Gram-negative bacteria partly rely on efflux pumps to facilitate growth under stressful conditions and to increase resistance to a wide variety of commonly used drugs. In recent years, Escherichia coli sequence type 131 (ST131) has emerged as a major cause of extraintestinal infection frequently exhibiting a multidrug resistance (MDR) phenotype.
KEYWORDS: efflux, efflux pump inhibitors, AcrB, E. coli, ST131
ABSTRACT
Gram-negative bacteria partly rely on efflux pumps to facilitate growth under stressful conditions and to increase resistance to a wide variety of commonly used drugs. In recent years, Escherichia coli sequence type 131 (ST131) has emerged as a major cause of extraintestinal infection frequently exhibiting a multidrug resistance (MDR) phenotype. The contribution of efflux to MDR in emerging E. coli MDR clones, however, is not well studied. We characterized strains from an international collection of clinical MDR E. coli isolates by MIC testing with and without the addition of the AcrAB-TolC efflux inhibitor 1-(1-naphthylmethyl)-piperazine (NMP). MIC data for 6 antimicrobial agents and their reversion by NMP were analyzed by principal-component analysis (PCA). PCA revealed a group of 17 MDR E. coli isolates (n = 34) exhibiting increased susceptibility to treatment with NMP, suggesting an enhanced contribution of efflux pumps to antimicrobial resistance in these strains (termed enhanced efflux phenotype [EEP] strains). Only 1/17 EEP strains versus 12/17 non-EEP MDR strains belonged to the ST131 clonal group. Whole-genome sequencing revealed marked differences in efflux-related genes between EEP and control strains, with the majority of notable amino acid substitutions occurring in AcrR, MarR, and SoxR. Quantitative reverse transcription-PCR (qRT-PCR) of multiple efflux-related genes showed significant overexpression of the AcrAB-TolC system in EEP strains, whereas in the remaining strains, we found enhanced expression of alternative efflux proteins. We conclude that a proportion of MDR E. coli strains exhibit an EEP, which is linked to an overexpression of the AcrAB-TolC efflux pump and a distinct array of genomic variations. Members of ST131, although highly successful, are less likely to exhibit the EEP.
INTRODUCTION
Increasing antibiotic resistance, especially in Gram-negative bacteria, is a growing health concern with significant socioeconomic implications (1). Mechanisms of antibiotic resistance are manifold and comprise such diverse elements as target mutations and vector-bound transfer of resistance genes, such as beta-lactamases. Another viable strategy is the overexpression of chromosomally encoded exporters, so-called efflux pumps (2). The Escherichia coli genome codes for several different efflux pumps, the physiological functions of which remain unknown in many cases. The most important and best understood efflux pump in E. coli is AcrAB (3, 4). This pump belongs to the class of the resistance-nodulation-cell division exporters (RND), closely related proteins that are found in virtually all Gram-negative bacteria (2). AcrB is a transport protein of the inner membrane which is linked to the outer membrane channel TolC via the membrane fusion protein AcrA. The substrate range of this pump system is extensive and includes antibiotics of diverse classes, organic solvents, dyes, bile salts, and fatty acids (2, 3). Because of its critical role in the homeostasis of the microbial cell, the expression of AcrAB is closely regulated. At the operon level, expression is mediated by the local repressor AcrR, while at the genomic scale, MarAR, SoxSR, and Rob form a well-characterized global regulatory network (5–7). Furthermore, AcrS, too, has been shown to efficiently repress the expression of AcrAB rather than that of AcrEF (8).
Efflux pumps contribute to antibiotic resistance development in Gram-negative bacteria. Selecting E. coli with fluoroquinolones rapidly yields AcrAB-overexpressing strains (9, 10). AcrB knockouts, on the other hand, lose a significant amount of their acquired and innate antibiotic resistance. This has been documented both in laboratory strains and in clinical isolates (11). One strategy to overcome microbial resistance to antibiotics may be the development of clinically safe and active efflux pump inhibitors (EPIs). Although, to date, no such substance is available, numerous agents that inhibit efflux pumps in vitro have been identified. The two best-characterized substances are phenylalanine–arginine–β-naphthylamide (PAN) and 1-(1-naphtylmethyl)-piperazine (NMP) (2, 12–14).
This study aimed to assess the contribution of efflux to antibiotic resistance in a panel of multidrug-resistant (MDR) E. coli clinical strains. We hypothesized that certain strains may be more reliant on efflux than others, a trait which could be called an enhanced efflux phenotype (EEP). We developed a method to identify strains exhibiting this phenotype via the proxy of EPI susceptibility. Furthermore, we demonstrate that the phenotypical differences are associated with distinct genomic mutations and can also be linked to typical patterns of expression of regulators.
RESULTS
MIC testing and PCA.
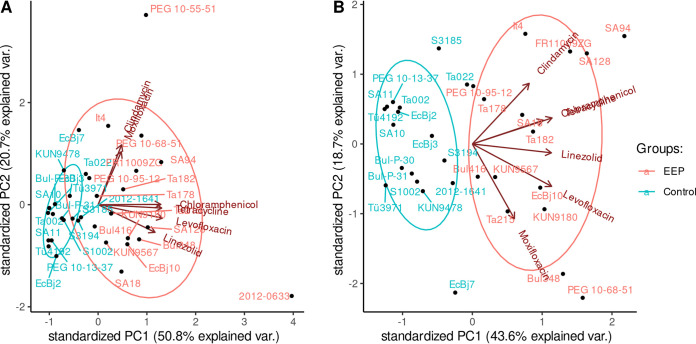
The MICs of a wide range of antibiotic substances with and without the addition of NMP (MICAB+NMPs and MICABs, respectively) were obtained for each strain (see Data Set S1 in the supplemental material). We selected six antibiotics (levofloxacin, moxifloxacin, clindamycin, chloramphenicol, linezolid, tetracycline), which are known to be efflux pump substrates, and analyzed the resulting MICAB/MICAB+NMP fractions by principal-component analysis (PCA) (Fig. 1). The first two principal components accounted for more than 70% of the variance. The already-distinct cluster in the third quadrant was further refined when we removed two outliers (strains PEG 10-55-51 and 2012-0633). All strains to the right of the origin (Fig. 1B) were considered to exhibit an enhanced efflux phenotype (EEP). A total of 17/34 strains were included in the EEP group. The remaining non-EEP MDR strains were included as a control group. In a second step, we obtained the MICs of mefloquine, carbonyl cyanide m-chlorophenylhydrazine (CCCP), and ethidium bromide (EtBr), which (i) are known efflux pump substrates, (ii) are not used for anti-infective treatment in clinical practice, and (iii) whose MICs are unlikely to be influenced by target mutations. As a sensitivity analysis, we then analyzed these MICs by PCA. Applying the aforementioned criteria, 12/17 strains were correctly identified to exhibit an enhanced efflux phenotype, and only 1/17 strains was incorrectly placed in the EEP group, yielding a sensitivity of 71% and a specificity of 94%, respectively (Fig. S1A). As negative control, a PCA of MICAB/MICAB+NMP fractions of streptomycin and gentamicin, which are drugs known not to be substrates of the AcrAB-TolC pump, was performed, yielding no discernible separation between the two groups (Fig. S1B). Additionally, we performed accumulation assays using Hoechst 33342 (Fig. 2). Reduction in dye accumulation as a proxy for efflux strength relative to that of knockout strain 3AG100ΔacrB was found to be greater in EEP strains than in control group strains, further corroborating our assumption that the increased susceptibility to EPIs in EEP strains is associated with increased efflux capacity.
FIG 1.
(A) PCA of all strains drawing on the x-fold MIC change after the addition of NMP for the listed antibiotics. PC, principal component; var., variance. (B) PCA omitting the outliers PEG 10-55-51 and 2012-0633 for enhanced clarity.
FIG 2.
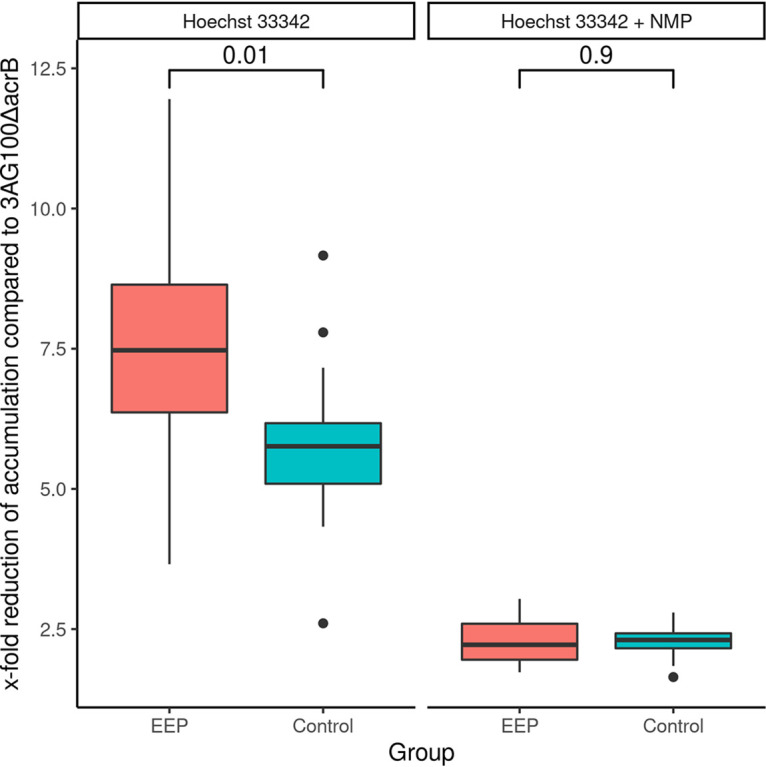
Efflux strengths estimated by x-fold reduction of Hoechst 33342 accumulation in EEP and control strains with and without NMP compared to that of the knockout strain 3AG100ΔacrS, with medians and interquartile ranges (IQR). Whiskers extend up to ±1.5 IQR, and dots represent outliers exceeding this range. No valid accumulation kinetics could be obtained for FR-11009_ZG. P values were calculated using Student's t test.
Sequence typing.
Sequence typing showed 13 sequence type 131 (ST131) strains (Table 1); 4 strains belonged to ST405, and 3 strains belonged to ST167. Interestingly, the distributions among the EEP and control strains were markedly different, with 12/17 strains in the control group belonging to ST131 and only 1/17 belonging to ST131 in the EEP group (P < 0.001, Fisher’s exact test). ST131 belonged to either the C1 nM27 or the C2 subclade, with the exception of one strain, which exhibited genomic traits of both subclades B and C2. Phylogenetic analysis confirmed the close relationship of ST131 strains (Fig. S3). However, non-ST131 strains in the control group were evenly distributed over the phylogenetic tree and did not exhibit closer relationship with the ST131 strains. Additionally, we recorded the material from which each isolate was obtained. However, no association with either sequence type or group membership was observed, with the majority of strains in both groups coming from urine samples.
TABLE 1.
Overview of included strains and mutations in efflux-relevant proteinsa
| Group | Strain | Origin | ST, subclade | Isolation source | Mutation(s) in: |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AcrR | MarA | MarR | SoxS | SoxR | Rob | AcrS | |||||
| E | PEG 10-68-51 | D/A/CH | ST1011 | Urine | No var | No var | G103S, D118N, Y137H | No var | No var | No var | D167N |
| 2012-0633 | Japan | ST1193 | Urine | No var | S127N | S3N, G103S, Y137H | No var | G74R, G121D | Q20H, A171S | E75D, Q213K, M220I | |
| KUN9180 | Japan | ST131, C1 nM27 | Blood | Frameshift | S127N | G103S, Y137H | No var | G74R | No var | E75D, Q213K, M220I | |
| FR-11009_ZG | D/A/CH | ST167 | Rectal swab | G28R | No var | No var | No var | A111T | No var | Deletion | |
| SA128 | Sudan | ST167 | Urine | Frameshift | No var | No var | No var | A111T | A201_L202insSb | No var | |
| SA94 | Sudan | ST167 | Urine | Frameshift | S127N | No var | No var | A111T | A201_L202insS | No var | |
| Bul416 | Bulgaria | ST361 | Drainage | P155L | No var | No var | No var | No var | No var | No var | |
| KUN9567 | Japan | ST361 | Blood | V29G | No var | A53E, G103S, Y137H | No var | T38S, G74R | No var | R141H, V153I, H212R, M220I | |
| PEG 10-95-12 | D/A/CH | ST361 | Urine | No var | No var | No var | No var | No var | No var | No var | |
| Bul348 | Bulgaria | ST405 | Urine | V29G, T213I, N214T | No var | K62R, G103S, Y137H | No var | T38S, G74R | No var | H27R, A146G, V153I, Q213K, M220I | |
| EcBj10 | France | ST405 | Abdominal cavity | V29G, T213I, N214T | No var | K62R, G103S, Y137H | No var | T38S, G74R | No var | H27R, A146G, V153I, Q213K, M220I | |
| It4 | Italy | ST405 | Urine | T213I, N214T, frameshift | No var | K62R, G103S, Y137H | No var | T38S, G74R | No var | H27R, A146G, V153I, Q213K, M220I | |
| Ta215 | Tanzania | ST405 | Blood | V29G, T213I, N214T | No var | K62R, G103S, Y137H | No var | T38S, G74R | No var | H27R, A146G, V153I, Q213K, M220I | |
| Ta182 | Tanzania | ST46 | Pus | No var | No var | G103S, Y137H | No var | No var | No var | No var | |
| SA18 | Sudan | ST648 | Urine | H115Y | No var | A53E, G103S, Y137H | No var | T38S, G74R | No var | R141H, V153I, H212R, M220I | |
| PEG 10-55-51 | D/A/CH | ST744 | Urine | No var | No var | Frameshift | No var | No var | No var | Q74*c | |
| Ta178 | Tanzania | ST940 | Pus | No var | No var | G103S, Y137H | No var | No var | No var | D167N | |
| S3194 | Sweden | ST10 | Urine | T213I, N214T | No var | G103S, Y137H | No var | G74R | No var | Deletion (39 bp) | |
| C | 2012-1641 | Japan | ST131, C1 nM27 | Urine | Frameshift | S127N | G103S, Y137H | No var | G74R | No var | E75D, Q213K, M220I |
| Bul-P-30 | Bulgaria | ST131, C1 nM27 | Urine | No var | S127N | G103S, Y137H | No var | G74R | No var | E75D, Q213K, M220I | |
| Bul-P-31 | Bulgaria | ST131, C1 nM27 | Wound | No var | S127N | G103S, Y137H | No var | G74R | No var | E75D, Q213K, M220I | |
| EcBj2 | France | ST131, C2 | Blood | No var | S127N | G103S, Y137H | No var | G74R | No var | E75D, Q213K, M220I | |
| EcBj7 | France | ST131, C2 | Urine | No var | S127N | G103S, Y137H | No var | G74R | No var | E75D, Q213K, M220I | |
| PEG 10-13-37 | D/A/CH | ST131, C1 nM27 | Blood | *216YP*c | S127N | G103S, Y137H | No var | G74R | No var | E75D, Q213K, M220I | |
| S3185 | Sweden | ST131, C2 | Urine | No var | S127N | G103S, Y137H | No var | G74R | No var | E75D, Q213K, M220I | |
| SA11 | Sudan | ST131, B/C2 | Urine | No var | S127N | G103S, Y137H | No var | G74R | I287T | E75D, Q213K, M220I | |
| Ta002 | Tanzania | ST131, C2 | Pus | No var | S127N | G103S, Y137H | No var | G74R | No var | E75D, Q213K, M220I | |
| Ta022 | Tanzania | ST131, C2 | Urine | No var | S127N | G103S, Y137H | No var | G74R | No var | E75D, Q213K, M220I | |
| Tü3971 | Turkey | ST131, C2 | Blood | No var | S127N | G103S, Y137H | No var | G74R | No var | E75D, Q213K, M220I | |
| Tü4192 | Turkey | ST131, C2 | Blood | No var | S127N | G103S, Y137H | No var | G74R | No var | E75D, Q213K, M220I | |
| SA10 | Sudan | ST2301 | Wound | No var | No var | No var | No var | A111T | No var | Deletion | |
| KUN9478 | Japan | ST354 | Sputum | No var | No var | G103S, Y137H | No var | T38S, G74R | No var | E131D, R141H, V153I, M220I | |
| S1002 | Greece | ST847 | Urine | No var | No var | G103S, Y137H | No var | No var | No var | D167N, T214A | |
| EcBj3 | France | ST90 | Blood | No var | No var | G103S, Y137H | No var | No var | No var | No var | |
E, enhanced efflux phenotype group; C, control group; D/A/CH, Germany, Austria, or Switzerland; ST, sequence type; No var, no variant.
A201_L202insS denotes an insertion of serine between A201 and L202.
Asterisks denote a stop codon.
Analysis of mutations.
Having established a phenotypical distinction between EEP and non-EEP strains, we analyzed the sequences of genes shown or presumed to be involved in efflux and efflux regulation. Significant differences between the EEP and control group were observed (Tables 1 and 2). These differences were most notable in the sequences of regulators of the AcrAB-TolC efflux pump system. Most mutations were in AcrR. The majority of these mutations (11/14) were in strains of the EEP group and included frameshift mutations presumably prohibiting regular gene function and mutations in the DNA binding region of AcrR (V29G and G28R) with very high SNAP2 scores (screening for nonacceptable polymorphisms). These mutations are likely to impair the DNA binding capacity of AcrR and have been shown to be associated with fluoroquinolone resistance (15–17). In the control group, only one strain had a frameshift mutation in AcrR, and there were no mutated DNA binding regions.
TABLE 2.
Frequency and predicted relevance of mutations in relevant efflux-regulating genes
| Gene | Mutation in proteina | SNAP2 scoreb | No. of mutations in: |
|
|---|---|---|---|---|
| EEP isolates (n = 17) | Control strains (n = 17) | |||
| acrR | G28R | 89 | 1 | 0 |
| V29G | 72 | 4 | 0 | |
| H115Y | 1 | 1 | 0 | |
| P155L | 31 | 1 | 0 | |
| T213I | –12 | 4 | 1 | |
| N214T | –54 | 4 | 1 | |
| *216YP* | 0 | 1 | ||
| Frameshift | 4 | 1 | ||
| marA | S127N | 60 | 3 | 12 |
| marR | S3N | –90 | 1 | 0 |
| A53E | –35 | 2 | 0 | |
| K62R | –66 | 4 | 0 | |
| G103S | –74 | 11 | 16 | |
| D118N | 10 | 1 | 0 | |
| Y137H | 12 | 11 | 16 | |
| Frameshift | 1 | 0 | ||
| soxR | T38S | –49 | 6 | 1 |
| G74R | −2 | 8 | 14 | |
| A111T | –13 | 3 | 1 | |
| G121D | 68 | 1 | 0 | |
| rob | Q20H | –92 | 1 | 0 |
| A171S | –96 | 1 | 0 | |
| A201_L202insSc | 2 | 0 | ||
| I287T | 26 | 0 | 1 | |
| acrS | H27R | –57 | 4 | 0 |
| E75D | –76 | 2 | 12 | |
| E131D | –33 | 0 | 1 | |
| R141H | −28 | 2 | 1 | |
| A146G | −56 | 4 | 0 | |
| V153I | –90 | 6 | 1 | |
| D167N | 33 | 2 | 1 | |
| H212R | −15 | 2 | 0 | |
| Q213K | −39 | 6 | 12 | |
| T214A | −38 | 0 | 1 | |
| M220I | 5 | 8 | 13 | |
| Q74* | 1 | 0 | ||
| Deletion | 1 | 2 | ||
Asterisks denote a stop codon.
SNAP2 scores were calculated as detailed in Materials and Methods. No prediction was possible for frameshifts, insertions, deletions, and stop codon mutations.
A201_L202insS denotes an insertion of serine between A201 and L202.
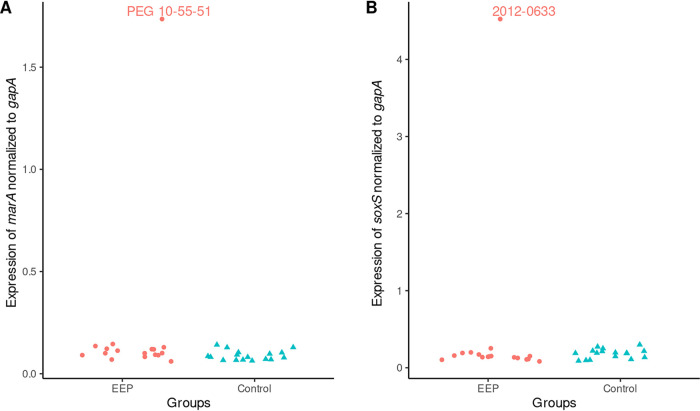
Analysis of marAR and soxSR revealed highly conserved sequences in the activators MarA and SoxS. In MarA, only one mutation, S127N, was found. This mutation concerns the last amino acid of the protein and seems to be closely associated with ST131 and related sequence types. In our collection, every member of ST131 exhibited this mutation. There were no mutations in SoxS. Conversely, both MarR and SoxR were found to harbor substantially more mutations than the respective activators. Most of these mutations had comparatively low SNAP2 scores. In marR, though, we found a frameshift mutation, presumably inactivating the MarR protein (strain 2012-0633). Similarly, we found one strain (PEG 10-55-51) exhibiting the G121D mutation in SoxR. This substitution has previously been described as a loss-of-function mutation by constitutive activation of the protein’s iron-sulfur center, rendering it incapable of suppressing the transcription of SoxS (18, 19). Interestingly, both of these strains, 2012-0633 and PEG 10-55-51, belonged to the EEP group and were shown to be outliers in the PCA described above, suggesting significantly increased susceptibility to EPIs.
Analysis of acrS, encoding another repressor of the AcrAB pump, yielded several loss-of-function mutations, namely, three deletions (S3194, FR-11009_ZG, SA10) and one codon conversion to a stop codon mutation (PEG 10-55-51). The deletions were due to transposases, the insertion of which in two strains (FR-11099_ZG, SA10) replaced the entire intergenic region between acrS and acrE as well as the first 191 bp of acrE, presumably abolishing the function of the AcrEF pump as well. The other observed mutations, receiving low to very low SNAP2 scores, were predicted to have little impact, with the exception of D167N, which was prevalent among EEP and control group strains. To further investigate the possible role of acrS loss-of-function mutations in an MDR background, we constructed the acrS-deficient strain 2012-0633ΔacrS by homologous recombination and introduced a low-copy-number plasmid (pACYC184) carrying acrS along with the promoter into the AcrS-deficient strains SA10 and FR11009ZG. However, neither 2012-0633ΔacrS nor the plasmid-complemented strains showed significant differences in MICs from those of their respective wild types (MIC in Data Set S1).
Analysis of additional efflux-associated genes (evgSA, baeSR, and emrR) did not yield any promising mutations, with the exception of a deletion of emrR in It4. This deletion, however, did not result in an overexpression of EmrAB (data not shown).
We also screened for acquired resistances via mobile genetic elements using ResFinder. Apart from tetracycline resistance genes TetA and TetB, which were present in all studied isolates, only one other known efflux-related, mobile genetic element was found, as one EEP strain (PEG 10-68-51) carried OqxA, a known multidrug efflux pump conferring resistance to ciprofloxacin, among others.
Gene expression data.
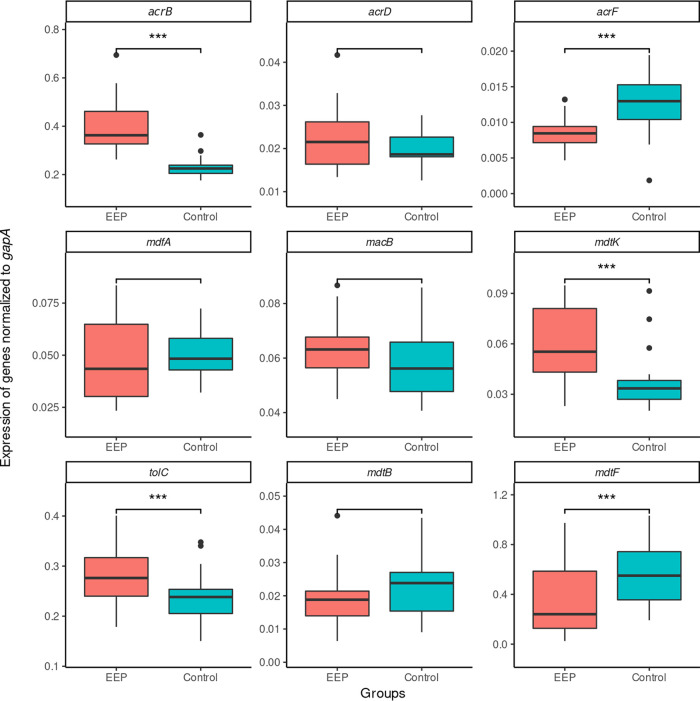
We then obtained quantitative reverse transcription-PCR (qRT-PCR) data for a panel of efflux-related proteins. There were significant differences between the two groups (Fig. 3 and Table 3). First, members of the EEP group showed an increased expression of all components belonging to the AcrAB-TolC pump system compared with that of the control strains. This was most noticeable for the expression of acrB, which had a highly significant result in an analysis of variance (ANOVA) (P < 0.001) and a large probabilistic index (PI) of 0.95 (95% confidence interval [95% CI], 0.81, 0.99) compared to that of the control group (Table 3) Although levels were lower overall, the expression of mdtK, too, was higher in the EEP group (P = 0.009; PI, 0.80; 95% CI, 0.61, 0.91). Second, the control strains exhibited a trend toward the overexpression of alternative pump proteins, especially mdtF, but acrF, too, was overexpressed compared to its level of expression in the EEP group, albeit at an overall lower level. There was no significant difference in expression of either marA or soxS between the two groups. However, for marA as well as soxS, there was one strain each that exhibited expression levels way above those of every other strain in our collection (Fig. 4). The corresponding strains exhibited the aforementioned loss-of-function mutations in their respective repressor genes (marR and soxR).
FIG 3.
Expression of genes normalized to the expression of gapA by group, with medians and interquartile ranges. Whiskers extend up to ±1.5 IQR, and dots represent outliers exceeding this range. *** denotes significant differences (P < 0.05) in the FDR-corrected ANOVA-type post hoc analysis.
TABLE 3.
Multivariate comparison of expression levels of efflux-related genes
| Gene | PI of EEP isolate vs control strain (95% CI)a | FDR Pb | Raw Pc | P (MANOVAd) |
|---|---|---|---|---|
| acrB | 0.95 (0.81, 0.99) | <0.001 | <0.001 | <0.001 |
| acrD | 0.58 (0.39, 0.75) | 0.503 | 0.447 | 0.227 |
| acrF | 0.24 (0.12, 0.43) | 0.018 | 0.006 | 0.006 |
| mdfA | 0.44 (0,27, 0,63) | 0.532 | 0.532 | 0.705 |
| macB | 0.60 (0,41, 0,76) | 0.408 | 0.317 | 0.317 |
| mdtK | 0.80 (0.61, 0.91) | 0.009 | 0.002 | 0.006 |
| tolC | 0.72 (0.50, 0.84) | 0.047 | 0.026 | 0.029 |
| mdtB | 0.39 (0.21, 0.56) | 0.408 | 0.285 | 0.280 |
| mdtF | 0.27 (0.13, 0.45) | 0.045 | 0.020 | 0.033 |
PI, probabilistic index. PIs greater than 0.56, 0.64, or 0.70 (and, conversely, below 0.44, 0.36, or 0.30) were deemed to denote a small, medium, or large relative effect, respectively. CI, confidence interval. PIs and corresponding CIs were calculated using nonparametric testing as described in Materials and Methods.
FDR, false-discovery rate. FDR-corrected P values are from analysis of variance (ANOVA)-type post hoc analyses.
Raw P values are from ANOVA-type post hoc analyses not accounting for multiple testing.
MANOVA, multivariate analysis of variance testing, reported for comparison.
FIG 4.
(A) Expression of marA normalized to that of gapA by group; (B) expression of soxS normalized to that of gapA by group. Red dots represent EEP strains, and green triangles represent control strains.
DISCUSSION
There is only limited and somehow conflicting information about the epidemiology of efflux overexpression and its clinical relevance in difficult-to-treat Gram-negative bacteria, such as MDR Enterobacterales, including E. coli clinical isolates (4, 20–24). In this study of MDR E. coli isolates, we screened for phenotypic traits indicating MDR efflux activity (i.e., MIC decrease by NMP) and used PCA to identify a cluster with enhanced MDR efflux. It was surprising to find ST131 isolates as some of the most successful MDR E. coli clones to be underrepresented among the group of isolates with enhanced efflux. The initial PCA results of clustering were largely confirmed after our sensitivity analysis and supported by the good correlation of group assignment and mutations in whole-genome sequencing (WGS) data.
The efflux capacity of ST131 E. coli has so far not been investigated in greater detail. In a study from Turkey, MarA expression was high in ST131 clinical isolates but not linked to fluoroquinolone resistance (25). Inactivation of the AcrAB-TolC pump in a clinical ST131 MDR isolate from Japan could restore sensitivity to a broad spectrum of antibiotics (11), but data showing a significantly lower organic solvent tolerance in ST131 E. coli isolates indicate a more limited role of MDR efflux in those strains (26). The reason for the probably reduced reliance on efflux activity for ST131 remains unclear. Since we were unable to find evidence pointing toward a relevant contribution of mobile genetic elements that ST131 E. coli isolates might lack and because of the AcrAB pump’s pivotal role even in these MDR strains, it seems unlikely that ST131 E. coli should lack crucial efflux genes. We speculate that in the extraintestinal habitat where ST131 strains are primarily successful, the selection pressure for efflux activity may be reduced compared to that in the gastrointestinal tract, and hence, occurrence of efflux overexpression is a rarer event. Also, the intrinsic fluoroquinolone resistance of ST131 isolates might lessen the need for increased efflux activity (27, 28).
Our analysis of the WGS data yielded high counts of mutations in efflux-regulating genes, and we believe that these data, if confirmed in a more extensive series, can be used to define an enhanced efflux genotype. Mutations in AcrR, MarR, and SoxR are well-known causes of acquired antibiotic resistance and have been documented in in vitro studies as well as in clinical isolates (15, 17, 19, 29–34). However, many of these mutations have uncertain functional relevance. Employing structural information and drawing on a systematic literature research, we attempted to ascertain the relevance of each individual mutation. Interestingly, most of the relevant mutations were found in the EEP group. This holds especially true for AcrR, where all but one inactivating mutation were found in the EEP group. It might be noted here as well that relevant mutations were far more common in AcrR than in either MarR or SoxR. This might be because loss-of-function mutations in the latter genes were associated with a drastically increased efflux activity, a condition that might not be beneficial under every circumstance. Loss-of-function mutations in MarR have previously been described in clinical isolates (29, 35) but were also associated with reduced fitness overall (35, 36). The findings in this study, however, provide further evidence that, at least in an MDR background, loss-of-function mutations in MarR and SoxR are viable paths for E. coli.
To the best of our knowledge, this is the first report of the occurrence of the constitutively SoxR-activating mutation G121D in a clinical E. coli isolate, which so far had been described only in laboratory strains and Salmonella spp. (18, 19, 37). Mutations in the effector proteins MarA and SoxS were rare, except for S127N in MarA. This mutation had a relatively high SNAP2 score, suggesting relevance, but given that this exchange concerns only the last amino acid of the protein, its relevance remains somewhat unclear. We are unaware of any other study reporting mutational data for AcrS from clinical isolates. Although loss-of-function mutations in AcrS may promote the derepression of AcrAB, we were unable to detect a distinct phenotype for AcrS knockout and knock-in strains in an MDR background. This might be due to the overall low expression of AcrS in both groups. Strains in the EEP group significantly differed from the remaining strains in levels of efflux pump gene expression. The higher levels of expression for all components of the AcrAB-TolC pump are in agreement with the findings of relevant mutations in genes regulating the expression of this MDR transporter, detailed above, and demonstrate its pivotal role.
A noteworthy finding was that in the control strains, we observed increased expression levels of alternative pump proteins. Although overexpression of AcrEF and MdtEF has been shown to be an alternative efflux strategy (38, 39), the significance of this increased expressions remains unclear. The overall very low level of AcrEF expression, even in the control group, renders the possibility of a significant contribution to the resistance profile unlikely. Another study found that overexpression of MdtEF facilitated growth under anaerobic conditions (39), but the bearing of this finding on fitness in extraintestinal habitats is not known. Finally, the higher expression levels of alternative pumps in the control group might be due to the fact that the marked overexpression of AcrAB-TolC components in the EEP group leaves little room for alternative transporters. Thus, it may be that these findings are due to a more balanced transporter interactome in the control group (40, 41).
A limitation of our work is the small number of isolates, which may not be representative; this prompted us to pursue nonparametric testing where feasible and prohibited large-scale genomic association studies. Nevertheless, results from our gene expression studies were additionally analyzed with parametric tests and found to be in good accordance with the results from nonparametric testing, which further increases confidence in their validity. Finally, our algorithm defined efflux capacity primarily via the proxy of EPI susceptibility. Although it is unlikely, since we found highly similar results using PAN (data not shown), this leaves room for the possibility of missing entire classes of efflux capacities immune to current EPIs.
In conclusion, we report a phenotypical approach to determining the efflux capacity of MDR E. coli. Although highly successful, ST131 E. coli isolates seem to rely less on efflux-mediated antibiotic resistance than do comparable MDR strains. Associated with the enhanced efflux phenotype, we find a distinct array of genomic variations and an overexpression of mainly the AcrAB-TolC pump.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
MDR clinical isolates were collected as part of an international cooperative project and were included in the present analysis if they met the following criteria: (i) a year of isolation not before 2010; (ii) an invasive nature (i.e., taken from blood, urine, abscess, or drainage fluid); (iii) in vitro resistance to ciprofloxacin, tetracycline (and/or doxy- and/or minocycline), co-trimoxazole, and at least one aminoglycoside; and (iv) production of extended-spectrum beta-lactamases (ESBLs). Bacteria were grown on Mueller-Hinton (MH) agar plates or in MH broth (Roth, Karlsruhe, Germany) at 37°C overnight, unless otherwise indicated.
Drugs and chemicals.
Drugs and chemicals were from Sigma (Taufkirchen, Germany), and 1-(1-naphthylmethyl)-piperazine (NMP) was from Chess (Mannheim, Germany).
Susceptibility testing and dye accumulation assays.
Microdilution MIC assays were conducted in MH broth according to CLSI guidelines using 96-well custom plates (Merlin, Bornheim-Hersel, Germany) in the absence and presence of NMP (100 mg/liter). The presence of an ESBL was confirmed using cefotaxime/cefotaxime and clavulanic acid strips with gradients of 0.25 to 16 and 0.016 to 1 mg/liter, respectively (Bestbion, Cologne, Germany) and by analyses of whole-genome sequencing (WGS) data (42; ResFinder, https://cge.cbs.dtu.dk/services/ResFinder/).
Intracellular accumulation of Hoechst 33342 was determined by fluorometry as described previously (43). Results were analyzed in comparison to dye accumulation in knockout strain 3AG100ΔacrB.
Mutational and gene expression analyses.
WGS data were generated according to a published protocol (44) and analyzed using CLC Genomics Workbench v 8.0.2 (Qiagen, CLC bio, Aarhus, Denmark). Chromosomal variants were detected by mapping reads to the reference genome from E. coli ATCC 25922 and to that from E. coli K-12 MG1655 (GenBank accession number CP009072.1 and NC_000913.3, http://www.ncbi.nlm.nih.gov/genbank/). Acquired resistance genes were detected by analyzing whole-genome sequencing data using the ResFinder-2.1 server (42; https://cge.cbs.dtu.dk//services/ResFinder/), and the multilocus sequence type (MLST) was determined using the MLST database (45; http://mlst.warwick.ac.uk/mlst/dbs/Ecoli/).
To assess the relevance of the identified mutations, we calculated the SNAP2 score (screening for nonacceptable polymorphisms) using a Web-based interface (46; https://rostlab.org/services/snap2web/). This algorithm makes use of a neuronal network trained with multiple protein databases and yields a score predicting the functional relevance of any mutation. SNAP2 scores range from −100 (very low probability of relevance) to +100 (very high probability of relevance).
For quantitative reverse transcription-PCR (qRT-PCR), RNA was isolated from cultures grown to an optical density at 600 nm (OD600) of 0.6 in Luria-Miller broth (Roth, Karlsruhe, Germany) starting with an OD600 of 0.05 and using the RNeasy Mini Plus kit (Qiagen, Hilden, Germany) as recommended by the manufacturer (with RNAprotect from Qiagen for RNA stabilization). Residual genomic DNA was eliminated with RNase-free DNase I from Qiagen. One microgram of RNA was reverse transcribed with the iScript cDNA kit (Bio-Rad, Munich, Germany), and PCR was conducted using the LightCycler fast-start DNA master SYBR green I kit (Roche, Grenzach-Wyhlen, Germany) with primer pairs for transporter and regulator genes (see Table S2 in the supplemental material). Expression was normalized to that of gapA and estimated with values from at least three independent RNA isolations.
Chromosomal gene inactivation.
Chromosomal gene deletion was conducted using the Quick & Easy E. coli gene deletion kit (Gene Bridges, Heidelberg, Germany). For mediating homologous recombination, a Red/ETCHL instead of Red/ETTET, plasmid was used. Correct insertion in acrS was verified by PCR and Sanger sequencing. For FRT-PGK-gb2-neo-FRT cassette amplification, including homology arms upstream and downstream of the insertion region in acrS, the oligonucleotides 5′-gctattttctcatcctgtgtcgaatatatttatttc-ctgaataattaatcAATTAACCCTCACTAAAGGGCGG-3′ and 5′-ctaaaactggttaactgtgacgaactgaattttcaggacagaatgtg-aatTAATACGACTCACTATAGGGCTC-3′ were used (uppercase letters indicate the priming region for the FRT-PGK-gb2-neo-FRT cassette).
Plasmid cloning.
The vector plasmid pACYC184 contains a p15a origin of replication and a chloramphenicol resistance cassette. After digestion with BamHI and XbaI, the acrS gene sequence from K-12-descendant 3AG100, including the promoter region, was cloned into the vector. Subsequently, strains were made electrocompetent and transformed via electroporation.
Phylogenetic analysis.
A taxonomic assignment check was done with Kraken v0.10.5-beta and the MiniKraken 4-GB database. Reads were mapped to reference genomeEC958/ST131 (GenBank accession no. HG941718) using smalt version 0.7.6 (https://www.sanger.ac.uk/science/tools/smalt-0). Single nucleotide polymorphisms (SNPs) were then filtered with GATK (https://gatk.broadinstitute.org/hc/en-us), and only SNPs with at least a 4-read coverage and present in >75% of reads were included. These variant-filtered files were then converted to a fasta file, where SNP sites and absent sites (N), compared to the reference genome, were replaced. All isolates were then combined to an alignment, and regions resembling mobile genetic elements were removed (https://github.com/andrewjpage/remove_blocks_from_aln). Sequence reads were assembled using SPAdes v3.11.1 (47; http://cab.spbu.ru/software/spades/), with kmer sizes 21, 33, 55, 77, 99, 109, and 123. Assemblies were then filtered to include only contigs with a minimum of 500 bp. MLST using the Achtman scheme was carried out with mlst v2.10 (https://github.com/tseemann/mlst) using the PubMLST website (https://pubmlst.org/) developed by Keith Jolley and sited at the University of Oxford.
After assembly and mapping, quality control (QC) was carried out. QC criteria are allocation of reads to the genus Escherichia, coverage of >30×, an appropriate genome size of ∼5 Mb, a number of contigs <500, the largest contig having >100,000 bp and an N50 of >100,000 bp, and identification of MLST alleles.
The core gene alignment was used for phylogenetic reconstruction with RAxML (48).
Statistical analysis.
All statistical analyses were performed using R (version 3.5.1) running RStudio (version 1.1.456). PCA was done using the packages stats and ggbiplot. Figures were created using the packages ggplot2 and cowplot. Multivariate analysis, employing a nonparametric approach, and calculation of PIs for individual variables were performed using the package npmv. The significance of the observed differences in gene expression levels between groups was analyzed using ANOVA-type testing, and correction for the false-discovery rate (FDR) was done according to the method of Benjamini and Hochberg (49). The PI was calculated in order to assess the relevance of the observed significant differences. In our design, the PI denotes the probability that a randomly chosen strain from one group exhibits a higher normalized expression level of the gene in question than a randomly chosen strain from the full cohort. PIs greater than 0.56, 0.64, or 0.70 (and conversely below 0.44, 0.36, or 0.30) were deemed to denote a small, medium, or large relative effect, respectively (50, 51). Confidence intervals for each PI were calculated according to the method of Newcombe using an Excel spreadsheet kindly provided by the author (52).
Supplementary Material
ACKNOWLEDGMENTS
We thank the following persons for their contribution of strains to this collection: Michael Kresken and Barbara Körber-Irrgang, Antiinfectives Intelligence, Rheinbach, Germany; Marie-Hélène Nicholas-Chanoine, Hôpital Beaujon, Clichy, France; Gunnar Kahlmeter and Hanna Odén Poulsen, Klinisk mikrobiologi, Centrallasarettet Växjö, Sweden; Yasufumi Matsumura, Kyoto University Graduate School of Medicine, Japan; Can Imirzalioglu, University Hospital Giessen and Marburg GmbH, Germany; Murat Akova, Hacettepe University Hospital, Ankara, Turkey; Mutasim E. Ibrahim, Abha National Polyclinic, Saudi Arabia; Stefania Stefani, Dipartimento di Scienze Bio-Mediche, Università degli Studi di Catania, Italy; and Rumyana Markovska, Department of Medical Microbiology, Medical University of Sofia, Bulgaria.
This work was supported in part by the Innovative Medicines Initiative (IMI) Joint Undertaking (project no. 115524 ND4BB Translocation [http://translocation.eu/], with contributions from the European Union seventh framework program and EFPIA companies).
We have no conflicts of interest to declare.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Marston HD, Dixon DM, Knisely JM, Palmore TN, Fauci AS. 2016. Antimicrobial resistance. JAMA 316:1193–1204. doi: 10.1001/jama.2016.11764. [DOI] [PubMed] [Google Scholar]
- 2.Li X-Z, Plésiat P, Nikaido H. 2015. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikaido H, Takatsuka Y. 2009. Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta 1794:769–781. doi: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nikaido H, Pagès J-M. 2012. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol Rev 36:340–363. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su C-C, Rutherford DJ, Yu EW. 2007. Characterization of the multidrug efflux regulator AcrR from Escherichia coli. Biochem Biophys Res Commun 361:85–90. doi: 10.1016/j.bbrc.2007.06.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma D, Alberti M, Lynch C, Nikaido H, Hearst JE. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol 19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 7.Jain K, Saini S. 2016. MarRA, SoxSR, and Rob encode a signal dependent regulatory network in Escherichia coli. Mol Biosyst 12:1901–1912. doi: 10.1039/c6mb00263c. [DOI] [PubMed] [Google Scholar]
- 8.Hirakawa H, Takumi-Kobayashi A, Theisen U, Hirata T, Nishino K, Yamaguchi A. 2008. AcrS/EnvR represses expression of the acrAB multidrug efflux genes in Escherichia coli. J Bacteriol 190:6276–6279. doi: 10.1128/JB.00190-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kern WV, Oethinger M, Jellen-Ritter AS, Levy SB. 2000. Non-target gene mutations in the development of fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother 44:814–820. doi: 10.1128/aac.44.4.814-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato T, Yokota S-I, Okubo T, Ishihara K, Ueno H, Muramatsu Y, Fujii N, Tamura Y. 2013. Contribution of the AcrAB-TolC efflux pump to high-level fluoroquinolone resistance in Escherichia coli isolated from dogs and humans. J Vet Med Sci 75:407–414. doi: 10.1292/jvms.12-0186. [DOI] [PubMed] [Google Scholar]
- 11.Schuster S, Vavra M, Schweigger TM, Rossen JWA, Matsumura Y, Kern WV. 2017. Contribution of AcrAB-TolC to multidrug resistance in an Escherichia coli sequence type 131 isolate. Int J Antimicrob Agents 50:477–481. doi: 10.1016/j.ijantimicag.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 12.Bohnert JA, Kern WV. 2005. Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob Agents Chemother 49:849–852. doi: 10.1128/AAC.49.2.849-852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kern WV, Steinke P, Schumacher A, Schuster S, von Baum H, Bohnert JA. 2006. Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Escherichia coli. J Antimicrob Chemother 57:339–343. doi: 10.1093/jac/dki445. [DOI] [PubMed] [Google Scholar]
- 14.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Blais J, Cho D, Chamberland S, Renau T, Leger R, Hecker S, Watkins W, Hoshino K, Ishida H, Lee VJ. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother 45:105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Dzink-Fox JL, Chen M, Levy SB. 2001. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother 45:1515–1521. doi: 10.1128/AAC.45.5.1515-1521.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson JS, Cobbold RN, Kyaw-Tanner MT, Heisig P, Trott DJ. 2010. Fluoroquinolone resistance mechanisms in multidrug-resistant Escherichia coli isolated from extraintestinal infections in dogs. Vet Microbiol 146:161–166. doi: 10.1016/j.vetmic.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Zayed AA-F, Essam TM, Hashem A-GM, El-Tayeb OM. 2015. 'Supermutators' found amongst highly levofloxacin-resistant E. coli isolates: a rapid protocol for the detection of mutation sites. Emerg Microbes Infect 4:e4. doi: 10.1038/emi.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koutsolioutsou A, Martins EA, White DG, Levy SB, Demple B. 2001. A soxRS-constitutive mutation contributing to antibiotic resistance in a clinical isolate of Salmonella enterica (serovar Typhimurium). Antimicrob Agents Chemother 45:38–43. doi: 10.1128/AAC.45.1.38-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vinué L, Corcoran MA, Hooper DC, Jacoby GA. 2015. Mutations that enhance the ciprofloxacin resistance of Escherichia coli with qnrA1. Antimicrob Agents Chemother 60:1537–1545. doi: 10.1128/AAC.02167-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davin-Regli A, Bolla J-M, James CE, Lavigne J-P, Chevalier J, Garnotel E, Molitor A, Pagès J-M. 2008. Membrane permeability and regulation of drug "influx and efflux" in enterobacterial pathogens. Curr Drug Targets 9:750–759. doi: 10.2174/138945008785747824. [DOI] [PubMed] [Google Scholar]
- 21.Li X-Z, Nikaido H. 2009. Efflux-mediated drug resistance in bacteria: an update. Drugs 69:1555–1623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cunrath O, Meinel DM, Maturana P, Fanous J, Buyck JM, Saint Auguste P, Seth-Smith HMB, Körner J, Dehio C, Trebosc V, Kemmer C, Neher R, Egli A, Bumann D. 2019. Quantitative contribution of efflux to multi-drug resistance of clinical Escherichia coli and Pseudomonas aeruginosa strains. EBioMedicine 41:479–487. doi: 10.1016/j.ebiom.2019.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paltansing S, Tengeler AC, Kraakman MEM, Claas ECJ, Bernards AT. 2013. Exploring the contribution of efflux on the resistance to fluoroquinolones in clinical isolates of Escherichia coli. Microb Drug Resist 19:469–476. doi: 10.1089/mdr.2013.0058. [DOI] [PubMed] [Google Scholar]
- 24.Morgan-Linnell SK, Becnel Boyd L, Steffen D, Zechiedrich L. 2009. Mechanisms accounting for fluoroquinolone resistance in Escherichia coli clinical isolates. Antimicrob Agents Chemother 53:235–241. doi: 10.1128/AAC.00665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atac N, Kurt-Azap O, Dolapci I, Yesilkaya A, Ergonul O, Gonen M, Can F. 2018. The role of AcrAB-TolC efflux pumps on quinolone resistance of E. coli ST131. Curr Microbiol 75:1661–1666. doi: 10.1007/s00284-018-1577-y. [DOI] [PubMed] [Google Scholar]
- 26.Johnson JR, Johnston B, Kuskowski MA, Sokurenko EV, Tchesnokova V. 2015. Intensity and mechanisms of fluoroquinolone resistance within the H30 and H30Rx subclones of Escherichia coli sequence type 131 compared with other fluoroquinolone-resistant E. coli. Antimicrob Agents Chemother 59:4471–4480. doi: 10.1128/AAC.00673-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicolas-Chanoine M-H, Bertrand X, Madec J-Y. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson JR, Porter SB, Thuras P, Johnson TJ, Price LB, Tchesnokova V, Sokurenko EV. 2015. Greater ciprofloxacin tolerance as a possible selectable phenotype underlying the pandemic spread of the H30 subclone of Escherichia coli sequence type 131. Antimicrob Agents Chemother 59:7132–7135. doi: 10.1128/AAC.01687-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato T, Yokota S-I, Uchida I, Okubo T, Usui M, Kusumoto M, Akiba M, Fujii N, Tamura Y. 2013. Fluoroquinolone resistance mechanisms in an Escherichia coli isolate, HUE1, without quinolone resistance-determining region mutations. Front Microbiol 4:125. doi: 10.3389/fmicb.2013.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yaqoob M, Wang LP, Kashif J, Memon J, Umar S, Iqbal MF, Fiaz M, Lu C-P. 2018. Genetic characterization of phenicol-resistant Escherichia coli and role of wild-type repressor/regulator gene (acrR) on phenicol resistance. Folia Microbiol (Praha) 63:443–449. doi: 10.1007/s12223-017-0579-7. [DOI] [PubMed] [Google Scholar]
- 31.Oethinger M, Podglajen I, Kern WV, Levy SB. 1998. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob Agents Chemother 42:2089–2094. doi: 10.1128/AAC.42.8.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fàbrega A, Martin RG, Rosner JL, Tavio MM, Vila J. 2010. Constitutive SoxS expression in a fluoroquinolone-resistant strain with a truncated SoxR protein and identification of a new member of the marA-soxS-rob regulon, mdtG. Antimicrob Agents Chemother 54:1218–1225. doi: 10.1128/AAC.00944-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kehrenberg C, Cloeckaert A, Klein G, Schwarz S. 2009. Decreased fluoroquinolone susceptibility in mutants of Salmonella serovars other than Typhimurium: detection of novel mutations involved in modulated expression of ramA and soxS. J Antimicrob Chemother 64:1175–1180. doi: 10.1093/jac/dkp347. [DOI] [PubMed] [Google Scholar]
- 34.Vinué L, Hooper DC, Jacoby GA. 2018. Chromosomal mutations that accompany qnr in clinical isolates of Escherichia coli. Int J Antimicrob Agents 51:479–483. doi: 10.1016/j.ijantimicag.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Praski Alzrigat L, Huseby DL, Brandis G, Hughes D. 2017. Fitness cost constrains the spectrum of marR mutations in ciprofloxacin-resistant Escherichia coli. J Antimicrob Chemother 72:3016–3024. doi: 10.1093/jac/dkx270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komp Lindgren P, Marcusson LL, Sandvang D, Frimodt-Møller N, Hughes D. 2005. Biological cost of single and multiple norfloxacin resistance mutations in Escherichia coli implicated in urinary tract infections. Antimicrob Agents Chemother 49:2343–2351. doi: 10.1128/AAC.49.6.2343-2351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen C, Choudhury A, Zhang S, Garst AD, Song X, Liu X, Chen T, Gill RT, Wang Z. 2020. Integrating CRISPR-enabled trackable genome engineering and transcriptomic analysis of global regulators for antibiotic resistance selection and identification in Escherichia coli. mSystems 5:e00232-20. doi: 10.1128/mSystems.00232-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olliver A, Vallé M, Chaslus-Dancla E, Cloeckaert A. 2005. Overexpression of the multidrug efflux operon acrEF by insertional activation with IS1 or IS10 elements in Salmonella enterica serovar typhimurium DT204 acrB mutants selected with fluoroquinolones. Antimicrob Agents Chemother 49:289–301. doi: 10.1128/AAC.49.1.289-301.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Xiao M, Horiyama T, Zhang Y, Li X, Nishino K, Yan A. 2011. The multidrug efflux pump MdtEF protects against nitrosative damage during the anaerobic respiration in Escherichia coli. J Biol Chem 286:26576–26584. doi: 10.1074/jbc.M111.243261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shuster Y, Steiner-Mordoch S, Alon Cudkowicz N, Schuldiner S. 2016. A transporter interactome is essential for the acquisition of antimicrobial resistance to antibiotics. PLoS One 11:e0152917. doi: 10.1371/journal.pone.0152917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuldiner S. 2018. The Escherichia coli effluxome. Res Microbiol 169:357–362. doi: 10.1016/j.resmic.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 42.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuster S, Kohler S, Buck A, Dambacher C, König A, Bohnert JA, Kern WV. 2014. Random mutagenesis of the multidrug transporter AcrB from Escherichia coli for identification of putative target residues of efflux pump inhibitors. Antimicrob Agents Chemother 58:6870–6878. doi: 10.1128/AAC.03775-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferdous M, Zhou K, Mellmann A, Morabito S, Croughs PD, de Boer RF, Kooistra-Smid AMD, Rossen JWA, Friedrich AW. 2015. Is Shiga toxin-negative Escherichia coli O157:H7 enteropathogenic or enterohemorrhagic Escherichia coli? Comprehensive molecular analysis using whole-genome sequencing. J Clin Microbiol 53:3530–3538. doi: 10.1128/JCM.01899-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hecht M, Bromberg Y, Rost B. 2015. Better prediction of functional effects for sequence variants. BMC Genomics 16(Suppl 8):S1. doi: 10.1186/1471-2164-16-S8-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 50.Acion L, Peterson JJ, Temple S, Arndt S. 2006. Probabilistic index: an intuitive non-parametric approach to measuring the size of treatment effects. Stat Med 25:591–602. doi: 10.1002/sim.2256. [DOI] [PubMed] [Google Scholar]
- 51.Kieser M, Friede T, Gondan M. 2013. Assessment of statistical significance and clinical relevance. Stat Med 32:1707–1719. doi: 10.1002/sim.5634. [DOI] [PubMed] [Google Scholar]
- 52.Newcombe RG. 2006. Confidence intervals for an effect size measure based on the Mann-Whitney statistic. Part 2: asymptotic methods and evaluation. Stat Med 25:559–573. doi: 10.1002/sim.2324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.