Abstract
Cell biology is immensely complex. To understand how cells work, we try to find patterns and suggest hypotheses to identify underlying mechanisms. However, it is not always easy to create a coherent picture from a huge amount of experimental data on biological systems, where the main players have multiple interactions or act in redundant pathways. In such situations, when a hypothesis does not lead to a conclusion in a direct way, theoretical modeling is a powerful tool because it allows us to formulate hypotheses in a quantitative manner and understand their consequences. A successful model should not only reproduce the basic features of the system but also provide exciting predictions, motivating new experiments. Much is learned when a model based on generally accepted knowledge cannot explain experiments of interest, as this indicates that the original hypothesis needs to be revised. In this Perspective, we discuss these points using our experiences in combining experiments with theory in the field of mitotic spindle mechanics.
The goal of anyone studying biology is to learn how life works, but for many students the choice of biology is reinforced by a desire to escape mathematics, physics, complex equations, and theoretical work. Yet research in biology often needs theoretical analysis. Theoretical modeling is valuable because it allows us to formulate our hypotheses in a rigorous manner and recognize their implications. Theory in cell biology has been the subject of thought-provoking reviews discussing different types of models as well as why and how to do theoretical modeling (Mogilner et al., 2006; Gunawardena, 2014; Möbius and Laan, 2015; Phillips, 2015; Tyson and Novák, 2015). In this essay, we illustrate the lessons that emerge from the interplay of theory and experiments using examples from spindle mechanics, emphasizing how theory is useful also when it cannot explain experiments and how it becomes especially valuable when it predicts unexpected behavior.
The mitotic spindle is a marvelous microtubule-based micromachine that segregates the genome from one cell into two equal parts destined to the future daughter cells (McIntosh, 2016). Spindle microtubules can be divided into three main classes according to their localization and function: kinetochore microtubules that bind the kinetochore, a protein complex at the centromere of each chromosome; overlap microtubules, which extend from the opposite spindle halves and overlap in the middle; and astral microtubules, which grow from the spindle pole toward the cell cortex. Nucleation, dynamics, and forces exerted by spindle microtubules are regulated by hundreds of microtubule-binding and other mitotic proteins, which have multiple mutual interactions. These complex biochemical interactions drive self-organization, a process where order arises from local interactions between initially disordered components, into a molecular machine that can generate large-scale forces to move the chromosomes (Pavin and Tolic´, 2016). Yet, despite the great amount of knowledge about the spindle, this complexity of interactions makes the mechanisms of spindle functioning still largely unclear. Precisely because of the complexity, theoretical modeling is helpful in testing hypotheses and identifying key mechanisms.
LESSON 1: A THEORETICAL MODEL SHOULD PROVIDE PREDICTIONS TO VALIDATE OR REFUTE IT
A theoretical model allows the formulation of hypotheses in a quantitative manner, and its solutions show the consequences of these hypotheses. A model provides predictions for various situations, which can be tested experimentally to support or refute the model. An area where theoretical modeling has been particularly useful for perceiving the effects of different mechanisms on system behavior is the balance of forces on chromosomes in metaphase. This central phase of spindle life is characterized by the neat alignment of kinetochores at the equatorial plane of the spindle (Figure 1A, left). Understanding the forces that establish and maintain this arrangement of kinetochores is key to the understanding of spindle formation as well as chromosome segregation.
FIGURE 1:
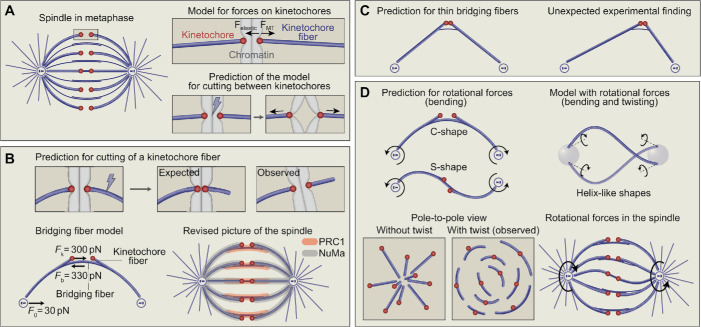
(A) Spindle in metaphase (left). Kinetochores are shown in red, microtubules and centrosomes in blue, and the rectangle marks the region around a pair of sister kinetochores that is enlarged on the right. A model for forces on kinetochores, Felastic and FMT (top right), predicts that upon laser cutting (bolt sign) of the centromere region, sister kinetochores will be pulled apart by kinetochore microtubules (bottom right). (B) The model from A also predicts that upon laser cutting of a kinetochore fiber, the kinetochores should get closer, but experiments showed that kinetochores can keep their distance and the kinetochore fiber stub rotates with its tip moving away from the spindle (top). In the bridging fiber model, tension on kinetochores, Fk, and compression at the pole, F0, are balanced by the compression in the bridging fiber, Fb (bottom left). Also shown is a simplified revised picture of the spindle in which overlap bundles act as bridges between sister kinetochore fibers (bottom right); localization of the cross-linkers PRC1 (orange) and NuMa (gray) is indicated. (C) The bridging fiber model predicts that if the bridging fiber is thinner, sister kinetochores will be closer and kinetochore fibers will be straight, giving the spindle a diamond-like shape (left). Experiments revealed an additional unexpected effect that thinner bridging fibers also lead to misaligned kinetochores (right). (D) The bridging fiber model predicts that if rotational forces (bending, curved arrows) act at the spindle poles in the opposite directions, the structure attains a C-shape, but if the direction of the rotational force is reversed at one pole, the structure curves into an S-shape (top left). A model with rotational forces (bending and twisting) predicts helix-like shapes of microtubule bundles (top right). Experimental test of this prediction by looking at the spindle from pole to pole: bundles without twist extend radially from the pole, whereas in the case with twist the bundles turn around the pole (bottom left). Rotational forces act in the spindle, twisting microtubule bundles into spiral shapes (bottom right).
Based on experiments that probed these forces (Rieder and Salmon, 1994), the first theoretical model assumed antagonistic forces acting on a chromosome: polar ejection forces exerted by microtubules that interact with chromosome arms and push them away from the pole, and poleward pulling forces exerted by kinetochore microtubules onto sister kinetochores, which are mechanically coupled by a spring (Joglekar and Hunt, 2002). Force-dependent kinetics of microtubule attachment to and detachment from the kinetochore is crucial to obtain the typical chromosome fluctuations around the spindle equator in this model. Subsequent models introduced force-dependent microtubule dynamics (Sprague et al., 2003; Gardner et al., 2005), motor proteins (Civelekoglu-Scholey et al., 2006), and more complex mechanisms (Liu et al., 2008; Gay et al., 2012; Civelekoglu-Scholey et al., 2013; Armond et al., 2015; Banigan et al., 2015; Mary et al., 2015; Gergely et al., 2016; Klemm et al., 2018).
A common assumption in these models is that kinetochore microtubules exert pulling forces on the kinetochore toward the spindle pole, whereas the elastic force exerted by the chromatin connecting the two sister kinetochores pulls the kinetochores together (Figure 1A, right). Thus, according to these models, removing the connection between sister kinetochores should result in kinetochores moving apart due to the pulling forces exerted by kinetochore microtubules. This prediction is in agreement with experiments in which the centromere region was ablated by a laser (Skibbens et al., 1995; Khodjakov and Rieder, 1996). Similarly, removing the microtubules should result in the movement of sister kinetochores toward each other, which is also consistent with experimental measurements (Waters et al., 1996). Thus, the central assumption of two antagonistic forces acting on the kinetochore is supported by experiments. This example illustrates how predictions of a theoretical model for different experimental situations can validate the model.
LESSON 2: THEORETICAL MODELS ARE USEFUL ALSO WHEN THEY CANNOT EXPLAIN EXPERIMENTS
Theoretical models sometimes cannot explain certain experimental observations because each model is a simplified picture of a complex system from the real world. Discrepancy between a model and experiments is not necessarily something bad, as the search for an explanation can motivate exciting new research and the development of new concepts.
In the model for force balance on a metaphase chromosome, the assumption of the antagonistic forces acting on the kinetochore implies that if a bundle of kinetochore microtubules, also called the kinetochore fiber, is cut by a laser, the pulling force exerted by this fiber will vanish and the elastic force of the chromatin will pull this kinetochore toward its sister (Figure 1B, top). During this movement, the microtubules will keep the same orientation as before the cut. However, such laser cutting experiments did not lead to the expected outcome. Cutting of the outermost kinetochore fiber about 2 µm away from the kinetochore resulted in sister kinetochores keeping their distance, but the microtubule stub that remained attached to the kinetochore after the cut changed its orientation as it rotated in the outward direction with respect to the spindle (Kajtez et al., 2016).
How can this result be explained? A possible explanation is that sister kinetochores are connected by something solid in addition to their connection via the soft chromatin. This solid connection could be made of microtubules. Indeed, nonkinetochore microtubules that extend along the kinetochore fiber, enter the region between sister kinetochores, and potentially interact with the sister (and other) kinetochore fiber have been observed in electron micrographs of spindles in human and PtK1 cells, plant endosperm, Xenopus egg extracts, and Drosophila S2 cells (Brinkley and Cartwright, 1971; McIntosh and Landis, 1971; Jensen, 1982; McDonald et al., 1992; Mastronarde et al., 1993; Ohi et al., 2003; Nixon et al., 2017; Strunov et al., 2018; O’Toole et al., 2020). In fluorescence microscopy images this microtubule bundle looks like a bridge between sister kinetochore fibers, which is why it is called bridging fiber (Kajtez et al., 2016; Tolic´, 2018). Laser cutting experiments showed that it moves together with the kinetochores and the kinetochore fiber stub after the cut, demonstrating that two kinetochore fibers and the bridging fiber make a self-sustained mechanical unit of the spindle that can segregate chromosomes (Kajtez et al., 2016; Vukusic et al., 2017). Microtubules within the bridging fiber are held together by the cross-linker PRC1 (Kajtez et al., 2016; Polak et al., 2017), which is the case not only in human cell lines but also in PtK2 cells (Suresh et al., 2020) and in mouse oocytes (So et al., 2019). Similarly, bridging fibers in metaphase bind a related central spindle protein, the kinesin-6 family member Subito, in mitotic cells and oocytes in Drosophila (Cesario et al., 2006; Das et al., 2018), and kinesin-14 family members in Caenorhabditis elegans oocytes (Mullen and Wignall, 2017).
The concept of a bridging fiber and its consequences for the force balance in the spindle were explored quantitatively by developing a theoretical model that includes the bridging fiber as a link between sister kinetochore fibers (Kajtez et al., 2016) (Figure 1B, bottom left). The aim of this model was to deduce the forces acting on kinetochores and within the bridging and kinetochore fiber. The shape of this mechanical unit consisting of sister kinetochore fibers and the bridging fiber is determined by these forces and the elastic properties of microtubule bundles. We cannot directly measure the forces within the bundles, but we can measure the shape and use it to determine the forces. This approach suggested that the bridging fiber splits from the kinetochore fiber 1–2 µm away from the kinetochore. The solution of the model also revealed the forces: The pole-proximal part of the kinetochore fiber is under compression of about 30 pN, and the kinetochore-proximal part is under tension of about 300 pN. This compression at the pole and the tension at the kinetochore are balanced by the compression in the central part of the bridging fiber of about 330 pN. Interestingly, the value of tension acting on the kinetochore is of the same order of magnitude as the force measured in the pioneering experiments by Nicklas (1983), where he used a flexible glass microneedle and found that a force of 700 pN is needed to stop the movement of a chromosome in anaphase. In agreement with this classical work and our estimate, a study using force sensors found that kinetochore fibers exert hundreds of pNs on the kinetochore (Ye et al., 2016).
How can the bridging fiber model be tested? If the model is right, then cutting of the kinetochore fiber at different locations will have a different result. If the cut is made far away from the kinetochore, the remaining fragment of the kinetochore fiber will stay linked to the bridging fiber, and thus the sister kinetochores will stay separated. On the contrary, a cut close to the kinetochore will disconnect the kinetochore fiber from the bridging fiber and the kinetochores will get closer. This prediction is unique to the bridging fiber model in comparison with previous models, and testing it is important for model validation as discussed in Lesson 1.
Several labs made systematic laser cutting of the kinetochore fiber at various distances from the kinetochore. We observed in human cells that the distance between sister kinetochores decreases to a larger extent when the cut is closer to the kinetochore (Kajtez et al., 2016; Milas and Tolic´, 2016), as predicted by the bridging fiber model, and a similar trend was found in Drosophila S2 cells (Maiato et al., 2017). A study on PtK2 cells went a step further by showing that this relationship holds not only for the outermost kinetochore fibers but also for the ones in the inner part of the spindle in PtK2 cells (Elting et al., 2017). Only the kinetochore pair whose fiber was cut showed relaxation, without effect on the neighboring kinetochores, supporting the picture in which two kinetochore fibers together with their bridging fiber are a self-sustained mechanical unit of the spindle. Moreover, depletion of the microtubule cross-linker PRC1 or inactivation of Eg5/kinesin-5 did not affect inter kinetochore relaxation after laser cutting, whereas depletion of the cross-linker NuMa led to a larger relaxation. Thus, a picture is emerging in which PRC1 cross-links the antiparallel microtubules within the bridging fiber together, while NuMa cross-links the parallel overlaps of kinetochore and bridging microtubules that extend from the same half of the spindle (Figure 1B, bottom right).
The story of the bridging fiber illustrates how a finding motivated by theoretical models that could not explain certain experiments initiated exciting discoveries. In general, this example shows how theory motivates new experiments, which can lead to new concepts and open up new ways of thinking about the system.
LESSON 3: EXPERIMENTS CAN BRING NEW FINDINGS THAT GO BEYOND THE PREDICTIONS OF THE MODEL DUE TO COMPLEXITY OF BIOLOGICAL SYSTEMS
Experiments that start as a test of a theoretical model sometimes reveal unexpected interesting effects that are outside of the scope of the model. In other words, perturbations designed to test the model can also perturb other important factors that are not described by the model, suggesting a link between the described and new factors. The model then needs to be extended by including this link.
One of the key predictions of the bridging fiber model is that weakening or removal of the bridging fibers will result in a decreased distance between sister kinetochores. Moreover, removal of the bridging fibers should lead to change in spindle shape from a round to a diamond-like outline due to straightening of kinetochore fibers (Figure 1C, left). To test these predictions, we designed an optogenetic approach, in which we remove the microtubule cross-linker PRC1 from the spindle to the cell membrane during metaphase (Jagrić et al., 2019). Acute PRC1 removal from an otherwise normally formed spindle resulted in substantially thinner bridging fibers. This was accompanied by sister kinetochores getting closer to each other and the spindle becoming more diamond-shaped, as the model predicted. In another approach, thinner or even absent bridging fibers were obtained by augmin depletion (Manenica et al., 2020), though this effect is not specific to bridging fibers as kinetochore fibers also become thinner (Zhu et al., 2008). In augmin-depleted cells, the kinetochores without a bridging fiber have a smaller interkinetochore distance and more straight shape of their kinetochore fibers when compared with the ones that have a bridging fiber (Manenica et al., 2020), providing further support for the model.
Remarkably, weakening of bridging fibers by optogenetic removal of PRC1 resulted in an unexpected finding that kinetochores are not well aligned at the spindle equator (Jagrić et al., 2019) (Figure 1C, right). The bridging fiber model does not make predictions related to kinetochore alignment because the model is static, whereas kinetochore alignment relies on microtubule dynamics. This example illustrates how experiments bring new findings that go beyond the predictions of the model because the biological system is much more complex than the model. To explore the role of the bridging fiber in kinetochore alignment, new phenomena should be included in the model, such as dynamics of kinetochore microtubules and sliding of bridging microtubules.
LESSON 4: THEORETICAL MODELS CAN LEAD TO NEW HYPOTHESES, DESIGN OF NEW EXPERIMENTS, AND DISCOVERY OF NEW BIOLOGY
Theoretical models can be used not only to explain experiments but also to lead the experiments based on new questions and hypotheses that are unique consequences of the model and would not be apparent without the model. Theories setting the stage for experiments are common in physics, but not so much in cell biology. In the spindle field, an exciting example is how a theoretical model led to the finding of spindle chirality.
We noticed that the bridging fiber model predicts dramatic changes in the shapes of microtubule bundles when the direction and type of force is changed. If rotational forces act at the spindle pole in the opposite direction, the bundle attains a C-shape. But if the direction of the rotational force is reversed at one pole, shapes resembling the letter “S” appear (Tolic´ et al., 2019) (Figure 1D, top left). This is the case for individual bundles. Can such C- and S-shaped bundles coexist within the same spindle? This is unlikely because the rotational forces at the spindle pole would need to act in opposite directions on different bundles.
We looked at real spindles in human cell lines and indeed observed both C- and S-shaped bundles within the same spindle. Even the usual confocal microscopy images of spindles show that C-shaped bundles are found in the outer region of the spindle, whereas the inner part contains bundles of various complex shapes including S-like profiles; see, for example, spindle images in Dick and Gerlich, 2013; Kajtez et al., 2016. Superresolution images obtained by using stimulated emission depletion (STED) microscopy clearly display C-shapes in the outer part and S-shapes together with more complex shapes in the inner part and show that the individual bundles extend almost from pole to pole (Novak et al., 2018). How can this be explained? One possibility is that opposing rotational forces act at different bundles at the same spindle pole, which is unlikely. Yet there is an alternative and more likely hypothesis: rotational forces that twist the bundles may exist in the spindle.
To explore this new hypothesis, we developed a physical model that incorporates both linear and rotational forces, including bending and twisting forces (Novak et al., 2018) (Figure 1D, top right). This model gives an interesting prediction that microtubule bundles should have a helix-like shape that extends in three dimensions, rather than planar C- and S-shapes. We tested this unusual prediction by looking at the spindles in a somewhat unusual way, from pole to pole, in order to be able to trace the three-dimensional contour of individual microtubule bundles (Figure 1D, bottom left). If the bundles in the spindle are arranged as meridians on Earth, then images of cross-sections of a vertically oriented spindle should show the bundles starting in a central spot at one pole, moving radially outward until the equatorial plane and then back toward the central spot at the other pole. Curiously, we found that the bundles do not extend in this manner but rotate around the spindle axis following a shape of a left-handed helix. The left-handedness of the bundles makes the whole spindle a chiral structure.
The discovery of spindle chirality is exciting because it shifts the view of spindle forces, which has traditionally been based only on linear forces (pushing and pulling), toward rotational forces (Figure 1D, bottom right). These rotational forces are likely caused by motor proteins such as Eg5/kinesin-5 (Novak et al., 2018), which not only slide but also rotate microtubules (Yajima et al., 2008). Yet how the rotations impact spindle chirality is not known. Moreover, the physiological significance of spindle chirality is still unclear. Although chirality may be merely a side effect of the action of torque-generating motors, the twisted shapes of microtubule bundles may contribute to spindle function, for example, by promoting physical separation of adjacent bundles or by allowing changes of spindle shape as a passive mechanical response to external forces. Thus, the finding of spindle chirality opens an exciting new area of research on the mechanisms and the biological roles of rotational forces in the spindle.
This last example shows how a theoretical model can motivate new scientific questions that emerge from the model, which would not be an obvious subject to investigate without the unique predictions of the model. These questions can guide new experimental approaches and lead to discovery of intriguing phenomena in cell biology.
Acknowledgments
Research in the Tolic´ and Pavin groups is supported by the European Research Council (ERC Synergy Grant, GA Number 855158, granted to I.M.T. and N.P.), the Croatian Science Foundation (HRZZ, projects PZS-2019-02-7653 granted to I.M.T. and IP-2019-04-5967 granted to N.P.), the QuantiXLie Center of Excellence, a project cofinanced by the Croatian Government and European Union through the European Regional Development Fund—the Competitiveness and Cohesion Operational Programme (Grant KK.01.1.1.01.0004). I.M.T. also acknowledges earlier support from the ERC (Consolidator Grant, GA Number 647077). We thank Ivana Šaric´ for the drawings and all group members for stimulating discussions. The ideas discussed in this paper were presented by I.M.T. at the ASCB|EMBO Meeting 2019 in Symposium 1: “Beyond Figure 7: Integrating modeling and experiment in cell biology.”
Abbreviation used:
- STED
stimulated emission depletion.
Footnotes
REFERENCES
- Armond JW, Harry EF, Mcainsh AD, Burroughs NJ (2015). Inferring the forces controlling metaphase kinetochore oscillations by reverse engineering system dynamics. PLoS Comput Biol 11, e1004607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banigan EJ, Chiou KK, Ballister ER, Mayo AM, Lampson MA, Liu AJ (2015). Minimal model for collective kinetochore-microtubule dynamics. Proc Natl Acad Sci USA 112, 12699–12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley BR, Cartwright J Jr (1971). Ultrastructural analysis of mitotic spindle elongation in mammalian cells in vitro. Direct microtubule counts. J Cell Biol 50, 416–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesario JM, Jang JK, Redding B, Shah N, Rahman T, McKim KS (2006). Kinesin 6 family member Subito participates in mitotic spindle assembly and interacts with mitotic regulators. J Cell Sci 119, 4770–4780. [DOI] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G, He B, Shen M, Wan X, Roscioli E, Bowden B, Cimini D (2013). Dynamic bonds and polar ejection force distribution explain kinetochore oscillations in PtK1 cells. J Cell Biol 201, 577–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G, Sharp DJ, Mogilner A, Scholey JM (2006). Model of chromosome motility in drosophila embryos: adaptation of a general mechanism for rapid mitosis. Biophys J 90, 3966–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Cesario J, Hinman AM, Jang JK, McKim KS (2018). Kinesin 6 regulation in drosophila female meiosis by the non-conserved N- and C- terminal domains. G3 (Bethesda) 8, 1555–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick AE, Gerlich DW (2013). Kinetic framework of spindle assembly checkpoint signalling. Nat Cell Biol 15, 1370–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elting MW, Prakash M, Udy DB, Dumont S (2017). Mapping load-bearing in the mammalian spindle reveals local kinetochore fiber anchorage that provides mechanical isolation and redundancy. Curr Biol 27, 2112–2112.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Pearson CG, Sprague BL, Zarzar TR, Bloom K, Salmon ED, Odde DJ (2005). Tension-dependent regulation of microtubule dynamics at kinetochores can explain metaphase congression in yeast. Mol Biol Cell 16, 3764–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay G, Courtheoux T, Reyes C, Tournier S, Gachet Y (2012). A stochastic model of kinetochore-microtubule attachment accurately describes fission yeast chromosome segregation. J Cell Biol 196, 757–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely ZR, Crapo A, Hough LE, McIntosh JR, Betterton MD (2016). Kinesin-8 effects on mitotic microtubule dynamics contribute to spindle function in fission yeast. Mol Biol Cell 27, 3490–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena J (2014). Models in biology: “accurate descriptions of our pathetic thinking.” BMC Biol 12, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagric´ M, Risteski P, Martincˇic´ J, Milas A, Tolic´ IM (2019). Optogenetic control of PRC1 reveals that bridging fibers promote chromosome alignment by overlap length-dependent forces. bioRxiv, 10.1101/865394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen CG (1982). Dynamics of spindle microtubule organization: kinetochore fiber microtubules of plant endosperm. J. Cell Biol 92, 540–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Hunt AJ (2002). A simple, mechanistic model for directional instability during mitotic chromosome movements. Biophys J 83, 42–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajtez J, Solomatina A, Novak M, Polak B, Vukusic K, Rudiger J, Cojoc G, Milas A, Sumanovac Sestak I, Risteski P, et al. (2016). Overlap microtubules link sister k-fibres and balance the forces on bi-oriented kinetochores. Nat Commun 7, 10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL (1996). Kinetochores moving away from their associated pole do not exert a significant pushing force on the chromosome. J Cell Biol 135, 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm AH, Bosilj A, Gluncic M, Pavin N, Tolic´ IM (2018). Metaphase kinetochore movements are regulated by kinesin-8 motors and microtubule dynamic instability. Mol Biol Cell 29, 1332–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Desai A, Onuchic JN, Hwa T (2008). An integrated mechanobiochemical feedback mechanism describes chromosome motility from prometaphase to anaphase in mitosis. Proc Natl Acad Sci USA 105, 13752–13757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Gomes A, Sousa F, Barisic M (2017). Mechanisms of chromosome congression during mitosis. Biology 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manenica M, Koprivec I, Štimac V, Simunic´ J, Tolic´ IM (2020). Augmin regulates kinetochore tension and spatial arrangement of spindle microtubules by nucleating bridging fibers. bioRxiv, 10.1101/2020.09.10.291740. [DOI] [Google Scholar]
- Mary H, Fouchard J, Gay G, Reyes C, Gauthier T, Gruget C, Pecreaux J, Tournier S, Gachet Y (2015). Fission yeast kinesin-8 controls chromosome congression independently of oscillations. J Cell Sci 128, 3720–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN, McDonald KL, Ding R, McIntosh JR (1993). Interpolar spindle microtubules in PTK cells. J Cell Biol 123, 1475– 1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KL, O’Toole ET, Mastronarde DN, McIntosh JR (1992). Kinetochore microtubules in PTK cells. J Cell Biol 118, 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR (2016). Mitosis. Cold Spring Harb Perspect Biol 8, a023218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Landis SC (1971). The distribution of spindle microtubules during mitosis in cultured human cells. J Cell Biol 49, 468–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milas A, Tolic´ IM (2016). Relaxation of interkinetochore tension after severing of a k-fiber depends on the length of the k-fiber stub. Matters 201603000025. 10.19185/matters.201603000025. [DOI] [Google Scholar]
- Möbius W, Laan L (2015). Physical and mathematical modeling in experimental papers. Cell 163, 1577– 1583. [DOI] [PubMed] [Google Scholar]
- Mogilner A, Wollman R, Marshall WF (2006). Quantitative modeling in cell biology: what is it good for? Dev Cell 11, 279–287. [DOI] [PubMed] [Google Scholar]
- Mullen TJ, Wignall SM (2017). Interplay between microtubule bundling and sorting factors ensures acentriolar spindle stability during C. elegans oocyte meiosis. PLoS Genet 13, e1006986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB (1983). Measurements of the force produced by the mitotic spindle in anaphase. J Cell Biol 97, 542–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon FM, Honnor TR, Clarke NI, Starling GP, Beckett AJ, Johansen AM, Brettschneider JA, Prior IA, Royle SJ (2017). Microtubule organization within mitotic spindles revealed by serial block face scanning electron microscopy and image analysis. J Cell Sci 130, 1845–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak M, Polak B, Simunic´ J, Boban Z, Kuzmic´ B, Thomae AW, Tolic´ IM, Pavin N (2018). The mitotic spindle is chiral due to torques within microtubule bundles. Nat Commun 9, 3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R, Coughlin ML, Lane WS, Mitchison TJ (2003). An inner centromere protein that stimulates the microtubule depolymerizing activity of a KinI kinesin. Dev Cell 5, 309– 321. [DOI] [PubMed] [Google Scholar]
- O’Toole E, Morphew M, McIntosh JR (2020). Electron tomography reveals aspects of spindle structure important for mechanical stability at metaphase. Mol Biol Cell 31, 184– 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavin N, Tolic´ IM (2016). Self-organization and forces in the mitotic spindle. Annu Rev Biophys 45, 279–298. [DOI] [PubMed] [Google Scholar]
- Phillips R (2015). Theory in biology: figure 1 or figure 7? Trends Cell Biol 25, 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak B, Risteski P, Lesjak S, Tolic´ IM (2017). PRC1-labeled microtubule bundles and kinetochore pairs show one-to-one association in metaphase. EMBO Rep 18, 217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED (1994). Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J Cell Biol 124, 223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Rieder CL, Salmon ED (1995). Kinetochore motility after severing between sister centromeres using laser microsurgery: evidence that kinetochore directional instability and position is regulated by tension. J Cell Sci 108(Pt 7), 2537–2548. [DOI] [PubMed] [Google Scholar]
- So C, Seres KB, Steyer AM, Mönnich E, Clift D, Pejkovska A, Möbius W, Schuh M (2019). A liquid-like spindle domain promotes acentrosomal spindle assembly in mammalian oocytes. Science 364, eaat9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague BL, Pearson CG, Maddox PS, Bloom KS, Salmon ED, Odde DJ (2003). Mechanisms of microtubule-based kinetochore positioning in the yeast metaphase spindle. Biophys J 84, 3529–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunov A, Boldyreva LV, Andreyeva EN, Pavlova GA, Popova JV, Razuvaeva AV, Anders AF, Renda F, Pindyurin AV, Gatti M, Kiseleva E (2018). Ultrastructural analysis of mitotic Drosophila S2 cells identifies distinctive microtubule and intracellular membrane behaviors. BMC Biol 16, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh P, Long AF, Dumont S (2020). Microneedle manipulation of the mammalian spindle reveals specialized, short-lived reinforcement near chromosomes. eLife 9, 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolic´ IM (2018). Mitotic spindle: kinetochore fibers hold on tight to interpolar bundles. Eur Biophys J 47, 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolic´ IM, Novak M, Pavin N (2019). Helical twist and rotational forces in the mitotic spindle. Biomolecules 9, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson JJ, Novák B (2015). Models in biology: lessons from modeling regulation of the eukaryotic cell cycle. BMC Biol 13, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukusic K, Bud¯a R, Bosilj A, Milas A, Pavin N, Tolic´ IM (2017). Microtubule sliding within the bridging fiber pushes kinetochore fibers apart to segregate chromosomes. Dev Cell 43, 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Skibbens RV, Salmon ED (1996). Oscillating mitotic newt lung cell kinetochores are, on average, under tension and rarely push. J Cell Sci 109, 2823–2831. [DOI] [PubMed] [Google Scholar]
- Yajima J, Mizutani K, Nishizaka T (2008). A torque component present in mitotic kinesin Eg5 revealed by three-dimensional tracking. Nat Struct Mol Biol 15, 1119–1121. [DOI] [PubMed] [Google Scholar]
- Ye AA, Cane S, Maresca TJ (2016). Chromosome biorientation produces hundreds of piconewtons at a metazoan kinetochore. Nat Commun 7, 13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Coppinger JA, Jang C-Y, Yates I, John R, Fang G (2008). FAM29A promotes microtubule amplification via recruitment of the NEDD1–γ-tubulin complex to the mitotic spindle. J Cell Biol 183, 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]