We conducted an updated analysis on yeast isolates causing fungemia in patients admitted to a tertiary hospital in Madrid, Spain, over a 13-year period. We studied 896 isolates associated with 872 episodes of fungemia in 857 hospitalized patients between January 2007 and December 2019.
KEYWORDS: fungemia, yeast, Candida, EUCAST, microsatellite genotyping, antifungal resistance, fluconazole, echinocandins
ABSTRACT
We conducted an updated analysis on yeast isolates causing fungemia in patients admitted to a tertiary hospital in Madrid, Spain, over a 13-year period. We studied 896 isolates associated with 872 episodes of fungemia in 857 hospitalized patients between January 2007 and December 2019. Antifungal susceptibility was assessed by EUCAST EDef 7.3.2. Mutations conferring azole and echinocandin resistance were further studied, and genotyping of resistant clones was performed with species-specific microsatellite markers. Candida albicans (45.8%) was the most frequently identified species, followed by the Candida parapsilosis complex (26.4%), Candida glabrata (12.3%), Candida tropicalis (7.3%), Candida krusei (2.3%), other Candida spp. (3.1%), and non-Candida yeasts (2.8%). The rate of fluconazole resistance in Candida spp. was 4.7%, ranging from 0% (C. parapsilosis) to 9.1% (C. glabrata). The overall rate of echinocandin resistance was 3.1%. Resistance was highly influenced by the presence of intrinsically resistant species. Although the number of isolates between 2007 and 2013 was almost 2-fold higher than that in the period from 2014 to 2019 (566 versus 330), fluconazole resistance in Candida spp. was greater in the second period (3.5% versus 6.8%; P < 0.05), while overall resistance to echinocandins remained stable (3.5% versus 2.4%; P > 0.05). Resistant clones were collected from different wards and/or time points, suggesting that there were no epidemiological links. The number of fungemia episodes has been decreasing over the last 13 years, with a slight increase in the rate of fluconazole resistance and stable echinocandin resistance. Antifungal resistance is not the cause of the spread of resistant clones.
INTRODUCTION
Candidemia is the most common fungal bloodstream infection in hospitalized patients. The overall pooled incidence rate in Europe is 3.88 episodes per 100,000 inhabitants and is associated with a crude mortality rate that ranges from 10% to 47% (1). Candida albicans remains the most commonly isolated species, although recent reports alert on the emergence of other Candida spp., non-Candida yeasts, echinocandin-resistant Candida glabrata isolates, or the multiresistant Candida auris (2).
Species-specific patterns may be used to try to predict antifungal susceptibility by identifying the species involved in the infection, which is generally sufficient due to intrinsically resistant isolates (3). However, detecting secondary resistance requires further antifungal susceptibility testing to monitor the resistance and guide empirical antifungal treatment. Therefore, periodic surveillance of isolates causing fungemia may provide further insight on the epidemiology and antifungal resistance. Nationwide fungemia studies have reported resistance rates ranging from 2.7% (Iceland) to 9.4% (Denmark) for fluconazole and 0% (Brazil and Iceland) to 15% (Portugal) for echinocandins (4–7). In Spain, resistance rates for fluconazole and echinocandins are 7% and 2%, respectively (8). Population-based studies are useful, although they may overlook interhospital differences (9).
In a previous study, we found low azole and echinocandin resistance rates in isolates causing fungemia obtained from patients admitted to our hospital (Hospital General Universitario Gregorio Marañón, Madrid, Spain) between January 2007 and December 2013 (10). In this study, we assessed the etiology and antifungal resistance of isolates collected between January 2014 and December 2019. We made an overall reassessment of the agents causing fungemia in patients admitted to a tertiary care hospital in Madrid (Spain) over a 13-year period, as well as their amphotericin B, azole, and echinocandin antifungal susceptibility using updated EUCAST clinical breakpoints, and the potential spread of resistant clones.
RESULTS
Epidemiology of species causing fungemia.
We studied 896 isolates causing 872 episodes of fungemia—single-species episodes (n = 848) and polyfungal episodes (n = 24)—in 857 patients. Isolates were from patients admitted to medical wards (30.2%), intensive care units (ICUs) (20.1%), surgical wards (15.7%), oncology-hematology (14.5%), neonatology (12.5%), and other wards (7%). Table 1 shows the distribution of species. C. albicans (45.8%) was the most frequently identified species, followed by the C. parapsilosis complex (26.4%), C. glabrata (12.3%), C. tropicalis (7.3%), C. krusei (2.3%), other Candida spp. (3.1%), and non-Candida yeasts (2.8%). In cases with polyfungal episodes, the following were isolated: C. albicans-C. glabrata (n = 11), C. albicans-C. parapsilosis (n = 4), C. glabrata-C. tropicalis (n = 2), and 1 case for each of the following combinations: C. albicans-C. dubliniensis, C. albicans-C. krusei, C. glabrata-C. parapsilosis, C. glabrata-C. metapsilosis, C. parapsilosis-C. guilliermondii, C. parapsilosis-C. metapsilosis, and C. tropicalis-C. krusei.
TABLE 1.
Species distribution of 896 isolates causing fungemia 857 patients
| Species | No. of isolates | % |
|---|---|---|
| C. albicansa | 410 | 45.8 |
| C. parapsilosis complexb | 237 | 26.4 |
| C. glabratac | 110 | 12.3 |
| C. tropicalis | 65 | 7.3 |
| C. krusei | 21 | 2.3 |
| C. guilliermondii complexd | 11 | 1.2 |
| C. dubliniensis | 8 | 0.9 |
| C. lusitaniae | 5 | 0.6 |
| C. kefyr | 2 | 0.2 |
| C. inconspicua | 1 | 0.1 |
| C. pelliculosa | 1 | 0.1 |
| Rhodotorula mucilaginosa | 7 | 0.8 |
| Trichosporon spp.e | 7 | 0.8 |
| Cryptococcus spp.f | 7 | 0.8 |
| Magnusiomyces capitatus | 2 | 0.2 |
| Arxula adeninivorans | 1 | 0.1 |
| Kodamaea ohmeri | 1 | 0.1 |
| Overall | 896g |
One isolate proved to be C. africana (0.11%).
Three isolates proved to be C. metapsilosis (0.33%); seven isolates proved to be C. orthopsilosis (0.78%).
One isolate proved to be C. nivariensis (0.11%).
Two isolates proved to be C. fermentati (0.22%).
T. inkin (n = 3 [0.33%]), T. asahii (n = 2 [0.22%]), T. dermatis (n = 1 [0.11%]), and T. asteroides (n = 1 [0.11%]).
C. neoformans (n = 5 [0.56%]) and C. deneoformans (n = 2 [0.22%]).
Forty-eight isolates caused 24 polyfungal episodes; species were distributed as follows: C. albicans (n = 17), C. glabrata (n = 15), C. parapsilosis (n = 8), C. tropicalis, (n = 3), C. krusei (n = 2), C. dubliniensis (n = 1), C. guilliermondii (n = 1), and C. metapsilosis (n = 1).
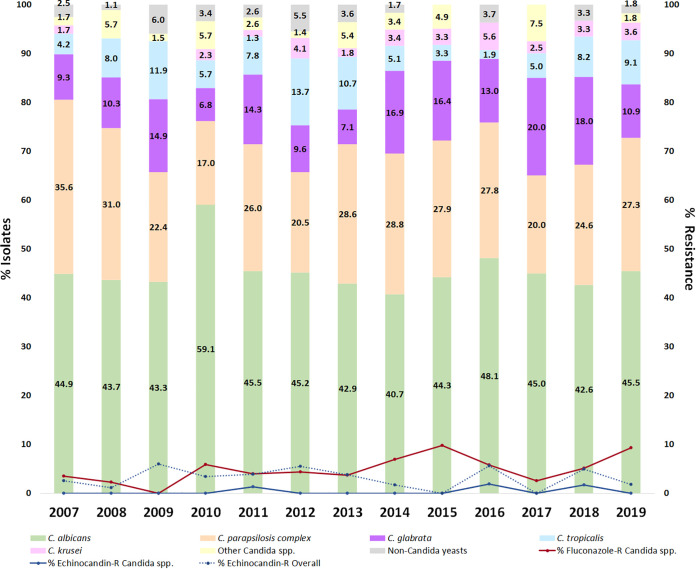
The number of episodes varied by year, ranging from 115 (2007) to 39 (2017). Figure 1 and Supplemental Table S1 show the percentage of isolates by individual species and year. C. albicans accounted for 40% to 48% of the isolates per year, except for 2010 (59%), whereas C. parapsilosis accounted for 17% to 36% of the isolates per year, although with a decreasing trend from initial years of the study period. In contrast, the proportion of other Candida spp. and non-Candida yeast isolates showed a spiked pattern. The distribution by species in different hospital locations is shown in Fig. S1 and Supplemental Table S2. With the exception of oncology-hematology, C. albicans was the dominant species, particularly in neonatology and other wards. In contrast, C. parapsilosis was more frequently detected in neonatology and oncology-hematology. The highest percentages of C. glabrata and C. tropicalis were found in medical wards and the ICU, respectively. C. krusei, Candida spp., and non-Candida yeasts were anecdotal in neonatology but notoriously frequent in oncology-hematology.
FIG 1.
Species distribution of isolates causing fungemia and rate of resistance to fluconazole and echinocandins during the study period.
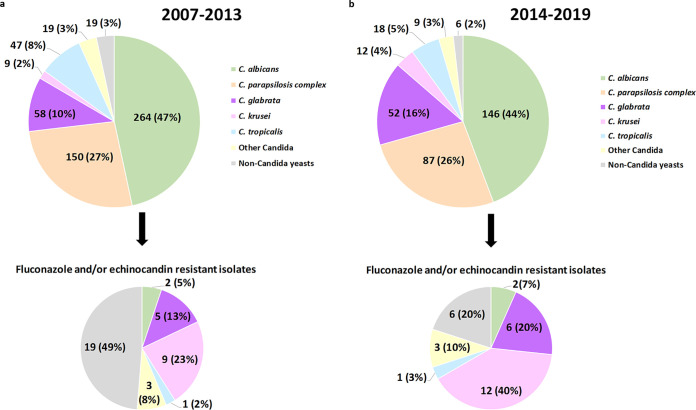
The number of isolates detected in the first period was almost twice that in the second period (n = 566 [63.2%] versus n = 330 [36.8%]). The proportions of C. albicans and C. parapsilosis isolates were similar in both subperiods, while those of C. glabrata and C. krusei significantly increased in the second period (10.2% versus 15.8% and 1.6% versus 3.6%, respectively; P < 0.05). Finally, the proportions of infections by C. tropicalis (8.3% versus 5.5%; P > 0.05), other Candida spp. (3.4% versus 2.7%; P > 0.05), and non-Candida yeasts (3.4% versus 1.8%; P > 0.05) decreased in the second period (Fig. 2).
FIG 2.
Species distribution of yeasts causing fungemia and fluconazole- and/or echinocandin-resistant isolates found in both subperiods.
Antifungal susceptibility testing and characterization of resistance.
Table 2 lists MIC distributions of the drugs tested. No resistance to amphotericin B was detected.
TABLE 2.
MIC distributions of studied antifungals versus isolates and rates of resistant/non-wild-type isolatesa
| Organism(s) and drug | No. of isolates at each MIC, in mg/liter |
No. of isolates (%) |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.001 | 0.002 | 0.004 | 0.008 | 0.016 | 0.03 | 0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | ≥64 | Resistant | Non-wild type | |
| C. albicans (n = 410) | |||||||||||||||||||
| Amphotericin B | — | — | 0 | 0 | 0 | 0 | 12 | 98 | 217 | 78 | 5 | 0 | 0 | 0 | — | — | — | 0 (0) | 0 (0) |
| Fluconazole | — | — | — | — | — | — | 28 | 239 | 125 | 12 | 2 | 0 | 0 | 1 | 0 | 0 | 3 | 4 (1.0) | 6 (1.5) |
| Voriconazole | 4 | 46 | 280 | 61 | 11 | 2 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 3b | — | — | — | 4 (1.0) | 6 (1.5) |
| Posaconazole | 1 | 4 | 83 | 205 | 81 | 27 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 3b | — | — | — | 4 (1.0) | 4 (1.0) |
| Isavuconazole | 25 | 239 | 120 | 16 | 6 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3b | — | — | — | ND | 4 (1.0) |
| Micafungin | 10 | 104 | 244 | 42 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | — | — | — | 0 (0) | 0 (0) |
| Anidulafungin | 13 | 178 | 190 | 19 | 9 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | — | — | — | 0 (0) | 0 (0) |
| C. parapsilosis complex (n = 237) | |||||||||||||||||||
| Amphotericin B | — | — | 0 | 0 | 0 | 0 | 4 | 20 | 105 | 101 | 7 | 0 | 0 | 0 | — | — | — | 0 (0) | 0 (0) |
| Fluconazole | — | — | — | — | — | — | 0 | 3 | 116 | 93 | 22 | 3 | 0 | 0 | 0 | 0 | 0 | 0 (0) | 0 (0) |
| Voriconazole | 0 | 0 | 16 | 125 | 78 | 15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | — | — | — | 0 (0) | 0 (0) |
| Posaconazole | 0 | 0 | 0 | 10 | 76 | 106 | 45 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | — | — | — | 0 (0) | 0 (0) |
| Isavuconazole | 0 | 33 | 104 | 83 | 16 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | — | — | — | ND | 0 (0) |
| Micafungin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 10 | 36 | 140 | 50 | 0 | 0 | — | — | — | 0 (0) | 0 (0) |
| Anidulafungin | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 14 | 53 | 153 | 13 | 0 | — | — | — | 0 (0) | 0 (0) |
| C. glabrata (n = 110) | |||||||||||||||||||
| Amphotericin B | — | — | 0 | 0 | 0 | 0 | 3 | 14 | 65 | 27 | 1 | 0 | 0 | 0 | — | — | — | 0 (0) | 0 (0) |
| Fluconazole | — | — | — | — | — | — | 0 | 0 | 0 | 0 | 0 | 5 | 37 | 49 | 9 | 2 | 8 | 10 (9.1) | 10 (9.1) |
| Voriconazole | 0 | 0 | 0 | 0 | 0 | 2 | 15 | 35 | 47 | 4 | 2 | 4 | 1 | 0 | — | — | — | ND | 5 (4.5) |
| Posaconazole | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 8 | 32 | 53 | 9 | 4 | 2 | 0 | — | — | — | ND | 6 (5.5) |
| Isavuconazole | 0 | 1 | 0 | 1 | 1 | 14 | 35 | 32 | 16 | 1 | 5 | 3 | 1 | 0 | — | — | — | ND | 10 (9.1) |
| Micafungin | 0 | 0 | 0 | 93 | 15 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | — | — | — | 0 (0) | 0 (0) |
| Anidulafungin | 0 | 0 | 0 | 0 | 41 | 64 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | — | — | — | 1 (0.9) | 1 (0.9) |
| C. tropicalis (n = 65) | |||||||||||||||||||
| Amphotericin B | — | — | 0 | 0 | 0 | 0 | 1 | 4 | 36 | 22 | 2 | 0 | 0 | 0 | — | — | — | 0 (0) | 0 (0) |
| Fluconazole | — | — | — | — | — | — | 0 | 10 | 37 | 14 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 (0) | 0 (0) |
| Voriconazole | 0 | 0 | 0 | 6 | 34 | 17 | 7 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | — | — | — | 0 (0) | 0 (0) |
| Posaconazole | 0 | 0 | 1 | 16 | 21 | 25 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | — | — | — | 0 (0) | 0 (0) |
| Isavuconazole | 0 | 3 | 28 | 31 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | — | — | — | ND | 0 (0) |
| Micafungin | 0 | 0 | 0 | 3 | 44 | 15 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | — | — | — | ND | 2 (3.1) |
| Anidulafungin | 0 | 0 | 0 | 15 | 38 | 8 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | — | — | — | 1 (1.5) | 1 (1.5) |
| C. krusei (n = 21) | |||||||||||||||||||
| Amphotericin B | — | — | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 17 | 3 | 0 | 0 | 0 | — | — | — | 0 (0) | 0 (0) |
| Fluconazole | — | — | — | — | — | — | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 8 | 7 | 21 (100) | 0 (0) |
| Voriconazole | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 9 | 8 | 0 | 0 | 0 | 0 | — | — | — | ND | 0 (0) |
| Posaconazole | 0 | 0 | 0 | 0 | 1 | 2 | 4 | 8 | 6 | 0 | 0 | 0 | 0 | 0 | — | — | — | ND | 0 (0) |
| Isavuconazole | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 12 | 6 | 1 | 0 | 0 | 0 | 0 | — | — | — | ND | 0 (0) |
| Micafungin | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | — | — | — | ND | 0 (0) |
| Anidulafungin | 0 | 0 | 0 | 0 | 0 | 14 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | — | — | — | 0 (0) | 0 (0) |
| Other Candida spp. (n = 28) | |||||||||||||||||||
| Amphotericin B | — | — | 0 | 0 | 0 | 2 | 6 | 10 | 6 | 4 | 0 | 0 | 0 | 0 | — | — | — | ND | ND |
| Fluconazole | — | — | — | — | — | — | 2 | 5 | 5 | 0 | 0 | 5 | 5 | 1 | 2 | 2 | 1 | 6 (21.4) | ND |
| Voriconazole | 0 | 0 | 3 | 8 | 2 | 0 | 6 | 4 | 3 | 1 | 0 | 0 | 1 | 0 | — | — | — | ND | ND |
| Posaconazole | 0 | 0 | 2 | 2 | 8 | 2 | 5 | 3 | 6 | 0 | 0 | 0 | 0 | 0 | — | — | — | ND | ND |
| Isavuconazole | 0 | 5 | 5 | 3 | 0 | 2 | 3 | 5 | 1 | 3 | 0 | 1 | 0 | 0 | — | — | — | ND | ND |
| Micafungin | 0 | 1 | 0 | 1 | 6 | 4 | 4 | 5 | 2 | 3 | 0 | 1 | 1 | 0 | — | — | — | ND | ND |
| Anidulafungin | 0 | 0 | 1 | 7 | 2 | 3 | 3 | 0 | 2 | 2 | 6 | 2 | 0 | 0 | — | — | — | ND | ND |
| Non-Candida yeasts (n = 25) | |||||||||||||||||||
| Amphotericin B | — | — | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 5 | 9 | 5 | 0 | 4b | — | — | — | ND | ND |
| Fluconazole | — | — | — | — | — | — | 0 | 0 | 0 | 2 | 5 | 1 | 5 | 1 | 1 | 2 | 8 | ND | ND |
| Voriconazole | 0 | 1 | 1 | 1 | 3 | 2 | 4 | 1 | 2 | 4 | 3 | 2 | 0 | 1b | — | — | — | ND | ND |
| Posaconazole | 0 | 0 | 0 | 2 | 3 | 5 | 5 | 3 | 4 | 3 | 0 | 0 | 0 | 0 | — | — | — | ND | ND |
| Isavuconazole | 0 | 1 | 1 | 1 | 5 | 2 | 6 | 1 | 2 | 3 | 1 | 2 | 0 | 0 | — | — | — | ND | ND |
| Micafungin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 21b | — | — | — | ND | ND |
| Anidulafungin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 2 | 20b | — | — | — | ND | ND |
Underlined values indicate non-wild-type isolates according to ECOFFs, and values in bold indicate resistant isolates (EUCAST breakpoint table v 10.0), with the exception of C. krusei, which was considered intrinsically resistant to fluconazole (30, 31). —, nontested antifungal concentration. ND, not done, as either breakpoints or ECOFFs were not available.
Isolates with MICs of ≥8 mg/liter.
Table 3 shows the characteristics of fluconazole-resistant Candida spp. isolates. Overall, 4.7% (n = 41) of isolates were fluconazole resistant (C. krusei [n = 21], C. glabrata [n = 10], C. albicans [n = 4], C. guilliermondii [n = 3], C. lusitaniae [n = 2], and C. inconspicua [n = 1]). Resistance rates in individual species ranged from 0% (C. parapsilosis) to 9.1% (C. glabrata). For the remaining triazoles, the overall rate of non-wild-type Candida species isolates was 1.3% for voriconazole (n = 11; C. albicans [n = 6] and C. glabrata [n = 5]), 1.1% for posaconazole (n = 10; C. albicans [n = 4] and C. glabrata [n = 6]), and 1.6% for isavuconazole (n = 14; C. albicans [n = 4] and C. glabrata [n = 10]). All isolates showing a non-wild-type phenotype to voriconazole, posaconazole, or isavuconazole were fluconazole non-wild type. The four fluconazole-resistant C. albicans isolates harbored mutations in ERG3 (V351A [n = 3], T293I V351A [n = 1]) and/or ERG11 (D116E/D E266D V488I/V [n = 1] and A114S Y257H G464S/G [n = 1]) and were non-wild type to the other azoles. The remaining two fluconazole non-wild-type C. albicans isolates also harbored mutations in ERG3 (V351A [n = 2]) and ERG11 (A114S Y257H [n = 2]). All fluconazole-resistant C. glabrata isolates exhibited cross-resistance to the four azoles (n = 5), posaconazole and isavuconazole (n = 1), or isavuconazole (n = 4). The updated fluconazole EUCAST breakpoints resulted in up to a 9% increase of the resistance rate in C. glabrata (compared to the 7% detected using the former 2018 breakpoints).
TABLE 3.
Characteristics of fluconazole-resistant isolates from patients with candidemia
| Isolate no. | Species | Fluconazole MIC (mg/liter) | ERG3-ERG 11 sequence | Ward | Date of isolation (day/mo/yr) | Genotype |
|---|---|---|---|---|---|---|
| 1 | C. albicansa | >64 | V351A-D116E/D E266D V488I/V | Digestive | 23/08/2010 | CA-170 |
| 2 | C. albicansa | 8 | V351A-A114S Y257H G464S/G | Neonatology ICU | 27/07/2011 | CA-276 |
| 3 | C. albicansa | >64 | T291I V351A-wild type | Internal medicine | 13/11/2019 | CA-313 |
| 4 | C. albicansa | >64 | V351A-wild type | Neurosurgery | 15/03/2015 | CA-413 |
| 5 | C. glabrataa | 64 | NDd | Oncology | 25/05/2018 | CG-01 |
| 6 | C. glabrataa | >64 | ND | Oncology | 08/11/2011 | CG-06 |
| 7 | C. glabratab | 64 | ND | Urology | 22/12/2008 | CG-09 |
| 8 | C. glabrataa | >64 | ND | ICU | 09/10/2007 | CG-13 |
| 9 | C. glabratab | 32 | ND | ICU | 12/01/2014 | CG-15 |
| 10 | C. glabratab | 64 | ND | Digestive | 13/08/2015 | CG-15 |
| 11 | C. glabrataa | 64 | ND | Emergency | 21/10/2007 | CG-23 |
| 12 | C. glabratab | 32 | ND | ICU | 26/01/2010 | CG-24 |
| 13 | C. glabrataa | >64 | ND | Surgery | 11/09/2015 | CG-24 |
| 14 | C. glabratac | 64 | ND | Surgery | 12/07/2019 | CG-27 |
| 15 | C. krusei | 32 | ND | Geriatrics | 02/01/2007 | ND |
| 16 | C. krusei | 16 | ND | Oncology | 27/11/2007 | ND |
| 17 | C. krusei | 32 | ND | ICU | 28/09/2010 | ND |
| 18 | C. krusei | 64 | ND | ICU | 27/12/2010 | ND |
| 19 | C. krusei | 32 | ND | ICU | 08/03/2011 | ND |
| 20 | C. krusei | 32 | ND | Oncology | 02/01/2012 | ND |
| 21 | C. krusei | 32 | ND | Digestive | 05/03/2012 | ND |
| 22 | C. krusei | 64 | ND | Nephrology | 29/05/2012 | ND |
| 23 | C. krusei | 64 | ND | Infectious diseases | 05/04/2013 | ND |
| 24 | C. krusei | 16 | ND | Digestive | 14/11/2014 | ND |
| 25 | C. krusei | 64 | ND | Onco/Hematoe | 24/11/2014 | ND |
| 26 | C. krusei | 32 | ND | Onco/Hemato | 24/04/2015 | ND |
| 27 | C. krusei | 16 | ND | Onco/Hemato | 16/07/2015 | ND |
| 28 | C. krusei | 32 | ND | ICU | 06/04/2016 | ND |
| 29 | C. krusei | 16 | ND | Oncology | 15/06/2016 | ND |
| 30 | C. krusei | 16 | ND | Onco/Hemato | 29/11/2016 | ND |
| 31 | C. krusei | 16 | ND | Oncology | 07/03/2017 | ND |
| 32 | C. krusei | 32 | ND | Internal medicine | 22/09/2018 | ND |
| 33 | C. krusei | 64 | ND | Onco/Hemato | 29/09/2018 | ND |
| 34 | C. krusei | 64 | ND | Intensive medicine | 17/03/2019 | ND |
| 35 | C. krusei | 64 | ND | Intensive medicine | 09/04/2019 | ND |
| 36 | C. guilliermondii | 16 | ND | Surgery | 10/10/2008 | ND |
| 37 | C. guilliermondii | 16 | ND | Digestive | 23/04/2013 | ND |
| 38 | C. guilliermondii | >64 | ND | Internal medicine | 31/01/2018 | ND |
| 39 | C. lusitaniae | 16 | ND | Onco/Hemato | 30/05/2010 | ND |
| 40 | C. lusitaniae | 32 | ND | Neurosurgery | 28/12/2014 | ND |
| 41 | C. inconspicua | 8 | ND | Traumatology | 22/01/2015 | ND |
Pan-azole-resistant isolate.
Cross-resistance among fluconazole and isavuconazole.
Cross-resistance among fluconazole, posaconazole, and isavuconazole.
ND, not done.
Onco/Hemato, Oncology and Hematology.
Table 4 presents the characteristics of echinocandin-resistant isolates. Only a few Candida species isolates were resistant to echinocandins (n = 3 [0.3%]; C. tropicalis [n = 2] and C. glabrata [n = 1]). FKS mutations were found in the three isolates, two of C. tropicalis (R656G/R FKS1 HS1 and S654F FKS1 HS1) and one of C. glabrata (F659S FKS2 HS1). No cross-resistance between azoles and echinocandins in Candida spp. was found. Non-Candida yeasts showed decreased fluconazole susceptibility and intrinsic echinocandin resistance. Pooled MICs against Candida spp. and non-Candida yeasts showed 3.1% (n = 28) echinocandin resistance.
TABLE 4.
Characteristics of echinocandin-resistant isolates from patients with candidemia
| Isolate no. | Species | MIC (mg/liter)a |
FKS mutation | Ward | Date on which species was isolated (day/mo/yr) | Genotype | |
|---|---|---|---|---|---|---|---|
| Micafungin | Anidulafungin | ||||||
| 1 | C. tropicalis | 0.125 | 0.03 | R656G/R FKS1 | ICU | 31/10/2011 | CT-033 |
| 2 | C. tropicalis | 2 | 2 | S654F FKS1 | Oncology and Hematology | 23/03/2018 | CT-165 |
| 3 | C. glabrata | 0.03 | 0.125 | F659S FKS2 | Digestive | 04/08/2016 | CG-024 |
Numbers in bold indicate resistance or non-wild-type isolates.
Rates of resistance to fluconazole (against Candida spp.) and echinocandins (in Candida spp. and non-Candida yeasts) fluctuated throughout the study period (Fig. 1). Fluconazole resistance rates in Candida spp. ranged from 0% (2009) to 9.8% (2015) and were highly influenced by the presence of species with intrinsic lower fluconazole susceptibility (C. glabrata and C. krusei), which accounted for 75.6% of all fluconazole-resistant Candida species isolates. Likewise, overall echinocandin resistance rates ranged between 0% (2015 and 2017) and 6.0% (2009) and were highly influenced by the presence of intrinsically echinocandin-resistant non-Candida species (89.3% of all echinocandin-resistant isolates).
Percentages of fluconazole (3.5% versus 6.8%; P < 0.05) and echinocandin (0.2% versus 0.6%; P > 0.05) resistance in Candida spp. were higher in the second period. However, pooled MICs against Candida spp. and non-Candida yeasts showed stable echinocandin resistance rates (3.5% versus 2.4%; P > 0.05) in the second period (Fig. 2).
Admission wards of patients with resistant isolates.
There was no relationship between patients infected with resistant Candida species isolates and specific hospital wards (Tables 3 and 4). The four fluconazole-resistant C. albicans isolates had different genotypes; this was also observed for the two echinocandin-resistant C. tropicalis isolates (Tables 3 and 4). In contrast, the 10 fluconazole-resistant C. glabrata isolates had eight genotypes; genotype CG-24 was detected in 3 isolates, 2 fluconazole-resistant isolates, and 1 echinocandin-resistant isolate. No epidemiological association was found between patients harboring C. glabrata clones since they were not admitted to the same wards or during the same period of candidemia onset.
DISCUSSION
Slight changes in the etiology and antifungal susceptibility in yeast species causing fungemia have been observed over time in our hospital. Despite the drop in the number of episodes since the beginning of the study, the rate of fluconazole resistance has increased, whereas resistance to echinocandin has remained stable. Moreover, resistance to fluconazole is not caused by the spread of resistant clones but is due to the presence of species showing intrinsic reduced susceptibility.
Population-based studies, aimed to assess the epidemiology of fungemia in large geographic areas, have proven C. albicans to be the main etiological agent. The second species in terms of frequency may be either C. parapsilosis (southern Europe), C. glabrata (northern Europe and North America), or C. tropicalis (Asia and Latin America) (2, 5–8, 11, 12). However, these types of studies may overlook interhospital nuances and differences that may be relevant to initiating empirical treatment while identifying the pathogen. Furthermore, periodic surveillance is also needed to follow epidemiological and antifungal resistance trends. We have been actively conducting monitoring studies with isolates causing fungemia at our hospital since 2007 (10). The current study is an update in which only incident isolates were included. We reassessed the data, dividing the study period into two subperiods, and applied the EUCAST 2020 updated fluconazole clinical breakpoints and epidemiological cutoffs (ECOFFs). The epidemiology of the examined species is in line with that reported in southern European hospitals (8, 11). The number of episodes decreased considerably (566 versus 330) between the two subperiods. This may explain why the implementation of educational campaigns, focused on catheter care and management, initiated in our hospital in 2010 led to a decrease in the number of fungemia infections (13). Health care workers’ knowledge on the guidelines for the prevention of catheter-related bloodstream infections was assessed before and after the program, and care bundles were implemented in adult and neonatal units, the pediatric ICU, and the internal medicine department (13–16). In the present study, no statistically significant epidemiological differences were observed, except for C. glabrata and C. krusei, for which increases were seen in the second period, in line with what has been reported elsewhere (5). C. glabrata is commonly found in elderly patients and among transplant recipients, whereas C. krusei is often isolated from patients with underlying hematological malignancies under fluconazole prophylaxis (6). C. albicans and C. parapsilosis are frequently associated with catheter-related candidemia. This may explain why the infection control campaign helped decrease the number of C. parapsilosis and C. albicans cases, while C. glabrata and C. krusei cases remained the same. On the other hand, rare Candida species and non-Candida yeasts showed a trend toward decrease over time, and C. auris was not detected in this study.
Antifungal resistance has been associated with poor outcome in patients with fungemia, as well as with higher hospital care costs (17). Echinocandin resistance is related to poor clinical response (18), whereas in cases of azole resistance, this is less clear (17). International guidelines recommend antifungal susceptibility testing in invasive Candida isolates, ideally using reference methods (3, 19). Given antifungal susceptibility species-specific patterns, correctly identifying the species is an invaluable tool to predict antifungal susceptibility and guide antifungal treatment (3). Nevertheless, antifungal susceptibility testing is necessary to detect either acquired fluconazole resistance in purportedly susceptible species such as C. albicans, C. tropicalis, and C. parapsilosis (9) or echinocandin resistance in species such as C. albicans, C. tropicalis, and C. glabrata (18, 20).
Fluconazole resistance is driven by species epidemiology, as shown in countries with a large number of C. glabrata- and C. krusei-related candidemia cases, which show high resistance rates (2). Globally, echinocandin resistance is generally low in most common Candida species, except for C. glabrata, for which increasing resistance rates have been reported in North America and northern Europe (2, 5, 18). In the multicenter CANDIPOP study carried out in Spain, a 6.9% rate of fluconazole resistance and low rates of echinocandin resistance were found (8). A later study, also conducted in Spain, showed lower rates of fluconazole resistance (3.4%) and full susceptibility to echinocandins (12). The rates of fluconazole resistance in Candida spp. (4.7%) and overall echinocandin resistance (3.1%) reported in this study are in line with those reported in the CANDIPOP study.
We observed an increase of fluconazole resistance over time, particularly due to the presence of C. glabrata and C. krusei. EUCAST antifungal susceptibility clinical breakpoint update testing affected only C. glabrata and fluconazole; the new breakpoint was lowered from ≥64 mg/liter to ≥32 mg/liter, implying that the rate of resistance in this species has increased (from 7% to 9%). Acquired resistance to azoles in C. albicans is due to efflux pumps or alterations in the ERG3 and ERG11 genes. We found polymorphisms in the ERG3 genes of the four azole-resistant C. albicans strains. However, the effects mutations in the ERG3 gene have on the acquisition of high-level azole resistance remain unclear (21). We further sequenced the ERG11 gene and detected previously reported mutations (D116E/D E266D V488I/V and A114S Y257H G464S/G) (22).
The echinocandin resistance communicated here is low and stable. We detected previously reported FKS gene mutations in our C. glabrata and C. tropicalis echinocandin-resistant isolates (23). The F659S substitution in C. glabrata isolates confers low-level resistance exclusively to anidulafungin (23–25). Amino acid substitutions in resistant C. albicans isolates at the S645 and R647 positions lead, respectively, to high MICs of echinocandins and slight MIC elevations but resistance exclusively to micafungin (26). The above-mentioned phenotypes in C. albicans were confirmed in this study for both echinocandin-resistant C. tropicalis isolates; furthermore, both S654F and R656G/R substitutions have been previously reported (27).
Monitoring resistant Candida clones may be helpful to avoid dissemination of resistance and prevent outbreaks (9). In our hospital, there is no spread of resistant C. albicans and C. tropicalis clones. Two C. glabrata clones infected 2 and 3, respectively, epidemiologically unrelated patients, suggesting the clones may have a limited capacity to cause outbreaks. Moreover, no association was found between resistant isolates and specific hospital locations. It is not uncommon to find clones involving patients who were either admitted to the same hospital but without a clear epidemiological link or admitted to different hospitals that in some cases were located in different countries (13, 28). The haploid species C. glabrata shows a relatively low genetic diversity compared to diploid Candida species such as C. albicans, although with sufficient typing discrimination; the two molecular methods mainly used for C. glabrata genotyping—multilocus sequence typing and microsatellite markers—correlate well (29). Clones without a clear epidemiological link may indicate the presence of common widespread clones or low typing resolution of the microsatellites (e.g., homoplasy, an intrinsic limitation of microsatellite typing analysis, leading to alleles with identical sizes but different sequences), or they may simply be a consequence of the lower diversity of a haploid species. Future studies using whole-genome sequencing will help decipher the role of epidemiologically unrelated clones.
We conclude that the etiology and antifungal susceptibility of the pathogens that cause fungemia have not significantly changed over time at our hospital. Antifungal resistance should be carefully monitored in order to check future trends.
MATERIALS AND METHODS
Definition of fungemia episode and study period.
We studied the incident isolates causing fungemia in patients admitted to Hospital General Universitario Gregorio Marañón (Madrid, Spain) between January 2007 and December 2019. A fungemia episode was defined as the presence of yeasts in blood cultures. The presence of two different species in a single patient was considered a polyfungal episode (both species found simultaneously in the same set of blood cultures) or different episodes (both species found in different sets of blood cultures).
Isolate identification and antifungal susceptibility testing.
Isolates were confirmed by amplification of the ITS1-5.8S-ITS2 region (10). Their in vitro antifungal susceptibilities to amphotericin B, fluconazole, voriconazole, and posaconazole (Sigma-Aldrich, Madrid, Spain), isavuconazole (Basilea Pharmaceutica, Basel, Switzerland), micafungin (Astellas Pharma, Inc., Tokyo, Japan), and anidulafungin (Pfizer Pharmaceutical Group, New York, NY) was assessed by the EUCAST EDef 7.3.2 broth dilution method using tissue-treated plates (CELLSTAR, 655180; Greiner bio-one, Frickenhausen, Germany) (30). Ranges of concentrations tested for each antifungal are shown in Table 2. C. parapsilosis ATCC 22019 and C. krusei ATCC 6258 were used as quality controls.
Isolates were categorized as resistant, susceptible, wild type, or non-wild type according to clinical breakpoints or tentative epidemiological cutoff values (ECOFFs) provided by EUCAST (31). Since there are no available clinical breakpoints for isavuconazole, we used tentative ECOFFs or wild-type upper limits previously proposed for C. albicans, C. parapsilosis, C. glabrata, C. tropicalis, and C. krusei isolates (32). Phenotypically resistant isolates were retested. The FKS1 and FKS2 genes were sequenced in anidulafungin- and/or micafungin-non-wild-type C. glabrata and C. tropicalis isolates (27). The ERG3 and ERG11 genes were sequenced in fluconazole non-wild-type C. albicans (33). Resistance/non-wild-type rates were calculated per isolate exclusively. In the absence of available clinical breakpoints/ECOFFs against non-Candida yeasts, resistance/non-wild-type rates were calculated either exclusively (azoles) in Candida species isolates or overall (echinocandins) in Candida spp. plus non-Candida yeasts given that echinocandins show systematically high MICs against non-Candida yeasts.
Proportions of species isolates and percentages of resistance found in the subperiods from 2007 to 2013 and 2014 to 2019 were compared (Epidat v.4.2; Consellería de Sanidade, Xunta de Galicia, Spain).
Genotyping.
Species-specific microsatellite markers were used to genotype C. albicans (CDC3, EF3, HIS3 CAI, CAIII, and CAVI), C. parapsilosis (CP1, CP4a, CP6, and B), C. glabrata (MTI, ERG3, GLM4, Cg4, Cg7, and Cg10), and C. tropicalis (Ctrm1, Ctrm10, Ctrm12, Ctrm21, Ctrm24, and Ctrm28) isolates showing resistance to any of the drugs tested (28, 34). We sized and scored the electrophoretic bands with GeneMapper version 4.0 (Applied Biosystems-Life Technologies Corporation, CA). The allelic composition was studied for each locus and converted to binary data by scoring the presence or absence of each allele. Genetic relationships between genotypes were clarified by constructing a minimum spanning tree using BioNumerics version 7.6 (Applied Maths, Sint-Martens-Latem, Belgium). Singletons were genotypes found only once. Isolates were considered genotypically identical when they showed the same alleles with all markers; identical genotypes were further defined as a match (found in serial samples of a given patient) or as a cluster (found in ≥2 patients). Different genotypes were codified as follows: CA-X (C. albicans), CP-X (C. parapsilosis), and CT-X (C. tropicalis), where X represents the internal code of the genotype in our collection.
Ethical considerations.
This study was approved by the Ethics Committee of Hospital Gregorio Marañón (CEIm; study no. MICRO.HGUGM.2020-011).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Dainora Jaloveckas (cienciatraducida.com) for editing assistance.
This study was supported by grants PI18/01155 from Fondo de Investigación Sanitaria (FIS; Instituto de Salud Carlos III, Plan Nacional de I+D+I 2013-2016). The study was cofunded by the European Regional Development Fund (FEDER) A Way of Making Europe.
P.E. (MS15/00115) is the recipient of a Miguel Servet contract supported by FIS. J.G. is a steady researcher contracted by Fundación para Investigación Sanitaria del Hospital Gregorio Marañón. J.D.-G. is supported by a predoctoral grant from FIS (FI19/00021). A.M. is supported by grants from the Ministerio de Ciencia, Innovación y Universidades (PEJ2018-004609-A) and from Fondo de Investigación Sanitaria (FI20/00089).
J.G. has received funds for participating at educational activities organized on behalf of Astellas, Gilead, MSD, Scynexis, and Biotoscana-United Medical; he has also received research funds from FIS, Gilead, Scynexis, and Cidara, outside the submitted work.
J.D.-G., methodology, formal analysis, writing – original draft preparation and review and editing, visualization; A.M., methodology; C.S.-C., data collection, resources (samples), writing – review and editing; E.R., data collection, resources (samples), writing – review and editing; P.M., data collection, resources (samples), writing – review and editing; P.E., conceptualization, project administration, data collection, supervision, validation, visualization, writing – original draft preparation and review and editing; J.G., conceptualization, project administration, data collection, supervision, validation, visualization, writing – original draft preparation and review and editing.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Koehler P, Stecher M, Cornely OA, Koehler D, Vehreschild M, Bohlius J, Wisplinghoff H, Vehreschild JJ. 2019. Morbidity and mortality of candidaemia in Europe: an epidemiologic meta-analysis. Clin Microbiol Infect 25:1200–1212. doi: 10.1016/j.cmi.2019.04.024. [DOI] [PubMed] [Google Scholar]
- 2.Pfaller MA, Diekema DJ, Turnidge JD, Castanheira M, Jones RN. 2019. Twenty years of the SENTRY Antifungal Surveillance Program: results for Candida species from 1997–2016. Open Forum Infect Dis 6:S79–S94. doi: 10.1093/ofid/ofy358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD. 2016. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 62:e1–e50. doi: 10.1093/cid/civ933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asmundsdottir LR, Erlendsdottir H, Gottfredsson M. 2013. Nationwide study of candidemia, antifungal use, and antifungal drug resistance in Iceland, 2000 to 2011. J Clin Microbiol 51:841–848. doi: 10.1128/JCM.02566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Astvad KMT, Johansen HK, Røder BL, Rosenvinge FS, Knudsen JD, Lemming L, Schønheyder HC, Hare RK, Kristensen L, Nielsen L, Gertsen JB, Dzajic E, Pedersen M, Østergård C, Olesen B, Søndergaard TS, Arendrup MC. 2017. Update from a 12-year nationwide fungemia surveillance: increasing intrinsic and acquired resistance causes concern. J Clin Microbiol 56:e01564-17. doi: 10.1128/JCM.01564-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faria-Ramos I, Neves-Maia J, Ricardo E, Santos-Antunes J, Silva AT, Costa-de-Oliveira S, Cantón E, Rodrigues AG, Pina-Vaz C. 2014. Species distribution and in vitro antifungal susceptibility profiles of yeast isolates from invasive infections during a Portuguese multicenter survey. Eur J Clin Microbiol Infect Dis 33:2241–2247. doi: 10.1007/s10096-014-2194-8. [DOI] [PubMed] [Google Scholar]
- 7.Doi AM, Pignatari ACC, Edmond MB, Marra AR, Camargo LFA, Siqueira RA, da Mota VP, Colombo AL. 2016. Epidemiology and microbiologic characterization of nosocomial candidemia from a Brazilian national surveillance program. PLoS One 11:e0146909. doi: 10.1371/journal.pone.0146909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guinea J, Zaragoza Ó, Escribano P, Martín-Mazuelos E, Pemán J, Sánchez-Reus F, Cuenca-Estrella M. 2014. Molecular identification and antifungal susceptibility of yeast isolates causing fungemia collected in a population-based study in Spain in 2010 and 2011. Antimicrob Agents Chemother 58:1529–1537. doi: 10.1128/AAC.02155-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mesini A, Mikulska M, Giacobbe DR, Del Puente F, Gandolfo N, Codda G, Orsi A, Tassinari F, Beltramini S, Marchese A, Icardi G, Del Bono V, Viscoli C. 2020. Changing epidemiology of candidaemia: increase in fluconazole-resistant Candida parapsilosis. Mycoses 63:361–368. doi: 10.1111/myc.13050. [DOI] [PubMed] [Google Scholar]
- 10.Marcos-Zambrano LJ, Escribano P, Sanchez C, Munoz P, Bouza E, Guinea J. 2014. Antifungal resistance to fluconazole and echinocandins is not emerging in yeast isolates causing fungemia in a Spanish tertiary care center. Antimicrob Agents Chemother 58:4565–4572. doi: 10.1128/AAC.02670-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guinea J. 2014. Global trends in the distribution of Candida species causing candidemia. Clin Microbiol Infect 20(Suppl 6):5–10. doi: 10.1111/1469-0691.12539. [DOI] [PubMed] [Google Scholar]
- 12.Nieto MC, Tellería O, Cisterna R. 2015. Sentinel surveillance of invasive candidiasis in Spain: epidemiology and antifungal susceptibility. Diagn Microbiol Infect Dis 81:34–40. doi: 10.1016/j.diagmicrobio.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 13.Escribano P, Sanchez-Carrillo C, Munoz P, Bouza E, Guinea J. 2018. Reduction in percentage of clusters of Candida albicans and Candida parapsilosis causing candidemia in a general hospital in Madrid, Spain. J Clin Microbiol 56:e00574-18. doi: 10.1128/JCM.00574-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guembe M, Pérez-Parra A, Gómez E, Sánchez-Luna M, Bustinza A, Zamora E, Carrillo-Álvarez A, Cuenca A, Padilla B, Martín-Rabadán P, Bouza E, GEIDI Study Group . 2012. Impact on knowledge and practice of an intervention to control catheter infection in the ICU. Eur J Clin Microbiol Infect Dis 31:2799–2808. doi: 10.1007/s10096-012-1630-x. [DOI] [PubMed] [Google Scholar]
- 15.Bouza E, Alcalá L, Muñoz P, Martín-Rabadán P, Guembe M, Rodríguez-Créixems M, GEIDI and the COMIC study groups . 2013. Can microbiologists help to assess catheter involvement in candidaemic patients before removal? Clin Microbiol Infect 19:E129–E135. doi: 10.1111/1469-0691.12096. [DOI] [PubMed] [Google Scholar]
- 16.Guembe M, Pérez-Granda MJ, Capdevila JA, Barberán J, Pinilla B, Martín-Rabadán P, Bouza E, NUVE Study Group . 2015. Nationwide study on the use of intravascular catheters in internal medicine departments. J Hosp Infect 90:135–141. doi: 10.1016/j.jhin.2015.01.024. [DOI] [PubMed] [Google Scholar]
- 17.Bassetti M, Vena A, Bouza E, Peghin M, Muñoz P, Righi E, Pea F, Lackner M, Lass-Flörl C. 2020. Antifungal susceptibility testing in Candida, Aspergillus and Cryptococcus infections: are the MICs useful for clinicians? Clin Microbiol Infect 26:1024–1033. doi: 10.1016/j.cmi.2020.02.017. [DOI] [PubMed] [Google Scholar]
- 18.Alexander BD, Johnson MD, Pfeiffer CD, Jiménez-Ortigosa C, Catania J, Booker R, Castanheira M, Messer SA, Perlin DS, Pfaller MA. 2013. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin Infect Dis 56:1724–1732. doi: 10.1093/cid/cit136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuenca-Estrella M, Verweij PE, Arendrup MC, Arikan-Akdagli S, Bille J, Donnelly JP, Jensen HE, Lass-Flörl C, Richardson MD, Akova M, Bassetti M, Calandra T, Castagnola E, Cornely OA, Garbino J, Groll AH, Herbrecht R, Hope WW, Kullberg BJ, Lortholary O, Meersseman W, Petrikkos G, Roilides E, Viscoli C, Ullmann AJ, ESCMID Fungal Infection Study Group . 2012. ESCMID guideline for the diagnosis and management of Candida diseases 2012: diagnostic procedures. Clin Microbiol Infect 18(Suppl 7):9–18. doi: 10.1111/1469-0691.12038. [DOI] [PubMed] [Google Scholar]
- 20.Grosset M, Desnos-Ollivier M, Godet C, Kauffmann-Lacroix C, Cazenave-Roblot F. 2016. Recurrent episodes of candidemia due to Candida glabrata, Candida tropicalis and Candida albicans with acquired echinocandin resistance. Med Mycol Case Rep 14:20–23. doi: 10.1016/j.mmcr.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowen LE, Sanglard D, Howard SJ, Rogers PD, Perlin DS. 2014. Mechanisms of antifungal drug resistance. Cold Spring Harb Perspect Med 5:a019752. doi: 10.1101/cshperspect.a019752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morio F, Loge C, Besse B, Hennequin C, Le Pape P. 2010. Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: new substitutions and a review of the literature. Diagn Microbiol Infect Dis 66:373–384. doi: 10.1016/j.diagmicrobio.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Arendrup MC, Perlin DS. 2014. Echinocandin resistance: an emerging clinical problem? Curr Opin Infect Dis 27:484–492. doi: 10.1097/QCO.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prigent G, Aït-Ammar N, Levesque E, Fekkar A, Costa JM, El Anbassi S, Foulet F, Duvoux C, Merle JC, Dannaoui E, Botterel F. 2017. Echinocandin resistance in Candida species isolates from liver transplant recipients. Antimicrob Agents Chemother 61:e01229-16. doi: 10.1128/AAC.01229-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castanheira M, Woosley LN, Messer SA, Diekema DJ, Jones RN, Pfaller MA. 2014. Frequency of fks mutations among Candida glabrata isolates from a 10-year global collection of bloodstream infection isolates. Antimicrob Agents Chemother 58:577–580. doi: 10.1128/AAC.01674-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernández-Silva F, Lackner M, Capilla J, Mayayo E, Sutton D, Castanheira M, Fothergill AW, Lass-Flörl C, Guarro J. 2014. In vitro antifungal susceptibility of Candida glabrata to caspofungin and the presence of FKS mutations correlate with treatment response in an immunocompromised murine model of invasive infection. Antimicrob Agents Chemother 58:3646–3649. doi: 10.1128/AAC.02666-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bordallo-Cardona MÁ, Marcos-Zambrano LJ, Sánchez-Carrillo C, Bouza E, Muñoz P, Escribano P, Guinea J. 2018. Resistance to echinocandins in Candida can be detected by performing the Etest directly on blood culture samples. Antimicrob Agents Chemother 62:e00162-18. doi: 10.1128/AAC.00162-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guinea J, Arendrup MC, Cantón R, Cantón E, García-Rodríguez J, Gómez A, Gómez E, Hare RK, Orden B, Sanguinetti M, Pemán J, Posteraro B, Ruiz-Gaitán AC, Parisi G, Da Matta DA, Lopes Colombo A, Sánchez-Carrillo C, Reigadas E, Munoz P, Escribano P. 2020. Genotyping reveals a high clonal diversity of Candida but widespread genotypes causing candidaemia at distant geographic areas. Front Cell Infect Microbiol 10:166. doi: 10.3389/fcimb.2020.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gabaldón T, Gómez-Molero E, Bader O. 2020. Molecular typing of Candida glabrata. Mycopathologia 185:755–764. doi: 10.1007/s11046-019-00388-x. [DOI] [PubMed] [Google Scholar]
- 30.Arendrup Mc Meletiadis J, Mouton JW, Lagrou K, Hamal P, Guinea J. 2020. EUCAST definitive document E.DEF 7.3.2. Method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Files/EUCAST_E_Def_7.3.2_Yeast_testing_definitive_revised_2020.pdf. [DOI] [PubMed]
- 31.Arendrup MC, Friberg N, Mares M, Kahlmeter G, Meletiadis J, Guinea J, Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST) . 2020. How to interpret MICs of antifungal compounds according to the revised clinical breakpoints v. 10.0 European committee on antimicrobial susceptibility testing (EUCAST). Clin Microbiol Infect 26:1464–1472. doi: 10.1016/j.cmi.2020.06.007. [DOI] [PubMed] [Google Scholar]
- 32.Marcos-Zambrano LJ, Gómez A, Sánchez-Carrillo C, Bouza E, Muñoz P, Escribano P, Guinea J. 2020. Isavuconazole is highly active in vitro against Candida species isolates but shows trailing effect. Clin Microbiol Infect 26:1589–1592. doi: 10.1016/j.cmi.2020.07.023. [DOI] [PubMed] [Google Scholar]
- 33.Liu JY, Shi C, Wang Y, Li WJ, Zhao Y, Xiang MJ. 2015. Mechanisms of azole resistance in Candida albicans clinical isolates from Shanghai, China. Res Microbiol 166:153–161. doi: 10.1016/j.resmic.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 34.Bordallo-Cardona M, Agnelli C, Gómez-Nuñez A, Sánchez-Carrillo C, Bouza E, Muñoz P, Escribano P, Guinea J. 2018. MSH2 gene point mutations are not antifungal resistance markers in Candida glabrata. Antimicrob Agents Chemother 63:e01876-18. doi: 10.1128/AAC.01876-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.