Antimicrobial peptides (AMPs) play an important role in the defense against pathogens by targeting and killing invading microbes. Some pathogenic bacteria have been shown to negatively regulate AMP expression, while several commensals may induce AMP expression.
KEYWORDS: Neisseria meningitidis, hBD2, Lactobacillus, NF-κB, A20, Neisseria, meningococcus
ABSTRACT
Antimicrobial peptides (AMPs) play an important role in the defense against pathogens by targeting and killing invading microbes. Some pathogenic bacteria have been shown to negatively regulate AMP expression, while several commensals may induce AMP expression. The expression of certain AMPs, such as human beta-defensin 2 (hBD2), can be induced via nuclear factor NF-κB, which, in turn, is negatively controlled by tumor necrosis factor alpha-induced protein 3 (TNFAIP3, or A20). In this work, we examined the expression of hBD1 and hBD2 during coincubation of pharyngeal epithelial cells with pathogenic Neisseria meningitidis and commensal lactobacilli. The Lactobacillus strains induced hBD2 expression in human pharyngeal cells, while the pathogen N. meningitidis did not. In coincubation experiments, meningococci were able to dampen the AMP expression induced by lactobacilli. We found that N. meningitidis induced the NF-κB inhibitor A20. Further, RNA silencing of A20 resulted in increased hBD2 expression after meningococcal infection. Since it is known that induction of A20 reduces NF-κB activity and thus hBD2 levels, meningococcal-mediated A20 induction could be a way for the pathogen to dampen AMP expression. Finally, treatment of N. meningitidis and lactobacilli with synthetic hBD2 reduced N. meningitidis viability more efficiently than Lactobacillus reuteri, explaining why maintaining low AMP levels is important for the survival of the pathogen.
INTRODUCTION
The epithelial surface constitutes the first point of contact with invading pathogens. Pathogen recognition results in the activation of a signaling cascade that ultimately recruits, directs, and amplifies the immune system. Numerous effector molecules are released, including cytokines, chemokines, and antimicrobial peptides (AMPs), which together contribute to eliminating the infection (1). AMPs are evolutionarily conserved components of the innate immune system and are found in vertebrates, insects, and plants (2). They exhibit a broad range of activities and, for this reason, are also known as host defense peptides. AMPs are capable of directly eliminating bacteria (3) but also display immune-stimulating properties by acting as chemoattractants (4) and altering the release of cytokines (5). The two main families of AMPs found in humans are cathelicidins and defensins. Beta-defensins, a subgroup of the defensin family, are characterized by a beta sheet structure stabilized by three disulfide bridges (6) and are expressed ubiquitously by epithelial surfaces throughout the human body (7–9). Human beta-defensin 1 (hBD1) is constitutively expressed (10), while hBD2 expression is variable and can be modulated by bacteria, bacterial components, or immune factors (7, 11, 12).
The induction of AMPs by various stimuli has been shown to be dependent on nuclear factor kappa B (NF-κB) (7, 13–15). While NF-κB is an important molecule involved in immune activation, it must be carefully regulated to avoid excessive inflammation (16). Tumor necrosis factor alpha-induced protein 3 (TNFAIP3; also known as A20) is a zinc finger protein involved in the regulation of NF-κB in a negative-feedback manner (17). A20 is induced by NF-κB-activating stimuli via NF-κB sites in the A20 promoter (18), which, in turn, results in reduced NF-κB activation. Termination of NF-κB signaling can occur through direct deactivation of upstream signaling molecules, for example, TNF, interleukin 17 (IL-17), Toll-like receptor 4 (TLR4), and nucleotide-binding oligomerization domain-containing protein 2 (NOD2) (17, 19–22).
An interesting discovery was made by Simanski et al., who showed that A20 signaling can be exploited by bacteria. Staphylococcus epidermidis, a commensal species of the human skin, was shown to induce A20 signaling, to dampen hBD2 expression in keratinocytes to allow permanent S. epidermidis colonization (23). Downregulation of AMPs has also been shown for pathogenic bacterial species (24, 25), suggesting that AMP modulation represents a bacterial evasion strategy. On the other hand, both commensal and pathogenic bacteria can also induce AMP expression, which can lead to elimination of infection, thereby representing a defense mechanism of the host (3). It is evident that AMP regulation is a complex process that is highly context dependent.
Neisseria meningitidis (meningococcus) is a Gram-negative bacterium colonizing the human nasopharyngeal mucosa. Approximately 5 to 20% of the human population carries the bacterium asymptomatically. However, in rare cases, N. meningitidis can cross the mucosal barrier and spread, which can cause life-threatening sepsis and/or meningitis (26, 27). Pathogenic bacteria coexist with commensals at the site of infection. Lactobacilli are Gram-positive bacteria, found as commensal species in various areas of the human body (28–30). Lactobacilli secrete lactic acid, regulate the environmental pH, and compete with and make it difficult for other (pathogenic) bacteria to grow and colonize the mucosal surface (31, 32). For example, it has been demonstrated that the presence of lactobacilli in nasopharyngeal epithelial cells reduces inflammation and necrotic cell death caused by N. meningitidis (33).
In this study, we sought to better understand how commensals and pathogens shape AMP responses. We compared two commensal lactobacillus strains isolated from human saliva with the human pathogen N. meningitidis for their ability to modulate hBD1 and hBD2 expression. We found that Lactobacillus induced hBD2 expression, while N. meningitidis did not. Coincubation of lactobacilli and N. meningitidis reduced hBD2 expression, indicating that N. meningitidis may influence the AMP-inducing properties of Lactobacillus or the host cell expression of hBD2. We found that N. meningitidis induced A20 expression, which is known to reduce NF-κB expression and thereby dampen the hBD2 response. Finally, we showed that treatment with synthetic hBD2 peptide was more lethal for N. meningitidis, explaining why it is important for the pathogen to maintain low levels of hBD2.
RESULTS
Expression of hBD2 is induced by lactobacilli but not by N. meningitidis.
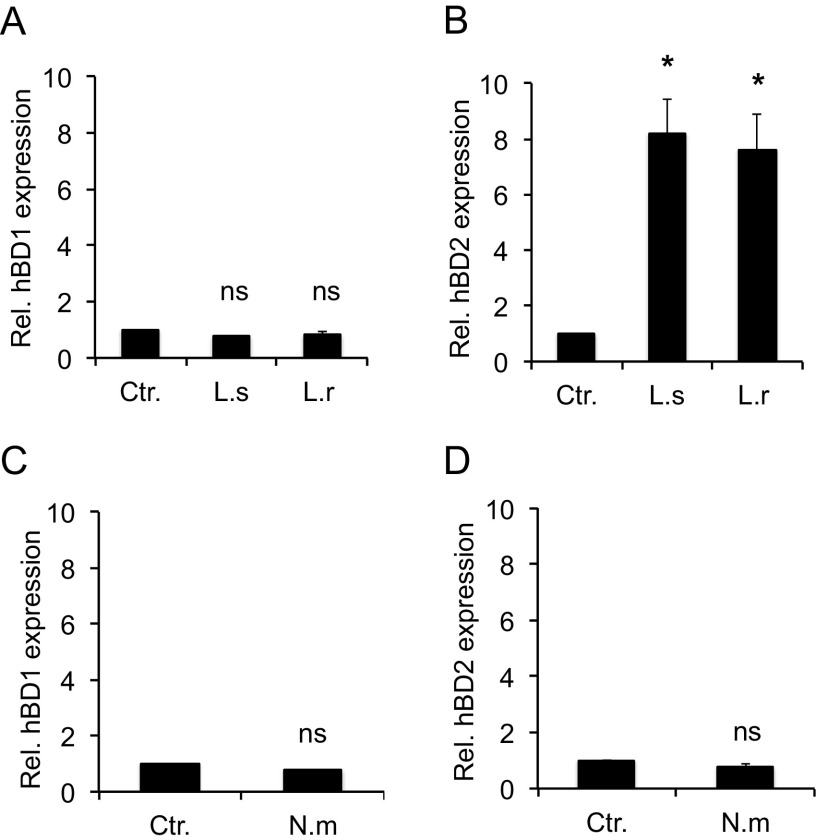
Previous studies have demonstrated that Lactobacillus can regulate the expression of human beta-defensins in human cells (13, 34, 35). To verify this, we incubated human pharyngeal epithelial cells with two different Lactobacillus species, Lactobacillus salivarius and Lactobacillus reuteri, and examined hBD1 and hBD2 gene expression. The cells were incubated with bacteria at a multiplicity of infection (MOI) of 100 for 6 h. At 2 h postincubation, cells were washed to remove unbound bacteria, and incubation was continued for an additional 4 h. In the presence of both Lactobacillus species, the expression of hBD1 remained unchanged compared to that of the control (Fig. 1A), while hBD2 expression was significantly induced (8-fold) (Fig. 1B). By testing different incubation times, we established that hBD2 gene expression peaked at 6 h (data not shown). These results are in agreement with a constitutive and inducible expression pattern of hBD1 and hBD2, respectively, reported for intestinal cells (7). Modulation of beta-defensin expression by N. meningitidis or during coincubation of lactobacilli and meningococci has thus far not been reported. Infection of pharyngeal epithelial cells with N. meningitidis did not induce expression of either hBD1 (Fig. 1C) or hBD2 (Fig. 1D). These data support the notion that epithelial cells are able to discriminate between commensals and pathogens by differential hBD2 modulation.
FIG 1.
Expression of hBD2 in response to Lactobacillus and N. meningitidis. Pharyngeal epithelial FaDu cells were incubated with two Lactobacillus species, L. salivarius and L. reuteri, at an MOI of 100 for 6 h, and the expression of the hBD1 (A) and hBD2 (B) genes was quantified using qPCR. FaDu cells were incubated with N. meningitidis at an MOI of 100 for 6 h, and the expression of the hBD1 (C) and hBD2 (D) genes was quantified using qPCR. The assays were performed in triplicate at least three times. Expression was normalized against the β-actin housekeeping gene and expressed as the fold change compared to the control. Data are represented as the mean values, with error bars representing the standard deviations. The assays were performed in triplicate at least three times. Significance was tested against the control. *, P < 0.05. ns, nonsignificant; Rel., relative; Ctr., control without bacteria; L.s, L. salivarius; L.r, L. reuteri; N.m, N. meningitidis.
Coincubation of L. reuteri with N. meningitidis dampens induction of hBD2 expression.
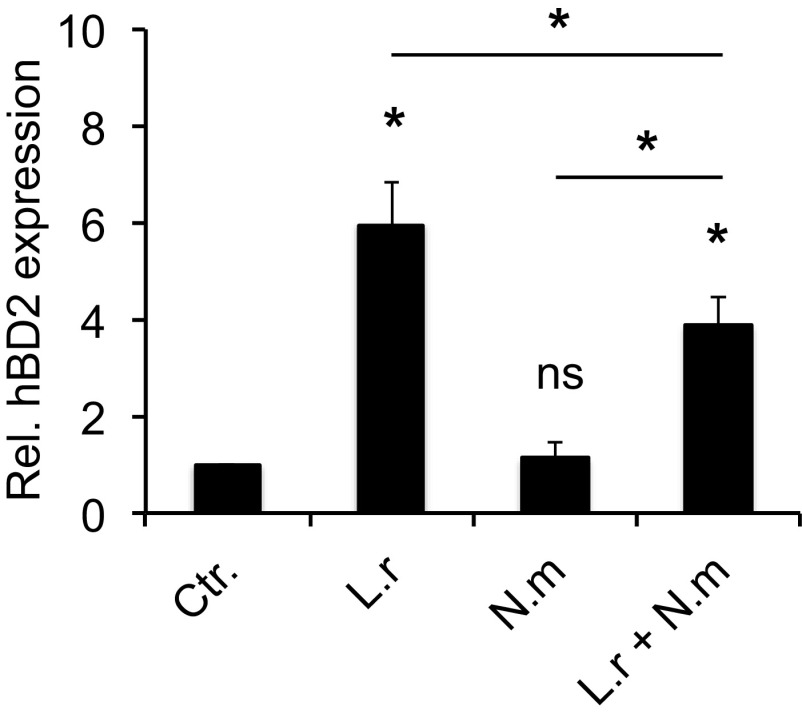
Since L. reuteri and L. salivarius induced hBD2 to similar levels, we continued experiments using L. reuteri. To mimic an environment where pathogenic and commensal bacteria are colocalized at the epithelial surface, we incubated epithelial cells simultaneously with L. reuteri and N. meningitidis. We found that the presence of N. meningitidis significantly dampened L. reuteri-induced hBD2 expression compared to that with L. reuteri alone (Fig. 2). Despite the dampening effect of N. meningitidis, there was still a significantly increased expression of hBD2 during coincubation compared to the control and N. meningitidis alone. These data show that N. meningitidis can reduce L. reuteri-mediated hBD2 expression.
FIG 2.
N. meningitidis affects L. reuteri-mediated hBD2 induction during coincubation. Pharyngeal epithelial cells were incubated with L. reuteri and N. meningitidis, either alone or coincubated, at an MOI of 100 for 6 h. Gene expression of hBD2 was quantified using qPCR. Expression was normalized against the β-actin housekeeping gene and expressed as the fold change compared to the control. Data are represented as the mean values, with error bars representing the standard deviations. The assay was performed in triplicate at least three times. Significance was tested against the control or as indicated. *, P < 0.05.
hBD2 induction by L. reuteri is independent of dose and levels of adhesion to host cells.
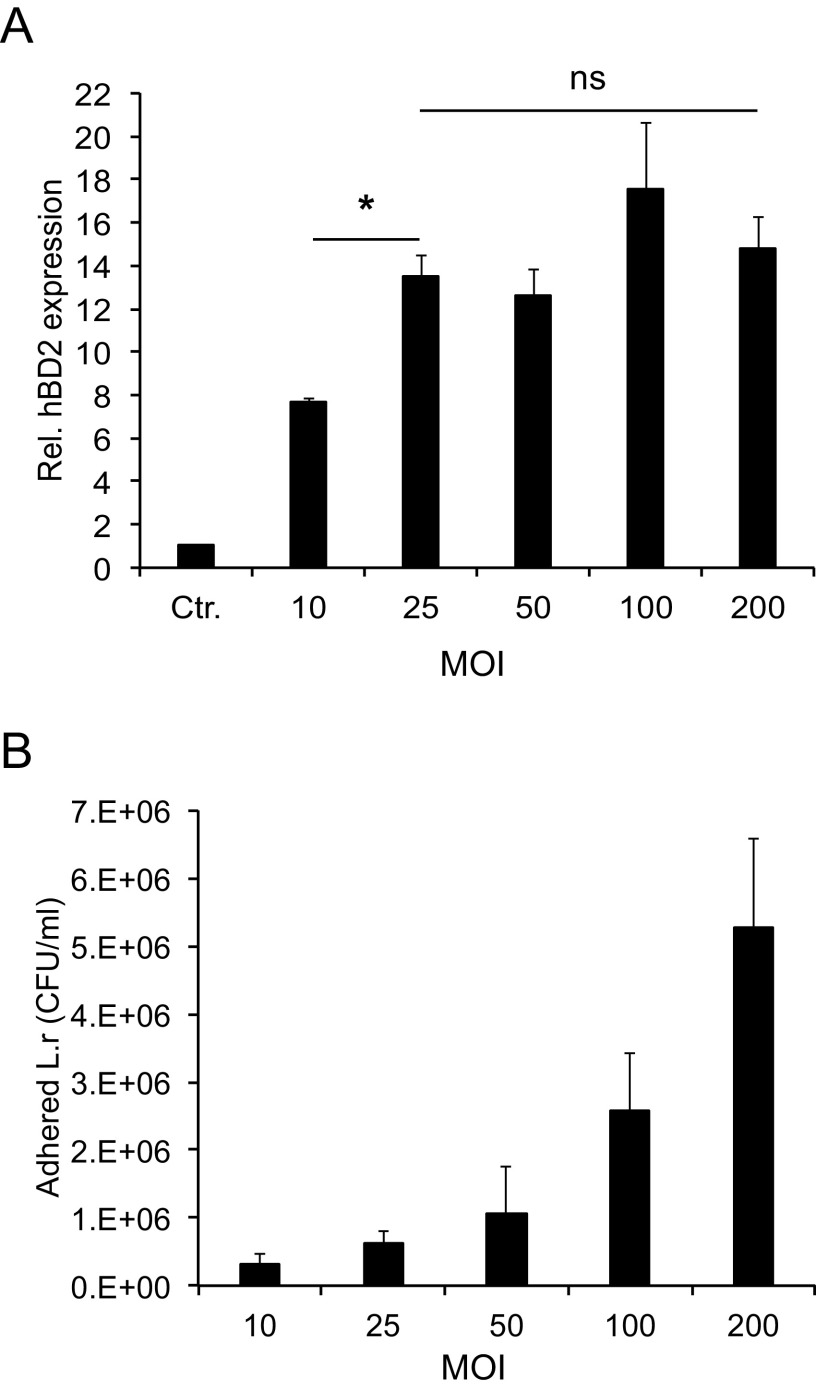
To determine whether modulation of hBD2 expression was correlated with the level of adhesion of bacteria to host cells, an adhesion assay at 6 h postincubation was performed. The attachment of L. reuteri to host cells was significantly reduced when the bacterium was coincubated with N. meningitidis (see Fig. S1A in the supplemental material). However, N. meningitidis adhesion to host cells remained unchanged in the presence or absence of L. reuteri (Fig. S1B). N. meningitidis, which did not induce hBD2, adhered approximately 4-fold better to host cells than did L. reuteri (Fig. S1A and B). The data suggest that the L. reuteri-induced hBD2 expression dampened by N. meningitidis may be due to reduced adhesion of L. reuteri to host cells. To investigate whether hBD2 expression depends on bacterial load, we incubated epithelial cells with L. reuteri or N. meningitidis at different MOIs. The expression of hBD2 remained uninduced by N. meningitidis regardless of the bacterial dose tested (Fig. S2). To examine whether the response to L. reuteri was dose dependent, we incubated with lactobacilli at a broader concentration range. Incubation of epithelial cells with L. reuteri at an MOI of 25, 50, 100, or 200 resulted in similar levels of hBD2 induced, but at an MOI of 10 the hBD2 level was lower (Fig. 3A). To investigate the correlation of hBD2 induction to the attachment level of L. reuteri, we performed an adherence assay at these different MOIs. Incubation with a higher bacterial load resulted in increased attachment of L. reuteri to epithelial cells (Fig. 3B). The data demonstrate that induction of hBD2 by L. reuteri at a range of MOIs from 25 to 200 is independent of the level of adhesion to host cells, as the induction of hBD2 at these bacterial concentrations remained constant (Fig. 3A). We suggest that to induce maximal hBD2 expression, L. reuteri must bind to host cells over a certain threshold value. Taken together, these data suggest that the N. meningitidis-mediated inhibition of L. reuteri hBD2 expression (Fig. 2) might not be a consequence of decreased attachment of L. reuteri but could have another explanation. An alternative possibility is that N. meningitidis triggers host cells to reduce hBD2 expression.
FIG 3.
L. reuteri-induced hBD2 expression. (A) Expression of the hBD2 gene in pharyngeal epithelial cells incubated with L. reuteri at an MOI of 10, 25, 50, 100, or 200 for 6 h. Expression was quantified using qPCR, normalized against the β-actin housekeeping gene and expressed as the fold change compared to the control. The assays were performed in triplicate at least three times. (B) Level of adherence of L. reuteri to pharyngeal epithelial cells. Epithelial cells were incubated with bacteria at an MOI of 10, 25, 50, 100, or 200 for 6 h, and bound bacteria were determined by plating. Data are represented as the mean values, with error bars representing the standard deviations. The assays were performed in triplicate at least three times. Significance was tested against the control or as indicated. *, P < 0.05.
hBD2 induction by L. reuteri requires live bacteria in direct contact with epithelial cells.
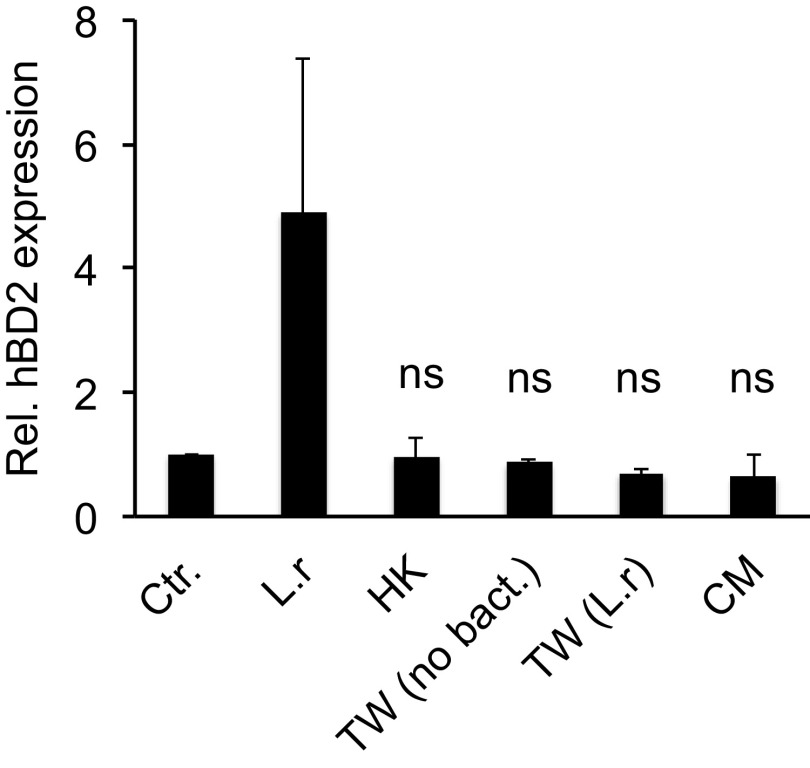
To determine whether induction of hBD2 expression requires an active interaction between L. reuteri and the host or if a released molecule mediates the effect, we incubated epithelial cells with heat-killed bacteria and bacteria that were physically separated from host cells. By using hanging cell culture Transwell filters, the bacteria and host cells were separated while allowing the diffusion of bacterially secreted factors. Incubation of epithelial cells with heat-killed bacteria did not result in the induction of hBD2 expression compared to the control (Fig. 4), suggesting that bacterial viability is important. Similarly, bacteria that were physically separated from host cells showed unchanged hBD2 expression compared to that of the control (Fig. 4), indicating that hBD2 induction is achieved by direct host-bacterium contact and that secreted factors are not involved. To verify this, we stimulated epithelial cells with L. reuteri conditioned medium (CM). CM did not trigger hBD2 expression compared to the control (Fig. 4). Taken together, these findings demonstrate that the induction of hBD2 expression is not mediated by a released component of L. reuteri; rather, live bacteria able to come in direct contact with host cells are necessary.
FIG 4.
hBD2 expression is induced by live lactobacilli in direct contact with host cells. Pharyngeal epithelial cells were incubated with live L. reuteri (L.r), L. reuteri that had been heat-killed (HK), or L. reuteri conditioned medium (CM). To separate bacteria from host cells, 0.4-μm hanging cell culture Transwell filters were used. Epithelial cells were maintained in the lower compartment, and live L. reuteri [TW (L.r)] or DMEM [TW (no bact)] were added to the upper compartment of the Transwell filters. After 6 h of incubation, the gene expression of hBD2 was quantified using qPCR. Expression was normalized against the β-actin housekeeping gene and expressed as the fold change compared to the control. Data are represented as the mean values, with error bars representing the standard deviations. The assay was performed in triplicate at least three times. Significance was tested against the control. *, P < 0.05.
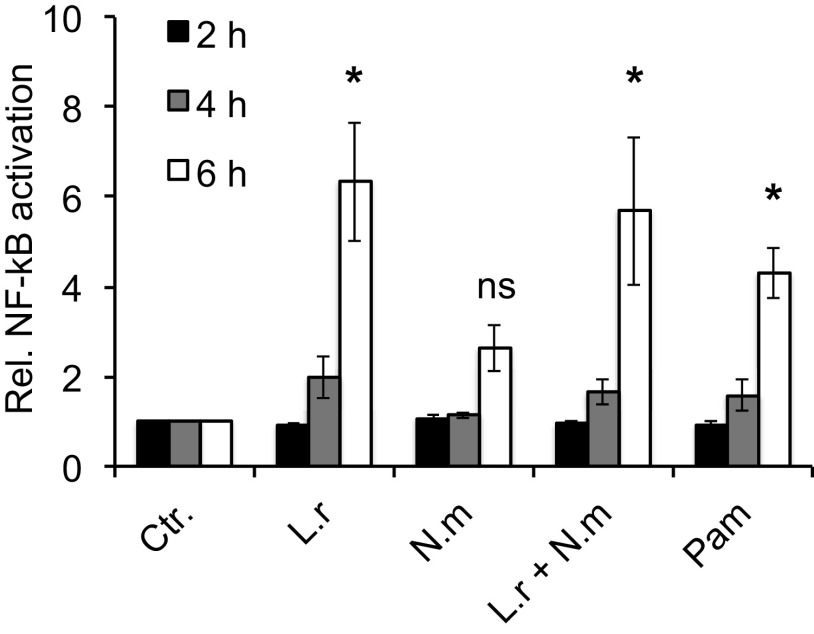
L. reuteri activates NF-κB signaling.
Modulation of hBD2 expression has previously been shown to occur through NF-κB signaling (7, 34). We therefore determined NF-κB activity during incubation with L. reuteri or N. meningitidis, both alone and coincubated. NF-κB activity was induced at 6 h of incubation in response to L. reuteri, alone or coincubated with N. meningitidis, relative to the control (Fig. 5). As a positive control, Pam3CSK4 was used. N. meningitidis did not significantly induce NF-κB activity. As a control, hBD2 expression was validated in transfected cells and showed a similar trend as nontransfected cells (Fig. S3). Further, in the presence of an NF-κB inhibitor L. reuteri no longer induced hBD2 expression (data not shown). These findings suggest that L. reuteri induces hBD2 expression through the NF-κB signaling pathway.
FIG 5.
NF-κB is activated by lactobacilli in a time-dependent manner. Pharyngeal epithelial cells were transfected with the inducible NF-κB reporter plasmid pNiFty and maintained for 2 days before incubation with L. reuteri and N. meningitidis, either alone or coincubated. Pam3CSK4 (Pam) stimulation was used as a positive control. Cell culture supernatants were collected at 2 h, 4 h, and 6 h postincubation. NF-κB activity is expressed relative to the control. Data are represented as the mean values, with error bars representing the standard deviations. The assay was performed in duplicate at least three times. Significance was tested against the control. *, P < 0.05.
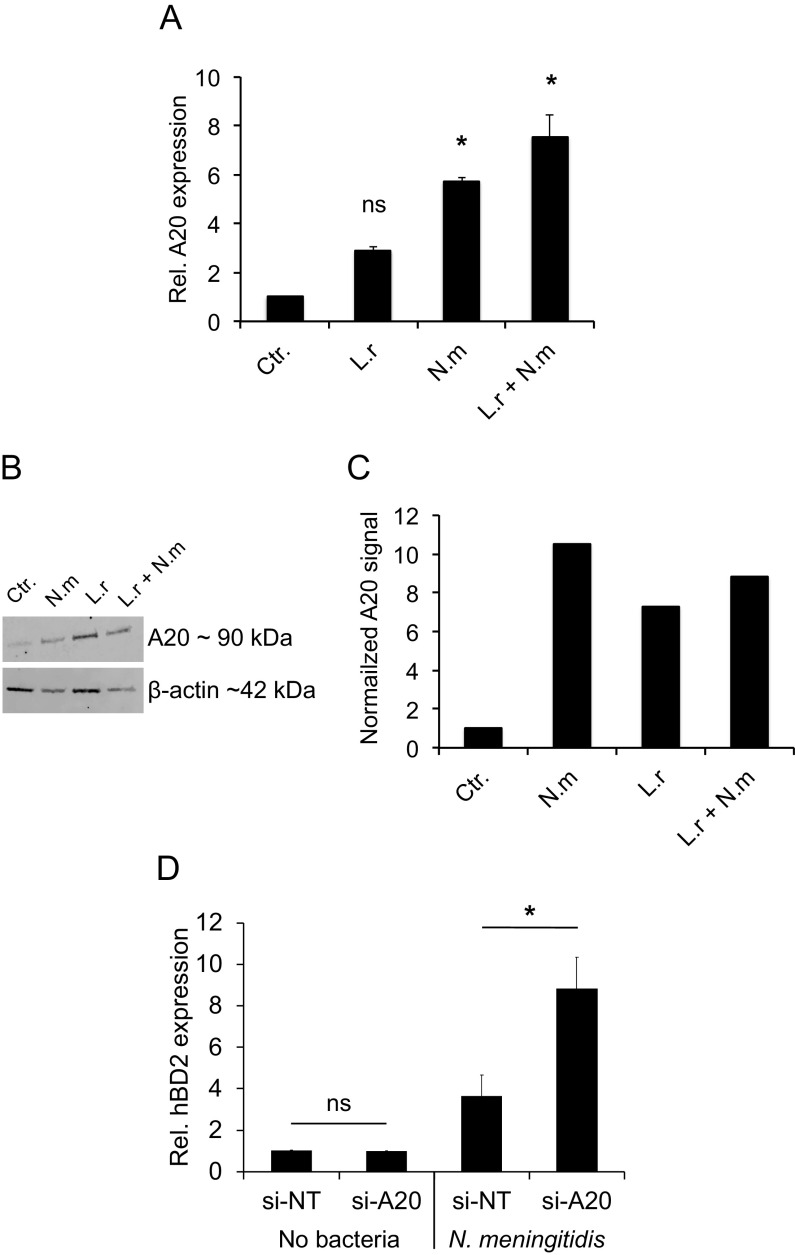
N. meningitidis induces A20 expression.
Simanski et al. recently showed that the skin commensal S. epidermidis is able to permanently colonize the skin by dampening host defenses, including hBD2, through induction of the host molecule A20 (23). A20 is an important signal molecule that negatively regulates NF-κB activation in the host to avoid excessive inflammation (17). Since we observed that NF-κB signaling is involved in L. reuteri-mediated induction of hBD2 expression, we hypothesized that N. meningitidis may also affect hBD2 in an A20-controlled manner. We therefore measured the expression of A20 in response to L. reuteri and N. meningitidis, both alone and coincubated. The expression of A20 in response to L. reuteri was not significantly induced compared to that of the control. However, N. meningitidis infection showed a significant induction of A20 expression (Fig. 6A). Coincubation of L. reuteri and N. meningitidis also induced A20 expression. After incubation with L. reuteri and N. meningitidis, the protein levels of A20 were assessed by Western blotting, which showed results consistent with the expression data (Fig. 6B and C). To investigate A20 expression in relation to bacterial load, we incubated epithelial cells with bacteria at MOIs of 50, 100, and 200. All three concentrations of L. reuteri induced A20 expression to similar levels (Fig. S4A), whereas A20 expression induced by N. meningitidis increased with increasing MOI (Fig. S4B). We next used small interfering RNA (siRNA) to silence A20 in epithelial cells to further verify the involvement of A20 in N. meningitidis-mediated hBD2 reduction. Silencing of A20 led to increased hBD2 expression (Fig. 6D). Thus, these findings suggest that N. meningitidis may exploit A20 signaling to dampen hBD2 expression.
FIG 6.
N. meningitidis induces A20 expression. Pharyngeal epithelial cells were incubated with L. reuteri and N. meningitidis, either alone or coincubated, at an MOI of 100 for 6 h. (A) Gene expression of A20 was quantified using qPCR. Expression was normalized against the β-actin housekeeping gene and expressed as the fold change compared to the control. Data are represented as the mean values, with error bars representing the standard deviations. The assay was performed in triplicate at least three times. Significance was tested against the control. (B) A20 protein level in cell lysates as determined by Western blotting and compared to the housekeeping protein β-actin. (C) Normalized A20 protein level relative to the control. (D) Pharyngeal epithelial cells were transfected with control siRNA (si-NT) or siRNA targeted against A20 (si-A20) for 24 h. The cells were then infected with N. meningitidis at an MOI of 100 for 6 h. Gene expression of hBD2 was quantified using qPCR, normalized against the β-actin housekeeping gene, and expressed as the fold change compared to the control. Data are represented as the mean values, with error bars representing the standard deviations. The assays were performed in triplicate at least three times. Significance was tested against the control or as indicated. *, P < 0.05.
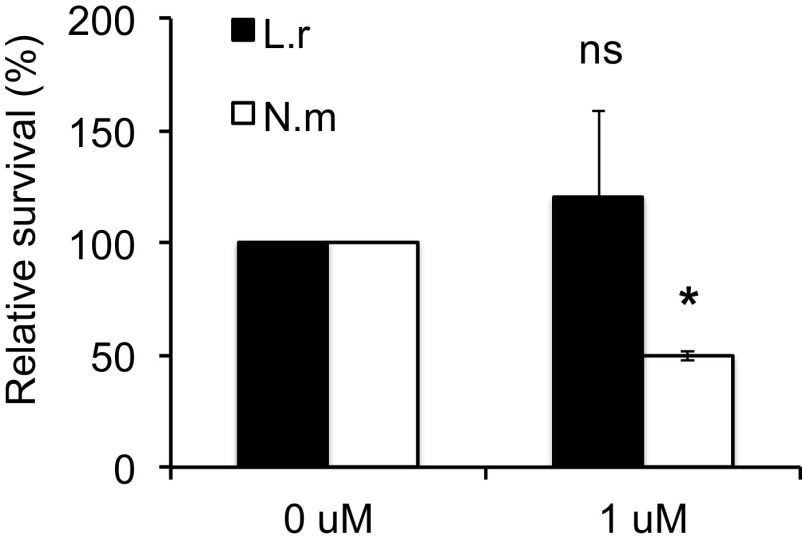
N. meningitidis is more susceptible to hBD2-mediated killing than is L. reuteri.
To understand why N. meningitidis dampens hBD2 expression in epithelial cells, we speculated that N. meningitidis might be susceptible to the killing activity of hBD2. It has been reported that the basal level of hBD2 in lung fluid is 0.5 μM (36), but in inflammatory circumstances it can be higher (37). We therefore used 1 μM hBD2 to compare the susceptibilities of N. meningitidis and L. reuteri to synthetic hBD2 peptide. Treatment of L. reuteri with hBD2 did not alter survival compared to that of the untreated control (Fig. 7). However, N. meningitidis treated with hBD2 showed a significant reduction in bacterial viability. Taken together, the results show that N. meningitidis dampening of hBD2 expression seems to correlate with a higher susceptibility to killing by hBD2 peptide.
FIG 7.
N. meningitidis is susceptible to hBD2-mediated killing. L. reuteri and N. meningitidis were grown to log phase, diluted in modified DMEM, and incubated with 1 μM hBD2 for 2 h. Viable counts were determined by plating. Survival is expressed relative to the untreated control. Data are represented as the mean survival, with error bars representing the standard deviations. The assay was performed in duplicate at least two times. Significance was tested against the untreated control. *, P < 0.05.
DISCUSSION
In this study, we compared the roles of commensal and pathogenic bacteria in the AMP response. In a natural environment, bacteria coexist and compete for nutrients and space (32). Both N. meningitidis and lactobacilli can colonize the oropharyngeal tract of humans and may influence AMP expression dissimilarly. Here, we demonstrate that pharyngeal epithelial cells discriminate between commensal and pathogenic bacteria by differential induction of hBD2. Expression of hBD2 was induced by the Lactobacillus species L. reuteri and L. salivarius. In contrast to lactobacilli, N. meningitidis was not able to induce hBD2 expression. We found that coincubation of L. reuteri together with N. meningitidis reduced hBD2 expression, suggesting that N. meningitidis dampens the response by acting on either lactobacilli or the host cells. We found that N. meningitidis acted on host cells by inducing A20 expression, which is known to reduce NF-κB activity and, in turn, lower hBD2 expression. Since hBD2 was found to be lethal to N. meningitidis, A20 induction might be an evasion mechanism for the pathogen to avoid attack by this AMP.
The closely related species N. gonorrhoeae has previously been shown to reduce the expression of cathelicidin LL-37 (25). Suppression of AMP-encoding genes can represent an escape mechanism to avoid elimination by the immune system and is likely an important feature for bacterial virulence. Previous studies of AMP modulation have also shown discrimination between commensal and pathogenic bacteria. However, in these studies, it was found that pathogenic bacteria induced hBD2 expression to higher levels than commensal bacteria and that coincubation resulted in an additive amplification of hBD2 expression (38). It is known that the modulation of the AMP response by bacteria depends on the bacterial species, the type of host cell, and the specific peptide involved; thus, AMP regulation appears complex and cannot be generalized (39).
We found that L. reuteri-induced hBD2 expression did not increase with increased attachment levels when incubated with an MOI of 25 and above. The data suggest that a minimum contact with host cells is required to induce hBD2 expression since additional adhesion of L. reuteri did not influence the hBD2 response. Furthermore, separating L. reuteri from host cells, using a Transwell filter, did not trigger hBD2 expression, supporting the notion that L. reuteri needs to be in direct contact with host cells. We also found that L. reuteri-conditioned medium did not induce hBD2 expression, indicating that hBD2 induction is not mediated by factors secreted or released by L. reuteri. A similar observation was made previously, where conditioned medium of S. epidermidis and Staphylococcus aureus was unable to induce hBD2 expression (38). The specific mechanism of how L. reuteri interacts with host cells and what components regulate hBD2 expression require further investigation.
Bekele et al. showed that coincubation of N. meningitidis with different Lactobacillus strains reduces inflammatory responses caused by the pathogen (33), suggesting that lactobacilli act in a protective manner toward the host. However, this protective ability was strain specific and dependent on inflammatory stimuli, and lactobacilli possessed adhesion-inhibitory effects against N. meningitidis. In contrast, the data in the current study demonstrated that the presence of N. meningitidis reduced the adhesion of L. reuteri to host cells during coincubation. It is possible that coincubation with meningococci reduced the available surface of host cells to L. reuteri; however, the attachment level of L. reuteri did not influence the hBD2 expression between MOIs of 25 and 200.
While AMPs have many benefits to the human host, their presence has been described as a double-edged sword: AMPs can protect against bacterial infection; however, exaggerated expression can lead to undesirable inflammatory responses (40). Regulation of inflammation is therefore imperative. A20 (or TNFAIP3) is an important host signal molecule that negatively regulates NF-κB activation, thereby limiting excessive inflammation. Although the role of A20 in innate immunity is still unclear, A20 dysregulation has been linked to autoimmune and inflammatory diseases (17). A recent study by Simanski et al. described the induction of A20 by commensal S. epidermidis. A20 attenuated IL-1β (a proinflammatory cytokine) and hBD2 expression, which may help S. epidermidis persist on the skin (23). We speculated that N. meningitidis dampens hBD2 induction by modifying A20 expression. Indeed, the results showed an increase in A20 expression during N. meningitidis infection, which was not found during incubation with L. reuteri. Thus, A20 expression was inversely related to NF-κB activation, as expected. Coincubation resulted in a somewhat higher induction of A20 than with bacteria alone, which did not coincide with the hBD2 induction previously observed during coincubation. However, protein levels of A20 matched hBD2 expression levels better. We found a direct link between meningococcus-mediated induction of A20 and dampening of hBD2 by RNA silencing of A20. Since N. meningitidis could dampen Lactobacillus-induced hBD2 expression, we hypothesized that meningococci might benefit from low hBD2 levels. Indeed, we found that N. meningitidis was killed by the hBD2 peptide, while L. reuteri was not. These results suggest that bacterial interference of A20 signaling may represent an escape mechanism for the pathogen.
Lactobacilli are often used as probiotic treatments. They exhibit numerous benefits, most importantly by inhibiting pathogen colonization and growth (41). Lactobacillus treatment of N. meningitidis-infected mice resulted in improved disease outcome and reduced bacterial dissemination (42). Pathogen-mediated downregulation of AMP expression can be successfully rebalanced by exogenous compounds such as butyrate, a short-chain fatty acid produced by the colon microbiota (24). Whether administration of lactobacilli represents a potential treatment for dysregulation of AMPs is an interesting prospect and requires further investigation. In this work we used N. meningitidis FAM20 of capsular serogroup C. In future studies it would be interesting to assess if meningococcal strains of different origins act the same and also if nonpathogenic Neisseria species affect hBD2 expression.
In conclusion, we show that host epithelial cells discriminate between commensal lactobacilli and pathogenic N. meningitidis: commensal lactobacilli induce hBD2 expression, while N. meningitidis dampens it. We suggest that the dampening of hBD2 is caused by bacterial exploitation of A20 signaling and that this represents a possible evasion mechanism to attenuate host immune responses to allow facilitated bacterial colonization and growth.
MATERIALS AND METHODS
Cell lines and culture conditions.
The human pharyngeal epithelial cell line FaDu (ATCC HBT43) was cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing GlutaMAX and pyruvate (Thermo Fisher Scientific) and supplemented with 10% heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich). Cells were maintained at 37°C and 5% CO2 in a humidified environment. Cells were seeded into 24-well tissue culture plates (Sarstedt) the day before experiments to form a monolayer overnight. Prior to incubation with bacteria, cells were washed two times with serum-free DMEM.
Bacterial strains and culture conditions.
Neisseria meningitidis serogroup C strain FAM20 has been described previously (43). Bacteria were grown on GC agar (Acumedia) supplemented with 1% Kellogg’s supplement (44) for 16 h at 37°C and 5% CO2 in a humidified environment. Lactobacillus salivarius LMG9477 and Lactobacillus reuteri FJ1 ATCC PTA5289, both isolated from human saliva, were grown on Rogosa agar (Oxoid) for 30 to 72 h at 37°C and 5% CO2 in a humidified environment. An overnight culture was grown in MRS broth (Oxoid) for 16 to 18 h at 37°C and 5% CO2 in a humidified environment. Lactobacillus strains were washed once in the appropriate medium before use in experiments.
Incubation of host cells with bacteria.
Epithelial FaDu cells were incubated with bacteria diluted in serum-free DMEM to a multiplicity of infection (MOI) of 10, 25 50, 100, or 200 for 6 h. During the incubation time, bacteria were allowed to adhere for 2 h, after which the cells were washed twice with serum-free DMEM to remove any unbound bacteria. After the last wash, serum-free DMEM was added, and the cells were incubated for an additional 4 h. In each experiment, the MOI was verified by plating. Following incubation, the cell lysate was collected for RNA isolation.
Pretreatment of bacteria.
To compare the effect of live and dead lactobacilli, a bacterial suspension at MOI 100 was incubated at 95°C for 5 min and added to epithelial cells for 6 h. Similar to incubation with whole bacteria, the cells were washed after 2 h, and the incubation was continued for an additional 4 h. The lack of viable bacteria in heat-killed (HK) suspensions was verified by plating. In each experiment, the MOI was verified by plating. Following incubation, the cell lysate was collected for RNA isolation.
Transwell filter experiments.
For experiments in which a separation between lactobacilli and epithelial cells was desired, 0.4-μm hanging cell culture Transwell inserts (Millipore) were used to prevent direct bacterial contact with host cells. Bacteria at an MOI of 100 or control serum-free DMEM was added to the upper compartment of the Transwell system for 6 h. Aliquots from the lower compartment were plated after incubation to verify that bacteria did not pass through the filter. At 2 h postincubation, the cells in the lower compartment were washed twice. The Transwell insert with bacteria was temporarily moved to the side to allow washing of cells. Following incubation, the cell lysate was collected for RNA isolation.
Conditioned medium.
Lactobacillus conditioned medium (CM) was prepared by incubating a suspension of L. reuteri at 3.5 × 107 CFU/ml, used for an MOI of 100, for 6 h at 37°C without shaking, to simulate experimental conditions as much as possible. The bacterial suspension was subsequently sterile filtered using 0.2-μm filters (Filtropur) to remove bacterial cells before addition to epithelial cells. The absence of bacteria was verified by plating. CM was incubated with FaDu epithelial cells for 6 h. Similar to incubation with whole bacteria, the cells were washed after 2 h, and then the incubation was continued for an additional 4 h with CM. After incubation the cell lysate was collected for RNA isolation.
qPCR analysis.
Epithelial cell lysate was collected following incubation of bacteria (or CM) with host cells. RNA was isolated using the RNeasy Plus kit (Qiagen) according to the manufacturer’s instructions. RNA yield and purity were determined using a NanoDrop 8000 UV-visible (UV-Vis) spectrophotometer (Thermo Fisher Scientific). Total RNA (0.2 μg) was reverse transcribed to cDNA using SuperScript VILO master mix (Invitrogen). Quantitative PCR was performed using a LightCycler 480 real-time PCR system (Roche) and the SYBR Green I Master kit (Roche). Expression of the β-actin, hBD1, hBD2, and A20 genes was detected using the primers listed in Table 1. The thermal cycling program was as follows: initial denaturation at 95°C for 10 min followed by amplification for 40 cycles with denaturation at 95°C for 10 s, annealing at 55°C for 20 s, and extension at 72°C for 20 s. The melting-curve analysis was as follows: 95°C for 5 s, 65°C for 1 min, and then increasing to 95°C at 0.08°C/s. The expression was normalized against the β-actin housekeeping gene. The expression levels were calculated using the comparative threshold cycle method (ΔΔCT method) and expressed as the fold change compared to that in control cells.
TABLE 1.
Primers used in this study
| Gene | Primer name | Sequence (5′–3′) |
|---|---|---|
| bAct | bAct_fwd | CATGCCATCCTGCGTCTGGACC |
| bAct_rev | ACATGGTGGTGCCGCCAGACAG | |
| hBD1 | hBD1_fwd | ATGGCCTCAGGTGGTAACTTTC |
| hBD1_rev | CACTTGGCCTTCCCTCTGTAAC | |
| hBD2 | hBD2_fwd | CCAGCCATCAGCCATGAGGGT |
| hBD2_rev | GGAGCCCTTTCTGAATCCGCA | |
| A20 | A20_fwd | GAGAGCACAATGGCTGAACA |
| A20_rev | TCCAGTGTGTATCGGTGCAT |
Bacterial adhesion to host cells.
Epithelial cells were incubated with N. meningitidis and L. reuteri at an MOI of 100 for 6 h and washed to remove unbound bacteria after 2 h. After incubation, host cells were washed 3 times with serum-free DMEM and treated with 1% saponin (Sigma-Aldrich) in serum-free DMEM for 10 min. Serial dilutions were plated on GC agar, and the adherence was determined from the viable counts.
NF-κB activity.
The activity of NF-κB was determined by an NF-κB-inducible reporter plasmid. Prior to transfection, epithelial cells were washed two times with serum-free DMEM. Epithelial cells at 80 to 90% confluence were transfected with 500 ng of the pNiFty-SEAP plasmid (InvivoGen) using 1 μl of Lipofectamine 2000 transfection reagent (Invitrogen). The cells were incubated at 37°C and 5% CO2 for 6 h, after which the cell culture medium was replaced with DMEM containing 10% FBS. The cells were further cultured for 2 days before incubation with N. meningitidis and L. reuteri. Pam3CSK4 (5 μg/ml; InvivoGen) was used as a positive control for NF-κB activation. Supernatants were collected after 2 h, 4 h, and 6 h of incubation. Quantification of the reporter plasmid was performed by incubating the supernatant with QUANTI-Blue (InvivoGen) for 6 h at 37°C and 5% CO2. The absorbance of the mixture was measured using a Spectramax i3x microplate reader (Molecular Devices) at 620 nm. NF-κB activity is expressed relative to that of the untreated control.
Western blot analysis.
Following incubation with N. meningitidis and L. reuteri at an MOI of 100 for 6 h, epithelial cells were washed twice with phosphate-buffered saline (PBS) and placed on ice. Cells were lysed with 200 μl of radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific) containing protease inhibitor cocktail cOmplete (Roche). Samples were mixed at a 2:1 ratio with sample buffer (63 mM Tris-HCl [pH 6.8], 25% glycerol, 1% SDS, 5% 2-mercaptoethanol) and boiled at 95°C for 5 min before loading of 20 µl on 4 to 20% Mini-PROTEAN TGX precast protein gels (Bio-Rad). After separation, proteins were transferred to an Immobilon-P polyvinylidene difluoride (PVDF) membrane (Millipore) using a semidry transfer system (Bio-Rad). The membrane was washed in water and blocked for 1 h in 5% skim milk powder (Sigma-Aldrich) in PBS at room temperature. The membrane was incubated overnight at 4°C with an antibody against A20 (Thermo Fisher Scientific; 59A426, 1:500 dilution) in 0.5% skim milk powder in PBS. After being washed three times with PBS, the membrane was incubated with IRDye800CW-conjugated goat anti-mouse antibody (LI-COR; 1:10,000 dilution) for 45 min at room temperature. Bands were visualized using an Odyssey IR scanner (LI-COR). The membrane was stripped with Restore stripping buffer (Thermo Fisher Scientific), blocked, incubated with an antibody against β-actin (Sigma-Aldrich; MAB1501, 1:2,000 dilution), washed, and incubated with the secondary antibody as described above.
RNA silencing.
Epithelial cells were seeded in 24-well plates 24 h prior to transfection. Transfection was carried out using HiPerfect transfection reagent (Qiagen) according to the manufacturer’s recommendation. Cells were washed twice with serum-free DMEM and then treated with SilenceSelect A20-targeting siRNA (ID s14259) and nontargeting siRNA (siRNA 4390844) as a control (Ambion; final concentration 10 nM). After 24 h of incubation with siRNA at 37°C and 5% CO2 in a humidified environment, cells were washed twice with serum-free DMEM to remove any extracellular siRNA. The cells were then used in infection experiments and for quantitative PCR (qPCR) analysis of hBD2 as described in the sections above. The efficiency of knockdown reached 25% was determined using qPCR.
Susceptibility to hBD2.
To determine the susceptibility of L. reuteri and N. meningitidis to hBD2, bacteria were grown to mid-log phase (optical density [OD] ≈ 0.6 and OD ≈ 0.4, respectively) in GC broth supplemented with 1% Kellogg’s supplement at 37°C and 5% CO2. Bacteria were diluted to 2 × 106 CFU/ml in modified DMEM (DMEM diluted 4× in sterile water) and mixed with synthetic hBD2 peptide (Innovagen). Fifty microliters of bacterial suspension was mixed with 10 μl of hBD2 (final concentration, 1 μM) and incubated in polypropylene tubes for 2 h at 37°C and 5% CO2 under shaking conditions. Sterile water instead of hBD2 served as the control. Bacterial viability was assessed by plating, and colonies were counted the next day. Bacterial survival is expressed relative to that of the untreated control.
Statistical analysis.
Testing of statistical significance was performed in GraphPad Prism (version 5). Two-tailed and unpaired Student’s t tests were used to compare differences between two groups. Analysis of variance (ANOVA) with Bonferroni’s post hoc test was used to compare differences between multiple groups. P values below 0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Swedish Research Council Dnr (2016-10279, 2019-01355) and the Swedish Cancer Society (grants to A.-B.J.).
The funders had no role in the study design, data collection, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Tauszig S, Jouanguy E, Hoffmann JA, Imler JL. 2000. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc Natl Acad Sci U S A 97:10520–10525. doi: 10.1073/pnas.180130797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 3.Hancock RE, Scott MG. 2000. The role of antimicrobial peptides in animal defenses. Proc Natl Acad Sci U S A 97:8856–8861. doi: 10.1073/pnas.97.16.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tjabringa GS, Ninaber DK, Drijfhout JW, Rabe KF, Hiemstra PS. 2006. Human cathelicidin LL-37 is a chemoattractant for eosinophils and neutrophils that acts via formyl-peptide receptors. Int Arch Allergy Immunol 140:103–112. doi: 10.1159/000092305. [DOI] [PubMed] [Google Scholar]
- 5.Mookherjee N, Hamill P, Gardy J, Blimkie D, Falsafi R, Chikatamarla A, Arenillas DJ, Doria S, Kollmann TR, Hancock RE. 2009. Systems biology evaluation of immune responses induced by human host defence peptide LL-37 in mononuclear cells. Mol Biosyst 5:483–496. doi: 10.1039/b813787k. [DOI] [PubMed] [Google Scholar]
- 6.Selsted ME, Tang YQ, Morris WL, McGuire PA, Novotny MJ, Smith W, Henschen AH, Cullor JS. 1993. Purification, primary structures, and antibacterial activities of beta-defensins, a new family of antimicrobial peptides from bovine neutrophils. J Biol Chem 268:6641–6648. doi: 10.1016/S0021-9258(18)53298-1. [DOI] [PubMed] [Google Scholar]
- 7.O’Neil DA, Porter EM, Elewaut D, Anderson GM, Eckmann L, Ganz T, Kagnoff MF. 1999. Expression and regulation of the human beta-defensins hBD-1 and hBD-2 in intestinal epithelium. J Immunol 163:6718–6724. [PubMed] [Google Scholar]
- 8.Ali RS, Falconer A, Ikram M, Bissett CE, Cerio R, Quinn AG. 2001. Expression of the peptide antibiotics human beta defensin-1 and human beta defensin-2 in normal human skin. J Invest Dermatol 117:106–111. doi: 10.1046/j.0022-202x.2001.01401.x. [DOI] [PubMed] [Google Scholar]
- 9.Harder J, Bartels J, Christophers E, Schroder JM. 2001. Isolation and characterization of human beta -defensin-3, a novel human inducible peptide antibiotic. J Biol Chem 276:5707–5713. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- 10.Zhao C, Wang I, Lehrer RI. 1996. Widespread expression of beta-defensin hBD-1 in human secretory glands and epithelial cells. FEBS Lett 396:319–322. doi: 10.1016/0014-5793(96)01123-4. [DOI] [PubMed] [Google Scholar]
- 11.Harder J, Bartels J, Christophers E, Schroder JM. 1997. A peptide antibiotic from human skin. Nature 387:861. doi: 10.1038/43088. [DOI] [PubMed] [Google Scholar]
- 12.Chadebech P, Goidin D, Jacquet C, Viac J, Schmitt D, Staquet MJ. 2003. Use of human reconstructed epidermis to analyze the regulation of beta-defensin hBD-1, hBD-2, and hBD-3 expression in response to LPS. Cell Biol Toxicol 19:313–324. doi: 10.1023/B:CBTO.0000004975.36521.c8. [DOI] [PubMed] [Google Scholar]
- 13.Schlee M, Harder J, Koten B, Stange EF, Wehkamp J, Fellermann K. 2008. Probiotic lactobacilli and VSL#3 induce enterocyte beta-defensin 2. Clin Exp Immunol 151:528–535. doi: 10.1111/j.1365-2249.2007.03587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDermott AM, Redfern RL, Zhang B, Pei Y, Huang L, Proske RJ. 2003. Defensin expression by the cornea: multiple signalling pathways mediate IL-1beta stimulation of hBD-2 expression by human corneal epithelial cells. Invest Ophthalmol Vis Sci 44:1859–1865. doi: 10.1167/iovs.02-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pistolic J, Cosseau C, Li Y, Yu JJ, Filewod NC, Gellatly S, Rehaume LM, Bowdish DM, Hancock RE. 2009. Host defence peptide LL-37 induces IL-6 expression in human bronchial epithelial cells by activation of the NF-kappaB signaling pathway. J Innate Immun 1:254–267. doi: 10.1159/000171533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. 2000. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science 289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catrysse L, Vereecke L, Beyaert R, van Loo G. 2014. A20 in inflammation and autoimmunity. Trends Immunol 35:22–31. doi: 10.1016/j.it.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Krikos A, Laherty CD, Dixit VM. 1992. Transcriptional activation of the tumor necrosis factor alpha-inducible zinc finger protein, A20, is mediated by kappa B elements. J Biol Chem 267:17971–17976. doi: 10.1016/S0021-9258(19)37138-8. [DOI] [PubMed] [Google Scholar]
- 19.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A. 2004. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol 5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 20.Garg AV, Ahmed M, Vallejo AN, Ma A, Gaffen SL. 2013. The deubiquitinase A20 mediates feedback inhibition of interleukin-17 receptor signaling. Sci Signal 6:ra44. doi: 10.1126/scisignal.2003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hitotsumatsu O, Ahmad RC, Tavares R, Wang M, Philpott D, Turer EE, Lee BL, Shiffin N, Advincula R, Malynn BA, Werts C, Ma A. 2008. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity 28:381–390. doi: 10.1016/j.immuni.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. 2004. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 23.Simanski M, Erkens AS, Rademacher F, Harder J. 2019. Staphylococcus epidermidis-induced interleukin-1 beta and human beta-defensin-2 expression in human keratinocytes is regulated by the host molecule A20 (TNFAIP3). Acta Derm Venereol 99:181–187. doi: 10.2340/00015555-3073. [DOI] [PubMed] [Google Scholar]
- 24.Islam D, Bandholtz L, Nilsson J, Wigzell H, Christensson B, Agerberth B, Gudmundsson G. 2001. Downregulation of bactericidal peptides in enteric infections: a novel immune escape mechanism with bacterial DNA as a potential regulator. Nat Med 7:180–185. doi: 10.1038/84627. [DOI] [PubMed] [Google Scholar]
- 25.Bergman P, Johansson L, Asp V, Plant L, Gudmundsson GH, Jonsson AB, Agerberth B. 2005. Neisseria gonorrhoeae downregulates expression of the human antimicrobial peptide LL-37. Cell Microbiol 7:1009–1017. doi: 10.1111/j.1462-5822.2005.00530.x. [DOI] [PubMed] [Google Scholar]
- 26.Pace D, Pollard AJ. 2012. Meningococcal disease: clinical presentation and sequelae. Vaccine 30(Suppl 2):B3–B9. doi: 10.1016/j.vaccine.2011.12.062. [DOI] [PubMed] [Google Scholar]
- 27.Christensen H, May M, Bowen L, Hickman M, Trotter CL. 2010. Meningococcal carriage by age: a systematic review and meta-analysis. Lancet Infect Dis 10:853–861. doi: 10.1016/S1473-3099(10)70251-6. [DOI] [PubMed] [Google Scholar]
- 28.Behera SS, Ray RC, Zdolec N. 2018. Lactobacillus plantarum with functional properties: an approach to increase safety and shelf-life of fermented foods. Biomed Res Int 2018:9361614. doi: 10.1155/2018/9361614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badet C, Thebaud NB. 2008. Ecology of lactobacilli in the oral cavity: a review of literature. Open Microbiol J 2:38–48. doi: 10.2174/1874285800802010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valeur N, Engel P, Carbajal N, Connolly E, Ladefoged K. 2004. Colonization and immunomodulation by Lactobacillus reuteri ATCC 55730 in the human gastrointestinal tract. Appl Environ Microbiol 70:1176–1181. doi: 10.1128/aem.70.2.1176-1181.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cadieux PA, Burton J, Devillard E, Reid G. 2009. Lactobacillus by-products inhibit the growth and virulence of uropathogenic Escherichia coli. J Physiol Pharmacol 60(Suppl 6):13–18. [PubMed] [Google Scholar]
- 32.Neal-McKinney JM, Lu X, Duong T, Larson CL, Call DR, Shah DH, Konkel ME. 2012. Production of organic acids by probiotic lactobacilli can be used to reduce pathogen load in poultry. PLoS One 7:e43928. doi: 10.1371/journal.pone.0043928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bekele T, Keith P, Adelina R, Vyvyan S, Victoria D. 2014. Oral Lactobacillus plantarum NCIMB 8825 inhibits adhesion, invasion and metabolism of Neisseria meningitidis serogroup B and affords anti-inflammatory and cytotoxic protection to nasopharyngeal epithelial cells. Adv Microbiol 4:81–93. doi: 10.4236/aim.2014.42013. [DOI] [Google Scholar]
- 34.Schlee M, Wehkamp J, Altenhoefer A, Oelschlaeger TA, Stange EF, Fellermann K. 2007. Induction of human beta-defensin 2 by the probiotic Escherichia coli Nissle 1917 is mediated through flagellin. Infect Immun 75:2399–2407. doi: 10.1128/IAI.01563-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krisanaprakornkit S, Kimball JR, Weinberg A, Darveau RP, Bainbridge BW, Dale BA. 2000. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect Immun 68:2907–2915. doi: 10.1128/iai.68.5.2907-2915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaller-Bals S, Schulze A, Bals R. 2002. Increased levels of antimicrobial peptides in tracheal aspirates of newborn infants during infection. Am J Respir Crit Care Med 165:992–995. doi: 10.1164/ajrccm.165.7.200110-020. [DOI] [PubMed] [Google Scholar]
- 37.Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, Ganz T, Gallo RL, Leung DY. 2002. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N Engl J Med 347:1151–1160. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- 38.Wanke I, Steffen H, Christ C, Krismer B, Gotz F, Peschel A, Schaller M, Schittek B. 2011. Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J Invest Dermatol 131:382–390. doi: 10.1038/jid.2010.328. [DOI] [PubMed] [Google Scholar]
- 39.Wassing GM, Bergman P, Lindbom L, van der Does AM. 2015. Complexity of antimicrobial peptide regulation during pathogen-host interactions. Int J Antimicrob Agents 45:447–454. doi: 10.1016/j.ijantimicag.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Lai Y, Gallo RL. 2009. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol 30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kemgang TS, Kapila S, Shanmugam VP, Kapila R. 2014. Cross-talk between probiotic lactobacilli and host immune system. J Appl Microbiol 117:303–319. doi: 10.1111/jam.12521. [DOI] [PubMed] [Google Scholar]
- 42.Belkacem N, Bourdet-Sicard R, Taha MK. 2018. Lactobacillus paracasei feeding improves the control of secondary experimental meningococcal infection in flu-infected mice. BMC Infect Dis 18:167. doi: 10.1186/s12879-018-3086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahman M, Kallstrom H, Normark S, Jonsson AB. 1997. PilC of pathogenic Neisseria is associated with the bacterial cell surface. Mol Microbiol 25:11–25. doi: 10.1046/j.1365-2958.1997.4601823.x. [DOI] [PubMed] [Google Scholar]
- 44.Kellogg DS, Jr, Cohen IR, Norins LC, Schroeter AL, Reising G. 1968. Neisseria gonorrhoeae. II. Colonial variation and pathogenicity during 35 months in vitro. J Bacteriol 96:596–605. doi: 10.1128/JB.96.3.596-605.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.