Abstract
The angiotensin-converting enzyme (ACE)-related carboxypeptidase, ACE-II, is a type I integral membrane protein of 805 amino acids that contains 1 HEXXH-E zinc binding consensus sequence. ACE-II has been implicated in the regulation of heart function and also as a functional receptor for the coronavirus that causes the severe acute respiratory syndrome (SARS). In this study, the potential of some flavonoids presents in propolis to bind to ACE-II receptors was calculated with in silico. Binding constants of ten flavonoids, caffeic acid, caffeic acid phenethyl ester, chrysin, galangin, myricetin, rutin, hesperetin, pinocembrin, luteolin and quercetin were measured using the AutoDock 4.2 molecular docking program. And also, these binding constants were compared to reference ligand of MLN-4760. The results are shown that rutin has the best inhibition potentials among the studied molecules with high binding energy − 8.04 kcal/mol, and it is followed by myricetin, quercetin, caffeic acid phenethyl ester and hesperetin. However, the reference molecule has binding energy of – 7.24 kcal/mol. In conclusion, the high potential of flavonoids in ethanolic propolis extracts to bind to ACE-II receptors indicates that this natural bee product has high potential for COVID-19 treatment, but this needs to be supported by experimental studies.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00203-021-02351-1.
Keywords: Coronavirus, COVID-19, Propolis, Flavonoids, ACE-II, Molecular docking
Introduction
Propolis is a natural mixture that honey bees collect from nature to protect their hives. Crude propolis is a highly viscous, slightly soluble mixture in water and best dissolved in 60–80% ethanol. Propolis has been an indispensable component of apitherapy for centuries and has recently been used as a food additive, or supplementary, under the name of traditional and complementary medicine (Anjum et al. 2019; Pobiega et al. 2019). Its composition varies according to the flora of the region where it is collected, but the majority of active ingredients of propolis comprise the family of polyphenols. Propolis is not consumed as raw, but their ethanolic and aqueous extracts are widely consumed in different formulations. It has been reported that polyphenolic agents such as gallic acid, caffeic acid, protocatechuic acid, chrysin, quercetin, rutin, galangin, kaempferol, hesperetin, pinocembrin, pinobanksin, apigenin, luteolin, daidzein, caffeic acid phenyl ester (CAPE) are the most active phenolic subtances of propolis samples and these secondary metabolites vary depending on the propolis source (Aliyazıcıoglu et al. 2013; Yang et al. 2013; Anjum et al. 2019).
Studies show that propolis extracts have high immunomodulatory effect and inhibition potential for some clinically important enzymes, such as urease, xanthine oxidase (XO), acetylcholinesterase (AChE), α-amylase, α-glucosidase (Baltas et al. 2016; Catchpole et al. 2018). In addition, in vivo and in vitro studies show that flavonoids, one of the active ingredients of propolis, have high potential for Angiotensin-Converting Enzyme (ACE) Inhibition (Hussain et al. 2018; Wang et al. 2018; Silveira et al. 2019).
The newly discovered SARS-CoV-2 was characterized as a beta-coronavirus and recognized as the seventh discrete coronavirus species capable of causing human disease. The disease caused by the virus is officially named Coronavirus Disease 2019 (COVID-19) by Word Health Organization (WHO). The emerged global epidemic spread rapidly with 122.524.424 confirmed cases and 2.703.620 deaths across 213 countries, areas and territories (COVID-19 situation Report WHO, 21 March 2021). Subsequent studies have shown that SARS-CoV-2 has been suggested to recognize human ACE-II more strongly than SARS-CoV, thereby increasing the ability to be transmitted from person to person (Wan et al. 2020).
ACE-II consists of 805 amino acids with a zinc binding motif (HEXXH-E) and is type I transmembrane glycoprotein with a single extracellular catalytic domain. ACE-II enzyme has been identified as a functional receptor for coronaviruses, including SARS-CoV and SARS-CoV-2 (Turner et al. 2004; Kuba et al. 2010). Thus, the SARS-CoV-2 spike protein was also estimated to have a strong binding affinity to human ACE-II. This similarity with SARS-CoV is important for proving that ACE-II is a functional SARS-CoV receptor in vitro (Li et al. 2003) and in vivo (Kuba et al. 2005) experiments.
According to the biochemical interaction analysis and crystal structure studies, it was found that the SARS-CoV envelope-anchored spike protein has a strong binding affinity to human ACE-II (Li et al. 2005). It is also known from the reports that residue 394 (Gln) in the SARS-CoV-2 receptor binding domain (RBD) corresponding to 479 residue in SARS-CoV is recognized by critical lysine 31 on the receptor of human ACE-II (Wu et al. 2012). Subsequent studies have shown that SARS-CoV-2 has been suggested to recognize human ACE-II more strongly than SARS-CoV, thereby increasing the ability to be transmitted from person to person (Wan et al. 2020). The spike protein first binds to the host's receptor, and then fuses viral and host membranes to mediate the coronavirus entering the host cells (Li 2016). A defined receptor binding site (RBD) of the SARS-CoV spike specifically recognizes the host receptor ACE-II (Li et al. 2003; Li 2015). Studies have shown that SARS-CoV RBD contains a core structure and a receptor binding motif (RBM) and that RBM binds to the outer surface of the claw-like structure of ACE-II (Li 2005). Therefore, ACE-II enzyme inhibition is important for treatments against these virus infections caused by SARS-CoV-2.
The aim of the study is to investigate the potential of ethanolic propolis extracts to bind to ACE-II receptors and compare with classical ACE-II inhibitors. For this reason, the study is based on Anatolian propolis composition which is analysed by HPLC–UV and to calculate in silico ACE-II binding constants of some flavanoids present in the propolis extract. In this molecular modeling for ACE-II (S, S)-2-{1-carboxy-2-[3-(3,5-dichloro-benzyl)-3h-imidazole-4-Yl]-ethylamino}-4-methyl-pentanoic acid (MLN-4760) is used as positive control.
Up to now, there are very limited studies about COVID-19 and most researchers focused primarily on clinical cases. However, to the authors' knowledge, no study has been made on inhibition of ACE-II known to be associated with COVID-19. Therefore, the paper will encourage further research about COVID-19 and candidate drug compounds.
Materials and methods
Materials
Raw propolis samples were obtained experienced beekeepers in 2018 from Black Sea Region, Turkey.
Chemicals
All phenolic standard for HPLC–UV analyses of gallic acid, protocathequic acid, p-OH benzoic acid, catechin, caffeic acid, syringic acid, epicatechin, p-coumaric acid, ferulic acid, rutin, myricetin, resveratrol, daidzein, luteolin, t-cinnamic acid, hesperetin, chrysin, pinocembrin, caffeic acid phenethyl ester (CAPE) were purchased from Sigma Chemical Co. (St Louis, MO, USA). All solvent for using mobile phases were analytical grade.
Preparation of propolis extracts
The raw propolis samples were frozen at − 20 °C and then grinded to powder. The following method was used to prepare the ethanolic propolis extract: 10 g powdered crude propolis was placed with 100 mL 70% ethanol in a glass flask and stirred with shaker (Heidolph Promax 2020, Schwabach, Germany) at room temperature for 24 h and then filtered with Whatman paper. The ethanolic filtrate is used for phenolic composition analyses of the ethanolic propolis extract.
Determination of phenolic profiles
In the study, 19 phenolic standards were used to high-performance liquid chromatography (HPLC) (Elite LaChrom Hitachi, Japan) with a UV detector. Separation was performed on a column with a reverse phase C18 column (150 mm × 4.6 mm, 5 μm; Fortis), in gradient solvent systems A (2% acetic acid in water) and solvent B (70:30, acetonitrile/water), which was sonicated before stirring and continuously degassed by the built-in HPLC system (Cakir et al. 2018; Malkoç et al. 2019). The flow rate was kept constant at 1 mL/min using gradient programming, starting the flow of the mobile phase as B (5%) to 3 min, gradually increasing (up to 15, 20, 25, 40 and 80% at 8, 10, 18, 25 and 35 min, respectively) and decreasing to 5% at 40 min, before being left for 10 min to equilibrate in the column. The standard phenolic substances chromatogram and a gradation graph are given in Figs. 1 and 2.
Fig. 1.
Chromatogram of nineteen phenolic compounds of HPLC–UV; (1): gallic acid, (2): protocathequic acid, (3): p-OH benzoic acid, (4): catechin, (5): caffeic acid, (6): syringic acid, (7): epicatechin, (8): p-coumaric acid, (9): ferulic acid, (10): rutin, (11): myricetin, (12): resveratrol, (13): daidzein, (14); luteolin, (15): t-cinnamic acid, (16): hesperetin, (17): chrysin, (18): pinocembrin, (19): Caffeic acid phenethyl ester (CAPE)
Fig. 2.
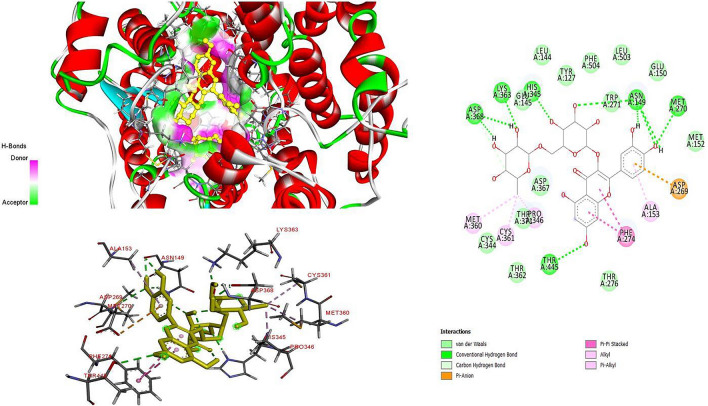
The two-dimension (2D) and three-dimension (3D) interaction analysis of ACE II with rutin
Molecular docking
In the study, some flavonoids detected in the ethanolic propolis extracts and they used as ligand (see in Supplementary File) for ACE-II receptors. The crystal structure of an ACE-II protein was downloaded from protein data bank web site (http://www.rcsb.org/pdb) (PDB ID: 1R4L: Resolution 3.00 Å). This crystal structure contains the inhibitory bound state of the extracellular metallopeptidase domain of ACE-II with MLN-4760 compound. Small compounds of flavonoids used in docking studies were obtained from ChEMBL as SDF form and hydrogen atoms were added to these 3D structures, geometrically cleaned, then converted to.pdb format with BIOVIA Discovery Studio (Dassault Systèmes BIOVIA 2017). Possible docking modes between compounds and the ACE-II enzyme were studied using the Autodock 4.2 (Morris et al. 2009) and Lamarckian genetic algorithm (LGA) was employed for docking simulations. The LGA for flexible ligand-receptor docking manages multiple degrees of freedom, allowing full flexibility of the ligand and partial flexibility of the receptor. Thus, it enables individual conformations to search their local conformational space, finding local minima, and then pass that knowledge to next generations. Therefore, in this study, we preferred the open-source AutoDock 4.2 program with LGA algorithm for semi-flexible docking.
The selected cavity is the binding site of reference inhibitor MLN-4760 [((S,S)-2-{1-carboxy-2-[3-(3,5-dichloro-benzyl)-3H-imidazol-4-YL]-ethylamino}-4-methyl-pentanoic acid)]. A grid box dimensions of 52, 34, and 47 points in x, y, and z directions were set with a grid spacing of 0.375 Å. The program was run for a total number of 100 Genetic algorithm runs. The default settings were applied for all other parameters. Results of the molecular docking described the affinity represented by docking score and binding interaction of each ligand on the interested protein target. The visualization of results was performed with the help of the BIOVIA Discovery Studio 2018 (Dassault Systèmes BIOVIA 2017).
Results and discussion
Phenolic composition of propolis extract
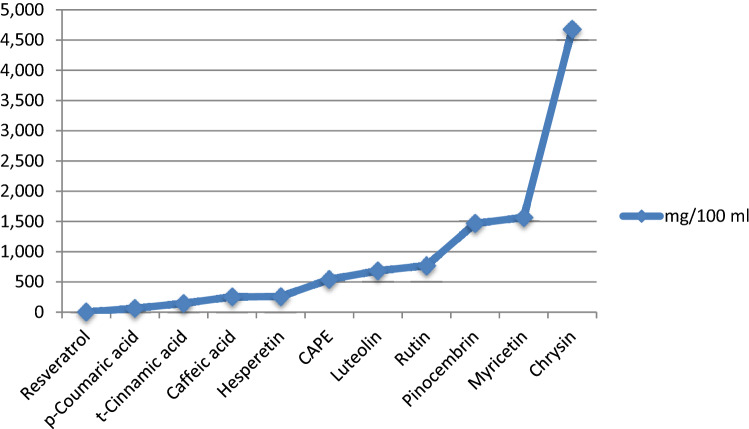
In this study, a standard HPLC–UV chromatogram prepared with nineteen phenolic standards including some phenolic acids and flavonoids is given in Fig. 1. The analysis data of the ethanolic propolis extract carried out according to this chromatogram are summarized in Table 1. Although the hydroxybenzoic acids and catechin derivatives of the propolis sample were found below determination limits, it was found to be rich in hydroxycinnamic acids and flavonoids. Among the hydroxy cinnamic acid derivatives, the caffeic acid phenyl ester is the highest amount of phenolic component in the sample and followed it caffeic acid and cinnamic acid. Ferulic acid could not be detected in the sample. Among the flavonoids subclass of flavonoids, the highest amount of myricetin was detected and rutin followed it.
Table 1.
Phenolic composition of propolis
| Group name | Compound name | mg/100 g |
|---|---|---|
| Hydroxybenzoic acids | Gallic acid | n.d |
| Protocateuic acid | n.d | |
| p-OH benzoic acid | n.d | |
| Syringic acid | n.d | |
| Catechin | Catechin | n.d |
| Epicatechin | n.d | |
| Hydroxycinnamic acids | Caffeic acid | 254.501 |
| p-Coumaric acid | 63.871 | |
| Ferulic acid | n.d | |
| t-Cinnamic acid | 145.455 | |
| CAPE | 541.213 | |
| Flavanones | Rutin | 770.970 |
| Myricetin | 1567.750 | |
| Flavones | Hesperetin | 258.010 |
| Chrysin | 4678.423 | |
| Pinocembrin | 1467.260 | |
| Isoflavons | Luteolin | 684.752 |
| Stilbands and Lignans | Daidzein | n.d |
| Resveratrol | 2.372 |
n.d Not detected
Among these three flavanons studied, chrysin is the most abundant compounds pinosembrin and hesperetin followed it. A smaller amount of flavone derivative of luteolin, was detected, while daidzein is not detected in the sample. Of all the studied compounds, chrysin and pinosembrin were detected as major flavonoids in the propolis sample.
Although composition of propolis varies according to the flora of the region where it is produced, these flavonoids were also reported in propolis samples of different countries samples (Chang et al. 2002; Can et al. 2015; Sun et al. 2015). There are many scientific studies showing that propolis, a natural bee product, is a very rich mixture of flavonoids and is an important agent of apitherapy. Polyphenolic profile of propolis varies according to the flora of the region and also caffeic acid, CAPE, rutin, quercetin, myricetin, kaempferol, hesperetin, naringin and naringenin, galangin are most active substances of Turkish propolis (Lu et al. 2004; Erdogan et al. 2011; Aliyazıcıoglu et al. 2013; Yang et al. 2013; Graikou et al. 2016). Barbarić et al. (2011) studied chemical composition of the ethanolic propolis extracts and determined ferulic acid, p-coumaric acid, caffeic acid, tectochrysin, galangin, pinocembrin, chrysin, apigenin, kaempferol, quercetin as phenolic compound in Croatia, Bosnia and Hercegovina and Macedonia propolis (Barbaric et al. 2011). Major compounds of red propolis samples from Brasilia were reported as luteolin (1.75 mg/g), naringenin (0.96 mg/g), kaempferol (0.59 mg/g), pinocembrin (0.41 mg/g) and biochanin A (0.39 mg/g) (Andrade et al. 2017). There are some differences between the findings because the chemical composition of propolis varies according to the geographical region, climate, environmental conditions and collection seasons (Sawaya et al. 2011; Lopez et al. 2014; Andrade et al. 2017). The findings show that propolis is a good source of phenolic substances. The literature states that propolis samples from different geographical origins have a good antioxidant antimicrobial, antifungal and antiviral (Avian influenza virus) activity (Pietta 2000; Pratsinis et al. 2010; Yildiz et al. 2014; Saral et al. 2016, 2019).
Binding affinity analysis for proteins and ligands with molecular docking
We focus here on the Anatolian propolis compounds used by people to treat infections against ACE-II with molecular docking methods. For this purpose, we calculated the binding energy and inhibition constant (Ki) for ACE-II and each compound by molecular docking analysis. The smaller binding energy and Ki value, the more tightly bound the ligand is, or the greater the binding affinity between ligand and protein. Accordingly, we found that rutin, myricetin, quercetin, CAPE and hesperetin have a better affinity against ACE-II enzyme than natural inhibitor MLN-4760 among the evaluated compounds, with low µM Ki and binding energy values (Table 2).
Table 2.
Summary of reference and flavonoids compounds aganist ACE II with binding energy and interacted residues in the ACE-II binding site
| Ligand Number | Ligand Name | Binding Energy (kcal/mol) | Ki (μM) | ACE II residues interacting with ligands |
|---|---|---|---|---|
|
MLN-4760 Referans ligand |
((S,S)-2-{1-Carboxy-2-[3-(3,5-dıchloro-benzyl)-3 h-ımıdazol-4-Yl]-ethylamıno}-4-methyl-pentanoic acid) | − 7.24 | Arg273, Phe274, His345, Pro346, Thr347, Ala348, Trp349, Thr371, His374, Glu375, His378, His401, Phe504, His505, Tyr510, Tyr515 | |
| 1 | Caffeic acid (3,4-dihyroxycinnamic acid) | − 5.53 | 88.12 | Arg273, His345, Pro346, Thr347, Ala348, His374, Glu375, His378, Phe504, His505, Tyr515 |
| 2 | Caffeic acid phenethyl ester (Caffeic acid 2-phenylethyl ester; β-Phenylethyl caffeate) (2-Phenylethyl (2E)-3-(3,4-dihydroxyphenyl)acrylate) | − 7.58 | 2.79 | Arg273, Phe274, His345, Pro346, Thr347, Ala348, Thr371, His374, Glu375, His378, Glu406, Phe504, His505, Tyr515, Arg518 |
| 3 | Chrysin (5,7-dihydroxyflavone) | − 7.17 | 5.58 | Tyr127, Ser128, Leu144, Glu145, Asn149, Val343, Cys344, His345, Pro346, Met360, Cys361, Thr362, Lys363, Asp368 |
| 4 | Galangin ( 3,5,7-trihydroxyflavone) | − 7.35 | 4.10 | Tyr127, Ser128, Leu144, Glu145, Trp271, Val343, Cys344, His345, Pro346, Met360, Cys361, Thr362, Lys363, Asp368, Phe504 |
| 5 | Myricetin (3,3′,4′,5,5′,7-hexahydroxyflavone) | − 7.59 | 2.74 | Arg273, Phe274, His345, Pro346, Thr347, Ala348, Thr371, His374, Glu375, His378, Glu406, Phe504, His505, Tyr515, Arg518 |
| 6 | Rutin (quercetin-3-rutinoside hydrate) | − 8.04 | 1.29 | Tyr127, Leu144, Glu145, Asn149, Glu150, Met152, Ala153, Asp269, Met270, Trp271, Phe274, Thr276, Cys344, His345, Pro346, Met360, Cys361, Lys363, Asp367, Asp368, Thr371, Thr445, Leu503, Phe504 |
| 7 | Hesperetin (3',5,7-trihydroxy-4'-methoxyflavanone) | − 7.45 | 3.46 | Arg273, Phe274, His345, Pro346, Thr347, Ala348, Trp349, His374, Glu375, His378, Glu402, Phe504, His505, Tyr510, Tyr515, Arg518 |
| 8 | Kaempferol (3,5,7-trihydroxy-2-(4-hydroxyphenyl)chromen-4-one) | − 7.23 | 5.05 | Arg273, Phe274, His345, Pro346, Thr347, Ala348, Thr371, His374, Glu375, His378, Glu406, Phe504, His505, Tyr510, Tyr515, Arg518 |
| 9 | Syringic acid (3,5-dimethoxy-4-hydroxybenzoic acid) | − 4.49 | 512.34 | Tyr127, Ser128, Leu144, Glu145, Asn149, Trp271, Val343, Cys344, His345, Pro346, Cys361, Phe504, |
| 10 | Pinocembrin (5,7-dihydroxy-2-phenyl-2,3-dihydro-4H-chromen-4-one) | − 7.16 | 5.61 | Tyr127, Ser128, Leu144, Glu145, Asn149, Val343, Cys344, His345, Pro346, Met360, Cys361, Thr362, Lys363, Asp368 |
| 11 | Luteolin (2-(3,4-dihydroxyphenyl)- 5,7-dihydroxy-4-chromenone) | − 7.29 | 4.52 | Tyr127, Ser128, Thr129, Leu144, Glu145, Asn149, Trp168, Val343, Cys344, His345, Pro346, Met360, Cys361, Thr362, Lys363, Asp368, Phe504 |
| 12 | Quercetin | − 7.58 | 2.79 | Arg273, Phe274, His345, Pro346, Thr347, Ala348, Thr371, His374, Glu375, His378, Glu406, Phe504, His505, Tyr515, Arg518 |
Furthermore, these compounds interacted with Phe274, Thr371, His345, Pro346 and Pro504 in ACE-II binding site. Especially, our in silico study showed that, rutin has the best binding affinity to the ACE-II enzyme (Binding energy: − 8.04 kcal/mol). This compound was observed to bind to the residues Asn149, Met270, His345, Lys363, Asp368 and Thr445 of ACE-II protein with the stronger hydrogen bond (Fig. 3). It can be suggested that these residues can contribute to the enhancement of ligand affinity for ACE-II enzyme. In addition, this compound has the pi-anion interaction with Arg269, pi-pi stacked interaction with Phe274, alkyl interaction with Pro346, Met360 and Cys361 and pi-alkyl interaction with Ala153 residues (Fig. 3).
Fig. 3.
Amounts of eleven phenolic compounds in mg/100 mL
Therefore, in this study, in silico effects of Anatolian propolis on ACE-II enzyme inhibition was investigated with the ten flavonoids as major substances. The results of this study showed that rutin, quercetin, CAPE, myricetin and hesperetin compounds effectively inhibit the ACE-II enzyme. These compounds can be clinically tested and used for the treatment disease role of ACE-II. Furthermore, Phe274, Thr371, His345, Pro346 and Pro504 are potential inhibitor targeting sites for the ACE-II enzyme. Based on this information, we propose guidelines to develop novel and specific inhibitors that target ACE-II enzyme.
Guerrero et al. (2012) experimentally demonstrated that some flavonoids have a relatively high inhibition potential for ACE-I. With the molecular docking studies, we have shown that some of these flavonoids inhibit ACE-II. ACE-I and ACE-II enzymes are metalloproteases, both of which contain similar zinc fingers (HEXXH) in their active sites. Molecular docking studies indicated that there are bond interactions between rutin and zinc finger residues of ACE-II. Because of similar active sites of ACE I and II, rutin may functionally bind both ACE I and II similar way.
It is revealed that COVID-19 binds to human Angiotensin-converting enzyme 2 (ACE-II) to enter the host cells. It is predicted that the rutin may compete with COVID-19 for ACE-II and have the potential to prevent or delay the entry of COVID-19 into the cell. In recent years, flavonoids have gained a great amount of interest with regards to their potential for cardiovascular protection. Many epidemiological studies associate an increased consumption of foods and beverages rich in flavonoids with a reduced risk of CVD death (Zhou et al. 2000; Joshipura et al. 2001; Kris-Etherton et al. 2002). Additionally, several of these flavonoids or their derivatives (i.e., diosmin, rutin and quercetin) are widely used as pharmaceutical agents for their vasoprotective properties (i.e., Daflon 500, cantaining flavonoid derivatives hesperedin and diosmin) (Muhammad et al. 2015). Therefore, rutin and other flavonoids used in this study can be used for prophylactic purposes as ACE-II inhibitors and competitor (Mishima et al. 2005; Maruyama et al. 2009).
Conclusion
This study shows that the high binding constants for the ACE-II receptors of flavanones in the ethanolic propolis extract make it a good competitive inhibitor and protective natural agents for the treatment of COVID-19. With the best binding affinity for ACE-II, rutin has the potential to compete with COVID-19 for ACE-II. In addition, it is predicted that COVID-19 may have a potential to prevent or delay its entry into the cell. However, this study should be supported with further in vivo studies and clinical trials.
Supplementary Information
Below is the link to the electronic supplementary material.
Declarations
Conflict of interest
No potential conflict of interest was reported by the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aliyazıcıoglu R, Sahin H, Erturk O, Ulusoy E, Kolayli S. Properties of phenolic composition and biological activity of propolis from Turkey. Int J Food Prop. 2013;16(2):277–287. doi: 10.1080/10942912.2010.551312. [DOI] [Google Scholar]
- Andrade JKS, Denadai M, de Oliveira CS, Nunes ML, Narain N. Evaluation of bioactive compounds potential and antioxidant activity of brown, green and red propolis from Brazilian northeast region. Food Res Int. 2017;101:129–138. doi: 10.1016/j.foodres.2017.08.066. [DOI] [PubMed] [Google Scholar]
- Anjum SI, Ullah A, Khan KA, Attaullah M, Khan H, et al. Composition and functional properties of propolis (bee glue): a review. Saudi J Biol Sci. 2019;26(7):1695–1703. doi: 10.1016/j.sjbs.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltas N, Yildiz O, Kolayli S. Inhibition properties of propolis extracts to some clinically important enzymes. J Enzyme Inhib Med Chem. 2016;31(sup1):52–55. doi: 10.3109/14756366.2016.1167049. [DOI] [PubMed] [Google Scholar]
- Barbarić M, Mišković K, Bojić M, Lončar MB, Smolčić-Bubalo A, et al. Chemical composition of the ethanolic propolis extracts and its effect on HeLa cells. J Ethnopharmacol. 2011;135(3):772–778. doi: 10.1016/j.jep.2011.04.015. [DOI] [PubMed] [Google Scholar]
- Cakir HE, Şirin Y, Kolayli S, Can Z. Validation methods for phenolic components with RP-HPLC-UV in various bee products. Apiterapi Ve Doğa Dergisi. 2018;1(1):13–19. [Google Scholar]
- Can Z, Yıldız O, Şahin H, Asadov A, Kolaylı S. Phenolic profile and antioxidant potential of propolis from Azerbaijan. Mellifera. 2015;15(1):16–28. [Google Scholar]
- Catchpole O, Mitchell K, Bloor S, Davis P, Suddes A. Anti-gastrointestinal cancer activity of cyclodextrin-encapsulated propolis. J Funct Foods. 2018;41:1–8. doi: 10.1016/j.jff.2017.12.023. [DOI] [Google Scholar]
- Chang CC, Yang MH, Wen HM, Chern JC. Estimation of total flavonoid content in propolis by two complementary colorimetric methods. J Food Drug Anal. 2002;10(3):178–182. [Google Scholar]
- Dassault Systèmes BIOVIA . Discovery studio modeling environment, release 2017. San Diego: Dassault Systèmes; 2017. [Google Scholar]
- Erdogan S, Ates B, Durmaz G, Yilmaz I, Seckin T. Pressurized liquid extraction of phenolic compounds from Anatolia propolis and their radical scavenging capacities. Food Chem Toxicol. 2011;49(7):1592–1597. doi: 10.1016/j.fct.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Graikou K, Popova M, Gortzi O, Bankova V, Chinou I. Characterization and biological evaluation of selected Mediterranean propolis samples. Is it a new type? LWT Food Sci Technol. 2016;65:261–267. doi: 10.1016/j.lwt.2015.08.025. [DOI] [Google Scholar]
- Guerrero L, Castillo J, Quiñones M, Garcia-Vallvé S, Arola L, et al. Inhibition of angiotensin-converting enzyme activity by flavonoids: structure-activity relationship studies. PLoS ONE. 2012;7(11):e49493. doi: 10.1371/journal.pone.0049493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain F, Jahan N, Rahman KU, Sultana B, Jamil S. Identification of hypotensive biofunctional compounds of coriandrum sativum and evaluation of their angiotensin-converting enzyme (ACE) inhibition potential. Oxid Med Cell Longev. 2018 doi: 10.1155/2018/4643736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshipura KJ, Hu FB, Manson JE, Stampfer MJ, Rimm E, et al. The effect of fruit and vegetable intake on risk for coronary heart disease. Ann Intern Med. 2001;134:1106–1114. doi: 10.7326/0003-4819-134-12-200106190-00010. [DOI] [PubMed] [Google Scholar]
- Kris-Etherton P, Keen C. Evidence that the antioxidant flavonoids in tea and cocoa are beneficial for cardiovascular health. Curr Opin Lipidol. 2002;13:41–49. doi: 10.1097/00041433-200202000-00007. [DOI] [PubMed] [Google Scholar]
- Kuba K, Imai Y, Rao S, Gao H, Guo F, et al. A crucial role of angiotensin converting enzyme 2 (ACE II) in SARS coronavirus—induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K, Imai Y, Ohto-Nakanishi T, Penninger J. Trilogy of ACE II: a peptidase in the renin–angiotensin system, a SARS receptor, and a partner for amino acid transporters. Pharmacol Ther. 2010;128(1):119–128. doi: 10.1016/j.pharmthera.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol. 2015;89:1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH, Moore MJ, Vasilieva N, Sui JH, Wong SK, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Li WH, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- López BGC, Schmidt EM, Eberlin MN, Sawaya AC. Phytochemical markers of different types of red propolis. Food Chem. 2014;146:174–180. doi: 10.1016/j.foodchem.2013.09.063. [DOI] [PubMed] [Google Scholar]
- Lu Y, Wu C, Yuan Z. Determination of hesperetin, cinnamic acid and nicotinic acid in propolis with micellar electrokinetic capillary chromatography. Fitoterapia. 2004;75(3–4):267–276. doi: 10.1016/j.fitote.2003.12.026. [DOI] [PubMed] [Google Scholar]
- Malkoç M, Çakır H, Kara Y, Can Z, Kolaylı S. Phenolic composition and antioxidant properties of Anzer honey from black sea region of Turkey. Uludağ Arıcılık Dergisi. 2019;19(2):143–151. doi: 10.31467/uluaricilik.602906. [DOI] [Google Scholar]
- Maruyama H, Sumitou Y, Sakamoto T, Araki Y, Hara H. Antihypertensive effects of flavonoids isolated from brazilian green propolis in spontaneously hypertensive rats. Biol Pharm Bull. 2009;32(7):1244–1250. doi: 10.1248/bpb.32.1244. [DOI] [PubMed] [Google Scholar]
- Mishima S, Yoshida C, Akino S, Sakamoto T. Antihypertensive effects of Brazilian propolis: identification of caffeoylquinic acids as constituents involved in the hypotension in spontaneously hypertensive rats. Biol Pharm Bull. 2005;28(10):1909–1914. doi: 10.1248/bpb.28.1909. [DOI] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad SA, Fatima N. In silico analysis and molecular docking studies of potential angiotensin-converting enzyme inhibitor using quercetin glycosides. Pharmacogn Mag. 2015;11(Suppl 1):S123. doi: 10.4103/0973-1296.157712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietta PG. Flavonoids as antioxidants. J Nat Prod. 2000;63(7):1035–1042. doi: 10.1021/np9904509. [DOI] [PubMed] [Google Scholar]
- Pobiega K, Kraśniewska K, Gniewosz M. Application of propolis in antimicrobial and antioxidative protection of food quality—a review. Trends Food Sci Technol. 2019;83:53–62. doi: 10.1016/j.tifs.2018.11.007. [DOI] [Google Scholar]
- Pratsinis H, Kletsas D, Melliou E, Chinou I. Antiproliferative activity of Greek propolis. J Med Food. 2010;13(2):286–290. doi: 10.1089/jmf.2009.0071. [DOI] [PubMed] [Google Scholar]
- Saral Ö, Yildiz O, Aliyazicioğlu R, Yuluğ E, Canpolat S, et al. Apitherapy products enhance the recovery of CCL4-induced hepatic damages in rats. Turk J Med Sci. 2016;46(1):194–202. doi: 10.3906/sag-1411-35. [DOI] [PubMed] [Google Scholar]
- Saral Ö, Kilicarslan M, Şahin H, Yildiz O, Dincer B. Evaluation of antioxidant activity of bee products of different bee races in Turkey. Turk J Vet Anim Sci. 2019;43(4):441–447. doi: 10.3906/vet-1901-3. [DOI] [Google Scholar]
- Sawaya ACHF, da Silva Cunha IB, Marcucci MC. Analytical methods applied to diverse types of Brazilian propolis. Chem Cent J. 2011;5(1):1–10. doi: 10.1186/1752-153X-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira MAD, Teles F, Berretta AA, Sanches TR, Rodrigues CE, et al. Effects of Brazilian green propolis on proteinuria and renal function in patients with chronic kidney disease: a randomized, double-blind, placebo-controlled trial. BMC Nephrol. 2019;20(1):140. doi: 10.1186/s12882-019-1337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Wu Z, Wang Z, Zhang H. Effect of ethanol/water solvents on phenolic profiles and antioxidant properties of Beijing propolis extracts. Evid Based Complement Altern Med. 2015 doi: 10.1155/2015/595393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner AJ, Hiscox JA, Hooper NM. ACE II: from vasopeptidase to SARS virus receptor. Trends Pharmacol Sci. 2004;25:291–294. doi: 10.1016/j.tips.2004.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7):e00127–e220. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sui X, Sui DJ, Yang P. Flavonoid extract from propolis inhibits cardiac fibrosis triggered by myocardial infarction through upregulation of SIRT1. Evid Based Complementary and Altern Med. 2018;27:4957573. doi: 10.1155/2018/4957573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KL, Peng GQ, Wilken M, Geraghty RJ, Li F. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J Biol Chem. 2012;287:8904–8911. doi: 10.1074/jbc.M111.325803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Yan QH, Ma JY, Wang Q, Zhang JW, et al. High performance liquid chromatographic determination of phenolic compounds in propolis. Trop J Pharm Res. 2013;12(5):771–776. [Google Scholar]
- Yildiz O, Karahalil F, Can Z, Sahin H, Kolayli S. Total monoamine oxidase (MAO) inhibition by chestnut honey, pollen and propolis. J Enzyme Inhib Med Chem. 2014;29(5):690–694. doi: 10.3109/14756366.2013.843171. [DOI] [PubMed] [Google Scholar]
- Zhou B, Yang L, Liu ZL. Antioxidative effects of green tea polyphenols on free radical initiated and photosensitized peroxidation of human low density lipoprotein. Chem Phys Lipid. 2000;106:53–63. doi: 10.1016/s0009-3084(00)00133-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.