Abstract
Objective:
We aimed to investigate the mechanism of the regulatory axis of miR-196b/AQP4 underlying the invasion and migration of lung adenocarcinoma (LUAD) cells.
Methods:
LUAD miRNA and mRNA expression profiles were downloaded from TCGA database and then differential analysis was used to identify the target miRNA. Target gene for the miRNA was obtained via prediction using 3 bioinformatics databases and intersection with the differentially expressed mRNAs searched from TCGA-LUAD. Then, qRT-PCR and western blot were used to validate the expression of miR-196b and AQP4. Dual-luciferase reporter assay was performed to confirm the targeting relationship between miR-196b and AQP4. Transwell assay was used to investigate the migration and invasion of LUAD cells.
Results:
MiR-196b was screened out by differential and survival analyses, and the downstream target gene AQP4 was identified. In LUAD, miR-196b was highly expressed while AQP4 was poorly expressed. Besides, overexpression of miR-196b promoted cell invasion and migration, while overexpression of AQP4 had negative effects. Moreover, the results of the dual-luciferase reporter assay suggested that AQP4 was a direct target of miR-196b. In addition, we also found that overexpressing AQP4 could suppress the promotive effect of miR-196b on cancer cell invasion and migration.
Conclusion:
MiR-196b promotes the invasion and migration of LUAD cells by down-regulating AQP4, which helps us find new molecular targeted therapies for LUAD.
Keywords: lung adenocarcinoma, miR-196b, AQP4, migration, invasion
Introduction
Lung cancer is regarded as the most common cause of cancer death worldwide, with a high rate of morbidity and mortality.1,2 Statistically, around 1,800,000 people are diagnosed with lung cancer each year, and their 5-year survival rate is as low as 4%-17%.3 Lung adenocarcinoma (LUAD), a typical histological type of non-small cell lung cancer (NSCLC), is common in females and non-smokers, accounting for 40% of all lung cancer cases.4-6 Despite the great advance in surgery, chemotherapy, radiotherapy and targeted therapy, LUAD is still apt to relapse and the 5-year overall survival rate is lower than 15%.7,8 Therefore, discovery of novel potential molecular mechanisms is of great significance in the effective diagnosis of LUAD and helps search for therapeutic targets.
MicroRNAs (miRNAs) are non-coding RNAs involved in the post-transcriptional regulation of gene expression by interacting with the 3′-untranslational regions (3′-UTRs) of target mRNAs.9,10 For example, suppression of miR-3941 expression in LUAD could reduce the apoptosis of cancer cells by targeting IGBP1.11 In addition, miR-374a and miR-320a could act as tumor suppressors through differentially down-regulated in LUAD tissue,12,13 while miR-222 and miR-214 had the negative effects.14,15 Studies have shown that miR-196b is up-regulated in NSCLC tissue and able to promote cell invasion.16 Moreover, miR-196b can act as a predictor in platinum-based chemotherapy in advanced NSCLC.17 Although miR-196b has been confirmed as an independent predictor for survival of LUAD patients,18 the specific mechanism still needs to be further studied.
Aquaporin-4 (AQP4), a membrane-bound protein that regulates water permeability, is expressed in skeletal muscle and epithelial cells in brain, lung, kidney, and gastrointestinal organs.19,20 AQP4 has been reported to be involved in cell migration by acting as an adhesion molecular.21,22 Warth, et al. found that the high expression of AQP4 in LUAD was correlated with great prognosis.23 However, the relationship between AQP4 and miR-196b has not been studied.
In the present study, we investigated the regulatory relationship between AQP4 and miR-196b in LUAD and discussed the corresponding mechanism underlying LUAD cell migration and invasion, thereby exploring novel approaches for LUAD diagnosis and therapy.
Materials and Methods
Bioinformatics Analysis
Expression data of miRNA (46 normal samples and 521 tumor samples) and mRNA (59 normal samples and 535 tumor samples) of LUAD were downloaded from TCGA database (https://cancergenome.nih.gov/). Differential analysis (|logFC|>2, padj < 0.01) was performed using “edgeR” package to screen out differentially expressed miRNAs (DEmiRNAs) and differentially expressed mRNAs (DEmRNAs). The FC (fold-change) and FDR (false discovery rate) values of all miRNAs and mRNAs are detailed in Attachment 1 and 2. Multiplicity correction was performed by applying the Benjamini-Hochberg method on the p-values to control the FDR. Then, miRDB (www.mirdb.org/), mirDIP (ophid.utoronto.ca/) and TargetScan (www.targetscan.org/) databases were used to predict downstream target genes for the target miRNA. An intersection was subsequently taken between the predicted genes and the DEmRNAs in TCGA-LUAD. For survival analysis, we set the median gene expression value of all samples as the critical value to divide tumor samples into high and low expression groups, and conducted coxph regression and log-rank test. Clinical characteristics of all samples obtained from TCGA-LUAD are presented in Attachment 3.
Cell Culture
Human normal lung fibroblasts IMR-90 (CCL-186) was cultured in Eagle’s Minimum Essential Medium (EMEM) containing 10% fetal bovine serum (FBS). Human LUAD cell lines NCI-H1568 (CRL-5876), NCI-H1563 (CRL-5875) and NCI-H2030 (CRL-5914) were cultured in RPMI-1640 medium with 10% FBS, appropriate 100 U/mL streptomycin and 100 U/mL penicillin. All the cells were incubated in an incubator with 5% CO2 at 37 °C. All the cells and mediums were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA).
Cell Transfection
LUAD cells in logarithmic phase were divided into 7 groups: NC mimic, miR-196b mimic, oe-NC, oe-AQP4, NC mimic+oe-NC, miR-196b mimic+oe-NC and miR-196b mimic+oe-AQP4. NC mimic, miR-196b mimic, oe-NC and oe-AQP4 (Ribo Bio, Guangzhou, China) were all transiently transfected into cells using the Lipofectamine® 2000 (Thermo Fisher Scientific, Inc., Waltham, USA), and the cells were cultured in corresponding mediums with 5% CO2 at 37 °C for 6 h. Then, the cells were continuously cultured for another 48 h with fresh mediums for subsequent experiments.
qRT-PCR
Total RNA was extracted from cells using TRIzol® regent (Invitrogen, Thermo Fisher Science, Inc.) and was reversely transcribed into complementary DNA (cDNA). qRT-PCR was performed on the ABI 7500 HT Fast Real-Time PCR System (Applied Biosystems, CA, USA) using the SYBR Green PCR Master Mix (Thermo Fisher Scientific). U6 and GAPDH were taken as internal references for detection of miR-196b and AQP4 mRNA expression levels by 2-ΔΔCT method. All the primers used were purchased from GeneCopoeia, and the sequences are as follows: miR-196b-F: 5′-TAGGTACCACTTTATCCCGTTCACCA-3′, miR-196b-R: 5′-ATCTCGAGGCAGGGAGAGAGGAATAA-3′; AQP4-F: 5′-GGTAAGTGTGGGACCTTTGTGT-3′, AQP4-R: 5′-CAAAGCAAAGGGAGAGAAC-3′; U6-F: 5′-GCCCATCTTGACCCGAAT-3′, U6-R: 5′ -AACGCTTCACGAATTTGCGT-3′; GAPDH-F: 5′-ACAGTCAGCCGCATCTTCTT-3′, GAPDH-R: 5′-ACGACCAAATCCGTTGACTC-3′. The experiment was repeated 3 times independently.
Western Blot
After 48 h of transfection, cells were rinsed with cold PBS 3 times. Whole cell lysate was used to lyse cells on ice for 10 min, and proteins contained were quantitated by the BCA protein assay kit (Thermo Fisher Scientific). Then, the protein samples were boiled at 95 °C for 10 min with 10 µL of loading buffer, and separated by SDS-PAGE at a voltage of 100 V. After electrophoresis, proteins were transferred onto nitrocellulose membrane, which was then blocked by 5% BSA/TBST for 1 h. Primary antibodies, including rabbit polyclonal antibody AQP4 (ab46182, 1 μg/mL, Abcam, China) and rabbit polyclonal antibody GAPDH (ab37168, 1 μg/mL, Abcam, China), were added onto the membrane for incubation overnight at 4 °C. On the following day, the membrane was firstly washed with 1 × TBST (pH7.4) 3 times, and then incubated with secondary antibody goat anti-rabbit IgG H&L (HRP) (ab205718, 1:10000, Abcam) at room temperature for 1 h. TBST was used to wash the membrane 3 times. Protein bands were visualized by ECL kit (ECL808-25, Biomiga, USA), and analyzed using the Image Pro Plus 6.0 software (Media Cybernetics, USA). The experiment was repeated 3 times.
Invasion Assay
A 24-well Transwell chamber (BD Biosciences, MD, USA) with 8 µm in aperture was used in the invasion assay and the migration assay followed. Around 2 × 104 cells were plated into the upper chamber that was pre-coated with Matrigel matrix (BD Bioscience). RPMI-1640 culture solution containing 10% FBS was added into the lower chamber. After 24 h of incubation at 37 °C, cells still in the upper chamber were wiped off by a cotton swab, and those invading into the lower chamber were fixed with methanol and stained by 0.1% crystal violet. Five visual fields were randomly selected by an inverted microscope to take pictures and count the cells. The experiment was repeated 3 times.
Migration Assay
LUAD cells in the logarithmic phase were starved for 24h. Next day, the cells were digested, centrifuged and resuspended by FBS-free RPMI-1640 medium to the cell concentration of 2 × 105 cells/mL. Totally 0.2 mL of cell suspension was seeded into the upper chamber, and 700 µL pre-cooled RPMI-1640 culture solution supplemented with 10% FBS was added into the lower chamber. After incubation in an incubator containing 5% CO2 at 37 °C for 24 h, cells in the upper chamber were wiped off by a cotton swab, and the migratory cells were fixed by methanol for 30 min and stained with crystal violet for 20 min. Then, the cells were rinsed and dried. Images of cells were captured under an inverted microscope, and 5 views were randomly selected for cell calculation.
Dual-Luciferase Reporter Assay
NCI-H2030 cells were seeded into 24-well plates at the density of 6 × 105 cells/well and incubated for 24 h. The mutant (MUT) and wild type (WT) 3′UTR of AQP4 mRNA were ligated into pmiRGLO (Promega Corp., WI, USA) to construct luciferase reporter vectors AQP4-WT and AQP4-MUT. 48 h later, miR-196b mimic or NC mimic and the luciferase reporter vectors were co-transfected into NCI-H2030 cells, respectively. Renilla luciferase expression vector pRL-TK (TaKaRa, Dalian, China) was taken as an internal reference. The transfected cells were cultured in RPMI-1640 medium with 10% FBS for 48 h, and the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) was applied to determine luciferase activities. The experiment was repeated 3 times.
Statistical Analysis
All data were analyzed by SPSS v.24.0 software (IBM Corp., NY, USA), and figures were plotted by the GraphPad Prism 6 software (GraphPad Software, Inc., CA, USA). All data were presented in the form of mean ± standard deviation (M ± SD). Comparisons between 2 groups were performed by t test. P < 0.05 was considered statistically significant.
Results
Expression Patterns of miR-196b and AQP4 in LUAD and Their Effect on Prognosis of LUAD Patients
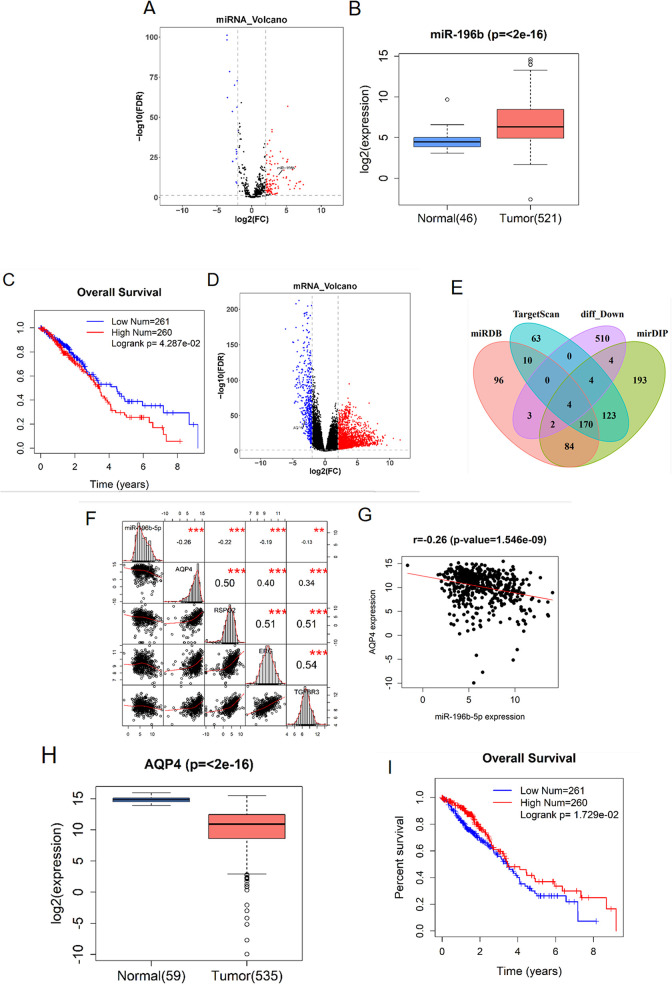
With the normal samples as control, 120 DEmiRNAs were obtained by “edgeR” differential analysis (Figure 1A). Among them, miR-196b has been reported in many literature as a biomarker of lung cancer.18,24,25 TCGA-LUAD data revealed that miR-196b expression in LUAD tissue was significantly higher than that in normal tissue (Figure 1B). Based on the survival analysis, it was found that high expression of miR-196b in LUAD tissue greatly affected prognosis and decreased survival time of patients (Figure 1C). To further study the downstream regulatory mechanism of miR-196b in LUAD, we firstly obtained 2,495 DEmRNAs via differential analysis from TCGA (Figure 1D) and then predicted the target genes of miR-196b by using miRDB, mirDIP and TargetScan databases. An intersection was made between the predicted genes and the down-regulated DEmRNAs, and 4 differential mRNAs with binding sites of miR-196b were obtained (Figure 1E). Pearson correlation analysis showed that there was a significant negative correlation between miR-196b and AQP4 (Figure 1F-G). Additionally, expression analysis based on TCGA data displaced that AQP4 expression in tumor tissue was remarkably lower by comparison with that in normal tissue (Figure 1H), and survival analysis showed that AQP4 had a significant effect on prognosis (Figure 1I). AQP4 has been proven to be a prognostic factor for a variety of human cancers in many studies.23,26,27 Combined with the above results of bioinformatics analysis and previous studies, we speculated that miR-196b may regulate the malignant progression of LUAD by targeting AQP4.
Figure 1.
Differential and survival analyses of miR-196b and AQP4 in LUAD tissue and cells. (A) Volcano plot of DEmiRNAs in TCGA-LUAD (the red dots represent differentially up-regulated genes and the blue dots refer to differentially down-regulated genes); (B) relative expression of miR-196b in tumor tissue (n = 521) and normal tissue (n = 46) in the TCGA-LUAD dataset; (C) survival significance of miR-196b; (D) volcano Plot of DEmRNAs in TCGA-LUAD (the red dots represent differentially up-regulated genes and the blue dots refer to differentially down-regulated genes); (E): Venn diagram of differentially down-regulated mRNAs in TCGA and predicted target genes on miRDB, mirDIP and TargetScan databases; (F) correlation analysis of miR-196b and 4 candidate target genes; (G) negative correlation between miR-196b and AQP4; (H) relative expression of AQP4 in tumor tissue (n = 535) and normal tissue (n = 59) in the TCGA-LUAD dataset; (I) survival significance of AQP4.
MiR-196b Is Up-Regulated in LUAD Cells and Promotes the Invasion and Migration of LUAD Cells
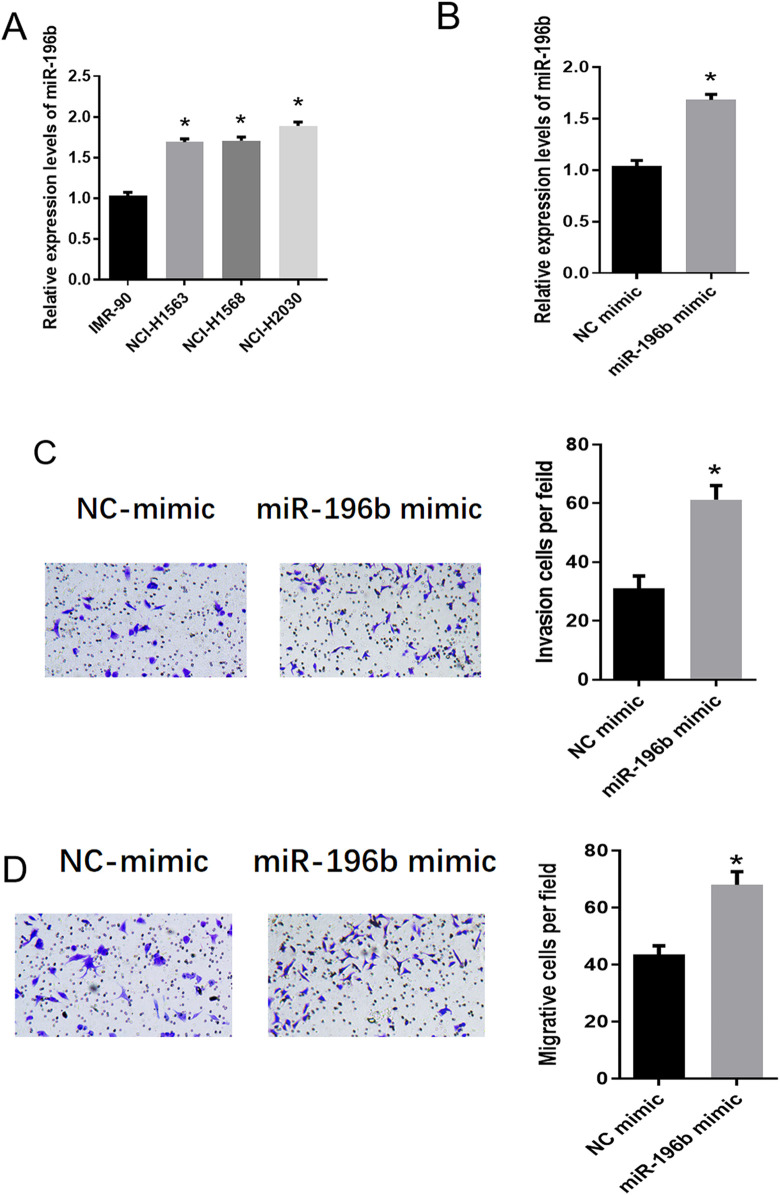
In order to verify the above hypothesis, we firstly detected the expression of miR-196b in human normal lung fibroblasts IMR-90 and LUAD cell lines (NCI-H1568, NCI-H1563 and NCI-H2030). qRT-PCR results showed that compared with IMR-90, LUAD cells had significantly up-regulated miR-196b, and the difference was the most significant in NCI-H2030 (Figure 2A). Therefore, NCI-H2030 cell line was selected for follow-up experiments. Then, we detected the transfection efficiency of miR-196b mimic by qRT-PCR. As shown in Figure 2B, the expression of miR-196b was significantly up-regulated after miR-196b mimic was transfected into cells, indicating a better transfection efficiency. Transwell experiments were performed to investigate the role of miR-196b in LUAD, suggesting that the numbers of invasive and migratory cells increased significantly after overexpressing miR-196b (Figure 2C-D). These results showed that the expression of miR-196b was significantly up-regulated in LUAD cells, which could promote cell invasion and migration.
Figure 2.
MiR-196b is up-regulated in LUAD cells and promotes the invasion and migration of cancer cells. (A) The expression of miR-196b in human normal lung fibroblasts IMR-90 and LUAD cells NCI-H1568, NCI-H1563 and NCI-H2030; (B) The expression of miR-196b in LUAD cell line NCI-H2030 after transfection with miR-196b mimic and NC mimic, respectively; (C) the effect of miR-196b overexpression on the invasion of NCI-H2030 cells; (D) the effect of miR-196b overexpression on the migration of NCI-H2030 cells. Note: “*” in Figure A refers to comparison with IMR-90, p < 0.05; “*” in Figure B, C, and D refer to comparison with NC mimic, p < 0.05.
AQP4 Is Down-Regulated in LUAD Cells and Inhibits the Invasion and Migration of LUAD Cells
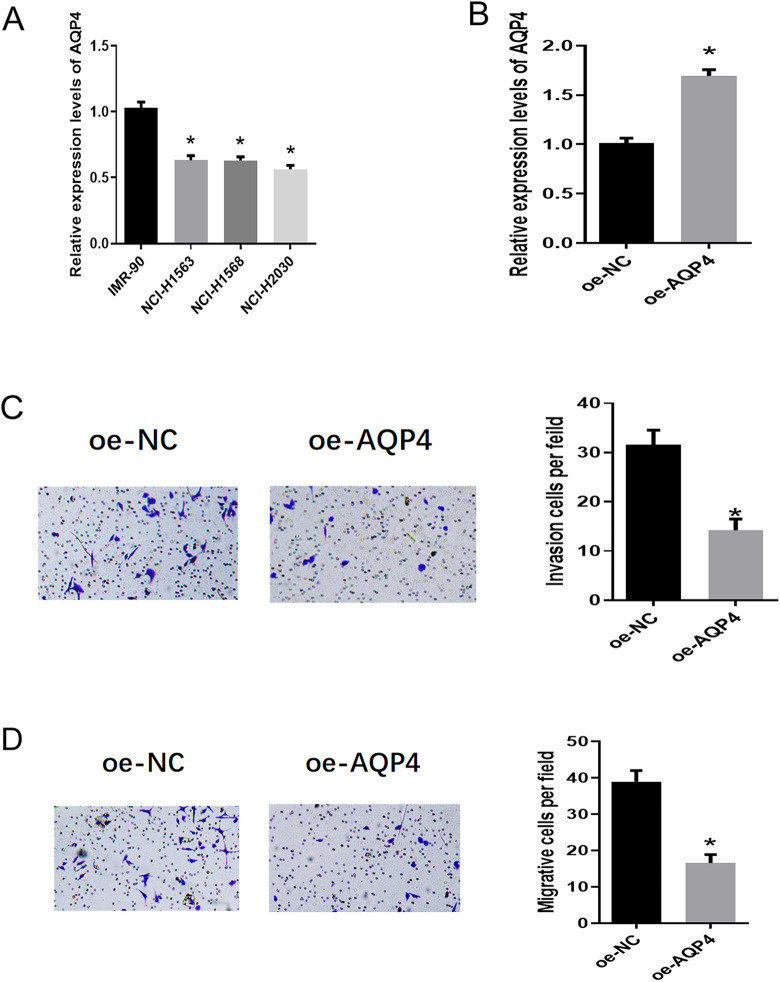
In order to explore the expression of AQP4 in LUAD cells and its effect on the biological function of LUAD cells, we used qRT-PCR to detect the expression of AQP4 in human normal lung fibroblasts IMR-90 and LUAD cell lines. We found that the mRNA expression of AQP4 was significantly down-regulated in LUAD cells, especially in NCI-H2030 cells (Figure 3A). NCI-H2030 cells were transfected with oe-NC or oe-AQP4, respectively, and qRT-PCR was performed to measure the transfection efficiency. As shown in Figure 3B, the expression of AQP4 was significantly up-regulated in the oe-AQP4 group. Moreover, the overexpression of AQP4 was found to greatly decrease the invasive and migratory abilities of NCI-H2030 cells as evidenced by Transwell assay (Figure 3C-D). These results suggested that the expression of AQP4 was down-regulated in LUAD cells, and overexpression of AQP4 could inhibit the invasion and migration of LUAD cells.
Figure 3.
AQP4 is down-regulated in LUAD cells and inhibits the invasion and migration of cancer cells. (A) The expression of AQP4 in human normal lung fibroblasts IMR-90 and LUAD cells NCI-H1568, NCI-H1563 and NCI-H2030; (B) the expression of AQP4 in LUAD cell line NCI-H2030 after transfection with oe-NC and oe-AQP4, respectively; (C) the effect of AQP4 overexpression on the invasion of NCI-H2030 cells; (D) the effect of AQP4 overexpression on the migration of NCI-H2030 cells. note: “*” in Figure A refers to comparison with IMR-90, p < 0.05; “*” in Figure B, C and D refer to comparison with NC mimic, p < 0.05.
Target Inhibition of AQP4 Expression by miR-196b
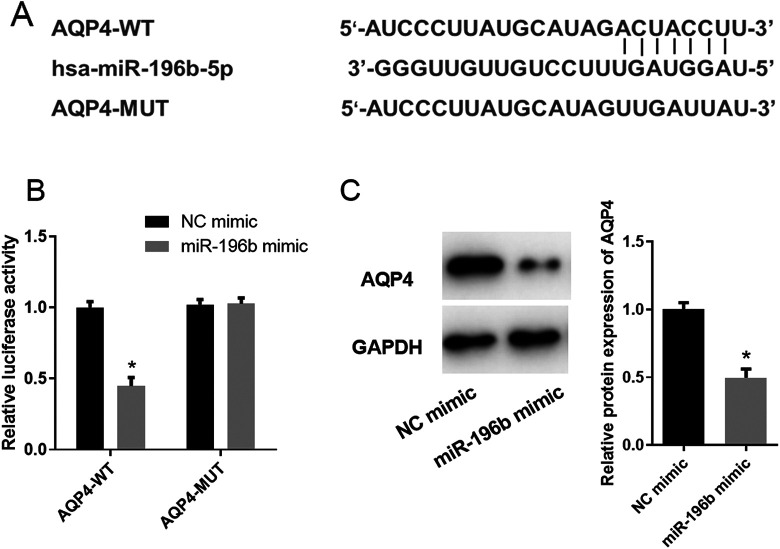
Previously, we used bioinformatics analysis to predict the target genes of miR-196b and found that AQP4 may be the direct target of miR-196b in LUAD (Figure 1F-G). The targeted binding sites of miR-196b on AQP4 mRNA 3′UTR were predicted by TargetScan website (Figure 4A) and verified by dual-luciferase assay. The result indicated that the luciferase activity was greatly decreased in the AQP4-WT group when miR-196b was overexpressed, while that in the AQP4-MUT group showed no significant difference (Figure 4B). This implied the targeting relationship between miR-196b and AQP4. In addition, western blot was further performed to investigate the effect of miR-196b overexpression on AQP4 protein expression, showing that AQP4 protein expression was significantly decreased upon miR-196b overexpression (Figure 4C). Collectively, miR-196b was verified to target and inhibit AQP4 expression.
Figure 4.
MiR-196b targets and inhibits the expression of AQP4. (A) Putative binding sites of miR-196b on AQP4 mRNA 3′UTR; (B) results of dual-luciferase reporter assay; (C) the effect of miR-196b overexpression on AQP4 protein expression. Note: “*” refers to comparison with NC mimic, p < 0.05.
MiR-196b Promotes the Invasion and Migration of LUAD Cells by Targeting AQP4
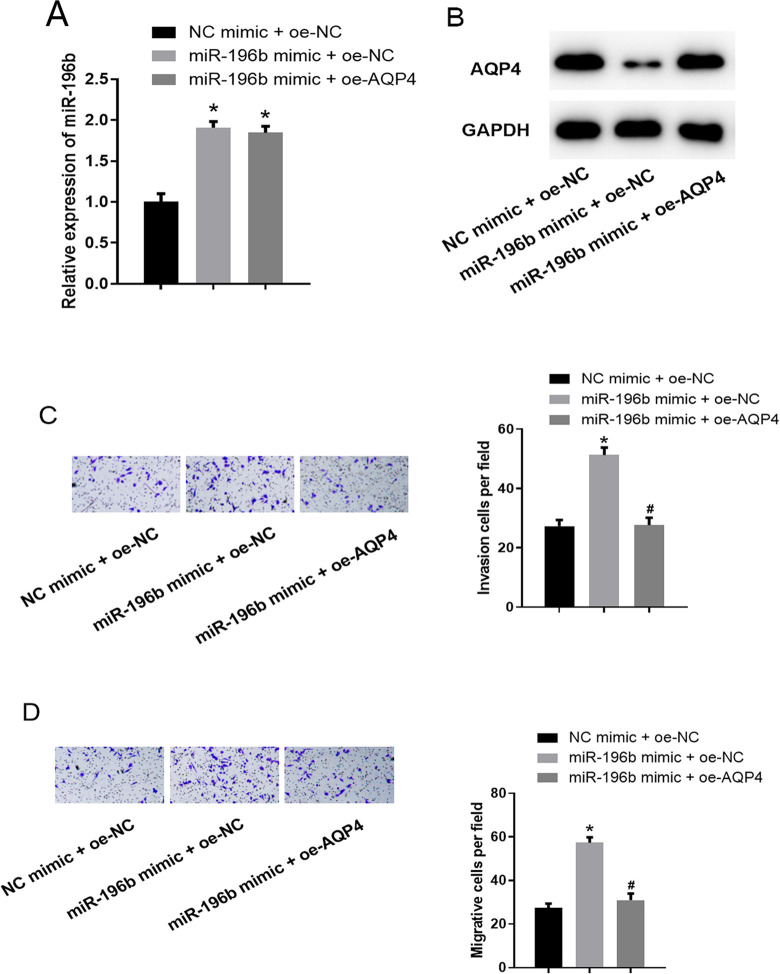
Considering the targeting relationship between miR-196b and AQP4 and their effects on the invasion and migration of LUAD cells, we designed a rescue experiment to explore the mechanism of miR-196b/AQP4 in LUAD. NC mimic + oe-NC, miR-196b mimic + oe-NC, miR-196b mimic + oe-AQP4 were transfected into NCI-H2030 cells, respectively. qRT-PCR was carried out for miR-196b expression detection and western blot was for AQP4 protein expression. As shown in Figure 5A-B, miR-196b was significantly up-regulated in the miR-196b mimic + oe-NC group by comparison with that in the NC mimic + oe-NC group, while AQP4 was down-regulated. Moreover, compared to the miR-196b mimic + oe-NC group, the expression of miR-196b in the miR-196b mimic + oe-AQP4 group showed no significant difference, while AQP4 expression was greatly up-regulated. Furthermore, results of the Transwell invasion and migration assays illustrated that cells in the miR-196b mimic + oe-NC group had much stronger invasive and migratory abilities compared to those in the NC mimic + oe-NC group. Meanwhile, with the presence of AQP4 overexpression, the promotive effect of miR-196b overexpression on the cancer cell invasion and migration was reversed (Figure 5C-D). These results confirmed that overexpression of AQP4 could reverse the inhibition of miR-196b on the invasion and migration of LUAD cells. In other words, miR-196b could inhibit the malignant progression of LUAD cells by targeting the expression of AQP4.
Figure 5.
Overexpression of AQP4 reverses the promotive effect of miR-196b on the invasion and migration of LUAD cells. (A) The expression of miR-196b in cells of each group (NC mimic + oe-NC, miR-196b mimic + oe-NC, miR-196b mimic + oe-AQP4); (B) the protein expression of AQP4 in cells of each group; (C): invasive ability of cells in each group; (D) migratory ability of cells in each group. Note: “*” refers to comparison with NC mimic + oe-NC group, p < 0.05; “#” refers to comparison with miR-196b mimic + oe-NC group, p < 0.05.
Discussion
LUAD is the most common subtype of lung cancer and one of the most aggressive and rapidly fatal tumor types, with an overall survival time less than 5 years.28,29 Although the diagnosis and therapies have been improved, patients with advanced LUAD still have poor prognosis, which is mainly attributed to the increase of distant metastasis after tumor resection.30 Therefore, it is important to find new functional genes and biomarkers responsible for tumor progression.
MiRNAs are frequently located at fragile sites and cancer-associated genomic regions, suggesting that miRNAs are involved in tumor malignant transformation.31,32 MiR-196b encoded in mammalian HOX clusters functions as a regulator of HOXB8 during vertebrate development.33,34 Recently, the abnormal expression of miR-196b in lung cancer has been reported. Ren, et al. found the positive effect of miR-196b on lung cancer development.35 Yu, et al. found that miR-196b was up-regulated in mesenchymal NSCLC cells and lung cancer tissues.16 Furthermore, studies made by Li HL revealed that overexpression of miR-196b promoted cell invasion and migration by down-regulating GATA6 in NSCLC.25 Despite the fact that miR-196b has been confirmed as an independent predictive factor for survival of LUAD patients,18 the specific role and molecular mechanism of miR-196b in LUAD have not been well studied. In the present study, we found that miR-196b was up-regulated in LUAD cells, which is consistent with previous studies. Moreover, the promotive effect of miR-196b overexpression on cell invasion and migration in LUAD was verified as well.
AQP4 has been reported to be down-regulated in breast cancer and gastric cancer,26 and its high expression shows a significant association with relapse-free survival (RFS).36 Warth et al. reported that high expression of AQP4 was beneficial for the prognosis of LUAD patients.23 Our study verified that AQP4 was lowly expressed in LUAD cells and overexpression of AQP4 could inhibit cancer cell invasion and migration, which are consistent with previous studies. In addition, we also revealed the targeting regulation relationship between miR-196b and AQP4 for the first time. MiR-196b could promote cell invasion and migration of LUAD by down-regulating AQP4, and such promotive effect could be suppressed by up-regulating AQP4.
In short, miR-196b was found to be up-regulated in LUAD cells and negatively correlated with AQP4 in the present study. MiR-196b could promote the invasion and migration of LUAD cells via targeting and down-regulating AQP4. Furthermore, miR-196b could function on the cell invasion and migration of LUAD through targeting AQP4, thereby affecting tumor progression. The study helps develop potential targets and provides novel approaches for LUAD targeted therapy.
Supplemental Material
Supplemental Material, Attachment1_miRNA_edgerOut for MiR-196b Promotes the Invasion and Migration of Lung Adenocarcinoma Cells by Targeting AQP4 by Xuhui Wu, Gongzhi Wu, Huaizhong Zhang, Xuyang Peng, Bin Huang, Mingjiang Huang, Jianyang Ding, Chaofan Mao and Congxiong Peng in Technology in Cancer Research & Treatment
Supplemental Material, Attachment2_mRNA_edgerOut for MiR-196b Promotes the Invasion and Migration of Lung Adenocarcinoma Cells by Targeting AQP4 by Xuhui Wu, Gongzhi Wu, Huaizhong Zhang, Xuyang Peng, Bin Huang, Mingjiang Huang, Jianyang Ding, Chaofan Mao and Congxiong Peng in Technology in Cancer Research & Treatment
Supplemental Material, Attachment3_clinical_characteristics for MiR-196b Promotes the Invasion and Migration of Lung Adenocarcinoma Cells by Targeting AQP4 by Xuhui Wu, Gongzhi Wu, Huaizhong Zhang, Xuyang Peng, Bin Huang, Mingjiang Huang, Jianyang Ding, Chaofan Mao and Congxiong Peng in Technology in Cancer Research & Treatment
Footnotes
Authors’ Note: Dr XH W and GZ W contributed to the study design. HZ Z conducted the literature search. XY P, B H, MJ H and JY D acquired the data. CF M and CX P wrote the article. XH W performed data analysis. XH W drafted. GZ W, HZ Z revised the article and gave the final approval of the version to be submitted. All authors read and approved the final manuscript. The data and materials in the current study are available from the corresponding author on reasonable request. Ethics approval and consent to participate is not applicable.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Congxiong Peng  https://orcid.org/0000-0002-2844-2287
https://orcid.org/0000-0002-2844-2287
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi:10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 3. Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. doi:10.1016/S0140-6736(16)30958-8 [DOI] [PubMed] [Google Scholar]
- 4. Bender E. Epidemiology: the dominant malignancy. Nature. 2014;513(7517):S2–S3. doi:10.1038/513S2a [DOI] [PubMed] [Google Scholar]
- 5. Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10(9):1243–1260. doi:10.1097/JTO.0000000000000630 [DOI] [PubMed] [Google Scholar]
- 6. Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers—a different disease. Nat Rev Cancer. 2007;7(10):778–790. doi:10.1038/nrc2190 [DOI] [PubMed] [Google Scholar]
- 7. Deng J, Deng H, Liu C, Liang Y, Wang S. Long non-coding RNA OIP5-AS1 functions as an oncogene in lung adenocarcinoma through targeting miR-448/Bcl-2. Biomed Pharmacother. 2018;98:102–110. doi:10.1016/j.biopha.2017.12.031 [DOI] [PubMed] [Google Scholar]
- 8. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 9. Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460(7255):642–646. doi:10.1038/nature08139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han H, Sun D, Li W, et al. A c-Myc-microRNA functional feedback loop affects hepatocarcinogenesis. Hepatology. 2013;57(6):2378–2389. doi:10.1002/hep.26302 [DOI] [PubMed] [Google Scholar]
- 11. Sato T, Shiba-Ishii A, Kim Y, et al. MiR-3941: a novel microRNA that controls IGBP1 expression and is associated with malignant progression of lung adenocarcinoma. Cancer Sci. 2017;108(3):536–542. doi:10.1111/cas.13148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu H, Liu Y, Shu XO, Cai Q. MiR-374a suppresses lung adenocarcinoma cell proliferation and invasion by targeting TGFA gene expression. Carcinogenesis. 2016;37(6):567–575. doi:10.1093/carcin/bgw038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lv Q, Hu JX, Li YJ, et al. MiR-320a effectively suppresses lung adenocarcinoma cell proliferation and metastasis by regulating STAT3 signals. Cancer Biol Ther. 2017;18(3):142–151. doi:10.1080/15384047.2017.1281497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun Q, Jiang CW, Tan ZH, et al. MiR-222 promotes proliferation, migration and invasion of lung adenocarcinoma cells by targeting ETS1. Eur Rev Med Pharmacol Sci. 2017;21(10):2385–2391. [PubMed] [Google Scholar]
- 15. Liu C, Luo J, Zhao YT, et al. TWIST1 upregulates miR-214 to promote epithelial-to-mesenchymal transition and metastasis in lung adenocarcinoma. Int J Mol Med. 2018;42(1):461–470. doi:10.3892/ijmm.2018.3630 [DOI] [PubMed] [Google Scholar]
- 16. Yu SL, Lee DC, Sohn HA, et al. Homeobox A9 directly targeted by miR-196b regulates aggressiveness through nuclear factor-kappa B activity in non-small cell lung cancer cells. Mol Carcinog. 2016;55(12):1915–1926. doi:10.1002/mc.22439 [DOI] [PubMed] [Google Scholar]
- 17. Szejniuk WM, Robles AI, McCulloch T, Falkmer UGI, Roe OD. Epigenetic predictive biomarkers for response or outcome to platinum-based chemotherapy in non-small cell lung cancer, current state-of-art. Pharmacogenomics J. 2019;19(1):5–14. doi:10.1038/s41397-018-0029-1 [DOI] [PubMed] [Google Scholar]
- 18. Li X, Shi Y, Yin Z, Xue X, Zhou B. An eight-miRNA signature as a potential biomarker for predicting survival in lung adenocarcinoma. J Transl Med. 2014;12:159. doi:10.1186/1479-5876-12-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ikeshima-Kataoka H. Neuroimmunological implications of AQP4 in astrocytes. Int J Mol Sci. 2016;17(8):1306. doi:10.3390/ijms17081306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Verkman AS, Phuan PW, Asavapanumas N, Tradtrantip L. Biology of AQP4 and anti-AQP4 antibody: therapeutic implications for NMO. Brain Pathol. 2013;23(6):684–695. doi:10.1111/bpa.12085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Papadopoulos MC, Verkman AS. Aquaporin water channels in the nervous system. Nat Rev Neurosci. 2013;14(4):265–277. doi:10.1038/nrn3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seifert G, Schilling K, Steinhäuser C. Astrocyte dysfunction in neurological disorders: a molecular perspective. Nat Rev Neurosci. 2006;7(3):194–206. doi:10.1038/nrn1870 [DOI] [PubMed] [Google Scholar]
- 23. Warth A, Muley T, Meister M, et al. Loss of aquaporin-4 expression and putative function in non-small cell lung cancer. BMC Cancer. 2011;11:161. doi:10.1186/1471-2407-11-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hamamoto J, Soejima K, Yoda S, et al. Identification of microRNAs differentially expressed between lung squamous cell carcinoma and lung adenocarcinoma. Mol Med Rep. 2013;8(2):456–462. doi:10.3892/mmr.2013.1517 [DOI] [PubMed] [Google Scholar]
- 25. Li H, Feng C, Shi S. MiR-196b promotes lung cancer cell migration and invasion through the targeting of GATA6. Oncol Lett. 2018;16(1):247–252. doi:10.3892/ol.2018.8671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thapa S, Chetry M, Huang K, et al. Significance of aquaporins’ expression in the prognosis of gastric cancer. Biosci Rep. 2018;38(3). doi:10.1042/BSR20171687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen Y, Gao F, Jiang R, et al. Down-regulation of AQP4 expression via p38 MAPK signaling in temozolomide-induced glioma cells growth inhibition and invasion impairment. J Cell Biochem. 2017;118(12):4905–4913. doi:10.1002/jcb.26176 [DOI] [PubMed] [Google Scholar]
- 28. Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res. 2016;5(3):288–300. doi:10.21037/tlcr.2016.06.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Denisenko TV, Budkevich IN, Zhivotovsky B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis. 2018;9(2):117. doi:10.1038/s41419-017-0063-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhan P, Zhang B, Xi GM, et al. PRC1 contributes to tumorigenesis of lung adenocarcinoma in association with the Wnt/beta-catenin signaling pathway. Mol Cancer. 2017;16(1):108. doi:10.1186/s12943-017-0682-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi:10.1073/pnas.0307323101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi:10.1038/nrc1997 [DOI] [PubMed] [Google Scholar]
- 33. Yu H, Lindsay J, Feng ZP, et al. Evolution of coding and non-coding genes in HOX clusters of a marsupial. BMC Genomics. 2012;13(1):251. doi:10.1186/1471-2164-13-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–596. doi:10.1126/science.1097434 [DOI] [PubMed] [Google Scholar]
- 35. Ren J, Fu J, Ma T, et al. LncRNA H19-elevated LIN28B promotes lung cancer progression through sequestering miR-196b. Cell Cycle. 2018;17(11):1372–1380. doi:10.1080/15384101.2018.1482137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu L, Ma N, Wang B, et al. Significant prognostic values of aquaporin mRNA expression in breast cancer. Cancer Manag Res. 2019;11:1503–1515. doi:10.2147/CMAR.S193396 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, Attachment1_miRNA_edgerOut for MiR-196b Promotes the Invasion and Migration of Lung Adenocarcinoma Cells by Targeting AQP4 by Xuhui Wu, Gongzhi Wu, Huaizhong Zhang, Xuyang Peng, Bin Huang, Mingjiang Huang, Jianyang Ding, Chaofan Mao and Congxiong Peng in Technology in Cancer Research & Treatment
Supplemental Material, Attachment2_mRNA_edgerOut for MiR-196b Promotes the Invasion and Migration of Lung Adenocarcinoma Cells by Targeting AQP4 by Xuhui Wu, Gongzhi Wu, Huaizhong Zhang, Xuyang Peng, Bin Huang, Mingjiang Huang, Jianyang Ding, Chaofan Mao and Congxiong Peng in Technology in Cancer Research & Treatment
Supplemental Material, Attachment3_clinical_characteristics for MiR-196b Promotes the Invasion and Migration of Lung Adenocarcinoma Cells by Targeting AQP4 by Xuhui Wu, Gongzhi Wu, Huaizhong Zhang, Xuyang Peng, Bin Huang, Mingjiang Huang, Jianyang Ding, Chaofan Mao and Congxiong Peng in Technology in Cancer Research & Treatment