Abstract
BACKGROUND:
Antimicrobial peptides (AMPs) are small proteins with potent antibacterial, antiviral, and antifungal activity. AMPs are ubiquitous among multicellular eukaryotes, with most plant and animal species expressing dozens of distinct AMP genes in epithelial tissues and in response to infection. The diversity and potency of AMPs make them attractive candidates for translational application, and several are already in clinical trials. However, if AMPs are to be used effectively and sustainably, it will be imperative to understand their natural biology and evolution in order to lessen the risk of collateral harm and avoid the resistance crisis currently facing conventional antibiotics.
ADVANCES:
For most of the past 25 years, the prevailing wisdom has been that AMPs are generally nonspecific and functionally redundant—largely interchangeable provided that they were produced quickly enough to a threshold that could contain infection. Support for this model was drawn from molecular evolutionary observations that AMP genes are rapidly duplicated and pseudogenized within and between species, often with little evolution at the level of the primary amino acid sequence. Furthermore, it was believed that the biochemical simplicity of AMPs reflected fundamentally irresistible modes of action, including permeabilization of the cell envelope through the formation of open pores, which was assumed to largely prevent bacterial evolution of resistance.
New evidence within the past 5 years, however, has begun to overturn that model. We now know that AMPs can exhibit remarkable levels of specificity and that some of the evolutionary degradation of AMP gene families may be adaptive. We are learning that genetic variability in AMPs, even at the level of single amino acids, can dramatically alter resistance to infection. There are now multiple documentations of convergent evolution of identical amino acid variants between species and of shared allelic diversity between species. It is increasingly clear that AMPs are highly functionally diversified and that they play roles in varied biological processes, including the regulation of symbiotic communities. It is also becoming apparent that bacteria can evolve resistance to AMPs, although the pharmacodynamics and mechanisms of killing of AMPs are much more favorable than those of conventional antibiotics for the prevention of resistance evolution.
OUTLOOK:
AMPs hold considerable promise for translational applications, but developing their potential will require more sophisticated foundational understanding. AMPs function synergistically in vivo, and emerging evidence indicates that their activities in biological contexts may not be fully captured with classical in vitro assays. Further development of mathematical approaches to study synergies will be required, especially for higher-order interactions, in order to rationally develop cocktails that have high efficacy at low concentrations. Synergies between AMPs and conventional antibiotics should be exploited to rescue drugs that are currently lost to resistance. AMPs should be mined from all domains of life: Although more than 3100 naturally occurring AMPs have been described from taxa representing the breadth of life on earth, almost 40% of AMPs under clinical trial are of human origin. This is potentially risky because any evolved resistance to those AMPs may result in collateral resistance to endogenous human immunity. The biochemical properties and pharmacodynamics of AMPs make them far more refractory to resistance evolution than conventional antibiotics, but care should still be taken to deploy them responsibly. Translational use of AMPs in clinical and other applied settings will be greatly enhanced by understanding how specific AMPs function in their natural contexts and how their evolutionary history may predict their future utility. If we combine the insights from the evolutionary diversification of the AMPs, their activity in the context of synergistic cocktails, and our growing understanding of how to limit resistance evolution, we may avoid repeating the mistakes that have resulted in the current crisis of antibiotic resistance.
Antimicrobial peptides (AMPs) are essential components of immune defenses of multicellular organisms and are currently in development as anti-infective drugs. AMPs have been classically assumed to have broad-spectrum activity and simple kinetics, but recent evidence suggests an unexpected degree of specificity and a high capacity for synergies. Deeper evaluation of the molecular evolution and population genetics of AMP genes reveals more evidence for adaptive maintenance of polymorphism in AMP genes than has previously been appreciated, as well as adaptive loss of AMP activity. AMPs exhibit pharmacodynamic properties that reduce the evolution of resistance in target microbes, and AMPs may synergize with one another and with conventional antibiotics. Both of these properties make AMPs attractive for translational applications. However, if AMPs are to be used clinically, it is crucial to understand their natural biology in order to lessen the risk of collateral harm and avoid the crisis of resistance now facing conventional antibiotics.
Graphical Abstract
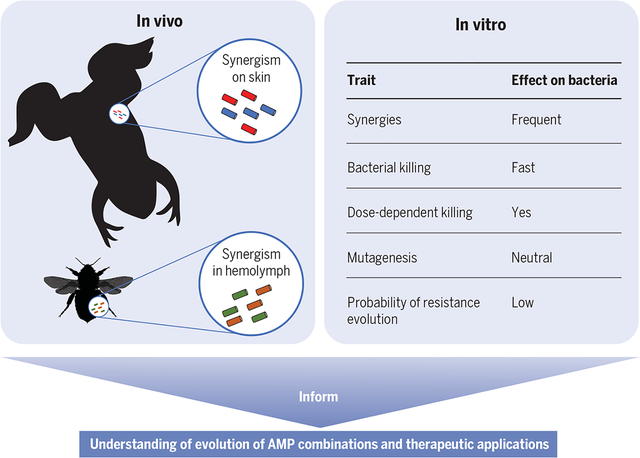
The combined insight from studying AMPs across the tree of life and the adaptive evolution of AMPs will inform their application and the understanding of AMPs in their natural context. In nature, AMPs are highly diverse, with most AMPs (more than 1000) described in Amphibia. They are released as synergistic cocktails in vivo. In vitro studies found that synergies are frequent and that other traits of AMPs result in a low probability of resistance evolution compared with conventional antibiotics.
Antimicrobial peptides (AMPs) are small proteins with antibacterial, antiviral, and antifungal activity. Sometimes referred to as “host-defense peptides,” AMPs are ubiquitous in the epithelial barriers and systemic induced defenses of multicellular eukaryotes (1). They are highly diverse within and across species, with most plant and animal genomes encoding 5 to 10 distinct AMP gene families that range in size from one to more than 15 paralogous genes. The diversity and potency of AMPs make them attractive targets for development as antimicrobial drugs (2) and surface antiseptics (3), and dozens of AMPs are currently being evaluated in clinical trials (4). More than 3100 AMPs have been described from varied plant and animal sources (5), diversified by rapid evolution between species as aptly illustrated by the diversity of AMPs in the most speciose groups of animals, the insects [several AMP families are specific to particular insect orders (6)].
The classically understood model of AMP efficacy (1) was that they exert microbial killing at threshold doses through simultaneous targeting of diverse aspects of microbial biology. Their inferred simplicity and redundancy were assumed to prevent evolution of resistance, and their lack of specificity was assumed to establish blanket protection against microbes. However, more recent findings are forcing a reevaluation of that model. New evidence indicates unexpectedly high degrees of specificity and synergisms among AMPs. We also now realize that resistance evolution is possible. Although some AMPs have evolved functions such as modulation of the immune system (7), deterrence of herbivores (8) and anticancer effects (9), we focus here on antimicrobial activity. However, additional functions can lead to complex and sometimes contradictory natural selection on AMP genes. We argue that it is essential to understand AMP biology in natural contexts before pursuit of translational application in order to maximize effectiveness and to avoid repeating the tragic mistakes of misuse that have led to widespread bacterial resistance to conventional antibiotics.
Classical perspective on AMPs
The basic design of antimicrobial peptides is simple (1). They are short peptides with a net-positive charge that attracts them to the generally negatively charged membranes of bacteria. The hydrophilic and hydrophobic amino acids of AMPs are structurally segregated to provide solubility in both aqueous and lipid-rich environments. The shortest AMPs may be as small as 15 to 20 amino acids in length, and even the largest are not more than ~150 amino acids (5). The biochemical simplicity of AMPs allows them to be easily evolved de novo, and certain three-dimensional structures, such as those of defensins, have independently evolved repeatedly across plants, insects, and vertebrate animals (10, 11).
The stereotypical mechanism of AMP action is to integrate into the bacterial cell membrane and disrupt its integrity, resulting directly or indirectly in cell lysis, although AMPs may also have more complex activities, including metabolic and translational inhibition (12, 13) and formation of nanonets (14, 15). Naturally cooccurring AMPs with distinct functions can synergize, as illustrated by an increasing number of studies showing that some AMPs permeabilize membranes to enable entry of other AMPs that have intracellular targets (16). For example, bumblebee hymenoptaecin opens pores in bacterial membranes that allow abaecin to enter and bind bacterial DnaK (17). In vertebrates, perforins form pores that allow lethal cationic cargo to reach the cytoplasm (18). Similarly, distinct AMPs may synergize to permeabilize bacterial membranes, as illustrated by the interaction between magainin 2 and PGLa (Fig. 1) (19). Eukaryotes can rapidly deploy multiple distinct classes of AMP simultaneously in response to challenge, in some cases up-regulating AMP gene expression several hundredfold within hours of infection, effectively killing microbes through simultaneous targeting of multiple critical cellular functions.
Fig. 1. Analyzing and depicting synergism.
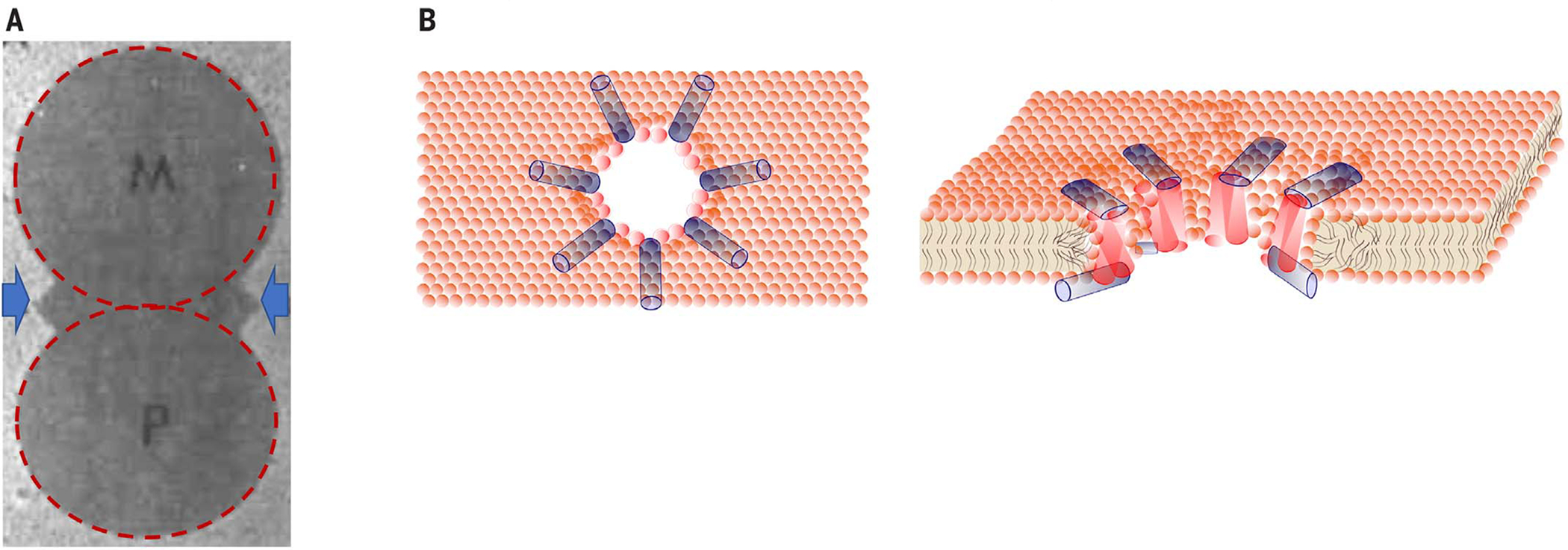
(A and B) Synergies between Magainin 2 (M) and PGLa (P) visualized with (A) a zone of inhibition assay and (B) a model of the interaction on the membrane. (A) Identical molar amounts of Magainin 2 and PGLa were applied to a freshly inoculated lawn of E. coli and photographed after a 24-hour incubation at 37°C. The sharp zones of bacterial killing reflect the steep concentration dependence of bactericidal activity. Arrows highlight the zones of activity resulting from synergy. (B) The synergistic interaction between PGLa and Magainin 2. Antiparallel PGLa dimers (red) span the membrane. Magainin 2 monomers (blue) lie on each surface of the membrane and contact each PGLa dimer tail to tail. [Redrawn from (19).]
These observations coalesced into a classical model of AMP function (1, 20, 21) that relied on three key interpretations of the data. First, it was presumed that host production of functionally diverse AMPs would result in more effective killing, in analogy to therapeutic application of multiple distinct antibiotics. Second, it was inferred that AMP dose was probably more important than specific peptide identity, making AMPs largely interchangeable provided that they were quickly produced above a threshold level that could rapidly overwhelm the infection. Last, the biochemically simple and highly efficient killing mechanism of even single AMPs was predicted to prevent bacterial evolution of resistance. These interpretations were fully consistent with strong up-regulation of AMP production in response to infection (22) and were superficially consistent with molecular evolutionary and comparative genomic data that showed rapid duplication, deletion, and pseudogenization of individual AMP genes while keeping total gene family size fairly constant (23–26), with little indication of adaptive amino acid diversification [(21), but also (27, 28)]. The classical model of interchangeable AMPs at threshold concentration was consistent with the data that were available over the roughly 20 years between 1994 and 2014. However, it is probably wrong.
Rethinking AMP function and evolution
The previously widespread belief that AMPs have generic, broad-spectrum activity has been recently challenged by new data. For example, in vivo disruption of individual AMP genes in the beetle Tenebrio molitor causes differential sensitivity to infection by the bacterium Staphylococcus aureus (29). In Drosophila melanogaster and its sister species Drosophila simulans, naturally occurring null alleles of the AMP gene Diptericin A cause acute sensitivity to infection by the bacterium Providencia rettgeri but not to other bacteria, including close relatives of P. rettgeri (Fig. 2) (30). Furthermore, a single polymorphic amino acid substitution in the Diptericin A peptide is sufficient to specifically alter resistance to P. rettgeri, and this susceptible mutation has arisen at least five independent times across the genus Drosophila (31). These findings suggest a previously unsuspected specificity in AMP activity.
Fig. 2. Small evolutionary changes matter.
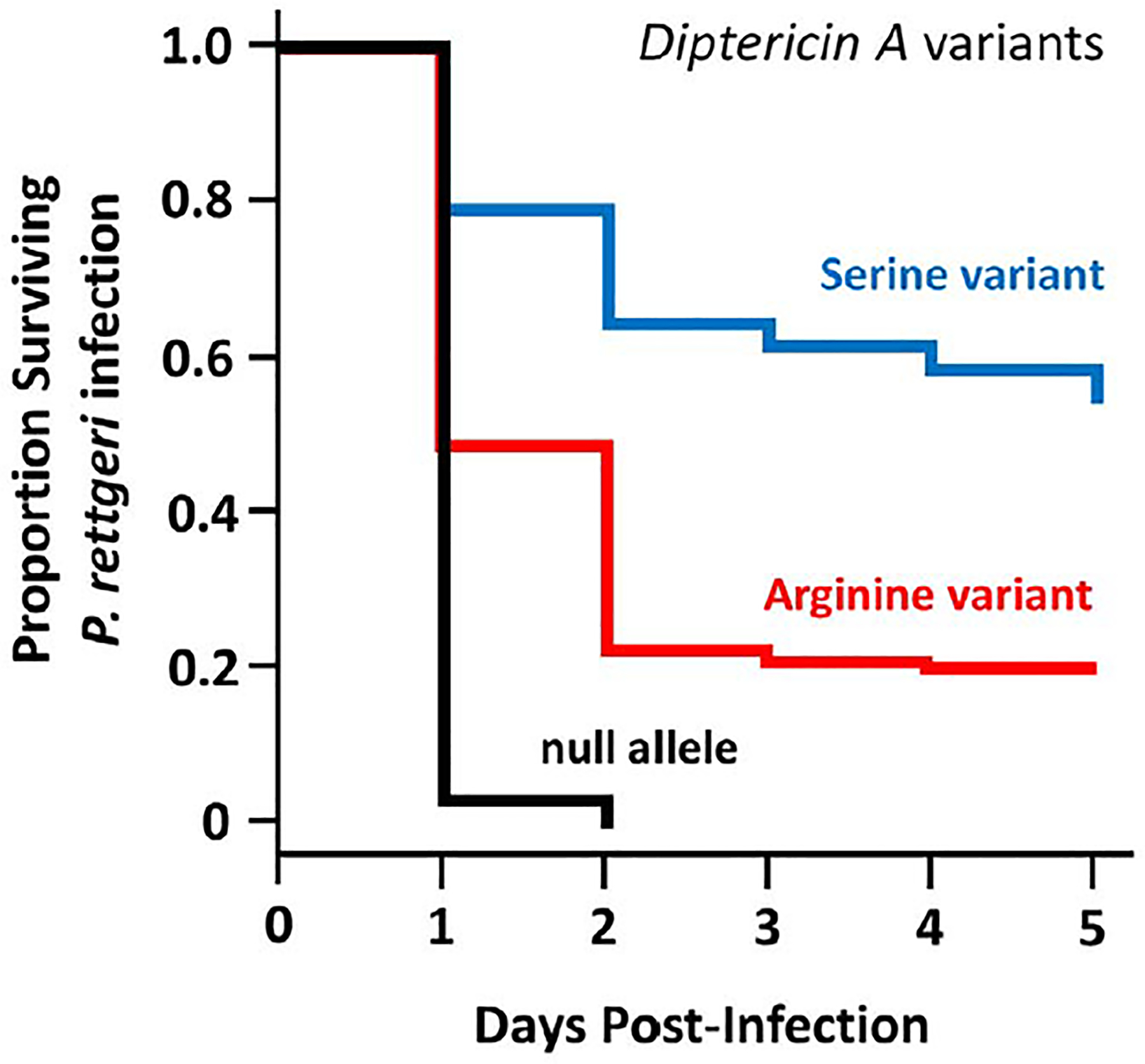
Small evolutionary changes in amino acid composition of AMPs can have major consequences for host survival during infection (30). In D. melanogaster, different alleles of Diptericin (arginine and serine) show pathogen-specific activity here against P. rettgeri. Lines of D. melanogaster with null alleles (black) show higher mortality than do lines homozygous for arginine (red). Lines homozygous for serine (blue) show the highest survival.
Expanding the work in D. melanogaster, Hanson and colleagues systematically deleted individual AMP genes and gene families and showed that distinct small subsets of the fruit fly AMP repertoire are wholly responsible for the control of diverse bacterial infections (32). Those observations were surprising because bacterial infections induce transcriptional expression of broad suites of AMP genes (33), but most of these appear to be functionally irrelevant to suppressing the pathogen in question. However, broad transcriptional induction makes sense in natural contexts, particularly if infections in nature are typically polymicrobial or if the host is unable to finely discriminate pathogens at the recognition stage (34). In these cases, comprehensive induction of AMPs at early stages of infection, including as a prophylactic response to wounding (33), would ensure activation of the subset with specific activity and would guarantee the most effective protection.
The high rate of duplication, deletion, pseudogenization, and de novo origin of AMP genes (23–26) had previously been interpreted as evidence that individual genes were superfluous if a threshold dose of AMPs was produced. However, the observations that laboratory mutants, as well as naturally occurring null alleles and point mutations, have profound and specific effects on resistance to infection are in complete contrast to the classical threshold model of interchangeable AMPs. An alternative interpretation is that the significantly elevated rates of gene duplication and diversification reflect adaptation to suites of microbes (35) and that AMP gene loss may be adaptive if the physiological costs of producing a given AMP outweigh its ecological benefit (36). An adaptive diversification model might predict that gene family expansions should be coupled with amino acid diversification of the encoded peptides. Consistent with this prediction, there are many reports of AMP gene family radiations coupled with sequence diversification, particularly in vertebrates (27, 28, 37). Furthermore, an adaptive diversification model might predict that newly duplicated genes would acquire distinct expression patterns, and this has been observed where it has been examined (38, 39).
Adaptation is also revealed in the parallel and/or convergent evolution of the same amino acid sequence in different species. Natural selection may promote evolutionary convergence in species that share ecological pressures, or adaptive maintenance of polymorphism within species if alternative alleles are more effective at killing specific pathogens that are commonly present at different times or places in the environment. Recent analyses have revealed a surprisingly high rate of convergent evolution in AMP genes sampled from organisms as diverse as mussels (40), birds (41), and multiple species of Drosophila (31). In some cases, multiple species are polymorphic for the same or similar alleles of AMP genes. For example, D. melanogaster and D. simulans have each independently evolved a Ser/Arg polymorphism at the same codon in Diptericin A, and the alternative alleles are demonstrably different in resistance to infection in both species (Fig. 2) (30). Such allele sharing has been interpreted as evidence that AMP polymorphism is adaptively maintained in populations. Allele sharing in AMP genes has been found in humans (42), frogs (43), passerine birds (41), waterfowl (44), codfish (45), mussels (40), and Drosophila (46), although typically without phenotypic analysis of the alternative alleles. These multiple examples may represent just the tip of the iceberg because population sequence samples from multiple species are required to detect shared polymorphisms, and sequences from constellations of related species are required to detect convergent evolutionary fixations, but these are rarely available. Additionally, molecular evolutionary signatures of adaptively maintained polymorphisms can be difficult to detect (47, 48), particularly in species with large population sizes (31).
Symbionts and coevolution
In addition to combatting infectious pathogens, AMPs are used to regulate bacterial symbionts and communities in the gut and other tissues (49). This is true even for organisms with fairly simple body plans, such as the cnidarian Hydra (50). In the mammalian gut, a growing body of work shows the importance of AMPs secreted by the Paneth cells in shaping the gut microbiota and hence determining healthy or pathological phenotypes (49). Many members of the gut microbiota display high intrinsic AMP resistance (51), suggesting that AMPs may be an important tool for establishment and maintenance of healthy communities. AMPs are also important for the regulation of highly co-adapted mutualists. For example, the weevil Sitophilus zeamais carries an obligate symbiont, Candidatus Sodalis pierantonius str. SOPE, in specialized cells called bacteriocytes. The symbiont shows hallmarks of tightly coevolved mutualism, including gene loss as it becomes more dependent on its partner (52). S. zeamais expresses one AMP, coleoptericin-A (ColA), exclusively in the bacteriocytes, and knocking down ColA expression results in symbiont escape into surrounding tissues (53). Similarly, the specialized plant nodules that harbor nitrogen-fixing symbionts in the leguminous plant Medicago truncatula express more than 700 peptides, many of which are cysteine-rich AMPs (54). The coevolved symbiont Sinorhizobium meliloti has evolved a peptidase that protects the symbionts against harmful effects of these AMPs and allows them to gain a competitive advantage over other microbes in that niche (54).
AMPs are additionally crucial to the well-documented example of bobtail squid, which obtains a bioluminescent bacterial symbiont, Vibrio fischeri, from the water column and sequesters it in a specialized light organ. The squid diurnally flushes its bacterial symbionts and then allows recolonization of the light organ from bacteria in the surrounding water. Specific colonization is ensured by AMPs that prevent V. fischeri from colonizing inappropriate tissues and that block colonization by undesirable bacteria. Specificity in this remarkable mutualism is further guaranteed by acidic mucus on the surface of the light organ that primes V. fischeri to become resistant to the AMPs and allows it to occupy this niche (55).
Sublethal concentrations of AMPs are deployed in certain host-symbiont interactions. For example, obligate microbial symbionts often undergo genome erosion (56) that makes them dependent on the host for exchange of metabolites. In many cases, genes that code for membrane transport are vulnerable to genome erosion. However, expression of AMPs at sublethal doses may substitute for the transport function by permeabilizing bacterial membranes without killing the cells in their symbiotic compartments (57). Similarly, low concentrations of AMP-like peptides that would be lethal at higher concentrations also stimulate terminal differentiation and metabolic specialization of the nitrogen-fixing bacteria in leguminous plants (58). How low concentrations of host AMPs influence bacterial physiology is poorly understood, and whether they contribute to bacterial evolution of resistance or induction of phenotypic resistance in other contexts is not known.
Synergism
Accumulating evidence shows marked functional synergism occurs among distinct AMPs (Fig. 1) (59–62). Synergism in vivo may reduce the chances of resistance evolving (63), especially by generalist pathogens that infect multiple hosts that express different combinations of AMPs. Yet, determining synergy from dose-response curves can be quite challenging, particularly because they are not linear. Currently, two main models are used to calculate synergies (64–66), and further development of these reference models will be crucial to our understanding of AMP interactions in both natural systems and drug applications, especially for higher-order interactions.
In vitro estimation of individual AMP activities may not reflect in vivo efficacy when synergisms among AMPs are common. For example, a recent report highlighted mismatches between the in vitro activities of the T. molitor AMPs Tenecin 1, 2, and 3, compared with infection resistance profiles observed when each of the genes that encode them was disrupted in vivo by means of RNA interference (29). Tenecin 2 showed no effect against S. aureus in vitro (67), but beetles deficient of Tenecin 2 suffered measurably increased mortality upon S. aureus infection. There are numerous reasons why in vitro assays may not reflect in vivo activities, including differences in local pH or salt concentrations, nutritional or osmotic stress on microbes, and synergisms among AMPs or between AMPs and other components of the immune system. This is an important problem because in vitro assays have been used as a tool for estimating the efficacy of particular AMPs against specific microbes, but results from experiments that do not accurately capture biological conditions may be misleading (20, 68).
Translational challenges for AMPs
Against the backdrop of accelerating antibiotic resistance (69, 70), antimicrobial peptides hold promise for use in clinical and veterinary settings. However, to effectively deploy AMPs and sustain their value, we need to learn from both the historical misuse of antibiotics (69, 70) and the evolutionary biology and natural pharmacology of AMPs.
Instead of reflexively relying on the current standard of testing minimum inhibitory concentration (MIC) of individual components in vitro (68), we need to understand and measure synergisms among AMPs in vivo so that we can exploit them effectively for microbial control at low concentrations. In addition, AMPs can synergize with conventional antibiotics (68, 71, 72), which raises the prospect that antibiotics that have been lost to resistance could be resurrected. For example, a recent study on multidrug-resistant Gram-negative bacteria showed strong synergisms between the antibiotic azithromycin—which showed no activity against Gram-negative bacteria in standard MIC tests—and the AMPs colistin and LL-37 (68). In another example, the AMP known as SAAP-148 was shown to be effective at killing drug-resistant bacteria even within biofilms in vivo on mouse skin (73). Host-directed therapies to boost natural AMP production can improve infection control. Application of compounds such as phenylbutyrate and aroylated phenylenediamines have been shown to boost LL-37 induction by 20- to 30-fold, and orally treated rabbits showed a decrease of bacterial load by up to five orders of magnitude (74). Synergisms can be synthetically generated very successfully. Chimeric peptide antibiotics that link polymyxin and murepavidin have been demonstrated to be active against multidrug-resistant Gram-negative ESKAPE pathogens (Enterococcus faecium, S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) (75). Combination therapies can be envisaged that integrate synthetic AMPs, stimulate natural AMPs, and deploy conventional antibiotics for the treatment of recalcitrant multidrug-resistant bacterial infections.
The low rates of evolved bacterial resistance to AMPs need to be maintained as peptide drugs are rolled out in the clinic. One route may be through collateral sensitivity (76), in which evolved resistance to one antibacterial renders bacteria sensitive to another. Collateral sensitivity to AMPs has been observed in Escherichia coli strains experimentally selected to be resistant to a variety of antibiotics (77). By contrast, independent studies in which S. aureus was selected for resistance to AMPs resulted in cross-resistance to several AMPs, including the peptide antibiotic Daptomycin, although not to other antibiotics (78, 79). Both collateral sensitivity and cross-resistance have been reported in multiple studies of E. coli selected for resistance against AMPs (80). Thus, active monitoring of target microbe populations and management of therapeutic AMP deployment will be essential.
The pharmacodynamics of AMPs reduces the probability of resistance evolution (Fig. 3) (81). Most AMPs interact with the bacterial cell surface and are not directly mutagenic, whereas many antibiotics can elevate bacterial mutation rates by triggering bacterial stress responses such as the SOS and rpoS pathways (82). AMPs kill faster than antibiotics—within minutes instead of hours (83)—allowing many fewer bacterial generations in which resistance could evolve. Because resistance to AMPs tends to be by nonspecific mechanisms, there may be fewer mutational routes by which resistance to AMPs can evolve (84) and lower likelihood of horizontal gene transfer that confers resistance (85). Perhaps most importantly, the concentration range of intermediate efficacy, in which resistance can evolve, is smaller for AMPs than for antibiotics. This phenomenon is captured by the Hill coefficient (the slope of the pharmacodynamic curve), which describes the window between concentrations that have no effect and concentrations that result in complete killing (Fig. 3A) (63). AMPs have high Hill coefficients, which means that there is a small window in which there is selective pressure on the bacteria to evolve resistance while they are still viable enough to do so. Combinations of AMPs have even higher Hill coefficients and hence reduce the risk of resistance evolution even further (81).
Fig. 3. Resistance evolution against AMPs.
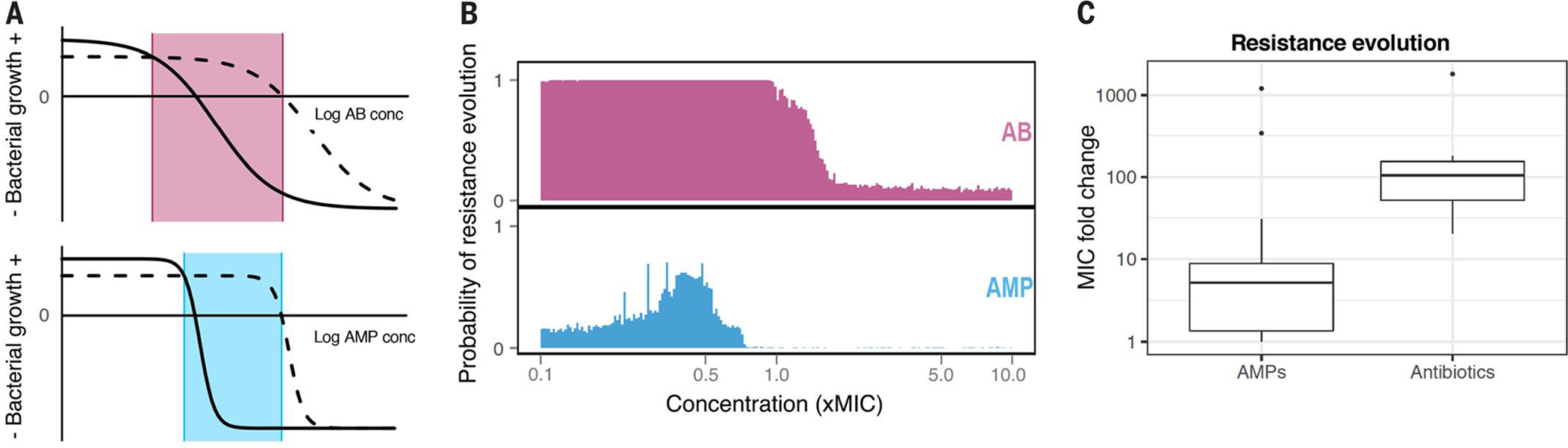
(A) Pharmacodynamics of AMPs differ from those of conventional antibiotics (63). The solid curved line depicts a susceptible bacterial strain, and the dashed line depicts a resistant strain. The respective MICs are shown at the intersections by the solid horizontal line. The Hill coefficient κ, depicting the slope, represents an antibiotic (top, κ = 1), and κ = 4 represents an AMP (bottom) [values are based on (63); typically κ values are higher for AMPs than for antibiotics]. The dose-response curve for the AMP is correspondingly steeper for an AMP, which results in a narrow mutant selection window (light blue) in which genetically resistant mutants are favored. (B) Combining the pharmacodynamical properties of AMPs and comparing them with those of antibiotics, computer simulations predict a lower probability of resistance evolution against AMPs compared with antibiotics. [Adapted from (81).] (C) Experimental resistance evolution of E. coli against 15 AMPs in vitro yields a significantly lower degree of resistance compared with the results for 12 antibiotics, with the exception of two AMPs. [Data are from (80).]
Another advantage of AMPs over antibiotics is that AMPs tend to be less stable than antibiotics and have much lower environmental persistence (86). This is partly a function of synthetic engineering of clinical antibiotics for extended shelf lives. The persistence of antibacterials in the environment at sublethal concentrations continues to select for antibiotic resistance and constitutes an important driver of antibiotic resistance evolution (69, 87). Because many antibiotics were originally evolved by bacteria and fungi for microbe-microbe warfare, soil bacterial communities harbor a rich antibiotic resistome (88). By contrast, presumably because of their typical provenance as defense molecules by multicellular eukaryotes, a recent study of soil microbes did not find high levels of preexisting AMP resistance (80).
Nevertheless, the early notion that bacteria are largely unable to evolve resistance to AMPs (1) has been refuted (89). Resistance to AMPs arises from a range of nonspecific mechanisms, including secretion of proteases and exopolymers, biofilm formation, and activation of efflux pumps. Tolerance of AMPs can be achieved through modification of the cell envelope, including its charge (90). However, a systematic study on resistance evolution against 12 individual AMPs in E. coli (Fig. 3C) (80) reported lower increases in MIC for AMPs than for antibiotics. The same study also showed that the physiological costs of evolving resistance to AMPs were lower than developing resistance to antibiotics. Other concerns, such as “bystander selection” for resistance in nontarget bacteria and phenotypic resistance through dormancy or persister states, may be as relevant for the usage of AMPs (91) as they have previously been shown for antibiotics (92, 93).
The most effective way to prevent widespread, generic resistance to AMPs is likely to be through the judicious application of synergistic and efficacious AMP cocktails that clear bacteria before resistant variants can emerge. In some cases, these cocktails could be carefully designed from a priori knowledge of the characteristics of individual components. However, the ease of synthesizing AMPs also means that random and highly diverse peptide mixtures can be evaluated against target microbes in controlled screens (94). Synthetic diversity of AMPs can also be achieved through random protein cleavage into short fragments that contain a high proportion of disorder-promoting amino acids (95). Although there has been concern that resistance could evolve more quickly against combinations (96), this argument is based on the assumption that resistance mechanisms are specific and therefore that several independent mutations are required for bacteria to evolve resistance. This may hold for antibiotic resistance but could be more tenuous for the generally nonspecific resistance to AMPs (90, 97).
Of greater concern, evolved resistance to therapeutically applied AMPs could result in undesirable cross-resistance to endogenous host AMPs (98). The only explicit test of this hypothesis has been carried out in the model insect Tenebrio (99, 100), in which some AMP-resistant S. aureus survived better in the host, albeit without showing increased virulence. However, similar experiments in a mammalian system or with human AMPs are lacking. This is particularly concerning given that at least 11 of the AMPs that are currently under clinical trial are of human origin (Fig. 4A). There is a much richer diversity of AMPs described from other animals and plants that could be therapeutically applied to humans (Fig. 4B) (5), and it may be wise to exploit those resources in order to limit the risk of evolved cross-resistance to endogenous host defense. Even the large diversity of known AMPs is a poor reflection of the diversity of biological sources that are potentially available, which is acutely illustrated by the fact that only about 16% of described AMPs are from arthropods despite arthropods constituting more than 60% of eukaryotic species on the planet. The challenge now is to infer from the selection exerted on a particular AMP whether it is a useful candidate for further experimental studies. Can we predict a given AMP’s potential utility in treatment from sequence alone, whether as drug or as hostdirected therapy?
Fig. 4. Evolution of AMPs and origin of AMPs as drugs.
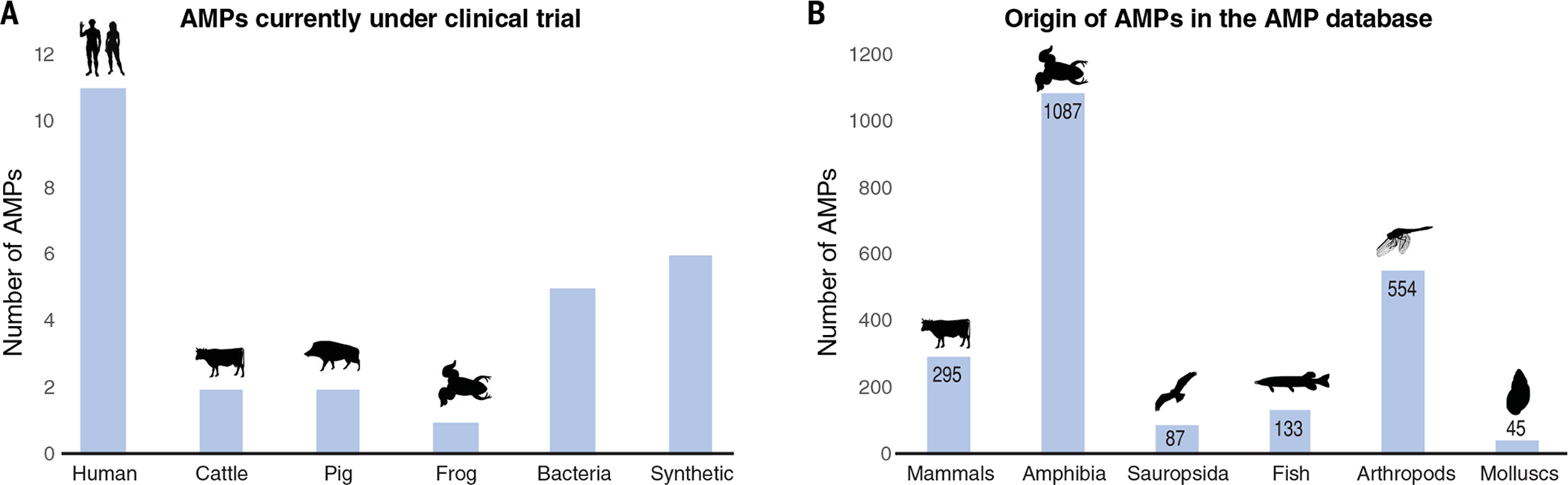
(A) The number of AMPs currently undergoing clinical trials [data are from (4)] and the organisms from which they are derived. (B) Relative representation of animal taxa in the antimicrobial peptide database (5). This representation is not correlated with the number of species in each of the groups because, for example, insects are by far the most species-rich taxon.
Conclusions
In 2002, Zasloff predicted, “If the story regarding diversity continues to unfold based on our current views, we might well discover that every species harbours a unique, specific collection of antimicrobial peptides, tuned to defend the organism against microorganisms that it will predictably encounter” (1). This prediction has been supported by the documentation of rapid evolutionary diversification of AMP families within and among species as well as by the recent realization that AMPs can be highly specific in their antimicrobial activities and in how they are naturally used by hosts. The pharmacodynamics of AMPs and the fact that multicellular hosts naturally deploy them in synergistic cocktails limit the probability of bacterial resistance evolution in nature. These same properties could be capitalized on for translation of AMPs into medicine and agriculture, informed by natural biology and evolution to avoid the mistakes that have resulted in the current crisis of antibiotic resistance.
ACKNOWLEDGMENTS
We thank J. Antonovics, S. Armitage, D. McMahon, and O. Judson for helpful feedback and D. Baeder, A. Rodriguez-Rojas, and C. Manthey for help with the figures.
Funding:
J.R. received funding from the European Research Council (EVOLchip) and the Deutsche Forschungsgemeinschaft (SFB 973). B.P.L. is supported by grant R01 AI141385 from the U.S. National Institutes of Health.
Footnotes
Competing interests: There are no competing interests.
REFERENCES AND NOTES
- 1.Zasloff M, Antimicrobial peptides of multicellular organisms. Nature 415, 389–395 (2002). doi: 10.1038/415389a; [DOI] [PubMed] [Google Scholar]
- 2.Czaplewski L et al. , Alternatives to antibiotics—A pipeline portfolio review. Lancet Infect. Dis 16, 239–251 (2016). doi: 10.1016/S1473-3099(15)00466-1; [DOI] [PubMed] [Google Scholar]
- 3.Forbes S et al. , Comparative surface antimicrobial properties of synthetic biocides and novel human apolipoprotein E derived antimicrobial peptides. Biomaterials 34, 5453–5464 (2013). doi: 10.1016/j.biomaterials.2013.03.087; [DOI] [PubMed] [Google Scholar]
- 4.Koo HB, Seo J, Antimicrobial peptides under clinical investigation. Pept. Sci 111, e24122 (2019). doi: 10.1002/pep2.24122 [DOI] [Google Scholar]
- 5.Wang G, Li X, Wang Z, APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res 44, D1087–D1093 (2016). doi: 10.1093/nar/gkv1278; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mylonakis E, Podsiadlowski L, Muhammed M, Vilcinskas A, Diversity, evolution and medical applications of insect antimicrobial peptides. Philos. Trans. R. Soc. London B Biol. Sci 371, 20150290 (2016). doi: 10.1098/rstb.2015.0290; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hancock REW, Haney EF, Gill EE, The immunology of host defence peptides: Beyond antimicrobial activity. Nat. Rev. Immunol 16, 321–334 (2016). doi: 10.1038/nri.2016.29; [DOI] [PubMed] [Google Scholar]
- 8.Campos ML, de Souza CM, de Oliveira KBS, Dias SC, Franco OL, The role of antimicrobial peptides in plant immunity. J. Exp. Bot 69, 4997–5011 (2018). doi: 10.1093/jxb/ery294; [DOI] [PubMed] [Google Scholar]
- 9.Deslouches B, Di YP, Antimicrobial peptides with selective antitumor mechanisms: Prospect for anticancer applications. Oncotarget 8, 46635–46651 (2017). doi: 10.18632/oncotarget.16743; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broekaert WF, Terras FR, Cammue BP, Osborn RW, Plant defensins: Novel antimicrobial peptides as components of the host defense system. Plant Physiol. 108, 1353–1358 (1995). doi: 10.1104/pp.108.4.1353; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shafee TMA, Lay FT, Phan TK, Anderson MA, Hulett MD, Convergent evolution of defensin sequence, structure and function. Cell. Mol. Life Sci 74, 663–682 (2017). doi: 10.1007/s00018-016-2344-5; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Florin T et al. , An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome. Nat. Struct. Mol. Biol 24, 752–757 (2017). doi: 10.1038/nsmb.3439; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagnon MG et al. , Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition. Nucleic Acids Res. 44, 2439–2450 (2016). doi: 10.1093/nar/gkw018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu H et al. , Human α-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science 337, 477–481 (2012). doi: 10.1126/science.1218831; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loth K et al. , The ancestral N-terminal domain of big defensins drives bacterially triggered assembly into antimicrobial nanonets. mBio 10, e01821–19 (2019). doi: 10.1128/mBio.01821-19; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rabel D et al. , Primary structure and in vitro antibacterial properties of the Drosophila melanogaster attacin C Pro-domain. J. Biol. Chem 279, 14853–14859 (2004). doi: 10.1074/jbc.M313608200; [DOI] [PubMed] [Google Scholar]
- 17.Rahnamaeian M et al. , Insect antimicrobial peptides show potentiating functional interactions against Gram-negative bacteria. Proc. Biol. Sci 282, 20150293 (2015). doi: 10.1098/rspb.2015.0293; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart SE et al. , The perforin pore facilitates the delivery of cationic cargos. J. Biol. Chem 289, 9172–9181 (2014). doi: 10.1074/jbc.M113.544890; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zerweck J et al. , Molecular mechanism of synergy between the antimicrobial peptides PGLa and magainin 2. Sci. Rep 7, 13153 (2017). doi: 10.1038/s41598-017-12599-7; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boman H, Marsh J, Goode J, Ed., Antimicrobial Peptides (Ciba Foundation Symposium) (John Wiley & Sons, 1994). [Google Scholar]
- 21.Lazzaro BP, Natural selection on the Drosophila antimicrobial immune system. Curr. Opin. Microbiol 11, 284–289 (2008). doi: 10.1016/j.mib.2008.05.001; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B, Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. U.S.A 98, 12590–12595 (2001). doi: 10.1073/pnas.221458698; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sackton TB et al. , Dynamic evolution of the innate immune system in Drosophila. Nat. Genet 39, 1461–1468 (2007). doi: 10.1038/ng.2007.60; [DOI] [PubMed] [Google Scholar]
- 24.Waterhouse RM et al. , Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 316, 1738–1743 (2007). doi: 10.1126/science.1139862; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramos-Onsins S, Aguadé M, Molecular evolution of the Cecropin multigene family in Drosophila: Functional genes vs. pseudogenes. Genetics 150, 157–171 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazzaro BP, Clark AG, Evidence for recurrent paralogous gene conversion and exceptional allelic divergence in the Attacin genes of Drosophila melanogaster. Genetics 159, 659–671 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semple CA, Gautier P, Taylor K, Dorin JR, The changing of the guard: Molecular diversity and rapid evolution of β-defensins. Mol. Divers 10, 575–584 (2006). doi: 10.1007/s11030-006-9031-7; [DOI] [PubMed] [Google Scholar]
- 28.Tennessen JA, Molecular evolution of animal antimicrobial peptides: Widespread moderate positive selection. J. Evol. Biol 18, 1387–1394 (2005). doi: 10.1111/j.1420-9101.2005.00925.x; [DOI] [PubMed] [Google Scholar]
- 29.Zanchi C, Johnston PR, Rolff J, Evolution of defence cocktails: Antimicrobial peptide combinations reduce mortality and persistent infection. Mol. Ecol 26, 5334–5343 (2017). doi: 10.1111/mec.14267; [DOI] [PubMed] [Google Scholar]
- 30.Unckless RL, Howick VM, Lazzaro BP, Convergent balancing selection on an antimicrobial peptide in Drosophila. Curr. Biol 26, 257–262 (2016). doi: 10.1016/j.cub.2015.11.063; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unckless RL, Lazzaro BP, The potential for adaptive maintenance of diversity in insect antimicrobial peptides. Philos. Trans. R. Soc. B 371, 20150291 (2016). doi: 10.1098/rstb.2015.0291; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanson MA et al. , Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach. eLife 8, e44341 (2019). doi: 10.7554/eLife.44341; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemaitre B, Reichhart J-M, Hoffmann JA, Drosophila host defense: Differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. U.S.A 94, 14614–14619 (1997). doi: 10.1073/pnas.94.26.14614; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tay WH, Chong KKL, Kline KA, Polymicrobial-host interactions during infection. J. Mol. Biol 428, 3355–3371 (2016). doi: 10.1016/j.jmb.2016.05.006; [DOI] [PubMed] [Google Scholar]
- 35.Peschel A, Sahl H-G, The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol 4, 529–536 (2006). doi: 10.1038/nrmicro1441; [DOI] [PubMed] [Google Scholar]
- 36.Hanson MA, Lemaitre B, Unckless RL, Dynamic evolution of antimicrobial peptides underscores trade-offs between immunity and ecological fitness. Front. Immunol 10, 2620 (2019). doi: 10.3389/fimmu.2019.02620; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mondragón-Palomino M, Stam R, John-Arputharaj A, Dresselhaus T, Diversification of defensins and NLRs in Arabidopsis species by different evolutionary mechanisms. BMC Evol. Biol 17, 255 (2017). doi: 10.1186/s12862-017-1099-4; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng X-J et al. , Gene expression divergence and evolutionary analysis of the drosomycin gene family in Drosophila melanogaster. J. Biomed. Biotechnol 2009, 315423 (2009). doi: 10.1155/2009/315423; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang K-Y, Wang X, Wan Q-H, Fang S-G, A crucial role of paralogous β-defensin genes in the Chinese alligator innate immune system revealed by the first determination of a Crocodilia defensin cluster. Dev. Comp. Immunol 81, 193–203 (2018). doi: 10.1016/j.dci.2017.11.018; [DOI] [PubMed] [Google Scholar]
- 40.Gosset CC, Do Nascimento J, Augé M-T, Bierne N, Evidence for adaptation from standing genetic variation on an antimicrobial peptide gene in the mussel Mytilus edulis. Mol. Ecol 23, 3000–3012 (2014). doi: 10.1111/mec.12784; [DOI] [PubMed] [Google Scholar]
- 41.Hellgren O, Sheldon BC, Buckling A, In vitro tests of natural allelic variation of innate immune genes (avian β-defensins) reveal functional differences in microbial inhibition. J. Evol. Biol 23, 2726–2730 (2010). doi: 10.1111/j.1420-9101.2010.02115.x; [DOI] [PubMed] [Google Scholar]
- 42.Cagliani R et al. , The signature of long-standing balancing selection at the human defensin β−1 promoter. Genome Biol. 9, R143 (2008). doi: 10.1186/gb-2008-9-9-r143; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tennessen JA, Blouin MS, Selection for antimicrobial peptide diversity in frogs leads to gene duplication and low allelic variation. J. Mol. Evol 65, 605–615 (2007). doi: 10.1007/s00239-007-9045-5; [DOI] [PubMed] [Google Scholar]
- 44.Chapman JR et al. , The evolution of innate immune genes: Purifying and balancing selection on β-defensins in waterfowl. Mol. Biol. Evol 33, 3075–3087 (2016). doi: 10.1093/molbev/msw167; [DOI] [PubMed] [Google Scholar]
- 45.Halldórsdóttir K, Árnason E, Trans-species polymorphism at antimicrobial innate immunity cathelicidin genes of Atlantic cod and related species. PeerJ 3, e976 (2015). doi: 10.7717/peerj.976; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chapman JR, Hill T, Unckless RL, Balancing selection drives the maintenance of genetic variation in Drosophila antimicrobial peptides. Genome Biol. Evol 11, 2691–2701 (2019). doi: 10.1093/gbe/evz191; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Charlesworth D, Balancing selection and its effects on sequences in nearby genome regions. PLOS Genet. 2, e64 (2006). doi: 10.1371/journal.pgen.0020064; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao Z, Przeworski M, Sella G, Footprints of ancient-balanced polymorphisms in genetic variation data from closely related species. Evolution 69, 431–446 (2015). doi: 10.1111/evo.12567; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bevins CL, Salzman NH, Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol 9, 356–368 (2011). doi: 10.1038/nrmicro2546; [DOI] [PubMed] [Google Scholar]
- 50.Franzenburg S et al. , Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc. Natl. Acad. Sci. U.S.A 110, E3730–E3738 (2013). doi: 10.1073/pnas.1304960110; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cullen TW et al. , Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 347, 170–175 (2015). doi: 10.1126/science.1260580; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oakeson KF et al. , Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol. Evol 6, 76–93 (2014). doi: 10.1093/gbe/evt210; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Login FH et al. , Antimicrobial peptides keep insect endosymbionts under control. Science 334, 362–365 (2011). doi: 10.1126/science.1209728; [DOI] [PubMed] [Google Scholar]
- 54.Arnold MFF et al. , Genome-wide sensitivity analysis of the microsymbiont Sinorhizobium meliloti to symbiotically important, defensin-like host peptides. mBio 8, e01060–17 (2017). doi: 10.1128/mBio.01060-17; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen F et al. , Bactericidal permeability-increasing proteins shape host-microbe interactions. mBio 8, e00040–17 (2017). doi: 10.1128/mBio.00040-17; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boscaro V et al. , Parallel genome reduction in symbionts descended from closely related free-living bacteria. Nat. Ecol. Evol 1, 1160–1167 (2017). doi: 10.1038/s41559-017-0237-0; [DOI] [PubMed] [Google Scholar]
- 57.Mergaert P, Kikuchi Y, Shigenobu S, Nowack ECM, Metabolic integration of bacterial endosymbionts through antimicrobial peptides. Trends Microbiol. 25, 703–712 (2017). doi: 10.1016/j.tim.2017.04.007; [DOI] [PubMed] [Google Scholar]
- 58.Van de Velde W et al. , Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327, 1122–1126 (2010). doi: 10.1126/science.1184057; [DOI] [PubMed] [Google Scholar]
- 59.Westerhoff HV et al. , Functional synergism of the magainins PGLa and magainin-2 in Escherichia coli, tumor cells and liposomes. Eur. J. Biochem 228, 257–264 (1995). doi: 10.1111/j.1432-1033.1995.00257.x; [DOI] [PubMed] [Google Scholar]
- 60.Yan H, Hancock REW, Synergistic interactions between mammalian antimicrobial defense peptides. Antimicrob. Agents Chemother 45, 1558–1560 (2001). doi: 10.1128/AAC.45.5.1558-1560.2001; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gueguen Y et al. , Oyster hemocytes express a proline-rich peptide displaying synergistic antimicrobial activity with a defensin. Mol. Immunol 46, 516–522 (2009). doi: 10.1016/j.molimm.2008.07.021; [DOI] [PubMed] [Google Scholar]
- 62.Marxer M, Vollenweider V, Schmid-Hempel P, Insect antimicrobial peptides act synergistically to inhibit a trypanosome parasite. Philos. Trans. R. Soc. London B Biol. Sci 371, 20150302 (2016). doi: 10.1098/rstb.2015.0302; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu G, Baeder DY, Regoes RR, Rolff J, Combination effects of antimicrobial peptides. Antimicrob. Agents Chemother 60, 1717–1724 (2016). doi: 10.1128/AAC.02434-15; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greco WR, Bravo G, Parsons JC, The search for synergy: A critical review from a response surface perspective. Pharmacol. Rev 47, 331–385 (1995). [PubMed] [Google Scholar]
- 65.Baeder DY, Yu G, Hozé N, Rolff J, Regoes RR, Antimicrobial combinations: Bliss independence and Loewe additivity derived from mechanistic multi-hit models. Philos. Trans. R. Soc. B 371, 20150294 (2016). doi: 10.1098/rstb.2015.0294; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Russ D, Kishony R, Additivity of inhibitory effects in multidrug combinations. Nat. Microbiol 3, 1339–1345 (2018). doi: 10.1038/s41564-018-0252-1; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chae J-H et al. , Purification and characterization of tenecin 4, a new anti-Gram-negative bacterial peptide, from the beetle Tenebrio molitor. Dev. Comp. Immunol 36, 540–546 (2012). doi: 10.1016/j.dci.2011.09.010; [DOI] [PubMed] [Google Scholar]
- 68.Lin L et al. , EBioMedicine 2, 690–698 (2015). doi: 10.1016/j.ebiom.2015.05.021; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holmes AH et al. , Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387, 176–187 (2016). doi: 10.1016/S0140-6736(15)00473-0; [DOI] [PubMed] [Google Scholar]
- 70.Laxminarayan R et al. , Antibiotic resistance-the need for global solutions. Lancet Infect. Dis 13, 1057–1098 (2013). doi: 10.1016/S1473-3099(13)70318-9; [DOI] [PubMed] [Google Scholar]
- 71.Petrosillo N, Ioannidou E, Falagas ME, Colistin monotherapy vs. combination therapy: Evidence from microbiological, animal and clinical studies. Clin. Microbiol. Infect 14, 816–827 (2008). doi: 10.1111/j.1469-0691.2008.02061.x; [DOI] [PubMed] [Google Scholar]
- 72.Vriens K et al. , Synergistic activity of the plant defensin HsAFP1 and caspofungin against Candida albicans biofilms and planktonic cultures. PLOS ONE 10, e0132701 (2015). doi: 10.1371/journal.pone.0132701; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Breij A et al. , The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med 10, eaan4044 (2018). doi: 10.1126/scitranslmed.aan4044; [DOI] [PubMed] [Google Scholar]
- 74.Ottosson H et al. , Potent inducers of endogenous antimicrobial peptides for host directed therapy of infections. Sci. Rep 6, 36692 (2016). doi: 10.1038/srep36692; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luther A et al. , Chimeric peptidomimetic antibiotics against Gram-negative bacteria. Nature 576, 452–458 (2019). doi: 10.1038/s41586-019-1665-6; [DOI] [PubMed] [Google Scholar]
- 76.Imamovic L, Sommer MOA, Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci. Transl. Med 5, 204ra132 (2013). doi: 10.1126/scitranslmed.3006609; [DOI] [PubMed] [Google Scholar]
- 77.Lázár V et al. , Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat. Microbiol 3, 718–731 (2018). doi: 10.1038/s41564-018-0164-0; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dobson AJ, Purves J, Kamysz W, Rolff J, Comparing selection on S. aureus between antimicrobial peptides and common antibiotics. PLOS ONE 8, e76521 (2013). doi: 10.1371/journal.pone.0076521; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kubicek-Sutherland JZ et al. , Antimicrobial peptide exposure selects for Staphylococcus aureus resistance to human defence peptides. J. Antimicrob. Chemother 72, 115–127 (2017). doi: 10.1093/jac/dkw381; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spohn R et al. , Integrated evolutionary analysis reveals antimicrobial peptides with limited resistance. Nat. Commun 10, 4538 (2019). doi: 10.1038/s41467-019-12364-6; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu G, Baeder DY, Regoes RR, Rolff J, Predicting drug resistance evolution: Insights from antimicrobial peptides and antibiotics. Proc. Biol. Sci 285, 20172687 (2018). doi: 10.1098/rspb.2017.2687; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rodríguez-Rojas A, Makarova O, Rolff J, Antimicrobials, stress and mutagenesis. PLOS Pathog. 10, e1004445 (2014). doi: 10.1371/journal.ppat.1004445; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fantner GE, Barbero RJ, Gray DS, Belcher AM, Kinetics of antimicrobial peptide activity measured on individual bacterial cells using high-speed atomic force microscopy. Nat. Nanotechnol 5, 280–285 (2010). doi: 10.1038/nnano.2010.29; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jochumsen N et al. , The evolution of antimicrobial peptide resistance in Pseudomonas aeruginosa is shaped by strong epistatic interactions. Nat. Commun 7, 13002 (2016). doi: 10.1038/ncomms13002; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kintses B et al. , Phylogenetic barriers to horizontal transfer of antimicrobial peptide resistance genes in the human gut microbiota. Nat. Microbiol 4, 447–458 (2019). doi: 10.1038/s41564-018-0313-5; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mathur D et al. , PEPlife: A repository of the half-life of peptides. Sci. Rep 6, 36617 (2016). doi: 10.1038/srep36617; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gullberg E et al. , Selection of resistant bacteria at very low antibiotic concentrations. PLOS Pathog. 7, e1002158 (2011). doi: 10.1371/journal.ppat.1002158; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Forsberg KJ et al. , The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111 (2012). doi: 10.1126/science.1220761; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Perron GG, Zasloff M, Bell G, Experimental evolution of resistance to an antimicrobial peptide. Proc. Biol. Sci 273, 251–256 (2006). doi: 10.1098/rspb.2005.3301; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Joo H-S, Fu C-I, Otto M, Bacterial strategies of resistance to antimicrobial peptides. Philos. Trans. R. Soc. B 371, 20150292 (2016). doi: 10.1098/rstb.2015.0292; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rodríguez-Rojas A, Baeder DY, Johnston P, Regoes RR, Rolff J, bioRxiv 802207 [Preprint] 19 February 2020; https://www.biorxiv.org/content/10.1101/802207v2. [Google Scholar]
- 92.Levin BR, Rozen DE, Non-inherited antibiotic resistance. Nat. Rev. Microbiol 4, 556–562 (2006). doi: 10.1038/nrmicro1445; [DOI] [PubMed] [Google Scholar]
- 93.Tedijanto C, Olesen SW, Grad YH, Lipsitch M, Estimating the proportion of bystander selection for antibiotic resistance among potentially pathogenic bacterial flora. Proc. Natl. Acad. Sci. U.S.A 115, E11988–E11995 (2018). doi: 10.1073/pnas.1810840115; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Topman S et al. , Random peptide mixtures as new crop protection agents. Microb. Biotechnol 11, 1027–1036 (2018). doi: 10.1111/1751-7915.13258; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Latendorf T et al. , Cationic intrinsically disordered antimicrobial peptides (CIDAMPs) represent a new paradigm of innate defense with a potential for novel anti-infectives. Sci. Rep 9, 3331 (2019). doi: 10.1038/s41598-019-39219-w; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pena-Miller R et al. , When the most potent combination of antibiotics selects for the greatest bacterial load: The smile-frown transition. PLOS Biol. 11, e1001540 (2013). doi: 10.1371/journal.pbio.1001540; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Koprivnjak T, Peschel A, Bacterial resistance mechanisms against host defense peptides. Cell. Mol. Life Sci 68, 2243–2254 (2011). doi: 10.1007/s00018-011-0716-4; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bell G, Gouyon P-H, Arming the enemy: The evolution of resistance to self-proteins. Microbiology 149, 1367–1375 (2003). doi: 10.1099/mic.0.26265-0; [DOI] [PubMed] [Google Scholar]
- 99.Dobson AJ, Purves J, Rolff J, Increased survival of experimentally evolved antimicrobial peptide-resistant Staphylococcus aureus in an animal host. Evol. Appl 7, 905–912 (2014). doi: 10.1111/eva.12184; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.El Shazely B, Urbański A, Johnston PR, Rolff J, In vivo exposure of insect AMP resistant Staphylococcus aureus to an insect immune system. Insect Biochem. Mol. Biol 110, 60–68 (2019). doi: 10.1016/j.ibmb.2019.04.017; [DOI] [PubMed] [Google Scholar]


