The malaria parasite Plasmodium falciparum contains the apicoplast organelle that synthesizes isoprenoids, which are metabolites necessary for posttranslational modification of Plasmodium proteins. We used fosmidomycin, an antibiotic that inhibits isoprenoid biosynthesis, to identify mechanisms that underlie the development of the parasite’s adaptation to the drug at sublethal concentrations.
KEYWORDS: apicoplast, fosmidomycin, prenylation, metabolomics, genome-scale metabolic model, Plasmodium falciparum, transcriptomics, metabolic modeling
ABSTRACT
The malaria parasite Plasmodium falciparum contains the apicoplast organelle that synthesizes isoprenoids, which are metabolites necessary for posttranslational modification of Plasmodium proteins. We used fosmidomycin, an antibiotic that inhibits isoprenoid biosynthesis, to identify mechanisms that underlie the development of the parasite’s adaptation to the drug at sublethal concentrations. We first determined a concentration of fosmidomycin that reduced parasite growth by ∼50% over one intraerythrocytic developmental cycle (IDC). At this dose, we maintained synchronous parasite cultures for one full IDC and collected metabolomic and transcriptomic data at multiple time points to capture global and stage-specific alterations. We integrated the data with a genome-scale metabolic model of P. falciparum to characterize the metabolic adaptations of the parasite in response to fosmidomycin treatment. Our simulations showed that, in treated parasites, the synthesis of purine-based nucleotides increased, whereas the synthesis of phosphatidylcholine during the trophozoite and schizont stages decreased. Specifically, the increased polyamine synthesis led to increased nucleotide synthesis, while the reduced methyl-group cycling led to reduced phospholipid synthesis and methyltransferase activities. These results indicate that fosmidomycin-treated parasites compensate for the loss of prenylation modifications by directly altering processes that affect nucleotide synthesis and ribosomal biogenesis to control the rate of RNA translation during the IDC. This also suggests that combination therapies with antibiotics that target the compensatory response of the parasite, such as nucleotide synthesis or ribosomal biogenesis, may be more effective than treating the parasite with fosmidomycin alone.
INTRODUCTION
Plasmodium falciparum possesses a nonphotosynthetic plastid organelle called the apicoplast, which it evolutionarily acquired through secondary endosymbiosis of a plastid-bearing red alga (1). Through the process of lateral gene transfer, most apicoplast genes migrated to the nuclear genome, which now encodes between 400 to 500 apicoplast-targeted proteins (2, 3). The apicoplast genome encodes fewer than 30 proteins, most of which are ribosome subunits (4). Previous work has shown that antibiotics that inhibit housekeeping functions of the apicoplast, such as DNA replication and transcription, cause the so-called delayed-death phenotype, in which the parasite successfully completes the first intraerythrocytic developmental cycle (IDC) but dies during the second IDC (5, 6). Specifically, it has been hypothesized that drug-induced delayed death begins with the inhibition of apicoplast housekeeping functions during the first IDC, which ultimately leads to irreversible apicoplast dysfunction and parasite death in the second cycle (1).
Previous studies have shown that the sole purpose of the apicoplast during the IDC is to synthesize isopentenyl diphosphate (IPP), which is involved in a wide range of functions, including vesicular trafficking (7), ubiquinone biosynthesis (8), protein glycosylation (9), and glycosylphosphatidylinositol (GPI) biosynthesis (9). Kennedy et al. (7) demonstrated that prenylation of Plasmodium proteins with a polyprenol (geranylgeraniol) is sufficient to rescue P. falciparum parasites undergoing drug-induced delayed death in the second IDC. However, these rescued parasites are still unable to survive the subsequent (third) IDC, suggesting that additional processes contribute to parasite death following disruption of IPP synthesis by a drug targeting apicoplast housekeeping functions.
In contrast to humans, which synthesize IPP via a mevalonate-dependent pathway (10), P. falciparum parasites synthesize IPP in the apicoplast through a nonmevalonate pathway. IPP synthesis via the latter pathway requires the metabolite 1-deoxy-d-xylulose 5-phosphate (DOXP) and the activity of DOXP reductoisomerase. Fosmidomycin, an antibiotic that selectively inhibits DOXP reductoisomerase, is effective in treating multidrug resistant parasites in vitro (11). Owing to recrudescent malaria infections (12), however, fosmidomycin is used clinically in combination with clindamycin, which results in highly efficacious treatment of children with P. falciparum malaria (13). The synergistic effect of combining the drugs is also observed under in vitro conditions (14); however, the mechanisms contributing to this synergy are not known. We hypothesize that by delineating and understanding how drug-induced adaption mechanisms allow the parasite to tolerate fosmidomycin, we will be able to provide a new rationale for selecting and expanding on possible alternative combination therapies.
To understand the impact of IPP depletion on P. falciparum blood-stage development, we first identified the fosmidomycin concentration that inhibited parasite growth by about 50% over one growth cycle. At a concentration of 1 μM, we collected transcriptomic and metabolomic data at multiple time points spanning a complete IDC. We then analyzed these data using a series of computational methods and models to identify global and stage-specific alterations in the parasite. Our analyses of the experimental transcriptomic data revealed alterations in genes involved in initiation and regulation of translation. To elucidate the metabolic adaptations of the parasite to fosmidomycin, we integrated the experimental transcriptomic and metabolomic data with a genome-scale metabolic model of P. falciparum (15–17). Our model-based analyses identified a reduction in lipid synthesis, an increase in polyamine metabolites, and marked perturbation of pH homeostasis in the treated parasites. Taken together, our analyses suggest that fosmidomycin-treated parasites regulate processes, such as nucleotide synthesis and ribosomal biogenesis, to control RNA translation as a survival mechanism. These results also provide a rationale for why combining fosmidomycin with antibiotics that target ribosomes (such as doxycycline, azithromycin, or clindamycin) may be more effective in treating malaria than fosmidomycin monotherapy (14).
RESULTS
In this section, we present the experimental multiomics data and metabolic network modeling results. In the first part, we describe the in vitro experiments that determined the fosmidomycin dose causing 50% growth inhibition at ∼7% parasitemia over one IDC. Next, we report our findings from the metabolomic and transcriptomic data analyses and describe their consequences for parasite physiology. Finally, we interpret the functional consequences of fosmidomycin-induced metabolomic and transcriptomic perturbations with the aid of a whole-genome metabolic network model of P. falciparum. Overall, these results identified the consequences of fosmidomycin-induced impairment in vesicular trafficking and possible metabolic adaptations that support P. falciparum survival under IPP deprivation (see Discussion).
Intraerythrocytic development of fosmidomycin-treated P. falciparum.
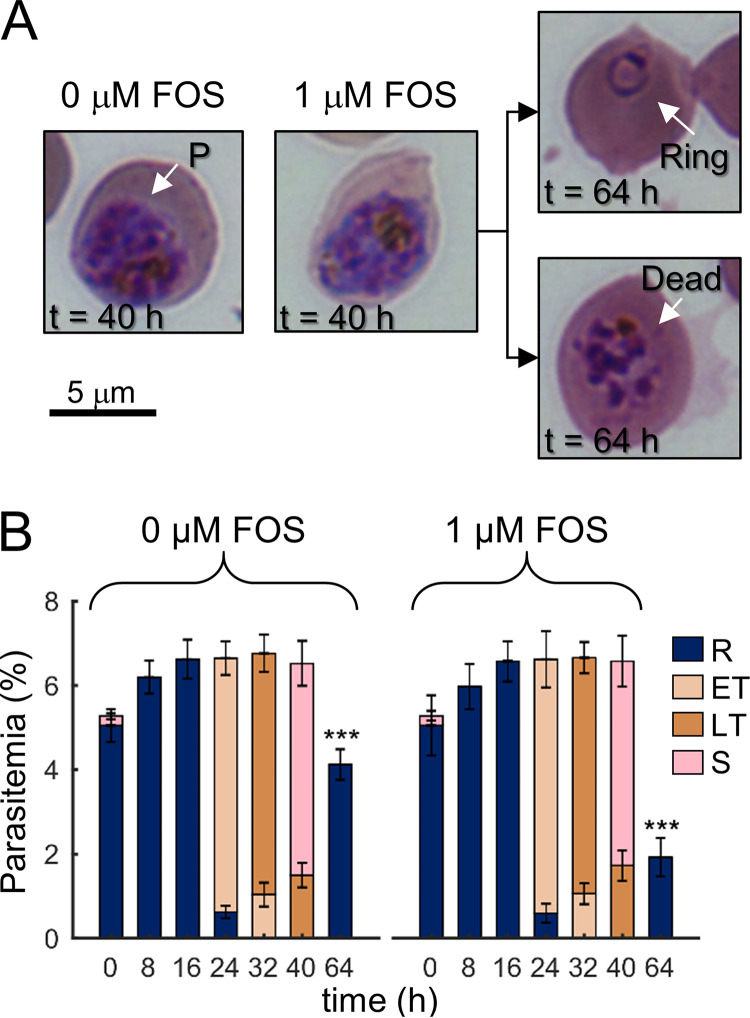
Fosmidomycin, which inhibits IPP synthesis in P. falciparum in the first IDC, prevents the prenylation of Plasmodium proteins that the parasite needs to complete the growth cycle (7). As noted above, however, additional processes are perturbed in parasites undergoing drug-induced delayed death (7). Moreover, because IPP supplementation is sufficient to prevent drug-induced delayed death indefinitely (1), we performed experiments under conditions that limited IPP synthesis to quantify the impact of IPP on Plasmodium processes. To determine the optimal fosmidomycin dose, we first conducted independent experiments in quadruplicate and at six different dosages of fosmidomycin ranging from 0.125 to 4 μM (see Fig. S1 in the supplemental material). Specifically, we treated synchronized ring-stage parasites at a given fosmidomycin concentration for 40 h and then transferred a tenth of the culture into drug-free medium to determine the viability of parasites to replicate in the second growth cycle. We found that a fosmidomycin concentration of 1 μM would allow us to assess the effects of the drug during the first IDC without prematurely killing the parasites. Figure 1A compares the morphologies of parasite-infected erythrocytes (iRBC) under untreated (0 μM) and treated (1 μM) conditions at 40 h of the IDC. Figure 1A also shows the morphologies of treated iRBC that progressed to the second IDC (t = 64 h, upper panel) and failed to progress to the second IDC (t = 64 h, lower panel). Under untreated and drug-treated conditions, the fractions of ring (R)-, early trophozoite (ET)-, late trophozoite (LT)-, and schizont (S)-stage parasites were similar at each sampled time point of the first IDC (from 0 up to 40 h, Fig. 1B). Nonetheless, parasite replication was reduced by ∼50% at 64 h (Fig. 1B), suggesting that significant metabolic perturbations had occurred during the 40-h treatment period and inhibited subsequent parasite development.
FIG 1.
Morphologies and stages of the intraerythrocytic developmental cycle (IDC) of P. falciparum under untreated (0 μM) and fosmidomycin-treated (1 μM) conditions. (A) Giemsa-stained images of erythrocytes infected with P. falciparum (denoted by P) at 40 h into the first IDC. At 40 h, no gross morphological changes were apparent in parasite-infected erythrocytes treated with fosmidomycin (0 μM versus 1 μM fosmidomycin), of which ∼50% progressed to the ring stage of the second IDC (64 h, upper panel). (B) Percentage of ring-, (early and late) trophozoite-, and schizont-stage parasites in the infected cultures. Fosmidomycin treatment did not alter parasite development over time, as indicated by the fractions of different parasite stages before division, i.e., 40 h. During the second IDC in the absence of the drug, approximately half of the ring population was nonviable at the 64-h time point. At each time point, we collected parasitemia data in quadruplicate from independent experiments with the error bars indicating the standard deviation from the mean percentage for a given parasite stage. Asterisks denote a statistically significant (P ≤ 0.01) reduction in the ring population after treatment compared to an untreated control. Abbreviations: ET, early trophozoite; FOS, fosmidomycin; LT, late trophozoite; R, ring; S, schizont.
Global analyses of metabolomic data.
Based on global metabolic profiling of parasite-infected cultures treated with 1 μM fosmidomycin, we identified 469 metabolites at six time points during the IDC (Data Set S1). To identify metabolic alterations due to fosmidomycin treatment, we compared these data with metabolomic data, quantifying 501 metabolites, from untreated cultures of the same Plasmodium strain during the IDC (18). We found that 368 (out of 469) metabolites were common with the metabolomic data from untreated cultures, while the remaining 101 (out of 469) metabolites were present exclusively in the fosmidomycin-treated cultures. To investigate whether fosmidomycin treatment caused production of any specific metabolic class, we categorized the metabolites present exclusively in treated (101 out of 469) and untreated (133 out of 501) cultures based on their respective pathways (see Fig. S2 in the supplemental material). We found that the number of metabolites detected in five out of seven metabolic classes were the same, suggesting that fosmidomycin treatment did not lead to production/consumption of specific metabolite classes. Hence, we focused on analyzing metabolites detected in both untreated and treated cultures.
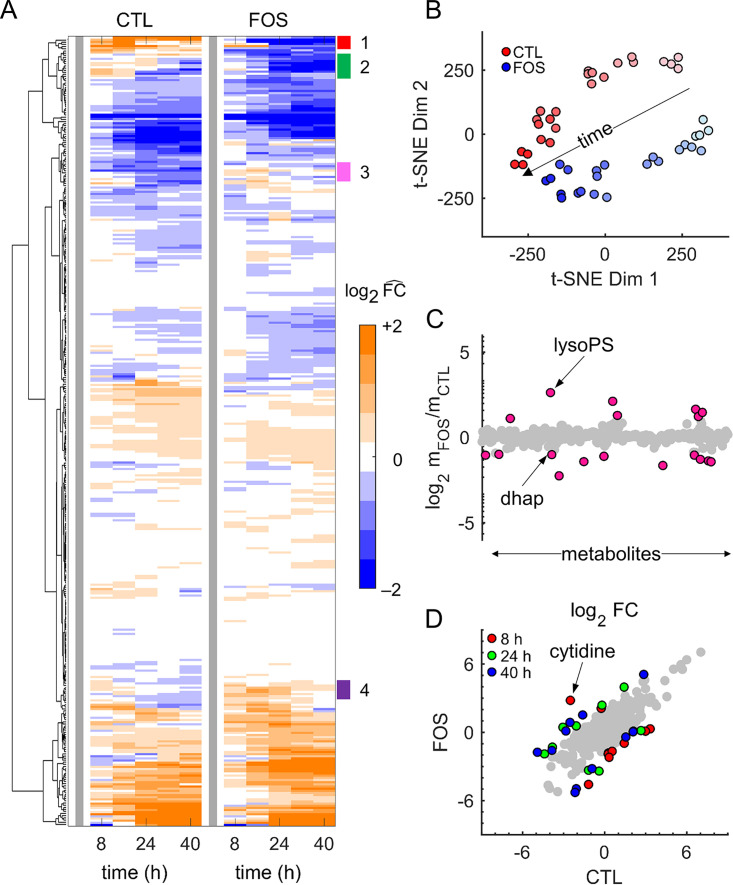
Figure 2A shows hierarchical clustering analysis of these 368 metabolites, in which we normalized the abundance of each metabolite from both untreated and treated cultures by its level at 0 h (thick gray line) to facilitate comparison between the two conditions. Although overall the metabolite profiles were largely similar, we identified four clusters (annotated 1 to 4) that showed differences among the detected metabolites in the treated condition. These metabolite categories included cofactor, glycolysis, purine, pyrimidine, and polyamine metabolites. Specifically, our analysis showed that abundance levels of cytidine, 2-phosphoglycerate, guanosine, adenylosuccinate, adenosine, dihydroxyacetone phosphate (DHAP), and vitamin B6 decreased relative to 0 h under the treated condition, while they increased under the untreated condition. By contrast, the abundance levels of phosphoenolpyruvate, adenosine 5′ diphosphate, and spermine increased relative to 0 h under the treated condition. Fig. S3 to S6 in the supplemental material show the temporal profiles of metabolites contained in clusters 1 to 4.
FIG 2.
Global analyses of metabolomic data. (A) Hierarchical clustering analysis of the averaged data from quadruplicate samples of untreated (CTL) and treated (FOS) parasite cultures. We normalized the data with respect to the corresponding condition results at 0 h (thick gray line) to facilitate temporal comparison between the two conditions. Overall, metabolite abundances in FOS cultures showed temporal alterations similar to those in CTL cultures. By contrast, the annotations (1 to 4) show metabolite clusters with different responses between CTL and FOS cultures. (B) Metabolomic data from quadruplicate samples of CTL and FOS cultures shown in a reduced dimensional representation. We used the t-distributed stochastic neighbor embedding (t-SNE) method (19) to perform dimension reduction. Markers with the lightest color represent data at 0 h, whereas those with the darkest color represent data at 40 h. The separation of CTL and FOS data along the time arrow captures the well-known observation that different parasite stages have different metabolic requirements (17). (C) Average abundance of metabolites quantified from parasite-infected erythrocyte (iRBC) cultures under CTL and FOS conditions. Pink markers denote a greater than (or equal to) 2-fold increase or decrease in the average abundance of a metabolite. (D) Fold change (FC) in abundance of metabolites quantified from uninfected erythrocyte and iRBC cultures under CTL (18) and FOS (this study) conditions. Metabolites for which the log2 FC between the conditions is greater than two are shown in red (8 h), green (24 h), and blue (40 h). All other metabolites are shown in gray. Abbreviations: dhap, dihydroxyacetone phosphate; lysoPS, lysophosphatidylserine.
To ascertain the impact of fosmidomycin treatment on the abundance levels of the quantified metabolites during the IDC, we used the t-distributed stochastic neighborhood embedding (t-SNE) method (19) to conduct a dimensional reduction and provide a simplified visual data representation. Figure 2B shows the separation of the data along two t-SNE dimensions that tracks to the dynamic evolution of P. falciparum parasite metabolism (17). Specifically, metabolomic data from treated cultures separated along the t-SNE dimensions with time in a manner similar to those of untreated cultures (Fig. 2B, arrow), suggesting that fosmidomycin did not cause any large-scale disturbances in parasite metabolism. To identify specific metabolites that showed significant alterations (>2-fold change on a log2 scale) during the IDC, we compared the average abundance of metabolites during the IDC between treated and untreated iRBC cultures (Fig. 2C). Seven metabolites yielded abundance levels more than 2-fold higher in treated relative to untreated cultures, while 11 yielded more than 2-fold lower abundance levels (Data Set S2). DHAP was among the 11 in the latter group. In contrast to our study, Kennedy et al. (7) found an increased abundance of DHAP in parasite cultures treated with indolmycin compared to that in untreated parasite cultures. In our study, we used fosmidomycin—a selective inhibitor of isoprenoid synthesis—whereas Kennedy et al. (7) used indolmycin, which disrupts isoprenoid synthesis by inhibiting RNA translation in the apicoplast. Therefore, this discrepancy in DHAP abundance may be a result of the different drugs that were used to probe isoprenoid synthesis in P. falciparum.
The iRBC cultures we used here consisted of ∼90% uninfected erythrocytes (see Materials and Methods). Given that the metabolism of uninfected erythrocytes (uRBC) changes during the duration of an IDC (18), we analyzed the metabolites in uRBCs cultured in parallel with iRBCs in the same medium containing fosmidomycin. We computed the fold change (FC) in the abundance levels of metabolites detected in iRBC cultures by normalizing them with respect to their abundance levels in the parallel uRBC cultures at each time point (Data Set S3). We performed this normalization to effectively subtract the uRBC background from the iRBC culture data, allowing us to focus on metabolic alterations caused by fosmidomycin treatment. Figure 2D shows the 25 distinct metabolites that were associated with a significant difference in FC between treated and untreated conditions at 8 h, 24 h, and 40 h of the IDC; 17 of these belong to the nucleotide or lipid class (Data Set S3, Main tab). For purine metabolites, FC values under fosmidomycin treatment were greater than three for those containing adenine (Data Set S3) at 0 and 8 h and relatively unchanged over time for all other purine metabolites, whereas FC values at all time points under the untreated condition were greater than four for the other purine metabolites and largely unchanged for the adenine-containing metabolites. By contrast, FC values for the pyrimidine metabolite cytidine under the treated condition were greater than three at all time points, whereas FC values for those under the untreated condition were lower. Together, these results suggest that fosmidomycin selectively alters purine and pyrimidine metabolism at different times during the IDC.
To identify metabolites significantly perturbed by fosmidomycin throughout the IDC, we next computed fold change (FCIDC) values for each metabolite using its average abundance in iRBC and uRBC cultures during the IDC. Table 1 lists metabolites that yielded a greater than 2-fold (log2 scale) difference in FCIDC magnitude under untreated and treated conditions. Of these, five belonged to the nucleotide class, of which cytidine—a precursor for pyrimidine nucleotides—showed higher abundance under the treated condition and lower abundance under the untreated condition. P. falciparum synthesizes pyrimidine metabolites de novo via a key enzyme (20), dihydroorotate dehydrogenase (DHODH), the activity of which requires ubiquinone. Each ubiquinone molecule has isoprenoid sidechains of various lengths (21), which ultimately determine DHODH activity (20). Therefore, the increased abundance of cytidine under fosmidomycin treatment may be a consequence of the inhibited isoprenoid synthesis in P. falciparum (22), which reduces synthesis of pyrimidine nucleotides, such as CTP. We have provided FCIDC values of all metabolites in Data set S4.
TABLE 1.
Fold change in average abundance of metabolites in infected cultures relative to that in uninfected erythrocyte cultures under untreated and fosmidomycin-treated conditions
| Main pathway | Subordinate pathway | Metabolite | FCIDC (SD)a |
|
|---|---|---|---|---|
| CTLb | FOS | |||
| Amino acid | Alanine, aspartate metabolism | N-Acetylasparagine | 1.63 (0.14) | 0.44 (0.03) |
| Carbohydrate | Glycolysis | 2-Phosphoglycerate | 0.29 (0.08) | 1.50 (0.30) |
| Pentose | Arabitol/xylitol | 0.22 (0.02) | 0.04 (0.01) | |
| Lipid | Lysophospholipid | 1-Oleoyl-GPS (18:1) | 1.49 (0.19) | 6.80 (1.10) |
| Monoacylglycerol | 1-Oleoylglycerol (18:1) | 2.59 (0.37) | 0.88 (0.07) | |
| Phospholipid | Phosphoethanolamine | 13.6 (1.14) | 3.87 (0.51) | |
| Sphingolipid | Lactosylceramide (d18:1/24:1) | 3.03 (0.20) | 1.01 (0.18) | |
| Nucleotide | Purine metabolism | Deoxyadenosine monophosphate | 0.74 (0.04) | 2.16 (0.27) |
| Guanine | 4.92 (0.89) | 1.17 (0.04) | ||
| Inosine | 1.30 (0.07) | 0.40 (0.02) | ||
| Xanthine | 3.83 (0.38) | 1.03 (0.03) | ||
| Pyrimidine metabolism | Cytidine | 0.48 (0.08) | 5.18 (0.42) | |
| Peptide | Dipeptide | Phenylalanylalanine | 0.52 (0.05) | 0.17 (0.02) |
Fold-change (FCIDC) values based on a comparison of the average abundance of a metabolite during the IDC in the iRBC culture relative to that in the uRBC culture. The standard deviation (SD) represents deviation from the mean of 10,000 bootstrap samples (see Materials and Methods).
bFCIDC values are from Tewari et al. (18). Abbreviations: CTL, untreated cultures; FOS, fosmidomycin-treated cultures; GPS, glycerophosphoserine; IDC, intraerythrocytic developmental cycle; iRBC, infected erythrocyte; uRBC, uninfected erythrocyte.
Global analyses of transcriptomic data.
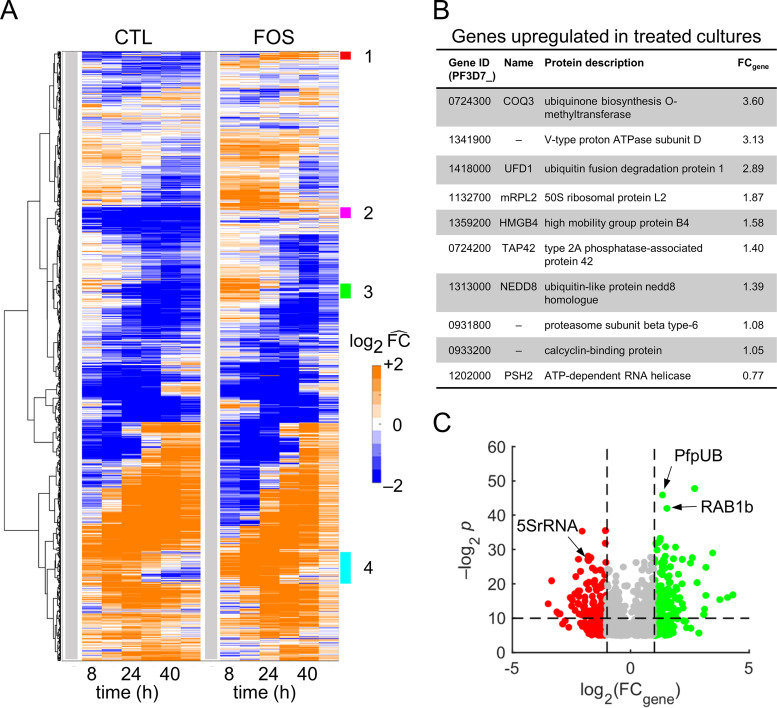
We performed global analyses of averaged transcriptomic data from quadruplicate samples of untreated and treated cultures using hierarchical clustering analyses (HCA) based on normalized expression levels of each gene during the IDC by its level at 0 h (denoted by ). Figure 3A shows the temporal profile of all Plasmodium genes as appearing largely unchanged in fosmidomycin-treated cultures, except for four clusters (annotated 1 to 4) that distinguish the transcriptomic responses. Clusters 1 and 2 primarily contained genes of unknown function, whereas clusters 3 and 4 primarily contained genes of known function (see Fig. S7 in the supplemental material). Most genes of known function belonged to the var, rifin, and stevor multigene families (Data Set S5, Sheets 3 and 4), which encode the highly variable surface proteins PfEMP1, RIFIN, and STEVOR (23), respectively. Although we are primarily concerned with mapping out the detailed physiological response to fosmidomycin, it is interesting to note that even a relatively benign drug stress is coupled to a change in expression of the surface variable protein. This does not impact the IDC progression per se but will affect the response of the host immune system. Importantly, cluster 4 contained genes related to isoprenoid synthesis and associated pathways, such as those of dolichol and terpenoid metabolism, that will affect the cell cycle progression.
FIG 3.
Global analyses of averaged transcriptomic data from quadruplicate samples of untreated and fosmidomycin-treated cultures of P. falciparum. (A) Hierarchical clustering analyses of gene-expression data from untreated (CTL) and treated (FOS) cultures. Each row corresponds to a Plasmodium gene. The gene-expression level, at a given time point, represents average of expression across quadruplicate samples. We normalized the average expression of each gene at each time point by its average expression at 0 h to allow comparison between the two conditions. These 0-h normalized values are denoted by . The vertical gray bar denotes the 0-h time point. We used a Euclidean distance measure of similarity to cluster genes with comparable temporal profiles under the two conditions. Overall, the temporal profile of all Plasmodium genes appeared largely unchanged in FOS cultures except for in four clusters (annotated 1 to 4). (B) Plasmodium genes temporally upregulated by fosmidomycin throughout the intraerythrocytic developmental cycle. We defined a temporally upregulated gene as one that showed 2-fold (or greater) expression levels relative to its 0-h level at all other sampled time points under the treated condition and within 25% of the 0-h level at all time points under the untreated condition. The table shows the 10 temporally upregulated genes with known or putative functions. The last column shows the average fold change in the expression level of the gene (FCgene) under treated versus untreated conditions. (C) Volcano plots of Plasmodium genes that significantly differed (q value < 0.1) under the treated condition. The horizontal dashed line denotes a P value of 0.01; the left and right vertical dashed lines correspond to FCgene values of 0.5 and 2.0, respectively. Green and red markers denote FCgene values of greater than 2.0 and less than 0.5, respectively. Abbreviations: 5SrRNA, gene encoding 5S rRNA; CTL, untreated; FOS, fosmidomycin-treated; PfpUB, gene encoding P. falciparum polyubiquitin; RAB1b, gene encoding Ras-related Rab1b protein.
P. falciparum develops resistance against fosmidomycin after upregulation of DOXP reductoisomerase (24) and methylerythritol phosphate cytidylyltransferase (25), which are enzymes of the isoprenoid biosynthesis pathway. We probed the entire transcriptome (5,851 genes) to further identify upregulated genes that could be involved in the development of resistance to fosmidomycin. Twenty-three genes showed an value of greater than or equal to 2 under the treated condition and within 25% of the expression level at 0 h under the untreated condition. Figure 3B lists the 10 genes with the greatest increase in average expression level with known or putative function that were temporally upregulated during the IDC. In particular, the last enzyme of the ubiquinone biosynthesis pathway, encoded by ubiquinone biosynthesis O-methyltransferase (COQ3) (26), yielded the largest fold change increase by a factor of 3.60. This suggests that the increased expression of COQ3 is a compensatory mechanism that counters the effects of IPP depletion on ubiquinone biosynthesis. Additional upregulated genes belonged to RNA translation and protein degradation processes.
To identify genes significantly altered by fosmidomycin, we further performed a two-way analysis of variance (ANOVA) for each of the 5,851 genes in the transcriptomic data from iRBC cultures, with treatment (0 μM or 1 μM fosmidomycin) and time (0, 8, 16, 24, 32, 40, or 48 h) as the between-subject factors. This identified significant alterations for 1,397 genes (P values ≤0.05 and q values <0.1; see Data Set S6). Figure 3C shows these significantly perturbed genes as a function of average fold change (FCgene) in expression due to fosmidomycin treatment. Among the significantly perturbed genes were Ras-related genes, such as RAB1b, which play a role in vesicular transport (27, 28) (Data Set S6). Because the addition of isoprenoid tails to proteins is an essential posttranslational modification for vesicular transport in P. falciparum (9), this result is a potential consequence of IPP synthesis inhibition due to fosmidomycin treatment. Additional highly altered expression levels were associated with P. falciparum genes encoding polyubiquitin (PfpUB) and 5S rRNA (5SrRNA) (Fig. 3C). PfpUB is essential for ubiquitylation, a posttranslational modification that not only tags proteins for proteasome-mediated protein degradation (29) but also has proteasome-independent functions, such as intracellular trafficking and ribosomal biogenesis (30). The 5SrRNA molecule binds with ribosomal protein L5, which interacts with the eukaryotic initiation factor 5A (eiF5A) (31), which, in turn, is involved in translational control (32) and protein synthesis (33). These results suggest that P. falciparum responds to sublethal fosmidomycin concentrations by altering processes that modulate the gene translation process.
Impact of fosmidomycin on the prenylome of P. falciparum.
Kennedy et al. (7) demonstrated that the disruption of protein prenylation is the proximal cause of delayed death in isoprenoid-deprived parasites. To quantify the effect of fosmidomycin on the genes encoding these proteins, we computed the FCgene of these genes and interpolated the time of peak gene-expression levels (tpeak) during the IDC under treated and untreated conditions (see Materials and Methods). Table 2 lists tpeak and FCgene values of Plasmodium genes that encode proteins prenylated by isoprenoids. The tpeak of genes encoding the SNARE protein (YKT6.2) and Rab1b protein, both of which are part of the vesicular transport pathway between the endoplasmic reticulum (ER) and Golgi apparatus (34, 35), differed between treated and untreated conditions by more than 30 h, with RAB1b having an FCgene value greater than three. Given that proteins incorrectly folded in the ER are ubiquitylated and shuttled to the proteasome for degradation (36, 37), the delay in the synthesis of genes encoding vesicular transport proteins and the increased expression of the gene encoding PfpUB (Fig. 3C) suggest that fosmidomycin treatment may lead to an increased abundance of misfolded proteins in the ER. Upon closer examination of genes involved in the ubiquitin-proteasome system of P. falciparum (see Table S1 in the supplemental material), we found that expression levels of genes encoding the proteasome and proteins that transport ubiquitylated proteins to proteasome were elevated after fosmidomycin treatment, suggesting that fosmidomycin-treated parasites deploy mechanisms to cope with misfolded proteins.
TABLE 2.
P. falciparum genes encoding proteins prenylated by isoprenoids
| Gene ID (PF3D7_) | Name | Protein description | tpeak (h of IDC)a |
FCgeneb (SD) | |
|---|---|---|---|---|---|
| CTL | FOS | ||||
| 1319100 | Conserved DUF544 protein, unknown function | 0 | 0 | 1.00 (0.16) | |
| 1437900 | HSP40 | HSP40, subfamily A, putative | 20 | 23 | 1.32 (0.21) |
| 0322000 | CYP19A | Peptidyl-prolyl cis-trans isomerase | 23 | 25 | 1.84 (0.54) |
| 1113100 | PRL | Protein tyrosine phosphatase | 0 | 0 | 1.03 (0.14) |
| 1231100 | RAB2 | Ras-related protein, Rab2 | 8 | 13 | 4.01 (4.22) |
| 0106800 | RAB5c | Ras-related protein, Rab5c | 35 | 35 | 2.21 (0.65) |
| 1320600 | RAB11a | Ras-related protein, Rab11a | 48 | 48 | 1.31 (0.32) |
| 1340700 | RAB11b | Ras-related protein, Rab11b | 36 | 37 | 0.93 (0.22) |
| 0807300 | RAB18 | Ras-related protein, Rab18 | 20 | 24 | 0.94 (0.37) |
| 0513800 | RAB1a | Ras-related protein, Rab1a | 28 | 31 | 0.98 (0.28) |
| 0512600 | RAB1b | Ras-related protein, Rab1b | 8 | 40 | 3.05 (0.76) |
| 0211200 | RAB5a | Ras-related protein, Rab5a | 30 | 34 | 0.80 (0.27) |
| 1310600 | RAB5b | Ras-related protein, Rab5b | 41 | 41 | 0.98 (0.17) |
| 1144900 | RAB6 | Ras-related protein, Rab6 | 39 | 41 | 1.34 (0.40) |
| 0903200 | RAB7 | Ras-related protein, Rab7 | 32 | 32 | 0.76 (0.10) |
| 0910600 | YKT6.1 | SNARE protein, putative | 48 | 48 | 1.47 (0.23) |
| 1324700 | YKT6.2 | SNARE protein, putative | 40 | 7 | 1.23 (0.40) |
Hour (tpeak) of intraerythrocytic developmental cycle (IDC) when the gene was maximally expressed.
The fold-change value (FCgene) of a gene was computed as the ratio of the average gene-expression level under the treated condition to that under the untreated condition. Abbreviations: CTL, untreated; FOS, fosmidomycin-treated; SD, standard deviation.
We also found that fosmidomycin substantially altered tpeak and FCgene of genes encoding amino acid (PF3D7_0515500), hexose (PF3D7_0204700), metal (PF3D7_0523800), and protein (PF3D7_0821800) transporters. For example, PF3D7_1341900 and PF3D7_0934500 had a more than 15 h delay in tpeak and an FCgene ≥2 under treated conditions. These two genes encode proton ATPases that parasites need to maintain an acidic pH in the food vacuole (38). Kennedy et al. (7) reported disruption of the food vacuole in the absence of isoprenoid synthesis. Because the cytosol of the parasite is less acidic than the food vacuole (pH 7.15 versus 5.18 [39]), a disruption of the vacuole would increase its pH via proton diffusion, requiring ATPases and/or transporters to maintain pH homeostasis. Table 3 lists substantially altered genes that encode ATPases and transporters of P. falciparum.
TABLE 3.
Genes encoding ATPases and transporters of P. falciparum
| Gene ID (PF3D7_) | Name | Protein description | tpeak (h of IDC)a |
FCgeneb (SD) | |
|---|---|---|---|---|---|
| CTL | FOS | ||||
| 0707400 | AAA family ATPase, putative | 15 | 31 | 3.29 (2.79) | |
| 1426500 | ABCG2 | ABC transporter G family member 2 | 0 | 39 | 0.81 (0.21) |
| 1037300 | ADT | ADP/ATP transporter on adenylate translocase | 13 | 23 | 0.87 (0.20) |
| 0515500 | Amino acid transporter, putative | 0 | 38 | 0.70 (0.22) | |
| 0523800 | Divalent metal transporter, putative | 0 | 6 | 1.88 (0.36) | |
| 0204700 | HT | Hexose transporter | 0 | 13 | 1.05 (0.38) |
| 0821800 | SEC61B | Protein transport protein, putative | 9 | 15 | 1.76 (0.55) |
| 0622300 | Vacuolar transporter chaperone, putative | 0 | 26 | 0.51 (0.11) | |
| 0721900 | V-type ATPase V0 subunit e, putative | 17 | 41 | 1.32 (0.33) | |
| 1354400 | V-type proton ATPase 21 kDa proteolipid subunit | 17 | 40 | 1.05 (0.13) | |
| 1341900 | V-type proton ATPase subunit D, putative | 0 | 16 | 3.13 (1.98) | |
| 0934500 | V-type proton ATPase subunit E, putative | 0 | 16 | 2.01 (0.98) | |
Hour (tpeak) of intraerythrocytic developmental cycle (IDC) when the gene was maximally expressed.
bThe fold-change value (FCgene) of a gene was computed as the ratio of the average gene-expression level under the treated condition to that under the untreated condition. Abbreviations: CTL, untreated; FOS, fosmidomycin-treated; SD, standard deviation.
Impact of fosmidomycin on the gene regulation machinery in P. falciparum.
During the IDC, P. falciparum executes a finely tuned gene-expression program (40), synthesizing specific genes at specific times. In response to stress, such as isoleucine deprivation (41) or fosmidomycin treatment, the parasite regulates gene expression via transcriptional, posttranscriptional, and translational mechanisms (42). We next quantified how fosmidomycin affects the posttranscriptional and translational machinery to allow the parasite to rapidly adjust gene expression in response to an environmental stressor.
Table 4 lists tpeak and FCgene values of genes that play a role in translational initiation (the rate-limiting step of translation [42]), specifically, those encoding eukaryotic initiation factors (eIFs), methyltransferases, and ribosome proteins. Both eIF2β and eIF2γ, which are part of the eIF2 complex (42), showed a delay in tpeak of 10 h (with FCgene ≥1.5) after drug treatment. In addition to interacting with partners of the translation complex, eIF2β also binds protein phosphatase 1 (43), a key regulator of gene transcription, protein synthesis, and cell division (44). DNA/RNA methylation is a key step of gene regulation requiring S-adenosylmethionine as the methyl donor. Methyltransferase genes associated with tRNA modification, mRNA 5′ capping, and ribosomal biogenesis showed a delay in tpeak after fosmidomycin treatment. Genes encoding ribosomal proteins also showed a substantial delay in tpeak (∼15 h) after fosmidomycin treatment, suggesting a decrease in the rate of RNA translation.
TABLE 4.
P. falciparum genes involved in translational initiation
| Gene ID (PF3D7_) | Name | Protein description | tpeak (h of IDC)a |
FCgeneb (SD) | |
|---|---|---|---|---|---|
| CTL | FOS | ||||
| Initiation factors | |||||
| 1010600 | eIF2β | Eukaryotic initiation factor 2 subunit beta | 13 | 23 | 1.53 (0.28) |
| 1410600 | eIF2γ | Eukaryotic initiation factor 2 subunit gamma | 6 | 18 | 2.29 (0.53) |
| 0918300 | EIF3F | Eukaryotic initiation factor 3 subunit F | 10 | 19 | 1.86 (0.51) |
| 0815600 | EIF3G | Eukaryotic initiation factor 3 subunit G | 0 | 12 | 1.94 (1.07) |
| 1468700 | eIF4A | Eukaryotic initiation factor 4A | 8 | 22 | 1.32 (0.24) |
| 1204300 | EIF5A | Eukaryotic initiation factor 5A | 18 | 24 | 1.48 (0.32) |
| Methyl group requiring enzymes | |||||
| 1415800 | KsgA1 | Dimethyladenosine transferase, putative | 13 | 20 | 1.47 (0.44) |
| 1313200 | MTFMT | Methionyl-tRNA formyltransferase, putative | 24 | 32 | 1.26 (1.01) |
| 1235500 | mRNA (N6-adenosine) methyltransferase, putative | 19 | 25 | 1.18 (0.14) | |
| 0525600 | mRNA (N7-guanine) methyltransferase | 24 | 47 | 1.18 (0.16) | |
| 1407500 | Multifunctional methyltransferase subunit TRM112 | 0 | 15 | 0.75 (0.18) | |
| 1406900 | RlmN | Radical SAM protein, putative | 22 | 42 | 1.03 (0.23) |
| 0622500 | RNA methyltransferase, putative | 9 | 15 | 0.68 (0.17) | |
| 1319300 | TRM1 | tRNA (guanine26-N2) dimethyltransferase | 11 | 16 | 0.75 (0.12) |
| 1343200 | tRNA guanosine-2′-O-methyltransferase, putative | 11 | 41 | 1.22 (0.24) | |
| 1120500 | tRNA nucleotidyltransferase, putative | 7 | 14 | 1.20 (0.49) | |
| Ribosomal proteins | |||||
| 0112300 | 18S ribosomal RNA | 4 | 15 | 0.95 (0.20) | |
| 1317800 | RPS19 | 40S ribosomal protein | 5 | 15 | 2.32 (1.28) |
| 0306900 | 40S ribosomal protein S23, putative | 0 | 15 | 1.39 (0.83) | |
| 0725800 | 5.8S ribosomal RNA | 0 | 39 | 0.91 (0.06) | |
| 1424400 | 60S ribosomal protein | 9 | 14 | 2.04 (0.65) | |
| 1338200 | RPL6 | 60S ribosomal protein | 7 | 15 | 3.05 (1.42) |
| API01800 | RPL16 | Apicoplast ribosomal protein L16 | 7 | 32 | 2.21 (1.31) |
| API00100 | RPS4 | Apicoplast ribosomal protein S4 | 0 | 31 | 0.62 (0.23) |
| 1457700 | Large ribosomal subunit nuclear export factor, putative | 0 | 15 | 1.41 (0.13) | |
Hour (tpeak) of intraerythrocytic developmental cycle (IDC) when the gene was maximally expressed.
The fold-change value (FCgene) of a gene was computed as the ratio of the average gene-expression level under the treated condition to that under the untreated condition. Abbreviations: CTL, untreated; FOS, fosmidomycin-treated; SD, standard deviation.
Translational repression could also allow the parasite to regulate translation in response to an external stressor. Table 5 lists genes involved in translation regulation and categorized into RNA-binding proteins, CNOT (carbon catabolite repression—negative on TATA-less) complex, mRNA decapping, and mRNA degradation functionalities. Among the genes encoding RNA-binding proteins that suppress translation, ALBA2 and CELF2 showed a delay in tpeak of greater than 15 h (with FCgene ≥1.5) after fosmidomycin treatment. The CNOT complex, which is the main structure involved in deadenylation (45, 46), controls mRNA degradation (47) and decapping (48) and, hence, regulates the rate of translation. Two subunits of the CNOT complex, CAF16 and CAF40, showed a delay in tpeak of greater than 6 h and FCgene values of 2 or greater after fosmidomycin treatment, which is a likely consequence of downregulation of the PUF2 gene (Table 5). Lindner et al. (49) found that downregulation of PUF2, which controls RNA metabolism, resulted in upregulation of genes of the CNOT complex (such as CAF16). After deadenylation, mRNA decays via either the 5′ to 3′ or the 3′ to 5′ pathway (50). The expression of genes involved in mRNA decapping that substantially changed after fosmidomycin treatment, such as LSM3 and LSM5, showed a delay in tpeak of 10 h or greater with an FCgene value of 5 or greater. No such perturbations occurred for mRNA degradation genes.
TABLE 5.
P. falciparum genes involved in translational regulation
| Gene ID (PF3D7_) | Name | Protein description | tpeak (h of IDC)a |
FCgeneb (SD) | |
|---|---|---|---|---|---|
| CTL | FOS | ||||
| RNA-binding proteins | |||||
| 1359400 | CELF1 | CUGBP Elav-like family member 1 | 11 | 15 | 1.13 (0.21) |
| 1409800 | CELF2 | CUGBP Elav-like family member 2, putative | 0 | 19 | 2.35 (1.65) |
| 1346300 | ALBA2 | DNA/RNA-binding protein Alba 2 | 0 | 48 | 1.68 (0.79) |
| 1006200 | ALBA3 | DNA/RNA-binding protein Alba 3 | 22 | 25 | 0.83 (0.15) |
| 0417100 | PUF2 | mRNA-binding protein PUF2 | 0 | 0 | 0.48 (0.27) |
| CNOT complex | |||||
| 0107200 | CCR4-3 | Carbon catabolite repressor protein 4, putative | 13 | 26 | 1.12 (0.66) |
| 1434000 | CAF16 | CCR4-associated factor 16, putative | 0 | 15 | 6.85 (5.67) |
| 1128600 | NOT2 | CCR4-NOT transcription complex subunit 2, putative | 0 | 29 | 0.96 (0.32) |
| 1235300 | NOT4 | CCR4-NOT transcription complex subunit 4, putative | 0 | 4 | 0.94 (0.14) |
| 1006100 | NOT5 | CCR4-NOT transcription complex subunit 5, putative | 5 | 10 | 1.34 (1.00) |
| 0507600 | CAF40 | Cell differentiation protein, putative | 8 | 14 | 1.88 (0.45) |
| mRNA decapping | |||||
| 1308900 | DCP2 | mRNA decapping enzyme 2, putative | 10 | 15 | 0.80 (0.13) |
| 0819900 | LSM3 | U6 snRNA-associated Sm-like protein LSm3, putative | 11 | 25 | 0.98 (0.22) |
| 1443300 | LSM5 | U6 snRNA-associated Sm-like protein LSm5, putative | 8 | 11 | 5.19 (4.48) |
| 1209200 | LSM7 | U6 snRNA-associated Sm-like protein LSm7, putative | 14 | 24 | 1.28 (0.36) |
| 0829300 | LSM8 | U6 snRNA-associated Sm-like protein LSm8, putative | 12 | 22 | 1.62 (0.55) |
| mRNA degradation | |||||
| 0209200 | MTR3 | 3′ exoribonuclease, putative | 8 | 11 | 1.50 (0.62) |
| 1106300 | 5′–3′ exoribonuclease 1, putative | 24 | 30 | 0.75 (0.25) | |
| 0720000 | CSL4 | Exosome complex component CSL4, putative | 8 | 12 | 0.75 (0.13) |
| 0909900 | Helicase SKI2W, putative | 0 | 5 | 1.24 (0.09) | |
aHour (tpeak) of intraerythrocytic developmental cycle (IDC) when the gene was maximally expressed.
The fold-change value (FCgene) of a gene was computed as the ratio of the average gene-expression level under the treated condition to that under the untreated condition. Abbreviations: CNOT, carbon catabolite repression—negative on TATA-less; CTL, untreated; FOS, fosmidomycin-treated; SD, standard deviation.
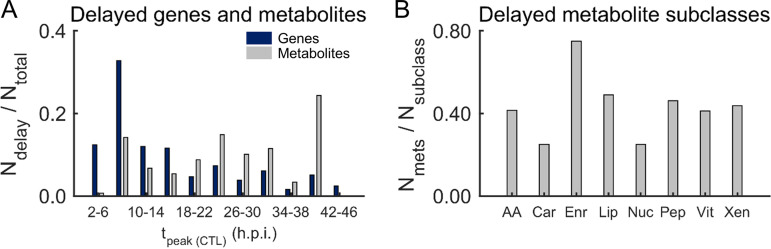
Overall, we found that fosmidomycin treatment increased and/or delayed the peak expression of genes involved in protein degradation, substrate transport, translational initiation, and translational regulation. Specifically, we found that fosmidomycin increased the tpeak by over 8 h for 1,271 genes (Data Set S7), of which ∼60% genes had peak expression levels at 0, 1, 47, or 48 h. Figure 4A (blue) shows genes that had a greater than 8 h delay in tpeak as a fraction of the total number of delayed genes with tpeak between 2 and 46 h. In Fig. 4A, we have omitted genes with tpeak at 0, 1, 47, and 48 h because these time points are identical, considering a delay ±4 h due to multiple magnetic-purifications steps (see Materials and Methods) and the period of a typical IDC, i.e., 48 h. In contrast to the delay in gene transcription, fosmidomycin increased the time of peak abundance level by more than 8 h for metabolites with peak abundance levels during the late stages of the IDC (Fig. 4A, gray). Further grouping of the delayed metabolites into subclasses showed that fosmidomycin primarily influenced energy metabolites, such as alpha-ketoglutarate, fumarate, and malate, which are part of the tricarboxylic acid (TCA) cycle in mitochondria (Fig. 4B). This metabolic remodeling may result from impaired ubiquinone metabolism, which depends on isoprenoid synthesis (51) that is inhibited due to fosmidomycin treatment. Overall, the gene-expression alterations preceded the metabolic alterations (Fig. 4A), presumably due to the gene-protein delay (52), making specific connections between the altered genes and associated metabolic pathway untenable. To investigate consequences of fosmidomycin-induced gene alterations on P. falciparum metabolism, we used a metabolic network model, incorporating the delay in gene expression and protein abundance of most Plasmodium genes (53).
FIG 4.
Estimated delay in peak gene expression and metabolite abundance in response to a sublethal dose of fosmidomycin. (A) Genes (blue) and metabolites (gray) with a delay in their peak expression (tpeak) and abundance of 8 h or more under treated versus untreated conditions. We normalized the number of delayed genes or metabolites (Ndelay) within each tpeak interval by the total number of delayed genes or metabolites (Ntotal) with tpeak between 2 and 46 h. The abscissa shows tpeak intervals used to count the number of delayed genes (blue) or metabolites (gray). There is no gray bar at “42 to 46” because the metabolomic data were only collected up to the 40-h time point. (B) Number of metabolites grouped by subclass, with a delay in abundance of 8 h or more under treated versus untreated conditions. We normalized the number of delayed metabolites (Nmets) by the total of number of metabolites in each subclass (Nsubclass). Abbreviations: AA, amino acids; Car, carbohydrates; CTL, untreated culture of parasite-infected erythrocytes; Enr, energy; h.p.i., hour post parasite-infection; Lip, lipids; Nuc, nucleotides; Pep, peptides; Vit, cofactor and vitamins; Xen, xenobiotics.
Quantifying the impact of fosmidomycin on the metabolism of P. falciparum.
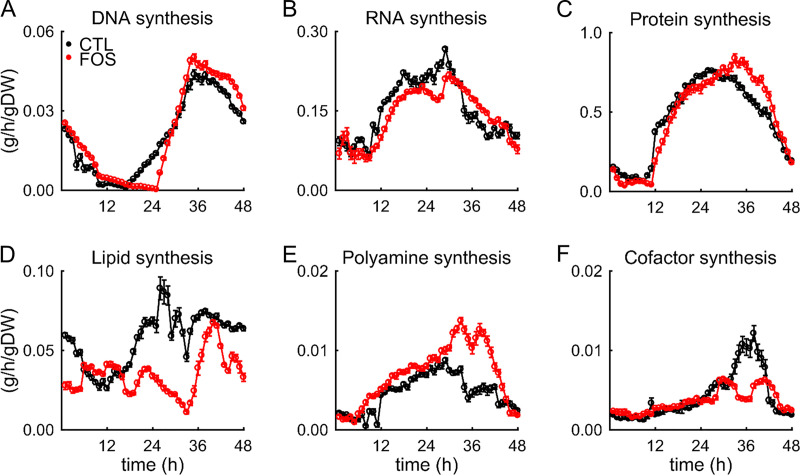
The major mechanism for controlling metabolic activity (54), as defined by the consumption and production of specific metabolites (metabolic fluxes), is gene regulation. Moxley et al. (55) demonstrated that combining mRNA levels with a metabolic network model enables a consistent approach to interpret changes in mRNA levels in relation to altered cellular metabolism. Here, we quantified the impact of fosmidomycin on metabolic fluxes using the transcriptomic data (presented above) and metabolic network modeling of P. falciparum (15). We integrated the metabolic fluxes during the IDC with the metabolic network to estimate the net synthesized amounts of several macromolecules and essential metabolites contributing to parasite growth in silico. Figures 5A, 5B, and 5C show simulated rates of DNA, RNA, and protein synthesis under untreated and fosmidomycin-treated conditions, respectively. In the presence of fosmidomycin, the rates of synthesis for lipids (Fig. 5D) and cofactors (Fig. 5F) were substantially lower than those in its absence. By contrast, the rate of polyamine synthesis increased under fosmidomycin treatment (Fig. 5E). These results indicate that in the presence of fosmidomycin, compensatory mechanisms maintain the rates of DNA, RNA, and protein synthesis but fail to synthesize the full complement of lipids.
FIG 5.
Rate of macromolecule synthesis in P. falciparum under untreated (black line) and treated (red line) conditions. Plots of synthesis rates are shown for (A) DNA, (B) RNA, (C) protein, (D) lipid, (E) polyamine, and (F) cofactor. Error bars indicate one standard deviation (SD) of 50 simulations computed after adding random Gaussian noise with a mean of zero and a SD of 5% to the transcriptomic data obtained under untreated and treated conditions. Abbreviations: CTL, untreated; FOS, treated; gDW, gram dry weight of the parasite.
Table 6 lists the total synthesized amount of essential metabolites under untreated and treated conditions. We numerically integrated the estimated rates of metabolic fluxes for essential metabolites and macromolecules over the 48-h IDC to estimate the total synthesized amount. In the presence of fosmidomycin, the synthesized amount of lipids was lowest among macromolecules, ∼1.5-fold lower than in its absence. Among essential metabolites, spermidine was the most synthesized, ∼2.3-fold higher in the presence of the drug than in its absence. Spermidine—a product of polyamine biosynthesis (56), which is part of the purine-recycling pathway used by most organisms to maintain adenine and methionine pools (57)—was higher under the treated condition (Fig. 5E). By contrast, phosphate was ∼6.6-fold lower under the treated condition; according to our metabolic network analysis, this occurred because of the metabolic remodeling that promotes the synthesis of purine-based nucleotides via polyamine biosynthesis, which requires phosphate to synthesize purine nucleotides.
TABLE 6.
Metabolite production/incorporation in untreated and fosmidomycin-treated parasites
| Metabolite | Total synthesized amount (mmol/gDW)a |
||
|---|---|---|---|
| Spermidine | 52.8 | 120.2 | 2.28 |
| Coenzyme A | 39.4 | 62.2 | 1.58 |
| Fe3+ | 32.3 | 46.2 | 1.43 |
| Nicotinamide adenine dinucleotide oxidized | 28.3 | 38.1 | 1.35 |
| Flavin adenine dinucleotide oxidized | 44.7 | 51.3 | 1.15 |
| Putrescine | 33.8 | 37.9 | 1.12 |
| Fe2+ | 47.0 | 47.0 | 1.00 |
| 47.4 | 47.1 | 0.99 | |
| 5,10-Methylenetetrahydrofolate | 61.6 | 60.7 | 0.98 |
| Thiamine diphosphate | 46.4 | 44.8 | 0.97 |
| S-Adenosylmethionine | 348.1 | 318.2 | 0.91 |
| 10-Formyltetrahydrofolate | 43.1 | 39.2 | 0.91 |
| Riboflavin | 55.3 | 47.0 | 0.85 |
| Ubiquinol | 46.1 | 38.5 | 0.83 |
| 259.7 | 200.9 | 0.77 | |
| 5,6,7,8-Tetrahydrofolate | 75.2 | 46.3 | 0.62 |
| Protoheme | 210.6 | 129.3 | 0.61 |
| Nicotinamide adenine dinucleotide phosphate | 122.5 | 47.40 | 0.39 |
| Pyridoxal 5-phosphate | 219.9 | 40.69 | 0.19 |
| 462.0 | 68.72 | 0.15 | |
| Macromolecule | Total synthesized amount (g/gDW) | Δm | |
| DNA | 0.9 | 1.0 | 1.16 |
| RNA | 6.6 | 5.1 | 0.78 |
| Protein | 21.2 | 15.8 | 0.74 |
| Phospholipid | 2.8 | 1.8 | 0.64 |
The total accumulated amount of a metabolite (or macromolecule) was computed by numerically integrating the synthesis rate (v) of each metabolite over the 48-h intraerythrocytic developmental cycle under control (CTL) or fosmidomycin (FOS)-treated conditions. Abbreviation: gDW, grams dry weight of the parasite.
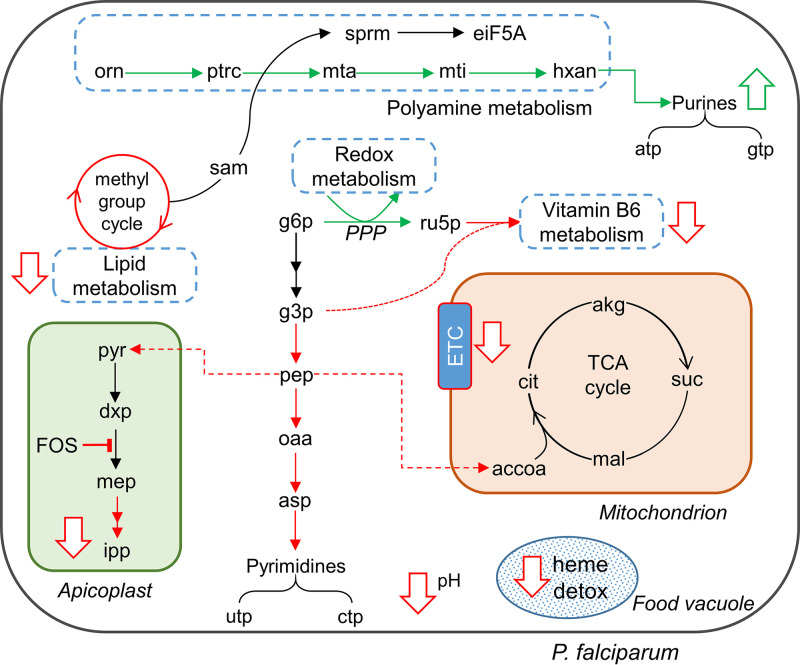
Figure 6 summarizes the findings of our integrated genome-scale metabolic network analysis. At the pathway level, our calculations predicted inhibition of heme detoxification, methyl-group reactions, pH homeostasis, isoprenoid synthesis, pyrimidine metabolism, the TCA cycle, and vitamin B6 metabolism. The inhibition of isoprenoid synthesis by fosmidomycin was expected (22). Moreover, the inhibition of heme detoxification, pH homeostasis, and vitamin B6 metabolism is consistent with work showing that isoprenoids are crucial for transporting hemoglobin into food vacuoles (7) and play a role in vitamin B6 biosynthesis (58). The metabolic network analysis predicted inhibition of the electron transport chain (ETC; Fig. 6, downward-pointing arrow in the mitochondrion) by fosmidomycin treatment (Δm of ubiquinol = 0.83; see Table 6). It also predicted a 10% reduction in the total accumulation of S-adenosylmethionine (Δm = 0.91; see Table 6), causing a reduction in the rate of lipid synthesis (Fig. 5D). At the same time, we saw activation of polyamine, purine, and redox metabolism under fosmidomycin treatment, wherein ATP and GTP metabolites were synthesized via polyamine synthesis using the purine-recycling pathway. It also predicted an increase in spermidine (denoted as sprm in Fig. 6) via the polyamine pathway. Spermidine—a polyamine metabolite—activates eiF5A (59), which in turn plays an important role in translation and is coupled to 5SrRNA (Fig. 3C).
FIG 6.
Metabolic alterations due to fosmidomycin (FOS) treatment predicted by genome-scale metabolic model simulations. FOS treatment causes an inhibition of isoprenoid synthesis, leading to a reduction in heme detoxification. The simulations suggest that the parasite enhances redox metabolism to counter increased oxidative stress, a consequence of reduced heme detoxification (60). Specifically, the parasite increased the flow of metabolites through the pentose phosphate pathway (PPP), reducing metabolic fluxes involved in the synthesis of pyrimidine metabolites and acetyl-CoA. In addition, the parasite increased flux through polyamine metabolism to generate the purine nucleotides ATP and GTP. This increased flux also activated a eukaryotic initiation factor, eiF5A, via spermidine (sprm) and hypusine (24). The simulations also predicted inhibition of the methyl-group cycle, reducing lipid synthesis under FOS treatment (Fig. 5D). Abbreviations: akg, alpha-ketoglutarate; asp, aspartate; cit, citrate; dxp, deoxy-xylulose phosphate; ETC, electron transport chain; g3p, glyceraldehyde 3-phosphate; g6p, glucose 6-phosphate; hxan, hypoxanthine; ipp, isopentenyl diphosphate; mal, malate; mep, methylerythritol phosphate; mta, methylthioadenosine; mti, methylthioinosine; oaa, oxaloacetate; orn, ornithine; pep, phosphoenolpyruvate; ptrc, putrescine; pyr, pyruvate; ru5p, ribulose 5-phosphate; sam, S-adenosylmethionine; suc, succinate.
Finally, consistent with the literature (7), our model predicted an inhibition of the heme polymerization reaction, a process that parasites deploy to detoxify heme radicals released by hemoglobin degradation (60). We found that the decrease in heme polymerization process was due to increased production of heme radicals and oxidized glutathione. To counter the increased oxidative stress, fosmidomycin-treated parasites enhanced flux through the pentose phosphate pathway, the main pathway for generating reducing equivalents for several antioxidant enzymes (61).
DISCUSSION
Most parasites of the phylum Apicomplexa synthesize isoprenoids via a nonmevalonate pathway that is absent in humans (2), making this pathway an attractive target for drug discovery. Several studies have investigated the effects of isoprenoid deprivation in the delayed death of P. falciparum during blood-stage development (1, 7, 9). In particular, Kennedy et al. (7) used indolmycin—an antibiotic that targets RNA translation in the apicoplast—to deplete isoprenoids in P. falciparum and investigate the mechanisms of delayed death. Unlike indolmycin, which depletes isoprenoids in the second IDC by causing apicoplast dysfunction, fosmidomycin is a fast-acting antibiotic that depletes isoprenoids and can cause death in the first IDC. Our approach of using a sublethal dose allowed us to study the effects of isoprenoid depletion without perturbing the function of the entire apicoplast. As the parasite completed the first IDC in the presence of fosmidomycin, we could characterize compensatory mechanisms and responses by monitoring time-dependent transcriptomic and metabolic alterations.
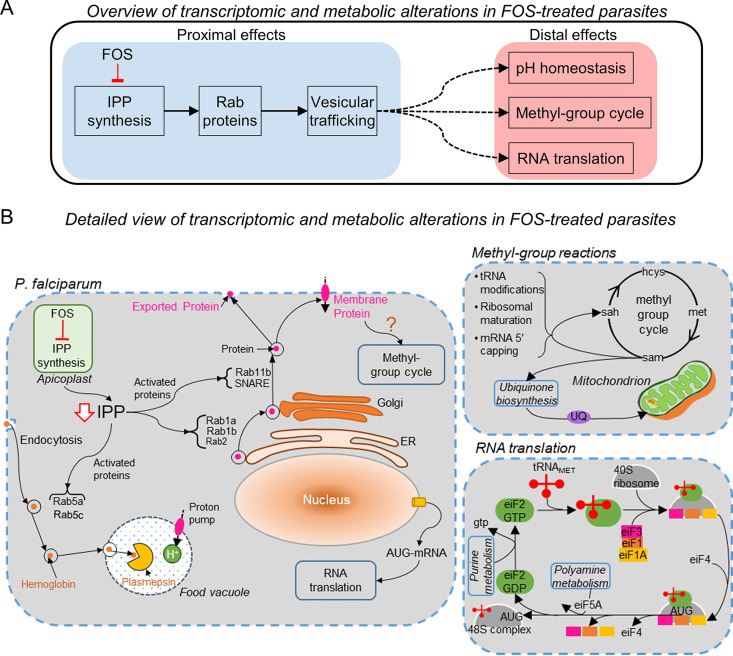
Figure 7A classifies our findings into proximal (blue-shaded region) and distal (red-shaded region) effects of fosmidomycin-induced isoprenoid depletion on P. falciparum. Figure 7B, on the other hand, provides a detailed view of our findings categorizing them into three major components: (i) intracellular effects of fosmidomycin-induced isoprenoid depletion on P. falciparum (left of the figure), (ii) consequences of impaired methyl-group cycling in the treated parasites (upper right), and (iii) consequences of altered metabolism on RNA translation (lower right). We expand on these topics in the text below.
FIG 7.
Transcriptomic and metabolic alterations in fosmidomycin-treated P. falciparum parasites. (A) An overview of proximal (direct) and distal (indirect) effects of fosmidomycin (FOS) on P. falciparum parasites. We know that FOS treatment directly inhibits synthesis of isoprenoids (IPP), which are essential for prenylation of proteins (Rab1a, Rab1b, etc.) involved in secretory and endocytic vesicular transport pathways. The blue-shaded region outlines the proximal effects of FOS treatment and the red-shaded region outlines the distal effects of FOS treatment identified in this study. (B) A detailed view of the proximal and distal processes altered in P. falciparum due to FOS treatment. We found that FOS substantially altered the expression levels of genes encoding Rab GTPases and SNARE proteins and caused a delay in peak expression of these genes. Our genome-scale metabolic model calculations predicted a delay in methyl-group cycle (upper right), which synthesizes S-adenosylmethionine (sam), the methyl-group donor for over 95% methyltransferase enzymes (70). We also found a delay in peak expression of methyltransferase genes associated with tRNA modification and ribosomal biogenesis, suggesting a probable delay in RNA translation due to FOS treatment (lower right). Our metabolic network model analysis predicted that P. falciparum increased the activity of enzymes in purine and polyamine metabolism (lower right), which support protein synthesis, to compensate for delays in RNA translation. Abbreviations: AUG, start codon of mRNA; eiF, eukaryotic initiation factor; hcys, homocysteine; IPP, isopentenyl diphosphate; met, methionine; NSF, N-ethylmaleimide sensitive factor; sah, S-adenosylhomocysteine; SNAP, soluble NSF attachment protein; SNARE, SNAP receptor; UQ, ubiquinone.
Effects of fosmidomycin-induced isoprenoid depletion on vesicular trafficking.
As the primary effect of fosmidomycin serves to diminish isoprenoid synthesis, this directly affects Rab GTPases, which require IPP for prenylation, and, hence, their ability to adopt their correct physiologic state and execute their proper function (Fig. 7A, blue-shaded region). These proteins regulate intracellular transport by acting as molecular switches that modulate the formation, tethering, and fusion of transport vesicles (28). Prenylation of Rab GTPases allows Rab proteins to modulate vesicular transport via retrograde transport (Rab5a and Rab5c), such as shuttling hemoglobin from the host erythrocyte to the food vacuole (Fig. 7B, left of the figure). This effect is observed in the study of Kennedy et al. (7), where parasites lacking isoprenoids could not transport hemoglobin to the food vacuole. We found that 1 μM fosmidomycin did significantly alter the average expression level for the gene encoding Rab5c ∼2-fold, whereas Rab5a mRNA levels did not change significantly (Table 2). This indicates that specific components of the hemoglobin transport mechanism are part of a transcriptionally regulated stress response.
Figure 7B (left of the figure) links prenylated Rab GTPases as key regulatory components in ER-Golgi protein transport (Rab1a, Rab1b, and Rab2) and in vesicle fusion (Rab11b and SNARE). In particular, Rab11b and SNARE mediate the fusion of vesicles containing carrier proteins with the plasma membrane (28, 62). We found that fosmidomycin altered the expression profiles of genes encoding these membrane fusion proteins, suggesting a probable alteration in the physiological function of associated Plasmodium carrier proteins. In support of these observations, we found that fosmidomycin caused a delay in transcription of genes encoding amino acid, ion, metal, and protein transporters (Table 3). Membrane transporters are integral for maintaining homeostasis of ions in the parasite, e.g., P. falciparum maintains a low level of protons in the cytosol and a relatively higher level of protons in the digestive food vacuole with the help of proton pumps (38). During the IDC, proton pumps are incorporated in the food vacuole via endocytic pathways that import hemoglobin in the vacuole (63). We found that genes encoding vacuolar proton pumps had a significant delay in peak expression and a greater than 2-fold increase in expression under treated conditions (Table 3). Moreover, since depletion of isoprenoids disrupts food vacuole integrity (7, 64), these perturbations may also result in altered pH homeostasis that was predicted by our integrated metabolic network analyses (Fig. 6).
Metabolic adaptations in P. falciparum due to fosmidomycin treatment.
To gain insight into fosmidomycin-induced metabolic adaptations in P. falciparum, we employed metabolic network analysis and identified two distinct alterations in the methionine-polyamine metabolic pathway. First, increased flux through the purine-recycling pathway allowed drug-treated parasites to maintain nucleotide and protein synthesis. Second, reduced methylation cycling affected the synthesis of ubiquinone and phosphocholine, a precursor for phosphatidylcholine and phosphatidylethanolamine (18). In support of our metabolic model predictions, we found an increase in adenine-based purine nucleotides (Data Set S3) and a decrease in phosphoethanolamine in fosmidomycin-treated parasite cultures (Table 1). Our analyses showed that increased synthesis of phosphatidylglycerol, the precursor of cardiolipin (3) (an inner mitochondrial membrane lipid that facilitates electron transport from ubiquinone to ETC complexes [65]), compensated for the reduced ubiquinone metabolism. As phosphatidylcholine and phosphatidylethanolamine jointly constitute ∼75 to 80% of P. falciparum membranes (18), and a reduction in their content is associated with growth defects and fewer progeny (66), the observed reduction in the number of ring-stage progeny could be a direct consequence of reduced phospholipid material available to support the complete complement of membranes.
Impact of altered metabolism on gene transcription.
Our metabolic network model analyses identified a delay in methyl-group reactions, all of which require S-adenosylmethionine as the methyl-group donor. The upper-right portion of Fig. 7B illustrates how, during the methyl-group donation process, S-adenosylmethionine (SAM) produces S-adenosylhomocysteine, which in turn is rapidly converted into the nontoxic homocysteine (67) and transported out of the infected erythrocytes via an amino acid transporter (26). Methyltransferase enzymes, which transfer the methyl group from S-adenosylmethionine to other substrates, play important roles in tRNA modification, ribosomal maturation, mRNA capping, and ubiquinone biosynthesis (67). As such, these processes are important for RNA translation (Fig. 7B, lower right). In support of our model predictions, we found a delay and/or increase in transcription of genes encoding membrane fusion proteins (Table 2), transporters (Table 3), methyltransferase enzymes (Table 4), and ribosomal proteins (Table 4).
Limitations of the study.
In this study, our aim was to characterize metabolic and transcriptomic alterations in P. falciparum treated with a sublethal dose of fosmidomycin. Toward this end, we first performed analyses of the multiomics data and discussed consequences of fosmidomycin treatment on P. falciparum. By analyzing the data, we were able to identify several processes that were changed substantially in fosmidomycin-treated parasites. However, with this approach, we could not ascertain whether these processes are specifically perturbed due to fosmidomycin treatment, e.g., a delay in translation presumably also occurs in the presence of isoleucine deprivation (41). Moreover, most gene-expression changes do not directly correlate with changes in metabolic abundances. Therefore, to gain a mechanistic insight and bridge the gap between gene-metabolic alterations, we employed a whole-genome metabolic network model of P. falciparum. This approach predicts alterations in metabolic enzyme activities based on the input data and explicit gene-protein reaction rules, which allows us to predict the metabolic state of P. falciparum as captured in the data and the reaction network. However, because this approach makes no assumption regarding the mechanisms of fosmidomycin action, the metabolic processes identified through model analyses are not directly influenced by fosmidomycin per se. Instead, the effect of fosmidomycin on these processes is likely indirect, being caused by isoprenoid deprivation and impaired vesicular trafficking, which may evoke multiple toxic effects in P. falciparum.
Summary.
Our results suggest that fosmidomycin causes metabolic and translational reprogramming, i.e., modulation of polyamine metabolism, methyl-group reactions, and delay of translation initiation in P. falciparum. These changes did not affect the duration and progression of the ring, trophozoite, and schizont stages during the first IDC. Although the primary effect of this adaptation was to allow the parasite to maintain short-term IDC viability, the ability to produce progeny was compromised by 50% as evident from the number of ring-stage parasites in a second drug-free IDC. Importantly, these progeny appeared fully viable with normal morphological characteristics. Because these compensatory mechanisms are inherent to the parasite and not a result of long-term drug exposure, the ability to produce even a radically reduced number of viable progeny may underlie the relapse of parasitemia that can occur in malaria patients receiving fosmidomycin monotherapy (12). These results also rationalize why clindamycin, an antibiotic inhibiting translation, when used in combination with fosmidomycin would be more effective than fosmidomycin monotherapy (14).
In contrast to Kennedy et al. (7), who identified the immediate cause of death in isoprenoid-deprived parasites, we identified compensatory adaptations that occurred as a consequence of administering a sublethal dosage of fosmidomycin during the IDC. We observed a broad range of physiological responses due to delays in transcription of genes encoding methyltransferase enzymes (tRNA and mRNA) and ribosomal proteins (5.8S, 18S, and 40S), which was coupled to a reduction in mitochondrial activity and delay of synthesis of TCA cycle metabolites. A specific metabolic adaptation occurred in polyamine metabolism in which a bifunctional enzyme—ornithine decarboxylase/S-adenosylmethionine decarboxylase—increased flux through spermidine synthase (purine-recycling pathway) and decreased flux through phosphoethanolamine N-methyltransferase (phosphatidylcholine metabolism). This enhanced purine-recycling and decreased activity of phosphatidylcholine metabolism allowed P. falciparum to maintain synthesis of purine nucleotides at the cost of suppressing synthesis of phospholipids, compromising membrane production, and reducing the ability to produce progeny. Thus, this detailed study of the sublethal response and survival adaptations provides a means to identify mechanisms used by the parasite to respond to and survive drug stress. In turn, these mechanisms can be hypothesized to underlie emerging drug resistance and resistant parasite strains and hence provide a rational approach to select combination therapies to block these adaptations.
MATERIALS AND METHODS
Parasite culture, fosmidomycin dose, and parasite viability.
We cultured P. falciparum NF54 parasites (gift from David Fidock, Columbia University) in human O-positive erythrocytes using methods and conditions described previously (16, 18). Briefly, we maintained parasite-infected erythrocytes in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, Gaithersburg, MD) supplemented with 20 mM HEPES, 12.5 μg/ml hypoxanthine, 25 μg/ml gentamicin, 0.5 μM R-lipoic acid, 0.3% sodium bicarbonate, and 0.5% AlbuMAX II (Life Technologies Inc., Carlsbad, CA).
Sample collection for metabolomic and transcriptomic profiling.
To collect samples for metabolomic and transcriptomic analyses, we passed 300 ml of synchronized parasite culture through a magnetically activated cell sorting (MACS) XS column (Miltenyi Biotec, Auburn, CA) to seed four flasks, serving as the source for quadruplicate measurements of transcriptomic and metabolomic data. Each flask had 75 ml of culture at 2% hematocrit (HCT). We monitored the parasite culture via blood smears until 0 to 2 h post-merozoite invasion, at which point we replaced the culture medium with fresh medium containing 1 μM fosmidomycin (time zero of the experiment). Beginning at time zero, we collected 10.5 ml of the resuspended parasite culture from each flask every 8 h until the 48-h time point and pelleted the erythrocytes by centrifugation at 400 × g for 5 min. Following aspiration of the media, we transferred 100 μl and 50 μl of the cell pellets to 1.5-ml tubes for metabolomic and transcriptomic analyses, respectively. We flash-froze the tubes in an ethanol/dry-ice bath and stored them at −80°C. We sent quadruplicate samples of the cell pellets collected at six time points (0, 8, 16, 24, 32, and 40 h) to Metabolon, Inc. (Durham, NC) for metabolite analysis and quadruplicate samples of the cell pellets collected at seven time points (0, 8, 16, 24, 32, 40, and 48 h) to the Johns Hopkins Genomic Analysis and Sequencing Core Facility for microarray analysis using an Agilent microarray chip (AMADID 037237; Santa Clara, CA). We have deposited the transcriptomic data to the National Center for Biotechnology Information Gene Expression Omnibus repository (GEO accession number: GSE159516). In parallel, we maintained four flasks with 50 ml of uninfected erythrocytes at 2% HCT in media containing fosmidomycin to serve as controls. Following the above-mentioned procedure, we collected, centrifuged, and aspirated 7 ml of uninfected culture from each flask. We transferred 100 μl of the cell pellets to 1.5-ml tubes, immediately flash-froze the tubes, and stored them at −80°C. We sent quadruplicate samples collected at six time points of these control cultures to Metabolon, Inc. for metabolomic analysis. We did not send the 48-h samples from control and parasite cultures to Metabolon, Inc. for metabolomic analyses. We have provided the metabolomic data as provided by Metabolon, Inc. in Data Set S8.
To evaluate the impact of fosmidomycin on parasite growth, we assessed parasitemia by a blood smear test at every time point. At 40 h, we transferred 20 μl of the pelleted erythrocytes from the four treatment flasks to 25 cm2 flasks and diluted them to 2% HCT with fresh blood and 5 ml of drug-free medium. We determined parasitemia in these diluted cultures by blood smear at 64 h.
Global analyses of metabolomic and transcriptomic data.
We performed HCA using the built-in MATLAB function “clustergram” and identified metabolites and genes that were differentially perturbed by fosmidomycin in iRBC cultures. We used Ward’s algorithm and the Euclidean distance metric to identify clusters of metabolites and genes. We quantile-normalized the metabolomic data from iRBC cultures maintained under untreated and treated conditions before performing the analysis. To perform t-SNE analysis, we downloaded the MATLAB implementation of the algorithm, publicly available on GitHub (https://lvdmaaten.github.io/software/). We reduced the metabolomic data into three dimensions and used six as the value of the “perplexity” variable. We used the bootstrap technique described previously (18) to compute average and temporal fold change values in metabolite abundance (or genes expression) due to fosmidomycin treatment.
To quantify the impact of fosmidomycin on gene transcription, we first fitted a smoothing spline to the transcriptomic profile under the two conditions using the built-in MATLAB function “fit.” Afterwards, we computed the hour of peak expression during the IDC (tpeak) using the fitted curve. We used the same procedure for estimating hour of peak metabolite abundance under untreated and treated conditions. We performed two-way ANOVAs on the transcriptomic data to identify genes that were significantly altered by fosmidomycin. We used the built-in MATLAB function “anova2” to perform this analysis. As the data were nonnormally distributed, we log2-transformed the data before performing the ANOVAs (68). To correct for multiple comparisons, we used the “mafdr” MATLAB function, which implements Storey’s method (69).
Simulating the effects of fosmidomycin on P. falciparum metabolism.
We used previously described methods (15, 16) to simulate the effects of drug treatment. Briefly, we integrated the data (transcriptomic and metabolomic), obtained under untreated and treated conditions, with a genome-scale metabolic network model of P. falciparum to estimate the rates of macromolecule synthesis under respective conditions. In line with our experimental observations, we assumed that treated parasites produced 50% less biomass than untreated parasites (Fig. 1B, 1 μM versus 0 μM fosmidomycin at 64 h). To compute the net synthesized amount of essential metabolites and macromolecules (Table 6), we numerically integrated the simulated enzyme fluxes using the MATLAB function “trapz,” which implements the trapezoidal method.
Data availability.
We have deposited the transcriptomic data to the National Center for Biotechnology Information Gene Expression Omnibus repository (GEO accession number: GSE159516).
Supplementary Material
ACKNOWLEDGMENTS
We thank Tatsuya Oyama for his comments on a previous version of the manuscript. The authors also thank Anne E. Jedlicka and Amanda Dziedzic at Johns Hopkins University’s Genomic Analysis and Sequencing Core Facility for transcript profiling. This work was primarily funded by the Network Science Initiative of the U.S. Army Medical Research and Development Command, Ft. Detrick, Maryland (Award W81XWH-15-C-0061; STP). This work also leveraged support by the National Institutes of Health R01 AI065853 (STP), training grant T32 AI007417 (KR), the Johns Hopkins Malaria Research Institute, and the Bloomberg Family Foundation. The opinions and assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the U.S. Army, the U.S. Department of Defense, or The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. (HJF). The manuscript has been approved for public release with unlimited distribution.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Yeh E, DeRisi JL. 2011. Chemical rescue of malaria parasites lacking an apicoplast defines organelle function in blood-stage Plasmodium falciparum. PLoS Biol 9:e1001138. doi: 10.1371/journal.pbio.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swift RP, Rajaram K, Liu HB, Dziedzic A, Jedlicka AE, Roberts AD, Matthews KA, Jhun H, Bumpus NN, Tewari SG, Wallqvist A, Prigge ST. 2020. A mevalonate bypass system facilitates elucidation of plastid biology in malaria parasites. PLoS Pathog 16:e1008316. doi: 10.1371/journal.ppat.1008316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mir SS, Biswas S, Habib S. 2011. Targeting apicoplast pathways in Plasmodium, p 163–186. In Becker K (ed), Apicomplexan parasites: molecular approaches toward targeted drug development. Wiley-Blackwell, Singapore. [Google Scholar]
- 4.Lim L, McFadden GI. 2010. The evolution, metabolism and functions of the apicoplast. Philos Trans R Soc Lond B Biol Sci 365:749–763. doi: 10.1098/rstb.2009.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodman CD, Su V, McFadden GI. 2007. The effects of anti-bacterials on the malaria parasite Plasmodium falciparum. Mol Biochem Parasitol 152:181–191. doi: 10.1016/j.molbiopara.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Dahl EL, Rosenthal PJ. 2007. Multiple antibiotics exert delayed effects against the Plasmodium falciparum apicoplast. Antimicrob Agents Chemother 51:3485–3490. doi: 10.1128/AAC.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kennedy K, Cobbold SA, Hanssen E, Birnbaum J, Spillman NJ, McHugh E, Brown H, Tilley L, Spielmann T, McConville MJ, Ralph SA. 2019. Delayed death in the malaria parasite Plasmodium falciparum is caused by disruption of prenylation-dependent intracellular trafficking. PLoS Biol 17:e3000376. doi: 10.1371/journal.pbio.3000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szkopinska A. 2000. Ubiquinone. Biosynthesis of quinone ring and its isoprenoid side chain. Intracellular localization. Acta Biochim Pol 47:469–480. doi: 10.18388/abp.2000_4027. [DOI] [PubMed] [Google Scholar]
- 9.Kennedy K, Crisafulli EM, Ralph SA. 2019. Delayed death by plastid inhibition in apicomplexan parasites. Trends Parasitol 35:747–759. doi: 10.1016/j.pt.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Odom AR. 2011. Five questions about non-mevalonate isoprenoid biosynthesis. PLoS Pathog 7:e1002323. doi: 10.1371/journal.ppat.1002323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weidemeyer C, Hintz M, Turbachova I, Eberl M, Zeidler J, Lichtenthaler HK, Soldati D, Beck E. 1999. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science 285:1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- 12.Lell B, Ruangweerayut R, Wiesner J, Missinou MA, Schindler A, Baranek T, Hintz M, Hutchinson D, Jomaa H, Kremsner PG. 2003. Fosmidomycin, a novel chemotherapeutic agent for malaria. Antimicrob Agents Chemother 47:735–738. doi: 10.1128/aac.47.2.735-738.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borrmann S, Issifou S, Esser G, Adegnika AA, Ramharter M, Matsiegui PB, Oyakhirome S, Mawili-Mboumba DP, Missinou MA, Kun JF, Jomaa H, Kremsner PG. 2004. Fosmidomycin-clindamycin for the treatment of Plasmodium falciparum malaria. J Infect Dis 190:1534–1540. doi: 10.1086/424603. [DOI] [PubMed] [Google Scholar]
- 14.Wiesner J, Henschker D, Hutchinson DB, Beck E, Jomaa H. 2002. In vitro and in vivo synergy of fosmidomycin, a novel antimalarial drug, with clindamycin. Antimicrob Agents Chemother 46:2889–2894. doi: 10.1128/aac.46.9.2889-2894.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tewari SG, Prigge ST, Reifman J, Wallqvist A. 2017. Using a genome-scale metabolic network model to elucidate the mechanism of chloroquine action in Plasmodium falciparum. Int J Parasitol Drugs Drug Resist 7:138–146. doi: 10.1016/j.ijpddr.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tewari SG, Rajaram K, Schyman P, Swift R, Reifman J, Prigge ST, Wallqvist A. 2019. Short-term metabolic adjustments in Plasmodium falciparum counter hypoxanthine deprivation at the expense of long-term viability. Malar J 18:86. doi: 10.1186/s12936-019-2720-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallqvist A, Fang X, Tewari SG, Ye P, Reifman J. 2016. Metabolic host responses to malarial infection during the intraerythrocytic developmental cycle. BMC Syst Biol 10:58. doi: 10.1186/s12918-016-0291-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tewari SG, Swift RP, Reifman J, Prigge ST, Wallqvist A. 2020. Metabolic alterations in the erythrocyte during blood-stage development of the malaria parasite. Malar J 19:94. doi: 10.1186/s12936-020-03174-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pezzotti N, Lelieveldt BPF, Van Der Maaten L, Hollt T, Eisemann E, Vilanova A. 2017. Approximated and user steerable tSNE for progressive visual analytics. IEEE Trans Vis Comput Graph 23:1739–1752. doi: 10.1109/TVCG.2016.2570755. [DOI] [PubMed] [Google Scholar]
- 20.Baldwin J, Farajallah AM, Malmquist NA, Rathod PK, Phillips MA. 2002. Malarial dihydroorotate dehydrogenase. Substrate and inhibitor specificity. J Biol Chem 277:41827–41834. doi: 10.1074/jbc.M206854200. [DOI] [PubMed] [Google Scholar]
- 21.Lee PC, Salomon C, Mijts B, Schmidt-Dannert C. 2008. Biosynthesis of ubiquinone compounds with conjugated prenyl side chains. Appl Environ Microbiol 74:6908–6917. doi: 10.1128/AEM.01495-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guggisberg AM, Amthor RE, Odom AR. 2014. Isoprenoid biosynthesis in Plasmodium falciparum. Eukaryot Cell 13:1348–1359. doi: 10.1128/EC.00160-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niang M, Yan Yam X, Preiser PR. 2009. The Plasmodium falciparum STEVOR multigene family mediates antigenic variation of the infected erythrocyte. PLoS Pathog 5:e1000307. doi: 10.1371/journal.ppat.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blasco B, Leroy D, Fidock DA. 2017. Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nat Med 23:917–928. doi: 10.1038/nm.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang B, Watts KM, Hodge D, Kemp LM, Hunstad DA, Hicks LM, Odom AR. 2011. A second target of the antimalarial and antibacterial agent fosmidomycin revealed by cellular metabolic profiling. Biochemistry 50:3570–3577. doi: 10.1021/bi200113y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ginsburg H, Abdel-Haleem AM. 2016. Malaria parasite metabolic pathways (MPMP) upgraded with targeted chemical compounds. Trends Parasitol 32:7–9. doi: 10.1016/j.pt.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Gisselberg JE, Zhang L, Elias JE, Yeh E. 2017. The prenylated proteome of Plasmodium falciparum reveals pathogen-specific prenylation activity and drug mechanism-of-action. Mol Cell Proteomics 16:S54–S64. doi: 10.1074/mcp.M116.064550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutagalung AH, Novick PJ. 2011. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev 91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lecker SH, Goldberg AL, Mitch WE. 2006. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol 17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 30.Mukhopadhyay D, Riezman H. 2007. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 31.Szymanski M, Barciszewska MZ, Barciszewski J, Erdmann VA. 1999. 5S ribosomal RNA data bank. Nucleic Acids Res 27:158–160. doi: 10.1093/nar/27.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaiser A. 2012. Translational control of eIF5A in various diseases. Amino Acids 42:679–684. doi: 10.1007/s00726-011-1042-8. [DOI] [PubMed] [Google Scholar]
- 33.Henderson A, Hershey JW. 2011. Eukaryotic translation initiation factor (eIF) 5A stimulates protein synthesis in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 108:6415–6419. doi: 10.1073/pnas.1008150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suazo KF, Schaber C, Palsuledesai CC, Odom John AR, Distefano MD. 2016. Global proteomic analysis of prenylated proteins in Plasmodium falciparum using an alkyne-modified isoprenoid analogue. Sci Rep 6:38615. doi: 10.1038/srep38615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. 2002. Molecular Biology of the Cell, 4th ed Garland Science, New York. [Google Scholar]
- 36.Li W, Ye Y. 2008. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci 65:2397–2406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung DW, Ponts N, Prudhomme J, Rodrigues EM, Le Roch KG. 2012. Characterization of the ubiquitylating components of the human malaria parasite’s protein degradation pathway. PLoS One 7:e43477. doi: 10.1371/journal.pone.0043477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saliba KJ, Allen RJ, Zissis S, Bray PG, Ward SA, Kirk K. 2003. Acidification of the malaria parasite’s digestive vacuole by a H+-ATPase and a H+-pyrophosphatase. J Biol Chem 278:5605–5612. doi: 10.1074/jbc.M208648200. [DOI] [PubMed] [Google Scholar]
- 39.Kuhn Y, Rohrbach P, Lanzer M. 2007. Quantitative pH measurements in Plasmodium falciparum-infected erythrocytes using pHluorin. Cell Microbiol 9:1004–1013. doi: 10.1111/j.1462-5822.2006.00847.x. [DOI] [PubMed] [Google Scholar]
- 40.Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol 1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Babbitt SE, Altenhofen L, Cobbold SA, Istvan ES, Fennell C, Doerig C, Llinas M, Goldberg DE. 2012. Plasmodium falciparum responds to amino acid starvation by entering into a hibernatory state. Proc Natl Acad Sci U S A 109:E3278–E3287. doi: 10.1073/pnas.1209823109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bennink S, Pradel G. 2019. The molecular machinery of translational control in malaria parasites. Mol Microbiol 112:1658–1673. doi: 10.1111/mmi.14388. [DOI] [PubMed] [Google Scholar]
- 43.Tellier G, Lenne A, Cailliau-Maggio K, Cabezas-Cruz A, Valdes JJ, Martoriati A, Aliouat EM, Gosset P, Delaire B, Freville A, Pierrot C, Khalife J. 2016. Identification of Plasmodium falciparum translation initiation eIF2beta subunit: direct interaction with protein phosphatase type 1. Front Microbiol 7:777. doi: 10.3389/fmicb.2016.00777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lenne A, De Witte C, Tellier G, Hollin T, Aliouat EM, Martoriati A, Cailliau K, Saliou JM, Khalife J, Pierrot C. 2018. Characterization of a protein phosphatase type-1 and a kinase anchoring protein in Plasmodium falciparum. Front Microbiol 9:2617. doi: 10.3389/fmicb.2018.02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webster MW, Chen YH, Stowell JAW, Alhusaini N, Sweet T, Graveley BR, Coller J, Passmore LA. 2018. mRNA deadenylation is coupled to translation rates by the differential activities of Ccr4-Not nucleases. Mol Cell 70:1089–1100. doi: 10.1016/j.molcel.2018.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hart KJ, Oberstaller J, Walker MP, Minns AM, Kennedy MF, Padykula I, Adams JH, Lindner SE. 2019. Plasmodium male gametocyte development and transmission are critically regulated by the two putative deadenylases of the CAF1/CCR4/NOT complex. PLoS Pathog 15:e1007164. doi: 10.1371/journal.ppat.1007164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, Tsujimoto M, Suzuki T, Katada T, Hoshino SI. 2007. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev 21:3135–3148. doi: 10.1101/gad.1597707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buschauer R, Matsuo Y, Sugiyama T, Chen YH, Alhusaini N, Sweet T, Ikeuchi K, Cheng J, Matsuki Y, Nobuta R, Gilmozzi A, Berninghausen O, Tesina P, Becker T, Coller J, Inada T, Beckmann R. 2020. The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science 368:eaay6912. doi: 10.1126/science.aay6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindner SE, Mikolajczak SA, Vaughan AM, Moon W, Joyce BR, Sullivan WJ, Jr, Kappe SH. 2013. Perturbations of Plasmodium Puf2 expression and RNA-seq of Puf2-deficient sporozoites reveal a critical role in maintaining RNA homeostasis and parasite transmissibility. Cell Microbiol 15:1266–1283. doi: 10.1111/cmi.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shock JL, Fischer KF, DeRisi JL. 2007. Whole-genome analysis of mRNA decay in Plasmodium falciparum reveals a global lengthening of mRNA half-life during the intraerythrocytic development cycle. Genome Biol 8:R134. doi: 10.1186/gb-2007-8-7-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cassera MB, Merino EF, Peres VJ, Kimura EA, Wunderlich G, Katzin AM. 2007. Effect of fosmidomycin on metabolic and transcript profiles of the methylerythritol phosphate pathway in Plasmodium falciparum. Mem Inst Oswaldo Cruz 102:377–383. doi: 10.1590/s0074-02762007000300019. [DOI] [PubMed] [Google Scholar]
- 52.Foth BJ, Zhang N, Chaal BK, Sze SK, Preiser PR, Bozdech Z. 2011. Quantitative time-course profiling of parasite and host cell proteins in the human malaria parasite Plasmodium falciparum. Mol Cell Proteomics 10:M110.006411. doi: 10.1074/mcp.M110.006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang X, Reifman J, Wallqvist A. 2014. Modeling metabolism and stage-specific growth of Plasmodium falciparum HB3 during the intraerythrocytic developmental cycle. Mol Biosyst 10:2526–2537. doi: 10.1039/c4mb00115j. [DOI] [PubMed] [Google Scholar]
- 54.Hoppe A. 2012. What mRNA abundances can tell us about metabolism. Metabolites 2:614–631. doi: 10.3390/metabo2030614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moxley JF, Jewett MC, Antoniewicz MR, Villas-Boas SG, Alper H, Wheeler RT, Tong L, Hinnebusch AG, Ideker T, Nielsen J, Stephanopoulos G. 2009. Linking high-resolution metabolic flux phenotypes and transcriptional regulation in yeast modulated by the global regulator Gcn4p. Proc Natl Acad Sci U S A 106:6477–6482. doi: 10.1073/pnas.0811091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramya TN, Surolia N, Surolia A. 2006. Polyamine synthesis and salvage pathways in the malaria parasite Plasmodium falciparum. Biochem Biophys Res Commun 348:579–584. doi: 10.1016/j.bbrc.2006.07.127. [DOI] [PubMed] [Google Scholar]
- 57.Ting LM, Shi W, Lewandowicz A, Singh V, Mwakingwe A, Birck MR, Ringia EA, Bench G, Madrid DC, Tyler PC, Evans GB, Furneaux RH, Schramm VL, Kim K. 2005. Targeting a novel Plasmodium falciparum purine recycling pathway with specific immucillins. J Biol Chem 280:9547–9554. doi: 10.1074/jbc.M412693200. [DOI] [PubMed] [Google Scholar]
- 58.Wanke M, Skorupinska-Tudek K, Swiezewska E. 2001. Isoprenoid biosynthesis via 1-deoxy-d-xylulose 5-phosphate/2-C-methyl-d-erythritol 4-phosphate (DOXP/MEP) pathway. Acta Biochim Pol 48:663–672. doi: 10.18388/abp.2001_3901. [DOI] [PubMed] [Google Scholar]
- 59.Park MH, Wolff EC. 2018. Hypusine, a polyamine-derived amino acid critical for eukaryotic translation. J Biol Chem 293:18710–18718. doi: 10.1074/jbc.TM118.003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Francis SE, Sullivan DJ, Jr, Goldberg DE. 1997. Hemoglobin metabolism in the malaria parasite Plasmodium falciparum. Annu Rev Microbiol 51:97–123. doi: 10.1146/annurev.micro.51.1.97. [DOI] [PubMed] [Google Scholar]
- 61.Fernandez-Marcos PJ, Nobrega-Pereira S. 2016. NADPH: new oxygen for the ROS theory of aging. Oncotarget 7:50814–50815. doi: 10.18632/oncotarget.10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beske O. 2000. Membrane fusion in a nutshell. Trends Cell Biol 10:514. doi: 10.1016/S0962-8924(00)01867-5. [DOI] [PubMed] [Google Scholar]
- 63.Abu Bakar N, Klonis N, Hanssen E, Chan C, Tilley L. 2010. Digestive-vacuole genesis and endocytic processes in the early intraerythrocytic stages of Plasmodium falciparum. J Cell Sci 123:441–450. doi: 10.1242/jcs.061499. [DOI] [PubMed] [Google Scholar]
- 64.Howe R, Kelly M, Jimah J, Hodge D, Odom AR. 2013. Isoprenoid biosynthesis inhibition disrupts Rab5 localization and food vacuolar integrity in Plasmodium falciparum. Eukaryot Cell 12:215–223. doi: 10.1128/EC.00073-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vos M, Geens A, Bohm C, Deaulmerie L, Swerts J, Rossi M, Craessaerts K, Leites EP, Seibler P, Rakovic A, Lohnau T, De Strooper B, Fendt SM, Morais VA, Klein C, Verstreken P. 2017. Cardiolipin promotes electron transport between ubiquinone and complex I to rescue PINK1 deficiency. J Cell Biol 216:695–708. doi: 10.1083/jcb.201511044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kilian N, Choi JY, Voelker DR, Ben Mamoun C. 2018. Role of phospholipid synthesis in the development and differentiation of malaria parasites in the blood. J Biol Chem 293:17308–17316. doi: 10.1074/jbc.R118.003213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trager W, Tershakovec M, Chiang PK, Cantoni GL. 1980. Plasmodium falciparum: antimalarial activity in culture of sinefungin and other methylation inhibitors. Exp Parasitol 50:83–89. doi: 10.1016/0014-4894(80)90010-7. [DOI] [PubMed] [Google Scholar]
- 68.McDonald JH. 2009. Handbook of biological statistics, 3rd ed, vol 2. Sparky House Publishing, Baltimore. [Google Scholar]
- 69.Storey JD. 2002. A direct approach to false discovery rates. J R Stat Soc Series B Stat Methodol 64:479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- 70.Lennard L, Wang LM. 2017. Methyltransferases, p 497–516. In Eaton DL (ed), Comprehensive Toxicology, vol 10. Elsevier, Oxford. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have deposited the transcriptomic data to the National Center for Biotechnology Information Gene Expression Omnibus repository (GEO accession number: GSE159516).