Abstract
Systemic lupus erythematosus (SLE) is a severe autoimmune disorder, the pathogenesis of which remains largely unknown. The present study aimed to investigate the role and mechanism of circular RNAs in the etiopathogenesis of SLE. CD4+ T cells in patients with SLE expressed higher levels of hsa_circ_0010957 compared with healthy individuals and was a good differentiator of the active from inactive SLE disease. It was also determined that hsa_circ_0010957 mediated microRNA (miR)-125b/STAT3 signaling and subsequent secretion of inflammatory cytokines interleukin (IL)-18, IL-6 and IL-17, which are important factors in the process of SLE. Hsa_circ_0010957 abrogated the proinflammatory effect of IL-6 via the blockade of STAT3 signaling. In conclusion, increased hsa_circ_0010957 may be involved in SLE pathogenesis via miR-125b/STAT3 signaling. Hsa_circ_0010957 promises to be a potential biomarker and therapeutic target for SLE.
Keywords: systemic lupus erythematosus, circular RNA, hsa_circ_0010957, microRNA-125b, STAT3
Introduction
Systemic lupus erythematosus (SLE) is a severe autoimmune disorder that affects several organs, such as the skin, kidney, heart, joints, and the central nervous system (1,2). Despite an imperfect understanding of the pathogenesis of SLE, significant gains have been made in recent years. One of the principal pathophysiological characteristics of SLE is the dysfunction of various immunocyte populations, including macrophages, dendritic cells, neutrophils and peripheral blood mononuclear cells (PBMCs) such as B cells and CD4+ T cells, resulting in changes in inflammatory and immune responses (3). Increased understanding of the pathogenesis of SLE can help to develop targeted immunotherapy in treating patients with SLE (4).
Circular RNAs (circRNAs) are a novel class of noncoding RNA molecules formed by a covalently closed loop structure without a 5′cap or 3′poly A tail; they are widely expressed in mammals (5). Growing evidence has shown that circRNAs play critical roles in the course of multiple diseases (6). Some studies have shown that circRNAs play important roles in SLE (7,8). CircRNAs can act as novel biomarkers for SLE; for example, circRNA_002453 might serve as a biomarker for diagnosing lupus nephritis (9); circPTPN22 may function as a potential activity indicator in SLE (10). CircRNAs such as circIBTK (11), hsa_circ_0045272 (12) and hsa_circ_0012919 (13) are also involved in SLE disease progression. Nevertheless, few studies have been done on circRNAs in SLE, and their roles in SLE are unclear.
Therefore, the present study investigated the expression and function of circRNAs in SLE. Hsa_circ_0010957 is encoded within the SEPN1 gene locus and is highly expressed in SLE-derived CD4+ T cells, as demonstrated by circRNA microarray analysis (13). The expression of hsa_circ_0010957 was verified in SLE-derived CD4+ T cells and healthy controls (HCs), and its function in SLE progression was investigated.
Materials and methods
Subjects
Thirty patients diagnosed with SLE (three males and twenty-seven females; mean age, 32.78±7.42) and twenty-five age- and sex-matched HCs (two males and twenty-three females; mean age, 33.12±7.31) were recruited from Jinhua Municipal Central Hospital. This study was approved by the Human Ethics Committee of Jinhua Municipal Central Hospital and written informed consents were obtained from all participants in this study. The diagnostic criteria for SLE were according to the 2012 Systemic Lupus International Collaborating Clinic (SLICC) revised criteria for classification of SLE (14). The SLEDAI scoring system was applied to assess disease activity, and SLEDAI score >4 was considered as active disease.
CD4+ T cell separation, culture and treatment
PBMCs were separated from peripheral venous blood using Ficoll-Paque (Sigma-Aldrich; Merck KGaA). CD4+ T cells were selected by Miltenyi beads (Miltenyi Biotec GmbH). Cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing 20% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) in an incubator at 37°C containing 5% CO2. CD4+ T cells were treated with interleukin (IL)-6 (10 ng/ml, 24 h; cat. no. HY-P7044, MedChemExpress) (15).
Transfection
Human T Cell Nucleofector kit (Lonza Group, Ltd.) was used to transfect hsa_circ_0010957 small interfering (si)RNAs (50 nM), microRNA (miR)-125b mimics (50 nM), miR-125b inhibitor (50 nM), STAT3 siRNA (50 nM) and corresponding negative control (NC) (50 nM) into CD4+ T cells. hsa_circ_0010957 siRNAs and NC siRNA, miR-125b mimics and NC mimics, miR-125b inhibitor and NC inhibitor were purchased from Shanghai GenePharma Co., Ltd. The mimic and inhibitor-NCs used in the present study were non-targeting sequences (sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′. Target sequences (5′→3′) of hsa_circ_0010957 siRNA: siRNA#1, AGAGAAGACTAACGTCCATCA; siRNA#2, GAAGACTAACGTCCATCACAT; siRNA#3, GACTAACGTCCATCACATCAA. STAT3 siRNA (cat. no. sc-29493) and control siRNA (cat. no. sc-37007) were purchased from Santa Cruz Biotechnology, Inc. Cells were harvested 48 h after transfection at 37°C, and transfection efficacy was then assessed by reverse transcription-quantitative PCR (RT-qPCR).
RT-qPCR
TRIzol LS reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to isolate total RNA from CD4+ T cells. cDNAs were synthesized with a PrimeScript RT-PCR kit (Takara Bio, Inc.; 37°C for 15 min and 85°C for 5 sec. RT-qPCR was performed with SYBR Premix Ex Taq (Takara Bio, Inc.) using the following thermocycling conditions: 95°C for 2 min, followed by 40 cycles of 95°C for 20 sec, 60°C for 30 sec and 72°C for 20 sec. Primers for hsa_circ_0010957 were: Forward, 5′-AGAGAAGACTAACGTCCATCACA-3′; reverse, 5′-TGGACGGGTCTTCAAAGGTG-3′. β-actin was used as an internal reference: Forward, 5′-GTGGCCGAGGACTTTGATTG-3′; reverse, 5′-CCTGTAACAACGCATCTCATATT-3′. miR-125b was detected using TaqMan Human MicroRNA assays (Applied Biosystems; Thermo Fisher Scientific, Inc.) and normalized to the U6 small nuclear RNA (U6 snRNA). The data were analyzed by 2−ΔΔCq method (16).
Enzyme-linked immunosorbent assay (ELISA)
IL-18, IL-6 and IL-17 concentrations were measured in the cell supernatants and serum of patients with SLE by ELISA using Human IL-18 ELISA kit (cat. no. ab215539; Abcam), human IL-6 ELISA kit (cat. no. ab46042; Abcam) and human IL-17 ELISA kit (cat. no. ab119535; Abcam), respectively.
Luciferase reporter assay
Wild-type and mutant sequences of hsa_circ_0010957 or STAT3 3′UTR containing miR-125b binding sites were synthesized and inserted into the downstream of firefly luciferase reporter vector pmirGLO (Promega Corporation). The luciferase and Renilla luciferase reporter vectors were co-transfected into CD4+ T cells with miR-125b mimics using Human T Cell Nucleofector kit (Lonza Group, Ltd.) according to the manufacturer's protocol. After 48 h culture, luciferase activity was quantified with a Dual Luciferase Reporter Assay kit (Promega Corporation) according to the manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase.
Western blotting
Total protein was extracted from CD4+ T cells using RIPA buffer (cat. no. P0013B; Beyotime Biotechnology) and quantified using the BCA Protein Assay Kit (Beyotime Biotechnology). Equal amounts of protein (20 µg/lane) were resolved on a 10% SDS-denaturing polyacrylamide gel and transferred to PVDF membranes. The membranes were subsequently blocked by 5% fat-free buffered milk for 2 h at room temperature and then incubated overnight at 4°C with antibodies to STAT3 (cat. no. ab119352; 1:2,000; Abcam), STAT3 (phospho Y705; cat. no. ab76315; 1:1,000; Abcam) and β-actin (cat. no. ab8226; 1:2,000; Abcam). The membranes were then washed with PBS-T solution (PBS with 0.1% Tween-20) and incubated with HRP-labeled Goat Anti-Rabbit IgG (cat. no. A0208; 1:1,000; Beyotime Institute of Biotechnology) and HRP-labeled Goat Anti-Mouse IgG (cat. no. A0216; 1:1,000; Beyotime Institute of Biotechnology) at room temperature for 2 h. Protein bands were visualized using a high sensitivity ECL chemiluminescence Kit (cat. no. P0018M; Beyotime Institute of Biotechnology). ImageJ software (version 1.8.0; National Institutes of Health) was used for densitometry.
Statistical analysis
Data analysis was performed using SPSS 19.0 statistical software (SPSS, Inc.). All experiments were performed in triplicate, and data are reported as mean ± standard deviation (SD). Differences between three or more groups were analyzed by one-way analysis of variance following Tukey's test. Differences between two groups were analyzed using paired (for paired groups) or unpaired (for unpaired groups) Student's t-test, when applicable. Non-parametric method was used to analyze the AUC (Area Under Curve) of receiver operating characteristic (ROC) curves. Spearman's analysis was used to test correlation. P<0.05 was considered to indicate a statistically significant difference.
Results
Hsa_circ_0010957 expression is increased in SLE and can serve as a biomarker
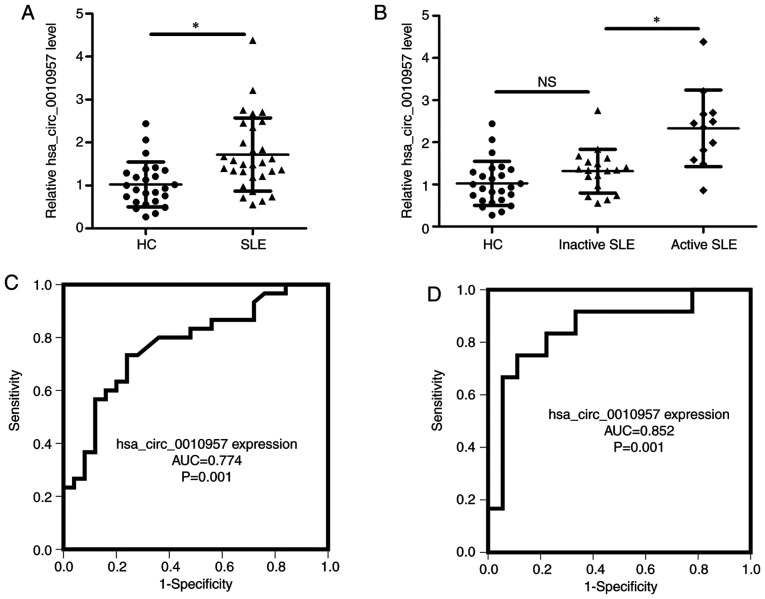
Previous microarray analyses have shown that hsa_circ_0010957 is upregulated in SLE-derived CD4+ T cells (12), but the level of hsa_circ_0010957 had not been verified by RT-qPCR. SLE-derived CD4+ T cells and cells derived from healthy volunteers demonstrated higher expression of hsa_circ_0010957 in SLE (Fig. 1A). There was also a difference in hsa_circ_0010957 expression between patients with active SLE and those with inactive disease (Fig. 1B), but there was no significant difference between patients with inactive disease and HCs (Fig. 1B). A receiver operating characteristic curve showed the diagnostic value of hsa_circ_0010957 for differentiating patients with SLE from HCs (Fig. 1C) and differentiating patients with active SLE from those with inactive disease (Fig. 1D). Thus, increased hsa_circ_0010957 expression might be a prospective biomarker for SLE.
Figure 1.
Hsa_circ_0010957 expression is increased in *P<0.05. NS, no significance and can serve as a biomarker. (A) Relative expression of has_circ_0010957 in CD4+ T cells from 30 patients with SLE and 25 HCs tested using reverse transcription-quantitative PCR. (B) Comparisons of relative has_circ_0010957 expression in CD4+ T cells from 25 HCs, 18 inactive and 12 active patients with SLE. (C and D) Receiver operating characteristics showing diagnosis value of hsa_circ_0010957 in differentiating patients with SLE from HCs (C) and differentiating active from inactive patients with SLE. (D) Data are shown as mean ± SD *P<0.05. NS, no significance; SLE, systemic lupus erythematosus; AUC, area under the curve; HCs, healthy controls.
Hsa_circ_0010957 absorbs miR-125b in SLE-derived CD4+ T cells
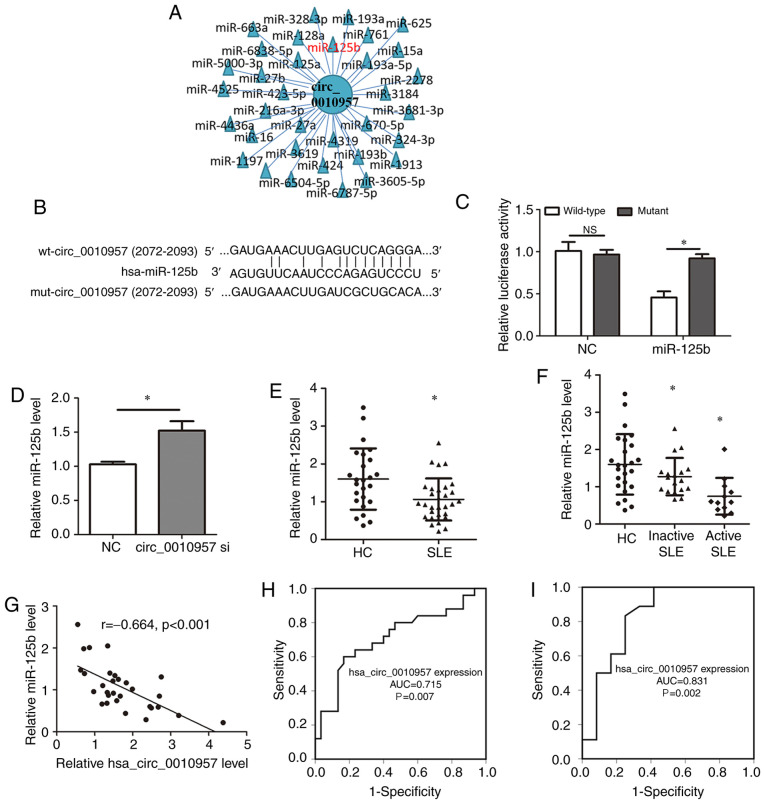
A common mechanism of circRNA function is to act as miRNA sponges to regulate target miRNA activity (17,18). Putative miRNA targets of hsa_circ_0010957 were searched using the StarBase database (http://starbase.sysu.edu.cn/index.php) (Fig. 2A) and identified miR-125b, which has previously been associated with SLE (19,20). Luciferase reporter assays were used to test the putative interaction (Fig. 2B). Overexpression of miR-125b decreased wild-type hsa_circ_0010957 activity but did not affect the mutant vector (Fig. 2C). Indeed, hsa_circ_0010957 knockdown led to increased miR-125b expression (Fig. 2D). miR-125b expression was also decreased in SLE-derived CD4+ T cells (Fig. 2E) and was lower in patients with active SLE compared with those with inactive disease (Fig. 2F). Moreover, miR-125b expression was negatively associated with hsa_circ_0010957 expression (Fig. 2G). Thus, hsa_circ_0010957 acts as a miR-125b sponge. It was also found that miR-125b is a good diagnostic marker for differentiating patients with SLE from HCs (Fig. 2H) and differentiating active SLE from inactive disease (Fig. 2I). These results suggested that decreased miR-125b expression in SLE might also be a prospective biomarker for the disease.
Figure 2.
Hsa_circ_0010957 absorbs miR-125b in SLE-derived CD4+ T cells. (A) StarBase database showing the potential target miRNAs of hsa_circ_0010957. (B) Potential binding site and the mutant site of hsa_circ_0010957 and miR-125b. (C) Luciferase assay showing overexpression of miR-125b decreased the luciferase activity of wide-type hsa_circ_0010957 vector, while not the mutant vector. (D) Relative miR-125b level was detected using RT-qPCR after knockdown of hsa_circ_0010957. (E) Relative miR-125b expression in CD4+ T cells from 30 patients with SLE and 25 HCs tested using RT-qPCR. (F) Comparisons of relative miR-125b expression in CD4+ T cells from 25 HCs, 18 inactive and 12 active patients with SLE. (G) Pearson's correlation analysis of hsa_circ_0010957 and miR-125b expression in CD4+ T cells of patients with SLE. (H and I) Receiver operating characteristic showing diagnosis value of miR-125b in differentiating patients with SLE from HCs (H) and differentiating active from inactive patients with SLE. (I) Data are shown as mean ± SD *P<0.05. NS, no significance; SLE, systemic lupus erythematosus; wt, wild-type; mut, mutant type; miR/miRNAs, microRNA; C, negative control; RT-qPCR, reverse transcription-quantitative PCR; HC, healthy control; AUC, area under the curve.
Hsa_circ_0010957 modulates inflammatory cytokines secretion via miR-125b
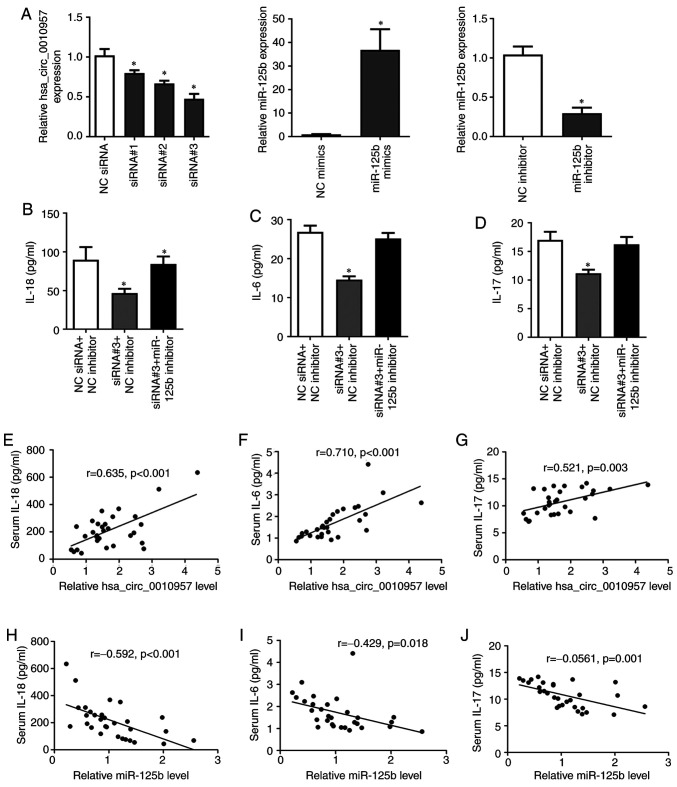
To investigate the biological role of hsa_circ_0010957 and miR-125b in SLE, first, we detected the overexpression or silence efficiency (Fig. 3A). Knockdown of hsa_circ_0010957 significantly reduced secretion of inflammatory cytokines IL-18, IL-6, and IL-17, while silencing miR-125b reversed these effects (Fig. 3B-D). We also observed a positive correlation between hsa_circ_0010957 and inflammatory cytokines expression (Fig. 3E-G) and a negative correlation between miR-125b and inflammatory cytokines (Fig. 3H-J). Therefore, we suggest hsa_circ_0010957 modulates inflammatory cytokine secretion via miR-125b.
Figure 3.
Hsa_circ_0010957 modulates inflammatory cytokines secretion via miR-125b. (A) Overexpression or silencing efficiency of hsa_circ_0010957 or miR-125b in CD4+ T cells of SLE detected by reverse transcription-quantitative PCR. (B-D) IL-18, IL-6 and IL-17 levels detected in cell supernatant after CD4+ T cells were transfected with hsa_circ_0010957 siRNAs and miR-125b inhibitor. (E-J) Pearson's correlation analysis between hsa_circ_0010957 or miR-125b expressions in CD4+ T cells of SLE with inflammatory cytokines in serum of patients with SLE. Data are shown as mean ± SD *P<0.05. SLE, systemic lupus erythematosus; miR, microRNA; siRNA, small interfering RNA; NC, negative control; IL, interleukin.
Hsa_circ_0010957 regulates activation of STAT3 signaling via miR-125b
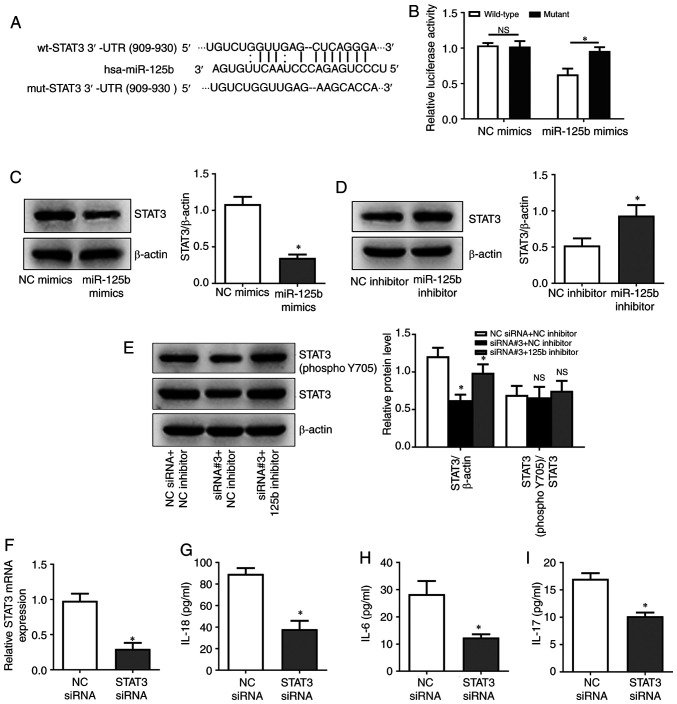
Through the microRNA.org database, STAT3 was identified as a potential target of miR-125b regulation (Fig. 4A). Luciferase reporter assays showed that miR-125b overexpression inhibited the activity of wild-type STAT3 3′-UTR but not a variant STAT3 3′-UTR (Fig. 4B). In addition, miR-125b overexpression led to decreased STAT3 protein expression, while miR-125b silencing had an inverse effect (Fig. 4C and D). The results showed that hsa_circ_0010957 siRNA decreased total and phosphorylation levels of STAT3 protein, and this effect can be rescued by miR-125b inhibition (Fig. 4E). These results indicated that hsa_circ_0010957 regulates STAT3 signaling activation via miR-125b. STAT3 was also successfully silenced in SLE-derived CD4+ T cells and showed that knockdown of STAT3 inhibited the expression of inflammatory cytokines IL-18, IL-6 and IL-17 (Fig. 4F-I). It was also found that hsa_circ_0010957 could regulate the activation of STAT3 signaling via miR-125b in CD4+ T cells from healthy controls (Fig. S1).
Figure 4.
Hsa_circ_0010957 regulates activation of STAT3 signaling via miR-125b. (A) Potential binding site and the mutant site of STAT3 3′UTR and miR-125b. (B) Luciferase assay showing overexpression of miR-125b decreased the luciferase activity of wide-type STAT3 3′UTR vector, while not the mutant vector. (C and D) STAT3 protein level after transfecting miR-125b mimics or inhibitor into CD4+ T cells of patients with SLE. (E) STAT3 and phosphorylated STAT3 protein level after transfecting hsa_circ_0010957 siRNA and miR-125b inhibitor into CD4+ T cells of patients with SLE. (F) Silence efficiency of STAT3 in CD4+ T cells of SLE detected by reverse transcription-quantitative PCR. (G-I) IL-18, IL-6 and IL-17 levels detected in cell supernatant after transfecting STAT3 siRNA into CD4+ T cells of patients with SLE. Data are shown as mean ± SD *P<0.05. NS, no significance; SLE, systemic lupus erythematosus; wt, wild-type; mut, mutant type; UTR, untranslated region; miR, microRNA; siRNA, small interfering RNA; NC, negative control; IL, interleukin.
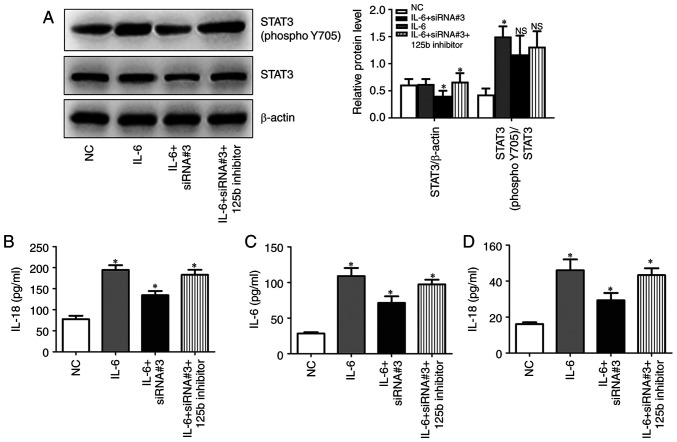
Hsa_circ_0010957 depletion blocks IL-6-induced activation of STAT3 signaling
Given that serum IL-6 levels are increased in SLE, and IL-6 is an established activator of STAT3 signaling, the role of hsa_circ_0010957 in IL-6/STAT3 signaling was explored. CD4+ T cells obtained from patients with SLE were incubated with IL-6 and then transfected with hsa_circ_0010957 siRNA and miR-125b inhibitor. Silencing hsa_circ_0010957 blocks IL-6-induced activation of STAT3 signaling via miR-125b (Fig. 5A). It was also observed that silencing hsa_circ_0010957 abolished IL-6-induced secretion of inflammatory cytokines (Fig. 5B-D).
Figure 5.
Hsa_circ_0010957 depletion blocks IL-6 induced activation of STAT3 signaling. (A) STAT3 and phosphorylated STAT3 protein levels when CD4+ T cells were treated with IL-6 and transfected with hsa_circ_0010957 siRNA and miR-125b inhibitor. (B-D) IL-18, IL-6 and IL-17 levels detected in cell supernatant when CD4+ T cells were treated with IL-6 and transfected with hsa_circ_0010957 siRNA and miR-125b inhibitor. Data are shown as mean ± SD *P<0.05. NS, no significance; IL, interleukin; miR, microRNA; siRNA, small interfering RNA; NC, negative control.
Discussion
In recent years, circRNAs have garnered attention as a new class of noncoding RNA. CircRNAs are potential biomarkers and therapeutic targets for many diseases; however, few studies have been done on circRNAs in SLE. The present study focused on hsa_circ_0010957, whose role in SLE was unknown.
CircRNAs are prospective biomarkers for many diseases, and it was demonstrated that hsa_circ_0010957 might serve as a potential biomarker for SLE. The present study found that hsa_circ_0010957 expression increases in SLE and is a good diagnostic indicator for differentiating patients with active SLE from inactive or no disease.
CircRNAs act as miRNA sponges, thus influencing their function (17,18). In the present study, it was found that hsa_circ_0010957 was a sponge for miR-125b in SLE. miR-125b is downregulated in SLE and inhibits autophagy by targeting UVRAG (19). Low expression of miR-125b, mainly in T cells, is inversely correlated with lupus nephritis and contributes to the pathogenesis of SLE (20). Here, it was shown that hsa_circ_0010957 modulates the expression of inflammatory cytokines IL-18, IL-6 and IL-17 via miR-125b. These cytokines are increased in SLE and play vital roles in disease progression (21–23).
Subsequently, it was shown that the hsa_circ_0010957/miR-125b axis activated STAT3 signaling via mediating STAT3 expression. STAT3, an essential element of the JAK-STAT signal pathway (24), was reported to participate in the pathogenesis of lupus-susceptible mice (25). Increased expression of STAT3 in SLE T cells promotes chemokine-mediated cell migration (26). It was found that knockdown of STAT3 inhibited the secretion of IL-18, IL-6 and IL-17 and concluded that hsa_circ_0010957/miR-125b influences the secretion of these inflammatory cytokines via regulating STAT3 signaling.
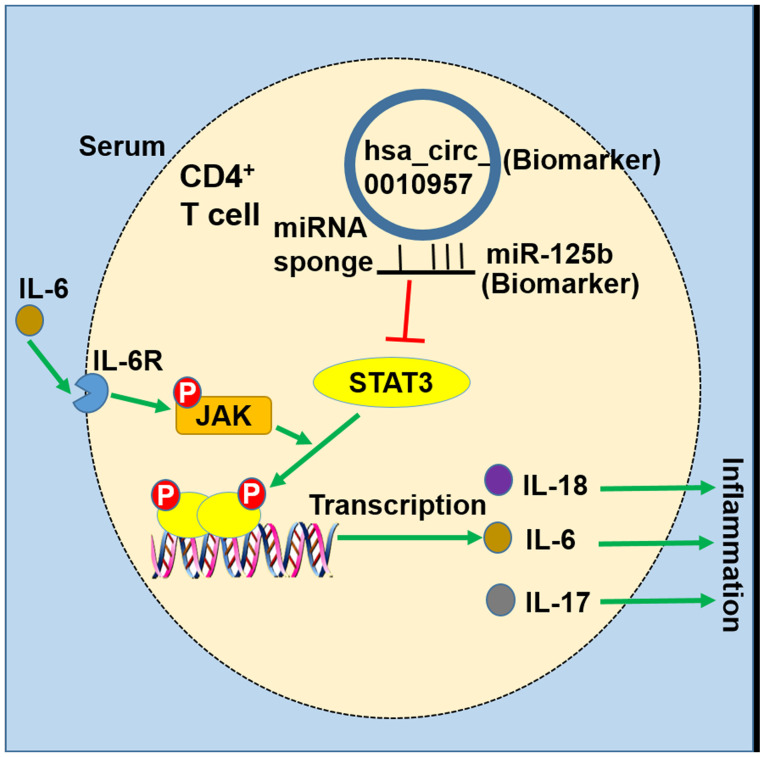
In addition, it was revealed that silencing hsa_circ_0010957 inhibited IL-6-induced inflammatory cytokines secretion through restraining STAT3 signaling. High IL-6 level presents in the serum of patients with SLE (19,20) and is responsible for the activation of STAT3 signaling (27). Another role of hsa_circ_0010957 in impeding the proinflammatory effect of IL-6 was also demonstrated. The proposed model of hsa_circ_0010957/miR-125b/STAT3 signaling in SLE is illustrated in Fig. 6.
Figure 6.
Schematic diagram exhibiting the hsa_circ_0010957/miR-125b/STAT3 signaling pathway in SLE. Upregulated hsa_circ_0010957 in CD4+ T cells of SLE is able to act as a sponge for miR-125b, and thereby activated the STAT3 signaling by suppressing miR-125b. Hsa_circ_0010957 in SLE is involved in the secretion of inflammatory cytokines of CD4+ T cells via regulating miR-125b/STAT3 signaling. SLE, systemic lupus erythematosus; miR/miRNA, microRNA; IL, interleukin.
In conclusion, the present findings show that the increased level of hsa_circ_0010957 in SLE is involved in the secretion of inflammatory cytokines of CD4+ T cells via regulating miR-125b/STAT3 signaling. Hsa_circ_0010957 and miR-125b can be used as a potential biomarker and therapeutic target for SLE.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
This work was supported by the Jinhua Public Welfare Technology Application Research Project (grant no. 2019-4-002).
Funding
This work was supported by the Jinhua Public Welfare Technology Application Research Project (grant no. 2019-4-002).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YZ, SH and HD conceived the study and performed the experiments. YW and XS analyzed and interpreted the data. YZ and SH confirm the authenticity of all the raw data. YZ and SH wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Human Ethics Committee of Jinhua Municipal Central Hospital (approval no. 2019-118) and written informed consents were obtained from all participants in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Miyagawa-Hayashino A, Yoshifuji H, Kitagori K, Ito S, Oku T, Hirayama Y, Salah A, Nakajima T, Kiso K, Yamada N, et al. Increase of MZB1 in B cells in systemic lupus erythematosus: Proteomic analysis of biopsied lymph nodes. Arthritis Res Ther. 2018;20:13. doi: 10.1186/s13075-018-1511-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fanouriakis A, Tziolos N, Bertsias G, Boumpas DT. Update οn the diagnosis and management of systemic lupus erythematosus. Ann Rheum Dis. 2021;80:14–25. doi: 10.1136/annrheumdis-2020-218272. [DOI] [PubMed] [Google Scholar]
- 3.Morel L. Immunometabolism in systemic lupus erythematosus. Nat Rev Rheumatol. 2017;13:280–290. doi: 10.1038/nrrheum.2017.43. [DOI] [PubMed] [Google Scholar]
- 4.Tsokos GC, Lo MS, Costa Reis P, Sullivan KE. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat Rev Rheumatol. 2016;12:716–730. doi: 10.1038/nrrheum.2016.186. [DOI] [PubMed] [Google Scholar]
- 5.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z, Yang T, Xiao J. Circular RNAs: Promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai CY, Shen CY, Liu CW, Hsieh SC, Liao HT, Li KJ, Lu CS, Lee HT, Lin CS, Wu CH, et al. Aberrant non-coding RNA expression in patients with systemic lupus erythematosus: Consequences for immune dysfunctions and tissue damage. Biomolecules. 2020;10:1641. doi: 10.3390/biom10121641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo Q, Li X, Fu B, Zhang L, Fang L, Qing C, Guo Y, Huang Z, Li J. Expression profile and diagnostic value of circRNAs in peripheral blood from patients with systemic lupus erythematosus. Mol Med Rep. 2021;23:1. doi: 10.3892/mmr.2020.11639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ouyang Q, Huang Q, Jiang Z, Zhao J, Shi GP, Yang M. Using plasma circRNA_002453 as a novel biomarker in the diagnosis of lupus nephritis. Mol Immunol. 2018;101:531–538. doi: 10.1016/j.molimm.2018.07.029. [DOI] [PubMed] [Google Scholar]
- 10.Miao Q, Zhong Z, Jiang Z, Lin Y, Ni B, Yang W, Tang J. RNA-seq of circular RNAs identified circPTPN22 as a potential new activity indicator in systemic lupus erythematosus. Lupus. 2019;28:520–528. doi: 10.1177/0961203319830493. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Zhang C, Wu Z, Chen Y, Shi W. CircIBTK inhibits DNA demethylation and activation of AKT signaling pathway via miR-29b in peripheral blood mononuclear cells in systemic lupus erythematosus. Arthritis Res Ther. 2018;20:118. doi: 10.1186/s13075-018-1618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li LJ, Zhu ZW, Zhao W, Tao SS, Li BZ, Xu SZ, Wang JB, Zhang MY, Wu J, Leng RX, et al. Circular RNA expression profile and potential function of hsa_circ_0045272 in systemic lupus erythematosus. Immunology. 2018;155:137–149. doi: 10.1111/imm.12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C, Wang X, Chen Y, Wu Z, Zhang C, Shi W. The down-regulation of hsa_circ_0012919, the sponge for miR-125a-3p, contributes to DNA methylation of CD11a and CD70 in CD4 T cells of systemic lupus erythematous. Clin Sci (Lond) 2018;132:2285–2298. doi: 10.1042/CS20180403. [DOI] [PubMed] [Google Scholar]
- 14.Petri M, Orbai AM, Alarcón GS, Gordon C, Merrill JT, Fortin PR, Bruce IN, Isenberg D, Wallace DJ, Nived O, et al. Derivation and validation of the systemic lupus international collaborating clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum. 2012;64:2677–2686. doi: 10.1002/art.34473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Zhao C, Zhang C, Mei X, Song J, Sun Y, Wu Z, Shi W. Increased HERV-E clone 4-1 expression contributes to DNA hypomethylation and IL-17 release from CD4+ T cells via miR-302d/MBD2 in systemic lupus erythematosus. Cell Commun Signal. 2019;17:94. doi: 10.1186/s12964-019-0416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 18.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 19.Cao W, Qian G, Luo W, Liu X, Pu Y, Hu G, Han L, Yuan L, Xiao A, Deng D. miR-125b is downregulated in systemic lupus erythematosus patients and inhibits autophagy by targeting UVRAG. Biomed Pharmacother. 2018;99:791–797. doi: 10.1016/j.biopha.2018.01.119. [DOI] [PubMed] [Google Scholar]
- 20.Luo X, Zhang L, Li M, Zhang W, Leng X, Zhang F, Zhao Y, Zeng X. The role of miR-125b in T lymphocytes in the pathogenesis of systemic lupus erythematosus. Clin Exp Rheumatol. 2013;31:263–271. [PubMed] [Google Scholar]
- 21.Mende R, Vincent FB, Kandane-Rathnayake R, Koelmeyer R, Lin E, Chang J, Hoi AY, Morand EF, Harris J, Lang T. Analysis of serum interleukin (IL)-1β and IL-18 in systemic lupus erythematosus. Front Immunol. 2018;9:1250. doi: 10.3389/fimmu.2018.01250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Y, Tao H, Gong Y, Chen F, Li C, Yang X. Changes of serum IL-6, IL-17, and complements in systemic lupus erythematosus patients. J Interferon Cytokine Res. 2019;39:410–415. doi: 10.1089/jir.2018.0169. [DOI] [PubMed] [Google Scholar]
- 23.Monzavi SM, Alirezaei A, Shariati-Sarabi Z, Afshari JT, Mahmoudi M, Dormanesh B, Jahandoost F, Khoshdel AR, Rezaie AE. Efficacy analysis of hydroxychloroquine therapy in systemic lupus erythematosus: A study on disease activity and immunological biomarkers. Inflammopharmacology. 2018;26:1175–1182. doi: 10.1007/s10787-018-0512-y. [DOI] [PubMed] [Google Scholar]
- 24.Zegeye MM, Lindkvist M, Fälker K, Kumawat AK, Paramel G, Grenegård M, Sirsjö A, Ljungberg LU. Activation of the JAK/STAT3 and PI3K/AKT pathways are crucial for IL-6 trans-signaling-mediated pro-inflammatory response in human vascular endothelial cells. Cell Commun Signal. 2018;16:55. doi: 10.1186/s12964-018-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pramanik R, Jørgensen TN, Xin H, Kotzin BL, Choubey D. Interleukin-6 induces expression of Ifi202, an interferon-inducible candidate gene for lupus susceptibility. J Biol Chem. 2004;279:16121–16127. doi: 10.1074/jbc.M313140200. [DOI] [PubMed] [Google Scholar]
- 26.Harada T, Kyttaris V, Li Y, Juang YT, Wang Y, Tsokos GC. Increased expression of STAT3 in SLE T cells contributes to enhanced chemokine-mediated cell migration. Autoimmunity. 2007;40:1–8. doi: 10.1080/08916930601095148. [DOI] [PubMed] [Google Scholar]
- 27.Johnson DE, O'Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15:234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.