Abstract
TRIM9 and TRIM67 are neuronally enriched E3 ubiquitin ligases essential for appropriate morphogenesis of cortical and hippocampal neurons and fidelitous responses to the axon guidance cue netrin-1. Deletion of murine Trim9 or Trim67 results in neuroanatomical defects and striking behavioral deficits, particularly in spatial learning and memory. TRIM9 and TRIM67 interact with cytoskeletal and exocytic proteins, but the full interactome is not known. Here we performed the unbiased proximity-dependent biotin identification (BioID) approach to define TRIM9 and TRIM67 protein–protein proximity network in developing cortical neurons and identified putative neuronal TRIM interaction partners. Candidates included cytoskeletal regulators, cytosolic protein transporters, exocytosis and endocytosis regulators, and proteins necessary for synaptic regulation. A subset of high-priority candidates was validated, including Myo16, Coro1A, MAP1B, ExoC1, GRIP1, PRG-1, and KIF1A. For a subset of validated candidates, we utilized total internal reflection fluorescence microscopy to demonstrate dynamic colocalization with TRIM proteins at the axonal periphery, including at the tips of filopodia. Further analysis demonstrated that the RNA interference–based knockdown of the unconventional myosin Myo16 in cortical neurons altered growth cone filopodia density and axonal branching patterns in a TRIM9- and netrin-1–dependent manner. Future analysis of other validated candidates will likely identify novel proteins and mechanisms by which TRIM9 and TRIM67 regulate neuronal form and function.
INTRODUCTION
Neuronal morphogenesis and function require coordinated cytoskeletal reorganization and plasma membrane expansion (McCormick and Gupton, 2020). Exocytosis leads to the addition of plasma membrane (Pfenninger, 2009; Gupton and Gertler, 2010; Winkle et al., 2014; Urbina and Gupton, 2020), whereas cytoskeletal reorganization molds the plasma membrane into the complex arborized neuronal shape. The functions of many cytoskeletal and membrane remodeling proteins are defined, yet how their functions are coordinated to promote cell shape change remains poorly understood. Further there is likely specialized machinery for the unique cell shape changes that occur in neurons, which achieve an elongated and polarized morphology in response to a host of guidance cues. Phylogenetic analysis suggested that the emergence of the class I TRIM (TRIpartitite Motif) E3 ubiquitin ligases and the emergence of neuronal-like cells appeared to have occurred contemporaneously (Boyer et al., 2018), and thus class I TRIMs may be well poised to regulate neuronal form and function.
The TRIM family is a subclass of RING-domain E3 ubiquitin ligases, which comprises more than 80 human members, some of which are brain-enriched and implicated in neuronal development and neurological disorders (Tocchini and Ciosk, 2015; Jin et al., 2017; Watanabe and Hatakeyama, 2017; George et al., 2018). TRIM ligases are characterized by an N-terminal RING ligase domain, zinc-finger B-Box domain(s), and coiled-coil multimerization domain (Meroni and Diez-Roux, 2005; Short and Cox, 2006). A variable C-terminal structure defines nine distinct classes of TRIM proteins and is often considered responsible for providing substrate specificity. Class I TRIMs contain a Cos domain that in some cases binds microtubules (MTs) (Short and Cox, 2006; Cox, 2012), an FN3 domain, and a SPRY domain. In vertebrates, six class I TRIMs are divided into three paralogue pairs, indicative of multiple gene duplication events (Short and Cox, 2006). In contrast, invertebrates have one or two class I TRIM proteins (Boyer et al., 2018).
Vertebrate class I TRIM pairs include TRIM1 and TRIM18, TRIM36 and TRIM46, and TRIM9 and TRIM67. Mutations in TRIM1 (MID2) and TRIM18 (MID1) are associated with X-linked disorders such as intellectual disability and Opitz G/BBB syndrome (Quaderi et al., 1997; Cainarca et al., 1999; De Falco et al., 2003; Geetha et al., 2014). TRIM36 regulates dorsal axis formation and cell cycle progression (Cuykendall and Houston, 2009; Miyajima et al., 2009). Mutations in TRIM36 are associated with anencephaly (Singh et al., 2017). TRIM46 establishes neuronal polarity and the axon initial segment (Van Beuningen et al., 2015; Harterink et al., 2019). TRIM9 and TRIM67 regulate neuronal morphological changes in response to the axon guidance cue netrin-1 (Winkle et al., 2014, 2016; Menon et al., 2015; Plooster et al., 2017; Boyer et al., 2020). TRIM9 localizes to Parkinsonian Lewy bodies and single nucleotide polymorphisms (SNPs) in TRIM9 may be associated with atypical psychosis (Tanji et al., 2010; Kanazawa et al., 2013). Paraneoplastic neurological syndromes and small-cell lung carcinoma are associated with TRIM9, TRIM67, and TRIM46 (van Coevorden-Hameete et al., 2017; Do et al., 2019). These findings demonstrate a role for class I TRIMs in neuronal form and function.
Loss of the single class I TRIM dTrim9 in Drosophila melanogaster and madd-2 in Caenorhabditis elegans leads to axon branching and guidance defects (Hao et al., 2010; Morikawa et al., 2011). Similar phenotypes occur with loss of netrin/unc-6 and its receptor, Frazzled/unc-40 (Kolodziej et al., 1996; Mitchell et al., 1996; Norris and Lundquist, 2011). Similarly, in vertebrates, loss of netrin-1 and its receptor, deleted in colorectal cancer (DCC) results in midline crossing defects such as agenesis of the corpus callosum (Serafini et al., 1996; Fazeli et al., 1997; Bin et al., 2015; Yung et al., 2015). dTrim9 and MADD2 interact with netrin receptors, Frazzled and UNC40 (Hao et al., 2010; Morikawa et al., 2011). Analogously, vertebrate TRIM9 and TRIM67 interact with DCC (Winkle et al., 2014; Boyer et al., 2018). However, unlike loss of Ntn1 or DCC, loss of murine Trim9 results in corpus callosum thickening, due at least partially to aberrant axon branching within the structure (Winkle et al., 2014). In contrast, loss of murine Trim67 results in thinning of the callosum (Boyer et al., 2018). These data suggest that both TRIM9 and TRIM67 function in axon projection downstream of netrin.
We have found that TRIM9 and TRIM67 mediate morphological changes downstream of netrin and DCC by regulating cytoskeletal dynamics and exocytosis (Winkle et al., 2014; Menon et al., 2015; Plooster et al., 2017; Urbina et al., 2018; Boyer et al., 2020). TRIM9 and TRIM67 interact with and modulate ubiquitination of the actin polymerase VASP to regulate growth cone dynamics and netrin-dependent axonal turning (Menon et al., 2015; Boyer et al., 2020). TRIM9-mediated ubiquitination of DCC is netrin-1 sensitive and regulates signaling, exocytosis, and axon branching. TRIM9 and TRIM67 regulate the frequency and mode of exocytosis, via interactions with exocytic t-SNAREs SNAP25 and SNAP47, respectively (Winkle et al., 2014; Urbina et al., 2018, 2020). TRIM9 and TRIM67 continue to modulate neuronal form and function in the mature animal. TRIM9 regulates morphogenesis of adult-born neurons including dendritic arborization, spine density, and localization in the hippocampus (Winkle et al., 2016). Loss of Trim67 results in hypotrophy of the hippocampus (Boyer et al., 2018). Along with these anatomical phenotypes, mice lacking Trim9 and Trim67 exhibit spatial learning and memory deficits (Winkle et al., 2016; Boyer et al., 2018), phenotypes typically associated with hippocampal dysfunction.
Together these studies indicate that TRIM9 and TRIM67 are important in orchestrating neuronal shape changes, yet the complete repertoire of their function is not known. Most E3 ubiquitin ligases ubiquitinate multiple substrates (Deshaies and Joazeiro, 2009), yet only a few substrates for TRIM9 and TRIM67 have been identified. Further, TRIM9 and TRIM67 interact with proteins without altering their ubiquitination. Therefore, identifying other potential interactors and substrates is critical to understanding the role of TRIM9 and TRIM67 in neuronal development. Here we utilized an unbiased proximity labeling proteomics approach (Roux et al., 2012) to identify candidate interacting partners and substrates. This yielded a number of high-priority interaction candidates poised to regulate the MT and actomyosin cytoskeletons, synaptic structure and maintenance, membrane remodeling, and cytosolic transport, processes that are critical to neuronal form and function. We validated the interaction of a subset of candidates with TRIM9 and TRIM67. Analysis of the validated interaction partner Myo16 showed that loss of this unconventional myosin affected growth cone filopodia density and axonal branching in a TRIM9- and netrin-1–dependent manner.
RESULTS
Proximity-dependent labeling identifies interaction partners of TRIM9 and TRIM67
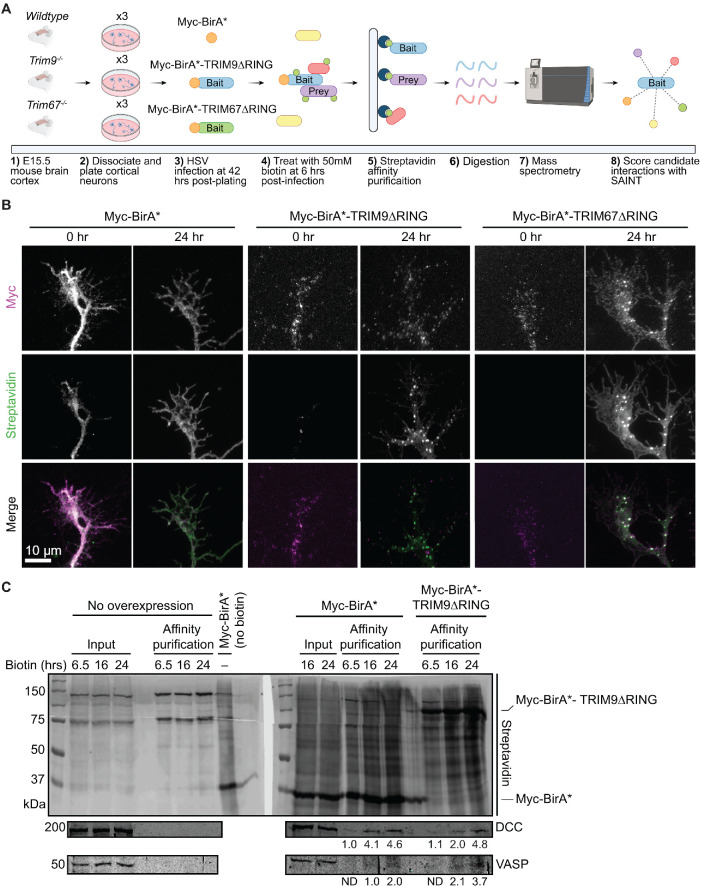
Unbiased identification of ligase interaction partners and substrates is difficult via standard coimmunoprecipitation (co-IP) techniques due to the transient nature of E3 ligase–substrate interactions (Iconomou and Saunders, 2016) and low abundance of ubiquitinated proteins. Proximity-dependent labeling approaches are an effective alternative strategy (Coyaud et al., 2015; Cloer et al., 2018). To identify candidate TRIM9 and TRIM67 interacting partners and substrates, we employed proximity-dependent labeling with the promiscuous biotin ligase (BirA*) attached to either TRIM9 or TRIM67. This approach exploits biotinylation of proteins within 10 nm of BirA* (Roux et al., 2012; Kim et al., 2014) and allows identification of transient interaction partners and substrates (Figure 1A). Further, we attached a Myc-BirA* to TRIM9 and TRIM67 lacking ligase domains (TRIM9∆RING and TRIM67∆RING) to minimize enrichment of biotinylated ubiquitin.
FIGURE 1:
BioID approach to identify candidate interaction partners of TRIM9 and TRIM67 in embryonic cortical neurons. (A) Graphical representation of the BioID approach. E15.5 cortical neurons were transduced with HSV carrying Myc-BirA*, Myc-BirA*TRIM9∆RING, or Myc-BirA*TRIM67∆RING, and media was supplemented with 50 μM biotin. Following cell lysis, biotinylated proteins were affinity purified, and enriched proteins were subjected to on-bead trypsinization. Candidate peptides were identified by mass spectrometry. (B) Images of axonal growth cones from control Myc-BirA*, Myc-BirA*-TRIM9∆RING, Myc-BirA*-TRIM67∆RING expressing neurons at 0 and 24 h post–biotin addition, stained with fluorescent streptavidin and anti-Myc antibody. Merged images show colocalized streptavidin and Myc signals. (C) Western blots of streptavidin affinity purification of biotinylated proteins from cortical neurons expressing Myc-BirA* or Myc-BirA*-TRIM9∆RING. Samples include inputs and affinity-purified samples from controls that do not express BirA* (6.5, 16, 24 h), input from neurons expressing Myc-BirA* but not supplemented with biotin, inputs (16, 24 h) and affinity-purified samples (6.5, 16, 24 h) from neurons expressing Myc-BirA* and supplemented with biotin, and affinity-purified samples from neurons expressing Myc-BirA*-TRIM9∆RING (6.5, 16, 24 h) and supplemented with biotin. Blots were probed for streptavidin, DCC, and VASP. Numbers below the anti-DCC blot represent DCC intensity values normalized to the intensity of the DCC band enriched in the affinity-purified samples (6.5 h) from neurons expressing Myc-BirA*. Numbers below the anti-VASP blot represent VASP intensity values normalized to the intensity of the VASP band enriched in the affinity-purified samples (16 h) from neurons expressing Myc-BirA*. VASP was not detected (N.D.) in affinity-purified samples (6.5 h) from neurons expressing Myc-BirA* or Myc-BirA*-TRIM9∆RING.
Because TRIM9 and TRIM67 are enriched in neurons, identification of interaction partners in neurons was critical. MycBirA* constructs were introduced to embryonic cortical neurons by herpes simplex virus (HSV) transduction. To ensure proper localization and function of constructs, we examined localization in neurons and biotinylation of known interaction partners. Myc-BirA*TRIM9∆RING and Myc-BirA*TRIM67∆RING localized to the axonal growth cone, primarily to the tips of filopodia and along the periphery of the growth cone (Figure 1B), indistinguishable from the localization of endogenous TRIM9 and TRIM67 (Winkle et al., 2014; Boyer et al., 2018, 2020). In contrast, Myc-BirA* exhibited an expected cytosolic distribution. In neurons supplemented with biotin, the TRIM9∆RING and TRIM67∆RING puncta (Myc-stained) overlapped with streptavidin signal (Figure 1B), indicating the BirA* was biotinylating proximal proteins. Previously identified TRIM9 substrates, VASP and DCC, were enriched by affinity purification from lysates of Trim9–/– neurons expressing Myc-BirA*TRIM9∆RING after biotin addition (Figure 1C). After 24 h of biotin addition, VASP and DCC were more enriched in Myc-BirA*TRIM9∆RING lysates than Myc-BirA*–expressing lysates. These results confirmed the feasibility of using an in vitro proximity-dependent biotin identification (BioID) approach with 24 h of biotin addition in developing neurons for the identification of candidate E3 ligase substrates and/or interacting partners.
Potential TRIM9 and TRIM67 interaction candidates affect cytoskeletal change, protein transport, and synaptic structure
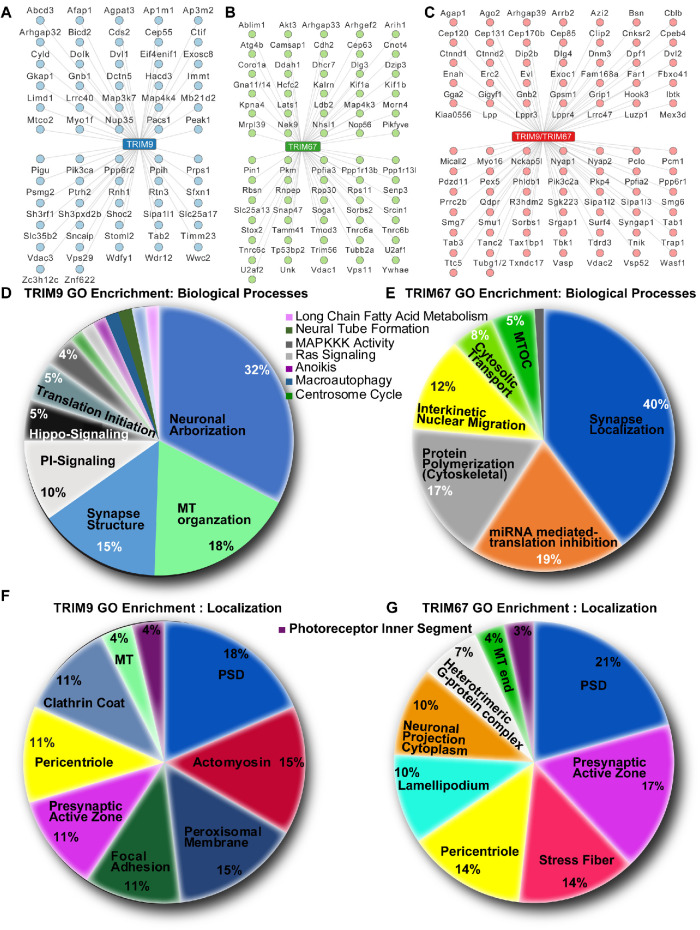
Biotinylated proteins were affinity purified from wild-type embryonic cortical neurons expressing Myc-BirA* (negative control), Trim9–/– neurons expressing Myc-BirA*TRIM9∆RING, and Trim67–/– expressing Myc-BirA*TRIM67ΔRING and analyzed using mass spectrometry. Totals of 2012 and 2315 potential interaction candidates were identified for TRIM9 and TRIM67, respectively (Supplemental Tables 1 and 2). The included proteins have a false discovery rate (FDR) ranging from 0.000 (high-confidence hits) through 0.7559 (low-confidence hits) based on enrichment over MycBirA* hits and variability across replicates. Of all TRIM9 and TRIM67 interaction candidates, 149 and 151, respectively, were classified as high-confidence interaction hits with FDR values ≤0.25 (Figure 2, A–C). Of these interaction candidates, 91 were common to both TRIM9 and TRIM67 (Figure 2C), approximately 60%. Known interaction partners, including all Enah/VASP family proteins: Enah (TRIM9 interaction FDR: 0, TRIM67 interaction FDR: 0), VASP (TRIM9 interaction FDR: 0, TRIM67 interaction FDR: 0), and Evl (TRIM9 interaction FDR: 0, TRIM67 interaction FDR: 0.0176), and the netrin receptor DCC (TRIM9 interaction FDR: 0.7559, TRIM67 interaction FDR: 0.7559) (Winkle et al., 2014; Menon et al., 2015; Boyer et al., 2018, 2020) were identified by the BioID approach.
FIGURE 2:
Candidate TRIM9 and TRIM67 interaction partners. (A–C) Potential high-confidence (FDR ≤ 0.259) interaction partners unique to TRIM9 (A), unique to TRIM67 (B), or shared (C). (D, E) Pie charts of biological processes enriched via GO classification of high-confidence TRIM9 and TRIM67 interaction partners, respectively. (F, G) Pie charts of cellular localization terms enriched via GO classification of high-confidence TRIM9 and TRIM67 interaction partners, respectively. Percentages for a particular GO term in D–G represent the number of gene IDs that hit against the total number of gene IDs annotated.
Gene ontology (GO) analysis of high-confidence candidates was performed using the ClueGO application in Cytoscape_3.7.2 (Shannon et al., 2003; Bindea et al., 2009). This approach identified enrichment of protein categories based on function (Figure 2, D and E) and cellular localization (Figure 2, F and G). Consistent with findings that TRIM9 regulates morphogenesis, TRIM9 interaction candidates included proteins regulating neuron projection arborization and localizing to the actomyosin and MT cytoskeletons and localization to clathrin coats, suggesting a possible role in endocytosis (Figure 2, D and F). Similarly, TRIM67 interaction candidates included proteins that regulated cytoskeletal polymerization and localized to cytoskeletal rich structures, including stress fibers, lamellipodium, the pericentriole, and neuronal projections (Figure 2, E and G). Although the proteomic analysis was performed in developing neurons before synaptogenesis, GO categorization for both TRIM9 and TRIM67 was enriched for proteins that localize to both the presynaptic region and post synaptic density as well as for proteins required for maintenance of synapse structures and synaptic localization of other proteins. This analysis suggested novel roles for TRIM9 and TRIM67 in signal transduction cascades and protein translation. For TRIM9 this included phosphatidylinositol-mediated signaling, hippo signaling, MAPKKK activity, anoikis, and translation initiation. Similarly, TRIM67 interaction candidates were enriched in cytosolic transport, mRNA-mediated inhibition of translation, interkinetic nuclear migration, and MAPKKK activity.
Validation of TRIM9 and TRIM67 interaction candidates
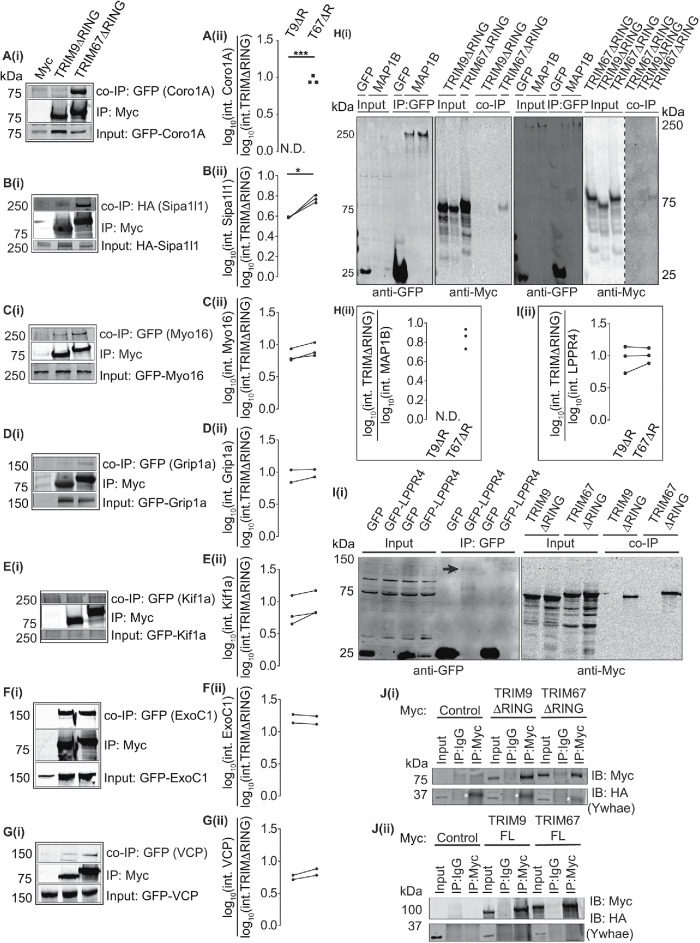
We focused validation efforts on candidates implicated in actin and MT regulation, membrane remodeling, and motor-based transport. Some high-FDR candidates were included in validation efforts because N-terminal tagging of BirA* may have affected the biotinylation of true C-terminal interacting partners, as previously observed (Redwine et al., 2017). Indeed the netrin-1 receptor DCC, which interacts directly with the C-terminal SPRY domain of TRIM9 (Winkle et al., 2014), and likely the SPRY domain of TRIM67 by homology, exhibited high FDR values (0.7559 for both TRIM9 and TRIM67, 1.05- and 1.26-fold enrichment over control in TRIM9 and TRIM67, respectively). To validate interactions, we used co-IP assays of Myc-tagged TRIM∆RING proteins and GFP- or HA-tagged candidate interactors coexpressed in HEK293 cells. This approach overcame issues of transient interactions, low abundance, and unvalidated antibodies to endogenous proteins. As a negative control, immunoprecipitation of an empty Myc construct was included in each assay. For the confirmed interactions described below, there was no detectable signal of candidate interactors coimmunoprecipitating with empty Myc (Figure 3, A–I).
FIGURE 3:
Validation of potential TRIM9 and TRIM67 interaction partners via co-IP assays. (A–G) Immunoblots showing co-IP of GFP- or HA-tagged proteins when immunoprecipitated using an anti-Myc antibody to enrich for Myc, Myc-TRIM9∆RING, or Myc-TRIM67∆RING. (A: Coro1A [n = 3], B: Sipa1l1 [n = 3], C: Myo16 [n = 3], D: GRIP1a [n = 2], E: Kif1a [n = 3], F: ExoC1 [n = 2], G: VCP [n = 2]). A(i) –G(i) show blots for inputs for the coimmunoprecipitated proteins, the immunoprecipitated Myc-TRIM9ΔRING or Myc-TRIM67ΔRING and co-IP proteins. A(ii) –G(ii) show individual datapoints of co-IP GFP- or HA-tagged proteins relative to Myc-TRIM9ΔRING (T9ΔR) or Myc-TRIM67ΔRING (T67ΔR). (H(i)) Immunoblot demonstrating co-IP of Myc-TRIM67∆RING, but not Myc-TRIM9∆RING with GFP-MAP1B. Two representative blots are included, one with GFP-control coexpressed with Myc-TRIM9∆RING and one with GFP-control coexpressed with Myc-TRIM67∆RING. H(ii) shows individual datapoints of co-IP Myc-TRIM67ΔRING (T67ΔR) relative to GFP-MAP1B (n = 3). Myc-TRIM9∆RING (T9ΔR) did not coprecipitate with GFP- MAP1B (N.D.). (Ii) Immunoblot showing Myc-TRIM9∆RING and Myc-TRIM67ΔRING co-IP with GFP-LPPR4 (PRG-1). The black arrow in the blot probed for GFP shows a faint GFP-LPPR4 band. Note that GFP-LPPR4 was not detected in inputs. I(ii) shows individual datapoints of coimmunoprecipitated Myc-TRIM9∆RING (T9ΔR) and Myc-TRIM67ΔRING (T67ΔR) relative to GFP-LPPR4 (n = 3). (J(i), (ii)) Immunoblots demonstrating that HA-Ywhae does not coimmunoprecipitate with Myc-TRIM∆RING (n = 3) or full-length constructs of TRIM9 and TRIM67 (n = 3). The white asterisks in J(i) denote a nonspecific band observed in all anti-Myc IP lanes. Datapoints were compared using an unpaired t test with Welch’s correction, *, P < 0.05, ***, P < 0.005.
Coro1a (TRIM9 FDR: 0.0027 and TRIM67 FDR: 0) is a type I coronin, a well-characterized actin regulator, and modulator of the signaling endosome (Suo et al., 2014, 2015; Martorella et al., 2017). Coro1a interacted with TRIM67∆RING, but not TRIM9∆RING (Figure 3A, i and ii). Sipa1l1, commonly called SPAR (spine-associated Rap GTPase-activating protein) (TRIM9 FDR: 0.0082, TRIM67 FDR: 0.5652), modulates actin dynamics in dendritic spines (Pak et al., 2001; Pak and Sheng, 2003; Hoe et al., 2009). Sipa1l1 interacted with both TRIM9∆RING and TRIM67∆RING, albeit with a higher affinity for TRIM67∆RING (Figure 3B, i and ii). Myo16 (TRIM9 FDR: 0.0855, TRIM67 FDR: 0.0855) is an unconventional myosin enriched in the developing brain that regulates actin dynamics via the wave regulatory complex (WRC), Arp2/3, and PI3K signaling (Patel et al., 2001; Yokoyama et al., 2011). FDR values suggested that Myo16 interacted with TRIM9∆RING and TRIM67∆RING; co-IP confirmed this (Figure 3C, i and ii).
Glutamate receptor interacting protein 1 (GRIP1; TRIM9 FDR: 0, TRIM67 FDR: 0) is a kinesin adaptor protein required for homeostatic AMPA receptor trafficking (Setou et al., 2002; Hoogenraad et al., 2005; Tan et al., 2015; Twelvetrees et al., 2019). GRIP1 interacted with both TRIM9∆RING and TRIM67∆RING (Figure 3D, i and ii). Kif1a (TRIM9 FDR: 0.271, TRIM67 FDR: 0.0587), a member of the kinesin 3 family, is a MT-based motor protein essential for the transport of synaptic vesicle precursors (Okada et al., 1995; Lo et al., 2011; Karasmanis et al., 2018; Guedes-Dias et al., 2019). Kif1a interacted with both TRIM67∆RING and TRIM9∆RING (Figure 3E, i and ii). ExoC1 (yeast homologue, Sec3, TRIM9 FDR: 0.0855, TRIM67 FDR: 0), a member of the octameric exocyst complex that plays a critical role in tethering vesicles to the plasma membrane before exocytosis (Wu and Guo, 2015; Kampmeyer et al., 2017; Yue et al., 2017; Liu et al., 2018), interacted similarly with TRIM9∆RING and TRIM67∆RING (Figure 3F, i and ii). Valosin-containing protein (VCP, TRIM9 FDR: 0.7599, TRIM67 FDR: 0.7599) is a clathrin-binding protein and member of the AAA+ ATPase family (Pleasure et al., 1993; Ritz et al., 2011). VCP interacted with both TRIM9∆RING and TRIM67∆RING (Figure 3G, i and ii).
MAP1B (TRIM9 interaction FDR: 0.7599, TRIM67 interaction FDR: 0.7599), a large MT-associated protein that plays a role in MT dynamics, neurite branching, and dendritic spine development (Ma et al., 2000; Li et al., 2006; Tortosa et al., 2011; Tymanskyj et al., 2012; Bodaleo et al., 2016), interacted with TRIM67∆RING, but not TRIM9∆RING (Figure 3H, i and ii). Two separate blots are shown to demonstrate the specificity of the interaction with TRIM67; one controlled for TRIM9∆RING (left) and one for TRIM67∆RING (right). LPPR4 (TRIM9 interaction FDR: 0, TRIM67 interaction FDR: 0.0738), commonly called PRG-1, is a member of the lipid phosphate phosphatase family. It is a transmembrane protein that functions as an ectoenzyme and promotes axonal growth by locally depleting lipid phosphates (Bräuer et al., 2003; Tokumitsu et al., 2010; Petzold et al., 2016). GFP-LPPR4 was detected on Western blots only after immunoprecipitation (Figure 3I). Myc-TRIM9∆RING and Myc-TRIM67∆RING were enriched in the GFP-LPPR4 immunoprecipitates, but not the GFP control (Figure 3I, i and ii). Ywhae (TRIM9 FDR: 0.5696, TRIM67 FDR: 0.2572), also called 14-3-3 epsilon, is brain-enriched and required for neurogenesis, neuronal differentiation, and migration (Toyo-Oka et al., 2003, 2014). A microduplication of the Ywhae gene locus is associated with autism spectrum disorders (Bruno et al., 2010). Ywhae did not interact with full-length or ∆RING variants of TRIM9 or TRIM67 (Figure 3J, i and ii). These co-IP experiments validated most (seven of eight) of the candidates tested as interaction partners, supporting the validity of the BioID approach for defining the interactome and suggesting that additional valid interaction partners await confirmation.
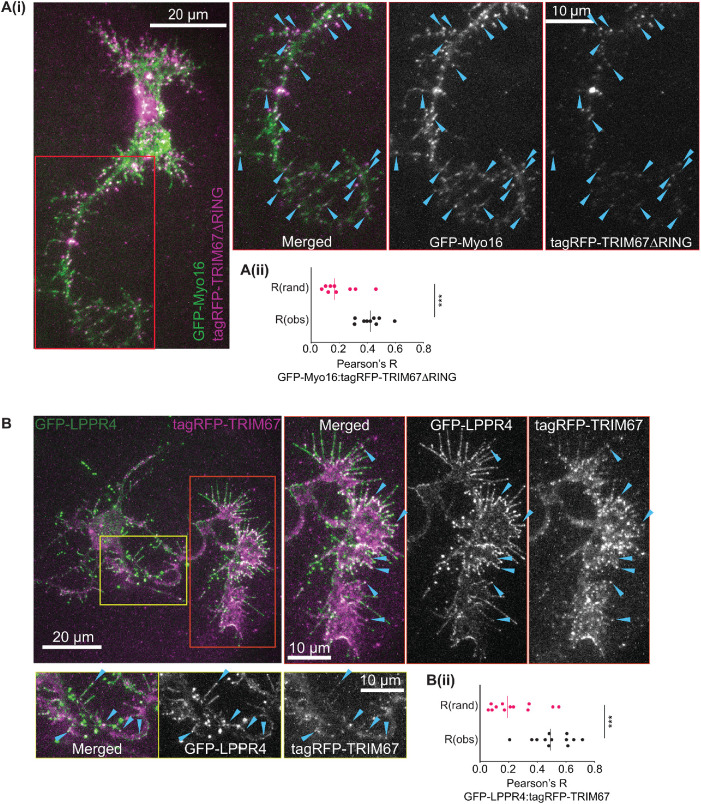
Live-cell total internal reflection fluorescence (TIRF) microscopy was performed to examine the colocalization of a subset of validated candidates at the basal surface of developing neurons. This revealed that GFP-Myo16 colocalized with tagRFP-TRIM67∆RING along the axon and at filopodia tips (Figure 4A, i, arrowheads, and ii). Similarly, GFP-LPPR4 colocalized with tagRFP-TRIM67 along filopodia tips along the axon and growth cone (Figure 4B, arrowheads; Figure 4Bii). Other interaction partners require investigation of colocalization.
FIGURE 4:
Myo16 and LPPR4 colocalize with TRIM67. (Ai) Representative images to demonstrate colocalization of GFP-Myo16 with tagRFP-TRIM67∆RING. Zoomed inset and the blue arrowheads show colocalization along the axon and in filopodia along the axon. (Aii) Pearson’s correlation of colocalization between GFP-Myo16 and tagRFP-TRIM67∆RING (Robs) compared with Fay-randomized controls (Rrand). (n = 9 cells from three biological replicates.) (Bi) Representative images to demonstrate colocalization of GFP-LPPR4 with tagRFP-TRIM67. Zoomed inset and the blue arrowheads show colocalization along the axon (yellow box) and at the axonal growth cone (red box). (Bii) Pearson’s correlation of colocalization between GFP-LPPR4 and tagRFP-TRIM67 (Robs) compared with Fay-randomized controls (Rrand). (n = 12 cells from four biological replicates.) Datapoints were compared using Student’s t test, ***, P < 0.005.
The Myo16 interaction is unique to TRIM9 and TRIM67
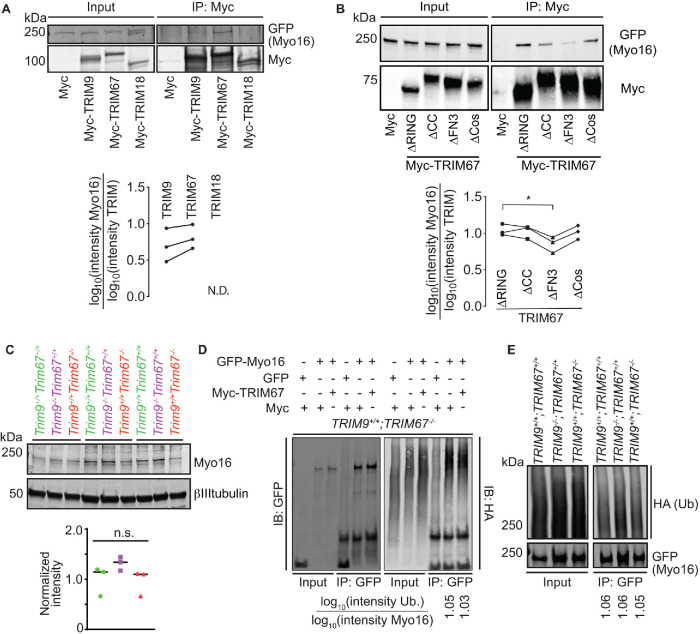
Myo16 expression peaks during neuronal development (Patel et al., 2001), where it regulates presynaptic organization in cerebellar Purkinje cells (Roesler et al., 2019). TRIM9 and TRIM67 are also expressed in the cerebellum and were recently identified as antibody targets in paraneoplastic cerebellar degeneration (Berti et al., 2002; Boyer et al., 2018; Do et al., 2019). Myo16 localizes along the length of the axon and to the tips of filopodia in hippocampal neurons (Patel et al., 2001). Similarly, endogenous TRIM9 and TRIM67 localize to the tips of filopodia, to the periphery of the axonal growth cone, and along the edges of the axon (Winkle et al., 2014; Boyer et al., 2018, 2020). Similar localization and expression patterns of Myo16, TRIM9, and TRIM67 motivated further investigation of the interaction with Myo16. To confirm that the interaction with Myo16 was specific to TRIM9 and TRIM67, we performed additional co-IP assays. Class I TRIMs share domain architecture; of the three class I paralogue pairs, TRIM1 and TRIM18 (MID1) share closest homology with TRIM9 and TRIM67 (Short and Cox, 2006; Carthagena et al., 2009). Whereas GFP-Myo16 precipitated with full-length Myc-TRIM9 and Myc-TRIM67, Myo16 failed to precipitate with Myc-TRIM18 (Figure 5A). Domain mutants of TRIM67 were employed to map the Myo16 interaction. The FN3 domain of TRIM67 was necessary for mediating the TRIM67/Myo16 interaction (Figure 5B). The FN3 domain of TRIM9 and TRIM67 are 75% identical, whereas they share less than 30% identity to the FN3 domain of other class I TRIMs (Supplemental Figure 1A).
FIGURE 5:
Myo16 interacts specifically with TRIM9 and TRIM67. (A) Immunoblot demonstrating GFP-Myo16 co-IP specifically with class I TRIM proteins, TRIM9 and TRIM67, but not TRIM18 (TRIM proteins used in this assay are full-length and Myc-tagged) (n = 3). Plot with individual datapoints of co-IP GFP-Myo16 relative to Myc-TRIM9 or Myc-TRIM67. GFP-Myo16 did not coprecipitate with Myc-TRIM18 (N.D.). (B) Representative immunoblot of TRIM67 domain deletion constructs (TRIM67∆RING, TRIM67∆CC, TRIM67∆FN3, TRIM67∆SPRY) IP demonstrating that the FN3 domain of TRIM67 is necessary for GFP-Myo16 and Myc-TRIM67 interaction (n = 3). Plot with individual datapoints of co-IPGFP-Myo16 relative to TRIM67 domain deletion constructs (∆RING, ∆CC, ∆FN3, ∆SPRY). (C) Immunoblot showing that endogenous Myo16 protein levels were not significantly different in cultured embryonic cortical neurons from wild type (Trim9+/+:Trim67+/+), Trim9–/–:Trim67+/+, Trim9+/+:Trim67–/– at 2 DIV. Quantification from three biological replicates. (D) Representative immunoblot demonstrating GFP (control) and GFP-Myo16 ubiquitination levels in TRIM67–/–HEK cells expressing either Myc or Myc-TRIM67. The blot was probed with anti-GFP and anti-HA antibodies to visualize Myo16 and ubiquitin, respectively. Myo16 ubiquitination status is not altered (n = 3). Ratio of log transformed intensities of HA-ubiquitin relative to immunoprecipitated GFP-Myo16 denoted below respective lanes. (E) Representative immunoblot demonstrating GFP-Myo16 ubiquitination levels in wild-type, TRIM9–/–;TRIM67+/+, TRIM9+/+;TRIM67–/–HEK cells. The blot was probed with anti-GFP and anti-HA antibodies to visualize Myo16 and ubiquitin, respectively. Myo16 ubiquitination status is not altered (n = 3). Ratio of log transformed intensities of HA-ubiquitin relative to immunoprecipitated GFP-Myo16 denoted below the respective lanes. Datapoints in A and B were analyzed using nonparametric analysis of variance (ANOVA) with Fisher’s Least Significant Difference (LSD) post-hoc correction, *, P < 0.05. Nonsignificant differences are not represented. Datapoints in C were analyzed using nonparametric ANOVA followed by the Benjamini–Hochberg procedure. Nonsignificant differences are not represented.
With evidence of the specificity of the Myo16 interaction with TRIM9 and TRIM67, we examined whether loss of these proteins affected Myo16. Myo16 protein levels in cortical neurons were not altered upon loss of Trim9 or Trim67 (Figure 5C). Because the interaction between TRIM67 and Myo16 appeared stronger, we hypothesized that TRIM67 may alter Myo16 ubiquitination. To test this, we performed denaturing immunoprecipitation-based ubiquitination assays of GFP-Myo16 from HEK293 cells in which TRIM67 had been removed via CRISPR/Cas9 genome editing (Boyer et al., 2020). GFP-Myo16 immunoprecipitation contained high-molecular-weight ubiquitin reactivity in TRIM67–/– HEK cells expressing Myc alone or Myc-TRIM67 (Figure 5D). In contrast, immunoprecipitation of GFP alone revealed no high-molecular-weight ubiquitin reactivity. These data indicated that Myo16 was ubiquitinated, but it was not TRIM67-dependent. Similar Myo16 ubiquitination was also observed in HEK293 cells in which TRIM9 had been deleted (Figure 5E). This further revealed that Myo16 ubiquitination levels were not different across the three genotypes. Together these data indicated that the Myo16 interaction with TRIM9 and TRIM67 was specific and mediated by the FN3 domain, but the interaction did not detectably alter Myo16 localization, protein levels, or ubiquitination.
Myo16 regulates netrin-1–dependent axonal branching and filopodial density at the axonal growth cone
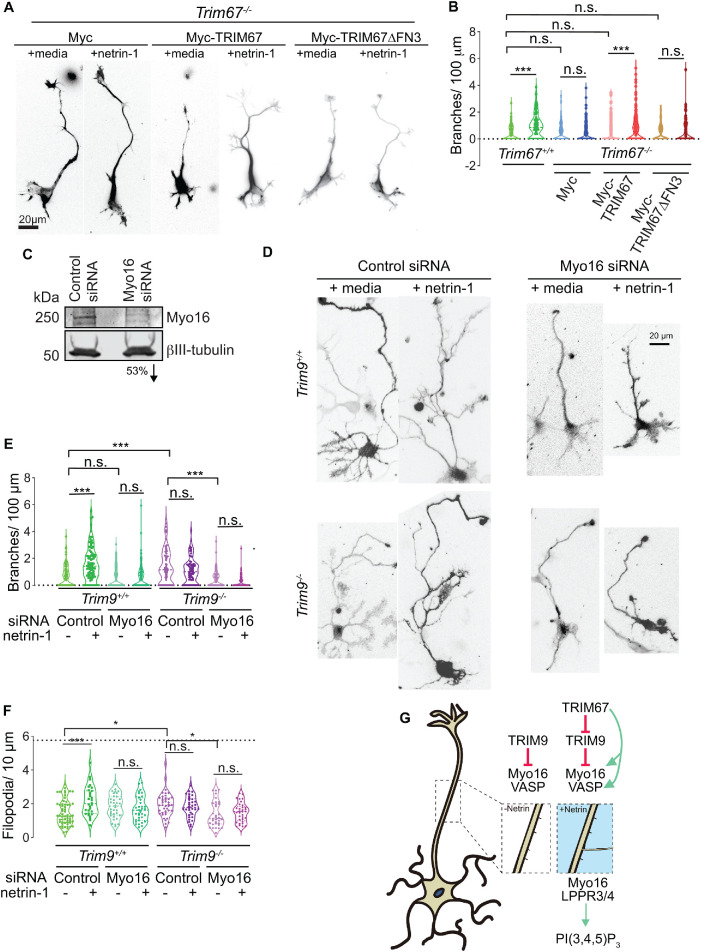
The axon guidance cue netrin-1 promotes axon branching in cortical neurons (Dent et al., 2004; Winkle et al., 2014). We have previously shown that deletion of Trim9 or Trim67 independently disrupts netrin-dependent axon branching (Winkle et al., 2014; Menon et al., 2015; Plooster et al., 2017; Boyer et al., 2020). In Trim9–/– neurons, basal levels of axon branching are two- to threefold higher but do not increase in response to netrin-1. In contrast, In Trim67–/– neurons, basal levels of axon branching are normal, but similarly do not increase in response to netrin-1 addition. Introduction of full-length TRIM67 into Trim67–/– neurons rescued netrin-dependent axon branching (Boyer et al., 2020). However, TRIM67∆FN3, which did not interact with Myo16 (Figure 5E), failed to rescue netrin-dependent branching (Figure 6, A and B), potentially implicating Myo16 in netrin-dependent axon branching. To test this hypothesis, Myo16 small interfering RNA (siRNA) was introduced into wild-type cortical neurons at day in vitro 0 (DIV 0), and Myo16 levels were reduced approximately 70% by 2 DIV (Figure 6C). Similar to Trim67 deletion, Myo16 knockdown did not alter basal axon branching levels, but abrogated netrin-dependent axonal branching (Figure 6, D and E). Axon length and progression of neuronal morphogenesis were not perturbed by Myo16 knockdown (Supplemental Figure 2). These data indicated that Myo16 was required for netrin-dependent axon branching. Because TRIM67 is known to interact with the netrin receptor DCC (Boyer et al., 2018) but yet there is no evidence that Myo16 interacts with DCC, we surmised that overexpression of Myo16 in Trim67–/– neurons was unlikely to rescue netrin sensitivity and netrin-dependent axon branching. We, however, hypothesized that the exuberant axon branching phenotype of Trim9–/– neurons (Winkle et al., 2014) may be dependent on Myo16. Here we confirmed previous data demonstrating that Trim9–/– cortical neurons showed elevated axon branching density (Figure 6, D and E). Reduction of Myo16 in Trim9–/– cortical neurons reduced axonal branch density (Figure 6, D and E), suggesting that Myo16 function was required for the aberrant branching phenotype in Trim9–/– neurons. However, axon branching still failed to increase in response to netrin.
FIGURE 6:
Knockdown of Myo16 inhibits netrin-dependent axonal branching. (A) Netrin-1–dependent axon branching assay and quantification in Trim67–/– neurons expressing Myc, Myc-TRIM67, or Myc-TRIM67∆FN3. The Myc and Myc-TRIM67 data were published in Boyer et al. (2020). Experiments with MycTRIM67∆FN3 were performed contemporaneously and not previously published. Staining for filamentous actin (phalloidin) and βIII tubulin were merged, to reveal neuronal morphology. (B) Quantification of axonal branching density shown as violin plots with individual datapoints from three biological replicates. n (cells) = 169 Myc, 177 Myc + netrin, 173 Myc-TRIM67, 166 Myc-TRIM67 + netrin, 87 Myc-TRIM67∆FN3, 83 Myc-TRIM67∆FN3 + netrin. (C) Representative immunoblot demonstrating effective knockdown of Myo16 at 2 DIV. A prominent Myo16 band that is immunoreactive at ∼250 kDa in the lane with control siRNA-treated lysate shows reduced levels in the lane with Myo16 siRNA-treated lysate. A 53% reduction in Myo16 levels is denoted below the corresponding lane. Average of ∼70% reduction in Myo16 levels was observed from two biological replicates. (D) Representative merged images of GFP (to reveal transfection with control or Myo16 siRNA) and βIII tubulin (to reveal cell morphology) in wild-type (Trim9+/+) and Trim9–/– cortical neurons at 3 DIV demonstrate branching phenotypes with or without 24 h of netrin treatment. (E) Quantification of axonal branching density shown as violin plots with individual datapoints from three biological replicates. n (cells) = 103 Trim9+/+ C siRNA, 92 Trim9+/+ C siRNA + netrin, 88 Trim9+/+ Myo16 siRNA, 105 Trim9+/+ Myo16 siRNA + netrin, 85 Trim9–/– C siRNA, 74 Trim9–/– C siRNA + netrin, 88 Trim9–/– Myo16 siRNA, 63 Trim9–/– Myo16 siRNA + netrin. (F) Quantification of filopodia density at the axonal growth cone at 2 DIV shown as violin plots with individual datapoints from two biological replicates. n (cells) = 59 Trim9+/+ C siRNA, 30 Trim9+/+ C siRNA + netrin, 34 Trim9+/+ Myo16 siRNA, 35 Trim9+/+ Myo16 siRNA + netrin, 41 Trim9–/– C siRNA, 34 Trim9–/– C siRNA + netrin, 33 Trim9–/– Myo16 siRNA, 33 Trim9–/– Myo16 siRNA + netrin. Nonparametric ANOVA with Tukey’s post-hoc correction was performed on all data sets. *, P < 0.05, ***, P < 0.005). (G) Model of TRIM-dependent regulation of axon branching in response to netrin. In the absence of netrin, TRIM9 reduces VASP and Myo16 function. In the presence of netrin-1, TRIM67 either directly promotes VASP and Myo16 function or does so via antagonizing TRIM9 to promote filopodia density and initiate axon branching. Myo16 in turn, along with LPPR3 and LPPR4, may locally promote increased PI(3,4,5)P3 levels in the plasma membrane.
Filopodia are required for axon branching response (Dent et al., 2004), and our previous work showed that netrin-dependent increases in growth cone filopodia density were also disrupted by deletion of Trim9 or Trim67 (Menon et al., 2015; Boyer et al., 2020). We thus investigated how Myo16 knockdown affected filopodia density after acute netrin addition. Myo16 knockdown in wild-type cortical neurons abrogated the netrin-dependent increase in filopodial densities at axonal growth cones (Figure 6F). Myo16 knockdown also reduced the aberrant elevated filopodial density of Trim9–/– cortical neurons (Figure 6F). These data confirm a novel role for Myo16 during axonal branching, growth cone filopodia density, and netrin responses.
DISCUSSION
Previously identified interacting partners and substrates suggested that TRIM9 and TRIM67 shared interaction partners, functioned in a yin–yang manner, and were poised to regulate the cytoskeletal machinery and membrane remodeling machinery that regulate neuronal morphogenesis, function, and connectivity. Here we used proximity biotinylation to identify previously unidentified candidate interaction partners. This revealed an overlapping interactome of proteins enriched in cellular process regulated by TRIM9 and TRIM67, including neuronal growth and arborization, cytoskeletal dynamics, and membrane remodeling. In addition, novel cellular processes such as synapse structure and maintenance were identified as potential TRIM9 and TRIM67 regulation points. We validated a subset of candidates implicated in cytoskeletal dynamics, membrane remodeling, and transport. One validated candidate, Myo16, plays a role in netrin-dependent axon branching, potentially in coordination with TRIM9 and TRIM67 (Figure 6G). Altogether this study demonstrates that TRIM9 and TRIM67 likely regulate neuronal form and function through a host of interaction partners and potential substrates and this interactome opens up an abundance of new avenues for investigation.
TRIM9 and TRIM67 regulate the neuronal actin and MT cytoskeletons
Our findings in Trim9–/– and Trim67–/– neurons revealed disruptions at multiple stages of neuronal morphogenesis, including filopodia stability, growth cone size and turning, axon branching, dendritic arborization, and dendritic spine density (Winkle et al., 2014, 2016; Menon et al., 2015; Plooster et al., 2017; Boyer et al., 2020). GO analysis of candidate interactors here bolsters the role of TRIM9 and TRIM67 function in regulating neuronal growth and guidance, with enrichment in neurite outgrowth regulation, and actin and MT cytoskeletal regulation, including actin and MT polymerization, as well as localization to multiple cytoskeletal structures, stress fibers, focal adhesion, MT ends, and lamellipodium. Here we found that the unconventional myosin, Myo16, interacts with both TRIM9 and TRIM67 and plays a critical role in netrin-dependent axon branching. TRIM67 structure/function results and rescue experiments in Trim9–/– neurons suggested Myo16 regulates netrin-dependent branching in coordination with TRIM9 and TRIM67. The mechanism of this coordinated response is not known. We did find that Myo16 was ubiquitinated, although we did not detect differences in the absence of TRIM9 or TRIM67. However, whether the same residues are ubiquitinated in all genotypes or whether TRIM9 and TRIM67 redundantly lead to Myo16 ubiquitination is not known. Our previous work found that TRIM9 and TRIM67 interact with each other (Boyer et al., 2018) and function in a yin–yang manner to mediate the nondegradative ubiquitination of VASP (Menon et al., 2015; Boyer et al., 2020). Nondegradative ubiquitination of VASP was essential for appropriate netrin-1–dependent axonal responses, such as filopodial stability at the axonal growth cone and axonal turning. Whether a similar antagonistic relationship exists between TRIM9, TRIM67, and Myo16 is an intriguing hypothesis. Similar questions regarding how other cytoskeletal regulators, like Coro1A and Sipa1l1, are involved in neuronal morphogenesis and regulated by TRIM proteins and downstream of netrin signaling remain to be answered.
Several tubulin isoforms and multiple MT-associated proteins and motors were identified, including MAP1B, DCX, and kinesins Kif1A and Kif1B. Class I TRIMs contain a Cos domain in their C-termini that has been implicated in MT binding (Short and Cox, 2006). Class I members TRIM18 (MID1) and TRIM46 have both been shown to interact with MTs and regulate MT organization (Van Beuningen et al., 2015; Wright et al., 2016). As MTs are a critical component in neuronal morphogenesis, investigation into how they are potentially regulated by TRIM9 and TRIM67 is warranted.
TRIM9 and TRIM67 regulate membrane remodeling
Developing neurons undergo large changes in plasma membrane surface area, driven by exocytosis. Previous work identified that TRIM9 interacts with and sequesters the exocytic t-SNARE SNAP25 from forming SNARE complexes, resulting in constrained constitutive exocytosis and axon branching in developing neurons (Winkle et al., 2014). This interaction was netrin sensitive, with netrin treatment increasing SNARE complex formation, exocytosis, and axon branching in a TRIM9-dependent manner. Strikingly, this increase in exocytosis primarily occurs in the axon, but how the localization of exocytic events occurs is unknown. Here we identified the exocyst component ExoC1 as a validated novel TRIM interactor. Whether this change in the localization of exocytic events involves vesicle tethering, such as the exocyst complex, is an intriguing hypothesis.
In contrast to TRIM9, TRIM67 regulates exocytic mode not frequency, promoting full vesicle fusion at the expense of kiss-and-run fusion (Urbina et al., 2020). The t-SNARE SNAP47 was also identified in the proteomic screen described here, specifically as a TRIM67 binding partner. As described elsewhere, we found that SNAP47 was partially responsible for TRIM67-mediated regulation of the exocytic mode in developing neurons, but additional proteins must also be involved. Potentially, other membrane-associated proteins like LPPR4 (Sigal et al., 2005) or known exocytic regulators like Munc18 and tomosyn, which we identified as candidate TRIM67 binding partners, may also regulate the exocytic mode downstream of TRIM67.
A potential role of TRIM9 and TRIM67 in modulating synapse morphology
Although BioID experiments were performed in neurons before synaptogenesis, we found an intriguing enrichment of proteins that localize to the postsynaptic density and the presynaptic active zone and modulate synapse formation and maintenance, such as validated interactors GRIP1, Myo16, VASP, PRG-1, and Kif1a, as well as yet to be validated candidates such as PSD95, BSN, PCLO, and SynGAP1. We previously demonstrated that deletion of Trim9 altered dendritic spine density in adult-born neurons of the dentate gyrus (Winkle et al., 2016), suggesting that TRIM9 may regulate synaptic connectivity. Adult-born hippocampal neurons are critical in spatial learning and memory, which was dramatically deficient in Trim9–/– mice (Winkle et al., 2016). Similarly, genetic deletion of Trim67 resulted in delayed spatial learning and memory and a loss of cognitive flexibility in the Morris Water Maze task (Boyer et al., 2018). Validation of proteins enriched in these categories will give us an insight into how TRIM9 and TRIM67 regulate synapse formation and function. Although how TRIM9 and TRIM67 function at the synapse and whether this is a developmental role or maintenance role in the adult is unknown, the array of BioID candidates provide multiple pathways for investigation. Many potential scenarios are plausible. This includes regulation of dendritic filopodia formation and spine maturation, based on the role of TRIM9 and TRIM67 in filopodia density and lifetime (Menon et al., 2015; Boyer et al., 2020). This could be through regulation of VASP or Myo16, both of which have been implicated in spine morphogenesis (Lin et al., 2010; Roesler et al., 2019). Alternatively, TRIM9 and TRIM67 may play a role in delivery or retrieval of receptors and membrane material (Winkle et al., 2014; Urbina et al., 2018, 2020) to the pre- or postsynaptic compartments via regulation of proteins such as GRIP1, VCP, or Kif1a.
Other roles for TRIM9 and TRIM67
TRIM67 function is implicated downstream of Ras in neuroblastoma cells (Yaguchi et al., 2012). Our studies indicate that both TRIM9 and TRIM67 are involved downstream of netrin and DCC. In the case of DCC, we found that TRIM9 plays a role in signaling. DCC was ubiquitinated in a TRIM9- and netrin-1–sensitive manner (Plooster et al., 2017). This ubiquitination resulted in reduced DCC phosphorylation levels, which constrained binding, autophosphorylation, and activation of focal adhesion kinase (FAK). GO analysis suggested that TRIM9 and TRIM67 may regulate additional signaling networks such as the MAP kinase pathway. Further, validated interactions with both Myo16 and LPPR4 implicate TRIM9 and TRIM67 in the phosphoinositol signaling pathway, which is critical to morphogenesis. Myo16, also known as NYAP3 (Neuronal tYrosine phosphorylated Adaptor for PI3K), promotes PI3K activity and increases levels of PI(3,4,5)P3 (Yokoyama et al., 2011). BioID also predicted that TRIM9 and TRIM67 interact with LPPR3 and LPPR4, which also increase membrane levels of PI(3,4,5)P3 levels, by antagonizing PTEN (Brosig et al., 2019). We speculate that netrin-dependent branches may involve TRIM-dependent changes in PI(3,4,5)P3 membrane levels, via regulation of Myo16, LPPR3, and LPPR4 (Figure 6G). Defining how TRIM proteins alter membrane levels of PI(3,4,5)P3 and what roles this plays in morphogenesis is critical. Regulation of translation initiation and microRNA (miRNA)-mediated inhibition of translation are some of the other categories enriched in the BioID candidates. Spatiotemporal regulation of protein translation is critical to the proper form and function of a neuron and regulates axonal branching and pathfinding, memory consolidation, and cognitive function (Yoon et al., 2009; Buffington et al., 2014; Wong et al., 2017). Recent evidence also suggests that there are receptor-specific interactomes that could serve as a hub for guidance cue–induced protein translation, such as in netrin-1/DCC signaling (Koppers et al., 2019). Deregulated protein translation has been attributed as the underlying cause of various neurodegenerative disorders (Kapur et al., 2017). Validation of these interacting partners remains to be completed. How these pathways are regulated by TRIM9 and TRIM67 and their participation in neuronal morphogenesis and function will be areas of future investigation.
Previous work together with this study provide evidence that TRIM9 and TRIM67 have multiple overlapping interaction partners and are poised to modulate a number of stages of neuronal morphogenesis, as well as neuronal function. Additional validation and investigations of the TRIM9/TRIM67 interactome are required. However, we propose that these ligases function as molecular rheostats to modulate a shared interactome that coordinates cytoskeletal dynamics and plasma membrane expansion. This coordination facilitates neuronal growth and guidance, essential for establishment of well-connected functional neural networks.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Animals
All mouse lines were on a C57BL/6J background and bred at the University of North Carolina with approval from the Institutional Animal Care and Use Committee. Timed pregnant females were obtained by placing male and female mice together overnight; the following day was designated as E0.5 if the female had a vaginal plug. Generation of Trim9–/– and Trim67–/– mice has been described previously (Winkle et al., 2014; Boyer et al., 2018).
Cortical neuron culture
E15.5 litters were removed from wild-type, Trim9–/–, and Trim67–/–pregnant dams by postmortem cesarean section. Dissociated cortical neuron cultures were prepared as described previously (Kwiatkowski et al., 2007). Briefly, cortices were microdissected and cortical neurons were dissociated with 0.25% trypsin for 20 min at 37°C, followed by quenching of trypsin with trypsin quenching medium (TQM; neurobasal medium supplemented with 10% fetal bovine serum [FBS] and 0.5 mM glutamine). After quenching, cortices were gently triturated in neurobasal medium supplemented with TQM. Dissociated neurons were then pelleted at 100 × g for 7 min. The pelleted cells were gently resuspended in serum free medium (SFM; neurobasal medium supplemented with B27 [Invitrogen]) and plated on coverslips or tissue culture plastic coated with 1 mg/ml poly-d-lysine (Sigma-Aldrich). For axon length and branching analysis, either neuronal growth media or 250 ng/ml netrin-1 in media was added to neuronal cultures at 2 DIV, followed by fixation with 4% paraformaldehyde (PFA) in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, pH = 6.9) after 24 h. For axonal growth cone filopodia density, 2 DIV neuronal cultures were treated with bath application of 600 ng/ml netrin-1 for 40 min followed by fixation with 4% PFA in PHEM buffer.
HEK293 cell culture
Wild-type and TRIM9–/–;TRIM67+/+HEK293 cells were obtained from Simon Rothenfußer (Klinikum der Universitat München, München, Germany) as previously described (Menon et al., 2015). TRIM9+/+;TRIM67–/–HEK293 cells were generated as previously described (Boyer et al., 2020). These cells were grown in DMEM with glutamine (Invitrogen), supplemented with 10% FBS (Hyclone) and maintained at 5% CO2/37°C.
Plasmids, antibodies, and reagents
Plasmids encoding Myc-TRIM9 full-length, Myc-TRIM67 full-length, Myc-TRIM9ΔRING, Myc-TRIM67∆RING, Myc-TRIM67∆CC, Myc-TRIM67∆Cos, Myc-TRIM67∆FN3, Myc-TRIM67∆Cos, and Myc-TRIM67∆SPRY constructs and tagRFP-tagged TRIM67 construct were previously described (Winkle et al., 2014; Boyer et al., 2018, 2020). The pDOWN Gateway entry vectors for expressing Myc-BirA*, Myc-BirA*TRIM9∆RING or Myc-BirA*TRIM67∆RING were constructed by VectorBuilder. HSV vectors for the BirA* constructs were generated by Rachael Neve, MIT Proteomics Core (currently at Gene Technology Core–Massachusetts General Hospital). The following plasmids were acquired: pDEST307-FLAG-PRG-1(LPPR4) (Herbert Geller, National Heart, Lung, and Blood Institute, National Institutes of Health [NIH]; PRG-1 was PCR amplified and recloned into an eGFP vector, eGFP-PRG-1), pML2 (EGFP-N1)–hCoro1A (James Bear, University of North Carolina, Chapel Hill), pEGFP-Myo16 (Anand Srivastava, Greenwood Genetic Center Foundation), VCP(wt)-EGFP (https://www.addgene.org/23971/, originally deposited by Nico Dantuma), Sipa1l1-HA (SPAR-HA) (Daniel Pak, Georgetown University), pEGFP-C3-Sec3 (GFP-ExoC1) (https://www.addgene.org/53755/, originally deposited by Channing Der), GFP-Kif1a (Juan Bonafacino, National Institute of Child Health and Human Development, NIH), GRIP1b-EGFP (Rick Huganir, Johns Hopkins University), pcDNA3.1-HA-14-3-3ε(Ywhae) (https://www.addgene.org/48797/, originally deposited by Huda Zhogbi), pCMV-6Myc-hMID1 (Timothy Cox, University of Washington), pEGFP-C1 MAP1B-GFP (Philips Gordon-Week, King’s College London), and GFP-hRas CAAX (Richard Cheney, University of North Carolina). Antibodies included goat polyclonal against DCC (Santa Cruz); rabbit polyclonal against VASP (Santa Cruz); mouse monoclonal against c-Myc (9E10); mouse monoclonal against mouse β-III-tubulin (Biolegend); mouse monoclonal against HA (12CA5) (Patrick Brennwald, University of North Carolina); rabbit polyclonal against Myo16 (Richard Cameron, Augusta University); rabbit polyclonal against Myo16 (Proteintech; 25104-1-AP); fluorescent secondary antibodies. Other fluorescently labeled reagents included streptavidin and phalloidin labeled with Alexa Fluor 561. Recombinant chicken myc-netrin-1 was concentrated from HEK293 cells (Serafini et al., 1996; Lebrand et al., 2004).
BioID , affinity purification, and peptide preparation
The negative control Myc-BirA* and Myc-BirA*TRIM9ΔRING (amino acids 132–710) or Myc-BirA*TRIM67ΔRING (amino acids 164–783) constructs were cloned into HSV vectors using Gateway Cloning (VectorBuilder and Rachael Neve, MIT Viral Vector Core). These vectors were packaged into Short Term HSV and driven under a IE 4/5 promoter. GFP expression is also driven in tandem downstream of a mCMV promoter. The two promoters are equivalent in the timing of expression and strength. GFP signal serves as a good indicator of transduction efficiency and timing of expression.
E15.5 cortical neurons from wild-type and Trim9–/– or Trim67–/– E15.5 litters were dissociated and plated on poly-d-lysine (PDL) (Sigma)-coated tissue culture dishes. Approximately 40 h postplating, neurons were infected with HSV (multiplicity of infection, MOI = 1.0) carrying either the negative control Myc-BirA*, Myc-BirA*TRIM9∆RING, or Myc-BirA*TRIM67∆RING. Six hours postinfection, cultures were supplemented with 50 μM biotin for 24 h. After incubation, cells were lysed using RIPA buffer (150 mM NaCl, 25 mM Tris-HCl, pH 7.5, 0.1% SDS, 1.0% NP-40, 0.25% deoxycholic acid, 2 mM EDTA, 10% glycerol, protease and phosphatase inhibitors). Lysates were treated with 0.5 ml benzonase/ml lysate with end-to-end rotation for 1 h at 4°C. Lysates were cleared by centrifugation, 16,000 × g for 30 min at 4°C. Biotinylated proteins were enriched using streptavidin-conjugated Sepharose beads (GE Healthcare Life Sciences). Beads were washed once with RIPA buffer, twice consecutively with TAP lysis buffer (50 mM HEPES [pH 8.0], 10% glycerol, 150 mM NaCl, 2 mM EDTA, and 0.1% NP-40), and three times consecutively with 50 mM ammonium bicarbonate. Enriched proteins were digested with 2.5 μg trypsin at 37°C for 16 h. Peptides were eluted using the RapiGEST SF Surfactant protocol (Waters). Eluted peptides were desalted using C18 columns followed by an ethyl acetate cleanup. Peptides were then stored at –20°C until analysis by mass spectrometry.
Mass spectrometry and data analysis
Reverse-phase nano-high-performance liquid chromatography (nano-HPLC) coupled with a nanoACQUITY ultraperformance liquid chromatography (UPLC) system (Waters Corporation, Milford, MA) was used to separate trypsinized peptides. Trapping and separation of peptides were performed in a 2 cm column (Pepmap 100; 3-m particle size and 100-Å pore size) and a 25-cm EASYspray analytical column (75-m inside diameter [i.d.], 2.0-m C18 particle size, and 100-Å pore size) at 300 nl/min and 35°C, respectively. Analysis of a 150-min gradient of 2–25% buffer B (0.1% formic acid in acetonitrile) was performed on an Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific). The ion source was operated at 2.4 kV, and the ion transfer tube was set to 300°C. Full MS scans (350–2000 m/z) were analyzed in the Orbitrap at a resolution of 120,000 and 1e6 AGC target. The MS2 spectra were collected using a 1.6 m/z isolation width and were analyzed by either the Orbitrap or the linear ion trap, depending on peak charge and intensity using a 3 s TopSpeed CHOPIN method. Orbitrap MS2 scans were acquired at 7500 resolution, with a 5e4 automatic gain control (AGC) and 22 ms maximum injection time after higher-energy collisional dissociation fragmentation with a normalized energy of 30%. Rapid linear ion trap MS2 scans were acquired using a 4e3 AGC, 250 ms maximum injection time after collision-induced dissociation 30 fragmentation. Precursor ions were chosen based on intensity thresholds (>1e3) from the full scan as well as on charge states (2–7) with a 30-s dynamic exclusion window. Polysiloxane 371.10124 was used as the lock mass.
For the data analysis all raw mass spectrometry data were searched using MaxQuant version 1.5.7.4. Search parameters were as follows: UniProtKB/Swiss-Prot mouse canonical sequence database (downloaded 1 February 2017), static carbamidomethyl cysteine modification, specific trypsin digestion with up to two missed cleavages, variable protein N-terminal acetylation and methionine oxidation, match between runs, and label-free quantification (LFQ) with a minimum ratio count of 2. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the data set identifier PXD021758. To rank candidate protein–protein interactions by likelihood of interaction, LFQ values for proteins identified in control and experimental conditions were input into SAINTq (version 0.0.4). Interactions were sorted by SAINT’s interaction probability, and a FDR threshold was assigned to each of the identified proteins.
Data representation and GO analysis
Cytoscape _3.7 open source bioinformatics interaction visualization software was used to visualize potential interaction candidates using a Prefuse Force Directed Layout in which the source node (TRIM9, TRIM67, or both) appears in the center of the network. GO enrichment analysis was performed using the ClueGo application available for download in Cytoscape_3.7.
Transfection of cortical neurons and HEK293 cells
For transfection of plasmids and siRNA, dissociated cortical neurons were resuspended in Lonza Nucleofector solution (VPG-1001) and electroporated with an Amaxa Nucleofector according to the manufacturer’s protocol. HEK293 cells were transfected using Polyplus jetPRIME reagent or Lipofectamine 2000 as per the manufacturer’s protocol.
Co-IP assays
For co-IP assays HEK293 cells were transfected with plasmids expressing tagged versions of the proteins-of-interest and Myc- or GFP-tagged TRIM9 or TRIM67 full-length or ∆RING constructs or Myc-tagged TRIM67 domain deletion constructs. Sixteen to eighteen hours posttransfection the cells were lysed using the immunoprecipitation (IP) buffer (10% glycerol, 1% NP-40, 50 mM Tris, pH 7.5, 200 mM NaCl, 2 mM MgCl2 and protease and phosphatase inhibitors). Cells were scraped from the dish and transferred into tubes and then centrifuged at 18,400 × g for 10 min. Approximately 500–1000 µg of protein per sample was used per co-IP. Myc-tagged proteins were enriched using anti-Myc (9E10) antibody. Antibody was incubated overnight with the lysate, following which Protein A–coupled agarose beads were added to the lysate–antibody mix. After 2 h of incubation the beads were precipitated, washed twice with IP buffer, and then boiled with 2× sample buffer. GFP-Trap beads (Chromotek) were used to enrich GFP-tagged proteins. The beads were incubated with the lysate for 1 h. The beads were then precipitated, washed twice with IP buffer, and then boiled with 2× sample buffer.
Ubiquitination assay
Wild-type, TRIM9–/–;TRIM67+/+, and TRIM9+/+;TRIM67–/– HEK cells were transfected with HA-Ub and GFP-Myo16, and in some cases Myc or Myc-TRIM67, using Lipofectamine 2000 and cultured for 24 h. These cells were treated with 10 μM MG132 for 4 h. The cells were then lysed in IP buffer (20 mM Tris-Cl, 250 mM NaCl, 3 mM EDTA, 3 mM EGTA (ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid), 0.5% NP-40, 1% SDS, 2 mM dithiothreitol, 5 mM NEM (N-ethylmaleimide), 3 mM iodoacetamide, protease and phosphatase inhibitors, pH = 7.3–7.4). For 5–6 million cells 270 µl of ubiquitin IP buffer was added and incubated on ice for 10 min. Cells were removed from the dish and transferred into tubes. Phosphate-buffered saline (PBS; 30 μl of 1×) was added and gently vortexed. Samples were boiled immediately for 20 min, until clear, and then centrifuged at 18,400 × g for 10 min. The boiled samples were diluted using IP buffer without SDS to reduce the SDS concentration to 0.1%. Immunoprecipitations were performed under conditions in which only covalent interactions were preserved with anti-GFP (mouse) antibody overnight at 4°C, following which Protein A agarose beads (Santa Cruz) were used to precipitate the antibody-bound complex.
Immunoblotting
The immunoprecipitated proteins from co-IP assays or ubiquitination assays or endogenous neuronal lysates were separated on reducing polyacrylamide gels and transferred to nitrocellulose membrane using standard procedures. The blots were probed with tag-specific or protein-specific primary antibodies followed by species-specific far-red conjugated secondary antibodies (Licor). Signal was detected using an Odyssey Imager (Licor). Western blots were analyzed using Li-Cor Image Studio Lite. Total fluorescence of labeled bands representing relative protein was measured. For all Western blot comparisons, the ratios of log transformed relative intensities (indicated for each blot) were analyzed using appropriate statistical tests as indicated in the corresponding figure legends.
Immunofluorescence
Neurons fixed with 4% PFA in PHEM buffer were permeabilized for 10 min in 0.1% Triton X-100. Permeabilized cells were blocked for 30 min in 10% donkey serum and stained with the indicated primary antibodies for 1 h at room temperature (RT). The primary antibodies were then washed off (4 × 5 min washes in 1× PBS), following which species-appropriate fluorescently labeled secondary antibodies or fluorescently labeled phalloidin were added and allowed to incubate for 1 h at RT. After the secondary antibodies/phalloidin was washed off, cells were mounted in a Tris/glycerol/n-propyl-gallate–based mounting medium and stored at 4°C till they were ready to be imaged.
Colocalization experiments
GFP-tagged Myo16 and tagRFP-TRIM67 constructs were expressed in Trim67–/– dissociated cortical neurons. Colocalization was observed using total internal reflection fluorescence (TIRF) microscopy. For live-cell TIRF and time-lapse imaging, cells were maintained at 37°C, 5% CO2, and humidity using a stage top incubator (Tokai Hit).
Microscopy and image acquisition
All immunofluorescence and live-cell images were collected on an Olympus IX81-ZDC2 inverted microscope. The following objective lenses were utilized: a UPLFLN 40×/1.39-NA differential interference contrast (DIC) objective (Olympus), UAPON 100×/1.49-NA DIC TIRF objective (Olympus). The microscope is equipped with an automated XYZ stage (Prior) and an Andor iXonelectron multiplying charge-coupled device. Cells for live imaging were maintained at 37°C and 5% CO2 using a TokaiHit on-stage cell incubator. Images were acquired using Metamorph acquisition software. Image analysis was performed in Fiji. Axon branches were defined as extensions off the primary axon (more than two times longer than other neurites) that were ≥20 µm in length. Pearson’s correlation of colocalization between GFP-tagged Myo16 or LPPR4 and tagRFP-tagged TRIM67 constructs was performed using the Colocalization Test plug-in for ImageJ with Fay randomization.
Supplementary Material
Acknowledgments
We thank Vong Thoon, Caroline Monkiewicz, Carey Hanlin, Janee Cadlett-Jete, and Natalia Riddick for mouse colony management. We thank Emily Cousins for help with troubleshooting the BioID approach and sample submission for mass spectrometric analysis. This work was supported by National Institutes of Health Grants R21MH108970 and R35GM135160 (S.L.G.).
Abbreviations used:
- BioID
proximity-dependent biotin identification
- BirA*
promiscuous bifunctional ligase/repressor BirA
- co-IP
coimmunprecipitation
- Coro1a
coronin, actin-binding protein 1A
- DCC
deleted in colorectal cancer
- ExoC1
exocyst complex component 1
- FDR
false discovery rate
- GO
gene ontology
- GRIP1
glutamate receptor interacting protein 1
- HSV
herpes simplex virus
- KIF1A
kinesin family member 1A
- MAP1B
microtubule-associated protein 1B
- MS
mass spectrometry
- MT
microtubule
- Myo16
myosin XVI
- PRG-1
phospholipid phosphatase related 4 (Plppr4)
- SNAP25
synaptosome-associated protein 25
- SNAP47
synaptosome-associated protein 47
- TIRF
total internal reflection fluorescence
- TRIM67
tripartite motif containing 67
- TRIM9
tripartite motif containing 9
- Ywhae
14-3-3 epsilon.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-10-0622) on December 30, 2020.
REFERENCES
- Berti C, Messali S, Ballabio A, Reymond A, Meroni G (2002). TRIM9 is specifically expressed in the embryonic and adult nervous system. Mech Dev 113, 159–162. [DOI] [PubMed] [Google Scholar]
- Bin JM, Han D, Lai Wing Sun K, Croteau LP, Dumontier E, Cloutier JF, Kania A, Kennedy TE (2015). Complete loss of netrin-1 results in embryonic lethality and severe axon guidance defects without increased neural cell death. Cell Rep 12, 1099–1106. [DOI] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J (2009). ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodaleo FJ, Montenegro-Venegas C, Henríquez DR, Court FA, Gonzalez-Billault C (2016). Microtubule-associated protein 1B (MAP1B)-deficient neurons show structural presynaptic deficiencies in vitro and altered presynaptic physiology. Sci Rep 6, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer NP, McCormick LE, Menon S, Urbina FL, Gupton SL (2020). A pair of E3 ubiquitin ligases compete to regulate filopodial dynamics and axon guidance. J Cell Biol 219, e201902088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer NP, Monkiewicz C, Menon S, Moy SS, Gupton SL (2018). Mammalian TRIM67 functions in brain development and behavior. ENeuro 5, 0186-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräuer AU, Savaskan NE, Kühn H, Prehn S, Ninnemann O, Nitsch R (2003). A new phospholipid phosphatase, PRG-1, is involved in axon growth and regenerative sprouting. Nat Neurosci 6, 572–578. [DOI] [PubMed] [Google Scholar]
- Brosig A, Fuchs J, Ipek F, Kroon C, Schrötter S, Vadhvani M, Polyzou A, Ledderose J, van Diepen M, Holzhütter HG, et al. (2019). The axonal membrane protein PRG2 inhibits PTEN and directs growth to branches. Cell Rep 29, 2028–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno DL, Anderlid BM, Lindstrand A, Van Ravenswaaij-Arts C, Ganesamoorthy D, Lundin J, Martin CL, Douglas J, Nowak C, Adam MP, et al. (2010). Further molecular and clinical delineation of co-locating 17p13.3 microdeletions and microduplications that show distinctive phenotypes. J Med Genet 47, 299–311. [DOI] [PubMed] [Google Scholar]
- Buffington SA, Huang W, Costa-Mattioli M (2014). Translational control in synaptic plasticity and cognitive dysfunction. Annu Rev Neurosci 37, 17–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cainarca S, Messali S, Ballabio A, Meroni G (1999). Functional characterization of the Opitz syndrome gene product (midin): evidence for homodimerization and association with microtubules throughout the cell cycle. Hum Mol Genet 8, 1387–1396. [DOI] [PubMed] [Google Scholar]
- Carthagena L, Bergamaschi A, Luna JM, David A, Uchil PD, Margottin-Goguet F, Mothes W, Hazan U, Transy C, Pancino G, et al. (2009). Human TRIM gene expression in response to interferons. PLoS One 4, e4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloer EW, Siesser PF, Cousins EM, Goldfarb D, Mowrey DD, Harrison JS, Weir SJ, Dokholyan NV, Major MB (2018). p62-dependent phase separation of patient-derived KEAP1 mutations and NRF2. Mol Cell Biol 38, e00644-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox TC (2012). The microtubule-associated C-I subfamily of TRIM proteins and the regulation of polarized cell responses. Adv Exp Med Biol 770, 105–118. [DOI] [PubMed] [Google Scholar]
- Coyaud E, Mis M, Laurent EMN, Dunham WH, Couzens AL, Robitaille M, Gingras AC, Angers S, Raught B (2015). BioID-based identification of skp cullin F-box (SCF)β-TrCP1/2 E3 ligase substrates. Mol Cell Proteomics 14, 1781–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuykendall TN, Houston DW (2009). Vegetally localized Xenopus trim36 regulates cortical rotation and dorsal axis formation. Development 136, 3057–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Falco F, Cainarca S, Andolfi G, Ferrentino R, Berti C, Rodríguez Criado G, Rittinger O, Dennis N, Odent S, Rastogi A, et al. (2003). X-linked opitz syndrome: novel mutations in the MID1 gene and redefinition of the clinical spectrum. Am J Med Genet 120A, 222–228. [DOI] [PubMed] [Google Scholar]
- Dent EW, Barnes AM, Tang F, Kalil K (2004). Netrin-1 and semaphorin 3A promote or inhibit cortical axon branching, respectively, by reorganization of the cytoskeleton. J Neurosci 24, 3002–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CAP (2009). RING domain E3 ubiquitin ligases. Annu Rev Biochem 78, 399–434. [DOI] [PubMed] [Google Scholar]
- Do LD, Gupton SL, Tanji K, Bastien J, Brugière S, Couté Y, Quadrio I, Rogemond V, Fabien N, Desestret V, et al. (2019). TRIM9 and TRIM67 are new targets in paraneoplastic cerebellar degeneration. Cerebellum 18, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazeli A, Dickinson SL, Hermiston ML, Tighe RV, Steen RG, Small CG, Stoeckli ET, Keino-Masu K, Masu M, Rayburn H, et al. (1997). Phenotype of mice lacking functional Deleted in colorectal cancer (Dcc) gene. Nature 386, 796–804. [DOI] [PubMed] [Google Scholar]
- Geetha TS, Michealraj KA, Kabra M, Kaur G, Juyal RC, Thelma BK (2014). Targeted deep resequencing identifies MID2 mutation for X-linked intellectual disability with varied disease severity in a large kindred from India. Hum Mutat 35, 41–44. [DOI] [PubMed] [Google Scholar]
- George AJ, Hoffiz YC, Charles AJ, Zhu Y, Mabb AM (2018). A comprehensive atlas of E3 ubiquitin ligase mutations in neurological disorders. Front Genet 9, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes-Dias P, Nirschl JJ, Abreu N, Tokito MK, Janke C, Magiera MM, Holzbaur ELF (2019). Kinesin-3 responds to local microtubule dynamics to target synaptic cargo delivery to the presynapse. Curr Biol 29, 268–282.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton SL, Gertler FB (2010). Integrin signaling switches the cytoskeletal and exocytic machinery that drives neuritogenesis. Dev Cell 18, 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao JC, Adler CE, Mebane L, Gertler FB, Bargmann CI, Tessier-Lavigne M (2010). The tripartite motif protein MADD-2 functions with the receptor UNC-40 (DCC) in netrin-mediated axon attraction and branching. Dev Cell 18, 950–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harterink M, Vocking K, Pan X, Soriano Jerez EM, Slenders L, Fréal A, Tas RP, van de Wetering WJ, Timmer K, Motshagen J, et al. (2019). Trim46 organizes microtubule fasciculation in the axon initial segment. J Neurosci 39, 4864–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe HS, Lee JY, Pak DTS (2009). Combinatorial morphogenesis of dendritic spines and filopodia by SPAR and α-actinin2. Biochem Biophys Res Commun 384, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad CC, Milstein AD, Ethell IM, Henkemeyer M, Sheng M (2005). GRIP1 controls dendrite morphogenesis by regulating EphB receptor trafficking. Nat Neurosci 8, 906–915. [DOI] [PubMed] [Google Scholar]
- Iconomou M, Saunders DN (2016). Systematic approaches to identify E3 ligase substrates. Biochem J 473, 4083–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Zhao H, Su G, Liu L, Li G, Xu L (2017). Tripartite motif-containing protein 33 (TRIM33) negatively regulates amyloid-β production by promoting proteasome-dependent degradation of BACE1. Int J Clin Exp Pathol 10, 5373–5378. [Google Scholar]
- Kampmeyer C, Karakostova A, Schenstrøm SM, Abildgaard AB, Lauridsen AM, Jourdain I, Hartmann-Petersen R (2017). The exocyst subunit Sec3 is regulated by a protein quality control pathway. J Biol Chem 292, 15240–15253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa T, Ikeda M, Glatt SJ, Tsutsumi A, Kikuyama H, Kawamura Y, Nishida N, Miyagawa T, Hashimoto R, Takeda M, et al. (2013). Genome-wide association study of atypical psychosis. Am J Med Genet Part B Neuropsychiatr Genet 162B, 679–686. [DOI] [PubMed] [Google Scholar]
- Kapur M, Monaghan CE, Ackerman SL (2017). Regulation of mRNA translation in neurons—a matter of life and death. Neuron 96, 616–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasmanis EP, Phan CT, Angelis D, Kesisova IA, Hoogenraad CC, McKenney RJ, Spiliotis ET (2018). Polarity of neuronal membrane traffic requires sorting of kinesin motor cargo during entry into dendrites by a microtubule-associated septin. Dev Cell 46, 204–218.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ (2014). Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci USA 111, 2453–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej PA, Timpe LC, Mitchell KJ, Fried SR, Goodman CS, Jan LY, Jan YN (1996). frazzled encodes a Drosophila member of the DCC immunoglobulin subfamily and is required for CNS and motor axon guidance. Cell 87, 197–204. [DOI] [PubMed] [Google Scholar]
- Koppers M, Cagnetta R, Shigeoka T, Wunderlich LCS, Vallejo-Ramirez P, Lin JQ, Zhao S, Jakobs MAH, Dwivedy A, Minett MS, et al. (2019). Receptor-specific interactome as a hub for rapid cue-induced selective translation in axons. eLife 8, e48718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski AV, Rubinson DA, Dent EW, van Veen JE, Leslie JD, Zhang J, Mebane LM, Philippar U, Pinheiro EM, Burds AA, et al. (2007). Ena/VASP is required for neuritogenesis in the developing cortex. Neuron 56, 441–455. [DOI] [PubMed] [Google Scholar]
- Lebrand C, Dent EW, Strasser GA, Lanier LM, Krause M, Svitkina TM, Borisy GG, Gertler FB (2004). Critical Role of Ena/VASP Proteins for Filopodia Formation in Neurons and in Function Downstream of Netrin-1. Neuron 42, 37–49. [DOI] [PubMed] [Google Scholar]
- Li W, Xia JT, Feng Y (2006). Microtubule stability and MAP1B upregulation control neuritogenesis in CAD cells. Acta Pharmacol Sin 27, 1119–1126. [DOI] [PubMed] [Google Scholar]
- Lin WH, Nebhan CA, Anderson BR, Webb DJ (2010). Vasodilator-stimulated phosphoprotein (VASP) induces actin assembly in dendritic spines to promote their development and potentiate synaptic strength. J Biol Chem 285, 36010–36020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Li X, Shen D, Novick P (2018). Two subunits of the exocyst, Sec3p and Exo70p, can function exclusively on the plasma membrane. Mol Biol Cell 29, 736–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KY, Kuzmin A, Unger SM, Petersen JD, Silverman MA (2011). KIF1A is the primary anterograde motor protein required for the axonal transport of dense-core vesicles in cultured hippocampal neurons. Neurosci Lett 491, 168–173. [DOI] [PubMed] [Google Scholar]
- Ma D, Himes BT, Shea TB, Fischer I (2000). Axonal transport of microtubule-associated protein 1B (MAP1B) in the sciatic nerve of adult rat: distinct transport rates of different isoforms. J Neurosci 20, 2112–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martorella M, Barford K, Winckler B, Deppmann CD (2017). Emergent role of coronin-1a in neuronal signaling. Vitam Horm 104, 113–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick LE, Gupton SL (2020). Mechanistic advances in axon pathfinding. Curr Opin Cell Biol 63, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Boyer NP, Winkle CC, McClain LM, Hanlin CC, Pandey D, Rothenfußer S, Taylor AM, Gupton SL (2015). The E3 ubiquitin ligase TRIM9 is a filopodia off switch required for netrin-dependent axon guidance. Dev Cell 35, 698–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroni G, Diez-Roux G (2005). TRIM/RBCC, a novel class of “single protein RING finger” E3 ubiquitin ligases. BioEssays 27, 1147–1157. [DOI] [PubMed] [Google Scholar]
- Mitchell KJ, Doyle JL, Serafini T, Kennedy TE, Tessier-Lavigne M, Goodman CS, Dickson BJ (1996). Genetic analysis of netrin genes in Drosophila: netrins guide CNS commissural axons and peripheral motor axons. Neuron 17, 203–215. [DOI] [PubMed] [Google Scholar]
- Miyajima N, Maruyama S, Nonomura K, Hatakeyama S (2009). TRIM36 interacts with the kinetochore protein CENP-H and delays cell cycle progression. Biochem Biophys Res Commun 381, 383–387. [DOI] [PubMed] [Google Scholar]
- Morikawa RK, Kanamori T, Yasunaga KI, Emoto K (2011). Different levels of the tripartite motif protein, anomalies in sensory axon patterning (Asap), regulate distinct axonal projections of Drosophila sensory neurons. Proc Natl Acad Sci USA 108, 19389–19394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris AD, Lundquist EA (2011). UNC-6/netrin and its receptors UNC-5 and UNC-40/DCC modulate growth cone protrusion in vivo in C. elegans. Development 138, 4433–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N (1995). The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell 81, 769–780. [DOI] [PubMed] [Google Scholar]
- Pak DTS, Sheng M (2003). Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science 302, 1368–1373. [DOI] [PubMed] [Google Scholar]
- Pak DTS, Yang S, Rudolph-Correia S, Kim E, Sheng M (2001). Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron 31, 289–303. [DOI] [PubMed] [Google Scholar]
- Patel KG, Liu C, Cameron PL, Cameron RS (2001). Myr 8, a novel unconventional myosin expressed during brain development associates with the protein phosphatase catalytic subunits 1α and 1γ1. J Neurosci 21, 7954–7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold S, Sommer B, Kröber A, Nitsch R, Schwegler H, Vogt J, Roskoden T (2016). NT-3 protein levels are enhanced in the hippocampus of PRG1-deficient mice but remain unchanged in PRG1/LPA2 double mutants. Neurosci Lett 612, 145–148. [DOI] [PubMed] [Google Scholar]
- Pfenninger KH (2009). Plasma membrane expansion: a neuron’s Herculean task. Nat Rev Neurosci 10, 251–261. [DOI] [PubMed] [Google Scholar]
- Pleasure IT, Black MM, Keen JH (1993). Valosin-containing protein, VCP, is a ubiquitous clathrin-binding protein. Nature 365, 459–462. [DOI] [PubMed] [Google Scholar]
- Plooster M, Menon S, Winkle CC, Urbina FL, Monkiewicz C, Phend KD, Weinberg RJ, Gupton SL (2017). TRIM9-dependent ubiquitination of DCC constrains kinase signaling, exocytosis, and axon branching. Mol Biol Cell 28, 2374–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaderi NA, Schweiger S, Gaudenz K, Franco B, Rugarli EI, Berger W, Feldman GJ, Volta M, Andolfi G, Gilgenkrantz S, et al. (1997). Opitz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp22. Nat Genet 17, 285–291. [DOI] [PubMed] [Google Scholar]
- Redwine WB, DeSantis ME, Hollyer I, Htet ZM, Tran PT, Swanson SK, Florens L, Washburn MP, Reck-Peterson SL (2017). The human cytoplasmic dynein interactome reveals novel activators of motility. eLife 6, e28257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz D, Vuk M, Kirchner P, Bug M, Schütz S, Bremer S, Lusk C, Baloh RH, Lee H, Glatter T, et al. (2011). Endolysosomal sorting by VCP and UBXD1. Nat Cell Biol 13, 1116–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler MK, Lombino FL, Freitag S, Schweizer M, Hermans-Borgmeyer I, Schwarz JR, Kneussel M, Wagner W (2019). Myosin XVI regulates actin cytoskeleton dynamics in dendritic spines of Purkinje cells and affects presynaptic organization. Front Cell Neurosci 13, 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini T, Colamarino SA, Leonardo ED, Wang H, Beddington R, Skarnes WC, Tessier-Lavigne M (1996). Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system. Cell 87, 1001–1014. [DOI] [PubMed] [Google Scholar]
- Setou M, Seog DH, Tanaka Y, Kanai Y, Takei Y, Kawagishi M, Hirokawa N (2002). Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature 417, 83–87. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short KM, Cox TC (2006). Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J Biol Chem 281, 8970–8980. [DOI] [PubMed] [Google Scholar]
- Sigal YJ, McDermott MI, Morris AJ (2005). Integral membrane lipid phosphatases/phosphotransferases: common structure and diverse functions. Biochem J 387, 281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Bhat VK, Tiwari A, Kodaganur SG, Tontanahal SJ, Sarda A, Malini KV, Kumar A (2017). A homozygous mutation in TRIM36 causes autosomal recessive anencephaly in an Indian family. Hum Mol Genet 26, 1104–1114. [DOI] [PubMed] [Google Scholar]
- Suo D, Park J, Harrington AW, Zweifel LS, Mihalas S, Deppmann CD (2014). Coronin-1 is a neurotrophin endosomal effector that is required for developmental competition for survival. Nat Neurosci 17, 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo X, Park J, Young S, Makita T, Deppmann CD (2015). Coronin-1 and calcium signaling governs sympathetic final target innervation. J Neurosci 35, 3892–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan HL, Queenan BN, Huganir RL (2015). GRIP1 is required for homeostatic regulation of AMPAR trafficking. Proc Natl Acad Sci USA 112, 10026–10031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji K, Kamitani T, Mori F, Kakita A, Takahashi H, Wakabayashi K (2010). TRIM9, a novel brain-specific E3 ubiquitin ligase, is repressed in the brain of Parkinson’s disease and dementia with Lewy bodies. Neurobiol Dis 38, 210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocchini C, Ciosk R (2015). TRIM-NHL proteins in development and disease. Semin Cell Dev Biol 47, 48–52. [DOI] [PubMed] [Google Scholar]
- Tokumitsu H, Hatano N, Tsuchiya M, Yurimoto S, Fujimoto T, Ohara N, Kobayashi R, Sakagami H (2010). Identification and characterization of PRG-1 as a neuronal calmodulin-binding protein. Biochem J 431, 81–91. [DOI] [PubMed] [Google Scholar]
- Tortosa E, Montenegro-Venegas C, Benoist M, Härtel S, González-Billault C, Esteban JA, Avila J (2011). Microtubule-associated protein 1B (MAP1B) is required for dendritic spine development and synaptic maturation. J Biol Chem 286, 40638–40648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyo-Oka K, Shionoya A, Gambello MJ, Cardoso C, Leventer R, Ward HL, Ayala R, Tsai LH, Dobyns W, Ledbetter D, et al. (2003). 14-3-3ε is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat Genet 34, 274–285. [DOI] [PubMed] [Google Scholar]
- Toyo-Oka K, Wachi T, Hunt RF, Baraban SC, Taya S, Ramshaw H, Kaibuchi K, Schwarz QP, Lopez AF, Wynshaw-Boris A (2014). 14-3-3ε and ζ regulate neurogenesis and differentiation of neuronal progenitor cells in the developing brain. J Neurosci 34, 12168–12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twelvetrees AE, Lesept F, Holzbaur ELF, Kittler JT (2019). The adaptor proteins HAP1a and GRIP1 collaborate to activate the kinesin-1 isoform KIF5C. J Cell Sci 132, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymanskyj SR, Scales TME, Gordon-Weeks PR (2012). MAP1B enhances microtubule assembly rates and axon extension rates in developing neurons. Mol Cell Neurosci 49, 110–119. [DOI] [PubMed] [Google Scholar]
- Urbina FL, Gomez SM, Gupton SL (2018). Spatiotemporal organization of exocytosis emerges during neuronal shape change. J Cell Biol 217, 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina FL, Gupton SL (2020). SNARE-mediated exocytosis in neuronal development. Front Mol Neurosci 13, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina FL, Menon S, Goldfarb D, Ben Major M, Brennwald P, Gupton SL (2020). TRIM67 regulates exocytic mode and neuronal morphogenesis via SNAP47. Cell Rep 34, 930404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beuningen SFB, Will L, Harterink M, Chazeau A, van Battum EY, Frias CP, Franker MAMM, Katrukha EA, Stucchi R, Vocking K, et al. (2015). TRIM46 controls neuronal polarity and axon specification by driving the formation of parallel microtubule arrays. Neuron 88, 1208–1226. [DOI] [PubMed] [Google Scholar]
- van Coevorden-Hameete MH, van Beuningen SFB, Perrenoud M, Will LM, Hulsenboom E, Demonet JF, Sabater L, Kros JM, Verschuuren JJGM, Titulaer MJ, et al. (2017). Antibodies to TRIM46 are associated with paraneoplastic neurological syndromes. Ann Clin Transl Neurol 4, 680–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Hatakeyama S (2017). TRIM proteins and diseases. J Biochem 161, 135–144. [DOI] [PubMed] [Google Scholar]
- Winkle CC, McClain LM, Valtschanoff JG, Park CS, Maglione C, Gupton SL (2014). A novel netrin-1-sensitive mechanism promotes local SNARE-mediated exocytosis during axon branching. J Cell Biol 205, 217–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkle CC, Olsen RHJ, Kim H, Moy SS, Song J, Gupton SL (2016). Trim9 deletion alters the morphogenesis of developing and adult-born hippocampal neurons and impairs spatial learning and memory. J Neurosci 36, 4940–4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HHW, Lin JQ, Ströhl F, Roque CG, Cioni JM, Cagnetta R, Turner-Bridger B, Laine RF, Harris WA, Kaminski CF, et al. (2017). RNA docking and local translation regulate site-specific axon remodeling in vivo. Neuron 95, 852–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KM, Du H, Dagnachew M, Massiah MA (2016). Solution structure of the microtubule-targeting COS domain of MID1. FEBS J 283, 3089–3102. [DOI] [PubMed] [Google Scholar]
- Wu B, Guo W (2015). The exocyst at a glance. J Cell Sci 128, 2957–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi H, Okumura F, Takahashi H, Kano T, Kameda H, Uchigashima M, Tanaka S, Watanabe M, Sasaki H, Hatakeyama S (2012). TRIM67 protein negatively regulates Ras activity through degradation of 80K-H and induces neuritogenesis. J Biol Chem 287, 12050–12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K, Tezuka T, Kotani M, Nakazawa T, Hoshina N, Shimoda Y, Kakuta S, Sudo K, Watanabe K, Iwakura Y, et al. (2011). NYAP: a phosphoprotein family that links PI3K to WAVE1 signalling in neurons. EMBO J 30, 4739–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon BC, Zivraj KH, Holt CE (2009). Local translation and mRNA trafficking in axon pathfinding. Results Probl Cell Differ 95, 852–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P, Zhang Y, Mei K, Wang S, Lesigang J, Zhu Y, Dong G, Guo W (2017). Sec3 promotes the initial binary t-SNARE complex assembly and membrane fusion. Nat Commun 8, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung AR, Nishitani AM, Goodrich LV (2015). Phenotypic analysis of mice completely lacking netrin 1. Development 142, 3686–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.