Abstract
Summary
A rich set of tools have recently been developed for performing genome-wide genotyping of tandem repeats (TRs). However, standardized tools for downstream analysis of these results are lacking. To facilitate TR analysis applications, we present TRTools, a Python library and suite of command line tools for filtering, merging and quality control of TR genotype files. TRTools utilizes an internal harmonization module, making it compatible with outputs from a wide range of TR genotypers.
Availability and implementation
TRTools is freely available at https://github.com/gymreklab/TRTools. Detailed documentation is available at https://trtools.readthedocs.io.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Tandem repeats (TRs) represent one of the largest sources of human genetic variation and are well known to affect many human phenotypes (Hannan, 2018). Improvements in sequencing technology and bioinformatics algorithms have led to the recent development of a rich set of tools for performing genome-wide analysis of TR variation (Bakhtiari et al., 2018; Dolzhenko et al., 2017; Kristmundsdottir et al., 2020; Mousavi et al., 2019; Willems et al., 2017). These tools take aligned sequencing reads as input and output Variant Call Format (VCF) files containing estimates of TR copy number at one or more genomic TRs. The resulting VCF files may be used for a wide variety of downstream applications. However, before doing so it is usually necessary to perform filtering, quality control (QC) and merging of files across samples. While utilities exist for performing such manipulations on VCF files containing SNP variants, these tools often do not handle multi-allelic TRs and are not designed to compute TR-specific statistics. Further, different TR genotypers use different allele annotations, complicating the use of downstream tools.
Here, we present TRTools, an open-source toolkit for performing analyses on TR genotypes. TRTools provides utilities for filtering, merging, comparing and performing QC on TR VCF files. It may be used to analyse either short tandem repeats (STRs; repeat units 1–6 bp) or variable number tandem repeats (VNTRs; repeat units >6 bp) collectively referred to here as TRs. It is currently compatible with five genotypers (GangSTR, HipSTR, ExpansionHunter, PopSTR2 and adVNTR, summarized in Supplementary Table S1) and can easily be extended to handle VCFs from additional tools.
2 Features and methods
TRTools consists of a suite of command-line utilities and a corresponding Python library for performing common operations on TR genotypes, including filtering, callset comparisons and other workflows. It parses VCF files using the PyVCF (Casbon, 2012) library and implements a ‘TR harmonizer’ module that converts VCF formats from each tool to a standardized representation (Supplementary Material). This harmonization step enables downstream operations to proceed agnostic of the original tool used to produce the genotypes. For all utilities described below, the --vcftype argument may be used to specify the genotyping tool used. If not specified, the type is automatically inferred. In the following sections, we summarize the current functionality available in TRTools. Utilities are summarized in Table 1. Each utility described below is available as a standalone command line tool within the TRTools package.
Table 1.
Summary of current TRTools utilities
| Command | Description |
|---|---|
| dumpSTR | Filter a TR genotype dataset |
| mergeSTR | Merge two or more VCFs generated by a TR genotyper |
| statSTR | Generate per-locus statistics from a VCF of TR genotypes |
| compareSTR | Compare two TR genotype call sets |
| qcSTR | Output QC plots for a TR genotype call set |
2.1 DumpSTR
dumpSTR is a tool for filtering TR VCF files. It performs call-level filtering (e.g. minimum call depth, minimum call quality) and locus-level filtering (e.g. minimum call rate or deviation from Hardy–Weinberg Equilibrium). dumpSTR is specially built to handle VCF FORMAT and INFO fields unique to TR genotypers. Unlike standard VCF filtering tools, it also computes locus-level metrics such as heterozygosity and Hardy–Weinberg Equilibrium based on TR allele lengths. It takes a VCF file as input and gives a new VCF with locus-level filters annotated in the FILTER column and call-level filters annotated in the FORMAT field for each call as output.
dumpSTR --vcf VCF --out OUTPREFIX \
[--vcftype={eh|gangstr|hipstr|popstr|advntr}] \
[filters]
2.2 MergeSTR
mergeSTR is a method for merging VCF files generated by TR genotyping methods. While methods for merging VCF files currently exist (Li, 2011), TR VCFs have unique characteristics that call for a specialized merging tool. TRs are often multi-allelic, and VCFs generated using different sample sets may contain different alternate allele sets. Further, existing tools may normalize TR alleles to remove redundant sequence when merging, which can interfere with downstream analysis of TR lengths. (For example, BCFtools (Li, 2011) normalizes REF=CAG, ALT=CAGCAG to REF=C, ALT=CAGC, which is not desirable in a TR analysis.) mergeSTR takes two or more VCFs generated by the same TR genotyper as input and a merged VCF file containing all of the samples included in the input VCFs as output.
mergeSTR --vcfs VCF1, VCF2[,…], VCFn \
--out OUTPREFIX
2.3 Statistics and QC utilities
TRTools provides a suite of statistics and QC utilities to allow fast high-level checks of TR runs.
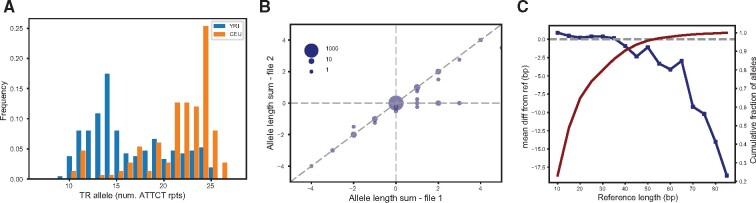
statSTR allows users to compute locus-level statistics on multi-sample TR VCFs, such as the mean allele length, allele frequency distributions and call rate. It outputs a tab-delimited file listing user-specified statistics for each TR. statSTR can also output plots of allele frequency distributions at specific TRs (Fig. 1a).
Fig. 1.
TRTools visualizations. (a) Allele frequency distribution at an example pentanucleotide TR output by statSTR based on GangSTR genotypes for two sample sets (YRI population consisting of Yorubans from Nigeria and CEU population of Northwestern European descent). (b) Example TR genotype comparison output by compareSTR. The plot compares genotypes (in terms of number of repeats difference from hg19) from HipSTR (x-axis) to those from ExpansionHunter (y-axis) on 5000 tetranucleotide TRs. Bubble sizes give the number of calls included in each point. (c) Example reference bias plot output by qcSTR using popSTR2 genotypes. The plot shows the average deviation of TR alleles called versus the reference length of the TR (in bp). The red line shows the cumulative percentage of allele calls below each reference length threshold
statSTR --vcf VCF --out OUTPREFIX [statistics]
compareSTR allows users to compare calls from two VCF files. These can be generated by the same or different tools. This allows users to compare calls across platforms or for different runtime options. Figure 1b shows an example plot created by compareSTR comparing two call sets.
compareSTR --vcf1 VCF1 --vcf2 VCF2 \
[--vcftype1 VCFTYPE] [--vcftype2 VCFTYPE] \
--out OUTPREFIX [options] \
qcSTR automatically generates plots for performing quality control of TR genotype datasets. For example, Fig. 1c shows a plot demonstrating an expected deletion bias at long alleles based on popSTR2 genotypes.
qcSTR --vcf VCF --out OUTPREFIX [options]
Additional use cases for each utility using output from each supported TR genotyping tool are provided in the TRTools documentation.
2.4 Python library for data analysis
To enable researchers to leverage TRTools features in their own custom tools, we have packaged it as a Python library. The underlying functionality for operations such as harmonizing VCF records across TR genotypers or performing string manipulations on TR sequences can be accessed by importing the library into a Python script.
import vcf, trtools.utils.tr_harmonizer as trh
reader = vcf. Reader(open(“my.vcf”))
vcftype = trh. InferVCFType(reader)
rec = reader.next()
trrecord = trh. HarmonizeRecord(vcftype, rec)
trrecord.GetAlleleFrequencies(uselength=True)
# {10: 0.2, 15: 0.8} dict of num. rpts.->freq
3 Discussion
QC and filtering are crucial steps for nearly any genome- or population-scale analysis. TRTools meets a pressing need for standardized tools for performing these tasks on TR datasets, which are not handled well by mainstream tools. This toolkit currently supports five major TR genotypers. It can easily be extended to additional TR genotyping methods for either short or long reads as long as they are compatible with the VCF standard and report precise repeat copy numbers. Improved handling of imprecise repeat copy numbers and more complex repeat sequences reported by error-prone long reads is a topic for future development. Finally, TRTools can incorporate additional utilities as the community continues to develop standards for TR analysis.
Supplementary Material
Acknowledgements
The authors thank Vineet Bafna, Mehrdad Bakhtiari, Jonghun Park and Snædis Kristmundsdottir for helpful comments and sharing VCF files. Whole genome sequencing data for individuals NA12881, NA12892, and NA12878 was obtained from dbGaP (phs001224.v1.p1).
Funding
This work was supported by the National Institutes of Health [R01HG010149, DP5OD024577].
Conflict of Interest: none declared.
Contributor Information
Nima Mousavi, Department of Electrical and Computer Engineering, University of California San Diego, La Jolla, 92093, USA.
Jonathan Margoliash, Department of Medicine, University of California San Diego, La Jolla, 92093, USA.
Neha Pusarla, Department of Bioengineering, University of California San Diego, La Jolla, 92093, USA.
Shubham Saini, Department of Computer Science and Engineering, University of California San Diego, La Jolla, 92093, USA.
Richard Yanicky, Department of Medicine, University of California San Diego, La Jolla, 92093, USA.
Melissa Gymrek, Department of Medicine, University of California San Diego, La Jolla, 92093, USA; Department of Computer Science and Engineering, University of California San Diego, La Jolla, 92093, USA.
References
- Bakhtiari M. et al. (2018) Targeted genotyping of variable number tandem repeats with adVNTR. Genome Res., 28, 1709–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casbon J. (2012) PyVCF – A Variant Call Format Parser for Python. Available from https://pyvcf.readthedocs.io/
- Dolzhenko E. et al. ; The US–Venezuela Collaborative Research Group. (2017) Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res., 27, 1895–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan A.J. (2018) Tandem repeats mediating genetic plasticity in health and disease. Nat. Rev. Genet., 19, 286–298. [DOI] [PubMed] [Google Scholar]
- Kristmundsdottir S. et al. (2020) popSTR2 enables clinical and population-scale genotyping of microsatellites. Bioinformatics, 36, 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics, 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi N. et al. (2019) Profiling the genome-wide landscape of tandem repeat expansions. Nucleic Acids Res., 47, e90–e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems T. et al. (2017) Genome-wide profiling of heritable and de novo STR variations. Nat. Methods, 14, 590–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.