Summary
With the recent approval of highly effective coronavirus disease 2019 (COVID-19) vaccines, functional and lasting immunity to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is currently under investigation as antibody levels in plasma were shown to decline during convalescence. Since the absence of antibodies does not equate to absence of immune memory, we evaluate the presence of SARS-CoV-2-specific memory B cells in convalescent individuals. Here, we report a longitudinal assessment of humoral immune responses on 32 donors up to 8 months post-symptom onset. Our observations indicate that anti-Spike and anti-receptor binding domain (RBD) immunoglobulin M (IgM) in plasma decay rapidly, whereas the reduction of IgG is less prominent. Neutralizing activity also declines rapidly when compared to Fc-effector functions. Concomitantly, the frequencies of RBD-specific IgM+ B cells wane significantly when compared to RBD-specific IgG+ B cells, which remain stable. Our results add to the current understanding of immune memory following SARS-CoV-2 infection, which is critical for secondary infection prevention and vaccine efficacy.
Keywords: coronavirus, COVID-19, SARS-CoV-2, Spike glycoproteins, RBD, antibodies, memory B cells, humoral responses, ADCC, neutralization
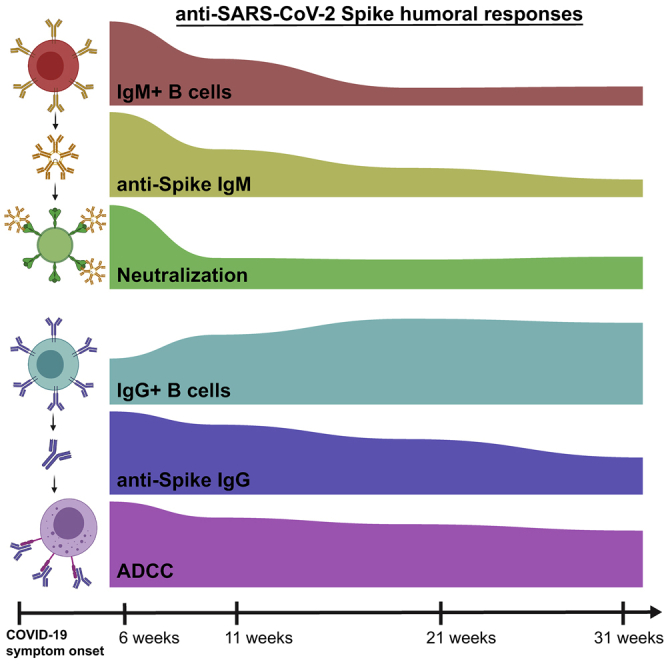
Graphical abstract
Highlights
Spike-specific IgM and IgA wane more rapidly than IgG after recovery
Fc-effector functions, but not neutralization, are sustained over time
SARS-CoV-2-specific B cell immunity persists despite overall antibody decline
In a cohort of SARS-CoV-2-recovered individuals, Anand et al. observe a concomitant decrease of IgM and neutralization although IgG and Fc-effector functions remain relatively stable. The presence and persistence of antigen-specific memory B cells in all individuals are encouraging with regards to long-term immunity.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the ongoing coronavirus disease 2019 (COVID-19) pandemic, is highly contagious and has infected close to a 100 million people worldwide and caused over 2 million deaths since its discovery. The dynamics and persistence of immune responses in individuals infected with SARS-CoV-2 is currently under needful investigation. Several studies with acute and convalescent COVID-19 patients have showed prompt induction of B and T cell responses upon infection, along with the detection of antigen-specific memory B and T cell responses several weeks into convalescence.1, 2, 3, 4, 5, 6 Additionally, antibodies (Abs) induced upon infection have been shown to protect from SARS-CoV-2 reinfection in animal models.7, 8, 9 Passive immunization using neutralizing monoclonal antibody treatments decreased viral loads in animal studies and in patients with COVID-19.10,11 The viral target of neutralizing antibodies is the highly immunogenic trimeric Spike (S) glycoprotein, which facilitates SARS-CoV-2 entry into host cells via its receptor-binding domain (RBD) that interacts with angiotensin-converting enzyme 2 (ACE-2).12,13
The evolution of overall antibody responses in convalescent individuals is being extensively analyzed, with studies showing that Ab titers and neutralization activity against Spike start decreasing during the first weeks after resolution of infection.3,6,14, 15, 16, 17, 18, 19, 20 Importantly, in addition to neutralizing viral particles, the antiviral activities of SARS-CoV-2-specific antibodies can expand to involve Fc-effector functions, including antibody-dependent cellular cytotoxicity (ADCC).21,22 Recent research has highlighted the importance of humoral immunity development and the ability of antibodies to carry out Fc-effector functions in decreasing mortality of patients exhibiting severe disease symptoms.23
In this study, we dissect multiple aspects of humoral immunity, including Fc-effector functions and antigen-specific B cells, longitudinally for up to 8 months post-symptom onset (PSO) in 32 convalescent individuals. Our findings aid in the understanding of durability of COVID-19 immunity, which is important in the context of secondary infections, vaccine efficacy, and herd immunity.
Results
SARS-CoV-2 RBD-specific and Spike-specific antibody levels in convalescent plasma decrease up to 8 months PSO
To monitor the evolution of antibody responses longitudinally, we analyzed serological samples from 32 convalescent individuals (Table 1) along with 10 pre-pandemic samples from uninfected individuals as experimental controls. The average age of the donors was 47 years old (range: 20–65 years), and samples were from 17 males and 15 females. Convalescent patients were sampled at four longitudinal time points between 16 and 233 days PSO: 6 weeks (16–95 days; median: 43 days); 11 weeks (48–127 days; median: 77 days); 21 weeks (116–171 days; median: 145 days); and 31 weeks (201–233 days; median: 218 days). Participants were tested positive for SARS-CoV-2 infection by reverse transcription PCR (RT-PCR) on nasopharyngeal swab specimens, had mild to moderate disease symptoms, and none of them were hospitalized. Convalescent participants were enrolled following two negative RT-PCR tests and a complete resolution of symptoms for at least 14 days before blood sampling.
Table 1.
Longitudinal SARS-CoV-2 convalescent cohort
| Group | N | Days after onset of symptoms (median; day range) | Age (median; age range) | Gender |
|
|---|---|---|---|---|---|
| Male (n) | Female (n) | ||||
| 6 weeks | 32 | 43 (16–95) | 47 (20–65) | 17 | 15 |
| 11 weeks | 28 | 77 (48–127) | 47 (20–65) | 16 | 12 |
| 21 weeks | 28 | 145 (116–171) | 48 (20–65) | 16 | 12 |
| 31 weeks | 13 | 218 (201–233) | 46 (20–65) | 9 | 4 |
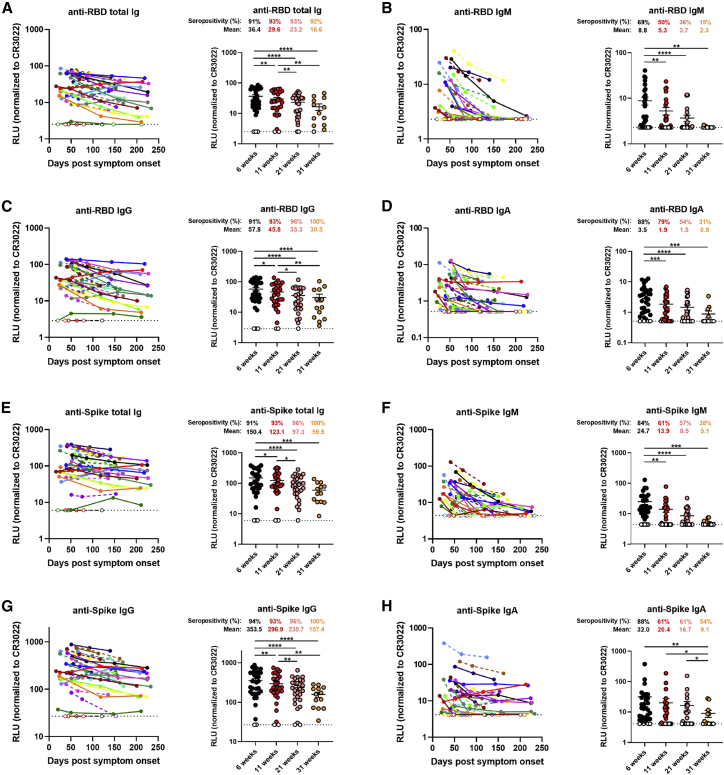
We began by evaluating the presence of RBD-specific antibodies by using a previously published enzyme-linked immunosorbent assay (ELISA) against the SARS-CoV-2 RBD antigen.16 In agreement with recent reports showing the waning of antibody levels in longitudinal convalescent plasma over time,14, 15, 16, 17 we observed that total RBD-specific immunoglobulin (Ig) levels, comprising IgG, IgM, and IgA, gradually decreased between 6 and 31 weeks after the onset of symptoms (Figure 1A). However, the percentage of convalescent individuals presenting detectable RBD-specific Ig levels remained stable, with a consistent seropositivity rate above 90% throughout the sampling time frame. Notably, 100% of the donors still had detectable IgG at the last time point, while IgM and IgA diminished more rapidly, with 85% and 69% of the donors having undetectable IgM and IgA levels, respectively, 31 weeks PSO (Figures 1B–1D and S1A). Because the RBD ELISA is limited to detect antibodies targeting only one domain of the Spike, we developed a high-throughput cell-based ELISA methodology to screen for antibodies recognizing the native full-length S protein on the cell surface. Human osteosarcoma (HOS) cells stably expressing the SARS-CoV-2 S glycoproteins were incubated with plasma samples, followed by the addition of secondary antibodies recognizing IgG, IgM, and/or IgA. We observed that 100% of the donors still had detectable S-specific total Ig and IgG in their plasma at 31 weeks PSO, whereas only 38% and 54% of the plasma samples tested positive for the presence of Spike-specific IgM and IgA, respectively (Figures 1E–1H and S1B). We confirmed this observation using a recently characterized flow-cytometry-based assay24 determining antibody binding to the full-length S protein on the surface of 293T cells (Figure S2A). The data obtained with both the cell-based ELISA and flow-cytometry techniques correlated significantly (r = 0.8120; p < 0.0001; Figure S2B).
Figure 1.
Decline of RBD- and Spike-specific antibodies in longitudinal convalescent plasma
(A–D) Indirect ELISA was performed using recombinant SARS-CoV-2 RBD protein and incubation with COVID-19+ plasma samples recovered between 6 and 31 weeks post-symptom onset. Anti-RBD antibody binding was detected using horseradish peroxidase (HRP)-conjugated (A) anti-human IgM+IgG+IgA, (B) anti-human IgM, (C) anti-human IgG, or (D) anti-human IgA. Relative light unit (RLU) values obtained with BSA (negative control) were subtracted and further normalized to the signal obtained with the anti-RBD CR3022 monoclonal antibody (mAb) present in each plate.
(E–H) Cell-based ELISA was performed using HOS cells expressing full-length SARS-CoV-2 Spike and incubation with COVID-19+ plasma samples recovered between 6 and 31 weeks post-symptom onset. Anti-Spike antibody binding was detected using HRP-conjugated (E) anti-human IgM+IgG+IgA, (F) anti-human IgM, (G) anti-human IgG, or (H) anti-human IgA. RLU values obtained with parental HOS (negative control) were subtracted and further normalized to the signal obtained with the CR3022 mAb present in each plate.
(Left panels) Each curve represents the normalized RLUs obtained with the plasma of one donor at every donation as a function of the days after symptom onset. (Right panels) Plasma samples were grouped in different time points post-symptom onset (6, 11, 21, and 31 weeks). Undetectable measures are represented as white symbols, and limits of detection are plotted. Error bars indicate means ± SEM. Statistical significance was tested using repeated-measures one-way ANOVA with a Holm-Sidak post-test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001).
Neutralizing and Fc-effector activities of antibodies present in convalescent plasma decrease at different rates over time
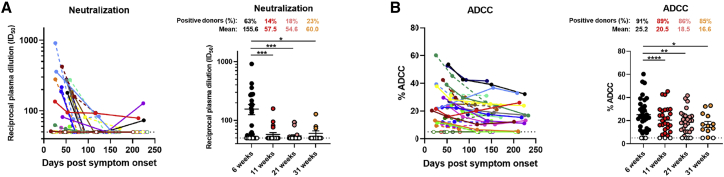
Recent studies have shown the importance of neutralizing antibodies in reducing viral load and preventing infection in animal models.10,25,26 Neutralizing monoclonal antibody cocktails also reduced viral load in COVID-19 patients.11 Neutralizing activity is often considered as a determining factor in convalescent plasma therapy, although its relative importance compared to Fc-effector activity is still unknown.27, 28, 29 Thus, we measured the capacity of convalescent plasma to neutralize pseudoviral particles carrying the SARS-CoV-2 Spike protein over time. Neutralizing antibody titers (inhibitory dilution [ID50]) were detected in 63% of the donors at 6 weeks PSO, while none of the uninfected controls had detectable neutralizing activity. Titers declined from 155.6 at 6 weeks to 60.0 at 31 weeks PSO, respectively, with 77% of donors having undetectable neutralization activity in their plasma at the last time point (Figure 2A). Of note, plasma from individuals that had undetectable neutralization activity at later time points started with lower peak titers at earlier time points. Because the depletion of IgM from plasma has been associated with loss of viral neutralization capacity30,31 and IgA has also been shown to dominate the early neutralizing response,32,33 the sharp decline in neutralization activity seen in this study is corroborated by the striking decrease in anti-Spike and anti-RBD IgM and IgA levels (Figures 1B, 1D, 1F, 1H, S1A, S1B, and S5).
Figure 2.
Neutralization and Fc-effector function activities in convalescent plasma decrease over time
(A) Pseudoviral particles coding for the luciferase reporter gene and bearing the SARS-CoV-2 S glycoproteins were used to infect 293T-ACE2 cells. Neutralizing activity was measured by incubating pseudoviruses with serial dilutions of COVID-19+ plasma samples recovered between 6 and 31 weeks post-symptom onset at 37°C for 1 h prior to infection of 293T-ACE2 cells. Neutralization half-maximal inhibitory serum dilution (ID50) values were determined using a normalized non-linear regression using GraphPad Prism software.
(B) CEM.NKr parental cells were mixed at a 1:1 ratio with CEM.NKr-Spike cells and were used as target cells. PBMCs from uninfected donors were used as effector cells in a fluorescence-activated cell sorting (FACS)-based ADCC assay. The graphs shown represent the percentages of ADCC obtained in the presence of COVID-19+ plasma samples recovered between 6 and 31 weeks post-symptom onset.
(Left panels) Each curve represents (A) the neutralization ID50 or (B) the percentages of ADCC obtained with the plasma of one donor at every donation as a function of the days after symptom onset. (Right panels) Plasma samples were grouped in different time points post-symptom onset (6, 11, 21, and 31 weeks). Undetectable measures are represented as white symbols, and limits of detection are plotted. Error bars indicate means ± SEM. Statistical significance was tested using repeated-measures one-way ANOVA with a Holm-Sidak post-test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001).
Fc-mediated effector functions of antibodies can contribute to the efficacy of immune response against SARS-CoV-2. Recent studies have examined Fc-mediated effector functions of antibodies elicited upon SARS-CoV-2 infection.21,34,35 Both the presence of IgG and their Fc-mediated effector activities have been linked to reduced severity of disease.23 Herein, we assessed the ability of plasma from convalescent donors to trigger ADCC responses over time. We developed an ADCC assay using a human T-lymphoid cell line resistant to NK-cell-mediated cell lysis (CEM.NKr) and stably expressing the full-length S protein on the cell surface as target cells. Peripheral blood mononuclear cells (PBMCs) from healthy individuals were used as effector cells. ADCC activity was measured by the loss of Spike-expressing GFP+ target cells (Figure S3). We observed that ADCC activity of convalescent plasma decreased gradually between 6 weeks and 31 weeks PSO (Figure 2B). However, this decline was modest when compared to the decrease in neutralization activity of plasma (Figure S1C), and 85% of the donors’ plasma still elicited substantial ADCC activity at the latest study time point. As expected, the levels of ADCC activity strongly correlated with the levels of anti-Spike IgG (Figure S5). The presence of Fc-mediated antibody effector functions up to 8 months PSO is corroborated with the presence of significant IgG levels.
RBD-specific memory B cells develop and remain stable up to 8 months PSO
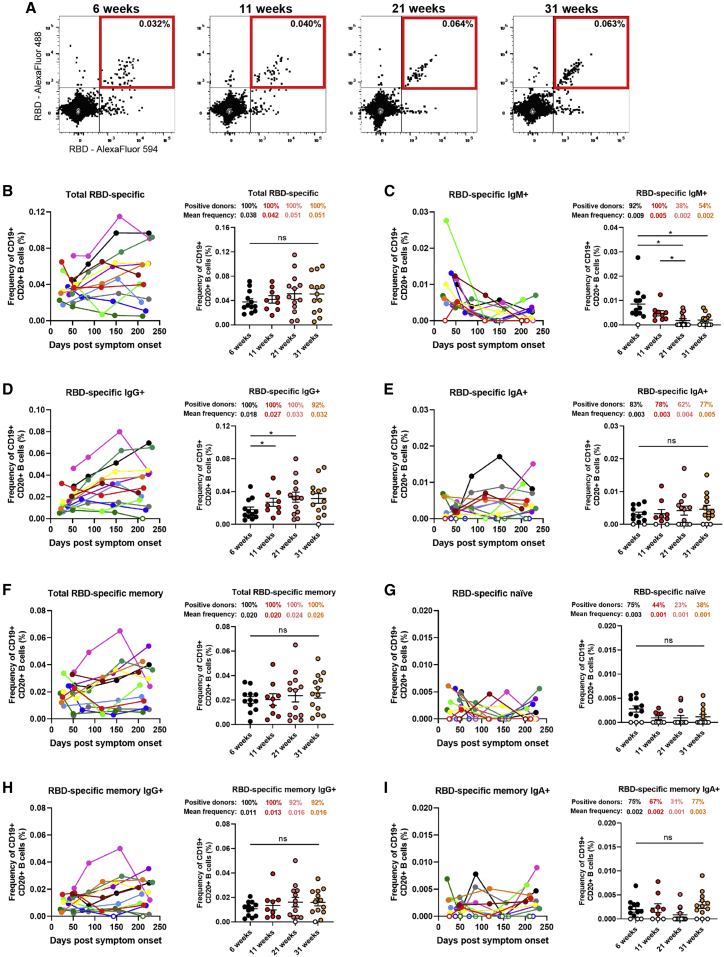
Recent studies on convalescent COVID-19 patients have indicated persistent antigen-specific memory B cell responses despite waning antibody levels.3,6 To monitor the circulating B cell compartment in our cohort of convalescent individuals, antigen-specific B cells were characterized by flow cytometry (identified as CD19+ CD20+). We focused on RBD-specific B cell responses to avoid selecting for pre-existing memory B cells cross-reactive with endemic coronaviruses.36 To identify distinct RBD-specific B cells, we used double discrimination with two recombinant RBD protein preparations labeled with fluorochromes Alexa Fluor 594 and Alexa Fluor 488, respectively. Detection of this double-positive population was specific because it was not detected in PBMCs from uninfected individuals (Figures S4A and S4B). RBD-specific B cells were detected 6 weeks PSO with a modest increase in mean frequency up to 31 weeks PSO (0.038%–0.051%; Figure 3B). Total RBD-specific B cells were evaluated for their differential surface Ig isotype expression, and we observed IgG+, IgM+, and IgA+ cells in 100%, 92%, and 83% of donors at 6 weeks PSO, respectively. Strikingly, the frequency of RBD-specific IgM+ B cells decreased significantly, with 46% of the donors having undetectable IgM+ B cells 31 weeks PSO (Figures 3C and S1D). Conversely, the frequency of RBD-specific IgG+ B cells significantly increased between 6 and 21 weeks PSO and remained stable up to 8 months PSO (Figure 3D); the detection of RBD-specific IgA+ B cells also persisted in 77% of the donors tested (Figures 3E and S1D). Furthermore, the total RBD-specific B cells were evaluated to distinguish memory and naive B cells (identified by using CD27 and CD21 markers; Figures S4C and S4D). Importantly, total RBD-specific memory B cells were detected in 100% of the donors and the mean frequency remained stable between 6 and 31 weeks PSO (0.020%–0.026%), while RBD-specific naive B cells were observed in lower proportions and modestly decreased over time (Figures 3F and 3G). Interestingly, the proportion of donors positive for RBD-specific naive B cells was reduced by half between the first and last time point, as also observed for RBD-specific IgM+ B cells, which is consistent with the decline in IgM levels observed between these groups. IgG+ RBD-specific memory B cells were detected in 100% of the donors, and the frequency of this population modestly increased up to 31 weeks PSO (Figure 3H). Meanwhile, the frequencies of IgA+ RBD-specific memory B cells were low but stable over the 8-month period (Figures 3G and 3I).
Figure 3.
RBD-specific memory B cells develop and persist up to 8 months post-symptom onset
(A) Flow cytometry plots of staining with fluorescent SARS-CoV-2 RBD probes on CD19+ CD20+ HLA-DR+ B cells. Samples from one representative convalescent donor are shown for 4 different time points post-symptom onset (6, 11, 21, and 31 weeks). All percentages shown represent the frequency of RBD-specific B cells on the total CD19+ CD20+ B cell population.
(B–G) Characterization of RBD-specific B cells was performed on longitudinal PBMC samples obtained from COVID-19+ convalescent individuals between 6 and 31 weeks post-symptom onset.
(B–E) Total (B) RBD-specific B cells were segregated by subsets based on cell surface expression of (C) IgM, (D) IgG, or (E) IgA BCR isotypes.
(F–I) Frequency of (F) total memory, (G) naive, (H) IgG+ memory, and (I) IgA+ memory B cells was determined based on CD21 and CD27 expression.
(Left panels) Each curve represents the frequency of a B cell subset on the total B cell population obtained with PBMCs from one donor at every donation as a function of the days after symptom onset. (Right panels) PBMC samples were grouped in different time points post-symptom onset (6, 11, 21, and 31 weeks). Error bars indicate means ± SEM. Statistical significance was tested using repeated-measures one-way ANOVA with a Holm-Sidak post-test (∗p < 0.05; ns, nonsignificant)
Evaluation of the relationship between different aspects of humoral immunity reveals the importance of IgG responses
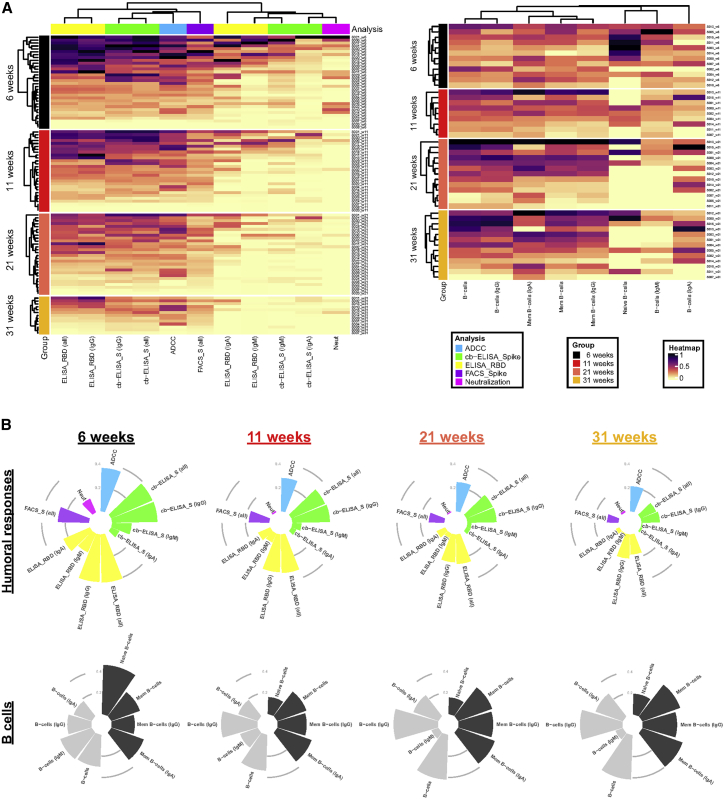
To examine interrelations between serological, immunological, and functional parameters assessed in this study, we performed comprehensive sets of correlation analyses. First, at each time point, the strongest immune response clusters were composed of total Ig and IgG binding levels and, thus, increased ADCC activity (Figure 4A, left panel). In contrast, weaker immune response clusters in plasma were formed by IgA and IgM levels and, subsequently, neutralization capacities. Overall, the responses decreased over the course of the 8-month period (Figure 4B). The inverse was observed with antigen-specific B cell frequencies (Figure 4B). Apart from naive B cells and the IgM fraction thereof, total RBD-specific B cells increased over the course of 8 months. At each time point, total IgG+ B cells and memory B cells (IgA+ and IgG+) constituted the most prominent responses (Figure 4A, right panel).
Figure 4.
Longitudinal patterns of B cell levels and humoral immune responses
(A) (Left panel) Heatmap of humoral immune responses normalized per parameter. Columns represent immune response parameters clustered based on similarity and grouped according to the provided color code. Rows represent IDs grouped according to study time point. IDs are clustered according to their immune response profiles within each time point. (Right panel) Heatmap of B cell levels with similar display as in left panel is shown.
(B) Circular bar plots represent averaged values of parameters at each time point.
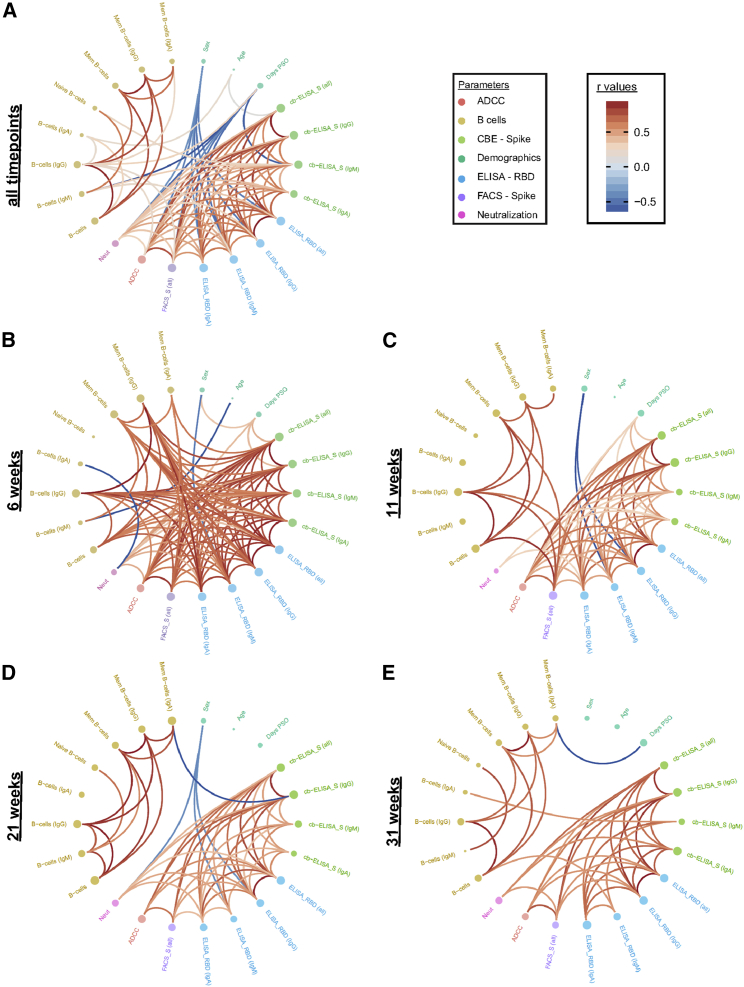
Additionally, at the earliest time point convalescent plasma was collected after symptom onset (6 weeks PSO), we observed a vast network of strong positive correlations among B cell populations, antibody levels, and antibody-mediated functional parameters (Figure 5B). Intriguingly, this was followed by a striking disconnect of associations between B cell frequencies and serological parameters at 11, 21, and 31 weeks PSO (Figures 5C–5E). This is caused by the concomitant diminution of antibody levels yet stabilization of antigen-specific B cells observed at these time points, suggestive of decreased antibody production by B cells after resolution of infection or the gradual replacement of Ig-secreting, short-lived plasma cell by memory B cells. When considering the collective datasets, prominent features were two subsequently dividing positive correlation clusters, the first among total and IgG+ RBD-specific B cells and the second among ADCC with total Ig and IgG binding responses. Furthermore, days PSO inversely correlated with neutralization as well as IgM- and IgA-specific responses (Figures 5A and S5).
Figure 5.
Longitudinal plasticity and separation of B cell and humoral correlation clusters
Edge bundling correlation plots where red and blue edges represent positive and negative correlations between connected parameters, respectively. Only significant correlations (p < 0.05) are displayed. Nodes are color coded based on the grouping of parameters according to the legend at the bottom. Node size corresponds to the degree of relatedness of correlations. Edge bundling plots are shown for correlation analyses using all data points (A) and datasets of individual time points, i.e., at 6 weeks (B), 11 weeks (C), 21 weeks (D), and 31 weeks (E).
Discussion
A better understanding of the type and longevity of immune responses following viral infections is critical to reveal immune mechanisms involved in protection from reinfection and protection by vaccination. Data regarding these important issues continue to be gathered for SARS-CoV-2. In this study, we have contributed to the current understanding by reporting on the evolution of the overall humoral immune responses in 101 blood samples obtained from 32 COVID-19 convalescent patients between 16 and 233 days PSO. Overall, we observed that IgM levels and the neutralizing capacity of plasma decreases rapidly, whereas IgG and Fc-effector activity are more sustained. Since disease severity has been shown to correlate with Ig titers and neutralization activity of plasma,37 the rapid decline of neutralizing activity in our cohort could be linked to the mild severity of symptoms observed by our donors. The antibody kinetics we observed herein are typical of those seen for other human coronavirus infections, with antibodies peaking 2–4 weeks PSO followed by a contraction phase.38 Furthermore, we show that COVID-19 patients generate RBD-specific memory B cells and IgG+ B memory cells that persist for over 8 months. This is similar to recent studies on the durability of SARS-CoV-2 immune responses showing that, despite decreases in antibody levels, Spike-specific IgG+ memory B cells are generated and maintained likely due to antigen persistence in convalescent individuals.3,5,6 Additionally, studies demonstrated enhanced cellular immunity that protects non-human primates from SARS-CoV-2 reinfection in the context of waning neutralizing antibodies.8 Thus, the decline of antibody levels does not negate the protective potential because of the importance of cellular responses against SARS-CoV-2 infection. This understanding is corroborated with a recent risk assessment carried out in a cohort of 43,000 convalescent individuals demonstrating that immunity elicited upon natural SARS-CoV-2 infection protects against reinfection with an efficacy of >90% for at least 7 months.39
Fc-mediated effector activity of antibodies has been recently shown to correlate with reduced disease severity and mortality.23 Antibodies capable of mediating Fc-dependent functions, such as antibody-dependent phagocytosis and ADCC, have been isolated from convalescent donors.40 Importantly, Fc-mediated effector activity was shown to contribute to the protection from SARS-CoV-2 infection in adapted mice and hamster models.9,21,34 Thus, our observations of the persistence of ADCC responses in convalescent plasma up to 8 months PSO suggest that Fc-mediated effector functions could play a vital role in protection from reinfection. Nevertheless, the heterogeneity of immune responses between individuals is and will be an important factor when evaluating the efficacy of immune responses upon re-exposure to SARS-CoV-2.
Recent studies comparing humoral immunity generated by mRNA vaccinees (mRNA-1273 or BNT162b2) and individuals recovered from natural infection observed similarities in antibody binding titers and plasma neutralization capacity.41 Furthermore, similar frequencies of RBD-specific memory B cells between vaccinees and infected individuals were also detected. Therefore, our results on the persistence of RBD-specific memory B cells up to 8 months after natural infection is reassuring with regards to long-term vaccine efficacy.
Limitations of study
A limitation of the current study is the relatively low number of individuals studied, which can affect the power of our statistical analyses. Longitudinal data from a larger sample size with varying clinical symptoms ranging from asymptomatic to critical cases as well as individuals below 20 years and over 65 years of age would be required for a more precise understanding of the overall immune durability against COVID-19. However, we note that results as obtained in this study are very similar to those observed in larger cross-sectional and longitudinal studies.3,5,6
Furthermore, although we successfully used our ADCC assay to show the importance of Fc-mediated effector functions of neutralizing Abs to protect from lethal SARS-CoV-2 challenges in K18-hACE2 transgenic mice42 and as an immune correlate for survival in acutely infected individuals,43 our assay uses cells constitutively expressing the Spike proteins and not bona fide SARS-CoV-2-infected cells.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Cross-reactive SARS-CoV-1 monoclonal antibody CR3022 | ter Meulen et al.44 | RRID: AB_2848080 |
| Alexa Fluor 647 AffiniPure Goat Anti-Human IgA + IgG + IgM (H+L) |
Jackson ImmunoResearch | Cat# 109-605-064; RRID: AB_2337886 |
| Goat anti-Human IgG Fc Cross-Adsorbed Secondary Antibody, HRP |
Invitrogen | Cat# A18823; RRID: AB_2535600 |
| Peroxidase AffiniPure Goat Anti-Human IgM, Fc5μ fragment specific | Jackson ImmunoResearch | Cat# 109-035-129; RRID: AB_2337588 |
| Peroxidase AffiniPure Goat Anti-Human Serum IgA, α chain specific | Jackson ImmunoResearch | Cat# 109-035-011; RRID: AB_2337580 |
| Peroxidase AffiniPure Goat Anti-Human IgA + IgG + IgM (H+L) |
Jackson ImmunoResearch | Cat# 109-035-064; RRID: AB_2337583 |
| BUV805 Mouse Anti-Human CD24 (Clone ML5) | BD Biosciences | Cat# 742010; RRID: AB_2871308 |
| BV421 Mouse Anti-Human IgG (Clone G18-145) | BD Biosciences | Cat# 562581; RRID: AB_2737665 |
| BUV737 Mouse Anti-Human IgM (Clone UCH-B1) | BD Biosciences | Cat# 748928; RRID: AB_2873331 |
| BV480 Mouse Anti-Human CD3 (Clone UCHT1) | BD Biosciences | Cat# 566105, RRID: AB_2739507 |
| BV480 Mouse Anti-Human CD56 (Clone NCAM16.2) | BD Biosciences | Cat# 566124; RRID: AB_2739525 |
| BV480 Mouse Anti-Human CD14 (Clone M5E2) | BD Biosciences | Cat# 746304; RRID: AB_2743629 |
| BV480 Mouse Anti-Human CD16 (Clone 3G8) | BD Biosciences | Cat# 566171; RRID: AB_2739510 |
| BV711 Mouse Anti-Human CD20 (Clone 2H7) | BD Biosciences | Cat# 563126; RRID: AB_2313579 |
| BV786 Mouse Anti-Human CD21 (Clone B-ly4) | BD Biosciences | Cat# 740969; RRID: AB_2740594 |
| BB700 Mouse Anti-Human HLA-DR (Clone G46-6) | BD Biosciences | Cat# 566480; RRID: AB_2744477 |
| APC-R700 Mouse Anti-Human CD27 (Clone M-T271) | BD Biosciences | Cat# 565116; RRID: AB_2739074 |
| APC anti-human CD19 (Clone SJ25C1) | BioLegend | Cat# 363006; RRID: AB_2564255 |
| PE Mouse Anti-Human IgA (Clone IS11-8E10) | Miltenyi Biotec | Cat# 130-113-476; RRID: AB_2733861 |
| Biological samples | ||
| Human Plasma from SARS-CoV-2-infected or uninfected donors | This paper | N/A |
| Peripheral blood mononuclear cells (PBMCs) from SARS-CoV-2 infected donors or uninfected donors | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco’s modified Eagle’s medium (DMEM) | Wisent | Cat# 319-005-CL |
| Roswell Park Memorial Institute 1640 medium (RPMI) | GIBCO Life Technologies | Cat# 11875093 |
| Penicillin/streptomycin | Wisent | Cat# 450-201-EL |
| Penicillin/streptomycin | GIBCO Life Technologies | Cat# 15140122 |
| Fetal bovine serum (FBS) | VWR | Cat# 97068-085 |
| Fetal bovine serum (FBS) | Seradigm | Cat# 1500-500 |
| Tris-buffered saline (TBS) | Thermo Fisher Scientific | Cat# BP24711 |
| Bovine Serum Albumin (BSA) | BioShop | Cat# ALB001.100 |
| Phosphate buffered saline (PBS) | Wisent | Cat# 311-010-CL |
| Dulbecco’s phosphate-buffered saline (DPBS) | GIBCO Life Technologies | Cat# 14190144 |
| Western Lightning Plus-ECL, Enhanced Chemiluminescence Substrate | Perkin Elmer Life Sciences | Cat# NEL105001EA |
| Tween20 | Thermo Fisher Scientific | Cat# BP337-500 |
| Puromycin dihydrochloride | Millipore Sigma | Cat# P8833 |
| Passive lysis buffer | Promega | Cat# E1941 |
| FreeStyle 293F expression medium | ThermoFisher Scientific | Cat# 12338002 |
| ExpiFectamine 293 transfection reagent | ThermoFisher Scientific | Cat# A14525 |
| Ni-NTA agarose | Invitrogen | Cat# R90110 |
| D-Luciferin potassium salt | Thermo Fisher Scientific | Cat# L2916 |
| LIVE/DEAD Fixable AquaVivid Cell Stain | Thermo Fisher Scientific | Cat# L34957 |
| Formaldehyde 37% | Thermo Fisher Scientific | Cat# F79-500 |
| HEPES (1 M) | GIBCO Life Technologies | Cat# 15630080 |
| Skim milk (non fat powder) | BioShop | Cat# SKI400.1 |
| eBioscience Cell Proliferation Dye eFluor 670 | ThermoFisher Scientific | Cat# 65-0840-85 |
| eBioscience Cell Proliferation Dye eFluor 450 | ThermoFisher Scientific | Cat# 65-0842-85 |
| Critical commercial assays | ||
| Alexa Fluor 594 Protein Labeling Kit | Invitrogen | Cat# A10239 |
| Alexa Fluor 488 Protein Labeling Kit | Invitrogen | Cat# A10235 |
| Experimental models: Cell lines | ||
| HEK293T human embryonic kidney cells | ATCC | Cat# CRL-3216; RRID: CVCL_0063 |
| FreeStyle 293F | ThermoFisher Scientific | Cat# R79007; RRID: CVCL_D603 |
| 293T-ACE2 | Prévost et al.16 | N/A |
| 293T-Spike | Anand et al.24 | N/A |
| Human osteosarcoma (HOS) | ATCC | Cat# CRL-1543; RRID: CVCL_0312 |
| HOS-Spike | This paper | N/A |
| CEM.NKr CCR5+ | NIH AIDS Reagent Program | Cat # 4376; RRID: CVCL_X623 |
| CEM.NKr-Spike | This paper | N/A |
| Recombinant DNA | ||
| pNL4.3 R-E- Luc | NIH AIDS Reagent Program | Cat# 3418 |
| pCG1-SARS-CoV-2 Spike | Hoffmann et al.13 | N/A |
| Lentiviral packaging plasmids (pLP1, pLP2) | Liu et al.45 | N/A |
| pSVCMV-IN-VSV-G | Lodge et al.46 | N/A |
| pLV-SARS-CoV-2 S C-GFPSpark tag | Sino Biological | Cat# VG40590-ACGLN |
| pcDNA3.1(+)-SARS-CoV-2 RBD | Beaudoin-Bussières et al.14 | N/A |
| Software and algorithms | ||
| FlowJo v10 | Tree Star | https://www.flowjo.com/ |
| GraphPad Prism v9 | GraphPad | https://www.graphpad.com/ |
| R v3 | R | https://www.r-project.org/ |
| RStudio v1 | RStudio | https://www.rstudio.com/ |
| Microsoft Excel v16 | Microsoft Office | https://www.microsoft.com/en-ca/microsoft-365/excel |
| Other | ||
| BD LSR II Flow Cytometer | BD Biosciences | N/A |
| TriStar LB 942 Microplate Reader | Berthold Technologies | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Andrés Finzi (andres.finzi@umontreal.ca).
Materials availability
All unique reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
All data are included in the manuscript. This study did not generate new or custom code.
Experimental model and subject details
Ethics statement
All work was conducted in accordance with the Declaration of Helsinki in terms of informed consent and approval by an appropriate institutional board. Convalescent plasmas were obtained from donors who consented to participate in this research project at CHUM (19.381). The donors met all donor eligibility criteria: previous confirmed COVID-19 infection and complete resolution of symptoms for at least 14 days.
Human subjects
The present study aims to determine the kinetics of the humoral response following SARS-CoV-2 infection. All available samples from adults recovered from SARS-CoV-2 infection collected at the time of the study were analyzed. No specific criteria such as number of patients (sample size), clinical or demographic were used for inclusion, beyond PCR confirmed SARS-CoV-2 infection in adults. A summary of demographic parameters for all individuals is included in Table 1. While all the participants were symptomatic at the time of the infection, none of them were hospitalized. The disease severity ranged from mild to moderate symptoms without any severe or critical cases.
Cell lines
293T human embryonic kidney cells and HOS cells (obtained from ATCC) were maintained at 37°C under 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) (Wisent) containing 5% fetal bovine serum (VWR) and 100 μg/ml penicillin-streptomycin (Wisent). CEM.NKr CCR5+ cells (NIH AIDS Reagent Program) were maintained at 37°C under 5% CO2 in RPMI (GIBCO) containing 10% fetal bovine serum (VWR) and 100 μg/ml penicillin-streptomycin (Wisent). 293T-ACE2 and 293T-SARS-CoV-2 Spike cell lines were previously reported16. For the generation of HOS and CEM.NKr CCR5+ cells stably expressing the SARS-CoV-2 Spike glycoproteins, transgenic lentiviruses were produced in 293T using a third-generation lentiviral vector system. Briefly, 293T cells were co-transfected with two packaging plasmids (pLP1 and pLP245), an envelope plasmid (pSVCMV-IN-VSV-G46) and a lentiviral transfer plasmid coding for a GFP-tagged SARS-CoV-2 Spike (pLV-SARS-CoV-2 S C-GFPSpark tag) (Sino Biological). Supernatant containing lentiviral particles was used to transduce HOS and CEM.NKr CCR5+ cells in presence of 5 μg/mL polybrene. The HOS and CEM.NKr CCR5+ cells stably expressing SARS-CoV-2 Spike (GFP+) were sorted by flow cytometry.
Method details
Plasma and antibodies
Plasma from SARS-CoV-2-infected and pre-pandemic uninfected donors were collected, heat-inactivated for 1h at 56°C and stored at −80°C until ready to use in subsequent experiments. Plasma from uninfected donors were used as negative controls and used to calculate the seropositivity threshold in our ELISA, cell-based ELISA, and flow cytometry assays. The RBD-specific monoclonal antibody CR302244 was used as a positive control in our ELISAs, cell-based ELISAs, and flow cytometry assays and was previously described14,16,24. Horseradish peroxidase (HRP)-conjugated antibodies able to detect all Ig isotypes (anti-human IgM+IgG+IgA; Jackson ImmunoResearch Laboratories, Inc.) or specific for the Fc region of human IgG (Invitrogen), the Fc region of human IgM (Jackson ImmunoResearch Laboratories, inc.) or the Fc region of human IgA (Jackson ImmunoResearch Laboratories, inc) were used as secondary antibodies to detect antibody binding in ELISA and cell-based ELISA experiments. Alexa Fluor-647-conjugated goat anti-human Abs able to detect all Ig isotypes (anti-human IgM+IgG+IgA; Jackson ImmunoResearch Laboratories, Inc.) were used as secondary antibodies to detect plasma binding in flow cytometry experiments.
Protein expression and purification
FreeStyle 293F cells (Thermo Fisher Scientific) were grown in FreeStyle 293F medium (Thermo Fisher Scientific) to a density of 1 × 106 cells/mL at 37°C with 8% CO2 with regular agitation (150 rpm). Cells were transfected with a plasmid coding for SARS-CoV-2 S RBD using ExpiFectamine 293 transfection reagent, as directed by the manufacturer (Thermo Fisher Scientific). One week later, cells were pelleted and discarded. Supernatants were filtered using a 0.22 μm filter (Thermo Fisher Scientific). The recombinant RBD proteins were purified by nickel affinity columns, as directed by the manufacturer (Invitrogen). The RBD preparations were dialyzed against phosphate-buffered saline (PBS) and stored in aliquots at −80°C until further use. To assess purity, recombinant proteins were loaded on SDS-PAGE gels and stained with Coomassie Blue.
Enzyme-Linked Immunosorbent Assay (ELISA)
The SARS-CoV-2 RBD ELISA assay used was recently described16. Briefly, recombinant SARS-CoV-2 S RBD proteins (2.5 μg/ml), or bovine serum albumin (BSA) (2.5 μg/ml) as a negative control, were prepared in PBS and adsorbed to plates (MaxiSorp Nunc) overnight at 4°C. Coated wells were subsequently blocked with blocking buffer (Tris-buffered saline [TBS] containing 0.1% Tween20 and 2% BSA) for 1h at room temperature. Wells were then washed four times with washing buffer (Tris-buffered saline [TBS] containing 0.1% Tween20). CR3022 mAb (50ng/ml) or a 1/250 dilution of plasma from SARS-CoV-2-infected or uninfected donors were prepared in a diluted solution of blocking buffer (0.1% BSA) and incubated with the RBD-coated wells for 90 minutes at room temperature. Plates were washed four times with washing buffer followed by incubation with the respective secondary Abs (diluted in blocking buffer, 0.4% BSA) for 1h at room temperature. The binding of CR3022 IgG was quantified with HRP-conjugated antibodies specific for the Fc region of human IgG. This was followed by four washes. HRP enzyme activity was determined after the addition of a 1:1 mix of Western Lightning oxidizing and luminol reagents (Perkin Elmer Life Sciences). Light emission was measured with a LB942 TriStar luminometer (Berthold Technologies). Signal obtained with BSA was subtracted for each plasma and was then normalized to the signal obtained with CR3022 mAb present in each plate. The seropositivity threshold was established using the following formula: mean of all COVID-19 negative plasma + (3 standard deviation of the mean of all COVID-19 negative plasma).
Cell-Based ELISA
Detection of the trimeric SARS-CoV-2 Spike at the surface of HOS cells was performed by cell-based enzyme-linked immunosorbent assay (ELISA). Briefly, parental HOS cells or HOS-Spike cells were seeded in 384-well plates (2.8 × 104 cells per well) overnight. Cells were blocked with blocking buffer (washing buffer [1.8 mM CaCl2, 1 mM MgCl2, 25 mM Tris (pH 7.5), and 140 mM NaCl] supplemented with 10mg/mL non-fat dry milk and 5mM Tris [pH 8.0]) for 30min. CR3022 mAb (1 μg/ml) or plasma from SARS-CoV-2-infected or uninfected donors (at a dilution of 1/250) were prepared in blocking buffer and incubated with the cells for 1h at room temperature. Respective HRP-conjugated secondary antibodies were then incubated with the samples for 45 min at room temperature. The binding of CR3022 IgG was quantified with HRP-conjugated antibodies specific for the Fc region of human IgG. For all conditions, cells were washed 6 times with blocking buffer and 6 times with washing buffer. HRP enzyme activity was determined after the addition of a 1:1 mix of Western Lightning oxidizing and luminol reagents (PerkinElmer Life Sciences). Light emission was measured with an LB 942 TriStar luminometer (Berthold Technologies). Signal obtained with parental HOS was subtracted for each plasma and was then normalized to the signal obtained with CR3022 mAb present in each plate. The seropositivity threshold was established using the following formula: mean of all COVID-19 negative plasma + (3 standard deviation of the mean of all COVID-19 negative plasma).
Cell surface staining and flow cytometry analysis
293T and 293T-Spike cells were mixed at a 1:1 ratio and stained with the anti-RBD CR3022 monoclonal Ab (5 μg/ml) or plasma (1:250 dilution). AlexaFluor-647-conjugated goat anti-human IgM+IgG+IgA Abs (1:800 dilution) were used as secondary antibodies. The percentage of transduced cells (GFP+ cells) was determined by gating the living cell population based on viability dye staining (Aqua Vivid, Invitrogen). Samples were acquired on a LSRII cytometer (BD Biosciences) and data analysis was performed using FlowJo v10.7.1 (Tree Star). The seropositivity threshold was established using the following formula: mean of all COVID-19 negative plasma + (3 standard deviation of the mean of all COVID-19 negative plasma).
ADCC assay
For evaluation of anti-SARS-CoV-2 antibody-dependent cellular cytotoxicity (ADCC), parental CEM.NKr CCR5+ cells were mixed at a 1:1 ratio with CEM.NKr. Spike cells. These cells were stained for viability (AquaVivid; Thermo Fisher Scientific, Waltham, MA, USA) and cellular dyes (cell proliferation dye eFluor670; Thermo Fisher Scientific) and subsequently used as target cells. Overnight rested PBMCs were stained with another cellular marker (cell proliferation dye eFluor450; Thermo Fisher Scientific) and used as effector cells. Stained target and effector cells were mixed at a ratio of 1:10 in 96-well V-bottom plates. Plasma from COVID+ or COVID- individuals (1/500 dilution) or monoclonal antibody CR3022 (1 μg/mL) were added to the appropriate wells. The plates were subsequently centrifuged for 1 min at 300xg, and incubated at 37°C, 5% CO2 for 5 hours before being fixed in a 2% PBS-formaldehyde solution. ADCC activity was calculated using the formula: [(% of GFP+ cells in Targets plus Effectors)-(% of GFP+ cells in Targets plus Effectors plus plasma/antibody)]/(% of GFP+ cells in Targets) x 100 by gating on transduced live target cells. All samples were acquired on an LSRII cytometer (BD Biosciences) and data analysis performed using FlowJo v10.7.1 (Tree Star). The specificity threshold was established using the following formula: mean of all COVID-19 negative plasma + (3 standard deviation of the mean of all COVID-19 negative plasma)
Virus neutralization assay
293T-ACE2 target cells were infected with single-round luciferase-expressing SARS-CoV-2 pseudoparticles in presence of convalescent plasma. Briefly, 293T cells were transfected by the calcium phosphate method with the lentiviral vector pNL4.3 R-E- Luc (NIH AIDS Reagent Program) and a plasmid encoding for SARS-CoV-2 Spike at a ratio of 5:4. Two days post-transfection, cell supernatants were harvested and stored at −80°C until use. 293T-ACE2 target cells were seeded at a density of 1 × 104 cells/well in 96-well luminometer-compatible tissue culture plates (Perkin Elmer) 24h before infection. Recombinant viruses in a final volume of 100 μL were incubated with the indicated plasma dilutions (1/50; 1/250; 1/1250; 1/6250; 1/31250) for 1h at 37°C and were then added to the target cells followed by incubation for 48h at 37°C; cells were lysed by the addition of 30 μL of passive lysis buffer (Promega) followed by one freeze-thaw cycle. An LB942 TriStar luminometer (Berthold Technologies) was used to measure the luciferase activity of each well after the addition of 100 μL of luciferin buffer (15mM MgSO4, 15mM KPO4 [pH 7.8], 1mM ATP, and 1mM dithiothreitol) and 50 μL of 1mM d-luciferin potassium salt (Thermo Fisher Scientific). The neutralization half-maximal inhibitory dilution (ID50) represents the plasma dilution to inhibit 50% of the infection of 293T-ACE2 cells by SARS-CoV-2 pseudoviruses.
Detection of antigen-specific B cells
To detect SARS-CoV-2-specific B cells, we conjugated recombinant RBD proteins with Alexa Fluor 488 or Alexa Fluor 594 (Thermo Fisher Scientific) according to the manufacturer’s protocol. Approximately 10 × 106 frozen PBMC from 13 convalescent donors were prepared in Falcon® 5ml-round bottom polystyrene tubes at a final concentration of 14 × 106 cells/mL in RPMI 1640 medium (GIBCO) supplemented with 10% of fetal bovine serum (Seradigm), Penicillin- Streptomycin (GIBCO) and HEPES (GIBCO). After a rest of 2h at 37°C and 5% CO2, cells were stained using Aquavivid viability marker (GIBCO) in DPBS (GIBCO) at 4°C for 20min. The detection of SARS-CoV-2-antigen specific B cells was done by adding the RBD probes to the following antibody cocktail: IgM BUV737, CD24 BUV805, IgG BV421, CD3 BV480, CD56 BV480, CD14 BV480, CD16 BV480, CD20 BV711, CD21 BV786, HLA DR BB700, CD27 APC R700 all from BD Biosciences; CD19 BV650 from Biolegend and IgA PE from Miltenyi. Staining was performed at 4°C for 30min and cells were fixed using 2% paraformaldehyde at 4°C for 15min. Stained PBMC samples were acquired on Symphony cytometer (BD Biosciences) and analyzed using FlowJo v10.7.1 (TreeStar). In each experiment, PBMC from unexposed donors (total of n = 9) were included to ensure consistent specificity of the assay.
Quantification and statistical analysis
Statistical analyses
Statistics were analyzed using GraphPad Prism version 9.0.0 (GraphPad, San Diego, CA). Every dataset was tested for statistical normality and this information was used to apply the appropriate (parametric or nonparametric) statistical test. P values < 0.05 were considered significant; significance values are indicated as ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001. Multiplicity adjustments of p values were performed with the Benjamini-Hochberg method in R and R Studio47,48 using the data.table and tidyverse packages.
Software scripts and visualization
Normalized heatmaps were generated using the complexheatmap, tidyverse, and viridis packages in R and RStudio47,48. Normalizations were done per parameter. IDs were grouped and clustered separately according to time point. Correlograms were generated using the corrplot and RColorBrewer packages in program R and RStudio using hierarchical clustering according to the first principal component (FPC). Circular barplots were generated in R and RStudio using the tidyverse package with averaged, normalized data per parameter and time point. Edge bundling graphs were generated in undirected mode in R and RStudio using ggraph, igraph, tidyverse, and RColorBrewer packages. Edges are only shown if p < 0.05, and nodes are sized according to the connecting edges’ r values. Nodes are color-coded according to groups of parameters. Area graphs were generated for the display of normalized time series. The plots were created in RawGraphs using DensityDesign interpolation and vertically un-centered values49.
Acknowledgments
The authors are grateful to the convalescent plasma donors who participated in this study. The authors thank the CRCHUM BSL3 and Flow Cytometry Platforms for technical assistance. We thank Dr. Stefan Pöhlmann and Dr. Markus Hoffmann (Georg-August University, Germany) for the plasmid coding for SARS-CoV-2 S glycoproteins and Dr. M. Gordon Joyce (US MHRP) for the monoclonal antibody CR3022. The following reagent was obtained through the NIH HIV Reagent Program, Division of AIDS, NIAID, NIH: CEM.NKR CCR5+ Cells, ARP-4376, contributed by Dr. Alexandra Trkola. The graphical abstract was prepared using images from BioRender.com. This work was supported by le Ministère de l’Économie et de l’Innovation du Québec, Programme de soutien aux organismes de recherche et d’innovation to A.F., by the Fondation du CHUM, and by the Fondation du CHU Sainte-Justine. This work was also supported by the American Foundation for AIDS Research (amfAR) to A.F. and D.E.K. This work was also supported by Canada’s COVID-19 Immunity Task Force (CITF), in collaboration with the Canadian Institutes of Health Research (CIHR) (grant VR2-173203); a CIHR foundation grant no. 352417 to A.F.; CIHR COVID-19 Rapid Research Funding to A.F., R.B., and P.B.; and by an Exceptional Fund COVID-19 from the Canada Foundation for Innovation (CFI) no. 41027 to A.F. and D.E.K. A.F. is the recipient of Canada Research Chair on Retroviral Entry no. RCHS0235 950-232424. V.M.-L. and P.B. are supported by FRQS Junior 1 salary awards. D.E.K. is a FRQS Merit Research Scholar. R.D. was supported by NIH grant R01 AI122953-05. S.P.A., J.P., and G.B.-B. are supported by CIHR fellowships. R.G. is supported by a MITACS Accélération postdoctoral fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
S.P.A., J.P., J.R., and A.F. conceived the study; S.P.A., J.P., M.N., G.B.-B., E.B.-R., J.R., D.E.K., and A.F. designed experimental approaches; S.P.A., J.P., M.N., G.B.-B., M.B., R.G., N.B., A.L., S.Y.G., C.B., G.G.-L., P.B., J.R., R.B., R.D., D.E.K., and A.F. performed, analyzed, and interpreted the experiments; R.D. performed statistical analyses; S.P.A., J.P., J.R., and G.G. contributed novel reagents; A.L., H.M., G.G.-L., L.G., C.M., V.M.-L., and C.T. collected and processed clinical samples; and S.P.A., J.P., R.D., and A.F. wrote the paper. Every author has read, edited, and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: May 5, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100290.
Contributor Information
Ralf Duerr, Email: ralf.duerr@nyulangone.org.
Daniel E. Kaufmann, Email: daniel.kaufmann@umontreal.ca.
Andrés Finzi, Email: andres.finzi@umontreal.ca.
Supplemental information
References
- 1.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartley G.E., Edwards E.S.J., Aui P.M., Varese N., Stojanovic S., McMahon J., Peleg A.Y., Boo I., Drummer H.E., Hogarth P.M. Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence. Sci. Immunol. 2020;5:eabf8891. doi: 10.1126/sciimmunol.abf8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wheatley A.K., Juno J.A., Wang J.J., Selva K.J., Reynaldi A., Tan H.X., Lee W.S., Wragg K.M., Kelly H.G., Esterbauer R. Evolution of immune responses to SARS-CoV-2 in mild-moderate COVID-19. Nat. Commun. 2021;12:1162. doi: 10.1038/s41467-021-21444-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., Trial Investigators REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 Spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beaudoin-Bussières G., Laumaea A., Anand S.P., Prévost J., Gasser R., Goyette G., Medjahed H., Perreault J., Tremblay T., Lewin A. Decline of humoral responses against SARS-CoV-2 Spike in convalescent individuals. MBio. 2020;11:e02590-20. doi: 10.1128/mBio.02590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perreault J., Tremblay T., Fournier M.J., Drouin M., Beaudoin-Bussières G., Prévost J., Lewin A., Bégin P., Finzi A., Bazin R. Waning of SARS-CoV-2 RBD antibodies in longitudinal convalescent plasma samples within 4 months after symptom onset. Blood. 2020;136:2588–2591. doi: 10.1182/blood.2020008367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prévost J., Gasser R., Beaudoin-Bussières G., Richard J., Duerr R., Laumaea A., Anand S.P., Goyette G., Benlarbi M., Ding S. Cross-sectional evaluation of humoral responses against SARS-CoV-2 Spike. Cell Rep. Med. 2020;1:100126. doi: 10.1016/j.xcrm.2020.100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., Elliott J., Hofmann C., Hausner M.A., Ferbas K.G., Tobin N.H., Aldrovandi G.M., Yang O.O. Rapid decay of anti-SARS-CoV-2 antibodies in persons with mild Covid-19. N. Engl. J. Med. 2020;383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K.J.A., Hemmings O., O’Byrne A., Kouphou N., Galao R.P. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pradenas E., Trinité B., Urrea V., Marfil S., Ávila-Nieto C., Rodríguez de la Concepción M.L., Tarrés-Freixas F., Pérez-Yanes S., Rovirosa C., Ainsua-Enrich E. Stable neutralizing antibody levels 6 months after mild and severe COVID-19 episodes. Med (N Y) 2021;2:313–320.e4. doi: 10.1016/j.medj.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan C.E.Z., Seah S.G.K., Chye D.H., Massey S., Torres M., Lim A.P.C., Wong S.K.K., Neo J.J.Y., Wong P.S., Lim J.H. The Fc-mediated effector functions of a potent SARS-CoV-2 neutralizing antibody, SC31, isolated from an early convalescent COVID-19 patient, are essential for the optimal therapeutic efficacy of the antibody. bioRxiv. 2020 doi: 10.1101/2020.10.26.355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dufloo J., Grzelak L., Staropoli I., Madec Y., Tondeur L., Anna F., Pelleau S., Wiedemann A., Planchais C., Buchrieser J. Asymptomatic and symptomatic SARS-CoV-2 infections elicit polyfunctional antibodies. Cell Rep. Med. 2021 doi: 10.1016/j.xcrm.2021.100275.p. Published online April 20, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zohar T., Loos C., Fischinger S., Atyeo C., Wang C., Slein M.D., Burke J., Yu J., Feldman J., Hauser B.M. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell. 2020;183:1508–1519.e12. doi: 10.1016/j.cell.2020.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anand S.P., Prévost J., Richard J., Perreault J., Tremblay T., Drouin M., Fournier M.J., Lewin A., Bazin R., Finzi A. High-throughput detection of antibodies targeting the SARS-CoV-2 Spike in longitudinal convalescent plasma samples. Transfusion. 2021 doi: 10.1111/trf.16318. Published online February 18, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rojas M., Rodríguez Y., Monsalve D.M., Acosta-Ampudia Y., Camacho B., Gallo J.E., Rojas-Villarraga A., Ramírez-Santana C., Díaz-Coronado J.C., Manrique R. Convalescent plasma in Covid-19: Possible mechanisms of action. Autoimmun. Rev. 2020;19:102554. doi: 10.1016/j.autrev.2020.102554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L., Zhang W., Hu Y., Tong X., Zheng S., Yang J., Kong Y., Ren L., Wei Q., Mei H. Effect of convalescent plasma therapy on time to clinical improvement in patients with severe and life-threatening COVID-19: a randomized clinical trial. JAMA. 2020;324:460–470. doi: 10.1001/jama.2020.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devasenapathy N., Ye Z., Loeb M., Fang F., Najafabadi B.T., Xiao Y., Couban R., Bégin P., Guyatt G. Efficacy and safety of convalescent plasma for severe COVID-19 based on evidence in other severe respiratory viral infections: a systematic review and meta-analysis. CMAJ. 2020;192:E745–E755. doi: 10.1503/cmaj.200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klingler J., Weiss S., Itri V., Liu X., Oguntuyo K.Y., Stevens C., Ikegame S., Hung C.-T., Enyindah-Asonye G., Amanat F. Role of IgM and IgA antibodies in the neutralization of SARS-CoV-2. medRxiv. 2020 doi: 10.1101/2020.08.18.20177303. [DOI] [Google Scholar]
- 31.Gasser R., Cloutier M., Prévost J., Fink C., Ducas É., Ding S., Dussault N., Landry P., Tremblay T., Laforce-Lavoie A. Major role of IgM in the neutralizing activity of convalescent plasma against SARS-CoV-2. Cell Rep. 2021;34:108790. doi: 10.1016/j.celrep.2021.108790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sterlin D., Mathian A., Miyara M., Mohr A., Anna F., Claër L., Quentric P., Fadlallah J., Devilliers H., Ghillani P. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021;13:eabd2223. doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Viant C., Gaebler C., Cipolla M., Hoffmann H.H., Oliveira T.Y., Oren D.A. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 2021;13:eabf1555. doi: 10.1126/scitranslmed.abf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schäfer A., Muecksch F., Lorenzi J.C.C., Leist S.R., Cipolla M., Bournazos S., Schmidt F., Maison R.M., Gazumyan A., Martinez D.R. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 2021;218:e20201993. doi: 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tso F.Y., Lidenge S.J., Poppe L.K., Peña P.B., Privatt S.R., Bennett S.J., Ngowi J.R., Mwaiselage J., Belshan M., Siedlik J.A. Presence of antibody-dependent cellular cytotoxicity (ADCC) against SARS-CoV-2 in COVID-19 plasma. PLoS ONE. 2021;16:e0247640. doi: 10.1371/journal.pone.0247640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song G., He W.T., Callaghan S., Anzanello F., Huang D., Ricketts J., Torres J.L., Beutler N., Peng L., Vargas S. Cross-reactive serum and memory B cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. bioRxiv. 2020 doi: 10.1101/2020.09.22.308965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X., Pan Z., Yue S., Yu F., Zhang J., Yang Y., Li R., Liu B., Yang X., Gao L. Disease severity dictates SARS-CoV-2-specific neutralizing antibody responses in COVID-19. Signal Transduct. Target. Ther. 2020;5:180. doi: 10.1038/s41392-020-00301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang A.T., Garcia-Carreras B., Hitchings M.D.T., Yang B., Katzelnick L.C., Rattigan S.M., Borgert B.A., Moreno C.A., Solomon B.D., Trimmer-Smith L. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat. Commun. 2020;11:4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abu-Raddad L.J., Chemaitelly H., Coyle P., Malek J.A., Ahmed A.A., Mohamoud Y.A., Younuskunju S., Ayoub H.H., Al Kanaani Z., Al Kuwari E. SARS-CoV-2 reinfection in a cohort of 43,000 antibody-positive individuals followed for up to 35 weeks. medRxiv. 2021 doi: 10.1101/2021.01.15.21249731. [DOI] [Google Scholar]
- 40.Natarajan H., Crowley A.R., Butler S.E., Xu S., Weiner J.A., Bloch E.M., Littlefield K., Wieland-Alter W., Connor R.I., Wright P.F. SARS-CoV-2 antibody signatures robustly predict diverse antiviral functions relevant for convalescent plasma therapy. medRxiv. 2020 doi: 10.1101/2020.09.16.20196154. [DOI] [Google Scholar]
- 41.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ullah I., Prévost J., Ladinsky M.S., Stone H., Lu M., Anand S.P., Beaudoin-Bussières G., Benlarbi M., Ding S., Gasser R. Live imaging of SARS-CoV-2 infection in mice reveals neutralizing antibodies require Fc function for optimal efficacy. bioRxiv. 2021 doi: 10.1101/2021.03.22.436337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brunet-Ratnasingham E., Anand S.P., Gantner P., Moquin-Beaudry G., Dyachenko A., Brassard N., Beaudoin-Bussières G., Pagliuzza A., Gasser R., Benlarbi M. Integrated immunovirological profiling validates plasma SARS-CoV-2 RNA as an early predictor of COVID-19 mortality. medRxiv. 2021 doi: 10.1101/2021.03.18.21253907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Z., Pan Q., Ding S., Qian J., Xu F., Zhou J., Cen S., Guo F., Liang C. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe. 2013;14:398–410. doi: 10.1016/j.chom.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 46.Lodge R., Lalonde J.P., Lemay G., Cohen E.A. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 1997;16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.R Studio Team . RStudio; 2015. RStudio: Integrated Development for R. [Google Scholar]
- 48.R Development Core Team . R Foundation for Statistical Computing; 2013. R: A language and environment for statistical computing. [Google Scholar]
- 49.Mauri M., Elli T., Caviglia G., Uboldi G., Azzi M. Proceedings of the 12th Biannual Conference on Italian SIGCHI Chapter. Association for Computing Machinery; 2017. RAWGraphs: a visualisation platform to create open outputs. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the manuscript. This study did not generate new or custom code.