Discovering new Gram-negative antibiotics has been a challenge for decades. This has been largely attributed to a limited understanding of the molecular descriptors governing Gram-negative permeation and efflux evasion.
KEYWORDS: efflux pumps, molecular descriptors, machine learning, Gram-negative bacteria
ABSTRACT
Discovering new Gram-negative antibiotics has been a challenge for decades. This has been largely attributed to a limited understanding of the molecular descriptors governing Gram-negative permeation and efflux evasion. Herein, we address the contribution of efflux using a novel approach that applies multivariate analysis, machine learning, and structure-based clustering to some 4,500 molecules (actives) from a small-molecule screen in efflux-compromised Escherichia coli. We employed principal-component analysis and trained two decision tree-based machine learning models to investigate descriptors contributing to the antibacterial activity and efflux susceptibility of these actives. This approach revealed that the Gram-negative activity of hydrophobic and planar small molecules with low molecular stability is limited to efflux-compromised E. coli. Furthermore, molecules with reduced branching and compactness showed increased susceptibility to efflux. Given these distinct properties that govern efflux, we developed the first efflux susceptibility machine learning model, called Susceptibility to Efflux Random Forest (SERF), as a tool to analyze the molecular descriptors of small molecules and predict those that could be susceptible to efflux pumps in silico. Here, SERF demonstrated high accuracy in identifying such molecules. Furthermore, we clustered all 4,500 actives based on their core structures and identified distinct clusters highlighting side-chain moieties that cause marked changes in efflux susceptibility. In all, our work reveals a role for physicochemical and structural parameters in governing efflux, presents a machine learning tool for rapid in silico analysis of efflux susceptibility, and provides a proof of principle for the potential of exploiting side-chain modification to design novel antimicrobials evading efflux pumps.
INTRODUCTION
Remarkably, no new classes of Gram-negative antibiotics of clinical importance have been discovered since the quinolones, dating back to the early 1960s (1). In the same period, Gram-negative superbugs resistant to existing antibiotics have become a pervasive public health concern (2). To address the growing need for new antibiotics, the World Health Organization (WHO) published a list of bacterial pathogens to guide research and development, highlighting Gram-negative pathogens as a critical priority (3). The Centers for Disease Control and Prevention recently revealed that Gram-negative pathogens were responsible for ∼50% of antibiotic-resistant microbial infections in the United States (4). A recent analysis of the clinical pipeline revealed 50 antibiotics in development, of which only 12 are active against some of the priority Gram-negative pathogens identified by the WHO (5, 6). Of these 12 agents, only murepavadin is considered new, with a novel pharmacophore, target, and mode of action. Most recently, and unfortunately, nephrotoxicity concerns have halted its development (5, 6). Thus, the clinical pipeline is currently devoid of new chemical matter to treat the most troublesome infections caused by Gram-negative pathogens. Herein, we examine one of the most important impediments to identifying leads for new antibiotics targeting Gram-negative bacteria, namely, the physicochemical and structural parameters that delineate compound efflux.
It is widely accepted among pundits of antibacterial drug discovery that intrinsic resistance mechanisms, the outer membrane barrier and active efflux pumps, have presented the greatest challenge to the development of new Gram-negative antibiotics (7–9). Indeed, there have been a growing number of calls for research to understand the physicochemical properties that facilitate compound entry and efflux avoidance (10–12). Inspiration comes from success correlating a drug’s physicochemical properties with its pharmacokinetics, where Lipinski mapped an ideal physicochemical space for orally available drugs, establishing the rule of five: molecular weight (MW) of (<500 g mol−1), lipophilicity (clogP) of ≤5, number of hydrogen bond donors of ≤10, and number of acceptors of ≤5 (13). Several analyses have shown, nevertheless, that antibacterial compounds are exceptional in this context. In one of the first reports applying these properties to trends in antimicrobials, O’Shea and Moser showed that Gram-negative antibiotics, on average, are slightly larger (MW of 414 g mol−1) and more hydrophilic (clogD of −2.8) than nonantibiotic drugs (338 g mol−1; clogD of 1.6) (14). When Gram-negative antibiotics were classified by their target location, those with a cytoplasmic target were smaller (254 to 465 g mol−1) and more hydrophobic (clogD of −1.4 to 1.1) than antibiotics with a periplasmic target (347 to 558 g mol−1; clogD of −5.1 to −1) (15). Although these studies suggest the potential of a physicochemical space for Gram-negative activity (14, 15), they include antibiotics that are susceptible to efflux pumps of the resistance-nodulation-division (RND) superfamily, composed of an inner membrane transporter, a periplasmic adaptor protein, and an outer membrane channel (16, 17). These tripartite pumps extrude a broad range of molecules from the inner membrane and periplasmic space to the extracellular milieu (17, 18). In an analysis of several high-throughput screening campaigns at AstraZeneca, Brown et al. showed that small (<300 g mol−1) and hydrophilic (clogD of <0) or very large (>700 g mol−1) and zwitterionic compounds were least susceptible to RND efflux pumps (19). Compared to the Moser analysis of the physicochemical space occupied by Gram-negative antibiotics (15), these results suggest that antimicrobials with a cytoplasmic target may be more susceptible to efflux than others. A recent study measuring compound accumulation in Gram-negative bacteria, however, has challenged some of these properties, showing that hydrophobic and primary amine-containing compounds could overcome the Gram-negative permeability barrier (20). Interestingly, these compounds were also rigid, with low globularity. Furthermore, Copper et al. (21) observed that properties governing antibiotic activity differ among Gram-negative bacteria: for example, electrostatic nature and surface area were key contributors to activity in Pseudomonas aeruginosa, while topology and atom or bond count governed activity in Escherichia coli. In all, these studies highlight the complexity of designing new Gram-negative antibacterial agents that evade efflux pumps, where no singular property can predict whether a molecule will be susceptible to efflux. In this context, both clogD and the pKa of some side-chain moieties decorating pyridopyrimidine inhibitors of biotin carboxylase of Gram-negative bacteria have been shown to impact their antibacterial activity and efflux susceptibility (22). These results suggest that “variable” properties such as hydrophobicity are best taken alongside other physicochemical properties in order to understand overall Gram-negative activity. As such, we hypothesize that combinations of properties could feasibly be derived from phenotypic chemical screens of efflux substrates and that efflux could be predicted in a similar manner to accumulation (20) by machine learning approaches.
In the work described herein, we have taken a novel approach that applies multivariate analysis, machine learning, and structure-based clustering of growth inhibitors of E. coli to identify molecular descriptors and structure-activity relationships that impact efflux susceptibility. The effort defines new physicochemical and structural parameters that contribute to the antibacterial activity and efflux susceptibility of small molecules targeting this model Gram-negative bacterium.
RESULTS
A screen for growth inhibitors of an efflux-deficient strain of E. coli.
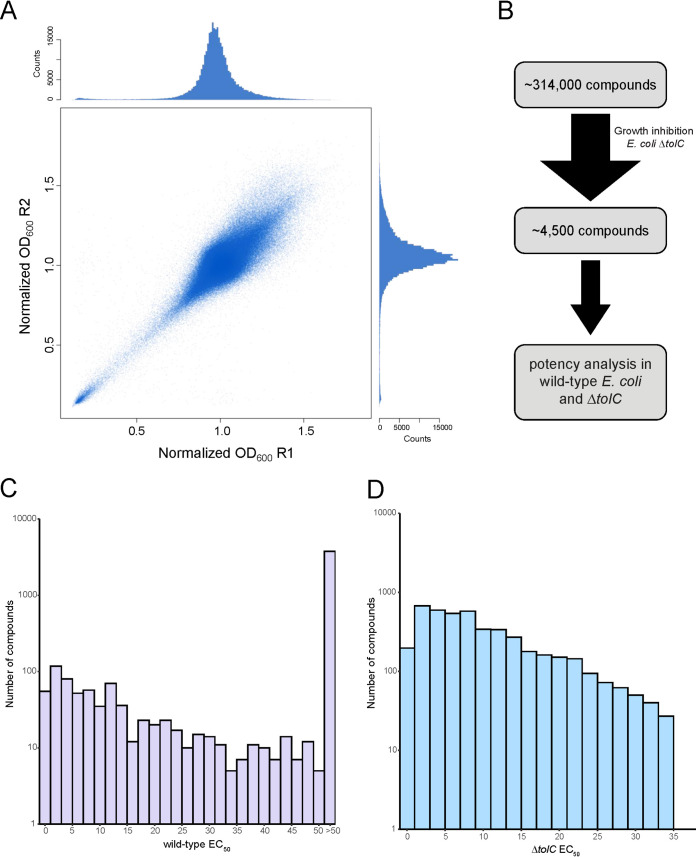
Our work began with a high-throughput screen to identify compounds with growth-inhibitory activity in the efflux-deficient E. coli ΔtolC mutant strain. The screen comprised ∼314,000 molecules, which were largely synthetic, and included a collection of ∼3,900 previously approved drugs and bioactive molecules (bioactive collection). The data were reproducible and defined a distinct collection of growth inhibitors in the primary screen (Fig. 1A). Indeed, ∼4,500 molecules (actives) exhibited strong growth inhibition (≤3σ below the mean [Fig. 1B]), of which 386 were from the bioactive collection. The potency (50% effective concentration [EC50]) of each active was measured in the wild-type E. coli strain and its ΔtolC mutant (Fig. 1C and D; see Fig S1 and Table S1 in the supplemental material). Of the 4,500 actives, a large fraction, some 84% (3,780/4,500), showed little activity at the highest concentration (50 μM) tested using wild-type E. coli (Fig. 1C), while the EC50 using the ΔtolC strain was less than 35 μM (Fig. 1D). This suggested that many of our actives were susceptible to efflux, where their antibacterial activity was abolished in the presence of efflux pumps. Among these molecules were antibiotics from the bioactive collection, which include β-lactams, macrolides, and other compounds, such as doxorubicin, known to have antibiotic activity and susceptibility to efflux (Table S1). This was an encouraging result from a data quality perspective given the previous literature on their susceptibility to efflux pumps in Gram-negative bacteria (18, 23–25).
FIG 1.
Primary small-molecule screen and potency analysis of actives. (A) Replicate plot of the primary screen of ∼314,000 small molecules in the efflux-deficient mutant strain E. coli BW25113 ΔtolC. Data were normalized as per Mangat et al. (47) and show good reproducibility. Density distributions are depicted in secondary plots for each of the two replicates. (B) From this primary screen, ∼4,500 actives exhibited strong growth inhibition (≤3σ below the mean [a molecule with a normalized mean OD value of ∼0.5 or less was deemed a growth inhibitor]) against the mutant strain E. coli BW25113 ΔtolC. The potency of these 4,500 actives was measured in wild-type E. coli BW25113 and the mutant strain E. coli BW25113 ΔtolC. A histogram shows the measured potency values obtained from dose-response analyses in (C) wild-type E. coli BW25113 and (D) the mutant strain E. coli BW25113 ΔtolC.
Hydrophobicity, level of saturation, and molecular stability contribute to Gram-negative activity in efflux-compromised E. coli.
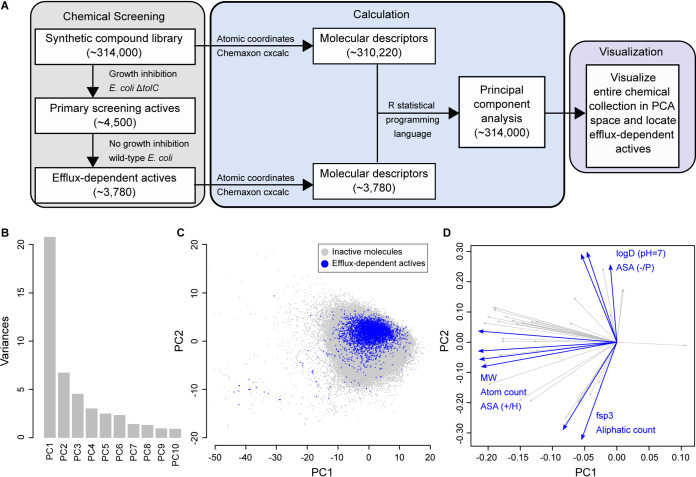
Next, we sought to identify the molecular descriptors for this set of 3,780 efflux-dependent active compounds. To this end, we calculated some 50 descriptors for all 314,000 compounds screened, which include efflux-dependent actives (Table S1), resulting in more than 15 million data points (Fig. 2A). To reduce the dimensionality of these data, we initially applied a principal-component analysis (PCA). The first three principal components of the PCA explained ∼63% of the variances between the molecular descriptors of efflux-dependent actives and those of non-growth-inhibitory (inactive) molecules in the ΔtolC mutant (Fig. 2B). Furthermore, the PCA identified a structural and physicochemical “pocket” that defines the antibacterial activity of the 3,780 efflux-dependent actives (Fig. 2C). The clustering of these actives near the origin (Fig. 2C) was largely due to molecular size, hydrophobicity, and molecular complexity, as observed by the eigenvectors in principal component 1 (PC1) and PC2 (Fig. 2D). Notably, PC1 uniquely comprised molecular size, which was represented by molecular weight (MW), the total atom count, and the solvent accessible surface area (ASA), as calculated by using the radius of the solvent. Particularly, ASA contained components of the surface area of partial positive charge (ASA+) and partial negative charge (ASA−). PC2, on the other hand, consisted of both hydrophobicity and molecular complexity. The former was represented by clogD (pH 7.0) and the ASA of polar atoms (ASA_P) and hydrophobic atoms (ASA_H), while the latter was described by the number of aliphatic atoms and the fraction of sp3 hybridized carbon atoms (Fsp3). Overall, the PCA results suggest that combinations of these nine molecular descriptors addressing molecular size, hydrophobicity, and molecular complexity contribute to Gram-negative activity in the ΔtolC mutant.
FIG 2.
Principal-component analysis of molecular descriptors of efflux-dependent actives and non-growth-inhibitory molecules from the primary screen. (A) Molecular descriptors for each efflux-dependent active and the remaining screened compounds were calculated. The former set and all non-growth-inhibitory molecules from the primary screen (inactive molecules) were visualized using a principal-component analysis (PCA). (B) Ten principal components (PC) explaining variances between descriptors of efflux-dependent actives and those of inactive molecules are shown. The first 3 principal components, PC1, PC2, and PC3, explain ∼63% of these variances. (C) For visualization, the first two principal component scores are presented, where efflux-dependent actives occupied a distinct chemical space (blue) among the inactive molecules (gray). (D) The loading eigenvectors for this PCA plot are shown. The molecular descriptors contributing the most to the scores of PC1 and PC2 are highlighted in blue: molecular weight (MW), atom count, accessible surface area (ASA) of partial negative charge (−), polar atoms (P), partial positive charge (+), hydrophobic atoms (H), the fraction of sp3 hybridized carbon atoms (Fsp3), and aliphatic count. Descriptors with lower contributions are shown in gray.
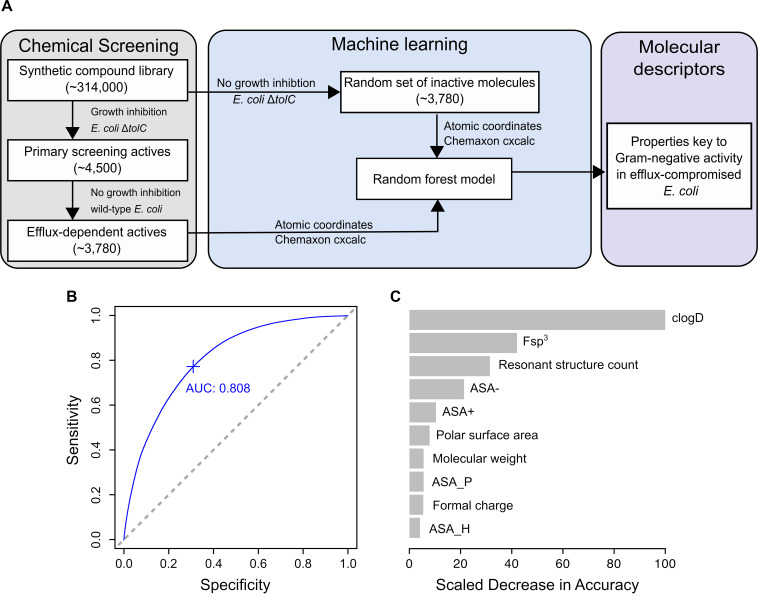
Given that the PCA highlighted some distinct molecular descriptors governing the antibacterial activity of efflux-dependent actives, we implemented a tree-based machine learning approach to quantify descriptors that restrict this activity to efflux-compromised E. coli. Herein, we developed a random forest classification model for the set of 3,780 efflux-dependent actives and 3,780 inactive molecules, randomly chosen from the primary screen (Fig. 3A). To eliminate bias from the PCA, all 50 molecular descriptors for each of these molecules were used to train the model, which achieved an area under the curve-receiver operating characteristic curve (AUC-ROC) of 0.808 (Fig. 3B; see Fig. S2 in the supplemental material). This illustrates a good measure of the model’s performance in classifying molecules with antibacterial activity in the ΔtolC mutant and those without antibacterial activity. The analysis revealed similar descriptors to that of the PCA (Fig. 2D and 3C); indeed, the model indicated that clogD (pH 7.0), Fsp3, and the resonant structure count (RSC) of a molecule were the top three factors driving Gram-negative activity of small molecules in efflux-compromised E. coli (Fig. 3C). Particularly, efflux-dependent actives were hydrophobic (clogD of 1 to 5), planar (Fsp3 of 0 to 0.5), and had low molecular stability (as measured by the resonant structure count at pH 7.0 of <4 [see Fig. S3 in the supplemental material]). Since these actives lost their antibacterial activity in efflux-proficient wild-type E. coli, these results suggest that the Gram-negative activity of molecules with such properties would be abolished due to efflux pumps. Of note, in the absence of clogD from the 50 molecular descriptors used to train this model, the model lost accuracy in classifying molecules with antibacterial activity in the ΔtolC mutant and those without activity (Fig. 3C). This highlights the importance of hydrophobicity as a physicochemical property that affects Gram-negative activity, regardless of efflux pumps.
FIG 3.
A machine learning approach identifies key molecular descriptors for Gram-negative activity of efflux-dependent actives. (A) Molecular descriptors for the 3,780 efflux-dependent actives and a random set of 3,780 inactive molecules (no growth inhibition in E. coli BW25113 ΔtolC) from the primary screen were used to train a random forest model to examine descriptors contributing to Gram-negative activity in efflux-compromised E. coli. (B) The area under the curve-receiver operating characteristic curve (AUC-ROC) plot for the random forest model is 0.808, showing a good distinction between efflux-dependent actives and inactive molecules. Sensitivity refers to the true-positive rate of the model, while specificity refers to its false-positive rate. (C) The top 10 molecular descriptors that reduce the model’s accuracy are shown, with clogD, Fsp3 (fraction of sp3 hybridized carbon atoms), and resonant structure count topping the list.
A random forest model predicts efflux-prone small molecules.
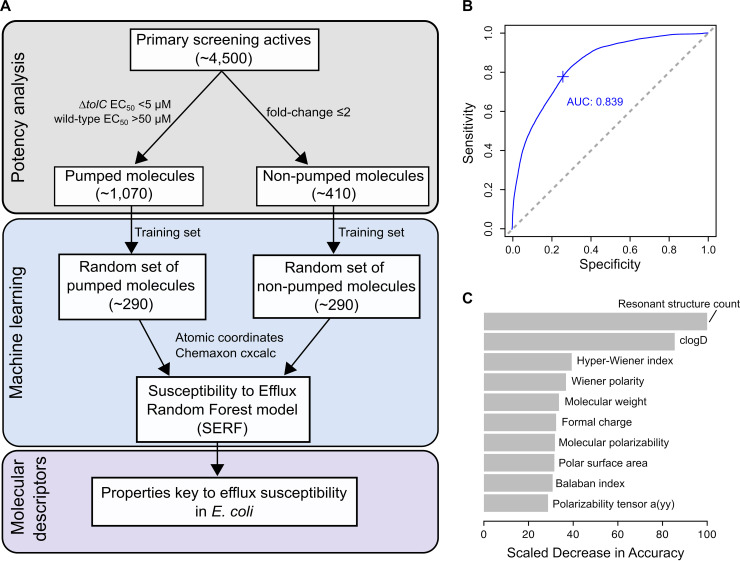
Consistent with the goal to understand efflux susceptibility in E. coli, we focused our analysis on determining key molecular descriptors that contribute to efflux susceptibility. First, we categorized the 4,500 actives into efflux-susceptible (pumped) molecules and unsusceptible (nonpumped) molecules based on their potency and calculated fold change in potency (wild-type EC50/ΔtolC EC50). Actives classified as pumped molecules were potent in the E. coli ΔtolC mutant (EC50 of <5 μM) and lacked antibacterial activity in the wild-type strain (EC50 of >50 μM), while those classified as nonpumped molecules showed insignificant changes in potency between both strains (fold change of ≤2) (Fig. 4A). The former set of molecules consisted of ∼1,070 actives, while the latter set constituted ∼410 actives. To ensure that efflux pumps largely limited the Gram-negative activity of pumped molecules, we measured their potency in the hyperpermeable strain E. coli WT-Pore (26). While some compounds exhibited antibacterial activity in permeabilized cells, pumped molecules were more potent in efflux-compromised E. coli (see Table S2 in the supplemental material), further highlighting their susceptibility to efflux pumps. As such, of the 1,480 actives categorized, a random set of ∼290 pumped molecules and ∼290 nonpumped molecules were chosen as a training set, where each molecule’s 50 descriptors were used to train a second random forest model, called Susceptibility to Efflux Random Forest (SERF) (Fig. 4A). This model achieved an AUC-ROC of 0.839 (Fig. 4B; see Fig. S4 in the supplemental material), indicating a good measure of classifying pumped molecules and nonpumped molecules. Here, we observed that along with hydrophobicity and molecular stability, molecular complexity was key to efflux susceptibility (Fig. 4C). Molecular complexity was represented by topological indices, namely, the hyper-Wiener index, Wiener polarity, and the Balaban index, that describe some aspects of molecular structure (27). Notably, the three descriptors largely governing efflux susceptibility were resonant structure count, clogD (pH 7.0), and the hyper-Wiener index (HWI), which describes molecular “branching” and “compactness” (27, 28). Molecules with a relatively low molecular stability (as measured by the resonant structure count at pH 7.0 of ≤6), hydrophobic nature (clogD of 1 to 5), and reduced branching and compactness (hyper-Wiener index of >6,000) showed increased susceptibility to efflux (see Fig. S5 in the supplemental material).
FIG 4.
The Susceptibility to Efflux Random Forest (SERF) model identifies molecular descriptors governing efflux. (A) The pipeline highlights the training set of compounds used to build the SERF model and identify key descriptors that contribute to efflux susceptibility. Based on the shown cutoffs for the 4,500 actives from the primary screen, ∼1,070 actives were pumped molecules and ∼410 actives were nonpumped molecules. For a random set of ∼290 pumped molecules and ∼290 nonpumped molecules, molecular descriptors for each of these compounds were used to train SERF and identify those contributing to efflux susceptibility in E. coli. (B) The area under the curve-receiver operating characteristic curve (AUC-ROC) plot for SERF is 0.839, showing a good distinction between pumped molecules and nonpumped molecules. Sensitivity refers to the true-positive rate of the model, while specificity refers to its false-positive rate. (C) The top 10 molecular descriptors that reduce the model’s accuracy are shown, with resonant structure count, clogD, and hyper-Wiener index accounting for the greatest impact on accuracy. “Polarizability tensor a(yy)” is the principal component of polarizability along the coordinate space a(yy).
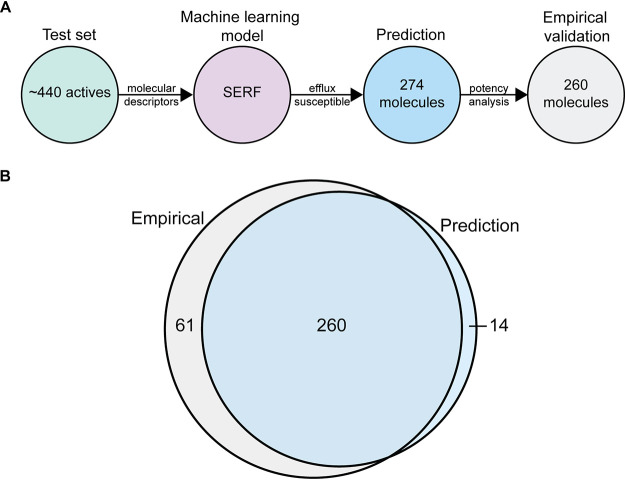
Following model development and optimization, we then assessed the predictive power of SERF to identify molecules susceptible to efflux. We used a test set of ∼440 actives (of 1,480) that were excluded from the training set, of which 274 were predicted by SERF to be efflux susceptible. We curated these 274 molecules and assessed their empirically measured potency in the wild-type and ΔtolC E. coli strains (Fig. 5A; see Table S3 in the supplemental material). Based on the chosen cutoff for pumped molecules (EC50 of <5 μM and wild-type EC50 of >50 μM), we observed that 260 of 274 (∼95%) predicted molecules were validated, illustrating a high accuracy in the model’s prediction (Fig. 5A). To determine the predictive power of SERF, we further assessed the potency of the remaining 166 molecules (of 440) in the wild-type and ΔtolC strains. Here, we noted 61 pumped molecules that were not reported by the model, revealing an overall 80% (260/321) predictive power in identifying efflux-susceptible molecules (Fig. 5B; see Table S4 in the supplemental material). In all, our analysis revealed that molecular stability and hydrophobicity, as well as molecular branching and compactness, contribute to efflux susceptibility. Furthermore, we designed the first random forest model (SERF) that analyzes the molecular descriptors of small molecules and identifies those susceptible to efflux. The code for SERF can be accessed online to test other small molecules for efflux susceptibility (https://github.com/sfrench007/serf).
FIG 5.
SERF identifies small molecules susceptible to efflux pumps. (A) Molecular descriptors of a test set of ∼440 actives (∼320 pumped molecules and ∼120 nonpumped molecules) that were excluded from the training set were used by SERF to identify molecules susceptible to efflux. SERF predicted some 274 molecules, of which 260 were validated by dose-response potency analysis. (B) The Venn diagram represents the accuracy and predictive power of SERF to identify efflux-susceptible molecules, as determined by empirical assessment and validation: 260 molecules were correct predictions, 14 molecules were incorrect predictions, and 61 molecules were not identified by SERF to be efflux susceptible.
Structure-activity relationship analyses reveal some structural modifications that impact efflux susceptibility.
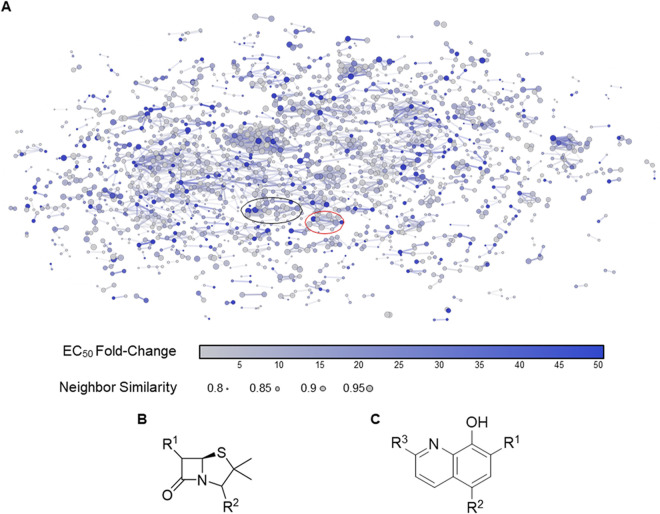
To analyze the chemical space occupied by our 4,500 actives, we applied structure-based clustering using structural fingerprints. This revealed ∼15 to 20 self-organized clusters, consisting on average of 15 or more molecules within a structural space (Fig. 6A). These clusters demonstrated distinct activity cliff regions that highlight how structural modifications within a compound series of molecules led to significant changes in biological activity (29). Some clusters were occupied by molecules known for their susceptibility to efflux, such as β-lactams and antifungals (Table 1; see Table S5 in the supplemental material). Notably, the β-lactam cluster consisted of 11 penicillin-type antibiotics that further divided into five semisynthetic β-lactamase-resistant penicillins (penicillins 1a to 4a and, 6a), four aminopenicillins (penicillins 8a, 9b, 10c, and 11a), and two ureidopenicillins (penicillins 5a and 7a) (Table 1). We assessed the local structure-activity relationship (SAR) of this β-lactam cluster, where we observed, as expected, that the penam was the common core among these penicillins (Fig. 6B and Table 1). Chemical modification of the acylamino side chain at the α-carbon of the penam largely contributed to this activity cliff (Table 1). Specifically, the amide group was decorated with a phenyl-isoxazolyl (penicillins 1a to 4a), naphthyl (penicillin 6a), or benzylamine (penicillins 5a, 7a, 8a, 9b, 10c, 11a, and 12a) moiety. The high fold change in the potency of penicillins 1a to 4a and 6a suggested that both phenyl-isoxazolyl and naphthyl moieties contribute to efflux susceptibility (Table 1). Substituting these moieties with more polar benzylamine groups, on the other hand, established antibacterial activity for most of the remaining penicillins in the wild-type strain (Table 1). This lowered the fold change in potency and the penicillin’s susceptibility to efflux (Table 1). Furthermore, compared to penicillins 1a to 4a and 6a, the lower hydrophobic character (clogP), an overall decrease in planar, or “flat,” nature (Fsp3), and increased molecular stability (as measured by the resonant structure count [RSC] at pH 7.0) of penicillins 8a, 9b, 10c, and 11a correlated with reduced efflux susceptibility. Notably, where the resonant structure count for penicillins 7a, 9b, and 11a remains constant, increased molecular compactness, as observed by lower hyper-Wiener indices (HWI [Table 1]), reduces efflux susceptibility as well. In all, these results suggest that the acylamino group at the α-carbon of penams and the overall hydrophobicity and reduced molecular complexity of penicillins contribute to their efflux susceptibility in E. coli.
FIG 6.
Structure-activity representation of the 4,500 actives from the primary screen. (A) Small molecules were clustered by structural similarity with fragment-based fingerprints in DataWarrior (50), using a force-directed method of clustering within the 2D space shown. Several clusters containing >15 molecules (neighbors) are seen from the primary screening data. The points are colored by a fold change in EC50 between wild-type E. coli strain BW25113 and the mutant strain BW25113 ΔtolC. The sizes of the points indicate the structural similarity between a compound and its immediate neighbors. The locations of the β-lactam cluster (black oval) and the hydroxyquinoline derivative cluster (red oval) are highlighted. Structures of the common core for the (B) β-lactam cluster and (C) the hydroxyquinoline derivative cluster are shown.
TABLE 1.
Structure, activity, and molecular descriptors of a β-lactam compound seriesa
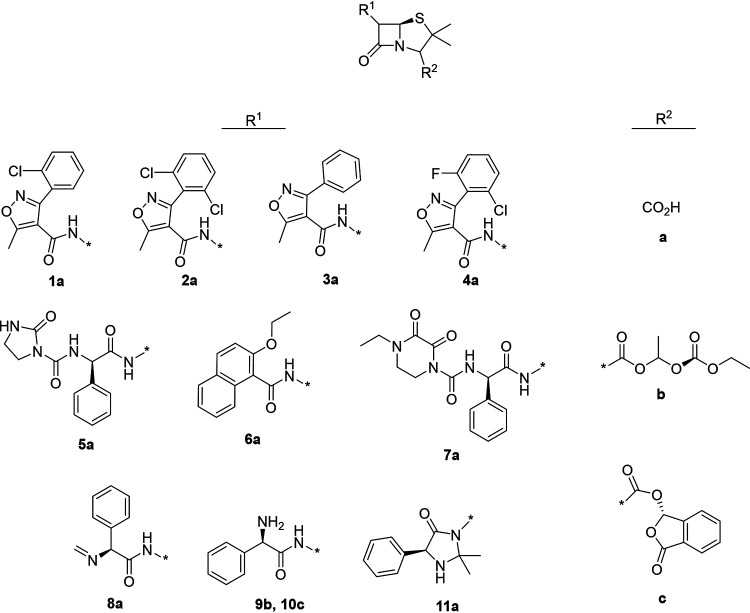 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cmpnd | Cmpnd name | EC50 (μM) for E. coli |
FC | MW (g mol−1) | clogP | PSA (Å2) | Fsp3 | RSC | HWI | |
| WT | ΔtolC | |||||||||
| 1a | Cloxacillin | >50 | 1.47 | >50 | 435.88 | 2.70 | 138 | 0.37 | 3 | 9,005 |
| 2a | Dicloxacillin | >50 | 2.88 | >34.7 | 470.32 | 3.31 | 138 | 0.37 | 3 | 9,877 |
| 3a | Oxacillin | >50 | 3.15 | >31.7 | 401.43 | 2.10 | 138 | 0.37 | 3 | 8,143 |
| 4a | Flucloxacillin | >50 | 3.91 | >25.6 | 453.87 | 2.80 | 138 | 0.37 | 3 | 9,877 |
| 5a | Azlocillin | >50 | 5.72 | >17.5 | 461.49 | 0.312 | 173 | 0.45 | 5 | 12,535 |
| 6a | Nafcillin | >50 | 10.5 | >9.49 | 414.47 | 3.12 | 121 | 0.38 | 2 | 8,612 |
| 7a | Piperacillin | 25.3 | 2.31 | 10.9 | 517.55 | −0.174 | 182 | 0.48 | 4 | 19,160 |
| 8a | Metampicillin | 14.9 | 1.82 | 8.19 | 361.41 | 0.542 | 124 | 0.41 | 5 | 5,716 |
| 9b | Bacampicillin | 30.7 | 7.13 | 4.31 | 465.51 | 1.29 | 163 | 0.52 | 4 | 15,846 |
| 10c | Talampicillin | 13.1 | 4.79 | 2.72 | 481.52 | 1.34 | 153 | 0.33 | 6 | 17,736 |
| 11a | Hetacillin | 4.18 | 1.57 | 2.67 | 389.46 | −0.234 | 115 | 0.53 | 4 | 6,164 |
Cmpd, compound; WT, wild type; FC, fold change; PSA, polar surface area; Fsp3, ratio of sp3 hybridized carbon atoms to total carbon atoms; RSC, resonant structure count; HWI, hyper-Wiener index.
Additionally, we identified a cluster occupied by six hydroxyquinoline derivatives (Fig. 6C), where the hydroxyquinoline core was decorated with different combinations of halogen atoms on its phenol and pyridine moieties (Table 2). Although these chemical changes seem minor, addition of a chlorine atom to the ortho and para positions of the phenol in compounds 12 and 13 abolished antibacterial activity in the wild type and maintained efflux susceptibility (Table 2). Reducing the number of chlorine atoms to one or none at these positions, however, established wild-type activity and decreased efflux susceptibility of compounds 14 to 17 (Table 2). Here, these modifications revealed that the addition of halogen atoms with low electronegativity to the phenol moiety improved whole-cell activity and reduced susceptibility to efflux (Table 2). Furthermore, decreasing the hydrophobic character (clogP of <3) reduced efflux susceptibility of compounds 15 to 17 (Table 2). Overall, these results highlighted modifications to the aromatic ring of hydroxyquinolines that affect their susceptibility to efflux.
TABLE 2.
Structure, activity, and molecular descriptors of a hydroxyquinoline compound seriesa
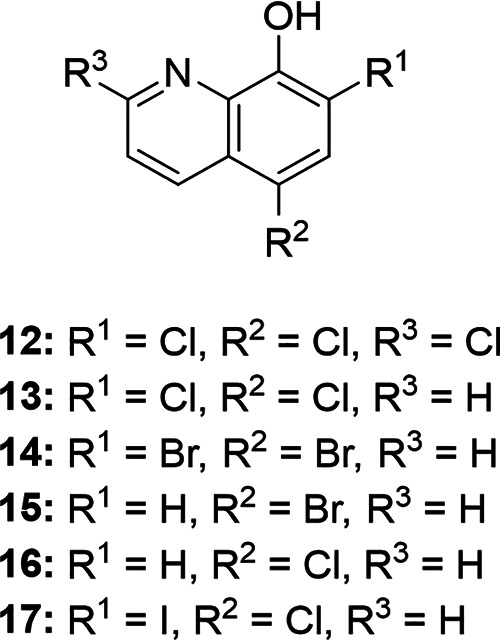 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cmpd | Cmpd name | EC50 (μM) for E. coli |
FC | MW (g mol−1) | clogP | PSA (Å2) | Fsp3 | RSC | HWI | |
| WT | ΔtolC | |||||||||
| 12 | ID-0366 | >50 | 0.644 | >50 | 248.49 | 3.55 | 33.1 | 0.00 | 3 | 623 |
| 13 | Chloroxine | >50 | 14.9 | >6.73 | 214.05 | 2.84 | 33.1 | 0.00 | 4 | 476 |
| 14 | Broxyquinoline | 3.72 | 0.168 | 22.1 | 302.95 | 3.08 | 33.1 | 0.00 | 4 | 476 |
| 15 | ID-0763 | 16.1 | 1.41 | 11.4 | 224.06 | 2.36 | 33.1 | 0.00 | 4 | 370 |
| 16 | Cloxyquin | 11.7 | 2.17 | 5.39 | 179.6 | 2.24 | 33.1 | 0.00 | 4 | 370 |
| 17 | Clioquinol | 2.64 | 0.560 | 4.72 | 305.5 | 2.67 | 33.1 | 0.00 | 4 | 476 |
Cmpd, compound; WT, wild type; FC, fold change; PSA, polar surface area; Fsp3, ratio of sp3 hybridized carbon atoms to total carbon atoms; RSC, resonant structure count; HWI, hyper-Wiener index.
The SAR and fold changes in potency available within the clusters highlighted above revealed that some chemical modifications impact both whole-cell activity and efflux susceptibility in E. coli. As such, structure-based clustering has the potential to provide some guidelines for optimal antibacterial activity and efflux evasion.
DISCUSSION
There is a need to understand compound penetration in Gram-negative bacteria, which consists of both outer membrane permeability and efflux evasion. There has been much progress in recognizing the properties that contribute to penetration of large chemical collections (14, 15, 19); however, the exact contribution of outer membrane permeability and efflux remains unclear. Herein, we addressed the contribution of efflux by using novel computational approaches to investigate the molecular descriptors of some 4,500 actives from a small-molecule screen in efflux-compromised E. coli. We applied principal-component analysis to visualize descriptors contributing to Gram-negative activity in the efflux-deficient E. coli ΔtolC strain and quantitatively assessed them using a machine learning approach. In doing so, we designed a machine learning model in order to identify descriptors governing efflux susceptibility of small molecules. The latter, referred to as Susceptibility to Efflux Random Forest (SERF), was further assessed for its predictive power to analyze the molecular descriptors of a set of small molecules and identify those susceptible to efflux pumps in silico. Notably, the model showed ∼95% accuracy in its prediction and had an overall predictive power of 80%. In all, this work presents the first large-scale study implementing PCA and machine learning in order to further resolve the contribution of efflux to compound penetration. Further, it provides the first machine learning tool (SERF) that identifies small molecules susceptible to efflux pumps in silico.
In this study, we identified some 4,500 actives from a phenotypic screen in the efflux-deficient E. coli ΔtolC strain. Dose-response potency analyses of these actives in the wild-type E. coli strain and ΔtolC mutant revealed that a majority (∼84%) lost antibacterial activity in the wild type (efflux-dependent actives), suggesting high efflux susceptibility. Among these compounds were conventional antibiotics, including β-lactams and macrolides, and other nonantibiotic drugs known to have cryptic antibacterial activity. These findings further validated our work since these compounds have been well recognized as efflux substrates in Gram-negative bacteria (18, 23–25).
A PCA of some 50 molecular descriptors for each of the efflux-dependent actives and non-growth-inhibitory molecules from the primary screen highlighted nine molecular descriptors that contribute to Gram-negative activity in efflux-compromised E. coli. These descriptors largely encompassed measures of molecular size, hydrophobicity, and molecular complexity. To quantify properties that contribute to Gram-negative activity and those that influence efflux susceptibility, we designed two random forest models. The first model addressed Gram-negative activity, highlighting that the antibacterial activity of hydrophobic and planar molecules with relatively low stability was limited to efflux-compromised E. coli. The second random forest model (SERF) assessed efflux susceptibility, revealing that hydrophobic molecules with reduced branching, compactness, and stability were most susceptible to efflux pumps. Overall, both random forest models suggest that hydrophobicity and molecular stability impact Gram-negative activity as well as efflux susceptibility. Furthermore, each model identified descriptors of molecular complexity that impact each of these aspects, where the level of saturation (Fsp3) governs antibacterial activity, potentially due to some influence from efflux pumps, while branching and compactness (hyper-Wiener index) uniquely affect efflux susceptibility.
The hydrophobic nature of efflux-susceptible Gram-negative antibacterials has been recognized by Brown et al. (19). Recent analyses of the physicochemical properties of some β-lactams, fluoroquinolones, and a subset of other antibiotics have shown that lipophilicity contributes to efflux susceptibility (21). Furthermore, O’Shea and Moser (14) have shown that Gram-negative antibiotics are more hydrophilic than other drugs. These studies validate our analyses, where we observed that the Gram-negative activity of hydrophobic molecules was abolished in efflux-proficient E. coli. The impact of molecular stability and molecular complexity on efflux susceptibility, however, provides novel insight into compound penetration. Particularly, planar, or “flat,” molecules with low molecular stability lost their Gram-negative activity in efflux-proficient E. coli, while “unbranched” and “elongated” molecules with low molecular stability showed increased susceptibility to efflux pumps. These molecular descriptors highlight ideal efflux substrates that may result in optimal binding to some efflux pumps in E. coli and their extrusion. Indeed, the outer membrane channel, TolC, is a component of many tripartite efflux pumps, including the well-characterized RND efflux pump AcrAB-TolC in E. coli (30). In addition to TolC, this multiple-component system consists of a periplasmic adaptor protein, AcrA, and an inner membrane transporter, AcrB (17). Cocrystallization of AcrB with some molecules revealed that it undergoes conformational cycling in order to bind and extrude molecules through TolC (18, 24). Hydrophilic molecules tend to bind to the upper “crevice” of the distal pocket, which is rich in hydrophilic and charged residues (30–32). On the other hand, hydrophobic molecules interact with the phenylalanine-rich hydrophobic trap located in the lower portion of the binding pocket (24, 33, 34). Molecular dynamics simulation studies have suggested that a tight interaction with the hydrophobic trap distorts the binding pocket and inhibits efflux, while a loose interaction facilitates binding and extrusion (30, 32, 35). As such, our analyses suggest that hydrophobic molecules that are planar, unbranched, and elongated may be ideal candidates for loose interactions with the hydrophobic trap in AcrB. Furthermore, decreasing the molecular stability of such molecules increases their reactivity, which may facilitate nonspecific binding and extrusion.
Additionally, we used the machine learning model SERF to predict, in silico, which antibacterials would be susceptible to efflux pumps. For a set of some 440 actives, SERF predicted that 274 molecules would be susceptible. Following empirical validation, we found that the prediction was ∼95% accurate, and the model showed good (80%) predictive power in identifying molecules susceptible to efflux. Overall, this highlights the potential of machine learning approaches to predict which compounds could be susceptible to efflux. Where novel approaches to study and overcome efflux are greatly required (10, 11, 36), SERF can be used as a tool to rapidly explore efflux susceptibility of small molecules in silico. As such, our machine learning model provides an opportunity to systematically develop and synthesize a novel library of Gram-negative antibacterial agents that could evade efflux pumps.
As for the structure-based clustering of the 4,500 actives, this analysis revealed ∼15 to 20 self-organized clusters, consisting of 15 or more molecules on average, within a self-organized structural space. We profiled the SAR within a cluster of β-lactams and hydroxyquinoline derivatives with uncharacterized modes of action. The β-lactam cluster consisted of penicillin-type antibiotics, including some five β-lactamase-resistant penicillins, four aminopenicillins, and two ureidopenicillins. Modifications to the acylamino moiety of these penicillins impacted their efflux susceptibility, where the phenyl-isoxazolyl and naphthyl moieties of β-lactamase-resistant penicillins showed high susceptibility to efflux. Substituting these moieties with polar benzylamine groups as well as decreasing the hydrophobicity of the antibiotic reduced efflux susceptibility of some aminopenicillins and ureidopenicillins. Moreover, increasing the molecular complexity of these antibiotics by reducing their “flat” nature as well as increasing their molecular stability and compactness allows for better efflux evasion. Overall, this analysis highlights that chemical substitutions along the acylamino group and changes in hydrophobicity and molecular complexity may be key to efflux susceptibility for penicillins. Indeed, earlier modifications of penicillins have shown that the β-lactamase-resistant penicillins are insensitive to hydrolysis by β-lactamases and susceptible to efflux (37–40). Further modifications largely focused on substitutions to the acylamino moiety (37, 41), which yielded the aminopenicillins and ureidopenicillins. Indeed, these penicillins have an extended activity spectrum that encompasses some Pseudomonas spp. (41, 42). Although some remain susceptible to efflux, these β-lactams have shown improved Gram-negative activity in efflux-proficient strains (23, 42). Of practical note, acylamino groups decorated with benzylamines could be used in medicinal chemistry efforts to assess for reduced efflux in other compound series.
The second cluster was composed of hydroxyquinoline derivatives exhibiting chemical modification to their aromatic rings. Substitution of chlorines (compounds 12 and 13) with halogen atoms decreasing in electronegativity (compounds 14 to 17) largely improved wild-type activity and decreased efflux susceptibility. A lower hydrophobic character within this compound series, also, seemed to contribute to improved activity in the wild type and reduced efflux. While hydrophobicity has been recognized for its impact on Gram-negative activity and efflux susceptibility (14, 15, 19), the hydroxyquinoline cluster revealed that decorating aromatic rings with different halogen atoms may lead to some changes in efflux susceptibility. In previous studies, compounds containing halogenated aromatic moieties have been shown to improve antibacterial activity in Gram-positive bacteria (43) and have been used as antibiotic adjuvants to overcome colistin resistance in some ESKAPE pathogens (i.e., Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) (44). Here, we highlight the prospect of such moieties to improve Gram-negative activity and reduce efflux susceptibility.
The empirical and computational approaches reported herein identified molecular descriptors and structural modifications that advance our understanding of efflux susceptibility. Our work here illustrates that physicochemical properties and chemical structures serve as guidelines for efflux, and combinations thereof generally define substrate quality. Leveraging these properties, we developed the first machine learning tool that can be used to assess efflux susceptibility of small molecules in silico. Further expansion of guidelines for efflux, however, will come from additional assessment of compound accumulation in Gram-negative bacteria, including those identified in this work.
Conclusion.
Efflux pumps are major contributors to the intrinsic resistance of many antibiotics in Gram-negative bacteria. These pumps have challenged the design of candidate molecules that would be otherwise efficacious in Gram-negative organisms. Understanding the molecular descriptors of small molecules susceptible to efflux pumps is important to overcome this challenge. Herein, we present a novel approach to determine the physicochemical and structural spaces occupied by such molecules. Multivariate analysis and a machine learning approach identified that hydrophobicity, the level of saturation, and molecular stability contribute to Gram-negative activity yet limit it to efflux-compromised E. coli. SERF, a second machine learning model, further revealed that along with hydrophobicity and molecular stability, the extent of molecular branching and compactness is a key factor that also governs efflux susceptibility. Additionally, SERF proved capable of predicting molecular efflux and is the first machine learning tool to identify small molecules susceptible to efflux pumps in silico. Furthermore, structure-activity relationship analyses revealed that some molecular side chains, and their associated physicochemical properties, serve as triggers for efflux. In all, the results of this work provide novel insight into the physicochemical properties and chemical structures governing efflux in Gram-negative bacteria, which can be used to guide the design of novel Gram-negative antimicrobials.
MATERIALS AND METHODS
Screen for antibacterial activity in the efflux-deficient E. coli ΔtolC strain.
The strains used in this study were E. coli BW25113 (wild type), the E. coli ΔtolC mutant (parent strain BW25113) (45), and GKCW102 (E. coli BW25113 WT-Pore, provided by H. Zgurskaya, University of Oklahoma) (26). In all experiments using wild-type E. coli and the ΔtolC mutant, a mid-log culture in M9 minimal medium was prepared as previously described (46) and used to prepare cells to a final working inoculum of ∼105 CFU/ml. In experiments using E. coli WT-Pore, cells were prepared as previously described (26), but with 0.5% arabinose and minimal medium. For the small-molecule screen, compounds were added to a 384-well assay plate to a final concentration of 10 μM. Molecules, dissolved in dimethyl sulfoxide (DMSO), were sourced from Enamine, ChemDiv, Asinex, ChemBridge, Maybridge, Sigma (Lopac), Prestwick, Biomol, and Microsource. To these plates, a final working inoculum of the E. coli ΔtolC strain was added to a final volume of 50 μl, and plates were incubated at 37°C for 18 h. All screens were performed in duplicate. Liquid handling was performed using a Beckman Coulter FXP laboratory automated workstation and an Echo acoustic dispenser (Labcyte). After incubation, absorbance (optical density at 600 nm [OD600]) of the 384-well assay plates was measured using a Perkin Elmer EnVision plate reader.
Analysis of screening data.
To reduce plate-to-plate variation, data from the OD600 measurements of each 384-well assay plate were rank ordered and the interquartile mean of each plate was calculated. Data were first normalized on a per plate basis as previously described (47). To account for positional effects, these data were further normalized by the interquartile mean of each well position as previously described (47). A cutoff at three standard deviations below the mean was established to determine actives, where any wells with values below this cutoff were considered actives.
Determination of antibacterial potency in dose.
The E. coli wild-type, ΔtolC mutant, and WT-Pore strains were grown and prepared as described above. These cells were then added to an assay plate containing half-log serial dilutions of one of the actives identified from the screen described above, where concentrations ranged from 50 μM to 0 μM. Assay plates were then incubated at 37°C for 18 h, and OD600 was measured using a Tecan plate reader (Infinite M1000). Growth at each exposed concentration was determined as G = Gi/G0, where Gi represents the growth in one of the wells exposed to the different concentrations of the tested compound and G0 represents the growth in the well that was not exposed to the tested compound. A value of 0 represents no growth on the plate, and 1 represents no growth inhibition. An EC50 dose-response curve was then fit to the data using the four-parameter dose-response model in GraphPad Prism in order to calculate the potency of a compound, based on the ratio of wild-type EC50 to ΔtolC EC50 (wild-type EC50/ΔtolC EC50).
Calculation and principal-component analysis of molecular properties.
Structures of all compounds in this study exist as MOL file coordinates within compiled SDF files. All structures had three-dimensional (3D) coordinates generated using cxcalc (ChemAxon), and the lowest-energy conformer was chosen as the basis for molecular property calculations. All calculations were done using cxcalc in a Linux terminal, with pH set to 7.0 where relevant. An initial principal-component analysis (PCA) was performed as a means of dimensionality reduction, in order to visualize the actives from the primary screen within the chemical space of the screened molecules. The R statistical programming language (48) was used here, with data appropriately scaled to eliminate bias from the different units and magnitudes of chemical descriptors. Code and data set examples, including cxcalc commands used to generate the chemical properties, can be found on GitHub (https://github.com/sfrench007/serf).
Machine learning and prediction of efflux susceptibility.
Further exploring the importance of the molecular descriptors specific to the 3,780 efflux-dependent actives, these molecules were used in a random forest machine learning approach to explore the properties key to Gram-negative activity in the efflux-deficient E. coli BW25113 ΔtolC strain. A binary (two-class) approach was used in creating a data set to train a random forest model. In this, the 3,780 efflux-dependent actives were classified as “true” and an equal number of randomly sampled inactive molecules from the screened collection were classified as “false.” This was performed in R, with the random forest learning done using the caret package (49). Molecular descriptors that were redundant based on a similarity matrix (>85% similarity) were removed from the model training set. The model was trained with 2,000 trees grown, 10 iterations, and a two-class summary function for “true” and “false” identifications. Validation was done through repeated random subsampling (70/30 split, training/test), with 10 repeats for each cross-validation. The number of properties randomly sampled when splitting at each tree node was incrementally increased and used to tune the optimal number of properties for the highest receiver operating characteristic (ROC) value. The ROC is a measure of how well the model can classify, in this case, Gram-negative active molecules. Upon training the model, molecular descriptors were ranked by their relative importance—importance in that removing that descriptor from the training set would result in incorrect classification and predictions. Frequency distributions for important descriptors were also examined, to visualize the separations, or shifts, between the two phenotypes used in the model.
To explore molecular descriptors contributing to efflux susceptibility, a second random forest model was created. This model used the same decision tree parameters as the previous model, this time comparing ∼290 pumped actives (EC50 of <5 μM and wild-type EC50 of >50 μM) and a random subset of ∼290 nonpumped actives (fold change in potency of ≤2). This random forest model, called Susceptibility to Efflux Random Forest (SERF), was tuned and evaluated in a similar manner to the first to identify descriptors of importance to efflux. A set of ∼440 actives was assessed by SERF to predict small molecules susceptible to efflux. All code and data set examples can be found on GitHub (https://github.com/sfrench007/serf).
Structure-activity relationship analysis.
Dose-response curves from primary screening actives (ΔtolC strain) were compared to dose-response curves of the wild-type strain. In this, EC50 values for each dose-response curve were calculated and compared to obtain a fold change value. These fold changes were tabulated along with chemical structure (as a MOL representation within an SDF file), and structural fingerprints were calculated in DataWarrior (50). The fingerprints were based on a fragment dictionary generated within DataWarrior, and a correlation matrix within the software compared all 4,507 actives in the data table. All active molecules are randomly placed within a 2D space, and force-directed clustering is applied (50) based on the correlation matrix, bringing like molecules together into groups. The result is a 2D structural representation of molecular connectivity, providing insights into the functional groups that define efflux-susceptible and efflux-evading molecules.
Clusters of actives in Fig. 6 were more closely examined by breaking down the molecules to common structural cores and R-groups. This was done using DataWarrior, which calculated the common core for each chemical analog within each group of molecules and generated a list of functional groups added to that core. Using this method, several structure-activity relationship tables were generated, indicating which functional groups had an impact on how well chemical analogs in each table were susceptible to efflux.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by a Foundation grant from the Canadian Institutes of Health Research (FRN 143215), infrastructure funding from the Canada Foundation for Innovation, a Research Excellence allocation from the Ontario Research Fund, and a Tier I Canada Research Chair award to E.D.B. In addition, S.S.E. was supported by an Ontario Graduate Scholarship and a Fred and Helen Knight Enrichment Award.
The high-throughput screen was performed in the Centre for Microbial Chemical Biology core facility at McMaster University. We thank Susan McCuskar for excellent technical assistance with the screen. We also thank Helen Zgurskaya for providing the E. coli porinated strain.
S.S.E. and E.D.B. conceived and designed the research. S.S.E., M.A.F., and G.K. performed the primary screen. S.S.E. and G.K. performed the dose-response experiments. S.S.E. analyzed the primary screen, dose-response, and SAR data. S.F. calculated the molecular descriptors for all screened molecules, generated the structure-activity representations, and wrote and developed the code for the machine learning models. S.S.E., S.F., and E.D.B. wrote the manuscript.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lewis K. 2020. The science of antibiotic discovery. Cell 181:29–45. doi: 10.1016/j.cell.2020.02.056. [DOI] [PubMed] [Google Scholar]
- 2.Brown ED, Wright GD. 2016. Antibacterial drug discovery in the resistance era. Nature 529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 3.WHO. 2017. Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 4.CDC. 2019. Antibiotic resistance threats in the United States, 2019. US Department of Health and Human Services, CDC, Atlanta, GA. [Google Scholar]
- 5.WHO. 2019. 2019 antibacterial agents in clinical development: an analysis of the antibacterial clinical development pipeline. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 6.Butler MS, Paterson DL. 2020. Antibiotics in the clinical pipeline in October 2019. J Antibiot (Tokyo) 73:329–364. doi: 10.1038/s41429-020-0291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 8.Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. 2015. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 9.Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. 2015. ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov 14:529–542. doi: 10.1038/nrd4572. [DOI] [PubMed] [Google Scholar]
- 10.Pew Charitable Trusts. 2017. Challenges in the discovery of Gram-negative antibacterials: the entry and efflux problem. Pew Charitable Trusts, Rockville, MD. [Google Scholar]
- 11.Pew Charitable Trusts. 2016. A scientific roadmap for antibiotic discovery. Pew Charitable Trusts, Rockville, MD. [Google Scholar]
- 12.Shore CK, Coukell A. 2016. Roadmap for antibiotic discovery. Nat Microbiol 1:16083. doi: 10.1038/nmicrobiol.2016.83. [DOI] [PubMed] [Google Scholar]
- 13.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. 2001. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 14.O'Shea R, Moser HE. 2008. Physicochemical properties of antibacterial compounds: implications for drug discovery. J Med Chem 51:2871–2878. doi: 10.1021/jm700967e. [DOI] [PubMed] [Google Scholar]
- 15.Reck F, Jansen JM, Moser HE. 2019. Challenges of antibacterial drug discovery. Arkivoc 2019:227–244. doi: 10.24820/ark.5550190.p010.955. [DOI] [Google Scholar]
- 16.Krishnamoorthy G, Leus IV, Weeks JW, Wolloscheck D, Rybenkov VV, Zgurskaya HI. 2017. Synergy between active efflux and outer membrane diffusion defines rules of antibiotic permeation into Gram-negative bacteria. mBio 8:e01172-17. doi: 10.1128/mBio.01172-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, Luisi BF. 2014. Structure of the AcrAB-TolC multidrug efflux pump. Nature 509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eicher T, Cha H-J, Seeger MA, Brandstätter L, El-Delik J, Bohnert JA, Kern WV, Verrey F, Grütter MG, Diederichs K, Pos KM. 2012. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc Natl Acad Sci U S A 109:5687–5692. doi: 10.1073/pnas.1114944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown DG, May-Dracka TL, Gagnon MM, Tommasi R. 2014. Trends and exceptions of physical properties on antibacterial activity for Gram-positive and Gram-negative pathogens. J Med Chem 57:10144–10161. doi: 10.1021/jm501552x. [DOI] [PubMed] [Google Scholar]
- 20.Richter MF, Drown BS, Riley AP, Garcia A, Shirai T, Svec RL, Hergenrother PJ. 2017. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature 545:299–304. doi: 10.1038/nature22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper SJ, Krishnamoorthy G, Wolloscheck D, Walker JK, Rybenkov VV, Parks JM, Zgurskaya HI. 2018. Molecular properties that define the activities of antibiotics in Escherichia coli and Pseudomonas aeruginosa. ACS Infect Dis 4:1223–1234. doi: 10.1021/acsinfecdis.8b00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrews LD, Kane TR, Dozzo P, Haglund CM, Hilderbrandt DJ, Linsell MS, Machajewski T, McEnroe G, Serio AW, Wlasichuk KB, Neau DB, Pakhomova S, Waldrop GL, Sharp M, Pogliano J, Cirz RT, Cohen F. 2019. Optimization and mechanistic characterization of pyridopyrimidine inhibitors of bacterial biotin carboxylase. J Med Chem 62:7489–7505. doi: 10.1021/acs.jmedchem.9b00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li XZ, Ma D, Livermore DM, Nikaido H. 1994. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as a contributing factor to beta-lactam resistance. Antimicrob Agents Chemother 38:1742–1752. doi: 10.1128/aac.38.8.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. 2006. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 25.Nakashima R, Sakurai K, Yamasaki S, Nishino K, Yamaguchi A. 2011. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature 480:565–569. doi: 10.1038/nature10641. [DOI] [PubMed] [Google Scholar]
- 26.Krishnamoorthy G, Wolloscheck D, Weeks JW, Croft C, Rybenkov VV, Zgurskaya HI. 2016. Breaking the permeability barrier of Escherichia coli by controlled hyperporination of the outer membrane. Antimicrob Agents Chemother 60:7372–7381. doi: 10.1128/AAC.01882-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Méndez-Lucio O, Medina-Franco JL. 2017. The many roles of molecular complexity in drug discovery. Drug Discov Today 22:120–126. doi: 10.1016/j.drudis.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Sheridan RP, Zorn N, Sherer EC, Campeau L-C, Chang CZ, Cumming J, Maddess ML, Nantermet PG, Sinz CJ, O'Shea PD. 2014. Modeling a crowdsourced definition of molecular complexity. J Chem Inf Model 54:1604–1616. doi: 10.1021/ci5001778. [DOI] [PubMed] [Google Scholar]
- 29.Sisay MT, Peltason L, Bajorath J. 2009. Structural interpretation of activity cliffs revealed by systematic analysis of structure-activity relationships in analog series. J Chem Inf Model 49:2179–2189. doi: 10.1021/ci900243a. [DOI] [PubMed] [Google Scholar]
- 30.Li X-Z, Plésiat P, Nikaido H. 2015. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takatsuka Y, Chen C, Nikaido H. 2010. Mechanism of recognition of compounds of diverse structures by the multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci U S A 107:6559–6565. doi: 10.1073/pnas.1001460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vargiu AV, Nikaido H. 2012. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc Natl Acad Sci U S A 109:20637–20642. doi: 10.1073/pnas.1218348109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bohnert JA, Schuster S, Seeger MA, Fähnrich E, Pos KM, Kern WV. 2008. Site-directed mutagenesis reveals putative substrate binding residues in the Escherichia coli RND efflux pump AcrB. J Bacteriol 190:8225–8229. doi: 10.1128/JB.00912-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakashima R, Sakurai K, Yamasaki S, Hayashi K, Nagata C, Hoshino K, Onodera Y, Nishino K, Yamaguchi A. 2013. Structural basis for the inhibition of bacterial multidrug exporters. Nature 500:102–106. doi: 10.1038/nature12300. [DOI] [PubMed] [Google Scholar]
- 35.Vargiu AV, Ruggerone P, Opperman TJ, Nguyen ST, Nikaido H. 2014. Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors. Antimicrob Agents Chemother 58:6224–6234. doi: 10.1128/AAC.03283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masi M, Réfregiers M, Pos KM, Pagès J-M. 2017. Mechanisms of envelope permeability and antibiotic influx and efflux in Gram-negative bacteria. Nat Microbiol 2:17001. doi: 10.1038/nmicrobiol.2017.1. [DOI] [PubMed] [Google Scholar]
- 37.Novick RP. 1962. Staphylococcal penicillinase and the new penicillins. Biochem J 83:229–235. doi: 10.1042/bj0830229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knowles JR. 1985. Penicillin resistance: the chemistry of beta-lactamase inhibition. Acc Chem Res 18:97–104. doi: 10.1021/ar00112a001. [DOI] [Google Scholar]
- 39.Drawz SM, Bonomo RA. 2010. Three decades of β-lactamase inhibitors. Clin Microbiol Rev 23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim SP, Nikaido H. 2010. Kinetic parameters of efflux of penicillins by the multidrug efflux transporter AcrAB-TolC of Escherichia coli. Antimicrob Agents Chemother 54:1800–1806. doi: 10.1128/AAC.01714-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walsh C. 2003. Antibiotics. American Society for Microbiology, Washington, DC. [Google Scholar]
- 42.Verbist L. 1979. Comparison of the activities of the new ureidopenicillins piperacillin, mezlocillin, azlocillin, and Bay k 4999 against Gram-negative organisms. Antimicrob Agents Chemother 16:115–119. doi: 10.1128/aac.16.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boudreau MA, Ding D, Meisel JE, Janardhanan J, Spink E, Peng Z, Qian Y, Yamaguchi T, Testero SA, O'Daniel PI, Leemans E, Lastochkin E, Song W, Schroeder VA, Wolter WR, Suckow MA, Mobashery S, Chang M. 2020. Structure-activity relationship for the oxadiazole class of antibacterials. ACS Med Chem Lett 11:322–326. doi: 10.1021/acsmedchemlett.9b00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barker WT, Chandler CE, Melander RJ, Ernst RK, Melander C. 2019. Tryptamine derivatives disarm colistin resistance in polymyxin-resistant Gram-negative bacteria. Bioorg Med Chem 27:1776–1788. doi: 10.1016/j.bmc.2019.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El Zahed SS, Brown ED. 2018. Chemical-chemical combinations map uncharted interactions in Escherichia coli under nutrient stress. iScience 2:168–181. doi: 10.1016/j.isci.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mangat CS, Bharat A, Gehrke SS, Brown ED. 2014. Rank ordering plate data facilitates data visualization and normalization in high-throughput screening. J Biomol Screen 19:1314–1320. doi: 10.1177/1087057114534298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ihaka R, Gentleman R. 1996. R: a language for data analysis and graphics. J Comput Graph Stat 5:299–314. doi: 10.2307/1390807. [DOI] [Google Scholar]
- 49.Kuhn M. 2008. Building predictive models in R using the caret package. J Stat Softw 28:1–26. doi: 10.18637/jss.v028.i05.27774042 [DOI] [Google Scholar]
- 50.Sander T, Freyss J, von Korff M, Rufener C. 2015. DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 55:460–473. doi: 10.1021/ci500588j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.