Abstract
The intrinsic dynamic nature of chromosomes is emerging as a fundamental component in regulating DNA transcription, replication, and damage-repair among other nuclear functions. With this increased awareness, reinforced over the last ten years, many new experimental techniques, mainly based on microscopy and chromosome conformation capture, have been introduced to study the genome in space and time. Owing to the increasing complexity of these cutting-edge techniques, computational approaches have become of paramount importance to interpret, contextualize, and complement such experiments with new insights. Hence, it is becoming crucial for experimental biologists to have a clear understanding of the diverse theoretical modeling approaches available and the biological information each of them can provide.
Current Opinion in Genetics and Development 2021, 67:25–32
This review comes from a themed issue on Genome architecture and expression
Edited by Gerd Blobel and Susan M Gasser
For a complete overview see the Issue and the Editorial
Available online 3rd December 2020
https://doi.org/10.1016/j.gde.2020.10.004
0959-437X/© 2020 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Introduction
In the last ten years, our understanding of the relationship between genome structure and function in eukaryotic cells has tremendously increased. Owing to the synergistic development of advanced microscopy [1, 2, 3, 4, 5] and high-throughput chromosome conformation capture (3C-based) techniques [6, 7, 8], it has been possible to characterize the various features of three-dimensional (3D) genome organization [9,10]. Briefly, at the nuclear scale, chromosomes occupy distinct territories with limited intermingling that has been proposed to impact gene regulation [11]. At the tens of megabases scale, chromatin segregates into spatial (A/B) compartments that are characterized by distinctive GC-content, gene density and diverse chromatin marks [7,8]. At the submegabase scale, genomes are partitioned into topologically associating domains (TADs), that are proposed to be the main functional and structural units of the 3D genome where enhancers and promoters colocalize. However, the latest experimental developments have revealed that many fundamental nuclear and cellular processes occur in a time-dependent dynamical context, prompting the advent of the so-called 4D nucleome [12,13].
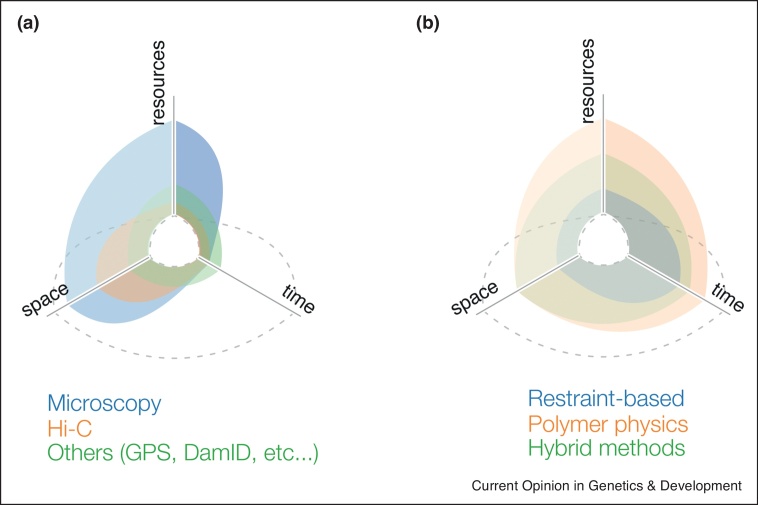
Biological processes happen in a wide range of time and spatial scales, which makes the concepts of genome structure and dynamics context-dependent. For example, in the fast dynamics and local length scales regime, gene transcription typically lasts within minutes. At intermediate scales, DNA replication, cell cycle and meiosis [14, 15, 16] span periods of hours, and involve the reorganization of entire chromosomes (hundreds of Mbs or ∼10 μm). Slower dynamics regulate, for example, cell differentiation and reprogramming [17,18••], lasting several days, and involving genome structural reorganization of both local and genome-wide scales. Notably, these phenomena need to be investigated by distinct experimental approaches each sensitive to a specific range of temporal and spatial scales (Figure 1a).
Figure 1.
Exploration map.
Radar chart displaying spanned areas of current experimental (a) and computational (b) approaches used to study the 4D nucleome. All axes are in arbitrary units. Vertical axis (‘resources’) indicates the required resources to execute the experiments/computations. Left axis (‘space’) indicates the coverage and depth of 3D space by either experiments or computation. Right axis (‘time’) indicates the coverage and depth of time by either experiments or computation. Dashed grey lines exemplifies a ‘perfect approach’ that requires very little resources but can provide the maximum insight in both space and time. Both the experimental and computational approaches have extensively charted the space dimension, but yet there is some work to do in unraveling the effects of the local scale on the global ones, and vice versa. In this sense, hybrid modeling has not yet exploited this to the fullest. The time axis has a great potential for further 4D nucleome modeling in parts of the exploration map still inaccessible to experiments. The resource dimension is currently the limiting factor, since both experiments and computation tend to use it at maximum. Experimental resource needs could be limited by, for example, reducing material requirements, as for instance a recently introduced low-input Hi-C technique [60]. As for computational resources, data-driven approaches are generally less demanding than top-down approaches, but the implementation of more efficient software may balance out this difference. For instance, bottom-up computational methods usually rely on few local force-fields and thus ought to be more computationally scalable than data-driven ones, given an efficient software implementation [34].
A crucial – and sometimes underappreciated – aspect of 4D nucleomics is the extensive development of computational tools that have been instrumental for reliable analysis, interpretation, and modeling of experimental data. To this end, data modeling encompasses two distinct, complementary strategies. On the one hand, data-driven or top-down approaches use experimental observations as input to generate 3D models representative of the data. However, the models represent more than a mere visualization as they often provide new insights into the structure-function relationships. In some cases [3], these models allow the integration of different datasets into unified models, disentangling possible similarities or incompatibilities between experiments. Data-driven 4D modeling usually covers the slow dynamics regime describing the large reorganization of genomes or of genomic regions at coarse genomic resolutions (tens of kbs) (Figure 1b). On the other hand, hypothesis-driven or bottom-up strategies build parametric, predictive models based on mechanistic hypotheses intuited from observations. By confronting model predictions of both genome structure and dynamics to experiments, the models allow to invalidate or consolidate the underlying assumed mechanisms and to propose novel experiments to further challenge them. Bottom-up approaches usually can treat both the fast dynamics regime describing short local genome kinetics at fine resolution and the slow dynamics regime describing chromosome reorganization at coarse genomic resolution (Figure 1b).
Taken together, these modeling achievements lead to an exploration map spanning dimensions of space, time and use of resources (Figure 1b). Here, we aim to review the recent efforts of charting new territories of this map to better characterise the 4D nucleome. Discussing exemplary applications, we highlight how modeling helped to add value to the experimental data providing novel biological insights otherwise inaccessible to experiments. We discuss criticalities and challenges, proposing feasible solutions which may drive the future developments of 4D nucleome modeling.
New insights from data-driven modeling
In recent years, experimental approaches (mainly 3C-based techniques) have offered an increasing number of time-resolved datasets, which aimed to study how the 3D genome architecture changes over time. An impressive range of temporal resolutions have been probed using these techniques, ranging from minutes [19,20], to hours [21••,22], to days [17,23••]. In some of these studies, data-driven 4D modeling has been used to convey an intuitive representation of complex dynamical behaviours of chromatin organization and nuclear shape [24], and to interpret the underlying features of the data [25,26], which enhanced our understanding of patterns not immediately observable in the raw representation of the data [21••,27••]. These hidden patterns often provide clues for further experimental exploration of the underlying biological system [21••,27••].
Data-driven techniques are based on four main methodological steps: (i) data collection; (ii) representation of the elementary genomic region; (iii) scoring of the possible structural conformations using the input data transformed into spatial restraints; and (iv) sampling the search space and ranking each conformation based on the satisfaction of the imposed restraints. The 3D models which optimally satisfy the input data-driven restraints are retained for further analysis [28].
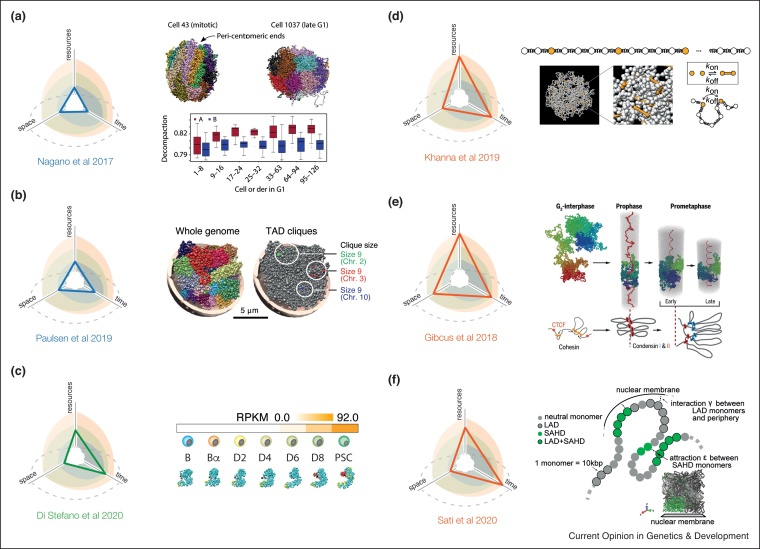
A common strategy to model the 4D nucleome is to consider each time-point separately, and generate 3D models whose characteristics can be used to explore the different ranges of temporal and spatial resolutions (Figure 2a). An illustrative example was provided by the Fraser and Tanay groups using single-cell Hi-C (scHi-C) experiments to characterize genome structure-dynamics during the cell cycle across thousands of individual cells [21••]. By in-silico inferred single-cell phasing, whole genome 3D models were generated representing the structural dynamics across stages of the cell cycle. From the modeling, the authors found that chromosomes rapidly decondense from a mitotic conformation during the progression of G1, yet with a more rapid decondensation of A (active/euchromatin) than B (inactive/heterochromatin) compartments. Further, the radial distribution of these compartments was found to be progressively established during G1, whereas long-range cis-contacts appeared earlier than the trans-compartment re-establishment.
Figure 2.
Illustration of selected 4D modeling studies.
On the left, each panel (a)–(f) shows the portion of the exploration map charted by the corresponding approach. These graphs illustrate the space and time dimensions one could explore given the available resources applying the various approaches. (a) Whole-genome models were obtained using single-cell Hi-C data at different cell-cycle phases [21••]. Models at different timepoints were crucial to unveil structural features only implicit in the Hi-C contact patterns such as the difference in decompaction speed between A (fast) and B (slow) during the G1 progression. (b) Center: Chrom3D whole-genome models in human adipose stem cells [18••] at the single-TAD level resolution. Each chromosome is indicated with a distinct color. Right: Three groups of TADs (cliques) in a repressive compartment predicted to be interacting with the nuclear lamina. (c) Trajectories of the Sox2 locus dynamics were simulated during the mouse B-to-iPSC reprogramming using the TADdyn tool [27••]. Upon expression activation in day 6 (D6), several regions (red beads) with enhancer characteristics (Open and Active chromatin) gather around the TSS of the locus (black bead) and form a 3D superenhancer hub. (d) Bead-and-spring polymer models with temporary crosslinking interactions were used in Ref. [41•] to study the structure and dynamics of the Igh locus in live mouse B-lymphocytes. The authors showed that the observed constrained motion of chromatin is consistent with a network of long-lived loops ensuring that the genomic region is ordered, but maintains enough fluidity. (e) 4D application of the loop extrusion model during mitotic chromosome folding [54••]. In particular, going from Prophase to Prometaphase, condensin II acts first by forming a helical scaffold of large adjacent loops (400 kbp), then condensin I further compacts these loops by folding them into shorter nested loops (80 kbp). (f) 3D organization of heterochromatin domains (SAHDs) in cycling and senescent human fibroblasts was modeled [23••] by polymer simulations accounting for the capacity of SAHDs to self-interact and to associate with the nuclear lamina. The slow 3D reorganization of SAHDs into large internal foci is consistent with a substantial weakening of lamina-SAHDs interactions mediated by HMGA-2.
More recently, Paulsen et al. used Hi-C and ChIP-seq of nuclear lamins (Lamin A/C and Lamin B1) to analyze genome structure dynamics during differentiation of human adipose stem cells into two distinct lineages [18••]. Using Chrom3D [29], the lamin-genome and Hi-C interactions were integrated to generate whole genome 3D models revealing a differentiation-coupled reinforcement of compartment compactification into a repressive state at the nuclear lamina (Figure 2b).
A recent alternative approach, called TADdyn [27••], integrates in a single trajectory chromosomal structural changes probed at discrete time-points along a biological process. This new hybrid (Figure 1b) approach allowed simulating gradual and smooth dynamical transitions between Hi-C experimental time-points by merging the methodological step of data-driven modeling with polymer-based representation of the chromatin fiber and molecular dynamics typically used in bottom-up approaches. TADdyn was used [27••] to study the structural changes of 21 genomic loci during the reprogramming of murine B cells to induced pluripotent stem cells. The simulations indicated the formation of 3D hubs harbouring enhancer-like regions around the transcription start site (TSS) of genes upon transcriptional activation. Similarly, these 3D super enhancers were found to disaggregate during gene silencing. For some genes (i.e. Sox2 and Nanog), the simulations also indicated the presence of a structural cage that embedded the TSS and confined its dynamics during gene expression (Figure 2c). These new types of simulations support the idea of local aggregation of active chromatin whose size correlates with the gene expression activity.
Deepening our understanding of mechanisms using modeling
Complementary to data-driven approaches, bottom-up modeling offers a set of quantitative frameworks to formalize, test and (in)validate mechanistic hypotheses on the dynamical processes driving the 4D nucleome. In particular, relying on computer simulations as their primary tool, these approaches use experimental data a priori to parameterize the models and a posteriori to validate the obtained results. The ultimate goal is to provide simple testable rules which can explain, in part, the complex nuclear architecture. While most of the current applications of bottom-up modeling aim at describing the average 3D organization of chromosomes [30], a plethora of new approaches are addressing how key molecular mechanisms affect the 4D nucleome.
To address the dynamical scales of the 4D nucleome, 3D polymer models parameterized with population-averaged 3D data have been used to predict and validate their consistency [31,32] mostly with experiments probing the fast dynamics of chromatin motion [33•,34, 35, 36, 37] and of chromatin-binding proteins [38,39]. An alternative perspective is to use 4D experimental data for inferring in parallel both the structure and dynamics of the genome [40,41•,42]. Khanna et al. tracked over minutes the motion of VH and DHJH segments at the Igh locus in live mouse B-lymphocytes. To explore the mechanistic origin of the measured dynamics, they built independent ensembles of models in different conditions and inferred the scenario capable of recapitulating quantitatively the data (Figure 2d). They found that polymer chains containing 5% of crosslinkable sites are consistent with the experimentally observed spatial confinement of the loci due to the formation of multiple transient loops. Tuning the bond lifetime of the simulated cross-linkings to 10 s, they recapitulated the relative constrained and subdiffusive motion of VH-DHJH segments, leading the system close to a liquid-to-gel transition.
A clear example of the potential of bottom-up approaches was their use to propose and demonstrate in silico that an active loop-extrusion model by SMC complexes [43•,44] impacts key structural elements of genome folding like, for example, the formation of TADs during interphase by cohesins [45,46]. These models and their predictions lead to the exciting development of many experiments corroborating directly in-vitro [47,48] and indirectly in-vivo [49, 50, 51] the loop extrusion mechanism. In terms of 4D folding, polymer modeling by the Mirny group demonstrated that loop extrusion by condensins is a main driver of the dynamical reorganization of chromosomes during mitosis [52,53,54••,55•]. Massive loading of condensins on chromatin leads to the full extrusion of the polymer into consecutive reinforced loops in early prophase, and drives the sister chromatids segregation in late-prophase (Figure 2e). In Gibcus et al. [54••], in combination with Hi-C experiments on synchronized chicken cells, bottom-up modeling of the loop extrusion mechanism established how condensins I and II time-coordinate during prometaphase to dramatically compact chromosomes (Figure 2e).
Bottom-up approaches have also been used to study slow dynamical behaviors, such as the 4D reorganization of the genome after biological cues or perturbations [23••,56•]. The general strategy is to start from a predictive mechanistic model of the 3D nuclear organization of wild-type or undifferentiated cells and to test different scenarios on how the cue or the perturbation will affect the model parameters. Then, by tracking in silico the dynamical changes of genome folding and by comparing with experimental data measured at different time-points during the reorganization, the more-likely scenario is inferred. An interesting example is provided by the recent investigation of the global rewiring of genome contacts during oncogene-induced senescence in human fibroblasts [23••]. Using microscopy and Hi-C, the authors monitored during several days the progressive reorganization of senescence-associated heterochromatin domains (SAHDs), that slowly detach from the nuclear periphery and segregate into large foci (SAHFs). A polymer model integrating attraction between SAHDs and the nuclear lamina and self-attraction between SAHDs (Figure 2f) suggested that the observed dynamics is consistent with a slow time relaxation of the chromosomes (senescent cells do not divide anymore) driven by phase-separation of SAHDs, combined with loss of interactions with the nuclear lamina. Interestingly, a similar change of affinity between heterochromatin and the lamina was associated with the slow dynamics of nuclear inversion observed during the differentiation of rod cells in nocturnal mammals using 4D bottom-up modeling [56•].
Challenges of the 4D nucleome modeling
Although modeling has yielded huge insights into the 4D nucleome, open challenges remain to be addressed.
Bottom-up approaches need an underlying model of the chromatin which can account for its average structural and dynamical physical properties, but yet can be efficiently simulated. In this respect, the community urges to reach a consensus on which polymer model should be used to describe this non-specific behaviour of the chromatin since several of its features, such as bending rigidity or chain crossability, may affect the resulting predictions [34]. Once defined, this null chromatin model should allow developing efficient multi-scale coarse-graining strategies to simulate both the local fast dynamics and the slow full genome motion over relevant time-scales within the same framework. Moreover, data-driven approaches would also benefit from this chromatin model, because it could be used to restrain genomic regions poorly characterized by the experimental information, for instance due to data sparsity.
Approaches addressing time-resolved experimental data [23••,27••], are in need of fine-grained observations that are consistent with the hypothesis of smooth dynamical structural chromosomal changes. If the temporal resolution of the data is too coarse, thereby excluding important chromosome structure transitions, those methods could fail to provide the best models to explain the biological observations. Thus, experimental methods need to be designed with a sufficiently resolved time-scale to answer the biological question at a reasonable experimental and computational price and, at the same time, comply with the underlying methodological hypothesis.
With the advent of novel experimental techniques [5,57], 4D modeling methods will remain a central component for unleashing the full potential of the data and revealing new biological insights. However, biases related to cell fixation, digestion, cross-linking, repetitive genome sequences, and probe hybridization would need to be properly handled during data processing and modeling. A substantial challenge for the 4D modeling community will be to integrate existing and new technologies into comprehensive 4D nucleome models by emphasizing their complementarity, while avoiding pitfalls related to technology-specific biases. A further challenge will also be to integrate multi-omics (transcriptomics and epigenomics) datasets seamlessly into the models to fully exploit all existing data.
Conclusions
Considering the exciting challenges we are facing, we propose possible ways forward for the community to continue uncovering new parts of our exploration map to deepen our understanding of the 4D nucleome.
Computational resources are a limiting factor in our ability to fully explore the ranges of spatial and temporal scales spanned by the exploration map. An effort to make computer code more efficient should be encouraged by including a wider range of software developers, by emphasizing good coding practices, and by sharing software early and often during the development process. Additionally, improved exploitation of multiprocessing via graphical processing units (GPUs) could drastically improve modeling efficiency and pave the way to explore even fast dynamics at large time scales.
A challenge is also posed to the experimental community to provide new techniques orthogonal to existing ones to significantly expand our coverage and depth, both in time and space. For instance, the development of high-throughput techniques measuring in live cells both structural and dynamical properties of many loci simultaneously will elucidate new aspects of genome structure-function relationship. In addition, we believe that more effort beyond current ones [58,59] should be spent to design experiments that address fundamental questions on chromatin structure, such as what is the elementary structure of the chromatin fiber? And, to what degree does this local folding depend on the DNA sequence itself, gene expression, and/or on the epigenetic marks?
Ultimately, to chart a larger space in the exploration map (Figure 1a and b), we believe that our efforts should not involve merely an improvement of our computational and experimental techniques, but also a restructuring of the community itself. As such, more dedicated communication channels between scientists with experimental skills and others with a more theoretical background (e.g. bioinformaticians and biophysicists) should be devised. We note that the 4D nucleome community has already greatly benefited from stable collaborations between experimental and theoretical labs, leading to outstanding scientific production [12,13]. However, in training our early stage researchers, the tight connections between computational and experimental efforts should be more highlighted and emphasized. Indeed, as the experimental techniques will become more complex, the correct interpretation of the data will rely on scientists well aware of the strengths of both experiments and modeling, and capable of synergistically applying both.
Author contributions
All authors wrote and read the manuscript.
Funding
M.A.M-R. acknowledges support from the European Union’s Seventh Framework Programme through the ERC (grant agreement 609989), European Union’s Horizon 2020 research and innovation programme (grant agreement 676556), and the Spanish Ministerio de Ciencia, Innovación y Universidades (BFU2017-85926-P). M.D.S. acknowledges support from the ORION project that has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement (grant agreement 741527). D.J. acknowledges Agence Nationale de la Recherche (ANR-18-CE12-0006-03, ANR-18-CE45-0022-01) for funding. CRG acknowledges support from ‘Centro de Excelencia Severo Ochoa 2013-2017’, SEV-2012-0208 and the CERCA Programme/Generalitat de Catalunya as well as support of the Spanish Ministry of Science and Innovation through the Instituto de Salud Carlos III and the EMBL partnership, the Generalitat de Catalunya through Departament de Salut and Departament d’Empresa i Coneixement, and the Co-financing with funds from the European Regional Development Fund (ERDF) by the Spanish Ministery of Science and Innovation coresponding to the Programa Opertaivo FEDER Plurirregional de España (POPE) 2014-2020 and by the Secretaria d'Universitats i Recerca, Departament d'Empresa i Coneixement of the Generalitat de Catalunya corresponding to the programa Operatiu FEDER Catalunya 2014-2020. Open Access fees paid from COST Action INC (CA18127), supported by COST (European Cooperation in Science and Technology).
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We apologize to those whose work could not be discussed due to space limitations. We thank all members of our labs for their extensive discussions and great insights.
Contributor Information
Marco Di Stefano, Email: marco.distefano@cnag.crg.es.
Marc A Marti-Renom, Email: martirenom@cnag.crg.eu.
References
- 1.Cremer T., Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 2.Boettiger A.N., Bintu B., Moffitt J.R., Wang S., Beliveau B.J., Fudenberg G. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature. 2016;529:418–422. doi: 10.1038/nature16496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nir G., Farabella I., Pérez Estrada C., Ebeling C.G., Beliveau B.J., Sasaki H.M. Walking along chromosomes with super-resolution imaging, contact maps, and integrative modeling. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bintu B., Mateo L.J., Su J.-H., Sinnott-Armstrong N.A., Parker M., Kinrot S. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science. 2018;362 doi: 10.1126/science.aau1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen H.Q., Chattoraj S., Castillo D., Nguyen S.C., Nir G., Lioutas A. 3D mapping and accelerated super-resolution imaging of the human genome using in situ sequencing. Nat Methods. 2020;17:822–832. doi: 10.1038/s41592-020-0890-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dekker J. Capturing chromosome conformation. Science. 2002:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 7.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonev B., Cavalli G. Organization and function of the 3D genome. Nat Rev Genet. 2016;17:772. doi: 10.1038/nrg.2016.147. [DOI] [PubMed] [Google Scholar]
- 10.Rowley M.J., Corces V.G. Organizational principles of 3D genome architecture. Nat Rev Genet. 2018;19:789–800. doi: 10.1038/s41576-018-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uhler C., Shivashankar G.V. Chromosome intermingling: mechanical hotspots for genome regulation. Trends Cell Biol. 2017;27:810–819. doi: 10.1016/j.tcb.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Dekker J., Belmont A.S., Guttman M., Leshyk V.O., Lis J.T., Lomvardas S. The 4D nucleome project. Nature. 2017;549:219–226. doi: 10.1038/nature23884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marti-Renom M.A., Almouzni G., Bickmore W.A., Bystricky K., Cavalli G., Fraser P. Challenges and guidelines toward 4D nucleome data and model standards. Nat Genet. 2018;50:1352–1358. doi: 10.1038/s41588-018-0236-3. [DOI] [PubMed] [Google Scholar]
- 14.Abramo K., Valton A.-L., Venev S.V., Ozadam H., Fox A.N., Dekker J. A chromosome folding intermediate at the condensin-to-cohesin transition during telophase. Nat Cell Biol. 2019;21:1393–1402. doi: 10.1038/s41556-019-0406-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H., Emerson D.J., Gilgenast T.G., Titus K.R., Lan Y., Huang P. Chromatin structure dynamics during the mitosis-to-G1 phase transition. Nature. 2019;576:158–162. doi: 10.1038/s41586-019-1778-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchal C., Sima J., Gilbert D.M. Control of DNA replication timing in the 3D genome. Nat Rev Mol Cell Biol. 2019;20:721–737. doi: 10.1038/s41580-019-0162-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stadhouders R., Vidal E., Serra F., Di Stefano B., Le Dily F., Quilez J. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat Genet. 2018;50:238–249. doi: 10.1038/s41588-017-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18••.Paulsen J., Liyakat Ali T.M., Nekrasov M., Delbarre E., Baudement M.-O., Kurscheid S. Long-range interactions between topologically associating domains shape the four-dimensional genome during differentiation. Nat Genet. 2019;51:835–843. doi: 10.1038/s41588-019-0392-0. [DOI] [PubMed] [Google Scholar]; By performing Hi-C and ChIP-seq of nuclear lamins the authors could model 4D genome structure at multiple time-points during a dual-lineage differentiation system. By applying Chrom3D modeling at each time-point, the authors identified that TADs engage in long-range interactions to reinforce a compact structure repositioned towards the nuclear periphery during the differentiation.
- 19.Ogiyama Y., Schuettengruber B., Papadopoulos G.L., Chang J.-M., Cavalli G. Polycomb-dependent chromatin looping contributes to gene silencing during Drosophila development. Mol Cell. 2018;71:73–88.e5. doi: 10.1016/j.molcel.2018.05.032. [DOI] [PubMed] [Google Scholar]
- 20.Hug C.B., Grimaldi A.G., Kruse K., Vaquerizas J.M. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell. 2017;169:216–228.e19. doi: 10.1016/j.cell.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 21••.Nagano T., Lubling Y., Várnai C., Dudley C., Leung W., Baran Y. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature. 2017;547:61–67. doi: 10.1038/nature23001. [DOI] [PMC free article] [PubMed] [Google Scholar]; By combining single-cell Hi-C (scHi-C) and structural modeling, the authors characterized the evolution of the 3D genome structure along the cell-cycle highlighting 4D features that were only implicit in the data, such as the progressive decondensations of A/B compartments and the establishment of chromosome radial positions approaching the G1 phase.
- 22.Beagan J.A., Pastuzyn E.D., Fernandez L.R., Guo M.H., Feng K., Titus K.R. Three-dimensional genome restructuring across timescales of activity-induced neuronal gene expression. Nat Neurosci. 2020;23:707–717. doi: 10.1038/s41593-020-0634-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23••.Sati S., Bonev B., Szabo Q., Jost D., Bensadoun P., Serra F. 4D genome rewiring during oncogene-induced and replicative senescence. Mol Cell. 2020;78:522–538.e9. doi: 10.1016/j.molcel.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using 4D polymer modeling, authors showed that the slow dynamical 3D re-organization of the genome during senescence is consistent with a loss of interactions of heterochromatin with the nuclear periphery combined to the capacity of heterochromatin to phase-separate.
- 24.Yang K.D., Damodaran K., Venkatachalapathy S., Soylemezoglu A.C., Shivashankar G.V., Uhler C. Predicting cell lineages using autoencoders and optimal transport. PLoS Comput Biol. 2020;16 doi: 10.1371/journal.pcbi.1007828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meluzzi D., Arya G. Computational approaches for inferring 3D conformations of chromatin from chromosome conformation capture data. Methods. 2019;181–182:24–34. doi: 10.1016/j.ymeth.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacKay K., Kusalik A. Computational methods for predicting 3D genomic organization from high-resolution chromosome conformation capture data. Brief Funct Genomics. 2020;19:292–308. doi: 10.1093/bfgp/elaa004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27••.Di Stefano M., Stadhouders R., Farabella I., Castillo D., Serra F., Graf T. Transcriptional activation during cell reprogramming correlates with the formation of 3D open chromatin hubs. Nat Commun. 2020;11 doi: 10.1038/s41467-020-16396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors introduced the TADdyn tool to merge in a modeling framework time-series Hi-C datasets at discrete time-points. 4D simulations of 21 loci during the B to iPSC reprogramming dynamics led to suggest the formation and disruption of 3D super enhancers as a structural motif capable of regulating gene expression.
- 28.Serra F., Di Stefano M., Spill Y.G., Cuartero Y., Goodstadt M., Baù D. Restraint-based three-dimensional modeling of genomes and genomic domains. FEBS Lett. 2015;589:2987–2995. doi: 10.1016/j.febslet.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Paulsen J., Sekelja M., Oldenburg A.R., Barateau A., Briand N., Delbarre E. Chrom3D: three-dimensional genome modeling from Hi-C and nuclear lamin-genome contacts. Genome Biol. 2017;18:21. doi: 10.1186/s13059-016-1146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tiana G., Giorgetti L. CRC Press; 2019. Modeling the 3D Conformation of Genomes. [Google Scholar]
- 31.Amitai A, Holcman D: Polymer physics of nuclear organization and function. doi: 10.1101/076661. [DOI]
- 32.Tortora M.M., Salari H., Jost D. Chromosome dynamics during interphase: a biophysical perspective. Curr Opin Genet Dev. 2020;61:37–43. doi: 10.1016/j.gde.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 33•.Di Pierro M., Potoyan D.A., Wolynes P.G., Onuchic J.N. Anomalous diffusion, spatial coherence, and viscoelasticity from the energy landscape of human chromosomes. Proc Natl Acad Sci U S A. 2018;115:7753–7758. doi: 10.1073/pnas.1806297115. [DOI] [PMC free article] [PubMed] [Google Scholar]; On the basis of a polymer model fitted to predict the 3D genome organization, this work reported how the same model without any additional constraints is compatible with the many experimental observations made on chromosome dynamics.
- 34.Ghosh S.K., Jost D. How epigenome drives chromatin folding and dynamics, insights from efficient coarse-grained models of chromosomes. PLoS Comput Biol. 2018;14 doi: 10.1371/journal.pcbi.1006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi G., Liu L., Hyeon C., Thirumalai D. Interphase human chromosome exhibits out of equilibrium glassy dynamics. Nat Commun. 2018;9 doi: 10.1038/s41467-018-05606-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shukron O., Piras V., Noordermeer D., Holcman D. Statistics of chromatin organization during cell differentiation revealed by heterogeneous cross-linked polymers. Nat Commun. 2019;10 doi: 10.1038/s41467-019-10402-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conte M., Fiorillo L., Bianco S., Chiariello A.M., Esposito A., Nicodemi M. Polymer physics indicates chromatin folding variability across single-cells results from state degeneracy in phase separation. Nat Commun. 2020;11:3289. doi: 10.1038/s41467-020-17141-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cortini R., Filion G.J. Theoretical principles of transcription factor traffic on folded chromatin. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04130-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amitai A. Chromatin configuration affects the dynamics and distribution of a transiently interacting protein. Biophys J. 2018;114:766–771. doi: 10.1016/j.bpj.2017.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arbona J.-M., Herbert S., Fabre E., Zimmer C. Inferring the physical properties of yeast chromatin through Bayesian analysis of whole nucleus simulations. Genome Biol. 2017;18:81. doi: 10.1186/s13059-017-1199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Khanna N., Zhang Y., Lucas J.S., Dudko O.K., Murre C. Chromosome dynamics near the sol-gel phase transition dictate the timing of remote genomic interactions. Nat Commun. 2019;10 doi: 10.1038/s41467-019-10628-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; Devising an interesting 4D bottom-up modeling approach, the authors showed that the relative constrained and subdiffusive motion of the VH and DHJH segments at the Igh locus can be recapitulated by a polymeric system with transient multiple loops.
- 42.Socol M., Wang R., Jost D., Carrivain P., Vaillant C., Le Cam E. Rouse model with transient intramolecular contacts on a timescale of seconds recapitulates folding and fluctuation of yeast chromosomes. Nucleic Acids Res. 2019;47:6195–6207. doi: 10.1093/nar/gkz374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Banigan E.J., Mirny L.A. Loop extrusion: theory meets single-molecule experiments. Curr Opin Cell Biol. 2020;64:124–138. doi: 10.1016/j.ceb.2020.04.011. [DOI] [PubMed] [Google Scholar]
- 44.Ghosh S.K., Jost D. Genome organization via loop extrusion, insights from polymer physics models. Brief Funct Genomics. 2020;19:119–127. doi: 10.1093/bfgp/elz023. [DOI] [PubMed] [Google Scholar]
- 45.Sanborn A.L., Rao S.S.P., Huang S.-C., Durand N.C., Huntley M.H., Jewett A.I. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fudenberg G., Imakaev M., Lu C., Goloborodko A., Abdennur N., Mirny L.A. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davidson I.F., Bauer B., Goetz D., Tang W., Wutz G., Peters J.-M. DNA loop extrusion by human cohesin. Science. 2019:1338–1345. doi: 10.1126/science.aaz3418. [DOI] [PubMed] [Google Scholar]
- 48.Kim Y., Shi Z., Zhang H., Finkelstein I.J., Yu H. Human cohesin compacts DNA by loop extrusion. Science. 2019:1345–1349. doi: 10.1126/science.aaz4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nora E.P., Goloborodko A., Valton A.-L., Gibcus J.H., Uebersohn A., Abdennur N. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 2017;169:930–944.e22. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rao S.S.P., Huang S.-C., Glenn St Hilaire B., Engreitz J.M., Perez E.M., Kieffer-Kwon K.-R. Cohesin loss eliminates all loop domains. Cell. 2017;171:305–320.e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwarzer W., Abdennur N., Goloborodko A., Pekowska A., Fudenberg G., Loe-Mie Y. Two independent modes of chromatin organization revealed by cohesin removal. Nature. 2017;551:51–56. doi: 10.1038/nature24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alipour E., Marko J.F. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 2012;40:11202–11212. doi: 10.1093/nar/gks925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goloborodko A., Imakaev M.V., Marko J.F., Mirny L. Compaction and segregation of sister chromatids via active loop extrusion. eLife. 2016;5 doi: 10.7554/eLife.14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54••.Gibcus J.H., Samejima K., Goloborodko A., Samejima I., Naumova N., Nuebler J. A pathway for mitotic chromosome formation. Science. 2018;359 doi: 10.1126/science.aao6135. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this synergistic experimental-modeling work, the authors provided mechanistic evidence for a condensin-mediated loop extrusion process leading to the progressive formation of mitotic chromosomes into compacted arrays of nested loops.
- 55•.Banigan E.J., van den Berg A.A., Brandão H.B., Marko J.F., Mirny L.A. Chromosome organization by one-sided and two-sided loop extrusion. eLife. 2020;9 doi: 10.7554/eLife.53558. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors provided an extensive, systematic analysis of the predictions of the loop-extrusion model: theoretical and computational bases, hypotheses on loop extrusion mechanism, contexts of applications, and experimental evidence driving its parameterization.
- 56•.Falk M., Feodorova Y., Naumova N., Imakaev M., Lajoie B.R., Leonhardt H. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature. 2019;570:395–399. doi: 10.1038/s41586-019-1275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Applying bottom-up modeling, the authors suggested that decreased attraction between heterochromatin and nuclear envelope may drive the slow dynamical repositioning of heterochromatin to the nuclear center during rod cells differentiation in nocturnal animals.
- 57.Barth R., Bystricky K., Shaban H.A. Coupling chromatin structure and dynamics by live super-resolution imaging. Sci Adv. 2020;6 doi: 10.1126/sciadv.aaz2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ricci M.A., Manzo C., García-Parajo M.F., Lakadamyali M., Cosma M.P. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell. 2015;160:1145–1158. doi: 10.1016/j.cell.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 59.Ou H.D., Phan S., Deerinck T.J., Thor A., Ellisman M.H., O’Shea C.C. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 2017;357 doi: 10.1126/science.aag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Díaz N., Kruse K., Erdmann T., Staiger A.M., Ott G., Lenz G. Chromatin conformation analysis of primary patient tissue using a low input Hi-C method. Nat Commun. 2018;9 doi: 10.1038/s41467-018-06961-0. [DOI] [PMC free article] [PubMed] [Google Scholar]