Abstract
Background
A dysregulated immune response is considered one of the major factors leading to severe COVID-19. Previously described mechanisms include the development of a cytokine storm, missing immunoglobulin class switch, antibody-mediated enhancement, and aberrant antigen presentation.
Objectives
To understand the heterogeneity of immune response in COVID-19, a thorough investigation of histomorphological patterns in regional lymph nodes was performed.
Materials and methods
Lymph nodes from the cervical, mediastinal, and hilar regions were extracted from autopsies of patients with lethal COVID-19 (n = 20). Histomorphological characteristics, SARS-CoV‑2 qRT-PCR, and gene expression profiling on common genes involved in immunologic response were analyzed.
Results
Lymph nodes displayed moderate to severe capillary stasis and edema, an increased presence of extrafollicular plasmablasts, mild to moderate plasmacytosis, a dominant population of CD8+ T‑cells, and CD11c/CD68+ histiocytosis with hemophagocytic activity. Out of 20 cases, 18 presented with hypoplastic or missing germinal centers with a decrease of follicular dendritic cells and follicular T‑helper cells. A positive viral load was detected by qRT-PCR in 14 of 20 cases, yet immunohistochemistry for SARS-CoV-2 N-antigen revealed positivity in sinus histiocytes of only one case. Gene expression analysis revealed an increased expression of STAT1, CD163, granzyme B, CD8A, MZB1, and PAK1, as well as CXCL9.
Conclusions
Taken together, our findings imply a dysregulated immune response in lethal COVID-19. The absence/hypoplasia of germinal centers and increased presence of plasmablasts implies a transient B‑cell response, implying an impaired development of long-term immunity against SARS-CoV‑2 in such occasions.
Keywords: CD8-positive T‑lymphocytes, Cytokine release syndrome, Germinal center, Immunohistochemistry, SARS-CoV‑2
Abstract
Hintergrund
Eine dysregulierte Immunantwort, z. B. in der Form eines Zytokinsturmes, einer Störung des Immunglobulinklassenwechsels, eines sog. antikörpervermitteltem Enhancements oder einer aberranten Antigenpräsentation wurde bereits in schweren Krankheitsverläufen von COVID-19 beschrieben.
Ziel der Arbeit
Zur Charakterisierung der COVID-19-Immunantwort wurde die Histomorphologie der Lymphknoten des pulmonalen Abflussgebietes untersucht.
Material und Methoden
Regionale Lymphknoten des pulmonalen Abflussgebiets wurden bei COVID-19-Autopsien asserviert (n = 20). Deren Histomorphologie, SARS-CoV-2-qRT-PCR sowie Genexpressionsanalysen von gängigen Genen der Immunantwort wurden berücksichtigt.
Ergebnisse
Histologisch zeigten sich ein mäßig- bis schwergradiges Ödem mit Kapillarostase, eine erhöhte Anzahl von extrafollikulären Plasmablasten, milde bis mäßige Plasmazytose, vermehrte CD8+-T-Zellen und CD11c/CD68+-Histiozyten mit Hämophagozytoseaktivität. Von 20 Fällen wiesen 18 hypoplastische oder fehlende Keimzentren sowie eine Verminderung der follikulären dendritischen Zellen und follikulären T‑Helferzellen auf. In 14 von 20 Fällen war der qRT-PCR-Nachweis von SARS-CoV‑2 positiv, jedoch zeigte sich nur bei einem einzigen Fall eine immunhistochemische Positivität für SARS-CoV-2-N-Antigene in Sinushistiozyten. In Genexpressionsanalysen war eine erhöhte Expression von STAT1, CD163, Granzym B, CD8A, MZB1 und PAK1, neben CXCL9 zu beobachten.
Diskussion
Die Befunde in den Lymphknoten deuten auf eine dysregulierte Immunantwort bei schweren COVID-19-Krankheitsverläufen hin. Insbesondere impliziert das Ausbleiben der Keimzentrumsreaktion und die vermehrte Präsenz von Plasmablasten eine nur transiente B‑Zellreaktion, welche die Entwicklung einer Langzeitimmunität infrage stellt.
Schlüsselwörter: CD8-positive T‑Lymphozyten, Zytokin-Freisetzungssyndrom, Keimzentrum, Immunohistochemie, SARS-CoV‑2
The year 2020 was markedly influenced by the coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Although the virus’ origin, cellular entry mechanisms, and epidemiology were rapidly elucidated [2, 18, 41], many fundamental immunopathological questions related to SARS-CoV‑2 remain unanswered.
In-situ analyses of viral pathophysiology thus remained at the level of small series and case reports [1, 30, 43], which were nonetheless instrumental in the understanding of this novel disease. For example, lymphopenia was one of the symptoms initially observed in cases with severe disease, its extent directly correlating with disease severity [16]. This was typically accompanied by an increased susceptibility to superimposed bacterial infections [30], but its morphological correlate in lymphoid tissues has not been fully elucidated.
Histomorphological patterns in lymph nodes [42] may provide valuable insights into the immunopathological mechanisms of disease. They reflect complex interactions between antigens, immune cells, signaling molecules (cytokines, chemokines, and growth factors), and the complement system [39]. Immunogenetics, such as gene polymorphisms of second messengers, receptors, and histocompatibility genes, often determines the severity and duration of the histomorphologically perceptible changes, as exemplified by sarcoidosis [14]. The most typical pattern of an adequate immune response is reflected by follicular hyperplasia with associated specific antibody production and establishment of an immunological memory [39]. Some other morphologic patterns also mirror signal transduction between messengers and cells. For instance, angiofollicular (Castleman/Castleman-like) hyperplasia is associated with chronic exposure to interleukin (IL)6 [12], while granulomas are associated with an overproduction of IL2, IL12, and tumor necrosis factor α (TNFα) [17]. Systemic lupus erythematosus, Kikuchi–Fujimoto disease, and toxoplasmosis show an increase in CD123/CD303+ plasmacytoid dendritic cells in lymph nodes due to the activation of Toll-like receptors 7 and 9 (TLR7, TLR9) and an overproduction of interferon γ (IFNγ) [20].
A somewhat less commonly considered histomorphological pattern in lymph nodes is the extrafollicular proliferation of B cells (B blasts) in the absence of follicular hyperplasia [5]. This is the morphological correlate of a rapid or primarily transient expansion of B cells without the formation of germinal centers. Histologically, lymph node architecture is intact; however, paracortical zones are replete with a polymorphic infiltrate of smaller blasts, centroblasts, immunoblasts, and—most instructively—plasmablasts. Immunophenotypic features of these blasts vary, showing heterogeneous positivity for CD20, CD30, CD38, CD79a, CD138, IRF4 (MUM1), and BLIMP1 [5]. They are polytypic for light chains and are 70–80% IgM+/CD27−/CD30−/CD79a+/CD138−, as they do not undergo an immunoglobulin class switch at the genetic level and do not experience a germinal center reaction at the physiological level. In 20–30% of cases, they are IgG+/CD27+/CD30±/CD79a−/CD138+, a likely immunophenotype of extrafollicularly activated marginal zone B cells without long-lasting B cell response [9]. A predominance of such extrafollicular B blasts would lead to an overarching production of low-affinity IgM antibodies without a lasting immunological memory. This pattern is predominantly observed in mucosal lymph nodes and splenic tissue.
This paper reviews current knowledge on immunopathological changes in COVID-19 and presents distinct histomorphological patterns of locoregional lymph nodes of COVID-19 lungs. These observations are discussed in the context of gene expression profiles and immunological phenomena in severe disease courses, as well as their potential role in the development of an effective immune response and immunological memory.
Lymph node (histo)pathology of COVID-19
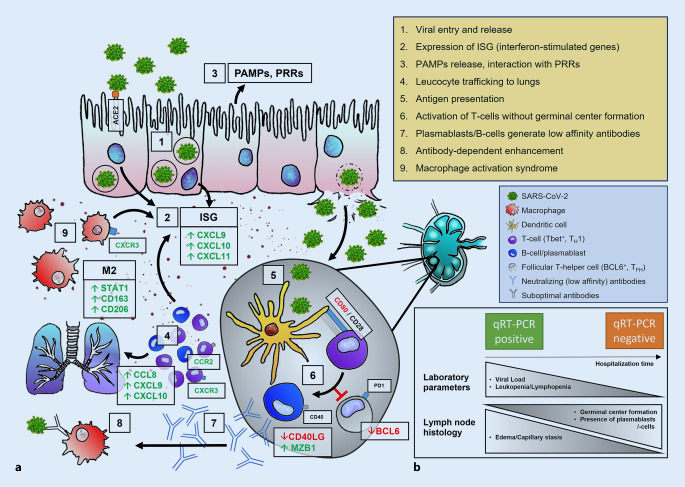
An overview of the major pathophysiological mechanisms of the immune response in COVID-19 is presented in Fig. 1a.
Fig. 1.
Immunopathology of SARS-CoV‑2 infection. a 1. Viral entry via ACE2, transcription of viral proteins and assembly, release of daughter virions. 2. Up-/dysregulation of ISG, cytokines, and chemokines. 3. Activation of PRRs by PAMPs. 4. Leukocyte trafficking into the lungs resulting in lymphopenia. 5. Antigen presentation in lymph nodes; activation of T cells (purple; e.g., by CTLA4/CD28) in a TH1 (Tbet+)-predominant milieu. 6. Inhibition of BCL6+ TFH cells (gray) disrupts germinal center formation, resulting in the activation of extrafollicular B blasts and, thus, an increase in plasmablasts (blue). 7. Release of specific (low-affinity) antibodies (blue) against SARS-CoV‑2. 8. Antibody-dependent enhancement. 9. Macrophage activation syndrome with predominance of an M2 phenotype including HLH and cytokine storm. b Temporal evolution of laboratory parameters and histology in COVID-19 lymph nodes. Short hospitalization time: high viral load, severe lymphopenia, absence of germinal centers accompanied by severe edema/capillary stasis and plasmablast accumulation. Longer hospitalization time: lower viral load, regressing lymphopenia, incipient (delayed) germinal center formation. ACE2 angiotensin-converting enzyme 2, HLH hemophagocytic lymphohistiocytosis, ISG interferon-stimulated gene, PAMPs pathogen-associated molecular patterns, PRRs pattern recognition receptors, RT-PCR reverse transcriptase polymerase chain reaction; green upregulated proteins, red downregulated proteins
Lymphopenia
Severe lymphopenia was one of the first observations among COVID-19 patients in early clinical case series [16]; its severity was shown to be the most important predictive factor of disease progression in a meta-analysis of 32 studies involving over 10,000 patients [26]. Pathophysiologically, lymphopenia could be the outcome of chemotactic dysregulation of CCL8, CCL20, and CXCL10, which were shown to be upregulated in COVID-19 lung tissue [1]. These molecules are responsible for chemotaxis and migration of (T) lymphocytes, possibly leading to extensive leukocyte trafficking into the lungs. Furthermore, lymphocytes express angiotensin-converting enzyme 2 (ACE2) [46], rendering possible direct cytopathic effects a further plausible explanation of lymphopenia.
Dysregulation of interferon-mediated signal transduction
IFN-mediated signal transduction plays a central role in the primary immune response to viral infections. Preliminary results of single-cell sequencing of peripheral blood mononuclear cells of COVID-19 patients revealed a phenotypic reconfiguration of immune cells with heterogeneous IFN-stimulated gene (ISG) signatures and downregulation of HLA class II genes, which was also associated with an increased plasmablast population [45]. Furthermore, ISG expression also appeared to be related to disease progression and viral load [34]. A subgroup of patients with early lethal disease showed high expression of ISG and cytokines, associated with high viral load and mild lung injury (ISGhigh) [30]. In contrast, another subgroup with prolonged disease showed decreased expression of ISGs (ISGlow) in association with severe diffuse alveolar damage, lower viral load, and increased infiltration of CD8+ cells and (M2-polarized) macrophages [34].
Dysregulation of antigen presentation
Gene expression analyses of COVID-19 lungs revealed downregulated expression of key costimulatory molecules of antigen presentation, such as TLR7, TLR9, and CD86 [1]. CD86 is expressed by antigen-presenting cells and binds to CD28 or CTLA4 expressed by T cells. Interaction between CD86 and CD28 subsequently stimulates a T cell response [39]. Other TLRs (TLR1, TLR4, TLR5) were also found to be downregulated in COVID-19. While TLR1 binds to peptidoglycans of Gram-positive bacteria, TLR4 interacts with lipopolysaccharides (LPS) of Gram-negative bacteria and is central for recognition of the SARS-CoV‑2 spike antigen [36]. Activation of TLR5 by flagellin stimulates the innate immune response against certain bacterial species [39]. This may explain why COVID-19 patients experience increased susceptibility towards bacterial superimposition.
Macrophage activation
Furthermore, the development of hemophagocytic lymphohistiocytosis (HLH) has been observed in COVID-19 patients [29, 38]. HLH typically is characterized by an uncontrolled macrophage activation syndrome, linked to impaired antigen presentation/recognition and excessive cytokine release (cytokine storm), but—to a limited and controlled extent—is known to be a typical reaction of the innate immune defense as a so-called pattern-recognition receptor (PRR)-associated response to pathogen-associated molecular patterns (PAMPs), specifically viral RNA [4, 30, 44]. HLH can be triggered by herpesviruses, especially EBV, but also by SARS-CoV‑1 and influenza viruses such as H5N1 and H1N1 [6, 15]. In accordance with this, COVID-19 tissue studies show increased expression of CCL2 and CCL7 [1], known to be chemotactic for macrophages, thus stressing the central role of the cytokine storm in COVID-19 pathogenesis and highlighting a potential therapeutic intervention in severe disease [25]. Furthermore, an upregulation of genes encoding complement activation and phagocytosis is observed in severe disease [48]. In this context, pediatric COVID-19 patients have been observed to develop multisystemic hyperinflammation, which phenotypically resembles Kawasaki syndrome [37]. In such cases, severe vascular and cardiac manifestation was observed, occasionally together with macrophage activation syndrome.
Dysregulation of the humoral immune response
In one of the first autopsy studies, an absence of germinal centers, an increased presence of plasmablasts, and intranodal capillary stasis were described in hilar lymph nodes and splenic parenchyma [30]. These histomorphological changes may be explained by a dysregulation of BCL6+ follicular T helper cells [21]which play an essential role in germinal center functionality. Additionally, early blockade of T helper cell differentiation, a predominance of T‑bet+ T helper 1 cells, and an extrafollicular accumulation of TNFα could be shown [21], corresponding to a loss of follicular B cells by flow cytometric analyses of peripheral blood from severely ill COVID-19 patients [21]. Interestingly, TLR4 and TLR5, which are both downregulated in COVID-19 as mentioned earlier, are also essential for the germinal center response as they activate the NF-κB signaling pathway via MYD88 [13].
The previously mentioned proliferation of plasmablasts in hilar lymph nodes in lethal COVID-19 infection could represent a morphological correlate of a dysregulation of immunoglobulin (Ig) class switching. This is supported by a markedly increased plasmablast population in patients with severe disease progression, whereas a robust adaptive immune response with clonally expanded CD8+ effector- or emory cells, at best, is observed in mild disease [23, 49]. Gene expression analyses of COVID-19 autopsy tissue also demonstrated downregulation of CD40LG [1], which is an essential link of communication between T and B cells and significantly influences B cell maturation [39]. A defect in CD40LG results in an absence of Ig class switching, which may favor preferential extrafollicular proliferation of B cells. As a reflection of the aforementioned two features, in-depth immunological analyses demonstrated a negative correlation between the number of memory B cells and COVID-19 symptom duration [33]; the number of these cells correlated with IgG1 and IgM against the SARS-CoV‑2 spike protein. This was also reflected in antibody titer measurements [32] and flow cytometric analyses, which showed oligoclonal plasmablast expansion (>30% of circulating B cells) [22] in cases with severe disease. This, along with other immunologic signatures that correlated with disease progression, allowed a biostatistical classification of three COVID-19 immune phenotypes with different risk profiles [28].
As a link to immunopathology, an incomplete humoral immune response with low-affinity, non-(sufficiently)-neutralizing, low-titer antibodies can lead to antibody-dependent enhancement. This is defined as the generation of suboptimal antibodies, which enable virus penetration into Fc/complement receptor-bearing monocytes, macrophages, and granulocytes [19]. Indeed, data indicate an interaction of anti-spike protein antibodies and macrophages that contributes significantly to lung injury in SARS-CoV‑1 [24]. Finally, spike protein reactivity was also shown to be present in over one third of SARS-CoV-2-naïve patients. This implies the presence of cross-reactive T cells, developed in the course of immunization against other coronaviruses and may explain the more robust immune response in some patient groups [4].
Methods
Tissue extraction and histology
Hilar, mediastinal, and cervical lymph nodes were acquired from autopsies of COVID-19 patients (n = 20). Staining protocols of histochemical and immunohistochemical assays (H&E, IgG, IgM, CD3, CD11c, CD20, CD79a, CD68, CD163, CD206, HLA-DR, SARS-CoV-2‑N antigen [Rockland 200-401-A50 rabbit polyclonal antibody, dilution 1:2000]) were performed in accordance with accredited standard operating procedure (SOP) protocols of the Institute of Pathology in Basel. Histologic characteristics (number of plasmablasts, plasma cells, edema/capillary stasis, presence of HLH and germinal centers) were evaluated using ordinal scales (0–3; Table 1).
Table 1.
Clinical, laboratory, and histopathological characteristics of qRT-PCR-positive and qRT-PCR-negative cases based on median viral load in lymph nodes
| qRT-PCR in lymph node | Total (n = 20) |
|||
|---|---|---|---|---|
| Positive (n = 14) |
Negative (n = 6) |
|||
| Clinical features/laboratory parameters (last value measured before death) | ||||
| Hospitalization time (days; median; IQR) | 6 (3–9) | 11 (4–30) | 8 (4–12) | |
| CRP (mg/dl; median; IQR) | 176 (100–271) | 315 (232–337) | 218 (144–280) | |
| Leucocytes (109/l; median; IQR) | 8.1 (6.3–11.4) | 16.0 (8.8–16.0) | 8.8 (7.4–13.8) | |
| Lymphocytes (109/l; median; IQR) | 0.7 (0.4–1.1) | 0.5 (0.4–0.96) | 0.7 (0.5–1.0) | |
| Neutrophilic granulocytes (109/l; median; IQR) | 6.7 (3.7–10.1) | 7.3 (7.26–10.6) | 6.8 (6.3–10.1) | |
| Histopathology and postmortem qRT-PCR | ||||
| Viral load, lungs (SARS-CoV‑2 genomes/106 RNaseP copies; median; IQR) | 2149 (57–60581) | 123 (0–365) | 146 (0–32218) | |
| Viral load, lymph nodes (SARS-CoV‑2 genomes/106 RNaseP copies; median; IQR) | 117 (21–10392) | 0 (0) | 21 (0–3100) | |
| Germinal centers (n, %) | Absent | 9 (64) | 3 (50) | 12 (60) |
| Scarce | 4 (29) | 2 (33) | 6 (33) | |
| Extensive | 1 (7) | 1 (17) | 2 (10) | |
| Plasmablasts (n, %) | Absent/scarce | 9 (64) | 3 (50) | 12 (60) |
| Moderate | 3 (21) | 3 (50) | 6 (33) | |
| Extensive | 2 (14) | 0 (0) | 2 (10) | |
| Plasma cells (n, %) | Absent/scarce | 12 (86) | 4 (66) | 16 (80) |
| Moderate | 2 (14) | 2 (33) | 4 (20) | |
| Extensive | 0 (0) | 0 (0) | 0 (0) | |
| Edema and capillary stasis (n, %) | Mild | 4 (29) | 0 (0) | 4 (20) |
| Moderate | 7 (50) | 6 (100) | 13 (65) | |
| Severe | 3 (21) | 0 (0) | 3 (15) | |
| Hemophagocytosis (n, %) | Absent | 5 (36) | 3 (50) | 8 (40) |
| Scarce | 6 (43) | 3 (50) | 9 (45) | |
| Extensive | 3 (21) | 0 (0) | 3 (15) | |
qRT-PCR quantitative reverse transcriptase polymerase chain reaction, CRP C‑reactive protein, IQR interquartile range
RT-PCR
All lymph nodes underwent RT-PCR examination to determine viral load. The protocol was described in our previous study [30].
Gene expression analyses
RNA was extracted by Forma HTG (HTG Molecular Diagnostics, Tucson, USA) from 10 µm thick untreated formalin-fixed paraffin-embedded (FFPE) blocks according to established protocols and processed and analyzed as described using the HTG EdgeSeq autoimmune assay [31].
Statistics
Statistical analyses were performed using SPSS version 25 (IBM, Armonk, NY, USA). All correlation analyses were performed using Spearman rho (ρ).
Results
Table 1 presents a summary of clinical and histological parameters of the 20 cases studied.
Clinical parameters and RT-PCR
In 14 out of 20 cases, qRT-PCR detected SARS-CoV‑2 RNA. Interestingly, in cases without detectable SARS-CoV‑2 RNA, hospitalization time was longer (6 vs. 11 days), suggesting a biphasic disease course with progressive viral elimination, similar to previously performed analyses of the same autopsy cohort ([34]; Fig. 1b). This is supported by a negative correlation of pulmonary viral load with hospitalization time (ρ = −0.776; p < 0.0001) and a positive correlation between pulmonary and lymph node viral load (ρ = 0.514; p = 0.035). Blood counts revealed a positive correlation between hospitalization time and absolute lymphocyte count (ρ = 0.582; p = 0.014) and a negative correlation with the absolute leucocyte count (ρ = −0.577; p = 0.015).
Histopathology
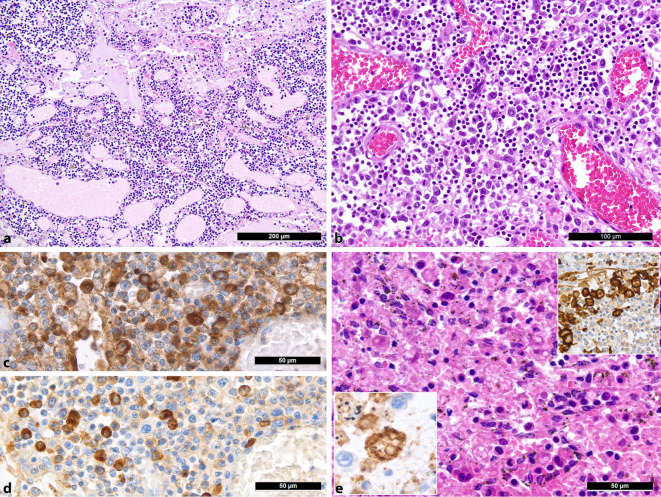
Major histologic lymph node features included moderate-to-severe edema and capillary stasis (Fig. 2a) in all cases, explained by acute right heart failure due to severe pulmonary disease. Further findings include a proliferation of extrafollicular B blasts, IgG- and IgM-positive plasmablasts in particular (Fig. 2b,c; 12/20), consistent with rapid or primarily transient B cell immune response bypassing the germinal center response as described above [5], with largely absent or hypoplastic/hypotrophic germinal centers or secondary follicles (12/20), including decreased follicular dendritic cells and follicular T helper cells. Correlation analyses showed a negative association between the presence of secondary follicles and viral load in the lungs (ρ = −0.645; p = 0.005) and CRP (secondary follicles = −0.522; p = 0.032), which, in conjunction with previous analyses of the same autopsy cohort [34], additionally underscores a delayed germinal center response. Furthermore, a discrete-to-moderate degree of plasmacytosis and a predominance of CD8+ T cells was observed, accompanied by numerous HLA-DR-, CD163-, and CD206-positive M2-polarized macrophages [24], CD11c- and CD68-positive histiocytes, and the presence of hemophagocytosis in the sinuses in 12/20 cases (Fig. 2d), consistent with the aforementioned phenomena of macrophage activation in COVID-19 [29, 38]; however, the full clinical picture of HLH presented itself in only one of the 20 patients examined here [47].
Fig. 2.
Histomorphological characteristics of regional lymph nodes of COVID-19 lungs. a Moderate to severe edema with capillary stasis (H&E; 50 ×). b Proliferation of extrafollicular plasmablasts (H&E; 200 ×). c, d IgG (c) and IgM (d) staining of plasmablasts (immunoperoxidase; 400 ×). e Hemophagocytosis in the sinus of a lymph node (H&E; 360 ×). Left inlet: positivity for CD11c in histiocytes undergoing hemophagocytosis (immunoperoxidase; 400 ×); right inlet: positivity for CD206, a specific marker of M2 polarization [48], in sinus histiocytes
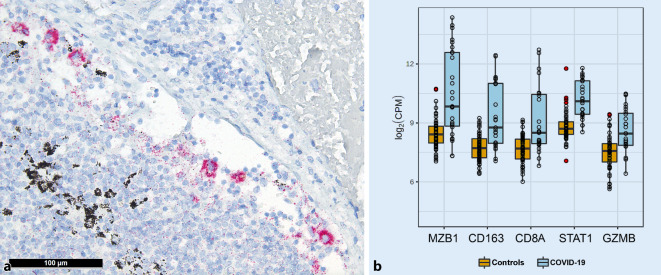
An increased number of sinus histiocytes with corresponding positive immunohistochemical detection of the nucleocapsid antigen of SARS-CoV‑2 in an elderly patient with a 30-day disease course may be interpreted as antibody-dependent enhancement, as further supported by observations from Martines et al. ([27]; Fig. 3a). Although the majority of lymph nodes examined illustrated imperceptible virus antigen levels as detected by immunohistochemistry, the lungs of the same patient showed a comparatively higher viral copy number than others (990 viral genome copies/106 human RNAseP copies in the lymph nodes and approximately 125000 in the lungs).
Fig. 3.
Immunohistochemistry detecting SARS-CoV‑2 and associated gene expression profiles in COVID-19. a Positivity in sinus histiocytes for SARS-CoV‑2 as detected by immunohistochemistry for the SARS-CoV-2 N-antigen (immunoperoxidase with 3‑amino-9-ethylcarbazole used as a chromogen; 200 ×). b Gene expression profile shows significantly increased expression of MZB1, CD163, CD8A, STAT1, and GZMB in COVID-19 compared to controls
Gene expression profiles
In line with the histomorphological patterns described above, gene expression profiles (Fig. 3b) showed increased expression of the following genes: STAT1 (central transcription factor in macrophage activation), CD163 (hemoglobin–haptoglobin complex receptor and marker of M2 macrophage polarization [24]), granzyme B (GZMB) but not perforin (consistent with the imbalance of both proteins in hemophagocytic lymphohistiocytosis [40]), CXCL9 and PAK1 (a chemokine and an enzyme important for the migration of cytotoxic T cells), CD8A (in line with the upregulation of PAK1), and, finally, MZB1 (marginal zone B and B1 cell-specific protein, which contributes to the composition and secretion of IgM and thus corresponds with the increased plasmablasts).
Discussion
The histomorphological patterns observed in lymph nodes appear to reflect the underlying immunopathology of lethal COVID-19. Our data imply a biphasic disease course, demonstrated by an initially high viral load and severe lymphopenia, which both eventually regress. Histologically, edema and plasmablast activation are evident in the lymph nodes of early lethal cases, followed by macrophage activation and a subtle germinal center reaction in cases with a longer disease duration (Fig. 1b), thus supporting this premise. These data are consistent with gene expression analyses performed on the same autopsy cohort [34], which unraveled a marked variance in ISG gene signature in patients with varying hospitalization time. A disruption of the IFN response is implied in severe COVID-19, as supported by the following:
All coronaviruses, especially SARS-CoV‑2, suppress the production and release of all three types of interferons [10].
Patients with severe COVID-19 show loss-of-function variants in TLR- and interferon-dependent genes or neutralizing antibodies to type I IFN (α and ω) [3, 49].
Dysregulation of type I IFN seems to be generally crucial for disease progression [7, 8].
-
The ISG response changes significantly as COVID-19 progresses and seems to influence different aspects of immunopathology [34].
This disruption of the IFN response may explain the dysregulation of BCL6+ follicular T helper cells [21], the rapid but poorly specific B cell immune response bypassing the germinal center reaction with plasmablasts generating low-affinity antibodies [5, 22, 45], the resultant antibody-dependent enhancement with macrophage (hyper)activation [19] with sinus histiocytosis and possibly HLH [29, 38], and M2 macrophage polarization [24, 34, 49]. This ultimately results in a highly pathogenic inflammatory monocyte-macrophage response that is well documented in SARS-CoV‑1 and MERS [7, 8, 24], leading to extensive organ damage [25, 48]. Antibody-dependent enhancement could also contribute to the in-situ positivity in monocytes/macrophages (Fig. 3a; [19]) and thus, by extension, systemic viral spread.
These complex immunopathological observations, illustrated in part by the histopathologic changes described in this study, could also explain the underlying challenges in efficient coronavirus vaccine development [11]. Furthermore, findings from in-situ studies of the lymphoid compartment in COVID-19 could provide valuable pharmacological approaches, such as TLR agonists [35], to increase the efficiency of vaccines under development, thus ensuring a durable immune response and memory.
Practical conclusion
The analysis of histopathological features in locoregional lymph nodes is indispensable for the understanding of COVID-19 pathophysiology. In particular, the absence of a germinal center response accompanied by plasmablast expansion and a lack of immunoglobulin class switching suggest an inefficient antibody response in patients with severe disease.
A dysregulation of interferon (IFN)-mediated signal transduction appears to be central to the immunopathology of COVID-19. Initial therapeutic approaches of IFN I‑interfering drugs (JAK inhibitors such as baricitinib or dexamethasone) are currently undergoing clinical trials (NCT04358614), while potential clinical studies involving TLR agonists are yet to be launched.
Further studies are needed to shed light on the underlying mechanisms of innate and humoral immune responses in mild and severe disease courses, as well as phenomena such as cross-reactivity of certain T cell subpopulations. This will potentially aid the development of an efficient vaccine.
Acknowledgments
Acknowledgements
This study was supported by the Botnar Research Centre for Child Health, BRCCH.
Abbreviations
- BLIMP-1/PRDM‑1
PR domain zinc finger protein 1
- CCL
Chemokine ligand
- CD
Cluster of differentiation
- COVID-19
Coronavirus disease 2019
- CTLA4
Cytotoxic T lymphocyte-associated protein 4
- CXCL9
Chemokine (C-X‑C motif) ligand 9
- EBV
Epstein–Barr virus
- Fc
Fragment constant
- HLA-DR
Human leukocyte antigen DR
- HLH
Hemophagocytic lymphohistiocytosis
- IFN
Interferon
- Ig
Immunoglobulin
- IL
Interleukin
- IRF4 (MUM-1)
Interferon regulatory factor 4
- ISG
interferon-stimulated genes
- LPS
Lipopolysaccharide
- MYD88
Myeloid differentiation primary response 88
- MUM‑1 (IRF4)
Multiple myeloma 1 protein
- MZB1
Marginal zone B and B1 cell-specific protein
- NF-κB
Nuclear factor kappa B
- PAK1
P21 (RAC1)-activated kinase 1
- PAMPs
Pathogen-associated molecular patterns
- PRRs
Pattern recognition receptors
- qRT-PCR
Quantitative reverse transcriptase polymerase chain reaction
- SARS-CoV-1/2
Severe acute respiratory syndrome coronavirus 1/2
- STAT1
Signal transducer and activator of transcription 1
- TLR
Toll-like receptor
- TNF‑α
Tumor necrosis factor alpha
Funding
Open access funding provided by University of Basel
Declarations
Conflict of interest
J.D. Haslbauer, M.S. Matter, A.K. Stalder, and A. Tzankov declare that they have no competing interests.
All procedures performed in studies involving human participants or on human tissue were in accordance with the ethical standards of the institutional and/or national research committee and with the 1975 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
The supplement containing this article is not sponsored by industry.
References
- 1.Ackermann M, Verleden SE, Kuehnel M, et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen KG, Rambaut A, Lipkin WI, et al. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastard P, Rosen LB, Zhang Q, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:423. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun J, Loyal L, Frentsch M, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020 doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 5.Brighenti A, Andrulis M, Geissinger E, et al. Extrafollicular proliferation of B cells in the absence of follicular hyperplasia: a distinct reaction pattern in lymph nodes correlated with primary or recall type responses. Histopathology. 2005;47:90–100. doi: 10.1111/j.1365-2559.2005.02173.x. [DOI] [PubMed] [Google Scholar]
- 6.Brisse E, Wouters CH, Andrei G, Matthys P. How viruses contribute to the pathogenesis of hemophagocytic lymphohistiocytosis. Front Immunol. 2017;8:1102. doi: 10.3389/fimmu.2017.01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Channappanavar R, Fehr AR, Vijay R, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-coV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Channappanavar R, Fehr AR, Zheng J, et al. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Invest. 2019;129:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chappell CP, Draves KE, Giltiay NV, Clark EA. Extrafollicular B cell activation by marginal zone dendritic cells drives T cell-dependent antibody responses. J Exp Med. 2012;209:1825–1840. doi: 10.1084/jem.20120774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu H, Chan JF-W, Wang Y, et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin Infect Dis. 2020;71:1400–1409. doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conte C, Sogni F, Affanni P, et al. Vaccines against coronaviruses: the state of the art. Vaccines (Basel) 2020;8:309. doi: 10.3390/vaccines8020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cronin DMP, Warnke RA. Castleman disease: an update on classification and the spectrum of associated lesions. Adv Anat Pathol. 2009;16:236–246. doi: 10.1097/PAP.0b013e3181a9d4d3. [DOI] [PubMed] [Google Scholar]
- 13.De Silva NS, Anderson MM, Carette A, et al. Transcription factors of the alternative NF-κB pathway are required for germinal center B-cell development. Proc. Natl. Acad. Sci. U. S. A. 2016;113:9063–9068. doi: 10.1073/pnas.1602728113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fingerlin TE, Hamzeh N, Maier LA. Genetics of sarcoidosis. Clin Chest Med. 2015;36:569–584. doi: 10.1016/j.ccm.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Griffin G, Shenoi S, Hughes GC. Hemophagocytic lymphohistiocytosis: an update on pathogenesis, diagnosis, and therapy. Best Pract Res Clin Rheumatol. 2020;34:101515. doi: 10.1016/j.berh.2020.101515. [DOI] [PubMed] [Google Scholar]
- 16.Guan W-J, Ni Z-Y, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helming L, Gordon S. Molecular mediators of macrophage fusion. Trends Cell Biol. 2009;19:514–522. doi: 10.1016/j.tcb.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwasaki A, Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol. 2020;20:339–341. doi: 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jegalian AG, Facchetti F, Jaffe ES. Plasmacytoid dendritic cells: physiologic roles and pathologic states. Adv Anat Pathol. 2009;16:392–404. doi: 10.1097/PAP.0b013e3181bb6bc2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaneko N, Kuo H-H, Boucau J, et al. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell. 2020;183:1–15. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuri-Cervantes L, Pampena MB, Meng W, et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci Immunol. 2020;5:1–15. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao M, Liu Y, Yuan J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 24.Liu L, Wei Q, Lin Q, et al. Anti–spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4:123–158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahmudpour M, Roozbeh J, Keshavarz M, et al. COVID-19 cytokine storm: the anger of inflammation. Cytokine. 2020;133:151–155. doi: 10.1016/j.cyto.2020.155151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malik P, Patel U, Mehta D, et al. Biomarkers and outcomes of COVID-19 hospitalisations: systematic review and meta-analysis. BMJ Evid Based Med. 2020 doi: 10.1136/bmjebm-2020-111536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martines RB, Ritter JM, Matkovic E, et al. Pathology and pathogenesis of SARS-coV-2 associated with fatal coronavirus disease, United States. Emerging Infect Dis. 2020;26:2005–2015. doi: 10.3201/eid2609.202095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathew D, Giles JR, Baxter AE, et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGonagle D, Sharif K, O’Regan A, Bridgewood C. The role of cytokines including interleukin-6 in COVID-19 induced pneumonia and macrophage activation syndrome-like disease. Autoimmun Rev. 2020;19:1025–1037. doi: 10.1016/j.autrev.2020.102537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menter T, Haslbauer JD, Nienhold R, et al. Post-mortem examination of COVID19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings of lungs and other organs suggesting vascular dysfunction. Histopathology. 2020;77:198–209. doi: 10.1111/his.14134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menter T, Hayoz S, Zucca E, et al. Immunomodulatory drugs may overcome the negative prognostic role of active Th17 axis in follicular lymphoma: evidence from the SAKK35/10 trial. Br J Haematol. 2020 doi: 10.1111/bjh.16876. [DOI] [PubMed] [Google Scholar]
- 32.Murphy P. Individuals with less severe manifestations of SARS-coV-2 infection may not develop long-lasting humoral immunity. Am J Clin Pathol. 2020 doi: 10.1093/ajcp/aqaa233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newell KL, Clemmer DC, Cox JB, et al. Switched and unswitched memory B cells detected during SARS-CoV-2 convalescence correlate with limited symptom duration. medRxiv. 2020;2020:1–29. doi: 10.1101/2020.09.04.20187724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nienhold R, Ciani Y, Koelzer VH, et al. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat Commun. 2020;11:5086. doi: 10.1038/s41467-020-18854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onofrio L, Caraglia M, Facchini G, et al. Toll-like receptors and COVID-19: a two-faced story with an exciting ending. Future Sci OA. 2020;6:605. doi: 10.2144/fsoa-2020-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park A, Iwasaki A. Type I and type III interferons—induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pouletty M, Borocco C, Ouldali N, et al. Paediatric multisystem inflammatory syndrome temporally associated with SARS-CoV-2 mimicking Kawasaki disease (Kawa-COVID-19): a multicentre cohort. Ann Rheum Dis. 2020;79:999–1006. doi: 10.1136/annrheumdis-2020-217960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prilutskiy A, Kritselis M, Shevtsov A, et al. SARS-coV-2 infection-associated hemophagocytic lymphohistiocytosis. Am J Clin Pathol. 2020;154:466–474. doi: 10.1093/ajcp/aqaa124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Punt J, Stranford S, Jones P, Owen J. Kuby immunology. 8. New York: Freeman, W.H. and Co; 2018. [Google Scholar]
- 40.Ramos-Casals M, Brito-Zerón P, López-Guillermo A, et al. Adult haemophagocytic syndrome. Lancet. 2014;383:1503–1516. doi: 10.1016/S0140-6736(13)61048-X. [DOI] [PubMed] [Google Scholar]
- 41.Sun J, He W-T, Wang L, et al. COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol Med. 2020;26:483–495. doi: 10.1016/j.molmed.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tzankov A, Dirnhofer S. A pattern-based approach to reactive lymphadenopathies. Semin Diagn Pathol. 2018;35:4–19. doi: 10.1053/j.semdp.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 43.Tzankov A, Jonigk D. Unlocking the lockdown of science and demystifying COVID-19: how autopsies contribute to our understanding of a deadly pandemic. Virchows Arch. 2020;477:331–333. doi: 10.1007/s00428-020-02887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Liu L. The membrane protein of severe acute respiratory syndrome coronavirus functions as a novel cytosolic pathogen-associated molecular pattern to promote beta interferon induction via a toll-like-receptor-related TRAF3-independent mechanism. mBio. 2016;7:01872–01815. doi: 10.1128/mBio.01872-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilk AJ, Rustagi A, Zhao NQ, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu H, Zhong L, Deng J, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12:1–5. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zellweger NM, Huber J, Tsakiris DA, Tzankov A, Gebhard CE, Siegemund M. Swiss Med Wkly. 2021;151:w20420. doi: 10.4414/smw.2021.20420. [DOI] [PubMed] [Google Scholar]
- 48.Zhang F, Gan R, Zhen Z, et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal Transduct Target Ther. 2020;5:1–11. doi: 10.1038/s41392-020-00263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Q, Bastard P, Liu Z, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:422. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]