Abstract
Pannexin 1 (PANX1) is a glycoprotein that forms large pore channels capable of passing ions and metabolites such as ATP for cellular communication. PANX1 has been implicated in many diseases including breast cancer and melanoma, where inhibition or deletion of PANX1 reduced the tumorigenic and metastatic properties of the cancer cells. We interrogated the effect of single amino acid changes in various PANX1 domains using naturally occurring variants reported in cancer patient tumors. We found that a previously reported variant (Q5H) is present in cancer cells, but was not different from the wild type (Q5) in glycosylation, trafficking, or channel function and did not affect cellular properties. We discovered that the Q5H variant is in fact the highly conserved ancestral allele of PANX1 with 89% of humans carrying at least one Q5H allele. Another mutated form Y150F, found in a melanoma patient tumor, prevented phosphorylation at Y150 as well as complex N-glycosylation while increasing intracellular localization. Sarcoma (SRC) is the predicted kinase to phosphorylate the Y150 residue, and its phosphorylation is not likely to be constitutive, but rather dynamically regulated. The Y150 phosphorylation site is the first one reported to play a role in regulating posttranslational modifications and trafficking of PANX1, with potential consequences on its large-pore channel structure and function in melanoma cells.
INTRODUCTION
Since its discovery in 2000 (Panchin et al., 2000), Pannexin 1 (PANX1 in human, Panx1 in rodents), a channel-forming glycoprotein that mediates the release of signaling molecules below 1 kDa in size, has been emerging as a key player in various physiological and pathological contexts ranging from blood pressure regulation (Locovei et al., 2006; Billaud et al., 2015), skin cell differentiation (Celetti et al., 2010; Penuela et al., 2014), facilitating neuronal cell death in ischemic conditions (Thompson et al., 2006; Bargiotas et al., 2012), apoptosis, and inflammation (Pelegrin and Surprenant, 2009; Chekeni et al., 2010; Qu et al., 2011) to regulating tumorigenic properties of cancers like melanoma (Penuela et al., 2012; Freeman et al., 2019). Depending on the cellular context and localization of the channel, PANX1 presents varying functions. Most studies report PANX1 to be localized at the cell surface where it is best characterized to function as an ATP release channel (Bao et al., 2004) while others report PANX1 to be found intracellularly within the endoplasmic reticulum (ER) acting as an ER calcium-leak channel (Vanden Abeele et al., 2006).
PANX1 is a tetra-spanning protein with intracellular N- and C-termini, with conserved cysteine residues within both extracellular loops (Baranova et al., 2004), and several domains that regulate PANX1 trafficking and channel function have been identified. The intracellular loop and C-terminus have been demonstrated to undergo both tyrosine and serine phosphorylation which has been linked to overall channel function. Disruption of the tyrosine phosphorylation sites, Y198 or Y308 (numbers differ between mouse Panx1 and human PANX1), either by mutagenesis or peptide inhibition interferes with Panx1 interactions with N-methyl-D-aspartate receptors (NMDARs) and α1-adrenergic receptors and affects channel function (Lohman et al., 2015; Poornima et al., 2015; Weilinger et al., 2016). The first extracellular loop regulates ATP-induced cholesterol-dependent internalization of the channel through its ATP-sensitive W74 residue (Boyce et al., 2015), whereas the second extracellular loop undergoes N-linked glycosylation on N254 (in mouse Panx1, N255 in human PANX1) which regulates cell-surface trafficking and intermixing with other pannexin members (Boassa et al., 2007; Penuela et al., 2009). Due to the differential N-glycosylation processes that PANX1 undergoes, the protein is present as three distinct species: the nonglycosylated Gly0 species, the high-mannose ER-resident Gly1 species, and the complex glycosylated Gly2 species which form in the Golgi apparatus (Boassa et al., 2007; Penuela et al., 2007). While all three species are capable of trafficking to the surface, Gly2 species tend to be more prevalent at the cell membrane (Penuela et al., 2009). PANX1 also undergoes S-nitrosylation at C40 of the N-terminus and C346 of the C-terminus and inhibits channel function (Lohman et al., 2012). Finally, the channel has been demonstrated to be regulated by the C-terminus in a “ball-and-chain” manner and proteolytic caspase cleavage of the C-terminus results in a constitutively active channel (Chekeni et al., 2010; Sandilos et al., 2012).
Only a few recent studies have analyzed the effects of common and rare naturally occurring PANX1 genetic variants. The first report associating a PANX1 mutation to a physiological disorder was a germline loss-of-function mutant, R217H that was reported in a young female with intellectual disability, short stature, sensorineural hearing loss, primary ovarian failure, and kyphosis (Shao et al., 2016). In another report, a somatic nonsense mutation, Q89*, was found to be enriched in two metastatic breast cancer cell lines. The truncated Q89* protein was found to enhance PANX1-mediated ATP release only when coexpressed with full-length PANX1 and was shown to enhance the metastatic efficiency of breast cancer cells by reducing deformation-induced purinergic receptor (P2Y)-mediated apoptosis in areas of microvasculature (Furlow et al., 2015). A recent report revealed PANX1 germline mutations that caused familial or sporadic female infertility by inducing oocyte cell death. They uncovered four germline mutations (K346E, C347S, Q392*, and p.21_23delTEP) that produced gain-of-function, hypo-glycosylated mutants consisting of predominantly Gly0 and Gly1 species (Sang et al., 2019). Finally, one variant that has been commonly reported is a missense variant, Q5H, which was shown to be associated with collagen-induced platelet reactivity in healthy subjects and displayed enhanced ATP release when ectopically expressed (Molica et al., 2015). The same group later showed that the variant was not associated with platelet reactivity in a larger cohort of cardiovascular patients under treatment (Stierlin et al., 2017). In a separate genome-wide association study (GWAS) identifying common variants associated with autism susceptibility risk, they found PANX1 to be implicated in cis using multiple single-nucleotide polymorphisms (SNPs) that included Q5H (Davis et al., 2012). Another GWAS study investigating associations of common pannexin variants with schizophrenia found no associations between Q5H and schizophrenia (Gawlik et al., 2016).
Analyses of these and other naturally occurring germline variants may provide further information about the protein domains they reside in and how they may affect functional and biochemical properties of PANX1. In this study, we interrogated three motifs of the PANX1 polypeptide using single amino acid changes and evaluated their role in PANX1 function. We introduced seven PANX1 naturally occurring variants reported in tumors of melanoma patients and human melanoma cells lines, ectopically expressing them into normal and cancer cell lines. These variants included Q5H, Y150F, G168E, T176I, H190Y, S239L, and Q264* that are in the N-terminus, intracellular loop, and second extracellular loop of the PANX1 polypeptide.
By probing these important motifs, we found that Q5H is equal to Q5 in channel function, glycosylation, and localization in human breast cancer cells. A higher allele prevalence of Q5H was demonstrated in global cohorts and has emerged to be the ancestral allele at this locus. Proteomic analyses also uncovered several other sites of posttranslational modifications (PTMs) in the PANX1 protein. We have uncovered Y150 as a new tyrosine phosphorylation site that when mutated can alter phosphorylation, N-glycosylation, and trafficking of the PANX1 channel protein.
RESULTS
Y150F alters normal PANX1 protein banding pattern indicative of glycosylation states
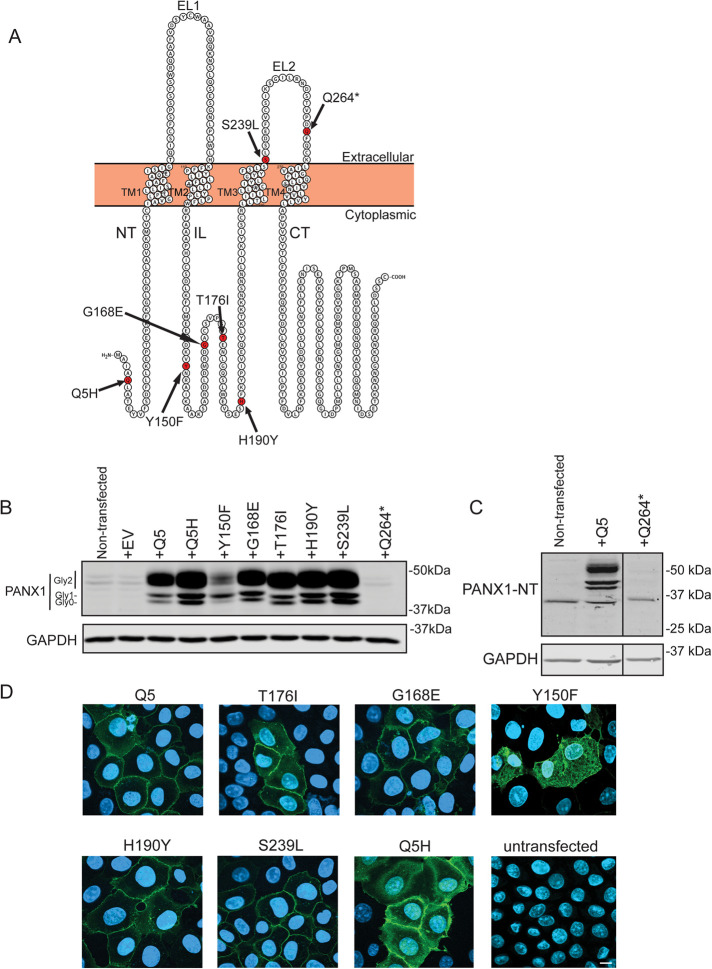
Seven PANX1 genetic variants identified in tumors of melanoma patients and several established cancer cell lines (Figure 1A; Supplemental Table S1), in addition to wild type (denoted as Q5 or WT hence forth), were assessed initially for their effects on PANX1 glycosylation and localization to investigate any changes that may result in altered PANX1 function. Differential N-glycosylation of PANX1 can occur and can be preliminarily inferred by observing the protein banding pattern of PANX1 consisting of Gly0, Gly1, and Gly2 species (Figure 1B) (Penuela et al., 2007).
FIGURE 1:
Impact of PANX1 mutations on protein banding and cell-surface localization in NRK cells. (A) Schematic of PANX1 polypeptide demonstrating the location of PANX1 variants Q5H, Y150F, G168E, T176I, H190Y, S239L, and Q264*. The schematic was generated using Protter (Omasits et al., 2014). NT, amino-terminus; TM, transmembrane; EL, extracellular loop; CT, carboxyl-terminus; IL, intracellular loop. (B) NRK cells were transfected to transiently overexpress selected PANX1 variants and PANX1 banding pattern was assessed 72 h posttransfection and compared with WT PANX1 (Q5). Immunoblotting with anti-PANX1-CT antibody revealed Y150F disrupts the normal banding pattern of PANX1, resulting in primarily Gly1 with minimal Gly0 and Gly2 species. EV, empty vector. (C) PANX1 protein was not detected in NRK cells transfected with Q264*-encoding vectors using two different antibodies against the C-Terminus or N-Terminus. The molecular weight of Q264* is expected to be approximately around 29 kDa. GAPDH was used as a loading control. Protein sizes are noted in kDa. (D) Immunofluorescence analyses revealed that most variants appeared to localize to the cell surface on ectopic expression in NRK cells, except for Y150F that had an increased intracellular profile. Bar: 10 µm.
Due to their relatively low Panx1 endogenous expression (Sanchez-Pupo et al., 2018) and large cytoplasm to visualize PANX1 localization, normal rat kidney (NRK) cells were used to transiently overexpress the selected PANX1 variants. Most of the variants, except for Y150F and Q264*, produced PANX1 banding patterns on immunoblots similar to Q5 (WT) consisting of Gly0, Gly1, and Gly2 (Figure 1B). In contrast, Y150F had lower levels of PANX1 protein and produced predominantly Gly1 species with reduced Gly2 and Gly0 species. The Q264* variant, which produces a truncated mutant lacking the last transmembrane domain (TM4) and the carboxyl-terminus (and thus required detection with an antibody targeting the amino-terminus), was not detected at the protein level (Figure 1C). We also expressed all the mutants in Hs575T cancer cells with endogenous PANX1 and observed similar banding patterns in Western blots with three different PANX1 antibodies (Supplemental Figure S1). Immunofluorescence analyses revealed that most PANX1 variants localized to the cell surface in NRK cells, with the exception of Y150F that presented increased intracellular localization (Figure 1D). As a result of Y150F altering the normal PANX1 banding pattern and localization, and Q5H being previously reported to enhance PANX1-mediated ATP release (Molica et al., 2015), we chose these two variants for further investigation.
Stable overexpression of Q5H does not affect cellular localization, dye uptake, cell growth, or migration relative to Q5 controls
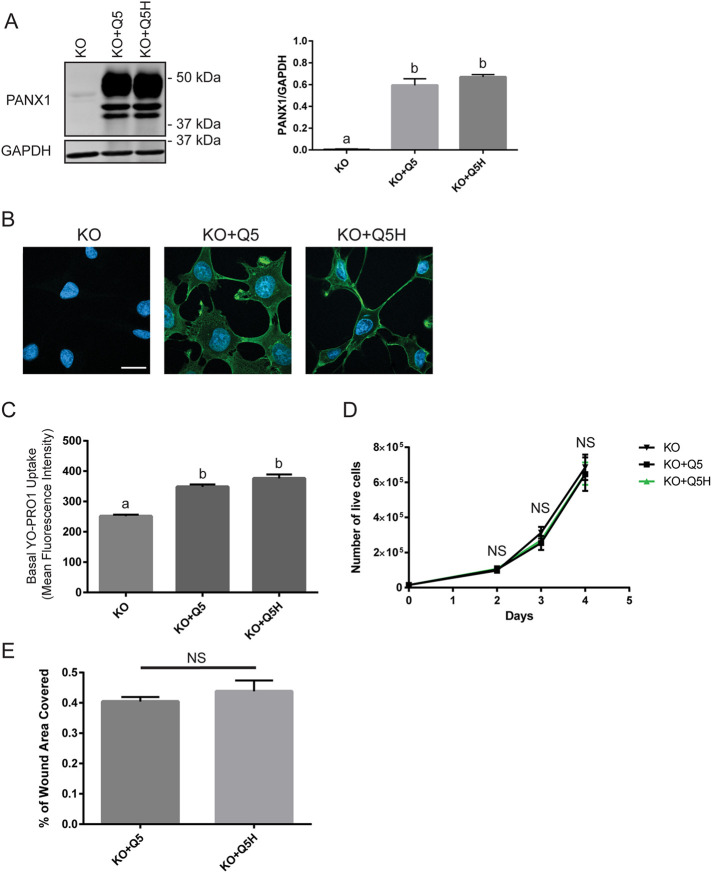
To assess if Q5H differs from Q5 regarding channel activity (as previously reported by Molica et al., 2015) and if it affects cancer cell properties when stably expressed, we generated stable clones of Hs578T(KO) overexpressing Q5 or Q5H at similar protein expression levels (Figure 2A). Immunofluorescence analyses demonstrate that Q5 and Q5H localize at the cell surface when stably expressed (Figure 2B). Channel activity, assessed with basal dye uptake of YOPRO1, did not differ significantly between Hs578T(KO) stably expressing Q5 or Q5H but both were significantly higher than nontransfected control (Figure 2C).
FIGURE 2:
Stable overexpression of Q5H in Hs578T(KO) cells does not differ in dye uptake, migration, or cell growth from Q5 controls. (A) Stable clones of Hs578T(KO) overexpressing Q5 or Q5H at similar protein levels were generated. GAPDH used as loading control. One-way ANOVA (letters denote significantly different groups). (B) Immunofluorescence analyses (PANX1, green) demonstrate that Q5 and Q5H both localize at the cell surface in stable clones. Nuclei are in blue. Scale bar: 20 µm. (C) Q5H did not differ significantly from Q5 in basal uptake of YO-PRO1 dye but both were significantly higher than nontransfected controls (KO). One-way ANOVA (letters depict significant differences). (D) Cell growth assay was performed by assessing the number of viable cells that excluded trypan-blue dye. Cell counts did not differ significantly between Hs578T(KO) stably overexpressing Q5 and Q5H and nontransfected controls. Two-way ANOVA (NS, not significant). (E) Cells were grown to confluence and a scratch was made on which the area cells migrated into after 15 h was recorded. Migration did not differ significantly between Hs578T(KO) stably overexpressing Q5 and Q5H. N = 3, paired t test (NS, not significant).
Furthermore, we characterized if the expression of Q5H affects migration and cell growth of breast cancer cells and found that they did not differ significantly between Hs578T(KO) stably overexpressing Q5 or Q5H (Figure 2, D and E).
Q5H has greater allele and homozygosity frequencies than Q5 in global cohorts
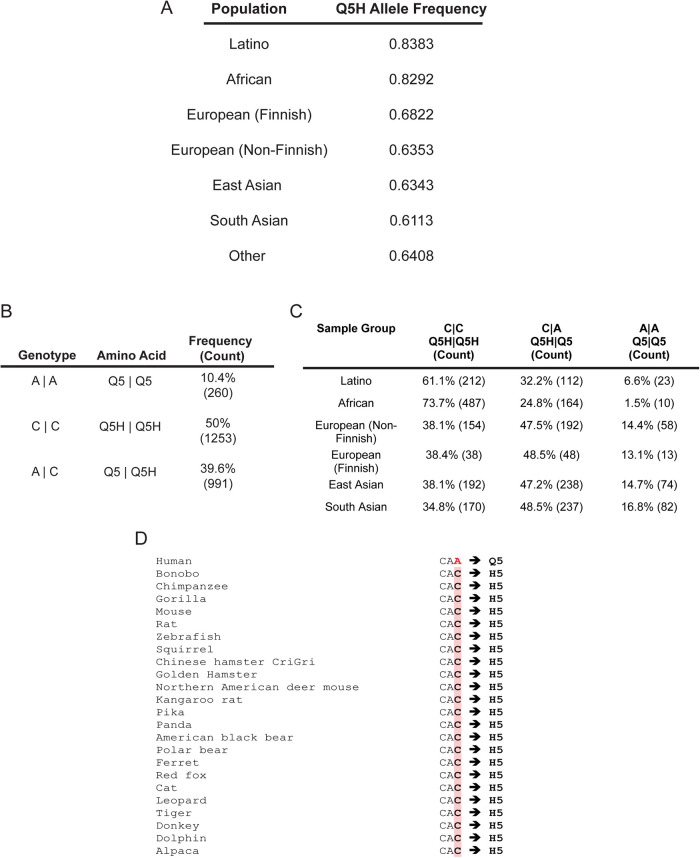
To assess the allele frequencies of Q5H and Y150F in global human cohorts, we obtained data from the ExAC Browser Exome Aggregation Consortium (Lek et al., 2016), an assembly of data from a variety of genome-sequencing endeavors that include 60,706 unrelated individuals sequenced for disease-specific and population-genetic studies. Within the ExAC database, the Y150F variant was not detected, but a Y150H missense variant was, albeit at a very low allele frequency of 8.24e-06 (Lek et al., 2016) corresponding to a single occurrence of this allele in a single heterozygous individual. In contrast, Q5H presented a higher allele frequency than Q5 in global cohorts with the highest allele frequency of 83.83% in Latino populations followed by African populations with an average of 82.92% (Figure 3A). Genotype frequencies of 1000 Genomes Project (phase 3) populations, a genome-sequencing initiative that assembled the genomes of 2504 individuals in 26 different populations across the world, were also assessed and revealed that approximately 89.6% of individuals possessed at least one copy of the Q5H allele, demonstrating its high prevalence in human populations (Figure 3, B and C).
FIGURE 3:
Higher Q5H allele frequency than Q5 in global cohorts. (A) Allele frequency of Q5H found in diverse populations (based on ancestry). Data were extracted from the ExAC Browser Beta. (B) The genotype frequency of individuals sequenced in the 1000 Genomes Project (phase 3) demonstrates that most individuals (∼89%) possess at least one allele copy of Q5H. (C) Genotype frequencies of Q5 and Q5H in different populations from the 1000 Genomes Project (phase 3) shows that the prevalence of each allele varies in different ethnic groups. (D) Ensembl phylogenetic context alignment revealed Q5H is a highly conserved site amongst vertebrate species with humans being the only species with Q5 (according to RefSeq NCBI).
Q5H is highly conserved among vertebrate species and is the ancestral allele of PANX1
The high prevalence of the Q5H allele globally indicated that Q5 could be a derived allele, one that arose through evolution as a result of a mutation, and points to Q5H as the ancestral allele. To determine if Q5H is the ancestral allele, we compared it to the allelic state of our last common ancestor, the chimpanzees, as an approximation (Spencer et al., 2006). By comparing the nucleotide codon encoding the fifth amino acid of PANX1 across 31 eutherian mammal species including the chimpanzees, we uncovered that the reference sequence of all assessed species, excluding humans, possessed nucleotide codons that only encoded for histidine (Figure 3D). Furthermore, some studies have noted that the highest frequency for ancestral alleles are found within African populations (Takahata et al., 2001), and African populations have the second highest Q5H allele frequency and highest percentage of Q5H homozygous individuals in the 1000 Genomes Project (Figure 3, A and C). These findings reveal that Q5H is not only the most representative variant of PANX1 within human populations but also the ancestral allele at this locus for PANX1.
Q5H and Y150F do not affect PANX1 channel currents
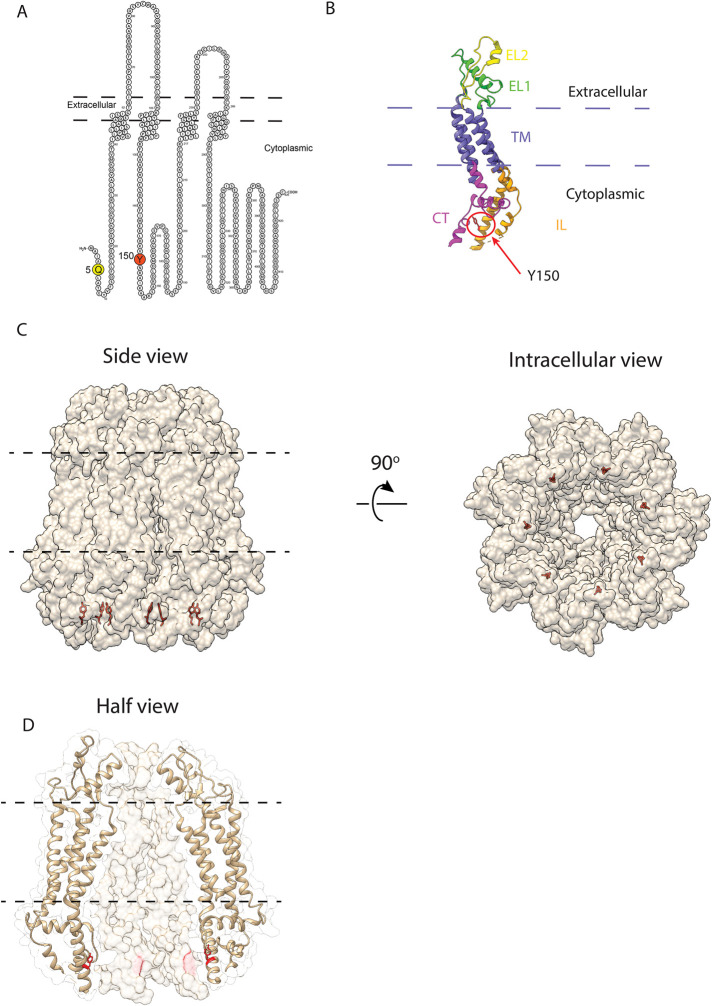
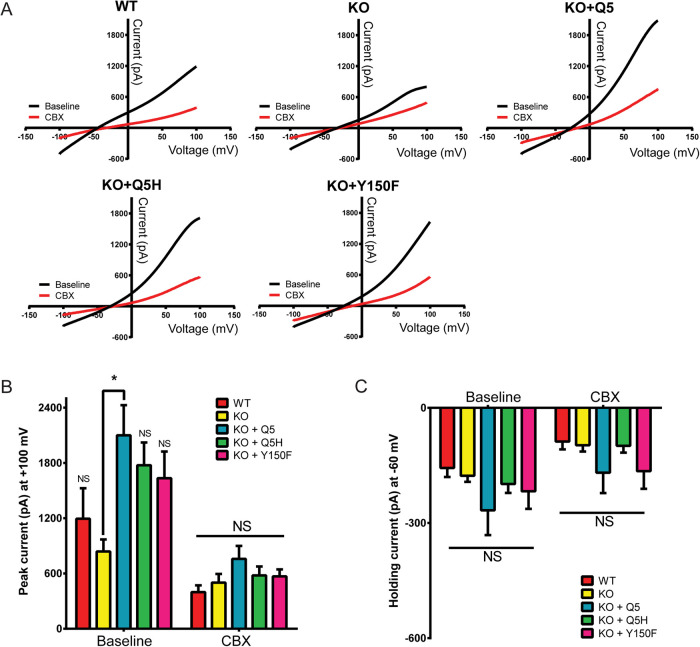
To assess the effects of Q5H and Y150F on PANX1 channel function, given their location in the NT and IL domains of the protein (Figure 4A), we used the recently published heptameric structure of PANX1 to model the possible location of Y150 in the monomeric 3D structure (Figure 4B) and in the heptameric channel structure (Figure 4, C and D). The model predicted Y150 to be in each PANX1 subunit within the inner linings of the pore but could not be used for Q5 since the structure of the NT region is not yet available. To evaluate any effects of the mutations in the ion conductance of the channel, we performed whole-cell patch-clamp electrophysiology on HEK293T cells transiently expressing PANX1. Electrophysiology recordings on HEK293T cells transiently transfected with Q5 and Q5H revealed no significant differences in current amplitude and no difference in carbenoxolone (CBX) sensitivity (Supplemental Figure S2, A and B). HEK293T cells transfected with Y150F experienced significant cell death and as a result were not included in electrophysiology recordings.
FIGURE 4:
Location of Q5 and Y150 residues in 2D and 3D structures of PANX1. (A) PANX1 2D polypeptide demonstrating the location of PANX1 Q5 and Y150 residues. (B) Monomeric 3D structure indicating the location of Y150 in the intracellular loop adjacent to a region (dashed line) not resolved in the 3D structure and in close proximity of a region (positions 322–329) of the carboxyl-terminus of the same monomer. NT, amino-terminus; TM, transmembrane; EL, extracellular loop; CT, carboxyl-terminus; IL, intracellular loop. The structure of the region corresponding to Q5 in the NT has not been published. (C, D) Solvent-excluded molecular surface and ribbon representations (transparency) of the side and intracellular views of the heptameric PANX1 channel showing the location of Y150 in each subunit within the inner linings of the pore. The hydroxyl-F side chain of Y150 is located inward pointing to the center of the channel. The 2D and 3D figures were generated using Protter (Omasits et al., 2014) and UCSF Chimera/ChimeraX (Pettersen et al., 2004), respectively. The cryo-EM 3D structure of PANX1 (PDB: 6m66) was used as a model for all the visualizations (Jin et al., 2020).
To eliminate all endogenous WT PANX1 that may interfere with the trafficking and glycosylation of the mutants, we generated two cancer cell lines engineered using CRISPR/Cas9 to delete the PANX1 gene and eliminate PANX1 expression (KO): the human triple-negative breast cancer cell, Hs578T(KO), and human melanoma cell line A375-P(KO). To assess the influence of Y150F on PANX1 channel currents, we selected the Hs578T(KO) cells, as moderate amounts of Y150F expression were noted to be well tolerated. No significant differences were observed among variants in the current amplitude recorded at positive (Figure 5, A and B) or negative potentials (Figure 5, A and C), apart from PANX1 KO versus Q5-expressing cells (Figure 5B). CBX sensitivity and reversal potentials were not significantly different among treatments (Figure 5). Therefore, under these conditions, Q5H and Y150F have similar electrophysiological properties relative to Q5.
FIGURE 5:
Q5H and Y150F do not affect PANX1 channel currents in Hs578T(KO) cells. (A) Averaged current-voltage relationships recorded in the presence or absence of PANX1 blocker CBX (red) in Hs578T (WT) and Hs578T (KO) cells expressing Q5, Q5H, or Y150F (n = 11 cells for each condition). (B) Summary of ramp currents recorded at +100 mV from WT, nontransfected KO and KO cells expressing Q5, Q5H, and Y150F (n = 11 cells per condition, *p < 0.05; NS, not significant; one-way ANOVA with means compared with KO). (C) Summary of currents recorded at –60 mV from WT, nontransfected KO, and KO cells expressing Q5, Q5H, and Y150F (n = 11 cells for each condition; NS, not significant; one-way ANOVA).
N-glycosylation is altered in Y150F variants
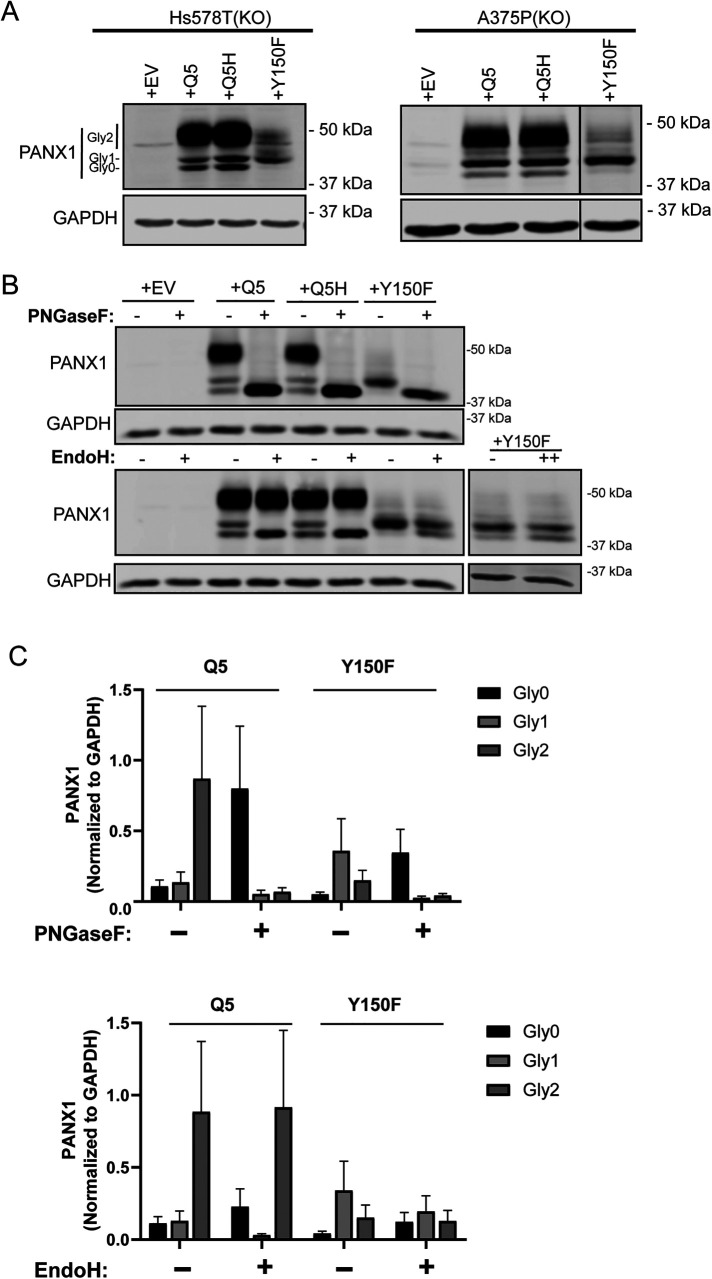
Using the two cancer cell lines engineered to eliminate all endogenous PANX1 expression (Hs578T[KO] and A375-P[KO]), we ectopically expressed Q5, Q5H, and Y150F in both cell types. The corresponding Western blots reproduced a PANX1 banding pattern (highly variable due to different levels of N-glycosylation), which was similar to the one seen in NRKs (Figure 1B). Y150F protein showed predominantly Gly1 with reduced Gly2 species (Figure 6A), while Q5 and Q5H had the typical PANX1 banding pattern with Gly 0,1, and 2.
FIGURE 6:
Y150F produces a Gly1 species that is partially resistant to EndoH deglycosylation. (A) Hs578T(KO) cells and A375P(KO) cells devoid of PANX1 were used to transiently overexpress EV, Q5, Q5H, and Y150F and assessed for changes in PANX1 banding pattern via immunoblotting. Western blot analyses revealed that Y150F disrupted the normal banding pattern of PANX1 in both cell lines, resulting in primarily Gly1 species. Gly0 (nonglycosylated), Gly1 (high mannose), Gly2 (complex glycosylation). (B) PNGase F digestion of protein lysates from Hs578T(KO) cells transiently overexpressing PANX1 variants and controls (N = 3). EndoH digestion of protein lysates from Hs578T(KO) transiently overexpressing PANX1 variants and controls resulted in a band shift of Gly1 species of Q5 and Q5H to Gly0 band. In contrast, Y150F was partially resistant to EndoH digestion, even when the amount of Endo H was doubled (++). GAPDH was used as a loading control. (C) Densitometric analysis of Western blots derived from deglycosylation experiments. Integrated intensity values were tested for normality with a Shapiro–Wilk test and converted -log10 for statistical analysis. Comparisons between treatments (+) and controls (–) were performed in each glyco-species individually using multiple t tests with the Holm–Sidak method. No statistical significance was observed (p > 0.05). Representative blots of N = 3.
The differential banding pattern of Y150F species in NRK, Hs578T(KO), and A375P(KO) indicated that Y150F may experience a different glycosylation process or other PTMs. To confirm that N-linked glycosylation is present throughout the various bands of Q5H and Y150F, we incubated protein samples from Hs578T(KO) cells overexpressing empty vector (EV), Q5, Q5H, and Y150F with PNGase F (Recombinant Flavobacterium meningosepticum PNGase F Protein deglycosydase), which cleaves all N-linked glycosylation which is first manifested in Gly1 species and maintained in complex Gly2 species of WT PANX1. This PNGase F treatment resulted in the shift of all bands to Gly0, indicating the presence of N-linked glycosylation in all bands above Gly0 (Figure 6B). Furthermore, we applied Endoglycosidase H (EndoH), a deglycosylation enzyme that cleaves only high-mannose glycosylation present in Gly1 species. Application of EndoH to EV, Q5, Q5H, and Y150F protein lysates resulted in complete digestion and downward shift of Gly1 band to Gly0 for Q5 and Q5H samples, but only a partial digestion of Gly1 species in Y150F lysates, even when applying a double dose of the EndoH enzyme (Figure 6B). Each band was quantified before and after deglycosylation (Figure 6C) showing the same pattern as the blots, but the changes were not statistically significant due to the high variability of the assays. This indicates that Y150F undergoes a different editing of its high-mannose species after its addition compared with controls.
Y150 is a tyrosine phosphorylation site of PANX1
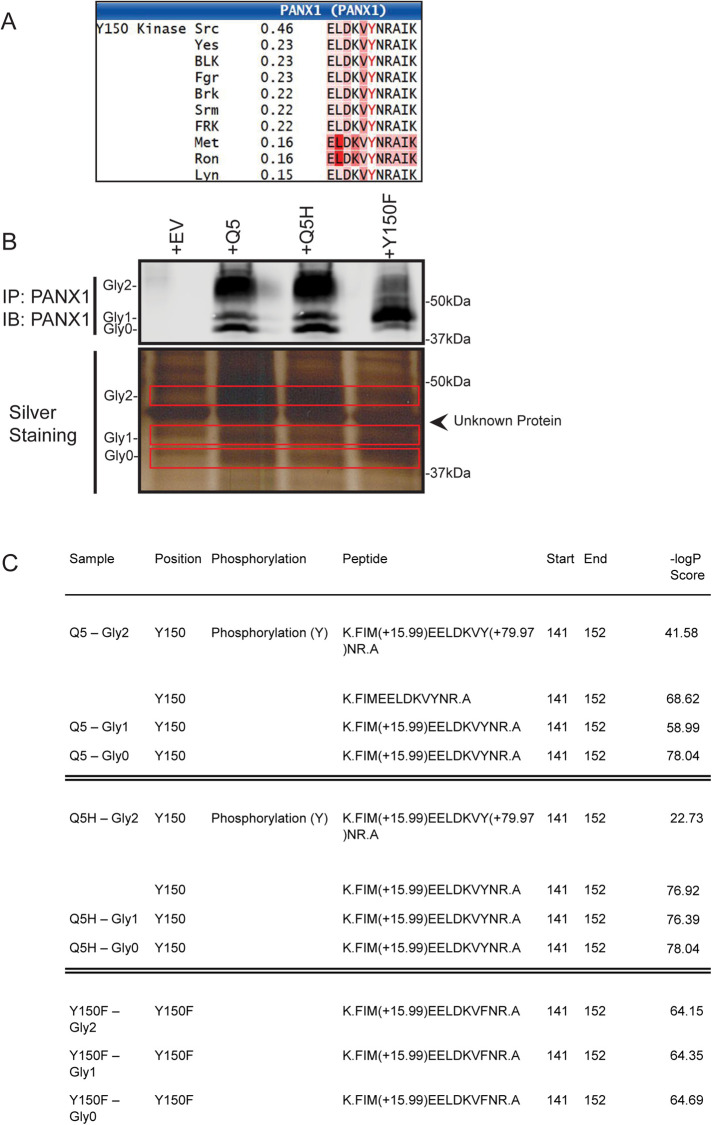
The consensus sequence around the Y150 site (146-LDKVYNRAI-154) was predicted to be recognized and phosphorylated as determined by the phosphorylation prediction software NetPhos 3.1 Server (Blom et al., 1999, 2004). NetworKIN server analyses predicted that the tyrosine 150 is likely phosphorylated by sarcoma (SRC) kinases (Figure 7A). To determine whether Y150 is indeed phosphorylated in Hs578T(KO) cells ectopically expressing PANX1, we immunoprecipitated (IP) PANX1 species from Hs578T(KO) cells overexpressing EV, Q5, Q5H, or Y150F and sent individual band samples from the different glycosylated species (Gly0, Gly1, and Gly2) for electrospray ionization (ESI) mass spectrometry (MS) to identify peptides containing phosphorylated and nonphosphorylated Y150 and peptides containing Y150F (Figure 7B; Supplemental Table S2).
FIGURE 7:
Y150 is a novel tyrosine phosphorylation site of PANX1. (A) Y150 is predicted to be a tyrosine-phosphorylation site by NetPhos3.1 Server. NetworKIN server predicted kinases of the Y150 site and the top candidate was SRC kinase (0.46 score). (B) PANX1 was IP from Hs578T(KO) transiently overexpressing EV, Q5, Q5H, or Y150F and bands from an SDS–PAGE gel representing Gly0, Gly1, and Gly2 of PANX1 were sent for ESI-MS processing and analysis. (C) Analyses of ESI LC MSMS data demonstrating identification of Y150-containing PANX1 peptides; –10logP Score: ions score, where P is the probability that the observed match is a random event; higher numbers identify the likelihood of a true match.
ESI analyses revealed the presence of the phosphorylated Y150-P residue only in Gly2 species of Q5 and Q5H variants, while the nonphosphorylated Y150 peptide was present in Gly0, Gly1, and Gly2 bands of both Q5 and Q5H samples. The Y150F peptide was detected in Gly0, Gly1, and Gly2 bands of Y150F IP lysates, but zero phosphorylated peptide at Y150 residue was detected in any of the Y150F bands (Figure 7C).
SRC kinase inhibition reduces Gly1 species and changes localization of PANX1
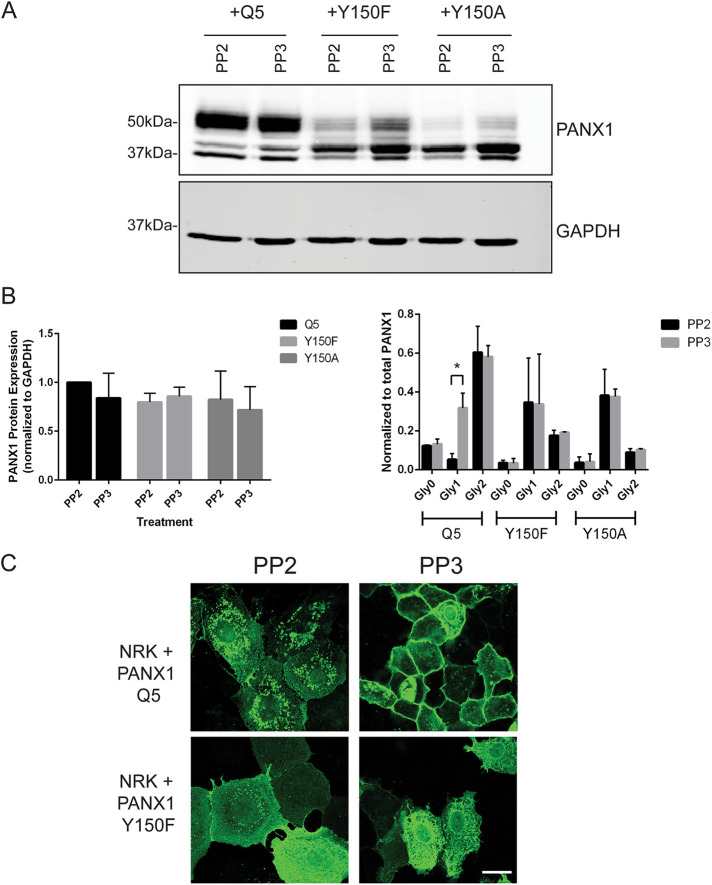
NRK cells ectopically expressing Q5, Y150F, or Y150A (nonphosphorylated mutants) were treated with the Src kinase inhibitor PP2 and its inactive analog PP3 (Figure 8). On treatment, the total levels of PANX1 protein did not change (Figure 8, A and B), but quantification of the individual bands (Gly0, Gly1, Gly2) demonstrated a significant effect of the drug in reducing the Gly1 band of Q5 PANX1, while no effect was observed with the Y150F and Y150A mutants (Figure 8B). Additionally, 24-h treatment with PP2 on NRK cells expressing Q5 PANX1 revealed a noticeable change in subcellular localization of the protein, with increased intracellular signal after immunofluorescence labeling with the anti-PANX1 antibody, while the localization of Y150F was not significantly altered (Figure 8C). In the control cells with the inactive analog PP3, the Q5 PANX1 localization was seen mostly at the cell surface as expected (Figure 8C).
FIGURE 8:
SRC kinase inhibition with PP2 reduces Gly1 species and decreases cell surface localization of PANX1. (A) NRK cells overexpressing PANX1 WT (Q5), Y150F or Y150A were treated with PP2 to inhibit Src kinases, and the inactive analog PP3 was used as a control. There was no decrease in total protein levels of PANX1 with PP2 treatment between mutants; however, kinase inhibition produced a significant decrease in the Gly1 species compared with control in PANX1 WT expressing cells (p < 0.05), with no effect on the nonphosphorylated mutants (Y150A and Y150F). (B) Protein levels were quantified from representative blots (N = 3) and a two-way ANOVA was performed (*p < 0.05). (C) NRK cells overexpressing PANX1 WT exhibited a more intracellular PANX1 pattern when treated with PP2 compared with the primarily cell surface localization seen in control PP3 cells.
Mutations at tyrosine Y150 change the banding profile and increase intracellular localization of PANX1 in normal and cancer cells
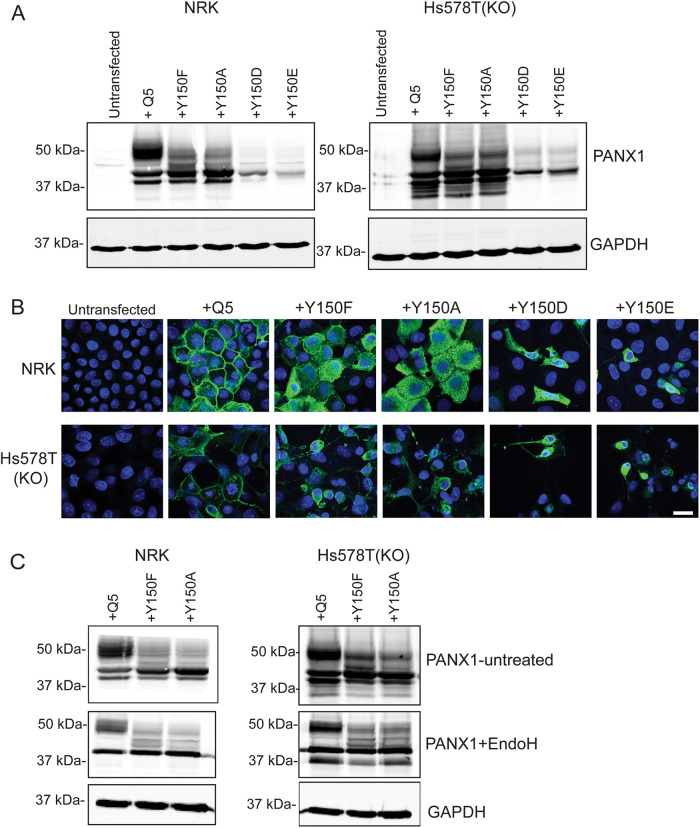
To assess the effect of different mutations on the Y150 site in both normal and cancer cells, we ectopically expressed the mutants in NRK and Hs578T-PANX1 KO cells. Similar patterns were observed in both cell types, as well as in mutants replacing the tyrosine with either phenylalanine (F) or alanine (A) (Figure 9A). Both nonphosphorylatable mutants (A and F) displayed increased intracellular localization in both cell types compared with Q5 (Figure 9B), with a subpopulation of the protein still visible at the cell surface. Lysates of Y150F- and Y150A-expressing cells showed the same pattern of partial resistance to EndoH digestion (Figure 9C), while the WT controls showed a complete digestion of Gly1 (high mannose) bands in the same assay.
FIGURE 9:
Impact of Y150 PANX1 mutations on protein banding and cell-surface localization in NRK and HS578T-PANX1-KO cells. (A) NRK and HS578T-PANX1 KO cells were transfected with 1 µg of DNA from either WT PANX1 (Q5), Y150 mutants unable to be phosphorylated (Y150F and Y150A) or Y150 phosphomimetics (Y150D and Y150E); 48 h post-transfection, all Y150 mutants disrupted the normal PANX1 banding pattern. Y150F and Y150A showed a loss of Gly2 and mainly Gly1 and Gly0. Y150D and Y150E mutants were toxic to the cells; however, protein that was able to be isolated demonstrated mainly Gly1 banding. (B) Immunoflourescence of all Y150 mutants 48 h post-transfection showed disruption of WT PANX1 localization within both NRK and HS578T-PANX1-KO cells. Y150F and Y150A demonstrate a mainly intracellular pattern of PANX1 compared with the cell surface localization of PANX1 WT. Y150D and Y150E mutants caused cell death, so localization is difficult to interpret. (C) EndoH digestion of protein lysates from NRK and Hs578T(KO) overexpressing WT PANX1 resulted in a shift for some of Gly2 and all the Gly1 species to the Gly0 band. Cells overexpressing Y150F and Y150A showed a partial shift of Gly1 to the Gly0 species, but a portion of Gly1 in these mutants remained partially resistant to EndoH digestion in both cell types (even in the presence of excess enzyme).
Interestingly, mutants carrying a phosphomimetic residue to represent constitutively phosphorylated Y150 sites (Y150D and Y150E) were toxic to the cells on transfection under the same conditions and resulted in lower PANX1 protein levels with mostly Gly0 and Gly1 bands (Figure 9A). Immunofluorescence of Y150D and Y150E in both NRK and Hs578T cells revealed intracellular localization of the mutant proteins and irregular cell morphology (Figure 9B). These findings suggest that phosphorylation at the Y150 site may need to be dynamically regulated rather than constitutively phosphorylated.
DISCUSSION
Given its widespread expression within the human body along with the wide range of signaling molecules it is permeable to, it is not surprising that PANX1 has been implicated in various pathological disorders such as ischemia, HIV infections, melanoma, and others (Thompson et al., 2006; Seror et al., 2011; Penuela et al., 2012; Freeman et al., 2019). While most studies explored the effects of altering expression or inhibition of PANX1, only a few investigations have explored the effects of coding variations on channel function in the context of disease (Furlow et al., 2015; Molica et al., 2015; Shao et al., 2016; Sang et al., 2019). This report studied the influence of two naturally occurring PANX1 variants, Q5H and Y150F, on the PTMs, trafficking, and function of PANX1. Overall, we uncovered a functionally important tyrosine phosphorylation site that may influence normal PANX1 N-glycosylation and trafficking and a highly prevalent ancestral PANX1 variant that, contrary to current knowledge, is the most common PANX1 variant found in humans.
What initiated our interest in Q5H was a previous report demonstrating that the Q5H variant was enriched in healthy Caucasian males with platelets that were hyperreactive to collagen (Molica et al., 2015). Interestingly, this study demonstrated that ectopic expression of Q5H in Chinese hamster ovary cells resulted in a greater basal and K+-activated ATP release than Q5 and controls (Molica et al., 2015). With the emergence of numerous reports demonstrating the tumor-promoting effects of PANX1 (Bao et al., 2012; Penuela et al., 2012; Furlow et al., 2015; Wei et al., 2015; Freeman et al., 2019), along with the finding that the Q5H variant was present in cancer patients (Zhang et al., 2011; Cerami et al., 2012; Gao et al., 2013) and cancer cell lines, we then developed the premise that Q5H may play a role in promoting tumorigenic properties. Contrary to our hypothesis, Q5H did not impact channel activity when expressed in breast cancer cells and HEK293T cells and did not affect growth or migration compared with Q5. However, it is important to note that Molica et al. (Molica et al., 2015) used basal and K+-activated ATP release to assess for PANX1 channel activity, whereas we employed basal dye uptake and examined voltage-dependent channel behavior recording the outward rectification of constitutively active currents by electrophysiology. The different stimuli used, voltage activation, and elevated extracellular K+ conditions have been previously proposed to promote different conformational shapes (Wang and Dahl, 2010; Wang et al., 2014) where the former involves the C-terminus lining the pore, but the latter does not. It has been suggested that elevated extracellular K+ conditions which favor ATP-permeability may induce a conformation similar to ATP-permeable connexin channels in which the N-terminus lines the pore of the channel (Bao et al., 2004; Dahl, 2018).
Our discovery that Q5H is a highly conserved and more prevalent ancestral allele than Q5, which is annotated in various genomic and protein sequencing databases to be the most representative allele, highlights the necessity to thoroughly analyze and confirm genomic sequences that are representative of the overall population. Ironically, when human PANX1 was fully sequenced in RefSeq in 2003 by Baranova et al. (Baranova et al., 2004; Accession No. AF398509.1), the PANX1 mRNA sequence contained the codon encoding for histidine in the fifth amino residue. However, currently the PANX1 mRNA sequence (Accession No. NM_015368.4) in RefSeq contains the codon that encodes for glutamine at the fifth residue. This has led to numerous studies over the years utilizing the derived allele that is less prevalent than the ancestral Q5H allele. As a result, future studies on human PANX1 need to take into consideration the two prevalent and divergent variants Q5 and Q5H (or rather, Q5 and H5), since it remains unknown how they may differ in other cellular contexts.
Only a couple of phosphorylation sites have been reported for PANX1 and they have an important role in functional regulation. With our proteomic analysis of PANX1, we found six additional sites of confirmed phosphorylation (S137, Y150, S159, S344, S357, T382) summarized alongside many other PTMs in Supplemental Table S2. We showed that when one of those phosphorylation sites, the Y150 residue, is mutated to F, it disrupts Y150 phosphorylation and affects normal N-glycosylation and trafficking of the protein. Although the phosphorylated and nonphosphorylated Y150 peptides were detected only in the Gly2 states of Q5 and Q5H, the spatiotemporal pattern of Y150 phosphorylation remains uncertain. It is possible that the phosphorylation happens in the Golgi as a checkpoint step between Gly1 and Gly2 modifications, but it is also possible that the phosphorylation of Y150 takes place at the cell surface. The resistance to EndoH deglycosylation acquired by Y150F and the reduction in Gly2 species production suggest that Y150 phosphorylation may facilitate a glycosylation pathway leading to complex glycosylation of the protein by either interacting with N-glycosyltransferases in the Golgi or by altering protein folding and/or stability. Disrupting its phosphorylation by mutating the residue to F led to the formation of a hypo-glycosylated species partially resistant to EndoH digestion. Therefore, this suggests that the Gly species found in Y150F may not be just high-mannose glycosylated but may experience another type of glycosylation modification such as hybrid glycosylation (Moremen et al., 2012). However, constitutive phosphorylation at the Y150 site (mimicked by Y150D and E) also disrupted glycosylation, indicating that a dynamic process of phosphorylation and dephosphorylation might be important for this regulatory step. Furthermore, Sang et al. (Sang et al., 2019) previously reported four hypo-glycosylated (Gly1) PANX1 mutants that increased oocyte cell death and induced increased expression of calreticulin, a chaperone protein that facilitates glycoprotein folding. This suggests that proper protein folding facilitates editing to complex glycosylation and that, in the case of Y150F, the lack of phosphorylation, or merely the mutation, may alter protein folding/stability and favor an alternate glycosylation-editing pathway. To test the possibility that the Y150F mutation could alter the structure of the protein, we modeled the potential effect of the Y to F substitution using the published heptameric structure of the PANX1 channel. Based on that prediction, Y150 can form hydrogen bonds with D327 that would be disrupted in the case of Y150F and D327, separated by a larger distance and too far to engage in hydrogen bonding interactions (Supplemental Figure S3). This could result in predicted structural changes on the heptameric channel, where a short helix (aa 325–336) may protrude outward in the Y150F mutant introducing a gap between adjacent chains (Supplemental Figure S4). We have seen that the ion conductance of the Y150F channel does not seem to change compared with WT (Figure 5), but we do not know the effects of this mutation on the PANX1 channel ability to pass larger metabolites like ATP, or its function as an intracellular channel. Based on the current heptameric PANX1 channel structure, we used protein modeling software to predict that the Y150F mutant pore channel would be straighter, with a progressively smaller pore diameter, and a narrowing in the pore structure when compared with Q5 PANX1 (Supplemental Figures S5 and S6). Unfortunately, due to their effects on cell health, we were not able to establish stable cell lines of Y150 mutants to perform additional functional assays and test if Y150F would have reduced permeability to larger metabolites.
Previous evidence of PANX1 tyrosine phosphorylation, specifically on Y198 and Y308 of mouse and rat Panx1, respectively, demonstrated phosphorylation required interactions with the Src family kinases (SFKs) (Iglesias et al., 2008; Lohman et al., 2012; Weilinger et al., 2012, 2016). Administration of interfering peptides against SFK consensus phosphorylation sites of Y198 and Y308 sites abolished Panx1 activation mediated by SFKs (Lohman et al., 2015; Weilinger et al., 2016). Furthermore, it has also been demonstrated that Y198 constitutive phosphorylation occurs at the cell surface and was dependent on SFK activity (DeLalio et al., 2019) and tyrosine phosphorylation of either Y198 or Y308 augmented channel activity, demonstrating that phosphorylation modulates Panx1 channel activity (Lohman et al., 2015; Weilinger et al., 2016). In this study, we show that once again, the most likely kinase responsible for Y150 phosphorylation is SRC kinase, as predicted by consensus sequence analysis and the effects of PP2 inhibition of SRC kinases on the WT PANX1, which did not affect the nonphosphorylatable mutants Y150F and Y150A. However, we need to consider that inhibiting SFKs would have many other effects on other PANX1 sites, as well as on other proteins in the cells that are regulated by SFKs. Given the localization and protein banding observed with the phosphomimetic mutants (Y150D and E), we propose that phosphorylation and dephosphorylation at the Y150 site need to be dynamically and precisely regulated for the PANX1 protein to be properly processed, trafficked, and posttranslationally modified by complex N-glycosylation.
Harboring a Y150F variant in the context of cancer can have different effects on the cancer cells depending on the expression level of the mutant. As aforementioned, only two cancer patients harbor mutations that disrupt the Y150 site, Y150F (melanoma) and Y150C (bladder cancer) mutations (Supplemental Table S1) (Cerami et al., 2012; Gao et al., 2013). In both cases, the allele frequency for the mutations is less than 0.5, suggesting heterozygosity with WT PANX1, which may produce effects different from those observed in this study when the mutant is coexpressed with the WT allele. However, we observed that the Y150F banding pattern was the same in the presence of endogenous PANX1 (Supplemental Figure S1). The melanoma patient who harbored Y150F in his tumor (TCGA-EE-A29E-06; RNA expression was confirmed on this tumor) is a white, 54-year-old male who presented only lymph node metastasis but no distant metastases. He has survived 63 mo after diagnosis without treatment, which is a good prognosis for a melanoma patient (cbioportal.org). Since only two patients possess a mutation at the Y150 site along with many other passenger mutations in other genes, it is not possible to establish a correlation between Y150 missense mutations and the disease status of these patients. Based on what we know about the Y150F mutation, we would predict a likely reduction of PANX1 large-pore channel function in those tumor cells.
In conclusion, by exploring various naturally occurring PANX1 variants from melanoma tumors and cells, we have uncovered two that revealed previously unknown characteristics of the widely expressed channel protein. The rare variant Y150F revealed a phosphorylation site that affects glycosylation and trafficking of PANX1, while the common variant Q5H had similar function to Q5 and unraveled itself as the ancestral allele. Q5H is also the more representative allele at this locus within human populations and should be included in future studies of PANX1.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cell culture
NRK cells (ATCC CRL-6509), human triple-negative breast cancer Hs578T (ATCC HTB-126) and human melanoma A375P (ATCC CRL-3224), A375-MA2 (ATCC CRL-3223), human melanoma 131/4-5B1(Cruz-Munoz et al., 2008), and human glioblastoma U87MG (ATCC HTB-14) cells were cultured in DMEM 1× (ThermoFisher) containing 4.5 g/l D-glucose, L-glutamine, 110 mg/l sodium pyruvate, 10,000 U of penicillin, 10 mg/ml streptomycin, and 10% fetal bovine serum (Wisent Institute). All cells were incubated at 37°C and 5% CO2. Cellular dissociation from culture plates was done using trypsin (0.25%, 1 mM EDTA 1×; ThermoFisher).
Generation of CRISPR/Cas9 knockout cells
PANX1 knockout cells were generated by CRISPR/Cas9 D10A as described in Ran et al. (2013). Briefly, cells were transfected with 1 µg each of pSpCas9n(BB)-2A-Puro (PX462) V2.0 and pSpCas9n(BB)-2A-GFP (PX461) (addgene.org) containing guide RNA sequences for human PANX1 in a 6-well plate. PANX1 gRNAs were designed with http://tools.genome-engineering.org (sequences GTTCTCGGATTTCTTGCTGA and CTCCGTGGCCAGTTGAGCGA). At 24 h post-transfection, cells were selected with 1 µg/ml Puromycin for 72 h. Following selection, cells underwent single colony selection via serial dilutions and were screened for PANX1 levels by Western blot. Plasmids were a gift from Feng Zhang (Addgene plasmid #48140 and #62987).
Plasmids and transfection
An expression vector encoding for WT PANX1 (Q5), pUNO1-hPANX1 was purchased from InvivoGen (San Diego, CA). PANX1 variant-encoding expression vectors (Q5H, Y150F, Y150A, Y150D, Y150E, G168E, T176I, H190Y, S239L, and Q264*) were generated by NorClone Biotech Industries (London, Ontario, Canada). For all experiments, cells were transfected with 1 μg of DNA (in 35-mm dishes) with expression vectors encoding for all the mutants using Lipofectamine 3000 (ThermoFisher) according to manufacturer’s instructions. Cells were analyzed for expression and functional analyses 48 and 72 h post-transfection. Hs578T(KO) clones stably overexpressing Q5 or Q5H were generated by blasticidin selection at 15 μg/ml for 1 wk and underwent single-cell colony selection by serial dilution.
Protein extraction and immunoblotting
Protein lysates from cells were extracted using IP buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40 (Igepal) (Honeywell Fluka, Seelze, Germany), 0.5% sodium deoxycholate, 1 mM sodium fluoride, 4 mM sodium orthovanadate, and one tablet of complete-mini EDTA-free protease inhibitor (Roche, Mannheim, Germany), or 1× RIPA buffer (Supplemental Figure S1). Protein concentration was quantified using the Pierce bicinchoninic acid assay (ThermoFisher Scientific); 30 μg of protein lysates were subjected to 10% SDS–PAGE, blocked with 3% bovine serum albumin (BSA) with 0.05% Tween-20 in 1× phosphate-buffered saline (PBS) or 1× Tris-buffered saline and immunoblotted with anti-human PANX1 antibody as previously described in Freeman et al. (2019) (1:1000, PANX1 CT-412; 0.35 μg/μl [ Shao et al., 2016]; 1:100 PANX1-EL2 [ Penuela et al., 2007, 2009]; 1:125, PANX1-NT ThermoFisher cat# 487900; anti-glyceraldehyde 3-phosphodehydrogenase [GAPDH] antibody [1:1000; Millipore Cat# MAB374, RRID: AB_2107445]). For detection, goat anti-rabbit IRDye-800CW and goat anti-mouse IRDye-680RD (LI-COR Biosciences, Lincoln, NE) were used as secondary antibodies at 1:10,000 dilutions and imaged using a LI-COR Odyssey infrared imaging system (LI-COR Biosciences). Western blot quantification and analysis was conducted using Image Studio Lite (LI-COR Biosciences).
Deglycosylation
Ten micrograms of whole-cell lysate from Hs578T-KO or NRK cells transiently overexpressing EV, Q5, Q5H, or Y150F, Y150A were incubated with either 500 or 1000 U of EndoH (after boiling the lysates; Cat. No. P0702S; New England BioLabs) or 1 U of PNGase F (Cat. No. P7367; Sigma-Aldrich) at 37°C according to the manufacturer’s protocol.
Immunofluorescence and confocal microscopy
Cells were grown on glass coverslips and were fixed 48–72 h posttransfection using ice-cold 8:2 methanol:acetone for 15 min at 4°C and blocked with 2% BSA-PBS. Coverslips were incubated with anti-human PANX1 antibody (1:500; PANX1 CT-412; 0.35 μg/μl), Hoechst 33342 (1:1000), and Alexa Fluor 488 goat anti-rabbit IgG (2 mg/ml, 1:700) and mounted using Airvol (Mowiol 4-88; Sigma Aldrich) prior to imaging. Immunofluorescence images were obtained using a Zeiss LSM 800 confocal microscope with a Plan-Apochromat 63×/1.40 Oil DIC objective (Carl Zeiss, Oberkochen, Germany). The laser lines used include 405 nm (Hoechst 33342) and 488 nm (Alexa488).
Whole-cell patch clamp recordings from HEK 293T cells
Whole-cell voltage-clamp recordings were performed at room temperature (20–22°C) using an Axon MultiClamp 700A amplifier and Digidata 1322A data acquisition system (Molecular Devices, Sunnyvale, CA). Patch electrodes were pulled using a Narishige two-stage puller (PP-83; Narishige, Greenvale, NY) from thin-walled borosilicate glass (TW150-F3; WPI, Sarasota, FL). Electrodes had a final resistance of 3–5 MΩ when filled with intracellular solution containing (in mM): 142 cesium gluconate, 10 HEPES, 2 MgCl2, 8 NaCl, pH 7.2 (adjusted with 1 M Cs-OH), and osmolarity between 290 and 295 mosmol/l. Standard extracellular solution (ECS) was composed of (in mM): 140 NaCl, 5.4 KCl, 25 HEPES, 33 glucose, 2 CaCl2, 1 MgCl2, pH of 7.4 (adjusted with 10N NaOH) and osmolarity between 300 and 305 mosmol/l. A computer-controlled, multibarreled perfusion system (SF-77B; Warner Institute, Hamden, CT) was used to exchange bath solutions from standard ECS to 100 µM CBX in ECS (Panx1 blocker). Transfected cells (identified by the presence of mCherry marker) were voltage-clamped at –60 mV and PANX1 currents were recorded by the application of voltage-ramps (±100 mV, 500 ms) to the membrane every 10 s. Current–voltage (I–V) relationships were constructed from these recordings. Currents were filtered at 2 kHz and sampled at 10 kHz, digitized, and acquired using pCLAMP 9.2 software (Molecular Devices). Data were collected from minimum three independent experiments and analyzed using Clampfit 10.7 software (Molecular Devices).
Cell growth assay
Fifteen-thousand cells were plated in a well of a 6-well culture plate at day 0. From day 2 to day 4 after cell plating, cells were dissociated with Trypsin (0.25%, 1 mM EDTA 1×; Life Technologies), and the number of trypan blue-excluded live cells were counted with an automated cell counter Cell Countess II (ThermoFisher) as described previously (Freeman et al., 2019).
Migration assay
Twenty-thousand cells were plated in each well of a 96-well culture plate coated with 0.01% poly-l-lysine (Millipore Sigma, Burlington, MA) and grown to confluence. A scratch wound was inflicted with a P200 pipette tip and serum DMEM was replaced with serum-free DMEM. Pictures were taken with a brightfield microscope within an hour of scratch and at 15 h after scratch. The area of migration was quantified with ImageJ software.
IP and silver staining
Protein lysates were collected 96 h post-transfection from Hs578T(KO) cells transfected with EV, Q5, Q5H, or Y150F on which confluency was reached; 1 mg of whole-cell protein lysate was incubated overnight at 4°C with Protein A/G beads that were cleaned twice with 1× Dulbecco-PBS (ThermoFisher) along with 10 μg of anti-human PANX1 antibody (PANX1 CT-412) cross-linked to Pierce Protein A/G-Agarose beads (Thermo Scientific). Subsequently, bound proteins underwent four washes with 500 μl of IP buffer. Beads were collected and resuspended in 2× Laemmli buffer with 10% β-mercaptoethanol, boiled for 5 min, and supernatant was collected after spinning down. Supernatant then was subjected to 10% SDS–PAGE and gel was stained with Pierce Silver Stain Kit (ThermoFisher Scientific) according to manufacturer’s protocol.
Peptide identification using MS
The peptides, obtained after Trypsin digest (Promega), were resuspended in 0.1% formic acid/99.9% water and loaded onto an ACQUITY UPLC Symmetry C18 NanoAcquity, 10K, 2G V/M, 180 μm × 20 mm, 100 A, 5 µm trapping column (Waters Corporation, Milford, MA) via a Waters NanoAcquity UPLC at a flow rate of 10 µl/min for 6 min using 99% buffer A (0.1% formic acid) and 1% buffer B (Acetonitrile + 0.1% formic acid). After trapping, the peptides were eluted onto the analytical column for separation using a 95-min run time. Flow was established at 300 nl/min for the ACQUITY UPLC Peptide BEH C18 nanoAcquity Column 10K psi, 130Am 1.7 µm × 25 mm which was held at 35°C. The gradient initial condition was 5% buffer B. Buffer B then increased to 40% over 60 min, then to 95% over 15 min, then to 5% over 2 min, and finally held at 5% for 13 min for re-equilibration to the initial condition. The LC system was directly connected to a NanoFlex (Thermo Electron, Waltham, MA) nanospray ionization source with a source voltage of 2.4 KV and was interfaced to an Orbitrap Elite, VelosPro MS. The MS was controlled by Xcalibur software (Thermo, v. 2.7.0) and operated in the data-dependent mode using an FT/IT/CID Top 20 scheme. The MS scan recorded the mass-to-charge ratios (m/z) of ions over the range of 400–1450 in FT (resolution of 120,000 at m/z 400), positive ion, profile, full MS mode using a lock mass (445.120025 m/z). The 20 most abundant multiply charged ions were automatically selected for subsequent collisional induced dissociation in ion trap mode (IT/CID) with an isolation width of 2.00 Da, rapid scan rate, centroid mode, with charge state filtering allowing only ions of +2, +3, and +4 charged states. Normalized collision energy was 35, and precursor ions were then excluded from further CID for 30 s.
Modified protein sequences for PANX1 (based on the RefSeq with single amino acid changes at Q5H, Y150F) were added to the Contaminants database (Uniprot, August 15th, 2018, 216 entries) and raw data were searched using PEAKS Studio 8.5 (build 20180507). Trypsin was selected with three missed cleavages and one nonspecific missed cleavage. Carbamidomethylation (C) was selected as a fixed modification and deamidation (NQ), oxidation (M), acetylation (Protein N-term), and phosphorylation (STY) were selected as variable modifications, with maximum seven variable modifications per peptide allowed. Parents mass error tolerance was set to 15 ppm and fragment mass error tolerance was set to 0.7 Da. FDR was estimated using a Target-Decoy. Another search was performed using the parameters as described above, only an S-nitrosylation was selected as a variable modification in addition to the above listed modifications. Also, the maximum number of variable modifications per peptide allowed was set to 6.
PP2 and PP3 treatments
NRK cells were grown in a 35-mm dish and transfected with 1 µg of each PANX1 encoding plasmid (WT, Y150F or Y150A). The following day (24 h post-transfection), cells were washed with PBS and treated with fresh media containing either 10 µM of PP2 or 10 µM PP3 control (Selleckchem, TX). Protein lysates for Western blots and coverslips for immunofluorescence were collected 24 h postdrug treatment and processed as described above.
Bioinformatics
Allele and genotype frequencies, in addition to population genetic distribution information for SNP rs1138800 encoding for Q5H, were extracted from Ensembl and ExAC databases. Clinical attributes of the TCGA SKCM cohort were downloaded from the cBioPortal database (Cerami et al., 2012; Gao et al., 2013). The results for TCGA SKCM cohort shown in this investigation are in whole based on data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Q5 and Q5H genome sequencing of cancer cell lines
Genomic DNA was extracted from cancer cell lines (A375-MA2, U87MG, BeWo, and 131/4-5B1) using PureLink Genomic DNA Mini Kit (Cat. No. K182001; ThermoFisher) according to the manufacturer’s instructions. Genomic DNA was converted to cDNA using High-Capacity cDNA Reverse Transcription Kit (Cat. No. 4368813; ThermoFisher) and primers spanning the N-terminus of PANX1 (forward: 5′-GGAAGCGCTTTGTTCCGC-3′, reverse: 5′-CCTCCCACAAACTTTGCCCTA-3′). cDNA products were sent for DNA sequencing at the Robarts Research Institute DNA Sequencing Facility (London, Ontario, Canada).
Statistical analyses
Statistical analyses were performed using GraphPad Prism software (version 8.0; San Diego, CA). Data error bars indicate mean ± standard error mean.
Protein structure modeling
The 2D and 3D figures were generated using Protter (Omasits et al. 2014) and UCSF Chimera/ChimeraX (Pettersen et al., 2004). The cryo-EM 3D structure of PANX1 (PDB: 6M66) was used as a model for all the visualizations (Jin et al., 2020) (Figure 4).
Sequence alignment and superposition were performed using Pymol (Alexander et al., 2011; Schiffrin et al., 2020). A Cryo-EM structure was generated with the Swiss Model homology modelling platform (Waterhouse et al., 2018) (Supplemental Figures S3 and S4). Pore visualization was done using PoreWalker (Pellegrini-Calace et al., 2009) (Supplemental Figure S5), and transmembrane output scores were estimated using CHEXVIS (Masood et al., 2015) (Supplemental Figure S6).
Supplementary Material
Acknowledgments
We thank Victoria Clarke for processing samples for ESI, and helpful discussions on the study. We also thank Jessica Esseltine for generating the gRNA used for CRISPR/Cas9 editing of PANX1. Funding for S.P. was provided from a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant RGPIN-2015-06794 and Canadian Institutes of Health Research Project Grant FRN 153112. Funding for G.B.G. was provided by NSERC Discovery Grant RGPIN-2015-03878.
Abbreviations used:
- A
alanine
- BSA
bovine serum albumin
- CBX
carbenoxolone
- CID
collisional induced dissociation
- ECS
extracellular solution
- EndoH
endoglycosidase H
- ER
endoplasmic reticulum
- ESI
electrospray ionization
- EV
empty vector
- F
phenylalanine
- GAPDH
glyceraldehyde 3-phosphodehydrogenase
- GWAS
genome-wide association study
- IP
immunoprecipitation
- MS
mass spectrometer
- NRK
normal rat kidney
- PANX1
pannexin 1 in human
- Panx1
pannexin 1 in rodents
- PBS
phosphate-buffered saline
- PTM
posttranslational modification
- SFK
Src family kinase
- SNP
single-nucleotide polymorphism
- SRC
sarcoma
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-10-0585) on January 6, 2021.
REFERENCES
- Alexander N, Woetzel N, Meiler J (2011). bcl::Cluster: A method for clustering biological molecules coupled with visualization in the Pymol Molecular Graphics System. IEEE Int Conf Comput Adv Bio Med Sci 2011, 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao BA, Lai CP, Naus CC, Morgan JR (2012). Pannexin1 drives multicellular aggregate compaction via a signaling cascade that remodels the actin cytoskeleton. J Biol Chem 287, 8407–8416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L, Locovei S, Dahl G (2004). Pannexin membrane channels are mechanosensitive conduits for ATP. FEBS Lett 572, 65–68. [DOI] [PubMed] [Google Scholar]
- Baranova A, Ivanov D, Petrash N, Pestova A, Skoblov M, Kelmanson I, Shagin D, Nazarenko S, Geraymovych E, Litvin O, et al. (2004). The mammalian pannexin family is homologous to the invertebrate innexin gap junction proteins. Genomics 83, 706–716. [DOI] [PubMed] [Google Scholar]
- Bargiotas P, Krenz A, Monyer H, Schwaninger M (2012). Functional outcome of pannexin-deficient mice after cerebral ischemia. Channels (Austin) 6, 453–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billaud M, Chiu YH, Lohman AW, Parpaite T, Butcher JT, Mutchler SM, DeLalio LJ, Artamonov MV, Sandilos JK, Best AK, et al. (2015). A molecular signature in the pannexin1 intracellular loop confers channel activation by the alpha1 adrenoreceptor in smooth muscle cells. Sci Signal 8, ra17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S (1999). Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294, 1351–1362. [DOI] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S (2004). Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633–1649. [DOI] [PubMed] [Google Scholar]
- Boassa D, Ambrosi C, Qiu F, Dahl G, Gaietta G, Sosinsky G (2007). Pannexin1 channels contain a glycosylation site that targets the hexamer to the plasma membrane. J Biol Chem 282, 31733–31743. [DOI] [PubMed] [Google Scholar]
- Boyce AK, Kim MS, Wicki-Stordeur LE, Swayne LA (2015). ATP stimulates pannexin 1 internalization to endosomal compartments. Biochem J 470, 319–330. [DOI] [PubMed] [Google Scholar]
- Celetti SJ, Cowan KN, Penuela S, Shao Q, Churko J, Laird DW (2010). Implications of pannexin 1 and pannexin 3 for keratinocyte differentiation. J Cell Sci 123, 1363–1372. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekeni FB, Elliott MR, Sandilos JK, Walk SF, Kinchen JM, Lazarowski ER, Armstrong AJ, Penuela S, Laird DW, Salvesen GS, et al. (2010). Pannexin 1 channels mediate ‘find-me’ signal release and membrane permeability during apoptosis. Nature 467, 863–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Munoz W, Man S, Xu P, Kerbel RS (2008). Development of a preclinical model of spontaneous human melanoma central nervous system metastasis. Cancer Res 68, 4500–4505. [DOI] [PubMed] [Google Scholar]
- Dahl G (2018). The Pannexin1 membrane channel: distinct conformations and functions. FEBS Lett 592, 3201–3209. [DOI] [PubMed] [Google Scholar]
- Davis LK, Gamazon ER, Kistner-Griffin E, Badner JA, Liu C, Cook EH, Sutcliffe JS, Cox NJ (2012). Loci nominally associated with autism from genome-wide analysis show enrichment of brain expression quantitative trait loci but not lymphoblastoid cell line expression quantitative trait loci. Mol Autism 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLalio LJ, Billaud M, Ruddiman CA, Johnstone SR, Butcher JT, Wolpe AG, Jin X, Keller TCST, Keller AS, Riviere T, et al. (2019). Constitutive SRC-mediated phosphorylation of pannexin 1 at tyrosine 198 occurs at the plasma membrane. J Biol Chem 294, 6940–6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman TJ, Sayedyahossein S, Johnston D, Sanchez-Pupo RE, O’Donnell B, Huang K, Lakhani Z, Nouri-Nejad D, Barr KJ, Harland L, et al. (2019). Inhibition of Pannexin 1 Reduces the Tumorigenic Properties of Human Melanoma Cells. Cancers (Basel) 11, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlow PW, Zhang S, Soong TD, Halberg N, Goodarzi H, Mangrum C, Wu YG, Elemento O, Tavazoie SF (2015). Mechanosensitive pannexin-1 channels mediate microvascular metastatic cell survival. Nat Cell Biol 17, 943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawlik M, Wagner M, Pfuhlmann B, Stober G (2016). The role of Pannexin gene variants in schizophrenia: systematic analysis of phenotypes. Eur Arch Psychiatry Clin Neurosci 266, 433–437. [DOI] [PubMed] [Google Scholar]
- Iglesias R, Locovei S, Roque A, Alberto AP, Dahl G, Spray DC, Scemes E (2008). P2X7 receptor-Pannexin1 complex: pharmacology and signaling. Am J Physiol Cell Physiol 295, C752–C760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q, Zhang B, Zheng X, Li N, Xu L, Xie Y, Song F, Bhat EA, Chen Y, Gao N, et al. (2020). Cryo-EM structures of human pannexin 1 channel. Cell Res 30, 449–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locovei S, Bao L, Dahl G (2006). Pannexin 1 in erythrocytes: function without a gap. Proc Natl Acad Sci USA 103, 7655–7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman AW, Leskov IL, Butcher JT, Johnstone SR, Stokes TA, Begandt D, DeLalio LJ, Best AK, Penuela S, Leitinger N, et al. (2015). Pannexin 1 channels regulate leukocyte emigration through the venous endothelium during acute inflammation. Nat Commun 6, 7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman AW, Weaver JL, Billaud M, Sandilos JK, Griffiths R, Straub AC, Penuela S, Leitinger N, Laird DW, Bayliss DA, Isakson BE (2012). S-nitrosylation inhibits pannexin 1 channel function. J Biol Chem 287, 39602–39612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masood TB, Sandhya S, Chandra N, Natarajan V (2015). CHEXVIS: a tool for molecular channel extraction and visualization. BMC Bioinformatics 16, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molica F, Morel S, Meens MJ, Denis JF, Bradfield PF, Penuela S, Zufferey A, Monyer H, Imhof BA, Chanson M, et al. (2015). Functional role of a polymorphism in the Pannexin1 gene in collagen-induced platelet aggregation. Thromb Haemost 114, 325–336. [DOI] [PubMed] [Google Scholar]
- Moremen KW, Tiemeyer M, Nairn AV (2012). Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol 13, 448–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits U, Ahrens CH, Muller S, Wollscheid B (2014). Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886. [DOI] [PubMed] [Google Scholar]
- Panchin Y, Kelmanson I, Matz M, Lukyanov K, Usman N, Lukyanov S (2000). A ubiquitous family of putative gap junction molecules. Curr Biol 10, R473–R474. [DOI] [PubMed] [Google Scholar]
- Pelegrin P, Surprenant A (2009). The P2X(7) receptor-pannexin connection to dye uptake and IL-1beta release. Purinergic Signal 5, 129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini-Calace M, Maiwald T, Thornton JM (2009). PoreWalker: a novel tool for the identification and characterization of channels in transmembrane proteins from their three-dimensional structure. PLoS Comput Biol 5, e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penuela S, Bhalla R, Gong XQ, Cowan KN, Celetti SJ, Cowan BJ, Bai D, Shao Q, Laird DW (2007). Pannexin 1 and pannexin 3 are glycoproteins that exhibit many distinct characteristics from the connexin family of gap junction proteins. J Cell Sci 120, 3772–3783. [DOI] [PubMed] [Google Scholar]
- Penuela S, Bhalla R, Nag K, Laird DW (2009). Glycosylation regulates pannexin intermixing and cellular localization. Mol Biol Cell 20, 4313–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penuela S, Gyenis L, Ablack A, Churko JM, Berger AC, Litchfield DW, Lewis JD, Laird DW (2012). Loss of pannexin 1 attenuates melanoma progression by reversion to a melanocytic phenotype. J Biol Chem 287, 29184–29193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penuela S, Kelly JJ, Churko JM, Barr KJ, Berger AC, Laird DW (2014). Panx1 regulates cellular properties of keratinocytes and dermal fibroblasts in skin development and wound healing. J Invest Dermatol 134, 2026–2035. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004). UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Poornima V, Vallabhaneni S, Mukhopadhyay M, Bera AK (2015). Nitric oxide inhibits the pannexin 1 channel through a cGMP-PKG dependent pathway. Nitric Oxide 47, 77–84. [DOI] [PubMed] [Google Scholar]
- Qu Y, Misaghi S, Newton K, Gilmour LL, Louie S, Cupp JE, Dubyak GR, Hackos D, Dixit VM (2011). Pannexin-1 is required for ATP release during apoptosis but not for inflammasome activation. J Immunol 186, 6553–6561. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pupo RE, Johnston D, Penuela S (2018). N-Glycosylation regulates Pannexin 2 localization but is not required for interacting with Pannexin 1. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandilos JK, Chiu YH, Chekeni FB, Armstrong AJ, Walk SF, Ravichandran KS, Bayliss DA (2012). Pannexin 1, an ATP release channel, is activated by caspase cleavage of its pore-associated C-terminal autoinhibitory region. J Biol Chem 287, 11303–11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Q, Zhang Z, Shi J, Sun X, Li B, Yan Z, Xue S, Ai A, Lyu Q, Li W, et al. (2019). A pannexin 1 channelopathy causes human oocyte death. Sci Transl Med 11. [DOI] [PubMed] [Google Scholar]
- Schiffrin B, Radford SE, Brockwell DJ, Calabrese AN (2020). PyXlinkViewer: A flexible tool for visualization of protein chemical crosslinking data within the PyMOL molecular graphics system. Protein Sci 29, 1851–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seror C, Melki MT, Subra F, Raza SQ, Bras M, Saidi H, Nardacci R, Voisin L, Paoletti A, Law F, et al. (2011). Extracellular ATP acts on P2Y2 purinergic receptors to facilitate HIV-1 infection. J Exp Med 208, 1823–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Q, Lindstrom K, Shi R, Kelly J, Schroeder A, Juusola J, Levine KL, Esseltine JL, Penuela S, Jackson MF, Laird DW (2016). A Germline Variant in the PANX1 Gene Has Reduced Channel Function and Is Associated with Multisystem Dysfunction. J Biol Chem 291, 12432–12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer CC, Deloukas P, Hunt S, Mullikin J, Myers S, Silverman B, Donnelly P, Bentley D, McVean G (2006). The influence of recombination on human genetic diversity. PLoS Genet 2, e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierlin FB, Molica F, Reny JL, Kwak BR, Fontana P (2017). Pannexin1 single nucleotide polymorphism and platelet reactivity in a cohort of cardiovascular patients. Cell Commun Adhes 23, 11–15. [DOI] [PubMed] [Google Scholar]
- Takahata N, Lee SH, Satta Y (2001). Testing multiregionality of modern human origins. Mol Biol Evol 18, 172–183. [DOI] [PubMed] [Google Scholar]
- Thompson RJ, Zhou N, MacVicar BA (2006). Ischemia opens neuronal gap junction hemichannels. Science 312, 924–927. [DOI] [PubMed] [Google Scholar]
- Vanden Abeele F, Bidaux G, Gordienko D, Beck B, Panchin YV, Baranova AV, Ivanov DV, Skryma R, Prevarskaya N (2006). Functional implications of calcium permeability of the channel formed by pannexin 1. J Cell Biol 174, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ambrosi C, Qiu F, Jackson DG, Sosinsky G, Dahl G (2014). The membrane protein Pannexin1 forms two open-channel conformations depending on the mode of activation. Sci Signal 7, ra69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Dahl G (2010). SCAM analysis of Panx1 suggests a peculiar pore structure. J Gen Physiol 136, 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46, W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Yang X, Shi X, Chen Y (2015). Pannexin1 silencing inhibits the proliferation of U87MG cells. Mol Med Rep 11, 3487–3492. [DOI] [PubMed] [Google Scholar]
- Weilinger NL, Lohman AW, Rakai BD, Ma EM, Bialecki J, Maslieieva V, Rilea T, Bandet MV, Ikuta NT, Scott L, et al. (2016). Metabotropic NMDA receptor signaling couples Src family kinases to pannexin-1 during excitotoxicity. Nat Neurosci 19, 432–442. [DOI] [PubMed] [Google Scholar]
- Weilinger NL, Tang PL, Thompson RJ (2012). Anoxia-induced NMDA receptor activation opens pannexin channels via Src family kinases. J Neurosci 32, 12579–12588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Baran J, Cros A, Guberman JM, Haider S, Hsu J, Liang Y, Rivkin E, Wang J, Whitty B, et al. (2011). International Cancer Genome Consortium Data Portal–a one-stop shop for cancer genomics data. Database (Oxford) 2011, bar026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.