To do this, we employed canonical correlation analysis (CCA), a well-studied statistical technique, which maximizes the correlation between the genetic and the geodemographic matrices by jointly finding linear combinations of the variables in each matrix. We used the top eight PCs of the genetic matrix as the results did not improve significantly, beyond that. We note that these eight PCs capture, collectively, 89% of the variance of the genetic matrix.
Running COGG-CCA on these inputs returns a statistically significant (supplementary fig. S7, Supplementary Material online) r2 equal to 0.94 () which is well above the obtained when COGG-CCA is run without including the sociolinguistic factors (see supplementary note, Supplementary Material online, for details).
Identifying the Features That Drive Population Structure within India
In order to formally investigate which of the nine features in the geodemographic matrix G contribute more in the optimization problem posed by COGG (eq. 2), we used the sparse approximation framework and the orthogonal matching pursuit (OMP) algorithm from applied mathematics (Natarajan 1995) (see supplementary note, Supplementary Material online). Running OMP on our data set, we obtain two sets of three features each, S1 and S2, for PC1 and PC2, respectively:
Plugging in S1 as the reduced feature space in COGG resulted in () for PC1 versus S1 and 0.85 () for PC2 versus S2. These values capture over 99% of the correlation returned by COGG when all the features in G are included. Membership to the AA and TB language groups which are identified among the top significant features correspond mostly to tribal nomadic hunter-gatherers dwelling in the hills and forests of Central East and North East India, respectively. Thus, the AA and TB language groups automatically capture SGC. On the other hand, membership to SGA, which is the other top significant feature that we identified, spans most of the IE and DR speakers found across Northern and Southern India. Thus, these three features appear to encompass most of the geographic, social, and linguistic diversity found in the Indian subcontinent and highlight their interplay.
Ethnic Groups Capturing Genetic Diversity across India
We developed a simple approach based on the RLS statistic (Alaoui and Mahoney 2015) (see Materials and Methods) to identify influential (from a genetic perspective) Indian populations which represent and capture the greatest portion of observed genetic diversity across India. Here, we analyzed the pan-Indian data set of 90 populations (details in Materials and Methods).
The RLS statistic highlights ethnic groups in the Indian subcontinent who either are quite distinct (e.g., underwent a founder event, or practiced endogamy and maintained isolation from other groups) or populations that show signs of admixture from distinctly different language families (table 1). Such populations create a mesh of complex layers of admixture across language and social barriers. We observe mostly SGB and SGC populations across all the language families in India encapsulate much of its genetic structure. Some of the highlighted populations are: 1) Great Andamanese and Jarawas from AND represent distinct ethnic groups and outliers with respect to mainland Indian populations (supplementary fig. S2B, Supplementary Material online). Great Andamanese are also linguistically divergent from Jarawa (Abbi 2009); 2) Vysyas, who underwent a founder event going back 100 generations, due to the strong imposition of endogamy (Reich et al. 2009); 3) Language isolates Vedda from Sri Lanka (Chaubey 2014); 4) Minicoy from Lakshadweep Archipelago with strong founder effects and diverse mixture due to the archipelago being a popular destination for maritime sailors (Samuel et al. 2009); 5) AA speaking Mundas who have Ancestral North and South Indian ancestry and an Ancestral Southeast Asian component (Tätte et al. 2019); 6) Manipuri Brahmins (TB_SGA) who show high shared ancestry with IE_SGA as well as TB_SGC (supplementary table S2, Supplementary Material online), since they are at the junction of the language families; and 7) TB speaking Changpas, who are seminomadic pastoralists dwelling in the high altitudes of Tibet and Ladakh in India.
Table 1.
Top Ten Significant Ethnic Groups in India Capturing the Genetic Structure of the Subcontinent as Reflected by the RLS Statistic.
| Population | State/Territory | Language Family | Social Group |
|---|---|---|---|
| Great Andamenese | Andaman and Nicobar Islands | Great Andamanese | SGC |
| Minicoy | Lakshadweep islands | IE | SGB |
| Vedda | Sri Lanka | IE | SGC |
| Vysya | Andhra Pradesh | DR | SGAa |
| Palliyar | Tamil Nadu | DR | SGC |
| Munda | Madhya Pradesh | AA | SGC |
| Changpas | Jammu and Kashmir | TB | SGC |
| Manipuri Brahmins | Manipur | TB | SGA |
| Meghawal | Rajasthan | IE | SGB |
| Jarawa | Andaman and Nicobar islands | Ongan | SGC |
Vysyas are classified as in between SGA and SGB; Moorjani et al. (2013).
Relationship between Sociolinguistic Groups
Our analyses using COGG clearly support the fact that language families and endogamy within social groups have played a significant role in shaping the genetic structure of the Indian subcontinent. Here, we further dissect the relationship between the endogamous social groups including the AND isolates (Thangaraj et al. 2003; Mondal et al. 2016) in order to highlight the cryptic relatedness among ethnic groups that COGG posits.
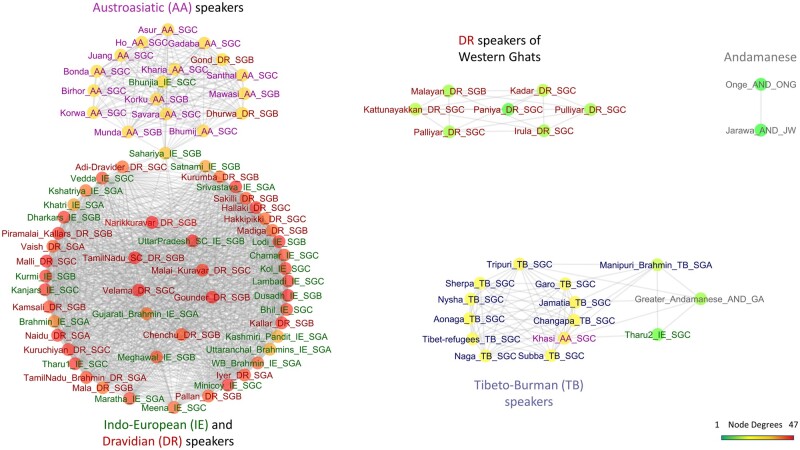
To better illustrate the intricacies in the relationships between the social groups in India, we constructed a network of all the 90 populations across India (fig. 2). The network was built as we have previously described (Paschou et al. 2014) based on weights that reflect shared ancestry (supplementary table S2, Supplementary Material online) as computed by meta-analysis of ADMIXTURE results (Alexander et al. 2009) (see Materials and Methods and supplementary note, Supplementary Material online, for details). The shared ancestry network, revealed four major clusters (i.e., 1. IE and DR, 2. AA, 3. TB, and 4. AND) and a few exceptions as outlined in detail below.
Fig. 2.
Network of 90 Indian populations (891 individuals) in the pan-Indian data set based on shared ancestry as defined by meta-analysis of ADMIXTURE results. Only the top 40% of edges (most related) populations are shown here (see Materials and Methods for details). The node labels are colored by their corresponding language groups as shown in figure 1.
IE and DR Populations across Social Groups
A cluster of IE and DR speakers across social groups resembling a nearly complete graph with over 60% of all possible edges was observed (fig. 2). This was further supported by a similar pattern of strong shared ancestry in outgroup f3 statistics (Patterson et al. 2012) using YRI from the 1000 Genomes data set as the outgroup (Auton et al. 2015) as well as in f3 tests for signs of admixture. We find that most IE and DR populations share more alleles with each other (supplementary fig. S8, Supplementary Material online) and are admixed with each other (supplementary table S3, Supplementary Material online). IE speakers share above 70% average ancestry with DR_SGA and DR_SGB (supplementary fig. S3B, Supplementary Material online) in the meta-analysis of ADMIXTURE. This supports the notion that there was mixture between IE and DR speakers across SGA and SGB around 1,900 to 4,200 years ago (Moorjani et al. 2013) and that the caste system originated in a “classless” seminomadic society, which became hierarchical with the knowledge of agriculture (Kosambi 1964; Majumder 2001). Furthermore, it provides a possible explanation for DR loanwords appearing in early Hindu texts which are not found in IE languages outside the Indian subcontinent (Mallory and Adams 1997; Witzel 2001; Moorjani et al. 2013). The high relatedness between SGA and SGC across IE and DR speakers barring a few exceptions (supplementary fig. S9, Supplementary Material online), also provides genetic evidence to the claim that although the caste system was formally defined and observed to be stringent, it was broken in some cases, allowing mixture between SGC and SGA (Thapar 2014).
AA Speakers Forming a Clique
Almost all AA populations from Central and East India tightly cluster together with fellow Central Indian groups such as Bhunjia (IE_SGC), Gonds (DR_SGB), and Sahariya (IE_SGB).
Clique of TB Speakers
TB speakers from North East India form a strongly connected cluster with the Khasis (AA speakers residing in North East India) who also clustered together with TB speakers in the scatter plot of the top two PCs (fig. 1B). The cluster also contains Manipuri Brahmins (TB_SGA), who are known to have significant admixture from IE_SGA and Tharus (IE_SGC) (Chaubey et al. 2014) from Tarai region in Nepal and eastern India (supplementary tables S3A and B, Supplementary Material online).
Isolated and Groups
The AND groups Jarawa and Onge diverge from the rest of the Indian populations. This has also been shown in (Thangaraj et al. 2003; Reich et al. 2009; Basu et al. 2016; Mondal et al. 2016). They belong to the Ongan language family which has a debatable connection with Austronesian languages (Blevins 2007), showing divergence from all language families in mainland India.
Populations outside Major Clusters
Above, we describe four major clusters each capturing the majority of individuals from different language groups: 1. The IE and DR cluster with 81% of IE and 69% of DR, 2. The AA cluster, capturing 93% of AA, 3. TB cluster with 73% of TB, and 4. a main AND cluster with 66% of AND populations. However, in each case, we also observed some exceptions revealing cryptic relatedness among ethnic groups which we outline here.
Few DR_SGC groups such as Kadar, Irula, Palliyar, and Paniya (which contain the lowest levels of Ancestral North Indian ancestry among Indian populations; Moorjani et al. 2013) formed a connected component, isolated from the main IE-DR cluster. They are hunter-gatherer populations dwelling in the forests of Western Ghats in Southern India, isolated from the rest of the DR_SGCs and very low shared ancestry with IE_SGC (supplementary fig. S9, Supplementary Material online).
The Gonds and Sahariyas are candidate mosaic Indian populations, which are also reflected by their location as bridge nodes between the AA and IE-DR cliques. They contain high AA, DR, and IE ancestry (supplementary figs. S8 and S9 and table S2, Supplementary Material online), which can be attributed to their central location in India (Chaubey et al. 2017) and their long history of exogamy.
We also found the Great Andamenese to be connected to TB speakers of North East India, rather than other AND populations. They share approximately 50% shared ancestry (supplementary table S2, Supplementary Material online) as well as showing strong shared genetic drift with respect to outgroup f3 statistics (supplementary fig. S9, Supplementary Material online). The Great Andamanese are known to be genetically divergent from other AND groups Jarawa and Onge (Thangaraj et al. 2003; Abbi 2009). To the best of our knowledge, this is the first observed interaction of the group to the rest of mainland Indian speakers based on autosomal markers and should be interpreted with caution due to small samples sizes of all groups involved. However, a study focused on the mitochondrial haplogroup M31 showed that with the exception of M31a1 (specific to AND), lineages M31a2, M31b, and M31c are prevalent in North East India and surrounding regions (Wang et al. 2011). The authors concluded with time estimation that the Andaman archipelago was likely settled by modern humans from North East India via the land-bridge connecting Andaman archipelago and Myanmar around Last Glacial Maximum (LGM) (Voris 2000; Clark et al. 2009).
The Mosaic of Indian Sociolinguistics in the Context of Eurasia
Indian populations from diverse sociolinguistic groups have different genetic affinities toward Eurasian populations. Outgroup f3 statistics between the sociolinguistic groups and European populations with YRI as outgroup, reveal greater shared genetic drift between IE speakers (across social groups) and DR_SGA with European and Middle Eastern populations (supplementary table S2, Supplementary Material online).
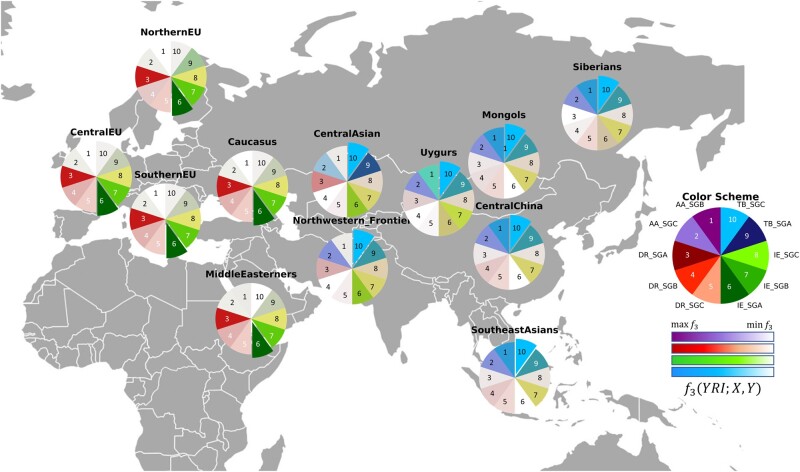
The East Asian populations have more shared drift with the TB speakers along with some affinity with AA speakers, which is in agreement with a previous study (Tätte et al. 2019). Our results clearly show two paths with a gradient of decreasing shared genetic drift from India and Eurasia: one from North East India toward China, Mongolia, and Siberia and the other from North West India toward Central Asia, Uygurs, Middle Easterners, and Europeans (fig. 3). This is concordant with our findings from network analysis with respect to connections with possible gateways to and from the Indian subcontinent (supplementary fig. S10, Supplementary Material online).
Fig. 3.
Shared genetic drift between 33 Indian populations (denoted by X) and 50 Eurasian/East Asian populations (denoted by Y) as estimated by f3 statistics with Yoruba as an outgroup f3 (YRI; X, Y). The darkest colors correspond to greatest portions of shared genetic drift with Indian populations. Full results can be found in supplementary table S4, Supplementary Material online.
Conclusion
India represents a country of great social and linguistic complexity. We established a quantitative deterministic and nonparametric framework called COGG, aiming to evaluate the relative contribution of language, social structure, and geography in shaping the Indian gene pool. COGG resulted in a dramatic increase in correlation between top PCs depicting genomic structure and the geodemographic factors that we investigated. We applied a feature selection algorithm to identify the most important factors shaping genomic structure in India, as well as a RLS statistic to highlight ethnic groups in India that best capture its diverse gene pool. Intriguingly, our study shows that spoken language seems to have been the major force bringing people together in India, across geographic and social barriers highlighting the need for population-specific studies.
We find evidence of wide mixture across all the social groups (tribal and nontribal) for IE speakers and across SGA and SGB for DR speakers. We also provide further support for broad admixture and a long contact between IE and DR speakers in India. Our analysis also identifies finer substructure and population relationships within Indian sociolinguistic groups as well as their relatedness with various Eurasian populations. Interestingly, we find stronger shared ancestry between the Great Andamenese with TB speakers of North East India than other mainland speakers, a relationship which is observed for the first time using autosomal markers.
The framework developed here in order to understand genetic structure within the Indian subcontinent can be applied more broadly to different populations to model the interaction between different factors that may have shaped genetic diversity. The possibility to correlate genomic background to geographic, social, and cultural differences opens new avenues for understanding how human history and mating patterns are translated into the genomic structure of extant human populations.
Materials and Methods
Study Design and Data Sets
We used PLINK1.9 (Chang et al. 2015) to assemble genome-wide data for 891 samples from 90 well-defined sociolinguistic groups (fig. 1A and supplementary table S1, Supplementary Material online) genotyped on 47,283 autosomal SNPs. These samples were collected from various sources (Reich et al. 2009; Chaubey et al. 2011; Metspalu et al. 2011; Moorjani et al. 2013; Basu et al. 2016) with the consent of the corresponding authors. We created subsets of this data set in order to construct an equal representation of social groups, language families, and geographical locations for this study and tested for correlation between genetics and geography along with sociolinguistic features. The normalized subset (see supplementary note, Supplementary Material online, for details) for which we have reported results on COGG, contains 368 samples from 33 populations genotyped on 47,283 SNPs (supplementary table S1B, Supplementary Material online). We converted all data to the same build (hg19) using LiftOver from the UCSC Genome Browser (Hinrichs 2006) before merging the data. Further quality control such as filtering out variants with missing call rates and minor allele frequency <0.05 was performed in PLINK1.9.
We merged 1,323 individuals across 50 populations from Eurasia and Southeast Asia, collected from various publicly available sources such as HGDP (Cann et al. 2002), the Estonian Biocenter (Behar et al. 2010; Yunusbayev et al. 2012, 2015; Di Cristofaro et al. 2013; Fedorova et al. 2013; Kovacevic et al. 2014; Raghavan et al. 2014), and the Allele Frequency Database (ALFRED) (Rajeevan et al. 2003) (supplementary table S1C, Supplementary Material online) with our normalized Indian data set to create a merged data set of 1,691 samples from 83 populations genotyped on 42,975 SNPs overlapping between all data sets.
PCA and LDA
We used TeraPCA (Bose et al. 2019) to perform PCA on our data sets after pruning for LD structure by setting –indep-pairwise 50 10 0.4 in PLINK1.9. We checked for outliers (using EIGENSTRAT’s; Price et al. 2006; outlier detection method) in the PCA plot (supplementary fig. S2A, Supplementary Material online) and removed three outliers, each one from TB speakers Jamatia, Tripuri, and Sherpa.
We implemented Rao’s discriminant analysis which is directly based on Fisher’s linear discriminant analysis (supplementary note, Supplementary Material online).
Mantel Tests
We computed pairwise FST distances between 33 Indian populations in the normalized data set using PLINK1.9. Thereafter, we computed the correlation between the FST and the distance matrix based on the geodemographic variables using the Mantel test function in Python’s scikit-bio package. We performed 10,000 permutations and estimated Spearman’s correlation, acknowledging the caveat of overestimation of P values obtained from the tests (Guillot and Rousset 2013).
COGG and Feature Selection Using OMP
Aimed to model genetic structure within India, COGG maximizes the correlation between the top two PCs (for more PCs, see CCA section in supplementary note, Supplementary Material online) and the geodemographic matrix which consists of nine variables (columns) corresponding to geographical coordinates (latitude and longitude), social groups, and language information encoded as indicator variables. COGG is explained in detail in New Approaches and supplementary note, Supplementary Material online.
On top of COGG, we used a greedy feature selection algorithm described in (Natarajan 1995) to select features of the geodemographic matrix G. We obtain two sets, S1 and S2 of the three most significant features from G, for PC1 and PC2, respectively. In short, it selects the column which results in the maximum r2 value from G and then projects G (and u) on the subspace perpendicular to the selected column in order to form (and ). We iterate the process until we have removed the required number of features from G (details in supplementary note, Supplementary Material online).
All the values returned by this method are statistically significant. When COGG was run with random permutations of the elements of S1 and S2, it returned negligible r2. We also considered all combinations of three feature sets and concluded that, out of all possible sets, only S1 and S2 return maximum correlation with PC1 and PC2, respectively.
Ridge Leverage Scores
We devised a simple method based on the RLS statistic in order to identify Indian populations that maximally contribute to the genetic diversity within the Indian subcontinent. We considered the genotype data, denoted by mean-centered (by SNPs) matrix where m is the number of individuals and n is the number of markers in the pan-Indian data set of 90 Indian populations (891 individuals) and 47,283 SNPs. Since we are interested in the median RLS statistic as the representative of a population, including groups of larger sample size would not introduce any bias, so there was no need for normalization. We also considered the mean-centered geodemographic matrix G. Our analysis procedure based on the RLS statistic has four steps:
We apply the RLS algorithm (supplementary note, Supplementary Material online) separately to the matrices Z and G to find their corresponding row RLSs, denoted by and , respectively, for .
We grouped the RLSs by populations to obtain a single score (median RLS) per group. If there are populations in the entire set of the Indian populations ( in this case), then we obtain RLSs in this manner, one per population ti, defined as the vectors and .
- Next, we compute an additive RLS for each population after normalizing the vectors obtained in the last step. This additive RLS highlights the significant rows (in our case, Indian populations), across both the genotype and geodemographic matrices Z and G. We define this consolidated additive RLS as,
Finally, we sort the entries of in descending order to obtain a set of representative populations.
Estimating Population Admixture and Meta-Analysis
We used the ADMIXTURE v1.22 software (Alexander et al. 2009) for all admixture analyses. Prior to running ADMIXTURE, we pruned for LD using PLINK1.9 by setting –indep-pairwise 50 10 0.8. We used 8-fold cross-validation (CV) to determine the optimal number of ancestral populations (K). We varied K between two and eight performing iterations until convergence for each value of K and selected the one with the lowest CV error.
We also performed a quantitative analysis (supplementary note, Supplementary Material online) of ADMIXTURE’s output as shown in (Stamatoyannopoulos et al. 2017). To compute the shared ancestry between populations X and Y, we create two matrices and containing the estimates from ADMIXTURE, where x and y are the numbers of samples in X and Y respectively. Thereafter, we project onto the subspace spanned by . In other words, we take the top p eigenvectors of and perform the following to find the shared ancestry between X and Y,
We compute the shared ancestry values for each K, by varying it from four to eight and report the mean shared ancestry across these ancestral components. Furthermore, we designed a color-coding scheme for better visualization. The highest and lowest shared ancestry correspond to black and white respectively, and all intermediate values follow a gradient from black to white.
Three Population Statistics
f 3 tests are conducted for checking whether a target population (Z) is admixed between two source populations (X and Y) or to measure the shared drift between two test populations (X and Y) from an outgroup (Z).
where pi is the allele frequency for a given site in population i (Patterson et al. 2012; Peter 2016) for a detailed exposition on f3 tests. We employ both these tests using ADMIXTOOLS (Patterson et al. 2012) to find signs of admixture and shared genetic drift within Indian populations as well as to find shared drift between Indian sociolinguistic groups and Eurasian populations using YRI as an outgroup. We set the significance thresholds for z-score as .
Network Analysis
To better visualize and understand the connection between the populations included in our study, we performed a network analysis where the nodes represent each of 90 Indian populations and the edge weights correspond to the mean shared ancestry computed by meta-analysis results of ADMIXTURE (varying K from four to eight), as shown in a previous study (Paschou et al. 2014). As we can have number of edges for an undirected graph with m nodes, we allow edges to the graph (fig. 2) until all the n populations (nodes) appear in the graph with their corresponding nearest neighbors (NN) sorted by decreasing edge weight (shared ancestry). Using this method with 3 NN, we obtained the top 40% of all edges for figure 2.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
Major part of this study was done when A.B. was a graduate student in the Department of Computer Science at Purdue University. We thank D. Reich, P. Moorjani, and P. P. Majumder for sharing genotypic data of 248 samples from Reich et al. (2009), 378 samples from Moorjani et al. (2013), and 367 samples from Basu et al. (2016), respectively. We also thank the editor and the reviewers for their thoughtful comments and suggestions which have helped us improve the manuscript. A.B., P.D. and P.P. were partially supported by National Science Foundation (NSF) 10001225, NSF 10001674, and NSF 10001390. A.B., D.E.P. and L.P. were supported by IBM.
Data Availability
Data used in this manuscript are available from the respective corresponding authors. Code for COGG and COGG-CCA is available here: https://github.com/aritra90/COGG.
References
- Abbi A. 2009. Is great Andamanese genealogically and typologically distinct from Onge and Jarawa? Lang Sci. 31(6):791–812. [Google Scholar]
- Alaoui AE, Mahoney MW.. 2015. Fast randomized kernel ridge regression with statistical guarantees. Proceedings of the 28th International Conference on Neural Information Processing Systems. Vol. 1. Cambridge (MA: ): MIT Press, NIPS’15. p. 775–783. [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ArunKumar G, Soria-Hernanz DF, Kavitha VJ, Arun VS, Syama A, Ashokan KS, Gandhirajan KT, Vijayakumar K, Narayanan M, Jayalakshmi M, et al. 2012. Population differentiation of southern Indian male lineages correlates with agricultural expansions predating the caste system. PLoS One 7(11):e50269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR.. 2015. A global reference for human genetic variation. Nature 526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M, Kivisild T, Watkins WS, Dixon ME, Ricker CE, Rao BB, Naidu JM, Prasad BVR, Reddy PG, Rasanayagam A, et al. 2001. Genetic evidence on the origins of Indian caste populations. Genome Res. 11(6):994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Mukherjee N, Roy S, Sengupta S, Banerjee S, Chakraborty M, Dey B, Roy M, Roy B, Bhattacharyya NP, et al. 2003. Ethnic India: a genomic view, with special reference to peopling and structure. Genome Res. 13(10):2277–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Sarkar-Roy N, Majumder PP.. 2016. Genomic reconstruction of the history of extant populations of India reveals five distinct ancestral components and a complex structure. Proc Natl Acad Sci U S A. 113(6):1594–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, Yunusbayev B, Metspalu M, Metspalu E, Rosset S, Parik J, Rootsi S, Chaubey G, Kutuev I, Yudkovsky G, et al. 2010. The genome-wide structure of the Jewish people. Nature 466(7303):238–242. [DOI] [PubMed] [Google Scholar]
- Blevins J. 2007. A long lost sister of proto-Austronesian?: proto-ongan, mother of Jarawa and Onge of the Andaman islands. Ocean Linguist. 46(1):154–198. [Google Scholar]
- Bose A, Kalantzis V, Kontopoulou EM, Elkady M, Paschou P, Drineas P.. 2019. Terapca: a fast and scalable software package to study genetic variation in tera-scale genotypes. Bioinformatics 35(19):3679–3683. [DOI] [PubMed] [Google Scholar]
- Bradburd GS, Ralph PL, Coop GM.. 2013. Disentangling the effects of geographic and ecological isolation on genetic differentiation. Evolution 67(11):3258–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmachari SK, Singh L, Sharma A, Mukerji M, Ray K, Roychoudhury S, Chandak GR, Thangaraj K, Habib S, Parmar D, et al. 2005. The Indian Genome Variation database (IGVdb): a project overview. Hum Genet. 118:1–11. [DOI] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, et al. 2002. A human genome diversity cell line panel. Science 296(5566):261–262. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Piazza A, Menozzi P, Mountain J.. 1988. Reconstruction of human evolution: bringing together genetic, archaeological, and linguistic data. Proc Natl Acad Sci U S A. 85(16):6002–6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ.. 2015. Second-generation plink: rising to the challenge of larger and richer datasets. GigaScience 4(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubey G. 2014. Language isolates and their genetic identity: a commentary on mitochondrial DNA history of Sri Lankan ethnic people: their relations within the island and with the Indian subcontinental populations. J Hum Genet. 59(2):61–63. [DOI] [PubMed] [Google Scholar]
- Chaubey G, Metspalu M, Choi Y, Mägi R, Romero IG, Soares P, Van Oven M, Behar DM, Rootsi S, Hudjashov G, et al. 2011. Population genetic structure in Indian austroasiatic speakers: the role of landscape barriers and sex-specific admixture. Mol Biol Evol. 28(2):1013–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubey G, Singh M, Crivellaro F, Tamang R, Nandan A, Singh K, Sharma VK, Pathak AK, Shah AM, Sharma V, et al. 2014. Unravelling the distinct strains of Tharu ancestry. Eur J Hum Genet. 22(12):1404–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubey G, Tamang R, Pennarun E, Dubey P, Rai N, Upadhyay RK, Meena RP, Patel JR, van Driem G, Thangaraj K, et al. 2017. Reconstructing the population history of the largest tribe of India: the Dravidian speaking Gond. Eur J Hum Genet. 25(4):493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zheng H, Bei J-X, Sun L, Jia W-h, Li T, Zhang F, Seielstad M, Zeng Y-X, Zhang X, et al. 2009. Genetic structure of the Han Chinese population revealed by genome-wide SNP variation. Am J Hum Genet. 85(6):775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark PU, Dyke AS, Shakun JD, Carlson AE, Clark J, Wohlfarth B, Mitrovica JX, Hostetler SW, McCabe AM.. 2009. The last glacial maximum. Science 325(5941):710–714. [DOI] [PubMed] [Google Scholar]
- Desai S, Dubey A.. 2012. Caste in 21st century India: competing narratives. Econ Polit Wkly. 46(11):40–49. [PMC free article] [PubMed] [Google Scholar]
- Di Cristofaro J, Pennarun E, Mazières S, Myres NM, Lin AA, Temori SA, Metspalu M, Metspalu E, Witzel M, King RJ, et al. 2013. Afghan Hindu Kush: where Eurasian sub-continent gene flows converge. PLoS One 8(10):e76748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drineas P, Lewis J, Paschou P.. 2010. Inferring geographic coordinates of origin for Europeans using small panels of ancestry informative markers. PLoS One 5(8):e11892–e11896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova SA, Reidla M, Metspalu E, Metspalu M, Rootsi S, Tambets K, Trofimova N, Zhadanov SI, Kashani BH, Olivieri A, et al. 2013. Autosomal and uniparental portraits of the native populations of Sakha (Yakutia): implications for the peopling of Northeast Eurasia. BMC Evol Biol. 13:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot G, Rousset F.. 2013. Dismantling the mantel tests. Methods Ecol Evol. 4(4):336–344. [Google Scholar]
- Hinrichs AS. 2006. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 34(90001):D590–D598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosambi D. 1964. The culture and civilisation of ancient India in historical outline. New Delhi: Vikas Publishing House Pvt. Ltd. [Google Scholar]
- Kovacevic L, Tambets K, Ilumäe AM, Kushniarevich A, Yunusbayev B, Solnik A, Bego T, Primorac D, Skaro V, Leskovac A, et al. 2014. Standing at the gateway to Europe – the genetic structure of Western Balkan populations based on autosomal and haploid markers. PLoS One 9(8):e105090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao O, van Duijn K, Kersbergen P, de Knijff P, Kayser M.. 2006. Proportioning whole-genome single-nucleotide–polymorphism diversity for the identification of geographic population structure and genetic ancestry. Am J Hum Genet. 78(4):680–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder PP. 2001. Indian caste origins: genomic insights and future outlook. Genome Res. 11(6):931–932. [DOI] [PubMed] [Google Scholar]
- Majumder PP. 2010. The human genetic history of South Asia. Curr Biol. 20(4):R184–R187. [DOI] [PubMed] [Google Scholar]
- Mallory J, Adams D.. 1997. Encyclopedia of Indo-European culture. London and Chicago: Fitzroy Dearborn. [Google Scholar]
- Metspalu M, Romero IG, Yunusbayev B, Chaubey G, Mallick CB, Hudjashov G, Nelis M, Mägi R, Metspalu E, Remm M, et al. 2011. Shared and unique components of human population structure and genome-wide signals of positive selection in South Asia. Am J Hum Genet. 89(6):731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal M, Casals F, Xu T, Dall’Olio GM, Pybus M, Netea MG, Comas D, Laayouni H, Li Q, Majumder PP, et al. 2016. Genomic analysis of Andamanese provides insights into ancient human migration into Asia and adaptation. Nat Genet. 48(9):1066–1070. [DOI] [PubMed] [Google Scholar]
- Moorjani P, Thangaraj K, Patterson N, Lipson M, Loh PR, Govindaraj P, Berger B, Reich D, Singh L.. 2013. Genetic evidence for recent population mixture in India. Am J Hum Genet. 93(3):422–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan VM, Patterson N, Moorjani P, Rohland N, Bernardos R, Mallick S, Lazaridis I, Nakatsuka N, Olalde I, Lipson M, et al. 2019. The formation of human populations in South and Central Asia. Science 365(6457):eaat7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan BK. 1995. Sparse approximate solutions to linear systems. SIAM J Comput. 24(2):227–234. [Google Scholar]
- Novembre J, Stephens M.. 2008. Interpreting principal component analyses of spatial population genetic variation. Nat Genet. 40(5):646–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olcott M. 1944. The caste system of India. Am Sociol Rev. 9(6):648–657. [Google Scholar]
- Paschou P, Drineas P, Yannaki E, Razou A, Kanaki K, Tsetsos F, Padmanabhuni SS, Michalodimitrakis M, Renda MC, Pavlovic S, et al. 2014. Maritime route of colonization of Europe. Proc Natl Acad Sci U S A. 111(25):9211–9216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschou P, Lewis J, Javed A, Drineas P.. 2010. Ancestry informative markers for fine-scale individual assignment to worldwide populations. J Med Genet. 47(12):835–847. [DOI] [PubMed] [Google Scholar]
- Pathak AK, Kadian A, Kushniarevich A, Montinaro F, Mondal M, Ongaro L, Singh M, Kumar P, Rai N, Parik J, et al. 2018. The genetic ancestry of modern Indus valley populations from northwest India. Am J Hum Genet. 103(6):918–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D.. 2012. Ancient admixture in human history. Genetics 192(3):1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BM. 2016. Admixture, population structure, and F-statistics. Genetics 202(4):1485–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D.. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 38(8):904–909. [DOI] [PubMed] [Google Scholar]
- Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, Moltke I, Rasmussen S, Stafford TW Jr, Orlando L, Metspalu E, et al. 2014. Upper Palaeolithic Siberian genome reveals dual ancestry of native Americans. Nature 505(7481):87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeevan H, Osier MV, Cheung KH, Deng H, Druskin L, Heinzen R, Kidd JR, Stein S, Pakstis AJ, Tosches NP, et al. 2003. ALFRED: the ALelle FREquency Database: update 2006. Nucleic Acids Res. 31(1):270–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L.. 2009. Reconstructing Indian population history. Nature 461(7263):489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Mahajan S, Gonzalez-Quevedo C, Blum MGB, Nino-Rosales L, Ninis V, Das P, Hegde M, Molinari L, Zapata G, et al. 2006. Low levels of genetic divergence across geographically and linguistically diverse populations from India. PLoS Genet. 2(12):2052–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhury S, Roy S, Basu A, Banerjee R, Vishwanathan H, Usha Rani MV, Sil SK, Mitra M, Majumder PP.. 2001. Genomic structures and population histories of linguistically distinct tribal groups of India. Hum Genet. 109(3):339–350. [DOI] [PubMed] [Google Scholar]
- Samuel PP, Krishnamoorthi R, Hamzakoya K, Aggarwal C.. 2009. Entomo-epidemiological investigations on chikungunya outbreak in the Lakshadweep islands, Indian Ocean. Indian J Med Res. 129(4):442–445. [PubMed] [Google Scholar]
- Schlebusch CM, Skoglund P, Sjödin P, Gattepaille LM, Hernandez D, Jay F, Li S, De Jongh M, Singleton A, Blum MGB, et al. 2012. Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science 338(6105):374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M, Oliveira M, Vieira D, Brandão A, Rito T, Pereira JB, Fraser RM, Hudson B, Gandini F, Edwards C, et al. 2017. A genetic chronology for the Indian subcontinent points to heavily sex-biased dispersals. BMC Evol Biol. 17(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal RR. 1991. Ancient movement patterns determine modern genetic variances in Europe. Hum Biol. 84:553–554. [DOI] [PubMed] [Google Scholar]
- Stamatoyannopoulos G, Bose A, Teodosiadis A, Tsetsos F, Plantinga A, Psatha N, Zogas N, Yannaki E, Zalloua P, Kidd KK, et al. 2017. Genetics of the Peloponnesean populations and the theory of extinction of the medieval Peloponnesean Greeks. Eur J Hum Genet. 25(5):637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tätte K, Pagani L, Pathak AK, Kõks S, Duy BH, Ho XD, Sultana GNN, Sharif MI, Asaduzzaman M, Behar DM, et al. 2019. The genetic legacy of continental scale admixture in Indian Austroasiatic speakers. Sci Rep. 9(1):3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangaraj K, Singh L, Reddy AG, Rao VR, Sehgal SC, Underhill PA, Pierson M, Frame IG, Hagelberg E.. 2003. Genetic affinities of the Andaman islanders, a vanishing human population. Curr Biol. 13(2):86–93. [DOI] [PubMed] [Google Scholar]
- Thapar R. 1990. A history of India. United Kingdom: Penguin Books. [Google Scholar]
- Thapar R. 2014. Can genetics help us understand Indian social history? Cold Spring Harb Perspect Biol. 6(11):a008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidyarthi LP, Rai BK.. 1977. The tribal culture of India. New Delhi: Concept Publishing Company. [Google Scholar]
- Voris HK. 2000. Maps of pleistocene sea levels in southeast Asia: shorelines, river systems and time durations. J Biogeogr. 27(5):1153–1167. [Google Scholar]
- Wang HW, Mitra B, Chaudhuri TK, Gounder Palanichamy M, Kong QP, Zhang YP.. 2011. Mitochondrial DNA evidence supports northeast Indian origin of the aboriginal Andamanese in the late Paleolithic. J Genet Genomics. 38(3):117–122. [DOI] [PubMed] [Google Scholar]
- Witzel M. 2001. Substrate languages in Old-Indo Aryan. Int J Dravidian Linguist. 30:1–94. [Google Scholar]
- Wooding S, Ostler C, Prasad BVR, Watkins WS, Sung S, Bamshad M, Jorde LB.. 2004. Directional migration in the Hindu castes: inferences from mitochondrial, autosomal and Y-chromosomal data. Hum Genet. 115(3):221–229. [DOI] [PubMed] [Google Scholar]
- Yunusbayev B, Metspalu M, Järve M, Kutuev I, Rootsi S, Metspalu E, Behar DM, Varendi K, Sahakyan H, Khusainova R, et al. 2012. The Caucasus as an asymmetric semipermeable barrier to ancient human migrations. Mol Biol Evol. 29(1):359–365. [DOI] [PubMed] [Google Scholar]
- Yunusbayev B, Metspalu M, Metspalu E, Valeev A, Litvinov S, Valiev R, Akhmetova V, Balanovska E, Balanovsky O, Turdikulova S, et al. 2015. The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia. PLoS Genet. 11(4):e1005068–e1005124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used in this manuscript are available from the respective corresponding authors. Code for COGG and COGG-CCA is available here: https://github.com/aritra90/COGG.