Abstract
Motivation
Recent advances in high-throughput long-read sequencers, such as PacBio and Oxford Nanopore sequencers, produce longer reads with more errors than short-read sequencers. In addition to the high error rates of reads, non-uniformity of errors leads to difficulties in various downstream analyses using long reads. Many useful simulators, which characterize long-read error patterns and simulate them, have been developed. However, there is still room for improvement in the simulation of the non-uniformity of errors.
Results
To capture characteristics of errors in reads for long-read sequencers, here, we introduce a generative model for quality scores, in which a hidden Markov Model with a latest model selection method, called factorized information criteria, is utilized. We evaluated our developed simulator from various points, indicating that our simulator successfully simulates reads that are consistent with real reads.
Availability and implementation
The source codes of PBSIM2 are freely available from https://github.com/yukiteruono/pbsim2.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
High-throughput DNA sequencing technology has markedly changed the style of biological research, from hypothesis-driven biology to data-driven biology. Notably, recent advances in long-read sequencers, including Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (Nanopore), have accelerated studies on the genome (Bowden et al., 2019; Chaisson et al., 2015; Dilthey et al., 2019; Jain et al., 2018; Korlach et al., 2017), epigenome (Simpson et al., 2017) and transcriptome (Weirather et al., 2017), among others (Mantere et al., 2019; van Dijk et al., 2018).
It is known that reads generated by long-read sequencers include more errors than those generated by short-read sequencers (e.g. Illumina HiSeq), and many tools and algorithms that specifically target long-read sequencers have been developed (Amarasinghe et al., 2020; Makałowski and Shabardina, 2019; Sedlazeck et al., 2018). However, in the development of tools/algorithms for long-read sequencers, it is generally difficult to evaluate those using real data. This is because real data that meets the necessary conditions cannot always be prepared; in addition, the true error information of real data is not easy to obtain. Therefore, simulators that generate reads with error information, such as alignments between reads and the reference sequences, are useful for the evaluation of new tools/algorithms (see Alosaimi et al., 2020; Escalona et al., 2016) for comprehensive reviews of read simulators.) Moreover, these simulators are useful for experimental design such as estimating the depth coverage required for genome assembly and variant detection. To make this possible, it is crucial to be able to properly simulate the characteristics of real reads, especially the characteristics of errors.
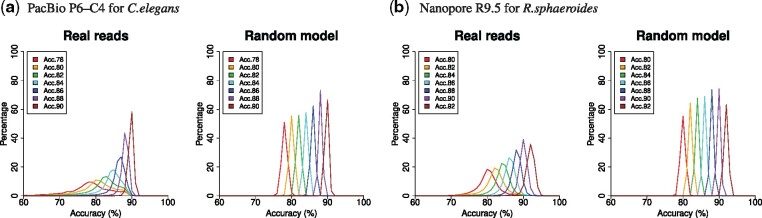
PacBio sequencers have lesser systematic (or context-specific) errors (e.g. errors in high- and low-GC regions and at homopolymer runs) than that of short-read sequencers, such as Illumina (Eid et al., 2009; Laehnemann et al., 2016; Ross et al., 2013). In contrast, it has been reported that PacBio reads have regional bias of error distribution within the reads, and very low-quality regions are sometimes observed (e.g. see Myers’ report, https://dazzlerblog.wordpress.com/2015/11/06/). Low-quality regions are caused by chimeras and undetected adapter sequences, as well as non-uniformity of errors. Figure 1 clearly shows the non-uniformity of quality scores, with the distributions of accuracy of 800 bp disjoint intervals in reads (Here, quality scores are used instead of actual errors, because it is difficult to obtain the true error information for reads, especially long reads. Note that the quality score is logarithmically related to error probability; Cock et al., 2010.). ‘Random models’ randomly generate quality scores according to real frequencies of quality scores, leading to a normal distribution of quality scores. When compared with random models, the distributions of real reads have broader accuracy ranges of 800 bp interval, especially for low read accuracy. Our previously developed simulator, PBSIM (Ono et al., 2013), employs a random model (Eid et al., 2009), and the reads generated by it are simpler and easier to handle than real reads; this is a problem when evaluating the tools/algorithms for long-read sequencers.
Fig. 1.
Non-uniformity of quality scores for real and simulated reads. After grouping reads by their accuracy, reads were segmented into 800 bp disjoint intervals, and accuracy of each interval was computed from quality scores. Each graph shows the distribution of averaged accuracy of 800 bp intervals, where colors of plotted lines represent read groups (e.g. ‘Acc.78’ refers to a read group with an accuracy of 77.5–78.4%). In random models, a house-made program randomly sampled quality scores according to the quality score distribution of each accuracy of real reads
Currently, there are several simulators that generate long reads (see Supplementary Table S1 for summary). With regard to simulation of low-quality regions, NanoSim (Yang et al., 2017) generates a set of read profiles from alignment-based analysis, and simulates low-quality regions using the profiles. PaSS (Zhang et al., 2019) adopts preset high error rates for both ends of the reads, to simulate low-quality regions. Badread (Wick, 2019) can introduce chimeras, adapter sequences, low-quality regions and low-complex repetitive sequences into simulated reads. However, there is still room for improvement in the simulation of the non-uniformity of errors (or quality scores).
To simulate the non-uniformity of quality scores, in this study, we developed a generative model for quality scores, based on a hidden Markov model (HMM) in combination with latest model selection criteria. Our computational experiments show that PBSIM2, the new version of PBSIM, simulates reads that have a tendency similar to real reads.
This article is organized as follows: In Section 2, after introducing a novel generative model for quality scores, we describe the detailed design of PBSIM2. In Section 3, we report comprehensive evaluations of PBSIM2 and related discussions. PBSIM2 newly added the function to simulate Nanopore reads, whereas it removed the function to simulate circular consensus sequencing (CCS also known as HiFi) reads. This is because the average accuracy of CCS exceeds 99%, which is outside the purpose of PBSIM to simulate error-prone reads. PBSIM2 is freely available from https://github.com/yukiteruono/pbsim2, and it will be useful for various studies using long reads.
2 Materials and methods
2.1 Datasets for long-read sequencers
In this study, we utilized various types of datasets for PacBio (seven datasets of CLR) and Nanopore sequencers (nine datasets), as summarized in Supplementary Tables S2 and S3, respectively.
2.2 Basic statistics of long reads
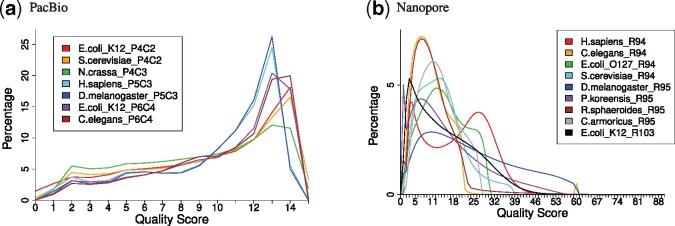
To learn the features of long reads, we obtained basic statistics, such as read length, accuracy distribution and quality score distribution, from the real reads in Supplementary Tables S2 and S3. As shown in Figure 2, in PacBio, quality score distributions are very similar within the same chemistry. Conversely, Nanopore has a wider range and more diverse distribution of quality scores than those of PacBio.
Fig. 2.
Quality score distributions of real reads. The frequency of quality scores was counted for each of the datasets in Supplementary Tables S2 and S3. Colors of plotted lines represent datasets. Dataset name is species (e.g. E.coli_K12) + chemistry (e.g. P4C2). The horizontal axis is PHRED33 quality score defined in terms of the estimated error probability (e.g. quality scores 4, 7 and 10 represent error probabilities of 40%, 20% and 10%, respectively; Cock et al., 2010)
Additionally, we conducted local alignments of real and simulated reads to reference sequences, and got error rates from the alignment results for several analyses. These local alignments were executed by LAST version 1047 (Kiełbasa et al., 2011). Alignments were filtered using last-map-probs. lastal was executed with parameters trained by last-train (Hamada et al., 2017) and ‘-m100 -j7’. lastdb, last-train, and last-map-probs were executed using the default parameters.
2.3 Generative model for quality scores
To construct a generative model for quality scores, we employed a HMM, which generates observed data from hidden states that follow the Markov model. Note that HMMs are utilized in many bioinformatics tools (e.g. Yoon, 2009). In our HMM, the emission probability distributions from each hidden state are provided by a categorical distribution, whose output is one of the quality scores. It should be emphasized that the parameters in categorical distribution with hidden states are different from each other.
In conventional HMM, the number of hidden states should be provided beforehand. In this study, we utilized HMM with the latest model selection criteria, called factorized information criteria (FIC-HMM; Hamada et al., 2015). This method is theoretically sound, enabling us to train not only parameters in HMM but also the number of hidden states (Fujimaki and Hayashi, 2012).
In this study, we adopted the model whose (lower bound of) FIC is maximum among five trials with different initial parameters, because FIC-HMM affects local optimal solutions in their training. The models were trained for each read accuracy of each chemistry (e.g. for 80% accuracy, training data comprise a read group with an accuracy of 79.5–80.4%). For read accuracy with insufficient training data, constant quality scores that match the accuracy were used.
2.4 Detailed design of PBSIM2
Given a reference sequence, PBSIM2 generates FASTQ file (Cock et al., 2010), including reads with quality scores, where the generative process is summarized as follows:
Determine read length according to the read length distribution in Section 2.4.1.
Determine read accuracy according to the read accuracy distribution in Section 2.4.2.
Generate quality scores of each position in the read using the generative model, which was trained for each read accuracy of each chemistry.
Sample a random position from the reference sequence and cut out a nucleotide sequence of the read length.
Introduce errors (substitution, insertion and deletion) into the nucleotide sequence according to a quality score at each position of the read and the ratio of error types as described in Section 2.4.3.
2.4.1 Read length distribution
On both PacBio and Nanopore sequencers, we utilized gamma distribution for read length, although log-normal distribution was employed in the previous version of PBSIM. This is because gamma distribution is more suitable than log-normal distribution for latest real datasets of both PacBio and Nanopore in our preliminary experiments (Supplementary Fig. S1). Note that DAZZ_DB/simulator ( https://github.com/thegenemyers/DAZZ_DB/blob/master/simulator.c), SimLoRD (Stöcker et al., 2016), and NPBSS (Wei and Zhang, 2018) employ log-normal distribution for PacBio; SiLiCO (Baker et al., 2016) employs log-normal distribution for PacBio, as well as gamma distribution for Nanopore; DeepSimulator1.5 (Li et al., 2020) employs beta, exponential and mixed gamma distribution for Nanopore; and Badread employs gamma distribution for both PacBio and Nanopore.
The distribution is defined by:
| (1) |
where shape and scale parameters (k and θ) are determined by averaged length and SD of reads in each dataset, respectively, which can be specified by the user as input parameters. PBSIM2 computes probability mass in each length, between the maximum and minimum length.
2.4.2 Read accuracy distribution
Both PacBio and Nanopore sequencers utilize exponential distributions for read accuracy, although normal distribution has been employed in the previous version of PBSIM. In other simulators, Badread employs beta distribution for both PacBio and Nanopore. Our preliminary experiments indicated that exponential distribution was more suitable than any other distribution for latest real datasets of both PacBio and Nanopore (Supplementary Fig. S2).
Precisely, we define read accuracy distribution by:
| (2) |
where
| (3) |
and the minimum and maximum of accuracy are determined by averaged accuracy of reads, which can be specified by the user as input parameters. PBSIM2 computes probability mass in each accuracy between the maximum and minimum accuracy.
2.4.3 Simulation of errors
A nucleotide sequence of a read is uniformly sampled from the reference sequence, and errors are introduced into the sequence as follows: For each position of the read, all error types (substitution, insertion and deletion) are introduced according to quality score at that position. In the previous version of PBSIM, deletion rate is uniform throughout all positions of every simulated read, but the latest datasets show that the rates of all error types are related to the quality scores (Supplementary Fig. S16). All error rates are calculated from quality scores and the ratio of error types given by the user. With regard to a deletion, there is no quality score for the deletion itself; thus, the quality score of the 5’neighbor is used. As in the previous version of PBSIM, half of the inserted nucleotides are chosen to be the same as their following nucleotides, and the other half are randomly chosen.
2.4.4 Sampling-based simulation
Sampling-based simulation implemented in PBSIM can also be used in PBSIM2. In this simulation, the length and quality scores of a read are randomly sampled from real data provided by the user. Subsequently, a nucleotide sequence is randomly extracted from the reference sequence, and errors are introduced in the same way as described in Section 2.4.3.
2.5 Execution of other simulators
To evaluate the ability of PBSIM2 to simulate the non-uniformity of real reads, we conducted simulations using other simulators and observed their non-uniformity. For NPBSS, we simulated PacBio CLR using the default error model. For PaSS, we simulated PacBio CLR using a prepared profile (sim.config). For LongISLND (Lau et al., 2016), we built models from real reads and simulated PacBio CLR using the models. For Badread, we built models from real reads and simulated PacBio CLR and Nanopore reads using the models. For DeepSimulator1.5, we simulated Nanopore fast5 using context-independent kmer pore model and basecalled using Guppy.
3 Result and discussion
3.1 CPU time and memory consumption
For each simulator, CPU time and maximum memory usage were measured for generating a total of 100 Mb of reads. NPBSS was executed on a Windows system equipped with Intel(R) Core(TM) CPU(i7-8565U@1.80 GHz). The others were executed on the National Institute of Genetics supercomputer system. The execution of DeepSimulator1.5 included basecalling by Guppy and checking read accuracy by Minimap2, and used ‘-c 8’ option (CPU number). Results are shown in Table 1. PBSIM is the fastest and consumes minimal memory, which enables users to simulate reads on their laptops.
Table 1.
CPU time and maximum memory for each simulator
| Simulator | CPU time (s) | Maximum memory (Gb) |
|---|---|---|
| PBSIM | 5 | 0.2 |
| PBSIM2 (this work) | 7 | 0.2 |
| LongISLND | 565 | 26.7 |
| NPBSS | 1024 | 0.1 |
| DeepSimulator1.5 | 113 344 | 15.3 |
| PaSS | 14 | 0.8 |
| Badread | 1498 | 3.5 |
Note: CPU time and maximum memory usage were measured for generating a total of 100 Mb of reads. NPBSS was executed on a Windows system equipped with Intel(R) Core(TM) CPU(i7-8565U@1.80 GHz). The others were executed on the National Institute of Genetics supercomputer system. The execution of DeepSimulator1.5 included basecalling by Guppy and assessing read accuracy by Minimap2; when using ‘-c 8’ option (CPU number), wall-clock time was 20 662 s.
3.2 Evaluation of a generative model of quality scores
To evaluate PBSIM2 that implemented a novel generative model of quality scores trained using FIC-HMM, we compared simulated reads of PBSIM2 with real reads in terms of non-uniformity of quality scores. PBSIM2 simulated reads with the same parameters (e.g. mean and SD of read length and accuracy) as real reads. We also evaluated simulated reads of Markov Model (MM), because in Nanopore sequencing, the raw current signal is mainly influenced by 5- or 6-mer that occupies the pore simultaneously (Rang et al., 2018), and Faucon et al. (2017) showed that the strongest feature for predicting the accuracy of each k-mer was the accuracy of neighboring k-mers, one step away. MM generates quality scores by first- and second-order MM (referred to as ‘1st-order MM’ and ‘2nd-order MM’, respectively) of transition probabilities of quality scores of real reads.
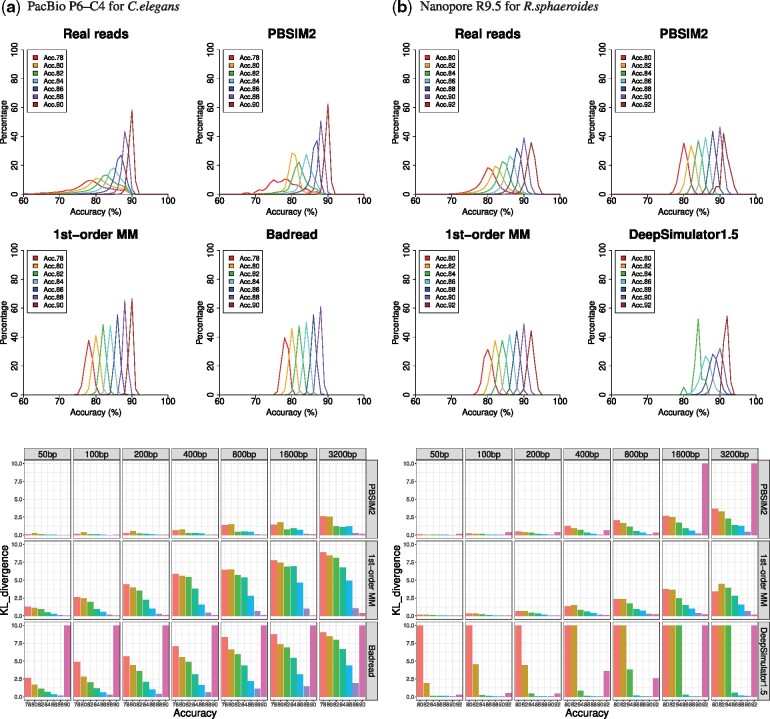
It is clear that the non-uniformity of simulated reads of PBSIM2 is sufficiently similar to that of real reads in both PacBio and Nanopore (Fig. 3 and Supplementary Figs S3 and S4 show graphs of all the interval sizes). Figure 3b also indicates that 1st-order MM is able to simulate the non-uniformity, as well as PBSIM2 in Nanopore. We utilized the Kullback–Leibler (KL) divergence for observing similarity between non-uniformity (see Fig. 3). For P (real distribution) and Q (simulated distribution), the KL divergence from Q to P is defined to be;
Fig. 3.
Simulation of non-uniformity of quality scores and evaluation by KL divergence. Each graph shows distributions of accuracy of 800 bp disjoint intervals in reads in the same way as Figure 1. Read groups (e.g. Acc.78) with insufficient data are not shown in the graphs. PBSIM2, the new version of PBSIM, generated reads using model-based simulation. ‘1st-order MM’, our in-house software tool, generated quality scores for each read group, by a 1st-order MM of transition probabilities of the quality score of real reads. Badread built a model and generated reads. DeepSimulator1.5 generated Nanopore fast5 using context-independent kmer pore model and basecalled using Guppy. KL divergence of distribution of accuracy of fixed size (50, 100, 200, 400, 800, 1600 and 3200 bp) intervals between real and simulated reads. Upper-limit value of KL divergence was 10
Figure 4 and Supplementary Figure S5 show that features of the transition probability matrix are clearly different between PacBio and Nanopore, and the transition ranges in Nanopore are narrower than those in PacBio. Thus, MM is successful in Nanopore. Furthermore Figures 5, 6 and Supplementary Figures S6 and S7 show that in FIC-HMM, the transition ranges of states in Nanopore are narrower than those in PacBio, and the emission ranges of states in Nanopore are narrower and simpler than those in PacBio. These observations in MM and FIC-HMM are consistent. We decoded training data for FIC-HMM into states using the Viterbi algorithm, and examined continuous length of state (e.g. if the same state is lined up five times in a row, the continuous length is 5). Supplementary Figure S8 shows that the continuous length of state in PacBio is longer than that in Nanopore. In both PacBio and Nanopore R9.5, 2nd-order MM was a slightly better simulation than 1st-order MM (Supplementary Figs S9–12). However, in Nanopore R10.3, they were almost the same (Supplementary Figs S13 and S14).
Fig. 4.
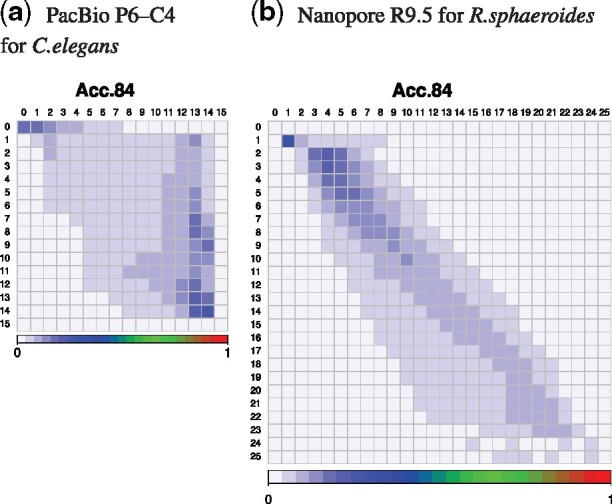
Transition probability matrixes of quality scores of real reads. The vertical and horizontal axes are PHRED33 quality scores defined in terms of the estimated error probability (e.g. quality score 4, 7 and 10 represent error probabilities of 40%, 20% and 10%, respectively; Cock et al., 2010). Quality scores on the vertical axis transition to ones on the horizontal axis. The sum of transition probabilities on each quality score of the vertical axis is 100%. These are matrixes of ‘Acc84’, denoting a read group whose accuracy is 83.5–84.4%. In Nanopore matrix, quality scores above 25 are not displayed
Fig. 5.
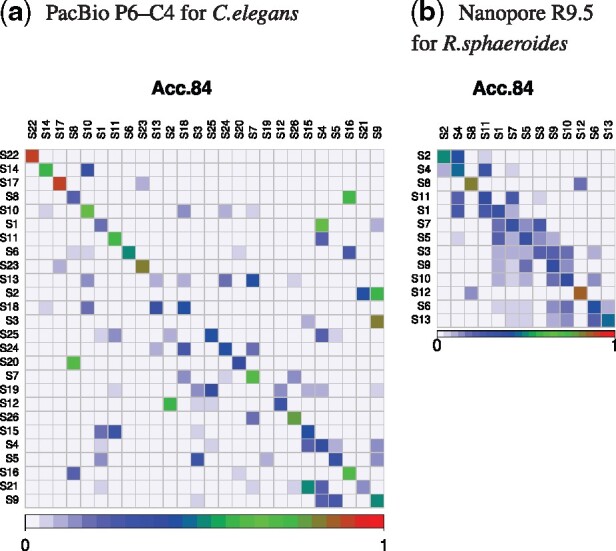
Transition probability matrixes of states of FIC-HMM. The vertical and horizontal axes represent states of FIC-HMM, which are sorted in order of increasing averaged quality score emitted by them. States on the vertical axis transition to ones on the horizontal axis. The sum of transition probabilities on each state of the vertical axis is 100%. These are matrixes of ‘Acc84’, a read group with an accuracy of 83.5–84.4%
Fig. 6.
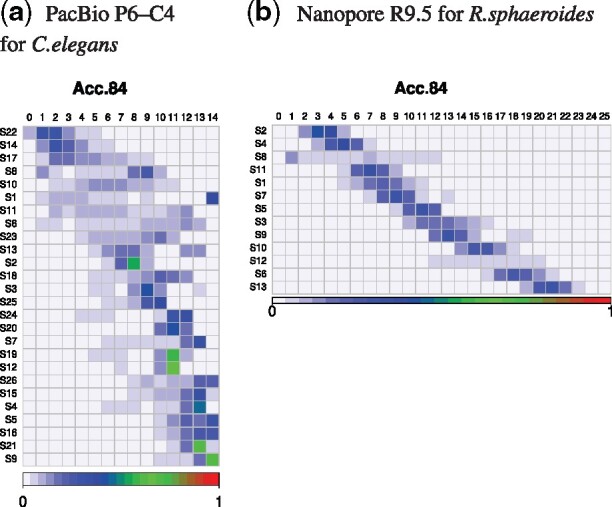
Emission probability matrixes of states of FIC-HMM. The vertical axis represents states of FIC-HMM, which are sorted in order of increasing averaged quality score emitted by them. The horizontal axis is PHRED33 quality score defined in terms of the estimated error probability (e.g. quality scores of 4, 7 and 10 represent error probability of 40%, 20% and 10%, respectively; Cock et al., 2010). States on the vertical axis emit quality scores on the horizontal axis. The sum of emission probabilities on each state of vertical axis is 100%. These are matrixes of ‘Acc84’, a read group with an accuracy of 83.5–84.4%. In the matrix of Nanopore, quality scores above 25 are not displayed
Supplementary Figures S9–14 also show comparisons with other long-read simulators. In simulation of PacBio reads, PBSIM2 is able to simulate the non-uniformity of real reads more than that of any other simulator (see Supplementary Fig. S10). Even in the simulation of Nanopore reads, PBSIM2 is one of the best simulators for overall read accuracy, but at 86–90% read accuracy of Supplementary Figure S12 and at 84–88% of Supplementary Figure S14, DeepSimulator1.5 is the best. However, DeepSimulator1.5 has narrow ranges of read accuracy.
3.3 Correlation between read length and accuracy
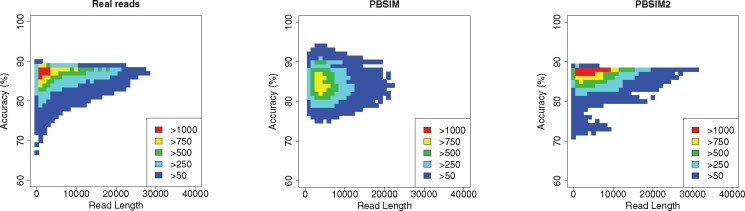
The previous version of PBSIM was unable to simulate realistic correlation between length and accuracy for each read (Stöcker et al., 2016; Wei and Zhang, 2018). As shown in Figure 7 and Supplementary Figure S15, PBSIM2 is able to simulate realistic correlations. This improvement was mainly due to change in read accuracy distribution, as mentioned in Section 2.4.2.
Fig. 7.
Correlation between read length and accuracy for each read of PacBio P6–C4 for E.coli K12 MG1655. Accuracy of each read was calculated from quality scores. PBSIM and PBSIM2 simulated reads with the same parameters (e.g. mean and SD of read length and accuracy) as real reads. Colors indicate the varying frequencies of each cell
3.4 Relationship between error rate and quality scores
In the previous version of PBSIM, the relationship between error rate and quality score deviated from the correct one with increasing quality score (Wei and Zhang, 2018). As shown in Supplementary Figure S16, the relationship is improved by changing the deletion rate, as mentioned in Section 2.4.3.
3.5 Nucleotide sequence-based error model
Using nucleotide sequence context (or k-mer)-based error models, generated from alignment-based analysis of real reads, several simulators are able to simulate features of real reads, such as error length (Faucon et al., 2017; Lau et al., 2016; Wick, 2019; Zhang et al., 2019). Thus, we investigated 6-mer error bias by analyzing alignments of reads to the reference sequences. As shown in Supplementary Figures S17 and S18, we observed that PacBio reads had small 6-mer bias, whereas Nanopore reads had significant 6-mer bias, which is consistent with a previous study (Faucon et al., 2017). It has been reported that homopolymers are difficult to be called accurately by base-callers; therefore, many deletions occur at homopolymers in read sequence of Nanopore (Rang et al., 2018). Concurrent with this report, we observed high deletion rate at homopolymers in Nanopore (see Supplementary Tables S4–6). We also observed that insertion and deletion (indels) were longer in Nanopore (R9.5) than those in PacBio (Supplementary Fig. S19a and b). Recently, the latest Nanopore chemistry, R10, has improved resolution of homopolymeric regions (Amarasinghe et al., 2020). Actually, indels are shorter in R10 than those in R9, and the indel length distribution becomes similar to that of PBSIM2 (Supplementary Fig. S19b and c), compared with PBSIM. In contrast, with regard to 6-mer error bias, R10 shows features similar to those of R9 (see Supplementary Fig. S18). Although it may be necessary to simulate 6-mer error bias, especially homopolymer-specific error, to simulate Nanopore reads accurately, this version of PBSIM2 does not address this issue because, as mentioned earlier, Nanopore R10 has improved for homopolymer, and basecalling software is improving for homopolymer basecalling (Wick et al., 2019).
3.6 Future directions
In addition to the low-quality regions, artifacts such as chimeras and adapter sequences are frequently observed in long reads (see Myers’ report, https://dazzlerblog.wordpress.com/2017/04/22/1344/). These errors are the major cause of poor genome assembly. Badread has previously simulated these errors, and we also plan to implement similar functions in the next version of PBSIM.
After PacBio Sequel sequencer, quality code is a fixed value and does not represent the actual error rate, so in this study, only RS II CLR was used as training data for quality scores. PBSIM2 is targeted at error-prone reads, so we are unsure if it can properly simulate HiFi reads. However, if a generative model of quality scores is created using the error information obtained from the alignment of reads to the reference sequences instead of the quality score, the latest PacBio Sequel II data can be used as the training data of FIC-HMM. Even though there are many problems, such as handling unaligned regions or regions where it is difficult to obtain accurate error information, including low-quality regions, learning alignment by FIC-HMM is expected to significantly improve the error model of long reads.
4 Conclusion
In this study, we proposed a novel simulator for long reads produced by PacBio and Nanopore sequencers, in which a novel generative model for quality scores is employed.
One of the novel points in this study was introducing a generative model of quality scores, based on a HMM with a model selection procedure. Our experiments showed that the generative model simulates quality scores that are more consistent with real reads of PacBio and Nanopore than other existing simulators.
Supplementary Material
Acknowledgements
We would like to extend our gratitude to Dr Martin Frith for his valuable comments on the article. Computations in this study were partially performed on the supercomputer systems at the ROIS National Institute of Genetics.
Funding
This work was supported, in part, by MEXT KAKENHI [grants JP24680031, JP16H05879 and JP20H00624 to M.H. and JP16H06279 and JP25240044 to M.H. and K.A.].
Conflict of Interest: none declared.
Contributor Information
Yukiteru Ono, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, University of Tokyo, Kashiwa 277-8561, Japan.
Kiyoshi Asai, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, University of Tokyo, Kashiwa 277-8561, Japan; Artificial Intelligence Research Center (AIRC), National Institute of Advanced Industrial Science and Technology (AIST), Tokyo 135–0064, Japan.
Michiaki Hamada, Department of Electrical Engineering and Bioscience, Faculty of Science and Engineering, Waseda University, Tokyo 169–8555, Japan; Computational Bio Big-Data Open Innovation Laboratory (CBBD-OIL), National Institute of Advanced Industrial Science and Technology (AIST), Tokyo 169–8555, Japan; Institute for Medical-oriented Structural Biology, Waseda University, Tokyo 162–8480, Japan; Graduate School of Medicine, Nippon Medical School, Tokyo 113–8602, Japan.
References
- Alosaimi S. et al. (2020) A broad survey of DNA sequence data simulation tools. Brief. Funct. Genomics, 19, 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe S.L. et al. (2020) Opportunities and challenges in long-read sequencing data analysis. Genome Biol., 21, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker E.A.G. et al. (2016) Silico: a simulator of long read sequencing in PacBio and Oxford Nanopore. BioRxiv, 076901. [Google Scholar]
- Bowden R. et al. (2019) Sequencing of human genomes with nanopore technology. Nat. Commun., 10, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson M.J. et al. (2015) Resolving the complexity of the human genome using single-molecule sequencing. Nature, 517, 608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock P.J. et al. (2010) The sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res., 38, 1767–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilthey A.T. et al. (2019) Strain-level metagenomic assignment and compositional estimation for long reads with Metamaps. Nat. Commun., 10, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J. et al. (2009) Real-time DNA sequencing from single polymerase molecules. Science, 323, 133–138. [DOI] [PubMed] [Google Scholar]
- Escalona M. et al. (2016) A comparison of tools for the simulation of genomic next-generation sequencing data. Nat. Rev. Genet., 17, 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucon P.C. et al. (2017) SNaResim: synthetic nanopore read simulator. In 2017 IEEE International Conference on Healthcare Informatics (ICHI), pp 338–344. IEEE, arXiv Preprint arXiv, and BioRxiv.
- Fujimaki R., Hayashi K. (2012) Factorized asymptotic Bayesian hidden Markov models. arXiv Preprint arXiv:1206.4679. [Google Scholar]
- Hamada M. et al. (2015) Learning chromatin states with factorized information criteria. Bioinformatics, 31, 2426–2433. [DOI] [PubMed] [Google Scholar]
- Hamada M. et al. (2017) Training alignment parameters for arbitrary sequencers with last-train. Bioinformatics, 33, 926–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M. et al. (2018) Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol., 36, 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiełbasa S.M. et al. (2011) Adaptive seeds tame genomic sequence comparison. Genome Res., 21, 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korlach J. et al. (2017) De novo PacBio long-read and phased avian genome assemblies correct and add to genes important in neuroscience research. BioRxiv, 103911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laehnemann D. et al. (2016) Denoising DNA deep sequencing data-high-throughput sequencing errors and their correction. Brief. Bioinform., 17, 154–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau B. et al. (2016) Longislnd: in silico sequencing of lengthy and noisy datatypes. Bioinformatics, 32, 3829–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. (2020) Deepsimulator1. 5: a more powerful, quicker and lighter simulator for nanopore sequencing. Bioinformatics, 36, 2578–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makałowski W., Shabardina V. (2019) Bioinformatics of nanopore sequencing. J. Hum. Genet., 65, 1–7. [DOI] [PubMed] [Google Scholar]
- Mantere T. et al. (2019) Long-read sequencing emerging in medical genetics. Front. Genet., 10, 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y. et al. (2013) PBSIM: PacBio reads simulator-toward accurate genome assembly. Bioinformatics, 29, 119–121. [DOI] [PubMed] [Google Scholar]
- Rang F.J. et al. (2018) From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biol., 19, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross M.G. et al. (2013) Characterizing and measuring bias in sequence data. Genome Biol., 14, R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlazeck F.J. et al. (2018) Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat. Rev. Genet., 19, 329–346. [DOI] [PubMed] [Google Scholar]
- Simpson J.T. et al. (2017) Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods, 14, 407–410. [DOI] [PubMed] [Google Scholar]
- Stöcker B.K. et al. (2016) SimLoRD: simulation of long read data. Bioinformatics, 32, 2704–2706. [DOI] [PubMed] [Google Scholar]
- van Dijk E.L. et al. (2018) The third revolution in sequencing technology. Trends Genet., 34, 666–681. [DOI] [PubMed] [Google Scholar]
- Wei Z.-G., Zhang S.-W. (2018) NPBSS: a new PacBio sequencing simulator for generating the continuous long reads with an empirical model. BMC Bioinformatics, 19, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirather J.L. et al. (2017) Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis. F1000Res., 6, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R. (2019) Badread: simulation of error-prone long reads. J. Open Source Softw., 4, 1316. [Google Scholar]
- Wick R.R. et al. (2019) Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol., 20, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. et al. (2017) NanoSim: nanopore sequence read simulator based on statistical characterization. GigaScience, 6, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon B.-J. (2009) Hidden Markov models and their applications in biological sequence analysis. Curr. Genomics, 10, 402–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. et al. (2019) PaSS: a sequencing simulator for PacBio sequencing. BMC Bioinformatics, 20, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.