Eradication of tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), has been a challenge due to its uncanny ability to survive in a dormant state inside host granulomas for decades. Mtb rewires its metabolic and redox regulatory networks to survive in the hostile hypoxic and nutrient-limiting environment, facilitating the formation of drug-tolerant persisters. Previously, we showed that protein kinase G (PknG), a virulence factor required for lysosomal escape, aids in metabolic adaptation, thereby promoting the survival of nonreplicating mycobacteria.
KEYWORDS: bacterial protein kinase, bacterial signal transduction, phosphorylation, Mycobacterium tuberculosis, drug discovery, latency, persisters, hypoxia, adjunct therapy, latent infection, persistence
ABSTRACT
Eradication of tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), has been a challenge due to its uncanny ability to survive in a dormant state inside host granulomas for decades. Mtb rewires its metabolic and redox regulatory networks to survive in the hostile hypoxic and nutrient-limiting environment, facilitating the formation of drug-tolerant persisters. Previously, we showed that protein kinase G (PknG), a virulence factor required for lysosomal escape, aids in metabolic adaptation, thereby promoting the survival of nonreplicating mycobacteria. Here, we sought to investigate the therapeutic potential of PknG against latent mycobacterium. We show that inhibition of PknG by AX20017 reduces mycobacterial survival in in vitro latency models such as hypoxia, persisters, and nutrient starvation. Targeting PknG enhances the bactericidal activity of the frontline anti-TB drugs in peritoneal macrophages. Deletion of pknG resulted in 5- to 15-fold-reduced survival of Mtb in chronically infected mice treated with anti-TB drugs. Importantly, in the Cornell mouse model of latent TB, the deletion of pknG drastically attenuated Mtb’s ability to resuscitate after antibiotic treatment compared with wild-type and complemented strains. This is the first study to investigate the sterilizing activity of pknG deletion and inhibition for adjunct therapy against latent TB in a preclinical model. Collectively, these results suggest that PknG may be a promising drug target for adjunct therapy to shorten the treatment duration and reduce disease relapse.
INTRODUCTION
Despite being a curable and preventable disease, tuberculosis (TB) continues to be among the leading causes of death due to infectious diseases. The broad-spectrum disease spans from active infection to dissemination to other organs, latency, reactivation, and transmission to other individuals. TB treatment involves 6 months of long multidrug chemotherapy, which leads to noncompliance resulting in treatment failure, higher infection relapse rates (7 to 13%), and the emergence of multidrug- to totally drug-resistant strains (1). Another major problem in the treatment of TB is the formation of persisters, metabolically quiescent nonreplicating cells that are highly recalcitrant to the current drug therapy. The vast majority of TB-infected individuals are able to contain the infection and thus remain asymptomatic (1). However, these latently infected individuals harbor a risk of reactivation of up to 5 to 15%, which increases upon immunodeficiency such as during HIV-TB coinfection or treatment with immunosuppressive agents (2, 3). In low-prevalence regions such as the United States, reactivation accounts for ∼47 to 84% of cases of active TB disease (4). The reactivation of TB can be averted by preventive therapy that includes either isoniazid (INH) monotherapy for 9 months or a combinatorial therapy with INH and rifampicin (RIF) for 12 weeks (5). INH is primarily effective against actively replicating bacteria, suggesting that Mycobacterium tuberculosis (Mtb) replicates during latent tuberculosis infection (LTBI), at least sporadically (6). The prolonged drug regimen has thus been suggested to reflect intervals of the nonreplicating phase, further indicating that cotargeting nonreplicating bacilli could effectively shorten therapy and reduce disease relapse. Thus, a priority for TB research is to identify the drivers of persisters and dormant cell formation and drugs that can effectively kill latent bacilli that are tolerant to the current regimen and, thus, shorten treatment.
Granulomas represent the pathological hallmark of TB. The low oxygen tension found inside the granulomas is regarded as one of the major host determinants associated with latency (7, 8). This is based on the observations that Mtb grown in vitro under hypoxic conditions switches to a nonreplicating lifestyle and is resistant to antibiotics (9). Detailed molecular characterization of the dynamic interplay between the host and bacterium factors that orchestrates the establishment of latent infection can help identify novel targets for chemotherapy. The two-component sensory kinases DosS and DosT enable Mtb to sense latency-associated signals such as O2 and NO concentrations and mediate adaptation through its cognate response regulator DosR (10–12). Additionally, proteins such as HspX, MosR, and Icl, etc., also promote survival under hypoxic conditions (13–15). In addition to the classical two-component signaling system, Mtb encodes 11 eukaryote-like serine-threonine protein kinases (STPKs) that facilitate its adaptation to the dynamic microenvironment of the host. Protein kinase G (PknG) is the sole cytosolic STPK present in Mtb, which harbors three domains: a rubredoxin domain at the N terminus, a central kinase domain, and tetratricopeptide domain at the C terminus (16, 17). PknG was shown to be secreted into the host cytosol and inhibit phagolysosomal fusion (18). However, the presence of PknG in nonpathogenic Mycobacterium smegmatis (Msm), very low secretory levels of PknG, and the absence of direct substrates in the host led to the speculation that the primary function of PknG is in the modulation of bacterial cellular metabolism (19). Consonantly, multiple groups reported definitive functions of PknG in the modulation of cellular events (20–25). In response to nutrient availability, PknG regulates central carbon and nitrogen metabolism through phosphorylation-dependent inhibition of GarA (Fig. 1a) (20–22, 26). The only other substrate of PknG characterized thus far is a ribosomal protein, L13 (24). PknG regulates NADH levels through the phosphorylation of L13 and is thus also involved in redox sensing and maintenance (24).
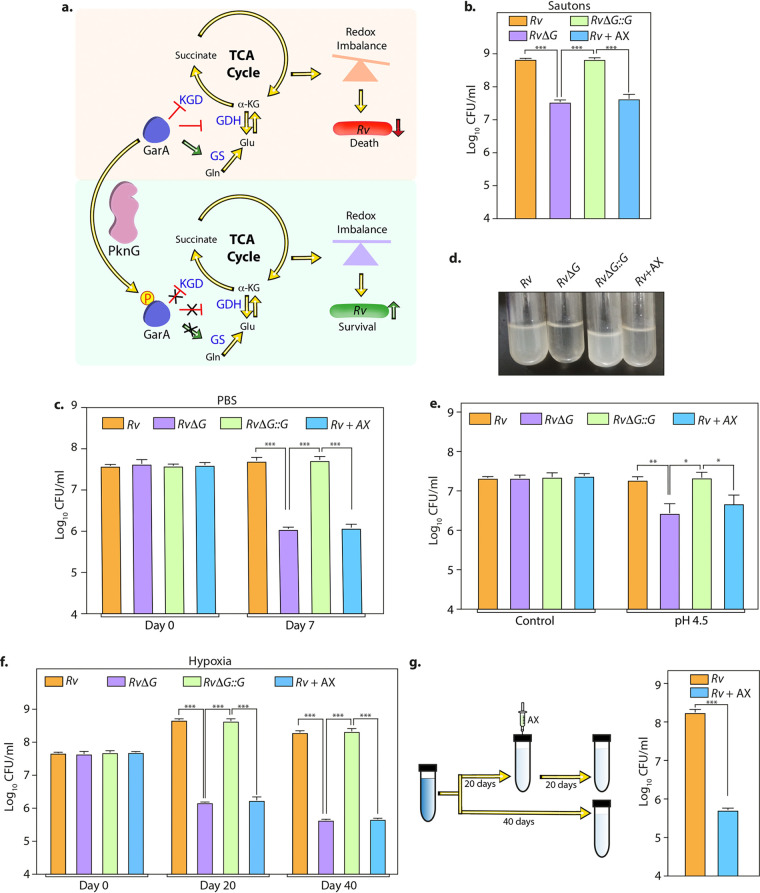
FIG 1.
AX inhibits Mtb survival in the in vitro models of latency. (a) Model illustrating the role of PknG during latency. GarA, a central metabolic regulator, inhibits α-ketoglutarate (α-KG) decarboxylase (KGD) and glutamate dehydrogenase (GDH) and activates glutamate synthase (GS). The phosphorylation of GarA by PknG alleviates the inhibition of KGD and GDH and the activation of GS. The PknG-GarA signaling axis is necessary for fine-tuning the tricarboxylic acid (TCA) cycle and glutamate metabolism, thus maintaining cellular redox homeostasis and abetting the survival of Mtb under latency-like conditions. (b to f) In the case of Rv plus AX (Rv+AX), 1 mM AX was added to Rv at the start of the experiment. (b) Rv, RvΔG, Rv+AX, and RvΔG::G were inoculated at an A600 of ∼0.1 in Sauton’s medium, and bacillary survival was enumerated at day 6. (c) Early-log-phase cultures were resuspended in PBS, and CFU were enumerated on day 7. (d) Pictorial representation of the growth of the indicated strains in acidified 7H9-ADC medium at day 7. (e) Single-cell suspensions of Rv, RvΔG, Rv+AX, and RvΔG::G were inoculated at an A600 of ∼0.1 in acidified 7H9-ADC medium, and CFU were enumerated on day 7. (f) Bacterial strains were inoculated at an A600 of ∼0.1 in 7H9-ADC medium containing methylene blue in tightly sealed tubes. CFU were enumerated on days 0, 20, and 40. (g) Schematic representation of the hypoxia experiment. Twenty days after the start of the experiment, 1 mM AX was injected using a fine needle, and tubes were resealed. A parallel group was left untreated (Rv). CFU were enumerated on day 40. Bars depict means ± SD (n = 3), representative of data from two biologically independent experiments. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Bacterial metabolism between actively replicating and dormant cells differs dramatically (14). A previous study from our group showed that while the PknG-GarA signaling pathway is dispensable for the in vitro growth of mycobacteria, it is crucial for survival during in vitro latency models such as hypoxia, persisters, and nutrient starvation (25). Infection of guinea pigs with the RvΔG (deletion mutant of pknG) strain resulted in drastically reduced formation of granulomas, repositories of dormant mycobacteria (25). We speculated that these mycobacteria that are not “walled off” would be more susceptible to anti-TB drugs. Since PknG kinase activity is involved in antibiotic tolerance (25, 27), adjunct therapy with a PknG inhibitor would target difficult-to-eradicate nonreplicating persisters. However, the role of PknG in abetting the survival of latent mycobacteria in vivo is not yet established.
Here, we tested our hypothesis that a combinatorial therapy of PknG inhibitors in conjunction with frontline anti-TB drugs would effectively kill both replicating and nonreplicating subpopulations. In line with this, we show that inhibition of PknG by its well-known inhibitor, AX20017 (AX), ameliorated the potency of first- and second-line TB drugs in both in vitro and ex vivo models. The deletion of pknG effectively reduced the drug-tolerant subpopulation and enhanced the efficacy of conventional anti-TB drugs in a murine model of TB infection. Importantly, we showed that the deletion of pknG leads to drastically lower disease relapse rates in the Cornell mouse model of latent TB. Collectively, our results showcase PknG as an attractive drug target for the treatment of latent TB.
RESULTS
AX inhibits Mtb survival in in vitro models of latency.
AX, a tetrahydrobenzothiophene compound, is an ATP competitive inhibitor that specifically inhibits PknG activity (16, 18). Treatment of macrophages with this inhibitor results in mycobacterial transfer to lysosomes, attenuating its intracellular survival. Even though multiple AX derivates and other compounds that inhibit PknG have been developed (28–34), we chose AX because it is highly specific and widely available, and its potency has been established by multiple groups (16, 18, 24). Since PknG is a nonessential protein, the addition of AX does not alter the growth of Mtb in vitro in 7H9-ADC medium (see Fig. S1 in the supplemental material). To examine the therapeutic potential of PknG, we first sought to test whether AX is able to inhibit PknG-mediated mycobacterial survival under in vitro latency-like conditions. Toward this, we assessed the growth profiles of the Mtb strains Rv, RvΔG, and RvΔG::G in nutrient-limited Sauton’s medium. While Rv and RvΔG::G were able to actively replicate, the deletion of PknG (RvΔG) resulted in the abrogation of growth. This is in congruence with a previous study that reported reduced growth of the pknG mutant under nutrient-limited conditions (35). The addition of AX reduced the survival of Rv and RvΔG::G by a log fold, nearly equivalent to the survival of RvΔG (Fig. 1b and Fig. S2a and b). Importantly, the addition of AX to RvΔG did not result in an additional decrease in survival, suggesting that AX specifically targets PknG inside the mycobacterial cells (Fig. S2a and b). Similarly, either deletion or inhibition of PknG compromised growth during starvation stress (Fig. 1c).
Mtb is an intracellular pathogen that resides mostly in the phagosome of macrophages. PknG-mediated cellular redox homeostasis helps Mtb combat multiple stresses, including acidic stress found inside phagosomes (25). To assess whether AX can efficiently inhibit survival under acidic conditions, we inoculated Rv, RvΔG, and RvΔG::G in acidified medium. In agreement with our previous report (25), RvΔG showed diminished survival compared with Rv and RvΔG::G. In a similar vein, treatment with AX also abrogated growth under acidic stress conditions (Fig. 1d and e). Next, we subjected mycobacterial strains to hypoxic stress according to a modification of Wayne’s model. Either the deletion or inhibition of PknG reduced the survival of Mtb by >2-log-fold (Fig. 1f and Fig. S2c). Importantly, the addition of AX to RvΔG had no additional effect, suggesting that AX specifically targets PknG even under hypoxic conditions (Fig. S2c). When a similar experiment was performed in the presence of various concentrations of AX, we observed that a similar impact could be achieved at 0.3 mM AX (Fig. S3a). Interestingly, the addition of even 0.03 and 0.1 mM AX also compromised the survival of Mtb under hypoxic conditions albeit not to a similar extent as the higher concentrations (Fig. S3a). However, we continued to use 1 mM AX for the subsequent in vitro experiments because (i) 1 mM AX does not impact mycobacterial survival under standard in vitro growth conditions (Fig. S1) and (ii) a similar concentration of AX has previously been used in other reports (24, 36). Mtb adapts to hypoxia in a biphasic manner. When the dissolved oxygen concentration drops below 1%, the bacilli enter microaerobic nonreplicating persistence (NRP) stage 1. When dissolved oxygen goes below 0.06% saturation, the organism enters NRP stage 2 and exhibits increased tolerance to INH (9). Thus, we next tested the ability of AX to inhibit mycobacterial survival in a preestablished hypoxia setup. Toward this, we injected the inhibitor 20 days after hypoxia and enumerated mycobacterial survival at 40 days. Interestingly, we found that the addition of AX attributed to an ∼2-log-fold reduction in mycobacterial survival (Fig. 1g). Together, these results suggest that PknG abets mycobacterial adaptation during NRP stage 2, and the addition of AX, unlike INH (37), can reduce the survival of dormant mycobacteria.
Inhibition of PknG suppresses the emergence of drug-resistant and -tolerant cells.
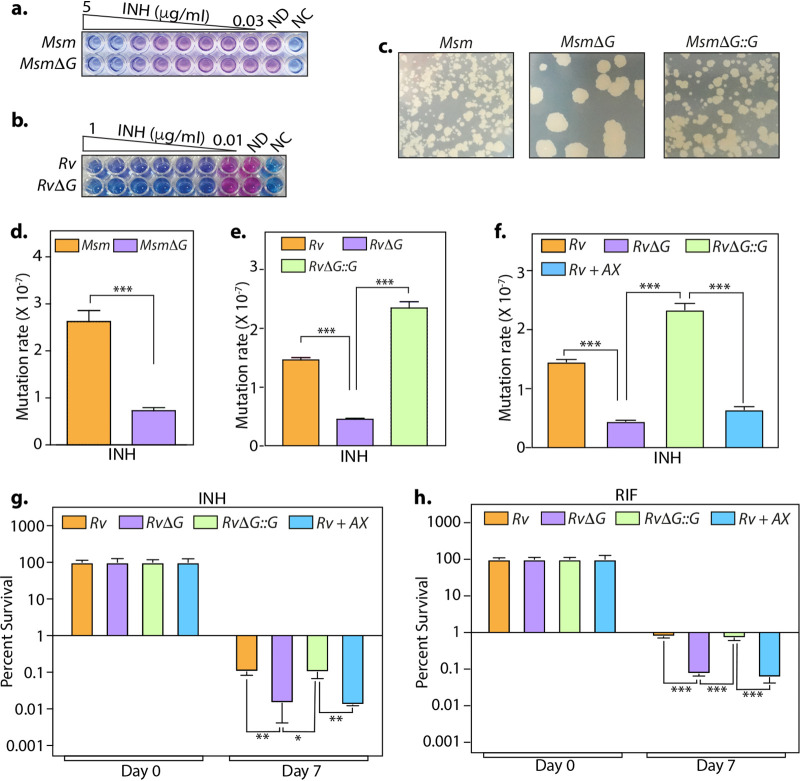
While nearly all frontline anti-TB antibiotics are active against actively replicating bacilli, they show limited efficacy against slowly replicating bacteria. Multiple reports link drug tolerance to inactive or slow metabolism, which in the case of Mtb echoes nonreplicating latent infection. Since PknG is necessary for the metabolic adaptation of Mtb in response to latency-like conditions (25), we speculated that deletion or inhibition of pknG would diminish bacillary survival in the presence of antibiotics. MICs of INH for parental and mutant strains were found to be similar in both Msm and Mtb (Fig. 2a and b). Subsequently, we quantified the mutation rate by enumerating the proportion of resistant mutants in the presence of INH. The deletion of pknG in either Msm or Mtb resulted in an ∼40 to 60% smaller resistor population (Fig. 2c to e). Importantly, the addition of 1 mM AX significantly reduced the mutation frequency to a similar extent (Fig. 2f). Analogous to RvΔG, the addition of 1 mM AX abated the persisters’ survival by ∼85% and ∼90% in the presence of INH and RIF, respectively (Fig. 2g and h). Moreover, a dose-response experiment wherein different concentrations of AX were used for the persister assay indicated that the addition of as little as 0.033 mM (30 times lower) also resulted in a significantly reduced survival of Mtb (Fig. S3b).
FIG 2.
Inhibition of PknG suppresses the emergence of drug-resistant and -tolerant cells. (a and b) The sensitivities of wild-type and pknG mutant Msm (a) and Mtb (b) strains to INH were determined with the help of an alamarBlue assay. ND and NC indicate no-drug and no-cell controls, respectively. (c) A total of 108 cells of Msm, MsmΔG, and MsmΔG::G were plated on 7H11-OADC plates containing 10 μg/ml isoniazid. (d and e) The antibiotic resistance frequency was determined by spotting 108 cells of Msm (d) and Mtb (e) strains on 7H11-OADC plates containing 100 μg/ml (d) and 10 μg/ml (e) INH, respectively. (f) Experiment performed as described above for panel e, with 1 mM AX added to Rv, where indicated. For panels d to f, the mutation rate is calculated as the number of colonies obtained on antibiotic-containing plates/number of colonies obtained on plain plates. The bar diagram represents means ± standard errors of the means (SEM) and is representative of results from a minimum of two independent biological replicates (n = 7). (g and h) Mtb strains were inoculated in 7H9-ADS medium containing 5 μg/ml INH (g) or 1 μg/ml RIF (h). Survival was monitored on days 0 and 7, and the survival obtained at day 0 was normalized to 100%. Percent survival at day 7 was calculated with respect to survival at day 0 for each strain. Data are represented as mean percent survival ± SD from one of three biologically independent experiments, each performed in triplicates (n = 3). *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Bedaquiline (BDQ) is among the recent anti-TB drugs used to treat multidrug-resistant bacteria. It binds to and induces large conformational changes in Mtb ATP synthase, resulting in a bactericidal effect (38–40). Importantly, BDQ has been shown to eradicate even the persistent bacterial population in mice (41, 42). Since both AX and BDQ target enzymes involved in energy metabolism, we sought to determine the effect of combinatorial therapy on both drug-resistant and -tolerant Mtb. Both the deletion and inhibition of PknG reduced the mutation rate of Mtb toward BDQ albeit not significantly (Fig. S4a). Importantly, the addition of AX or the deletion of pknG resulted in ∼93% and 95% decreases in persister survival in the presence of BDQ (Fig. S4b). Collectively, these data demonstrate the ability of AX to suppress the formation of drug-resistant and -tolerant cells with different antibiotic treatments, highlighting its potential utility in eliminating latent mycobacterial reservoirs.
Adjunct therapy with AX20017 reduces drug tolerance inside murine macrophages.
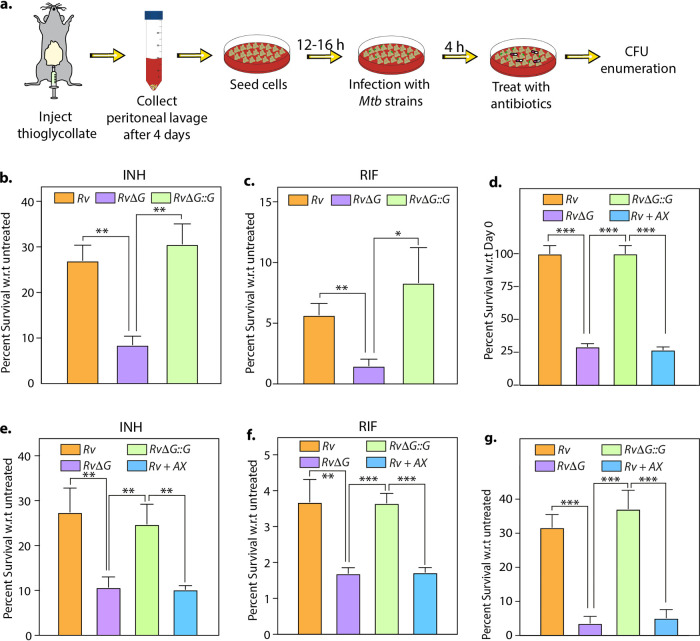
Intrigued by the AX potency to reduce drug tolerance in vitro, we sought to evaluate its efficacy within the host. Toward this, we infected peritoneal macrophages with the Rv, RvΔG, and RvΔG::G strains. After 4 h of infection, we added antibiotics and examined bacillary survival at 48 h postinfection (p.i.) (Fig. 3a). As depicted in Fig. 3b and c, compared with Rv and RvΔG::G, RvΔG showed an additional 30 to 40% reduction in survival in the presence of INH and RIF, indicating that PknG assists Mtb’s ability to tolerate anti-TB antibiotics even in the host (Fig. 3b and c). Next, we added AX in conjunction with INH and RIF to test its ability to inhibit PknG-mediated drug tolerance in the infected host. In agreement with previous reports (18, 24), the addition of AX in the absence of any additional antibiotic was able to reduce the intracellular survival of Mtb (Fig. 3d). Importantly, in the presence of INH, RIF, and BDQ, Mtb survival was further reduced by an additional ∼30%, 40%, and 80%, respectively, upon treatment with AX (Fig. 3e to g). Taken together, these results suggest that the addition of AX can reduce the drug-tolerant Mtb population within the infected macrophages.
FIG 3.
Adjunct therapy with AX20017 lowers drug tolerance inside murine macrophages. (a) Schematic representation of the peritoneal macrophage infection experiment. (b and c) Peritoneal macrophages were infected with Rv, RvΔG, and RvΔG::G at a multiplicity of infection (MOI) of 1:10. After 4 h, 0.5 μg/ml INH (b) or RIF (c) was added to the infected macrophages. At 48 h postinfection, macrophage cells were lysed, and bacillary survival was examined. CFU obtained for untreated infected macrophages at 48 h postinfection were normalized to 100%. The mean percent intracellular survival (±SD) (n = 3) of the bacterial strain in the treated macrophages was calculated with respect to (w.r.t) the corresponding untreated cells for that strain. (d to g) The same experiment as in panels b and c except with an additional Rv-infected sample, wherein 25 μM AX was added at 4 h postinfection (Rv+AX). Infected macrophages were either left untreated (d) or treated with 0.5 μg/ml INH (e), 0.5 μg/ml RIF (f), or 1 μg/ml BDQ (g). The mean percent intracellular survival (±SD) (n = 3) of the bacterial strain in the treated macrophages was calculated with respect to the corresponding untreated cells for that strain. Data are representative of results from two biologically independent experiments, each performed in triplicates. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
PknG abets drug tolerance in an infected murine model of tuberculosis.
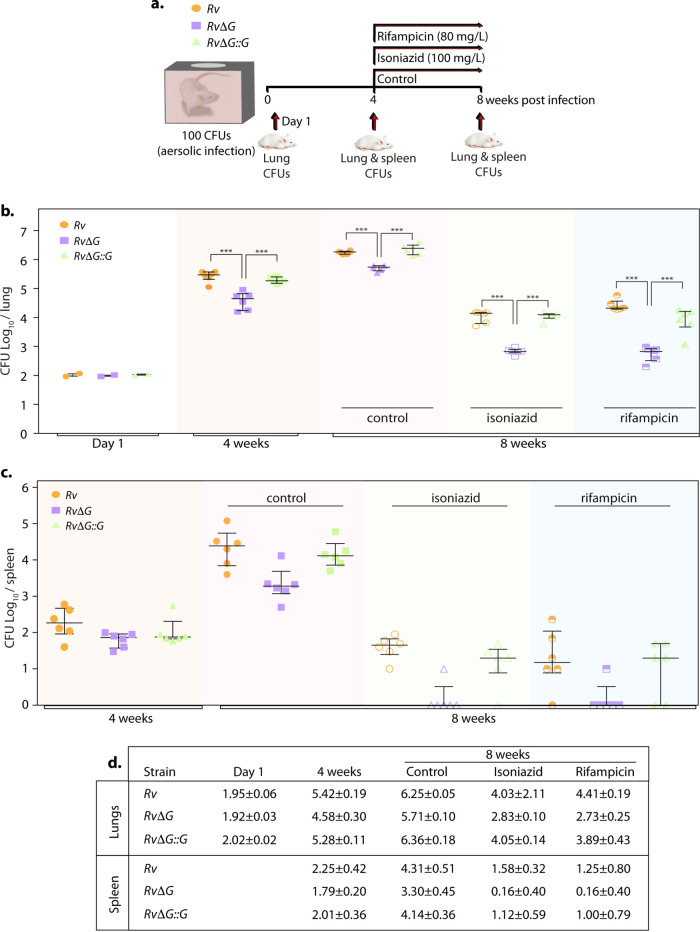
To further validate these findings, we infected mice with the Rv, RvΔG, and RvΔG::G strains, and CFU were enumerated at day 1 and weeks 4 and 8 p.i. in the lungs and spleen of infected animals (Fig. 4a). Bacillary survival in the lungs of infected animals at day 1 indicated equal deposition. After 4 weeks postinfection, RvΔG showed attenuated survival compared with Rv and RvΔG::G (Fig. 4b). In line with our previous guinea pig infection data (25), the magnitude of the difference between Rv and RvΔG decreased at 8 weeks postinfection (Fig. 4b). These results further corroborate an earlier report wherein the RvΔG mutant showed compromised survival in infected mice, more so at an earlier time point (35). Mice infected with RvΔG::G had CFU similar to those of mice infected with Rv, indicating that the complemented strain was able to restore the phenotype of the parental strain. Dissemination of RvΔG to the spleen was also attenuated in comparison with Rv and RvΔG::G at 8 weeks (Fig. 4c). Importantly, the survival of RvΔG was more compromised in the presence of INH and RIF in both the lungs and spleen. The relative survival of RvΔG with respect to Rv in the lungs of infected mice was 28.94% ± 6.18% (mean ± standard deviation [SD]) in the untreated control. Importantly, upon INH treatment, the relative survival of RvΔG with respect to Rv was 5.93% ± 1.39%. Similarly, upon RIF treatment, the relative survival of the mutant was found to be 2.16% ± 1.02% compared with Rv. Complementation of the mutant with pknG (RvΔG::G) restored the survival defects observed in the mutant in both the absence and presence of antibiotics. These results demonstrate that in the absence of pknG, the pathogen displays an additional ∼5- and 13-fold hypersensitivity to INH and RIF treatment, respectively.
FIG 4.
PknG abets drug tolerance in a murine model of tuberculosis infection. (a) Schematic representation of the murine infection experiment. Mice (n = 6 per group at each time point) were infected with Rv, RvΔG, and RvΔG::G. After the establishment of infection for 4 weeks, antibiotics (INH or RIF) were added to drinking water for the next 4 weeks. A parallel group was left untreated (control). Bacillary survival was assessed at day 1 and weeks 4 and 8. (b and c) Bacillary loads in lungs (b) and spleen (c) of infected mice at 4 and 8 weeks postinfection (n = 6). Each data point represents the log10 CFU of an infected animal in individual organs, and the error bar depicts the SD for each group. *, P < 0.05; **, P < 0.005; ***, P < 0.0005. (d) Table depicting mean log10 CFU ± SD in the lungs and spleen of infected animals at the indicated time points.
Deletion of PknG suppresses reactivation and lowers disease relapse rates in antibiotic-treated mice.
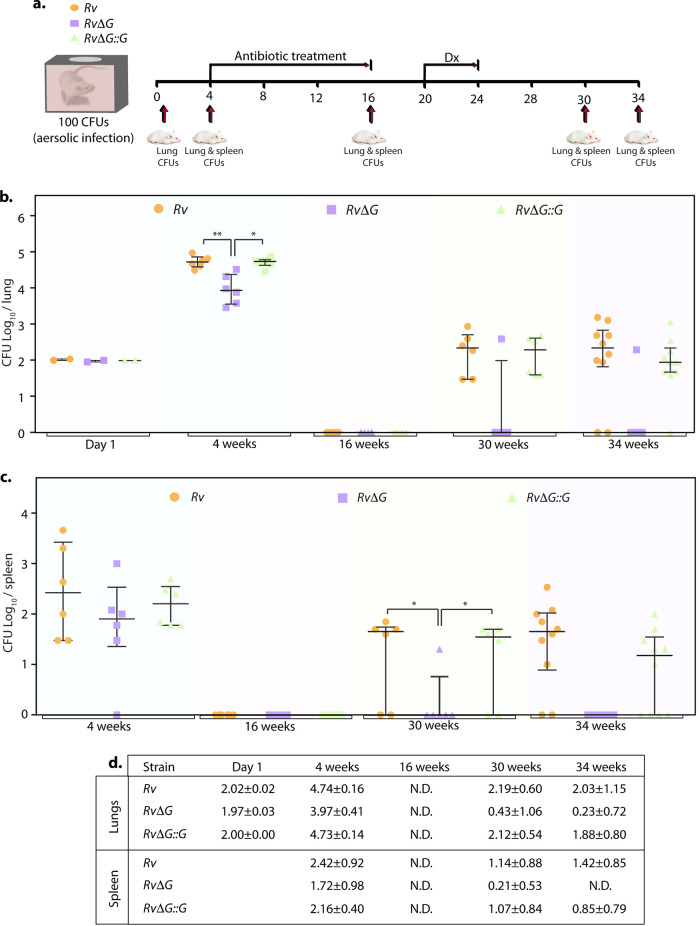
To deduce the importance of PknG in latent Mtb survival, we employed the modified Cornell model of infection (43). This model attempts to mimic a human-like latent infection by eliminating actively replicating bacilli with antibiotic treatment. Toward this, mice were infected with the Rv, RvΔG, and RvΔG::G strains, and after 4 weeks, mice were treated with INH and RIF for 12 weeks to establish latency. Four weeks after the establishment of latency, dexamethasone was administered for 4 weeks to suppress mouse immunity, which would allow the reactivation of latent viable bacilli. Eight and 12 weeks after the completion of dexamethasone treatment, CFU were enumerated (at the 30th and 34th weeks postinfection) (Fig. 5a).
FIG 5.
PknG is required for the resuscitation of Mtb in the modified Cornell mouse model of infection. (a) Schematic representation of the modified Cornell mouse model of tuberculosis infection experiment. Mice (n = 6 to 10 per group for each time point/strain) were infected with Rv, RvΔG, and RvΔG::G. After the establishment of infection for 4 weeks, antibiotics (INH and RIF) were added to drinking water for 12 weeks to eliminate actively replicating bacilli. Following 4 weeks of rest, dexamethasone (Dx) was injected for 4 weeks for immune suppression. Bacillary survival was examined at day 1 and weeks 4, 16, 30, and 34. (b and c) Bacillary loads in lungs (b) and spleen (c) of infected mice at the indicated time points postinfection (n = 6 to 10). Each data point represents log10 CFU of an infected animal in individual organs, and the error bar depicts the SD for each group. *, P < 0.05; **, P < 0.005; ***, P < 0.0005. N.D., not determined. (d) Table depicting mean log10 CFU ± SD in the lungs and spleen of infected animals at the indicated time points.
CFU counts were indistinguishable at day 1 postinfection, indicating equal deposition. At 4 weeks postinfection, the survival of RvΔG was compromised compared to Rv and RvΔG::G in both the lungs and spleen of infected mice (Fig. 5b and c). After 12 weeks of combinatorial therapy with INH and RIF, no viable CFU were found in the lungs and spleen of mice infected with either of the strains, indicating that all the actively replicating bacilli were eliminated. Eight weeks after immunosuppression, disease relapse rates were determined by the percentage of mice that showed the presence of mycobacterial colonies in the entire homogenates of lungs, spleen, or both. Examination of bacillary survival after immunosuppression clearly demonstrated the resurgence of both the Rv and RvΔG::G strains in almost all infected animal lungs (Fig. 5b) (30th and 34th weeks). The splenic bacillary loads of Rv- and RvΔG::G-infected mice observed at the 30th and 34th weeks also indicate the revival of these strains (Fig. 5c). In contrast, only 1 out of 6 and 10 RvΔG-infected mice at the 30th and 34th weeks, respectively, showed the presence of viable bacteria in the lungs (Fig. 5b). A similar trend was observed when bacillary survival in the spleen was evaluated (Fig. 5c). As indicated, the relapse noted for RvΔG-infected mice was 10% compared with 80% and 90% observed for Rv- and RvΔG::G-infected mice, respectively (Table 1). The resuscitated bacteria were found to be drug susceptible, as previously suggested (44), indicating that PknG is crucial for drug tolerance in vivo. Importantly, the data reinforce the above-described in vitro, ex vivo, and in vivo findings (Fig. 1 to 4) showing that PknG is essential for the survival of latent mycobacteria.
TABLE 1.
Number of infected mice per group in which viable CFU were found in either the lungs or spleen, both the lungs and spleen, or neither of them at the 30th and 34th weeks postinfectiona
| Wk | Strain | No. of mice with viable CFU/total no. of mice |
% relapse |
||||
|---|---|---|---|---|---|---|---|
| Lungs only | Spleen only | Both organs | Neither organ | Lungs | Spleen | ||
| 30 | Rv | 6/6 | 4/6 | 4/6 | 0/6 | 100 | 66.6 |
| RvΔG | 1/6 | 1/6 | 1/6 | 5/6 | 16.6 | 16.6 | |
| RvΔG | 6/6 | 4/6 | 4/6 | 0/6 | 100 | 66.6 | |
| 34 | Rv | 8/10 | 8/10 | 8/10 | 2/10 | 80 | 80 |
| RvΔG | 1/10 | 0/10 | 0/10 | 9/10 | 10 | 0 | |
| RvΔG | 9/10 | 6/10 | 6/10 | 1/10 | 90 | 60 | |
Percent relapse depicts the percentage of animals in which viable CFU were found in the lungs or spleen.
DISCUSSION
Treatment of TB involves two stages: (i) an initial intensive 2-month multidrug course aimed at eliminating actively replicating bacilli and (ii) a 4-month continuation phase involving INH and RIF to eradicate latent/persistent bacilli. Even though frequently used antimycobacterial drugs show profound bactericidal activity in vitro, the chemotherapeutic regimen spans over months. Inside host macrophages, Mtb faces a diverse array of stresses such as oxygen deprivation, low pH, and nutrient limitation (8, 45–47). In response to these stresses in vitro, the bacilli enter a nonreplicating, metabolically quiescent state (48). During the chronic phase of infection in mice, the average population doubling time of Mtb increases to 1,676 h (∼70 days) (49). Since most anti-TB antibiotics act on actively replicating cells (50), the compromised potency of antibiotics in vivo has been attributed to a nonreplication state and the concomitantly reduced metabolic activity of the bacilli resulting in drug tolerance (50). This further indicates that directly targeting these nonreplicating bacilli might be key to shortening anti-TB regimens.
Wayne’s model of hypoxia exposes the bacilli to progressively decreasing oxygen concentrations in vitro (37), which can be used to recapitulate the microaerobic conditions found within necrotic host granulomas (9). Metronidazole, a drug that is active only under anoxic conditions (51, 52), was found to prevent reactivation of latent Mtb infection in macaques (6). Furthermore, the deletion mutant of hspX, a dormancy-associated antigen, has also been shown to shorten antibiotic treatment and decrease disease relapse rates in mice (53). Our results show that inhibition of PknG by AX also reduces bacillary survival in in vitro latency models such as starvation and hypoxia (Fig. 1). Importantly, PknG aids the survival of Mtb even during NRP stage 2 (Fig. 1). This is in congruence with previous reports from multiple groups, including ours, that demonstrated increased expression of PknG specifically during later stages of hypoxia (25, 54). This suggests that the potential usage of AX in clinical settings wherein bacilli persist in preestablished hypoxic lesions would enhance Mtb clearance (Fig. 6).
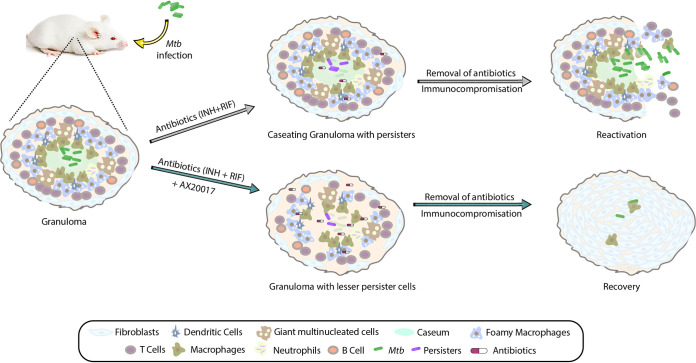
FIG 6.
Model depicting the therapeutic potential of AX20017. In response to Mtb infection, the host immune system contains the bacilli inside a granuloma. A granuloma is an organized and compact bunch of immune cells such as macrophages, multinucleated giant cells, dendritic cells, T cells, and B cells. Depending on the severity of the infection, granulomas can be of various kinds. The bacterium exploits this niche as its “safe haven” to survive and disseminate, leading to disease progression. The microaerophilic conditions found inside the granulomas drive cells of Mtb toward its nonreplicating antibiotic-tolerant persister phase. PknG helps Mtb adapt to hypoxic conditions by waning its metabolism. Deletion of pknG or its inhibition reduces Mtb’s ability to survive under in vivo latency-like conditions and attenuates its ability to thrive upon antibiotic stress. Hence, adjunct therapy with a PknG inhibitor could potentially suppress reactivation and lower the disease relapse rate and, hence, the transmission of the disease. The proof of principle for the interventions depicted in the schematic has been established with the help of various in vitro, ex vivo, and in vivo models of latency in this study.
RvΔG fails to persist in the presence of antibiotics due to its inability to phosphorylate GarA and L13, necessary for maintaining cellular redox poise (24, 25, 55). AX binds to the ATP binding pocket and inhibits the kinase activity of PknG (16). Even though PknG was reported to contribute to intrinsic antibiotic resistance of mycobacteria in a previous report (27), we did not observe any significant difference in the MIC of INH upon the deletion of pknG. We found that the deletion or inhibition of PknG reduces the survival of the drug-tolerant population both in vitro and within the host cells (Fig. 2 and 3). Hence, the addition of a PknG inhibitor to the therapeutic regimen would aid in the clearance of nonreplicating persisters. Since treatment with AX results in lower resistance to other antibiotics, its addition to anti-TB therapy is likely to suppress the formation of antibiotic-resistant cells (Fig. 2). Treatment with AX in this study serves as a proof of principle to demonstrate the therapeutic potential of PknG inhibitors in eliminating latent TB. We believe that potent inhibitors of PknG that directly target the substrate binding cleft or ATP binding pocket (such as AX and its derivates) would result in lower survival rates of dormant mycobacterium.
Unlike in vitro models of dormancy, in vivo models involve a vast array of host immune components, which are crucial for LTBI establishment. Granulomas are thought to restrict mycobacterial growth by limiting the oxygen supply, nutrients, and localized acidic pH (8). Previously, using a guinea pig model, we found that the absence of pknG results in reduced granuloma formation (25), which could be due to its failure to provide persistent chronic stimuli required for stable granuloma formation. Here, we sought to investigate the bactericidal activity of a PknG inhibitor given in combination with the frontline regimen in chronically infected mice. However, as the pharmacokinetics of AX is not established in the murine TB infection model, we used RvΔG as the alternate model for in vivo experiments. We show that the deletion of pknG enhances the bactericidal activity of INH and RIF in a murine model of chronic TB infection (Fig. 4). Importantly, in the Cornell mouse model of latent TB, the deletion of pknG drastically attenuated Mtb’s ability to resuscitate after antibiotic treatment. The absence of pknG reduced bacterial CFU to near sterility and significantly reduced disease relapse rates (10%) compared with those of mice infected with the wild-type (80%) and complemented (90%) strains (Fig. 5). Notably, this is the first study demonstrating the therapeutic potential of PknG in the 38-week mouse model of TB chemotherapy.
The major caveat of our proof-of-concept study is the use of a murine model, which, unlike guinea pigs, rabbits, and nonhuman primates, does not form human-like granulomas with caseous necrotic centers that contain a hypoxic environment (8). However, the absence of a similar Cornell in vivo latency TB model in guinea pigs restricted our choice. Since PknG is important for the virulence but not the survival of Mtb, we speculate that there would be a lower evolutionary burden on the bacilli to develop resistance. Importantly, this study lends additional support to the notion that a drug targeting nonreplicating bacteria can be effective in the treatment of the disease. However, preclinical studies that focus on the pharmacokinetics, standardization of the dosage, and optimization of the route of PknG inhibitors would be required. Collectively, we propose that PknG is a promising drug target that can be exploited in combination with conventional chemotherapeutic regimens to shorten the treatment duration and reduce disease relapse.
MATERIALS AND METHODS
Growth conditions and in vitro stresses.
Mtb wild-type (Rv), knockout (RvΔG), and complemented (RvΔG::G) strains were used in this study, as described previously (25, 35). The strains were grown at 37°C with shaking at 100 rpm in Middlebrook 7H9 broth supplemented with 10% ADC (albumin, dextrose, NaCl, and catalase), 0.2% glycerol (Sigma), and 0.05% Tween 80 (Sigma) or 7H11 agar supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC) and 0.2% glycerol. Kanamycin (Sigma), hygromycin (Sigma), and AX20017 (AX) (Calbiochem) were used at 25 μg/ml, 100 μg/ml, and 264.34 μg/ml (24), respectively. To evaluate the effect of AX on the in vitro growth of Mtb, mid-log-phase cultures (A600 of ∼0.6 to 0.8) were inoculated at an A600 of ∼0.1 in Sauton’s medium and at an A600 of ∼0.05 in 7H9-ADC medium. At the indicated time points, aliquots were withdrawn to measure the A600, and serial dilutions were plated to enumerate CFU.
To test susceptibility to acidic and starvation stress, mid-log-phase mycobacterial strains were washed with phosphate-buffered saline (PBS) containing 0.05% Tween 80 (PBST), and a single-cell suspension was inoculated at an A600 of ∼0.05 into 7H9 medium acidified to pH 4.5 or in PBST, respectively. CFU were enumerated on days 0 and 7. To assess viability after hypoxic stress, bacterial strains were inoculated with a headspace of 15% at an A600 of ∼0.1 in 7H9-ADC medium containing 1.5 μg/ml methylene blue (colorimetric redox indicator of dissolved oxygen) in 1-ml glass high-performance liquid chromatography (HPLC) vials. The caps of the tubes were sealed with multiple layers of parafilm, and the culture was left at 37°C with intermittent manual mixing to allow uniform hypoxia establishment. AX20017 was either added to the culture at the start of the experiment or injected on day 20 with a fine 27.5-gauge needle. CFU were enumerated on days 0, 20, and 40.
MIC analysis by an alamarBlue assay.
A 96-well plate was filled with 100 μl 7H9-ADS (albumin, dextrose, NaCl) medium without Tween 80. One hundred microliters of twice the maximum concentration of the test drug was added to the first well of three consecutive rows (for technical replicates) of the 96-well plate. The drug was serially diluted across the column by adding 100 μl of the drug from the first well to a consecutive one. One hundred microliters of Rv or RvΔG in 7H9-ADS medium at an A600 of ∼0.01 was added to each well. For controls, in one row, the drug was not added (no drug [ND]), and in the other, bacterial cells were replaced with medium only (no cells [NC]). The plate was sealed with parafilm and incubated at 37°C for 5 days, without shaking. After incubation, 20 μl of 0.25% filtered resazurin (prepared in MilliQ water) was added to each well, and the plate was further incubated until color development.
Persisters and antibiotic resistance.
To enumerate persisters, log-phase cultures of bacterial strains were diluted to an A600 of ∼0.05 in 7H9-ADS medium containing 5 μg/ml isoniazid (INH), 1 μg/ml rifampicin (RIF), or 1.75 μg/ml bedaquiline (BDQ) and incubated at 37°C for 7 days. The antibiotic resistance frequency was determined by plating 108 cells on 7H11 plates containing 10 μg/ml INH, 1 μg/ml RIF, or 1.75 μg/ml BDQ. The initial bacillary load was calculated by plating dilutions of the culture on 7H11 plates. The mutation rate was calculated as the number of resistant colonies obtained on antibiotic-containing plates divided by the CFU obtained in the absence of antibiotics.
Isolation and infection of peritoneal macrophages.
A 4% thioglycolate solution (Hi-Media) was injected into the peritoneal cavity of BALB/c mice. At 96 h postinjection, cells were isolated and seeded in 48-well plates in RPMI 1640 (Life Technologies, Inc.) containing 10% heat-inactivated fetal bovine serum (Gibco). Cells were maintained at 37°C in a humidified (5% CO2) incubator. The next day, cells were washed with RPMI 1640 medium and infected with a single-cell suspension of mycobacterial strains. At 4 h postinfection, medium was aspirated to remove extracellular bacteria, and fresh medium containing 0.5 μg/ml INH, 0.5 μg/ml RIF, 1 μg/ml BDQ, or 6.6 μg/ml AX was added. CFU were enumerated at 48 h postinfection.
Infection experiment in mice.
BALB/c mice (4 to 6 weeks old of either sex) were housed in individually ventilated cages at the Tuberculosis Aerosol Challenge Facility (TACF) at the International Centre for Genetic Engineering and Biotechnology (New Delhi, India) and cared for according to established animal ethics and guidelines. Mice (n = 6 at a minimum, per time point) were infected by the aerosol route using the Madison aerosol chamber (University of Wisconsin, Madison, WI), which was precalibrated to deliver ∼80 to 150 bacilli/mouse. Two mice for each strain were sacrificed at 24 h postexposure to assess bacterial deposition. To enumerate bacterial loads in the lungs and spleens at various time points as indicated, organs were removed aseptically and homogenized in 1 ml sterile normal saline. The homogenates were thereafter serially diluted and plated on 7H11 agar (containing polymyxin B, amphotericin B, nalidixic acid, trimethoprim, and azlocillin [PANTA] to prevent contamination) in triplicates to assess the bacillary load. For the antibiotic resistance model, after 4 weeks of infection, the mice were treated with either 100 mg/liter INH or 80 mg/liter RIF in drinking water (every 2 to 3 days) for 4 weeks. One of the parallel groups was left untreated as a control. Bacillary survival was assessed at 4 and 8 weeks p.i.
For the modified Cornell model, following 4 weeks of infection, mice were treated with 100 mg/liter INH and 60 mg/liter RIF administered in drinking water for 12 weeks in order to eliminate actively replicating bacilli and establish latency (56). Four weeks after the cessation of antibacterial treatment, dexamethasone was administered intraperitoneally (10 mg/kg of body weight in PBS) every alternate day for 4 weeks. Bacillary survival was assessed at day 1 to examine equal deposition, at week 4 to evaluate the establishment of infection, at week 16 to determine the establishment of latency, and at weeks 30 and 34 to assess reactivation.
Statistical analysis.
Student’s t test was used to determine statistical significance. Results were plotted with GraphPad Prism and modified with Adobe Illustrator. P values of >0.05 were considered statistically significant (*, P < 0.05; **, P < 0.005; ***, P < 0.0005).
Ethics statement.
The animal experimental protocol was approved by the Animal Ethics Committee of the National Institute of Immunology, New Delhi, India (IAEC numbers 409/16 and 462/18), according to the guidelines issued by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), Government of India.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yossef Av-Gay for sharing the Mtb pknG mutant strain. We thank the ICGEB and their staff for access to and help in their Tuberculosis Aerosol Central Facility (TACF).
M.Z.K. is supported by a research associate fellowship from the SERB (CRG/2018/001294). The study was supported by a core grant provided by the National Institute of Immunology, DBT grant BT/PR13522/COE/34/27/2015, SERB grant CRG/2018/001294, and J. C. Bose fellowship JCB/2019/000015 to V.K.N.
M.Z.K. designed and performed the experiments. V.K.N. supervised the design and acquired funding. M.Z.K. and V.K.N. wrote the manuscript.
We declare no competing interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Mitchison DA. 2005. Shortening the treatment of tuberculosis. Nat Biotechnol 23:187–188. doi: 10.1038/nbt0205-187. [DOI] [PubMed] [Google Scholar]
- 2.Getahun H, Matteelli A, Chaisson RE, Raviglione M. 2015. Latent Mycobacterium tuberculosis infection. N Engl J Med 372:2127–2135. doi: 10.1056/NEJMra1405427. [DOI] [PubMed] [Google Scholar]
- 3.Comstock GW, Livesay VT, Woolpert SF. 1974. The prognosis of a positive tuberculin reaction in childhood and adolescence. Am J Epidemiol 99:131–138. doi: 10.1093/oxfordjournals.aje.a121593. [DOI] [PubMed] [Google Scholar]
- 4.Lin PL, Flynn JL. 2010. Understanding latent tuberculosis: a moving target. J Immunol 185:15–22. doi: 10.4049/jimmunol.0903856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Getahun H, Matteelli A, Abubakar I, Aziz MA, Baddeley A, Barreira D, Den Boon S, Borroto Gutierrez SM, Bruchfeld J, Burhan E, Cavalcante S, Cedillos R, Chaisson R, Chee CB-E, Chesire L, Corbett E, Dara M, Denholm J, de Vries G, Falzon D, Ford N, Gale-Rowe M, Gilpin C, Girardi E, Go U-Y, Govindasamy D, Grzemska M, Harris R, Horsburgh CR, Jr, Ismayilov A, Jaramillo E, Kik S, Kranzer K, Lienhardt C, LoBue P, Lonnroth K, Marks G, Menzies D, Migliori GB, Mosca D, Mukadi YD, Mwinga A, Nelson L, Nishikiori N, Oordt-Speets A, Rangaka MX, Reis A, Rotz L, Sandgren A, Sane Schepisi M, et al. 2015. Management of latent Mycobacterium tuberculosis infection: WHO guidelines for low tuberculosis burden countries. Eur Respir J 46:1563–1576. doi: 10.1183/13993003.01245-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin PL, Dartois V, Johnston PJ, Janssen C, Via L, Goodwin MB, Klein E, Barry CE, III, Flynn JL. 2012. Metronidazole prevents reactivation of latent Mycobacterium tuberculosis infection in macaques. Proc Natl Acad Sci U S A 109:14188–14193. doi: 10.1073/pnas.1121497109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saunders BM, Cooper AM. 2000. Restraining mycobacteria: role of granulomas in mycobacterial infections. Immunol Cell Biol 78:334–341. doi: 10.1046/j.1440-1711.2000.00933.x. [DOI] [PubMed] [Google Scholar]
- 8.Via LE, Lin PL, Ray SM, Carrillo J, Allen SS, Eum SY, Taylor K, Klein E, Manjunatha U, Gonzales J, Lee EG, Park SK, Raleigh JA, Cho SN, McMurray DN, Flynn JL, Barry CE, III. 2008. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun 76:2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wayne LG, Hayes LG. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun 64:2062–2069. doi: 10.1128/IAI.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leistikow RL, Morton RA, Bartek IL, Frimpong I, Wagner K, Voskuil MI. 2010. The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J Bacteriol 192:1662–1670. doi: 10.1128/JB.00926-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park HD, Guinn KM, Harrell MI, Liao R, Voskuil MI, Tompa M, Schoolnik GK, Sherman DR. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol 48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar A, Toledo JC, Patel RP, Lancaster JR, Jr, Steyn AJ. 2007. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci U S A 104:11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayuri, Bagchi G, Das TK, Tyagi JS. 2002. Molecular analysis of the dormancy response in Mycobacterium smegmatis: expression analysis of genes encoding the DevR-DevS two-component system, Rv3134c and chaperone alpha-crystallin homologues. FEMS Microbiol Lett 211:231–237. doi: 10.1016/S0378-1097(02)00684-5. [DOI] [PubMed] [Google Scholar]
- 14.Eoh H, Rhee KY. 2013. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 110:6554–6559. doi: 10.1073/pnas.1219375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abomoelak B, Hoye EA, Chi J, Marcus SA, Laval F, Bannantine JP, Ward SK, Daffe M, Liu HD, Talaat AM. 2009. mosR, a novel transcriptional regulator of hypoxia and virulence in Mycobacterium tuberculosis. J Bacteriol 191:5941–5952. doi: 10.1128/JB.00778-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scherr N, Honnappa S, Kunz G, Mueller P, Jayachandran R, Winkler F, Pieters J, Steinmetz MO. 2007. Structural basis for the specific inhibition of protein kinase G, a virulence factor of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 104:12151–12156. doi: 10.1073/pnas.0702842104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koul A, Choidas A, Tyagi AK, Drlica K, Singh Y, Ullrich A. 2001. Serine/threonine protein kinases PknF and PknG of Mycobacterium tuberculosis: characterization and localization. Microbiology (Reading) 147:2307–2314. doi: 10.1099/00221287-147-8-2307. [DOI] [PubMed] [Google Scholar]
- 18.Walburger A, Koul A, Ferrari G, Nguyen L, Prescianotto-Baschong C, Huygen K, Klebl B, Thompson C, Bacher G, Pieters J. 2004. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science 304:1800–1804. doi: 10.1126/science.1099384. [DOI] [PubMed] [Google Scholar]
- 19.Khan MZ, Kaur P, Nandicoori VK. 2018. Targeting the messengers: serine/threonine protein kinases as potential targets for antimycobacterial drug development. IUBMB Life 70:889–904. doi: 10.1002/iub.1871. [DOI] [PubMed] [Google Scholar]
- 20.O’Hare HM, Duran R, Cervenansky C, Bellinzoni M, Wehenkel AM, Pritsch O, Obal G, Baumgartner J, Vialaret J, Johnsson K, Alzari PM. 2008. Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol Microbiol 70:1408–1423. doi: 10.1111/j.1365-2958.2008.06489.x. [DOI] [PubMed] [Google Scholar]
- 21.Rieck B, Degiacomi G, Zimmermann M, Cascioferro A, Boldrin F, Lazar-Adler NR, Bottrill AR, Le Chevalier F, Frigui W, Bellinzoni M, Lisa MN, Alzari PM, Nguyen L, Brosch R, Sauer U, Manganelli R, O’Hare HM. 2017. PknG senses amino acid availability to control metabolism and virulence of Mycobacterium tuberculosis. PLoS Pathog 13:e1006399. doi: 10.1371/journal.ppat.1006399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharyya N, Nkumama IN, Newland-Smith Z, Lin L-Y, Yin W, Cullen RE, Griffiths JS, Jarvis AR, Price MJ, Chong PY, Wallis R, O’Hare HM. 2018. An aspartate-specific solute-binding protein regulates protein kinase G activity to control glutamate metabolism in mycobacteria. mBio 9:e00931-18. doi: 10.1128/mBio.00931-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen L, Walburger A, Houben E, Koul A, Muller S, Morbitzer M, Klebl B, Ferrari G, Pieters J. 2005. Role of protein kinase G in growth and glutamine metabolism of Mycobacterium bovis BCG. J Bacteriol 187:5852–5856. doi: 10.1128/JB.187.16.5852-5856.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolff KA, de la Pena AH, Nguyen HT, Pham TH, Amzel LM, Gabelli SB, Nguyen L. 2015. A redox regulatory system critical for mycobacterial survival in macrophages and biofilm development. PLoS Pathog 11:e1004839. doi: 10.1371/journal.ppat.1004839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan MZ, Bhaskar A, Upadhyay S, Kumari P, Rajmani RS, Jain P, Singh A, Kumar D, Bhavesh NS, Nandicoori VK. 2017. Protein kinase G confers survival advantage to Mycobacterium tuberculosis during latency-like conditions. J Biol Chem 292:16093–16108. doi: 10.1074/jbc.M117.797563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner T, Andre-Leroux G, Hindie V, Barilone N, Lisa M-N, Hoos S, Raynal B, Vulliez-Le Normand B, O’Hare HM, Bellinzoni M, Alzari PM. 2019. Structural insights into the functional versatility of an FHA domain protein in mycobacterial signaling. Sci Signal 12:eaav9504. doi: 10.1126/scisignal.aav9504. [DOI] [PubMed] [Google Scholar]
- 27.Wolff KA, Nguyen HT, Cartabuke RH, Singh A, Ogwang S, Nguyen L. 2009. Protein kinase G is required for intrinsic antibiotic resistance in mycobacteria. Antimicrob Agents Chemother 53:3515–3519. doi: 10.1128/AAC.00012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen D, Ma S, He L, Yuan P, She Z, Lu Y. 2017. Sclerotiorin inhibits protein kinase G from Mycobacterium tuberculosis and impairs mycobacterial growth in macrophages. Tuberculosis (Edinb) 103:37–43. doi: 10.1016/j.tube.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Gil M, Grana M, Schopfer FJ, Wagner T, Denicola A, Freeman BA, Alzari PM, Batthyany C, Duran R. 2013. Inhibition of Mycobacterium tuberculosis PknG by non-catalytic rubredoxin domain specific modification: reaction of an electrophilic nitro-fatty acid with the Fe-S center. Free Radic Biol Med 65:150–161. doi: 10.1016/j.freeradbiomed.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santhi N, Aishwarya S. 2011. Insights from the molecular docking of withanolide derivatives to the target protein PknG from Mycobacterium tuberculosis. Bioinformation 7:1–4. doi: 10.6026/97320630007001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hegymegi-Barakonyi B, Székely R, Varga Z, Kiss R, Borbély G, Németh G, Bánhegyi P, Pató J, Greff Z, Horváth Z, Mészáros G, Marosfalvi J, Erōs D, Szántai-Kis C, Breza N, Garavaglia S, Perozzi S, Rizzi M, Hafenbradl D, Ko M, Av-Gay Y, Klebl BM, Orfi L, Kéri G. 2008. Signalling inhibitors against Mycobacterium tuberculosis—early days of a new therapeutic concept in tuberculosis. Curr Med Chem 15:2760–2770. doi: 10.2174/092986708786242886. [DOI] [PubMed] [Google Scholar]
- 32.Singh N, Tiwari S, Srivastava KK, Siddiqi MI. 2015. Identification of novel inhibitors of Mycobacterium tuberculosis PknG using pharmacophore based virtual screening, docking, molecular dynamics simulation, and their biological evaluation. J Chem Inf Model 55:1120–1129. doi: 10.1021/acs.jcim.5b00150. [DOI] [PubMed] [Google Scholar]
- 33.Szekely R, Waczek F, Szabadkai I, Nemeth G, Hegymegi-Barakonyi B, Eros D, Szokol B, Pato J, Hafenbradl D, Satchell J, Saint-Joanis B, Cole ST, Orfi L, Klebl BM, Keri G. 2008. A novel drug discovery concept for tuberculosis: inhibition of bacterial and host cell signalling. Immunol Lett 116:225–231. doi: 10.1016/j.imlet.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Kidwai S, Bouzeyen R, Chakraborti S, Khare N, Das S, Priya Gosain T, Behura A, Meena CL, Dhiman R, Essafi M, Bajaj A, Saini DK, Srinivasan N, Mahajan D, Singh R. 2019. NU-6027 inhibits growth of Mycobacterium tuberculosis by targeting protein kinase D and protein kinase G. Antimicrob Agents Chemother 63:e00996-19. doi: 10.1128/AAC.00996-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cowley S, Ko M, Pick N, Chow R, Downing KJ, Gordhan BG, Betts JC, Mizrahi V, Smith DA, Stokes RW, Av-Gay Y. 2004. The Mycobacterium tuberculosis protein serine/threonine kinase PknG is linked to cellular glutamate/glutamine levels and is important for growth in vivo. Mol Microbiol 52:1691–1702. doi: 10.1111/j.1365-2958.2004.04085.x. [DOI] [PubMed] [Google Scholar]
- 36.Houben EN, Walburger A, Ferrari G, Nguyen L, Thompson CJ, Miess C, Vogel G, Mueller B, Pieters J. 2009. Differential expression of a virulence factor in pathogenic and non-pathogenic mycobacteria. Mol Microbiol 72:41–52. doi: 10.1111/j.1365-2958.2009.06612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wayne LG, Diaz GA. 1967. Autolysis and secondary growth of Mycobacterium tuberculosis in submerged culture. J Bacteriol 93:1374–1381. doi: 10.1128/JB.93.4.1374-1381.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koul A, Dendouga N, Vergauwen K, Molenberghs B, Vranckx L, Willebrords R, Ristic Z, Lill H, Dorange I, Guillemont J, Bald D, Andries K. 2007. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat Chem Biol 3:323–324. doi: 10.1038/nchembio884. [DOI] [PubMed] [Google Scholar]
- 39.Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 40.Guo H, Courbon GM, Bueler SA, Mai J, Liu J, Rubinstein JL. 2021. Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline. Nature 589:143–147. doi: 10.1038/s41586-020-3004-3. [DOI] [PubMed] [Google Scholar]
- 41.Hu Y, Pertinez H, Liu Y, Davies G, Coates A. 2019. Bedaquiline kills persistent Mycobacterium tuberculosis with no disease relapse: an in vivo model of a potential cure. J Antimicrob Chemother 74:1627–1633. doi: 10.1093/jac/dkz052. [DOI] [PubMed] [Google Scholar]
- 42.Zhang T, Li SY, Williams KN, Andries K, Nuermberger EL. 2011. Short-course chemotherapy with TMC207 and rifapentine in a murine model of latent tuberculosis infection. Am J Respir Crit Care Med 184:732–737. doi: 10.1164/rccm.201103-0397OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dhillon J, Dickinson JM, Sole K, Mitchison DA. 1996. Preventive chemotherapy of tuberculosis in Cornell model mice with combinations of rifampin, isoniazid, and pyrazinamide. Antimicrob Agents Chemother 40:552–555. doi: 10.1128/AAC.40.3.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alnimr AM. 2015. Dormancy models for Mycobacterium tuberculosis: a minireview. Braz J Microbiol 46:641–647. doi: 10.1590/S1517-838246320140507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Voss JJ, Rutter K, Schroeder BG, Su H, Zhu Y, Barry CE, III. 2000. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc Natl Acad Sci U S A 97:1252–1257. doi: 10.1073/pnas.97.3.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohde K, Yates RM, Purdy GE, Russell DG. 2007. Mycobacterium tuberculosis and the environment within the phagosome. Immunol Rev 219:37–54. doi: 10.1111/j.1600-065X.2007.00547.x. [DOI] [PubMed] [Google Scholar]
- 47.Pandey AK, Sassetti CM. 2008. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci U S A 105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayes CS, Low DA. 2009. Signals of growth regulation in bacteria. Curr Opin Microbiol 12:667–673. doi: 10.1016/j.mib.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munoz-Elias EJ, Timm J, Botha T, Chan WT, Gomez JE, McKinney JD. 2005. Replication dynamics of Mycobacterium tuberculosis in chronically infected mice. Infect Immun 73:546–551. doi: 10.1128/IAI.73.1.546-551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gomez JE, McKinney JD. 2004. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinb) 84:29–44. doi: 10.1016/j.tube.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Wayne LG, Sramek HA. 1994. Metronidazole is bactericidal to dormant cells of Mycobacterium tuberculosis. Antimicrob Agents Chemother 38:2054–2058. doi: 10.1128/aac.38.9.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klinkenberg LG, Sutherland LA, Bishai WR, Karakousis PC. 2008. Metronidazole lacks activity against Mycobacterium tuberculosis in an in vivo hypoxic granuloma model of latency. J Infect Dis 198:275–283. doi: 10.1086/589515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu Y, Liu A, Menendez MC, Garcia MJ, Oravcova K, Gillespie SH, Davies GR, Mitchison DA, Coates AR. 2015. HspX knock-out in Mycobacterium tuberculosis leads to shorter antibiotic treatment and lower relapse rate in a mouse model—a potential novel therapeutic target. Tuberculosis (Edinb) 95:31–36. doi: 10.1016/j.tube.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 54.Gopinath V, Raghunandanan S, Gomez RL, Jose L, Surendran A, Ramachandran R, Pushparajan AR, Mundayoor S, Jaleel A, Kumar RA. 2015. Profiling the proteome of Mycobacterium tuberculosis during dormancy and reactivation. Mol Cell Proteomics 14:2160–2176. doi: 10.1074/mcp.M115.051151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen L. 2016. Antibiotic resistance mechanisms in M. tuberculosis: an update. Arch Toxicol 90:1585–1604. doi: 10.1007/s00204-016-1727-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCune RM, Feldmann FM, Lambert HP, McDermott W. 1966. Microbial persistence. I. The capacity of tubercle bacilli to survive sterilization in mouse tissues. J Exp Med 123:445–468. doi: 10.1084/jem.123.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.