A total of 15 Candida auris isolates from the SENTRY antimicrobial surveillance program between 2006 and 2019 were combined with 21 isolates from other collections for the evaluation of antifungal susceptibility and synergy against anidulafungin plus voriconazole or isavuconazole using the checkerboard method. Surveillance isolates were analyzed for genetic relatedness and resistance mechanisms.
KEYWORDS: Candida auris, azoles, echinocandins, synergy testing
ABSTRACT
A total of 15 Candida auris isolates from the SENTRY antimicrobial surveillance program between 2006 and 2019 were combined with 21 isolates from other collections for the evaluation of antifungal susceptibility and synergy against anidulafungin plus voriconazole or isavuconazole using the checkerboard method. Surveillance isolates were analyzed for genetic relatedness and resistance mechanisms. Applying the tentative statistical epidemiological cutoff values and the Centers for Disease Control tentative breakpoints, 32/36 isolates were resistant to fluconazole, 5/36 were resistant to amphotericin B, 5/36 were non-wild-type (NWT) to anidulafungin, 3/36 were NWT to micafungin, and 1/36 and 10/36 were NWT to isavuconazole and voriconazole, respectively. Of these, 10 isolates were multidrug resistant, which means that these isolates were resistant to 2 antifungal classes. Synergy or partial synergy was noted in 5/36 and 22/36, respectively, of the isolates with the combination of anidulafungin plus voriconazole, and 11/36 and 19/36 isolates, respectively, for the combination of anidulafungin plus isavuconazole. Multilocus sequence type (MLST) analysis of the 15 SENTRY isolates demonstrated that the isolates from the US were genetically related to, but different from, isolates from Latin America (Panama and Colombia) and Germany. Single nucleotide polymorphism (SNP) analysis showed that the 15 SENTRY isolates belonged to the described international clades and had associated Erg11 alterations, including 11 isolates displaying K143R, one displaying F126L, and one displaying Y501H alterations and a fluconazole MIC result of ≥64 mg/liter. Resistance mechanisms were not observed in the two isolates displaying fluconazole MIC values at 4 and 16 mg/liter. Isavuconazole displayed activity and greater synergy when tested with anidulafungin than seen with anidulafungin plus voriconazole against the C. auris clinical isolates that displayed resistance phenotypes.
INTRODUCTION
Candida auris is an emerging opportunistic fungal pathogen characterized by the ability to survive on inanimate surfaces, demonstrate prolonged colonization of infected and uninfected patients, transmit from patient to patient in hospital and long-term-care facilities, and express resistance to multiple classes of antifungal agents (1–5). Following its initial detection in 2009, C. auris spread worldwide, causing serious infection and death in hospitalized patients in over 30 countries and 6 continents (3, 6–10). Difficulties in the accurate identification of C. auris, coupled with its resistance to fluconazole (as high as 100%) and other azoles (28% resistance to voriconazole), often accompanied by decreased susceptibility to amphotericin B and the echinocandins (30% and 3 to 7% resistance, respectively) have complicated the clinical approach to prevention and therapy of infections of this unique species (10–14). The three classes of systematically active antifungal agents are azoles, echinocandins, and polyenes. Approximately 18 to 41% of isolates have been found to be resistant to two different classes and 4% of isolates are resistant to all three classes (1, 7, 9, 15–18).
The application of molecular methods to the characterization of C. auris provided important insights into the introduction and spread of this organism through geographic regions as well as its mechanisms of resistance to the azoles and echinocandins (1, 10–13, 15, 17). Specifically, whole-genome sequencing (WGS) has documented four geographic genetic clades: South Asia (clade I), East Asia (clade II), South Africa (clade III), and South America (clade IV) (3, 5, 7, 15, 17). Substantial interclade variation and intraclade homogeneity exist, suggesting that C. auris emerged independently in each region (7, 15). C. auris-related infections in other parts of the world, such as the United States (US) and the United Kingdom (UK), have been caused by strains that are genetically related to these clades (5, 7).
Resistance to at least two different classes of antifungal agents is categorized as exhibiting a multidrug-resistant (MDR) phenotype. MDR among C. auris clinical isolates led investigators to characterize the mechanisms of resistance to azoles and echinocandins (10–13, 19). Thus far, molecular characterization of C. auris demonstrates that, as with Candida albicans, resistance to fluconazole and other azoles is mediated by mutations in the gene ERG11 and potentially augmented by the overexpression of CDR1 efflux pumps (19, 20). Resistance to the echinocandins is acquired post exposure to these agents and is associated with a mutation in FKS1 (12, 14). Mutations in the ERG11 gene are strongly associated with the geographic clades: F126T, South African clade (clade III); Y132F, South American clade (clade IV); and Y132F or K143F, South Asian clade (clade I) (7, 10, 14, 19). Echinocandin resistance in C. auris isolates is mediated through limited mutations in FKS1, usually encoding S639P or S639F (12). Isolates with these FKS1 mutations have been detected in several different regions (12).
The MDR profile of C. auris severely limits its therapeutic options. Given the relatively low frequency of resistance to the echinocandins, this class of agents has been recommended as the first-line option for treatment of clinical infection pending the results of antifungal susceptibility testing (3, 21, 22). This recommendation is complicated by the observation that resistance to the echinocandin class may emerge in patients that have been treated with an echinocandin (14). In the interest of optimizing the therapeutic approach to infections with this organism, some investigators have proposed an approach that uses combinations of antifungal agents (3, 5, 23).
The goal of combination therapy is to employ agents with different mechanisms of action to achieve a synergistic interaction, thus increasing the likelihood of therapeutic success and limiting the emergence of resistance to employed agents (24, 25). Presently, there are six published in vitro studies that examined the activity of antifungal drug combinations against MDR C. auris (26–31). The investigated combinations include an echinocandin (caspofungin or micafungin) plus an azole (fluconazole or voriconazole) (28); flucytosine plus amphotericin B, voriconazole, or micafungin (26); flucytosine plus amphotericin B, azoles (isavuconazole, itraconazole, posaconazole, or voriconazole), or echinocandins (anidulafungin, caspofungin, or micafungin) (29); sulfamethoxazole plus an azole (fluconazole, itraconazole, or voriconazole) (27); an antileishmanial agent, miltifosine, plus either amphotericin B or fluconazole (30); and an HIV protease inhibitor, lopinavir, plus azoles (fluconazole, itraconazole, and voriconazole) (31). In each instance, either synergy or indifference/additivity was observed for the drug combinations when these combinations were tested against a limited number (10 to 15 isolates, most of which were South Asian clade I or from the CDC AR Collection) of C. auris isolates. No instances of antagonism were detected in the studies. Arguably, the most effective combination was micafungin plus voriconazole, as synergy was observed against all 10 MDR isolates of C. auris belonging to the South Asian clade (28).
In the present study, we examined the in vitro activity of isavuconazole and voriconazole tested alone and in combination with anidulafungin against a collection of 36 C. auris isolates obtained from the SENTRY antifungal surveillance program (15 isolates), the antimicrobial resistance (AR) bank from the Centers for Disease Control and Prevention (CDC; 10 isolates), and 11 isolates of C. auris from Kenya that were previously received at JMI Laboratories (32). In addition to reviewing the antifungal combination studies, we established the phenotypic resistance profile of these isolates by testing individual comparator antifungal agents (caspofungin, micafungin, fluconazole, and amphotericin B), employing whole-genome sequencing (WGS) to characterize the mechanisms of resistance to azoles and echinocandins, and determining the clades represented in the 15 isolates from the SENTRY collection.
RESULTS
Activity of isavuconazole and comparator agents against C. auris.
The activity of isavuconazole, echinocandins, other azoles, and amphotericin B when tested alone against the 36 C. auris isolates are listed in Table 1. Isavuconazole MIC values ranged from ≤0.008 mg/liter to 4 mg/liter, with a mode of 0.5 mg/liter and a MIC50/MIC90 of 0.5/1 mg/liter. Using the tentative epidemiological cutoff value (ECV or ECOFF) of ≤1 mg/liter proposed by Arendrup et al. (33), all but one of these isolates were classified as WT to isavuconazole. By comparison, the activity of voriconazole was within ±2-fold (MIC50/MIC90, 1/2 mg/liter; 26/36 inhibited by ≤1 mg/liter [S]). Fluconazole had limited activity against these isolates. Only 4 isolates were susceptible to fluconazole when applying the CDC tentative breakpoint of <32 mg/liter (Table 1).
TABLE 1.
Antifungal activity against Candida auris isolates using the CLSI reference broth microdilution method
| Organism group (no. of isolates) and drug | No. (cumulative %) of isolates inhibited at MIC (mg/liter) of: |
MIC50 | MIC90 | % S using CDC tentative breakpointsa | % WT applying ECV/ECOFFb | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.008 | 0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | >64 | |||||
| All Candida auris isolates (36) | |||||||||||||||||||
| Anidulafungin | 0 (0.0) | 1 (2.8) | 0 (2.8) | 4 (13.9) | 26 (86.1) | 5 (100.0) | 0.5 | 1 | 100.0 | 86.1 | |||||||||
| Caspofungin | 0 (0.0) | 1 (2.8) | 2 (8.3) | 14 (47.2) | 10 (75.0) | 7 (94.4) | 2 (100.0) | 0.25 | 0.5 | 100.0 | |||||||||
| Micafungin | 0 (0.0) | 2 (5.6) | 7 (25.0) | 24 (91.7) | 3 (100.0) | 0.25 | 0.25 | 100.0 | 91.7 | ||||||||||
| Fluconazole | 0 (0.0) | 1 (2.8) | 2 (8.3) | 0 (8.3) | 1 (11.1) | 0 (11.1) | 1 (13.9) | 31 (100.0) | >64 | >64 | 11.1 | 11.1 | |||||||
| Isavuconazole | 2 (5.6) | 0 (5.6) | 1 (8.3) | 8 (30.6) | 4 (41.7) | 2 (47.2) | 13 (83.3) | 5 (97.2) | 0 (97.2) | 1 (100.0) | 0.5 | 1 | 97.2 | ||||||
| Voriconazole | 1 (2.8) | 1 (5.6) | 1 (8.3) | 1 (11.1) | 0 (11.1) | 0 (11.1) | 3 (19.4) | 19 (72.2) | 7 (91.7) | 2 (97.2) | 0 (97.2) | 1 (100.0) | 1 | 2 | 72.2 | ||||
| Amphotericin B | 0 (0.0) | 1 (2.8) | 30 (86.1) | 5 (100.0) | 1 | 2 | 86.1 | 100.0 | |||||||||||
| Candida auris from the SENTRY program (15) | |||||||||||||||||||
| Anidulafungin | 0 (0.0) | 3 (20.0) | 11 (93.3) | 1 (100.0) | 0.5 | 0.5 | 100.0 | 93.3 | |||||||||||
| Caspofungin | 0 (0.0) | 2 (13.3) | 9 (73.3) | 2 (86.7) | 2 (100.0) | 0.12 | 0.5 | 100.0 | |||||||||||
| Micafungin | 0 (0.0) | 3 (20.0) | 11 (93.3) | 1 (100.0) | 0.25 | 0.25 | 100.0 | 93.3 | |||||||||||
| Fluconazole | 0 (0.0) | 1 (6.7) | 0 (6.7) | 1 (13.3) | 0 (13.3) | 1 (20.0) | 12 (100.0) | >64 | >64 | 13.3 | 13.3 | ||||||||
| Isavuconazole | 0 (0.0) | 1 (6.7) | 2 (20.0) | 0 (20.0) | 0 (20.0) | 8 (73.3) | 4 (100.0) | 0.5 | 1 | 100.0 | |||||||||
| Voriconazole | 0 (0.0) | 1 (6.7) | 1 (13.3) | 0 (13.3) | 0 (13.3) | 1 (20.0) | 11 (93.3) | 1 (100.0) | 1 | 1 | 93.3 | ||||||||
| Amphotericin B | 0 (0.0) | 12 (80.0) | 3 (100.0) | 1 | 2 | 80.0 | 100.0 | ||||||||||||
| Candida auris from Kenya (11) | |||||||||||||||||||
| Anidulafungin | 0 (0.0) | 1 (9.1) | 9 (90.9) | 1 (100.0) | 0.5 | 0.5 | 100.0 | 90.9 | |||||||||||
| Caspofungin | 0 (0.0) | 1 (9.1) | 0 (9.1) | 4 (45.5) | 6 (100.0) | 0.25 | 0.25 | 100.0 | |||||||||||
| Micafungin | 0 (0.0) | 1 (9.1) | 3 (36.4) | 6 (90.9) | 1 (100.0) | 0.25 | 0.25 | 100.0 | 90.9 | ||||||||||
| Fluconazole | 0 (0.0) | 6 (100.0) | >64 | >64 | 0.0 | 0.0 | |||||||||||||
| Isavuconazole | 0 (0.0) | 5 (45.5) | 3 (72.7) | 1 (81.8) | 2 (100.0) | 0.12 | 0.5 | 100.0 | |||||||||||
| Voriconazole | 0 (0.0) | 2 (18.2) | 6 (72.7) | 3 (100.0) | 1 | 2 | 72.7 | ||||||||||||
| Amphotericin B | 0 (0.0) | 11 (100.0) | 1 | 1 | 100.0 | 100.0 | |||||||||||||
| Candida auris from the CDC AR bank (10) | |||||||||||||||||||
| Anidulafungin | 0 (0.0) | 1 (10.0) | 0 (10.0) | 0 (10.0) | 6 (70.0) | 3 (100.0) | 0.5 | 1 | 100.0 | 70.0 | |||||||||
| Caspofungin | 0 (0.0) | 1 (10.0) | 2 (30.0) | 5 (80.0) | 2 (100.0) | 0.5 | 1 | 100.0 | |||||||||||
| Micafungin | 0 (0.0) | 1 (10.0) | 1 (20.0) | 7 (90.0) | 1 (100.0) | 0.25 | 0.25 | 100.0 | 90.0 | ||||||||||
| Fluconazole | 0 (0.0) | 1 (10.0) | 1 (20.0) | 0 (20.0) | 0 (20.0) | 0 (20.0) | 0 (20.0) | 8 (100.0) | >128 | >128 | 20.0 | 20.0 | |||||||
| Isavuconazole | 2 (20.0) | 0 (20.0) | 0 (20.0) | 1 (30.0) | 1 (40.0) | 1 (50.0) | 3 (80.0) | 1 (90.0) | 0 (90.0) | 1 (100.0) | 0.25 | 1 | 90.0 | ||||||
| Voriconazole | 1 (10.0) | 1 (20.0) | 0 (20.0) | 0 (20.0) | 0 (20.0) | 0 (20.0) | 0 (20.0) | 2 (40.0) | 3 (70.0) | 2 (90.0) | 0 (90.0) | 1 (100.0) | 2 | 4 | 40.0 | ||||
| Amphotericin B | 0 (0.0) | 1 (10.0) | 7 (80.0) | 2 (100).0 | 1 | 2 | 80.0 | 100.0 | |||||||||||
CDC tentative breakpoints are available at https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html.
Echinocandins were active against all isolates (36/36) tested using the CDC tentative breakpoints of ≥2mg/liter for caspofungin and ≥4 mg/liter for anidulafungin and micafungin. When the more conservative ECV/ECOFF values proposed by Arendrup et al. (33) were applied, 31/36 of the isolates were inhibited by ≤0.5 mg/liter of anidulafungin and 33/36 were inhibited by ≤0.25 mg/liter of micafungin. Whereas all isolates were classified as WT to amphotericin B using the ECV/ECOFF value of ≤2 mg/liter, 31/36 were inhibited at the CDC breakpoint of ≤1 mg/liter (Table 1).
Among the 3 collections tested, 13/15 of the isolates from the SENTRY program, 8/10 from the CDC AR panel, and all isolates from Kenya (6/6) were resistant to fluconazole (MIC ≥32 mg/liter; Table 1). All isolates from the SENTRY (15/15) and Kenya (11/11) collections were inhibited by ≤1 mg/liter of isavuconazole, compared to 9/10 for the CDC panel. Voriconazole was more active against isolates from SENTRY (14/15 inhibited by ≤1 mg/liter [S]) compared to those from Kenya (8/11 inhibited by ≤1 mg/liter [S]) and the CDC (4/10 inhibited by ≤1 mg/liter [S]) (Table 1). Whereas micafungin showed similar activity against isolates from all three collections (9/10 [CDC], 10/11 [Kenya], and 14/15 [SENTRY] inhibited by ≤0.25 mg/liter [WT]), anidulafungin was more active against isolates from SENTRY (14/15 inhibited by ≤0.5 mg/liter [WT]) and Kenya (10/11 inhibited by ≤0.5 mg/liter [WT]) than against those isolates from the CDC panel (7/10 inhibited by ≤0.5 mg/liter [WT]). All isolates from Kenya (11/11) were susceptible to amphotericin B at the CDC breakpoint of ≤1 mg/liter, whereas only 12/15 and 8/10 of isolates from the SENTRY and CDC collections, respectively, were inhibited at this cutoff value.
Frequency of MDR C. auris.
We assessed the extent of MDR in the 36-isolate collection by applying the CDC breakpoints for fluconazole (≥32 mg/liter) and amphotericin B (≥2 mg/liter) as well as the tentative ECV/ECOFFs for isavuconazole (>1 mg/liter), voriconazole (>1 mg/liter), and anidulafungin (>0.5 mg/liter) to determine the frequency of resistance or NWT strains and assess the degree of cross-resistance (azole-azole) or co-resistance (azole-echinocandin or azole-amphotericin B) in the collection. Overall, 32/36 isolates were resistant to fluconazole. Among the fluconazole-resistant population (n = 32 isolates), 9 were NWT to voriconazole, 5 were NWT to anidulafungin, 5 were NWT to amphotericin B, and none were NWT to isavuconazole. In total, 10 isolates were resistant to 2 agents of different classes as follows: 5 were resistant to fluconazole and amphotericin B (all were WT to anidulafungin) and 5 were resistant to fluconazole and NWT to anidulafungin (all were susceptible [WT] to amphotericin B). The frequency of MDR strains was 10/36 for the total collection. There were no isolates that were resistant to 3 different classes of antifungal agents.
Assessment of synergy between anidulafungin and isavucoazole or voriconazole.
A synergistic (∑FIC ≤0.5) interaction with anidulafungin was noted in 11/36 isolates with isavuconazole and 5/36 isolates with voriconazole (Tables 2 and 3). Partial synergy (∑FIC >0.5 to <1.0) was observed for 19/36 isolates and 22/36 of the isolates tested with the isavuconazole/anidulafungin combination and voriconazole/anidulafungin combination, respectively. Additivity (∑FIC 1.0) was observed in 2/36 of the isolates and 4/36 of the isolates tested with isavuconazole/anidulafungin and voriconazole/anidulafungin, respectively. Indifference (∑FIC >1.0 to <4.0) was noted with 4 and 3 of the isolates tested with isavuconazole/anidulafungin and voriconazole/anidulafungin, respectively. Antagonism (∑FIC ≥4.0) was observed for 1 isolate with the combination of voriconazole and anidulafungin (Tables 2 and 3).
TABLE 2.
Synergy results testing anidulafungin plus Isavuconazole for each Candida auris isolate
| Isolate source and collection no. |
MIC (mg/liter) |
ΣFIC resultsa |
|||
|---|---|---|---|---|---|
| Isavuconazole | Anidulafungin | Isavuconazole/anidulafungin | ΣFIC | Interpretation | |
| SENTRY program | |||||
| 512195 | 0.06 | 0.25 | 0.015/0.12 | 0.75 | Partial Synergy |
| 805548 | 0.5 | 0.5 | 0.03/0.25 | 0.56 | Partial Synergy |
| 871779 | 1 | 1 | ≤0.008/0.5 | 0.51 | Partial Synergy |
| 899807 | 0.5 | 0.5 | 0.06/0.25 | 0.63 | Partial Synergy |
| 968647 | 0.5 | 0.5 | 0.25/0.25 | 1.0 | Additivity |
| 968648 | 0.5 | 0.5 | 0.06/0.25 | 0.63 | Partial Synergy |
| 1042982 | 0.5 | 0.25 | ≤0.008/0.25 | 1.02 | Indifference |
| 1057395 | 0.5 | 0.5 | ≤0.008/0.25 | 0.52 | Partial Synergy |
| 1081160 | 1 | 0.5 | 0.03/0.25 | 0.53 | Partial Synergy |
| 1081169 | 0.5 | 0.5 | 0.015/0.25 | 0.53 | Partial Synergy |
| 1081171 | 1 | 0.5 | 0.03/0.25 | 0.53 | Partial Synergy |
| 1081172 | 1 | 0.5 | 0.06/0.25 | 0.56 | Partial Synergy |
| 1087877 | 0.06 | 0.5 | ≤0.008/0.12 | 0.38 | Synergy |
| 1087878 | 0.03 | 0.25 | ≤0.008/0.12 | 0.75 | Partial Synergy |
| 1089308 | 0.5 | 0.5 | 0.03/0.25 | 0.56 | Partial Synergy |
| Kenya collaboration | |||||
| 1 | 0.12 | 0.5 | 0.03/0.12 | 0.50 | Synergy |
| 2 | 0.06 | 0.5 | 0.015/0.12 | 0.50 | Synergy |
| 4 | 0.12 | 0.5 | 0.03/0.12 | 0.50 | Synergy |
| 5 | 0.5 | 0.5 | 0.06/0.12 | 0.38 | Synergy |
| 8 | 0.5 | 0.5 | 0.12/0.12 | 0.50 | Synergy |
| 10 | 0.06 | 0.5 | ≤0.008/0.25 | 0.63 | Partial Synergy |
| 12 | 0.25 | 0.5 | 0.06/≤0.03 | 0.31 | Synergy |
| 15 | 0.06 | 0.5 | 0.03/0.12 | 0.75 | Partial Synergy |
| 17 | 0.12 | 1 | 0.015/0.25 | 0.38 | Synergy |
| 19 | 0.06 | 0.12 | 0.06/≤0.03 | 1.25 | Indifference |
| 22 | 0.06 | 1 | ≤0.008/0.25 | 0.38 | Synergy |
| CDC antimicrobial resistance collection | |||||
| 0381 | ≤0.008 | 0.06 | ≤0.008/≤0.03 | 1.5 | Indifference |
| 0382 | 4 | 0.5 | ≤0.008/0.12 | 0.25 | Synergy |
| 0383 | 0.06 | 1 | ≤0.008/0.25 | 0.38 | Synergy |
| 0384 | 0.12 | 0.5 | ≤0.008/0.25 | 0.57 | Partial Synergy |
| 0385 | 1 | 0.5 | 0.25/0.25 | 0.75 | Partial Synergy |
| 0386 | 0.5 | 1 | 0.03/0.5 | 0.56 | Partial Synergy |
| 0387 | ≤0.008 | 0.5 | ≤0.008/≤0.03 | 1.06 | Indeterminate |
| 0388 | 0.5 | 0.5 | 0.25/0.25 | 1.0 | Additivity |
| 0389 | 0.5 | 0.5 | 0.03/0.25 | 0.56 | Partial Synergy |
| 0390 | 0.25 | 0.5 | 0.03/0.25 | 0.63 | Partial Synergy |
ΣFIC, sum of the fractional inhibitory concentrations.
TABLE 3.
Synergy results for anidulafungin plus voriconazole for each Candida auris isolate
| Isolate source and collection no. | MIC (mg/liter) |
ΣFIC resultsa |
|||
|---|---|---|---|---|---|
| Voriconazole | Anidulafungin | Voriconazole/anidulafungin | ΣFIC | Interpretation | |
| SENTRY program | |||||
| 512195 | 0.5 | 0.5 | 0.015/0.25 | 0.53 | Partial synergy |
| 805548 | 1 | 0.5 | 0.12/0.25 | 0.63 | Partial synergy |
| 871779 | 1 | 1 | 0.015/0.25 | 0.27 | Synergy |
| 899807 | 1 | 0.5 | 0.25/0.25 | 0.75 | Partial synergy |
| 968647 | 1 | 0.5 | 0.25/0.25 | 0.75 | Partial synergy |
| 968648 | 1 | 0.5 | 0.5/0.25 | 1.0 | Additivity |
| 1042982 | 1 | 0.25 | ≤0.008/0.25 | 1.01 | Indifference |
| 1057395 | 1 | 0.5 | 0.06/0.25 | 0.56 | Partial synergy |
| 1081160 | 1 | 0.5 | 0.25/0.25 | 0.75 | Partial synergy |
| 1081169 | 1 | 0.5 | 0.06/0.25 | 0.56 | Partial synergy |
| 1081171 | 2 | 0.5 | 0.12/0.25 | 0.56 | Partial synergy |
| 1081172 | 1 | 0.5 | 0.25/0.25 | 0.75 | Partial synergy |
| 1087877 | 0.06 | 0.25 | 0.03/0.12 | 1.0 | Additivity |
| 1087878 | 0.03 | 0.12 | 0.03/≤0.03 | 1.25 | Indifference |
| 1089308 | 1 | 0.5 | 0.06/0.25 | 0.56 | Partial synergy |
| Kenya collaboration | |||||
| 1 | 2 | 0.5 | 0.03/0.25 | 0.52 | Partial synergy |
| 2 | 0.5 | 0.5 | 0.03/0.25 | 0.56 | Partial synergy |
| 4 | 1 | 0.5 | 0.25/0.12 | 0.50 | Synergy |
| 5 | 0.5 | 0.5 | 0.06/0.25 | 0.63 | Partial synergy |
| 8 | 1 | 0.5 | 0.06/0.25 | 0.56 | Partial synergy |
| 10 | 2 | 0.5 | 0.06/0.25 | 0.53 | Partial synergy |
| 12 | 1 | 0.25 | 0.25/0.12 | 0.75 | Partial synergy |
| 15 | 1 | 0.5 | 0.12/0.25 | 0.63 | Partial synergy |
| 17 | 2 | 1 | ≤0.008/≥2 | 2.0 | Indifference |
| 19 | 1 | 0.25 | 0.06/0.12 | 0.56 | Partial synergy |
| 22 | 1 | 1 | 0.25/0.12 | 0.37 | Synergy |
| CDC antimicrobial resistance collection | |||||
| 0381 | ≤0.008 | 0.06 | ≤0.008/≤0.03 | 1.5 | Indifference |
| 0382 | >8 | 0.5 | 0.015/0.12 | 0.25 | Synergy |
| 0383 | 2 | 1 | 0.25/0.25 | 0.38 | Synergy |
| 0384 | 2 | 0.5 | ≤0.008/≥2 | 4.0 | Antagonism |
| 0385 | 4 | 0.5 | ≤0.008/0.5 | 1.0 | Additivity |
| 0386 | 4 | 1 | 0.06/0.5 | 0.52 | Partial synergy |
| 0387 | 0.015 | 0.5 | ≤0.008/0.25 | 1.0 | Additivity |
| 0388 | 1 | 0.5 | 0.25/0.25 | 0.75 | Partial synergy |
| 0389 | 2 | 0.5 | 0.03/0.25 | 0.52 | Partial synergy |
| 0390 | 1 | 0.5 | 0.12/0.25 | 0.63 | Partial synergy |
ΣFIC, sum of the fractional inhibitory concentrations.
The frequency of synergy or partial synergy between isavuconazole or voriconazole plus anidulafungin was highest among the isolates from Kenya (10/11 for both) compared to the SENTRY and CDC isolates tested with both combinations. A total of 7/10 and 6/10 of the CDC AR isolates displayed synergy or partial synergy with isavuconazole/anidulafungin and voriconazole/anidulafungin, respectively, and 13/15 of the SENTRY Program isolates displayed synergy or partial synergy for isavuconazole plus anidulafungin. The frequency of synergy/partial synergy was slightly lower for isolates from the SENTRY program for the combination of voriconazole plus anidulafungin (11/15 isolates).
While the combination of either isavuconazole or voriconazole and anidulafungin generally resulted in a favorable interaction (synergy, partial synergy, or additivity), the MIC of the echinocandin in each combination decreased by only about 2- to 4-fold (33/36 [isavuconazole plus anidulafungin] and 32/36 [voriconazole plus anidulafungin]) (Tables 2 and 3). Notably, of the 5 isolates for which the anidulafungin MIC tested alone was > 0.5 mg/liter (NWT), the MIC was reduced to ≤0.5 mg/liter (WT) for all isolates in the combination with isavuconazole and for 4/5 isolates in the combination with voriconazole (Tables 2 and 3). In contrast, the azole MIC results showed a greater decrease when tested in combination, with results 4- to 256-fold lower for isavuconazole (29/36 isolates) and voriconazole (31/36) (Tables 2 and 3). Although there was only 1 isolate that was NWT to isavuconazole (MIC, 4 mg/liter), the MIC in combination was decreased by 256-fold (MIC, 0.008 mg/liter) (Table 2). There were 10 isolates that were resistant to voriconazole, with MIC values ≥2 mg/liter. For 9/10 isolates, the voriconazole MICs were reduced in combination by 2- to 256-fold, resulting in a change from resistant to susceptible using the CDC breakpoint of ≤1 mg/liter (Table 3).
Among the 10 MDR isolates, 5 were resistant to fluconazole and amphotericin B and 5 were resistant to fluconazole and resistant/NWT to anidulafungin. Against the fluconazole/amphotericin B-resistant isolates, the combination of anidulafungin and isavuconazole showed partial synergy for 4 isolates and additivity for 1 isolate, whereas the combination of anidulafungin and voriconazole showed partial synergy for all 5 tested isolates. Against the fluconazole-resistant and anidulafungin-resistant/NWT isolates, the combination of anidulafungin and isavuconazole was synergistic for 3, indifferent for 1, and additive for 1 isolate. Importantly, the combination of anidulafungin and voriconazole showed antagonism for 1 of the fluconazole/anidulafungin-resistant isolates, synergy for 2, and partial synergy for 2 isolates.
Genetic characterization of C. auris isolates from the SENTRY program.
Alterations of the genes ERG11, ERG3, CDR1, and UPC2, which are associated with azole resistance, and FKS, which encodes the target for the echinocandins, are displayed in Table 4.
TABLE 4.
Mutations in genes associated with resistance to the azoles and echinocandins detected among the Candida auris isolates from the SENTRY program
| Study yr | Country, State (hospital [H] 1 or 2) | MIC (mg/liter) |
Amino acid alterationsa |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Anidulafungin | Fluconazole | Isavuconazole | Voriconazole | CDR1 | ERG11 | ERG3 | FKS1 | UPC2 | ||
| 2009 | Germany | 0.5 | >64 | 0.06 | 0.5 | S21L | S74L, A25L, F126L | WT | S906L | S36L, S89L, S237L, S376L, S436L, S456L |
| 2013 | NY (H1), USA | 0.5 | >128 | 0.5 | 1 | S21L, V704L | S74L, K143R | WT | S906L | S36L, S89L, S237L, S376L, S436L, S456L |
| 2014 | Colombia | 1 | 64 | 1 | 1 | S21L, H771R | S74L, K177R, N335S, E343D, Y501H | S58T | S906L | S36L, S89L, I223M, S237L, S376L, S436L, S456L |
| 2015 | NJ, USA | 0.5 | >128 | 0.5 | 1 | S21L, V704L | S74L, K143R | WT | S906L | S36L, S89L, S237L, S376L, S436L, S456L |
| 2016 | NY (H2), USA | 0.5 | >128 | 0.5 | 1 | S21L, V704L | S74L, K143R | WT | S906L | S36L, S89L, S237L, S376L, S436L, S456L |
| 2016 | NY (H2), USA | 0.5 | >128 | 0.5 | 1 | S21L, V704L | S74L, K143R | WT | S906L | S36L, S89L, S237L, S376L, S436L, S456L |
| 2017 | NY (H1), USA | 0.25 | >128 | 0.5 | 1 | S21L, V704L | S74L, K143R | WT | S906L | S36L, S89L, S237L, S376L, S436L, S456L |
| 2018 | NJ, USA | 0.5 | >128 | 0.5 | 1 | S21L, V704L | S74L, K143R | WT | S906L | S36L, S89L, S376L, S436L, S456L |
| 2018 | NY (H2), USA | 0.5 | >128 | 1 | 1 | S21L, V704L | S74L, K143R | WT | S906L | S36L, S237L, S89L, S376L, S436L, S456L |
| 2018 | NY (H2), USA | 0.5 | >128 | 0.5 | 1 | S21L, V704L | S74L, K143R | WT | S906L | S36L, S89L, S237L, S376L, S436L, S456L |
| 2018 | NY (H2), USA | 0.5 | >128 | 1 | 2 | S21L, V704L | S74L, K143R | WT | S906L | S36L, S89L, S237L, S376L, S436L, S456L |
| 2018 | NY (H2), USA | 0.5 | >128 | 1 | 1 | S21L, V704L | S74L, K143R | WT | S906L | S36L, S89L, S237L, S376L, S436L, S456L |
| 2018 | Panama | 0.25 | 16 | 0.06 | 0.06 | S21L | S74L, K177R, N335S, E343D | S58T | S906L | S36L, S89L, I223M, S237L, S376L, S436L, S456L |
| 2018 | Panama | 0.25 | 4 | 0.03 | 0.03 | S21L | S74L, K177R, N335S, E343D | S58T | S906L | S36L, S89L, I223M, S237L, S376L, S436L, S456L |
| 2019 | NJ, USA | 0.5 | >128 | 0.5 | 1 | S21L, V704L | S74L, K143R | WT | S906L | S36L, S89L, S237L, S376L, S436L, S456L |
Mutations in boldface have previously have been shown to cause resistance.
The SENTRY program isolates displaying elevated fluconazole MIC values (≥32 mg/liter) harbored the mutations in the ERG11 gene that were described in the literature as associated with azole resistance. These mutations included F126L, detected in one isolate from Germany; K143R, detected among 11 isolates from the US; and Y501H, detected in one isolate from Colombia. Two isolates from Panama had lower fluconazole MIC values (4 and 16 mg/liter) and did not harbor the mutations previously described to cause azole resistance.
None of the isolates analyzed had the S639P or S639F alterations in FKS that have been reported to result in echinocandin resistance. Only one of the SENTRY isolates tested as NWT to anidulafungin (MIC 1 mg/liter) and it did not harbor a mutation in FKS. Lastly, the mutations observed in CDR1 and UPC2 have not been described in the literature to be associated with antifungal resistance.
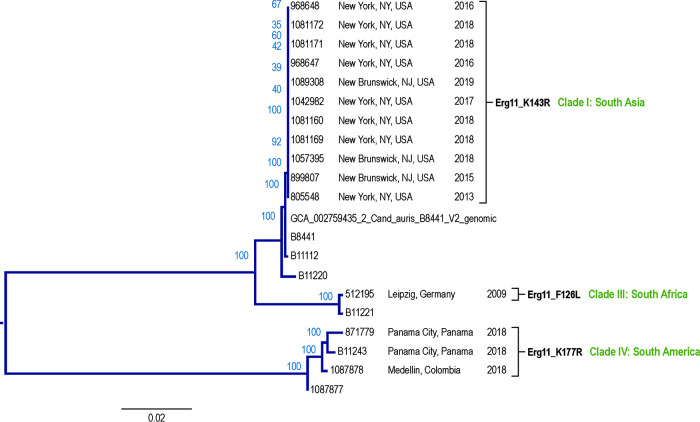
MLST- and SNP-based phylogenetic analysis grouped the isolates from the SENTRY program into three main groups: (i) three isolates from Latin America (clade III, South America), (ii) one isolate from Germany (clade II, South Africa), and (iii) 11 isolates from the USA (clade I, South Asia) (Fig. 1). The isolates from Latin America (one from Colombia and two from Panama) carried the same ERG11 alterations (K177R, N335S, and E343D), Erg3 amino acid substitutions (S58T), and one additional UPC2 change that other isolates did not carry (I223M; Table 3). The fluconazole-resistant isolate from Colombia differed from the fluconazole-susceptible isolates from Panama, as it harbored additional amino acid changes in Erg11 (Y501H) and a unique mutation in CDR1 (encoding H771R). The grouping of isolates by both methods was concordant.
FIG 1.
Phylogenetic tree of Candida auris isolates collected as part of the SENTRY program.
In contrast, isolates from the US carried the ERG11 alteration K143R, which is the same ERG11 alteration prevalent in the South Asia clade, and no alterations in ERG3. The single isolate from Germany carried none of these signature alterations. Like the South Africa clade, this isolate carried an alteration in ERG11 at position F126 (F126L).
DISCUSSION
Several recent reviews have provided extensive documentation of the global epidemiology, antifungal resistance profiles, and the mechanism of resistance to azoles and echinocandins for C. auris isolates from infected and colonized patients throughout the world (3–5, 8–10, 15, 23). The results of the present study support these observations for three distinct collections of isolates. Overall, we found most isolates were resistant to fluconazole (32/36), with cross-resistance detected for voriconazole (10/36) but not isavuconazole. Although not approved for the treatment of candidiasis, isavuconazole exhibited a WT MIC profile, with 35 of the 36 isolates inhibited at the tentative ECV/ECOFF of ≤1 mg/liter (Table 1). As with previous reports, we found 5/36 isolates were resistant to amphotericin B and 5/36 isolates were NWT to one or more echinocandins (9, 10).
In the literature, between 0.0% and 40.7% of C. auris isolates express an MDR phenotype and <4% were resistant to three or more classes of agents (1, 7, 9, 15–18). These findings vary according to the geographic origin and/or the predominant clade of the isolates tested (10, 12, 13, 19). Among the 36 isolates tested in the present study, 10 were shown to exhibit an MDR profile, whereas 5 were resistant to fluconazole and amphotericin B and 5 were resistant/NWT to fluconazole and anidulafungin. None of the isolates were resistant to 3 or more agents. This MDR profile prompted concern that the optimal therapy for a C. auris infection may be difficult to achieve, raising questions concerning the role of antifungal combination therapy (3, 5, 23). To this end, there have been six different studies of the in vitro activity of various agents tested in combination against collections of MDR C. auris (26–30). These combination studies included combinations of azoles and echinocandins; flucytosine with azoles, echinocandins, and polyenes; sulfa drugs or an antileishmanial agent with amphotericin B, azoles, or echinocandins; and an HIV protease inhibitor with azoles (26–31). For most combinations, the results were judged to be additive or indifferent, with a few combinations showing synergy against small numbers (10 to 15 isolates, most South Asia clade 1 or the CDC AR collection) of isolates. These results are consistent with other reports of combinations of azoles and amphotericin B used against the Candida species. For example, in clinical studies that compared fluconazole alone versus fluconazole combined with amphotericin B in treatment of candidemia, nonneutropenic patients studied showed higher rates of and more rapid fungemia clearance when treated with the combination of azole and amphotericin B (34).
Although isavuconazole has been shown to be active against C. auris (33), it has only been evaluated in combination with flucytosine against 15 isolates from the South Asian clade, with largely indifferent results (29). Notably, none of these studies have shown antagonism to any drug combination. Perhaps the most encouraging results were seen in the combination of azoles with echinocandins, as micafungin plus voriconazole produced synergy against a panel of 10 MDR isolates of C. auris (28).
As shown in Tables 2 and 3, the combination of anidulafungin and either isavuconazole or voriconazole resulted in synergy or partial synergy for most of the isolates tested in this study. The combination of anidulafungin and isavuconazole resulted in greater synergy/partial synergy (30/36) compared to anidulafungin and voriconazole (27/36). In contrast to previous studies, we observed antagonism between anidulafungin and voriconazole for one MDR (resistant to fluconazole and anidulafungin) isolate of C. auris.
In vitro studies on antifungals showed that combinations can broaden the coverage, increase the fungicidal effect, and reduce the risk of antifungal resistance development. Combined antifungal agents also can display synergistic activity with decreased toxicity (35).
Antagonism can be the result of several different mechanisms. For example, both antifungals may have direct action on the same target site, thereby decreasing the availability of one another. In addition, the modification of a target in the fungal cell by one antifungal agent can make that target less susceptible to exposure to a second antifungal. This modification happens in the interaction between the azoles and amphotericin B, as the azoles prevent the synthesis of ergosterol, thereby making amphotericin B inactive (36).
Thus far, clinical or in vivo evidence supporting combination therapy in infections with C. auris is lacking, save for a study showing that the combination of sulfamethoxazole and voriconazole enhanced survival of Caenorhabditis elegans infected with C. auris by nearly 70% (27). Regardless, given that C. auris is often an MDR organism, combination therapy is intriguing and should be evaluated further in vivo. The overall activity of isavuconazole alone or in combination with an echinocandin such as anidulafungin is promising. Additional evaluation is encouraged as a potential option for treatment of C. auris infections.
The SENTRY antifungal surveillance program has been active on a global scale since 1997 (37), with identification to species level confirmed by molecular sequencing or matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) since 2006. Between 2006 and 2019, a total of 16,273 isolates of Candida, consecutively collected from invasive infections, were characterized at JMI Laboratories. Of these, 15 (0.09%) isolates were identified as C. auris. Among these isolates, one was collected in 2009 (Germany), six were collected between 2013 and 2017 (five from the US and one from Colombia), seven were collected in 2018 (two from Panama and five from US), and one was collected in 2019 (US) (Table 3). The US isolates were collected from two institutions in New York (eight isolates) and one in New Jersey (three isolates). Characterization of the SENTRY isolates by WGS confirmed these identifications and showed these isolates represented three previously defined clades: South Asia (clade I, 11 US isolates), South America (clade IV; three total, including Panama [two isolates] and Colombia [one isolate]), and South Africa (clade III; one German isolate). MLST analysis revealed that the isolates from the US were genetically related to but different from isolates from Latin America (Panama and Colombia) and Germany (Fig. 1). All US isolates were resistant to fluconazole and harbored the K143R mutation in ERG11. As noted by Healey et al. (19), the isolates originating from Colombia and Panama (South American clade) all contained the same three ERG11 substitutions (K177R, N335S, and E343D), in contrast to the isolates from NY/NJ (South Asia clade) and Germany (South African clade). Notably, the isolate from Colombia expressed resistance to fluconazole and differed from the fluconazole-susceptible isolates from Panama by additional substitutions in ERG11 (Y501H) and CDR1 (H771R) (Table 3). The Y501H mutation was described previously in a fluconazole-resistant isolate from Colombia (19). Healey et al. (19) examined the role of these Erg11 substitutions in resistance to fluconazole by expressing the Colombian ERG11 WT allele (K177R, N335S, and E343D) and the allele encoding the Y501H substitution in Saccharomyces cerevisiae. Neither of these alleles resulted in the expression of fluconazole resistance in S. cerevisiae, indicating that these alleles do not impact resistance. It is possible that Y501H may contribute to reduced susceptibility to azoles in C. auris, but this conclusion would be dependent on an increase in ERG11 expression or efflux (19, 20). The CDR1 mutation (H771R) detected in this study has not been described to result in resistance to azoles.
There are two limitations that the present study shares with the six previous studies of antifungal drug combinations against C. auris. First, the total number of isolates tested was small (although greater than the numbers tested in the other studies), and second, aside from fluconazole and voriconazole, the number of isolates resistant to azoles and echinocandins was limited. The one isolate from the SENTRY program that was NWT to anidulafungin did not harbor a mutation in FKS as determined by WGS (Table 4). Regardless, this study found that isavuconazole had a high level of activity against the clinical isolates of C. auris, irrespective of resistance to fluconazole and other azoles or the presence of resistance mutations in ERG11. The combination of anidulafungin and isavuconazole resulted largely in synergy or partial synergy greater than the results seen with the combination of anidulafungin and voriconazole. Although no antagonism was seen with the combination of isavuconazole and anidulafungin, antagonism was observed for anidulafungin and voriconazole in one MDR isolate of C. auris. The importance of this finding should be confirmed by additional tests of this combination using in vivo models. WGS, applied to the 15 isolates of C. auris from the 2006 to 2019 SENTRY program, revealed three previously established distinct clades, each harboring the characteristic clade mutation in ERG11. Also, these data indicate that the increase in C. auris worldwide was captured by the SENTRY program, which collects invasive fungal isolates annually. The MDR nature of C. auris negatively impacts the therapy of infections, suggesting that combination antifungal therapy may be warranted. The combination of anidulafungin and isavuconazole should be explored further in vivo.
MATERIALS AND METHODS
Fungal isolates.
A total of 36 isolates of C. auris were included in the study. The 15 isolates from the SENTRY program were collected during 2006 to 2019 and comprised 0.09% of 16,273 Candida species isolates consecutively collected from invasive infections. Most of these isolates were from New York (NY; 8 isolates) and New Jersey (NJ; 3 isolates), while 1 isolate each was from Germany and Colombia and 2 isolates were from Panama. The 10 isolates from Kenya all were from patients with candidemia sent to JMI Laboratories (North Liberty, IA, USA) for identification and antifungal susceptibility testing (32). The 10 isolates from the CDC antimicrobial resistance (AR) bank were from a panel of isolates supplied by the CDC. Isolates from the SENTRY program and isolates from Kenya were identified to the species level using either nucleic acid sequencing or matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) analysis (Bruker Daltonics, Bremen, Germany).
Antifungal susceptibility testing.
All isolates were tested by the broth microdilution (BMD) method as described by the Clinical and Laboratory Standards Institute (CLSI) documents M27 and M60 (38, 39). The range of antifungal agent concentrations tested were 0.002 to 4 mg/liter for the echinocandins, isavuconazole, itraconazole, and voriconazole; 0.06 to 4 mg/liter for amphotericin B; and 0.008 to 128 mg/liter for fluconazole. There are no clinical breakpoints established by CLSI or the European Committee for Antimicrobial Susceptibility Testing (EUCAST) for any of the systemically active antifungal agents and C. auris. Tentative ECV/ECOFF values have been established according to CLSI and EUCAST (10, 33). Using the CLSI method of setting the cutoff at the theoretical MIC distribution at 97.5% of the wild type (WT) population and the ECOFF finder program for ECV determination, tentative ECVs have been proposed for the CLSI BMD method for itraconazole (0.25 to 0.5 mg/liter), isavuconazole (1 mg/liter), micafungin (0.25 mg/liter), anidulafungin (0.5 mg/liter), and amphotericin B (2 mg/liter) (33). In addition to these tentative statistical ECV/ECOFF values, the CDC has also published tentative resistant breakpoints for fluconazole (≥32 mg/liter), voriconazole (≥2 mg/liter), amphotericin B (≥2 mg/liter), anidulafungin (≥4 mg/liter), caspofungin (≥2 mg/liter), and micafungin (≥4 mg/liter) (https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html).
Synergy testing of antifungal combinations was performed with various concentrations of anidulafungin (range, 0.03 to 2 mg/liter) combined with isavuconazole (range, 0.008 to 8 mg/liter) or voriconazole (range, 0.008 to 8 mg/liter) using a checkerboard grid analysis following the CLSI BMD method (40).
Drug combination interactions were calculated algebraically by determining the fractional inhibitory concentration (FIC), as previously described (38). The sum of the FICs (ΣFIC) was calculated as follows: ΣFIC = FICA + FICB. FICA was calculated as the MIC of drug A in the combination divided by the MIC of drug A alone, and FICB equals the MIC of drug B in the combination divided by the MIC of drug B alone. The interpretation of ΣFIC was as follows: synergy, ≤0.5; partial synergy, >0.5 to <1.0; additivity, 1.0; indifference, >1.0 to <4.0; antagonism, ≥4.0 (39). The indifferent category was used when the result for one agent tested alone demonstrated an off-scale (≤ or ≥) MIC result.
Quality control (QC) was performed as recommended in documents M27 (39) and M60 (41) using Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258, and both strains were tested simultaneously according to M27 (39) and M60 (41) guidelines. Acceptable MIC ranges for the QC strains were published by CLSI in the document M60 (41).
Genetic characterization of C. auris isolates from the SENTRY program.
As described previously, C. auris isolates from the SENTRY program were submitted to further testing using whole-genome sequencing (WGS) analysis of resistance mechanisms and genetic relatedness (42). Sequences were compared to published C. auris sequences. DNA regions including FKS1, ERG3, ERG11, CDR1, and UPC2 were compared to sequences available in the literature. MLST analysis was performed based on ITS, D1/D2, RPB1, and RPB2 sequences as described by Prakash et al. (43). Additional SNP-based phylogenetic analysis was performed (https://doi.org/10.1038/nmeth.4285) by comparing the study isolates to the international clades described by Lockhart et al. (7).
ACKNOWLEDGMENTS
We acknowledge SJ Ryan Arends and Cecilia G. Carvalhaes for reviewing the synergy data and the SENTRY program participants, as well as Rodney Adam (Kenya collaboration), for contributing the isolates used in this study.
This study was performed by JMI Laboratories and supported by Pfizer Inc., which included funding for services related to preparing the manuscript.
JMI Laboratories contracted to perform services in 2019 for Achaogen, Inc., Albany College of Pharmacy and Health Sciences, Allecra Therapeutics, Allergan, AmpliPhi Biosciences Corp., Amicrobe Advanced Biomaterials, Amplyx, Antabio, American Proficiency Institute, Arietis Corp., Arixa Pharmaceuticals, Inc., Astellas Pharma Inc., Athelas, Basilea Pharmaceutica Ltd., Bayer AG, Becton, Dickinson and Company, bioMérieux SA, Boston Pharmaceuticals, Bugworks Research Inc., CEM-102 Pharmaceuticals, Cepheid, Cidara Therapeutics, Inc., CorMedix Inc., DePuy Synthes, Destiny Pharma, Discuva Ltd., Falk Pharma GmbH, Emery Pharma, Entasis Therapeutics, Eurofarma Laboratorios SA, US Food and Drug Administration, Fox Chase Chemical Diversity Center, Inc., Gateway Pharmaceutical LLC, GenePOC Inc., Geom Therapeutics, Inc., GlaxoSmithKline plc, Harvard University, Helperby, HiMedia Laboratories, F. Hoffmann-La Roche Ltd., ICON plc, Idorsia Pharmaceuticals Ltd., Iterum Therapeutics plc, Laboratory Specialists, Inc., Melinta Therapeutics, Inc., Merck & Co., Inc., Microchem Laboratory, Micromyx, MicuRx Pharmaceuticals, Inc., Mutabilis Co., Nabriva Therapeutics plc, NAEJA-RGM, Novartis AG, Oxoid Ltd., Paratek Pharmaceuticals, Inc., Pfizer, Inc., Polyphor Ltd., Pharmaceutical Product Development, LLC, Prokaryotics Inc., Qpex Biopharma, Inc., Roivant Sciences, Ltd., Safeguard Biosystems, Scynexis, Inc., SeLux Diagnostics, Inc., Shionogi and Co., Ltd., SinSa Labs, Spero Therapeutics, Summit Pharmaceuticals International Corp., Synlogic, T2 Biosystems, Inc., Taisho Pharmaceutical Co., Ltd., TenNor Therapeutics Ltd., Tetraphase Pharmaceuticals, Theravance Biopharma, University of Colorado, University of Southern California-San Diego, University of North Texas Health Science Center, VenatoRx Pharmaceuticals, Inc., Viosera Therapeutics, Vyome Therapeutics Inc., Wockhardt, Yukon Pharmaceuticals, Inc., Zai Lab, and Zavante Therapeutics, Inc.
There are no speakers’ bureaus or stock options to declare.
REFERENCES
- 1.Adams E, Quinn M, Tsay S, Poirot E, Chaturvedi S, Southwick K, Greenko J, Fernandez R, Kallen A, Vallabhaneni S, Haley V, Hutton B, Blog D, Lutterloh E, Zucker H, Candida auris Investigation Workgroup. 2018. Candida auris in healthcare facilities, New York, USA, 2013-2017. Emerg Infect Dis 24:1816–1824. doi: 10.3201/eid2410.180649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bidaud AL, Chowdhary A, Dannaoui E. 2018. Candida auris: an emerging drug resistant yeast—a mini-review. J Mycol Med 28:568–573. doi: 10.1016/j.mycmed.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Forsberg K, Woodworth K, Walters M, Berkow EL, Jackson B, Chiller T, Vallabhaneni S. 2019. Candida auris: the recent emergence of a multidrug-resistant fungal pathogen. Med Mycol 57:1–12. doi: 10.1093/mmy/myy054. [DOI] [PubMed] [Google Scholar]
- 4.Hata DJ, Humphries R, Lockhart SR, College of American Pathologists Microbiology Committee. 2020. Candida auris: an emerging yeast pathogen posing distinct challenges for laboratory diagnostics, treatment, and infection prevention. Arch Pathol Lab Med 144:107–114. doi: 10.5858/arpa.2018-0508-RA. [DOI] [PubMed] [Google Scholar]
- 5.Jeffery-Smith A, Taori SK, Schelenz S, Jeffery K, Johnson EM, Borman A, Manuel R, Brown CS, Candida auris Incident Management Team. 2018. Candida auris: a review of the literature. Clin Microbiol Rev 31:e00029-17. doi: 10.1128/CMR.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Cassia Orlandi Sardi J, Silva DR, Soares Mendes-Giannini MJ, Rosalen PL. 2018. Candida auris: epidemiology, risk factors, virulence, resistance, and therapeutic options. Microb Pathog 125:116–121. doi: 10.1016/j.micpath.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhodes J, Fisher MC. 2019. Global epidemiology of emerging Candida auris. Curr Opin Microbiol 52:84–89. doi: 10.1016/j.mib.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Snyder GM, Wright SB. 2019. The epidemiology and prevention of Candida auris. Curr Infect Dis Rep 21:19. doi: 10.1007/s11908-019-0675-8. [DOI] [PubMed] [Google Scholar]
- 10.Lockhart SR. 2019. Candida auris and multidrug resistance: defining the new normal. Fungal Genet Biol 131:103243. doi: 10.1016/j.fgb.2019.103243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaabane F, Graf A, Jequier L, Coste AT. 2019. Review on antifungal resistance mechanisms in the emerging pathogen Candida auris. Front Microbiol 10:2788. doi: 10.3389/fmicb.2019.02788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kordalewska M, Lee A, Park S, Berrio I, Chowdhary A, Zhao Y, Perlin DS. 2018. Understanding echinocandin resistance in the emerging pathogen Candida auris. Antimicrob Agents Chemother 62:e00238-18. doi: 10.1128/AAC.00238-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kordalewska M, Perlin DS. 2019. Identification of drug resistant Candida auris. Front Microbiol 10:1918. doi: 10.3389/fmicb.2019.01918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y, O'Brien B, Leach L, Clark A, Bates M, Adams E, Ostrowsky B, Quinn M, Dufort E, Southwick K, Erazo R, Haley VB, Bucher C, Chaturvedi V, Limberger RJ, Blog D, Lutterloh E, Chaturvedi S. 2020. Laboratory analysis of an outbreak of Candida auris in New York from 2016 to 2018: impact and lessons learned. J Clin Microbiol 58:e01503-19. doi: 10.1128/JCM.01503-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chow NA, Gade L, Tsay SV, Forsberg K, Greenko JA, Southwick KL, Barrett PM, Kerins JL, Lockhart SR, Chiller TM, Litvintseva AP, US Candida auris Investigation Team. 2018. Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect Dis 18:1377–1384. doi: 10.1016/S1473-3099(18)30597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chowdhary A, Prakash A, Sharma C, Kordalewska M, Kumar A, Sarma S, Tarai B, Singh A, Upadhyaya G, Upadhyay S, Yadav P, Singh PK, Khillan V, Sachdeva N, Perlin DS, Meis JF. 2018. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother 73:891–899. doi: 10.1093/jac/dkx480. [DOI] [PubMed] [Google Scholar]
- 17.Escandon P, Chow NA, Caceres DH, Gade L, Berkow EL, Armstrong P, Rivera S, Misas E, Duarte C, Moulton-Meissner H, Welsh RM, Parra C, Pescador LA, Villalobos N, Salcedo S, Berrio I, Varon C, Espinosa-Bode A, Lockhart SR, Jackson BR, Litvintseva AP, Beltran M, Chiller TM. 2019. Molecular epidemiology of Candida auris in Colombia reveals a highly related, countrywide colonization with regional patterns in Amphotericin B resistance. Clin Infect Dis 68:15–21. doi: 10.1093/cid/ciy411. [DOI] [PubMed] [Google Scholar]
- 18.Eyre DW, Sheppard AE, Madder H, Moir I, Moroney R, Quan TP, Griffiths D, George S, Butcher L, Morgan M, Newnham R, Sunderland M, Clarke T, Foster D, Hoffman P, Borman AM, Johnson EM, Moore G, Brown CS, Walker AS, Peto TEA, Crook DW, Jeffery KJM. 2018. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med 379:1322–1331. doi: 10.1056/NEJMoa1714373. [DOI] [PubMed] [Google Scholar]
- 19.Healey KR, Kordalewska M, Jimenez Ortigosa C, Singh A, Berrio I, Chowdhary A, Perlin DS. 2018. Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob Agents Chemother 62:e01427-18. doi: 10.1128/AAC.01427-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rybak JM, Doorley LA, Nishimoto AT, Barker KS, Palmer GE, Rogers PD. 2019. Abrogation of triazole resistance upon deletion of CDR1 in a clinical isolate of Candida auris. Antimicrob Agents Chemother 63:e00057-19. doi: 10.1128/AAC.00057-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenters N, Kiernan M, Chowdhary A, Denning DW, Peman J, Saris K, Schelenz S, Tartari E, Widmer A, Meis JF, Voss A. 2019. Control of Candida auris in healthcare institutions: outcome of an International Society for Antimicrobial Chemotherapy expert meeting. Int J Antimicrob Agents 54:400–406. doi: 10.1016/j.ijantimicag.2019.08.013. [DOI] [PubMed] [Google Scholar]
- 22.Tsay S, Kallen A, Jackson BR, Chiller TM, Vallabhaneni S. 2018. Approach to the investigation and management of patients with Candida auris, an emerging multidrug-resistant yeast. Clin Infect Dis 66:306–311. doi: 10.1093/cid/cix744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corsi-Vasquez G, Ostrosky-Zeichner L. 2019. Candida auris: what have we learned so far? Curr Opin Infect Dis 32:559–564. doi: 10.1097/QCO.0000000000000603. [DOI] [PubMed] [Google Scholar]
- 24.Fohrer C, Fornecker L, Nivoix Y, Cornila C, Marinescu C, Herbrecht R. 2006. Antifungal combination treatment: a future perspective. Int J Antimicrob Agents 27 Suppl 1:25–30. doi: 10.1016/j.ijantimicag.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee PK, Sheehan DJ, Hitchcock CA, Ghannoum MA. 2005. Combination treatment of invasive fungal infections. Clin Microbiol Rev 18:163–194. doi: 10.1128/CMR.18.1.163-194.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bidaud AL, Botterel F, Chowdhary A, Dannaoui E. 2019. In vitro antifungal combination of flucytosine with amphotericin B, voriconazole, or micafungin against Candida auris shows no antagonism. Antimicrob Agents Chemother 63:e01393-19. doi: 10.1128/AAC.01393-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eldesouky HE, Li X, Abutaleb NS, Mohammad H, Seleem MN. 2018. Synergistic interactions of sulfamethoxazole and azole antifungal drugs against emerging multidrug-resistant Candida auris. Int J Antimicrob Agents 52:754–761. doi: 10.1016/j.ijantimicag.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 28.Fakhim H, Chowdhary A, Prakash A, Vaezi A, Dannaoui E, Meis JF, Badali H. 2017. In vitro interactions of echinocandins with triazoles against multidrug-resistant Candida auris. Antimicrob Agents Chemother 61:e01056-17. doi: 10.1128/AAC.01056-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Brien B, Chaturvedi S, Chaturvedi V. 2020. In vitro evaluation of antifungal drug combinations against multidrug-resistant Candida auris isolates from New York outbreak. Antimicrob Agents Chemother 64:e02195-19. doi: 10.1128/AAC.02195-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Y, Totten M, Memon W, Ying C, Zhang SX. 2019. In vitro antifungal susceptibility of the emerging multidrug-resistant pathogen Candida auris to miltefosine alone and in combination with amphotericin B. Antimicrob Agents Chemother 64:e02063-19. doi: 10.1128/AAC.02063-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eldesouky HE, Salama EA, Lanman NA, Hazbun TR, Seleem MN. 2020. Potent synergistic interactions between lopinavir and azole antifungal drugs against emerging multidrug-resistant Candida auris. Antimicrob Agents Chemother 65:e00684-20. doi: 10.1128/AAC.00684-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adam RD, Revathi G, Okinda N, Fontaine M, Shah J, Kagotho E, Castanheira M, Pfaller MA, Maina D. 2019. Analysis of Candida auris fungemia at a single facility in Kenya. Int J Infect Dis 85:182–187. doi: 10.1016/j.ijid.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 33.Arendrup MC, Prakash A, Meletiadis J, Sharma C, Chowdhary A. 2017. Comparison of EUCAST and CLSI reference microdilution MICs of eight antifungal compounds for Candida auris and associated tentative epidemiological cutoff values. Antimicrob Agents Chemother 61:e00485-17. doi: 10.1128/AAC.00485-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rex JH, Pappas PG, Karchmer AW, Sobel J, Edwards JE, Hadley S, Brass C, Vazquez JA, Chapman SW, Horowitz HW, Zervos M, McKinsey D, Lee J, Babinchak T, Bradsher RW, Cleary JD, Cohen DM, Danziger L, Goldman M, Goodman J, Hilton E, Hyslop NE, Kett DH, Lutz J, Rubin RH, Scheld WM, Schuster M, Simmons B, Stein DK, Washburn RG, Mautner L, Chu TC, Panzer H, Rosenstein RB, Booth J, National Institute of A, Infectious Diseases Mycoses Study Group. 2003. A randomized and blinded multicenter trial of high-dose fluconazole plus placebo versus fluconazole plus amphotericin B as therapy for candidemia and its consequences in nonneutropenic subjects. Clin Infect Dis 36:1221–1228. doi: 10.1086/374850. [DOI] [PubMed] [Google Scholar]
- 35.Johnson MD, MacDougall C, Ostrosky-Zeichner L, Perfect JR, Rex JH. 2004. Combination antifungal therapy. Antimicrob Agents Chemother 48:693–715. doi: 10.1128/aac.48.3.693-715.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheven M, Scheven C, Hahn K, Senf A. 1995. Post-antibiotic effect and post-expositional polyene antagonism of azole antifungal agents in Candida albicans: dependence on substance lipophilia. Mycoses 38:435–442. doi: 10.1111/j.1439-0507.1995.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 37.Pfaller MA, Diekema DJ, Turnidge JD, Castanheira M, Jones RN. 2019. Twenty years of the SENTRY Antifungal Surveillance Program: results for Candida species from 1997–2016. Open Forum Infect Dis 6:S79–S94. doi: 10.1093/ofid/ofy358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfaller MA, Rhomberg PR, Messer SA, Castanheira M. 2015. In vitro activity of a Hos2 deacetylase inhibitor, MGCD290, in combination with echinocandins against echinocandin-resistant Candida species. Diagn Microbiol Infect Dis 81:259–263. doi: 10.1016/j.diagmicrobio.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 39.Clinical and Laboratory Standards Institute. 2017. M27Ed4; reference method for broth dilution antifungal susceptbility testing of yeasts. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 40.Clinical and Laboratory Standards Institute. 2018. M23Ed5; development of in vitro susceptibility testing criteria and quality control parameters, 5th edition. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 41.Clinical and Laboratory Standards Institute. 2017. M60Ed1; performance standards for antifungal susceptibility testing of yeasts. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 42.Castanheira M, Deshpande LM, Davis AP, Rhomberg PR, Pfaller MA. 2017. Monitoring antifungal resistance in a global collection of invasive yeasts and moulds: application of CLSI epidemiological cutoff values and whole genome sequencing analysis for detection of azole resistance in Candida albicans. Antimicrob Agents Chemother 61:e00906-17. doi: 10.1128/AAC.00906-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prakash A, Sharma C, Singh A, Kumar Singh P, Kumar A, Hagen F, Govender NP, Colombo AL, Meis JF, Chowdhary A. 2016. Evidence of genotypic diversity among Candida auris isolates by multilocus sequence typing, matrix-assisted laser desorption ionization time-of-flight mass spectrometry and amplified fragment length polymorphism. Clin Microbiol Infect 22:277.e1-277.e9. doi: 10.1016/j.cmi.2015.10.022. [DOI] [PubMed] [Google Scholar]