Abstract
Coat protein complex II (COPII) factors mediate cargo export from the endoplasmic reticulum (ER), but bulky collagens and lipoproteins are too large for traditional COPII vesicles. Mammalian CTAGE5 and TANGO1 have been well characterized individually as specialized cargo receptors at the ER that function with COPII coats to facilitate trafficking of bulky cargoes. Here, we present a genetic interaction study in zebrafish of deletions in ctage5, tango1, or both to investigate their distinct and complementary potential functions. We found that Ctage5 and Tango1 have different roles related to organogenesis, collagen versus lipoprotein trafficking, stress-pathway activation, and survival. While disruption of both ctage5 and tango1 compounded phenotype severity, mutation of either factor alone revealed novel tissue-specific defects in the building of heart, muscle, lens, and intestine, in addition to previously described roles in the development of neural and cartilage tissues. Together, our results demonstrate that Ctage5 and Tango1 have overlapping functions, but also suggest divergent roles in tissue development and homeostasis.
INTRODUCTION
Secretory proteins are trafficked by coat protein complex II (COPII) vesicles that form at specialized sites of the endoplasmic reticulum (ER) called ER-exit sites (ERES; Bonifacio and Glick, 2004; Lee et al., 2004). These sites contain conserved proteins such as SEC12, SEC16, SEC13, SEC31, SEC23, SEC24, and SAR1, among others that are important for COPII-mediated trafficking (Sato & Nakano, 2005; Watson et al., 2006; Townley et al., 2008; Kim et al., 2012; Saito et al., 2014; Maeda et al., 2017). Some bulky molecules are too large to fit into traditional COPII vesicles, which are 60–90 nm (Miller and Schekman, 2013; Malhotra and Erlman, 2015). Recently, a transport system using specialized cargo receptors, the CTAGE5 (MEA6)/TANGO1(MIA3) family, has been described for large molecules such as collagens, chylomicrons, and very–low density lipoproteins (VLDLs; Saito et al., 2009; Saito et al., 2011; Wilson et al., 2011; Maeda et al., 2016; Maiers et al., 2016; Santos et al., 2016; Rios-Barrera et al., 2017).
Evolution has created significant diversity in the CTAGE5/TANGO1 protein families. The human CTAGE5 and TANGO1 family is transcribed at the MIA2 and MIA3 gene loci, respectively. The MIA2 locus transcribes MIA2, CTAGE5, and TALI isoforms and the MIA3 locus transcribes TANGO1S (short) and TANGO1L (long) isoforms (reviewed in McCaughey and Stephens, 2019). Human and mouse studies have suggested that CTAGE5 and TANGO1L are expressed ubiquitously, whereas MIA2 and TALI expression are more limited, based on tissue transport requirements (Pitman et al., 2011; Santos et al., 2016). TANGO1S was only recently characterized (Maeda et al., 2016) and its expression across tissues is still unknown. Ensemble transcript analysis revealed that the zebrafish mia2 gene locus only transcribes ctage5 (mia2-205) and the mia3 gene locus transcribes tango1s (mia3-201) and tango1l (mia3-202; Supplemental Figure S1). The simplified protein diversity of Ctage5/Tango1 in zebrafish compared with humans allows investigation of their complementary and divergent functions. In the current study, we specifically investigate deletion of ctage5 and tango1l separately and together.
Members of the CTAGE5/TANGO1 family form a ring-like structure at the base of ERES vesicles and promote their expansion to accommodate large cargo (Lui et al., 2017; Raote et al., 2017; Reynolds et al., 2019) or potentially facilitate defined tubular connections for transfer of material to the cis-Golgi (Kurokawa et al., 2014; Raote and Malhotra, 2019; Raote et al., 2020). In vitro studies have suggested some redundant functions of the CTAGE5/TANGO1 family for transport of some collagens (Maeda et al., 2016), but unique functions for trafficking others (Santos et al., 2016). The families may also act cooperatively for transport of other large molecules such as VLDLs and chylomicrons (Santos et al., 2016). In vivo studies of TANGO1/CTAGE5 family members have been relatively limited, likely because in mice Tango1 deletion is perinatally lethal (Wilson et al., 2011) and Ctage5 deletion is essential for early embryonic development (Wang et al., 2016). Tango1l deletion caused buildup of multiple collagen subtypes in chondrocytes, fibroblasts, endothelial cells, and mural cells (Wilson et al., 2011). Conditional knockout of Ctage5 in neurons of mice show that in addition to secretory components, nonsecretory protein trafficking is also affected (Zhang et al., 2018). Similarly, Drosophila studies have shown that secretion of multiple extracellular proteins including mucins, laminins, and perlecan is disrupted after loss of Tango1 (Petley-Ragan et al., 2016; Liu et al., 2017; Rios-Barrera et al., 2017; Reynolds et al., 2019). The specific mechanisms underlying the broad disruption of ER trafficking are still being investigated. Of significance, however, studies of Ctage5 and Tango1 deletions within a single animal model have not been reported, highlighting a need to investigate their potential redundancies or unique functions in different cell types.
When trafficking dynamics is altered at the endoplasmic reticulum, stress pathways within the cell can be activated (Walter and Ron, 2011). This coordinated response has three major branches: the ATF6, PERK, and XBP1 pathways. After TANGO1 knockdown, the ER is distended (Wilson et al., 2011; Petley-Ragan et al., 2016; Rios-Barrerra et al., 2017; Maiers et al., 2016) and XBP1 and ATF4 ER stress pathways are activated, but ATF6 was not investigated (Maiers et al., 2016; Petley-Ragan et al., 2016). CTAGE5 KO can also cause ER stress in pancreatic islets (Fan et al., 2017). With buildup of material at the ER and ER stress, autophagy is also commonly activated (Kabir et al., 2018). After loss of TANGO1 in HEPG2 or Caco-2 cells, APOB accumulates intracellularly and is targeted for lysosomal degradation (Santos et al., 2016). Furthermore, CTAGE5 was shown as essential for starvation-induced autophagosome formation in HeLa cells, acting at the ERES for membrane donation (Ge et al., 2017). The role of TANGO1, however, was not investigated in that study. Interestingly, noncanonical autophagy can directly remove collagen from ERESs to maintain homeostasis (Omari et al., 2018). Cumulatively, the literature suggests that protein trafficking and membrane donation functions of Ctage5 and Tango1 could be closely related to stress pathway activation and cellular homeostasis (McCaughey and Stephens, 2019).
Zebrafish have been used previously to study COPII proteins, including the feelgood (creb3l2; Melville et al., 2011), and crusher (sec23; Lang et al., 2006) mutants, but neither Ctage5 nor Tango1 has been investigated in this model system. In this study, we generated ctage5 and tango1l mutant zebrafish and characterized phenotypes in the single- as well as double-homozygous mutant animals. We find that like other COPII pathway mutants, ctage5 and tango1 mutant zebrafish are small and have decreased survival. Overall, our studies reveal that Ctage5 and Tango1 have complementary, but also divergent roles in craniofacial development, collagen and lipoprotein trafficking, and in activation of cellular stress pathways.
RESULTS
Morphogenesis and survival are differentially affected by ctage5 or tango1l deletion
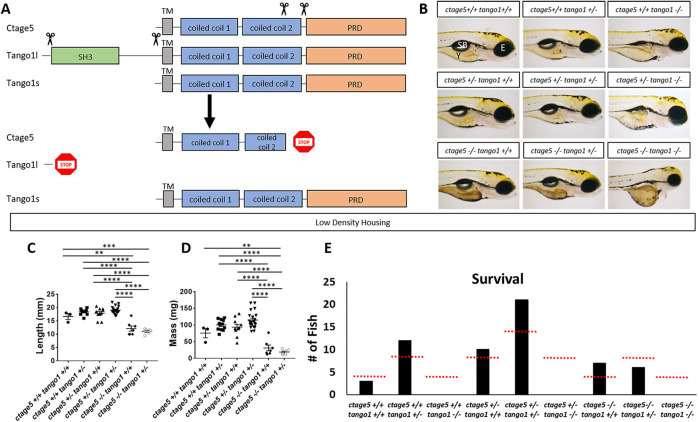
To investigate how disruption of ctage5 and tango1 affects zebrafish development and survival, large portions of genomic coding sequence were deleted with Crispr/Cas9, eliminating critical functional domains (Figure 1A). At the mia2 gene locus, transcripts of the ctage5 mutant variant were decreased twofold compared with wild-type ctage5 mRNA (Supplemental Figure S2). For the mia3 gene locus, the deletion specifically eliminates the genomic region that transcribes and translates the unique luminal portion of tango1l, sparing the tango1s transcript. Primers designed to measure tango1s mRNA did not reveal a change between wild-type and tango1l mutant embryos, suggesting that Tango1s protein could still be made (Supplemental Figure S2). Furthermore, our analysis indicated there was no compensatory up-regulation of ctage5 or tango1 expression with deletion of the other gene (Supplemental Figure S2). Therefore, our study is designed to investigate deletion of ctage5 or tango1l (subsequently referred to as tango1).
FIGURE 1:
ctage5 and tango1 mutations affect size and survival of zebrafish: (A) Schematic showing CRISPR guide RNA cut sites (scissors) and the estimated final truncated proteins. (B) Representative brightfield images of all combinations of ctage5 and tango1 mutations in 4dpf zebrafish larvae. Y = yolk, SB = swim bladder, E = eye. (C) Length (one-way ANOVA, F = 38.6, p < 0.0001), (D) mass (one-way ANOVA, F = 28.22, p < 0.0001) and (E) survival (χ2 = 24.59, p < 0.05, n = 59) measurements of all combinations of ctage5 and tango1 mutant zebrafish at 2 mo raised in a low-density environment (about 10 fish per tank). Red-dotted lines in E represent expected survival, and black bars are actual survival. ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
During embryogenesis, ctage5 mutants look normal except for having darker yolk (Figure 1B). When raised at low rearing density (10 fish per tank), ctage5 mutants are shorter (Figure 1C) and have decreased mass (Figure 1D) but can survive to expected numbers (Figure 1E). When raised at a high density (30 fish per tank), however, mutants have reduced survival (Supplemental Figure S3B). Regardless of rearing density, most ctage5 mutants die by 1 year, while heterozygous and wild-type siblings show normal survival past 3 years. In contrast to ctage5 mutant embryos, tango1 mutants are significantly shorter, have obvious craniofacial defects, as the mouth fails to protrude past the eyes, and do not survive to adulthood regardless of rearing density (Figure 1, B and E; Figure S3A). The ctage5;tango1 double mutants have similar, but more severe phenotypes compared with single mutants and also fail to survive to adulthood (Figure 1, B and E). Together, these results suggest that ctage5 and tango1 mutations differentially affect zebrafish during development and homeostasis, pointing to potential differences in cellular function.
Craniofacial and other morphological defects in tango1 mutants are accentuated with loss of ctage5
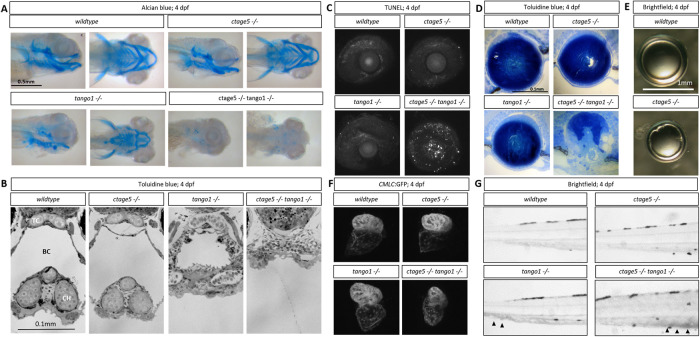
Mutations in COPII trafficking pathway components commonly cause craniofacial defects (Lang et al., 2006; Townley et al., 2008; Melville et al., 2011). For example, in mice, loss of Tango1 causes dwarfed embryos with defects in cartilage formation and bone mineralization (Wilson et al., 2011). Alcian blue staining confirmed that like mice, zebrafish tango1 mutant embryos have defects in cartilage formation at 4dpf (Figure 2A). ctage5;tango1 double mutants have a near-complete absence of cartilage staining (Figure 2A). Histology also revealed severe craniofacial defects in tango1 mutant embryos that were exacerbated by ctage5 deletion (Figure 2B). Interestingly, ocular lens defects exemplified by increased cell death and a degenerative lens were only observed in ctage5;tango1 double mutant embryos (Figure 2, C and D). Although ctage5 mutant lenses appear normal during development, by 2 months lens epithelial defects were observed (Figure 2E), suggesting a lens homeostasis role for Ctage5. Cardiac chamber dysgenesis was another unique phenotype to ctage5;tango1 double mutants. While both ctage5 and tango1 mutants showed normal heart development, severe cardiac structural changes were apparent by 4dpf when these genes were disrupted in tandem (Figure 2F). However, blood pooling in the caudal vein was noticed in both tango1 single and ctage5;tango1 double mutant larvae, suggesting that tango1 mutation could be driving cardiovascular defects, which only become obvious with the additional deletion of ctage5 (Figure 2G). These results extend previous in vivo observations demonstrating that Tango1 plays a critical role in morphogenesis of multiple tissues. However, Ctage5 also acts in many developmental processes, as phenotypes observed in tango1 mutants are enhanced by ctage5 deletion.
FIGURE 2:
Craniofacial and other morphological defects in tango1 mutants are accentuated by ctage5 deletion. (A) Representative lateral (left) and ventral (right) images of Alcian blue–stained 4dpf larvae. (B) Representative images of plastic-sectioned and toluidine blue–stained 4dpf larvae cropped to show craniofacial alterations. TC = trabeculae, BC = buccal cavity, CH = ceratohyal cartilage. (C) Representative maximum intensity projection images showing TUNEL staining of 4dpf lens and sclera. (D) Representative images of plastic-sectioned and toluidine blue–stained 4dpf lens. (E) Representative images of lens dissected from 3-mo-old zebrafish. (F) Representative images showing ventricle-enriched cardiac myosin light chain (CMLC:GFP) expression. (G) Representative brightfield image of the trunk for each genotype. Arrowheads detail blood pooling in the caudal vein.
Large-molecule trafficking defects differ between ctage5 and tango1 mutants
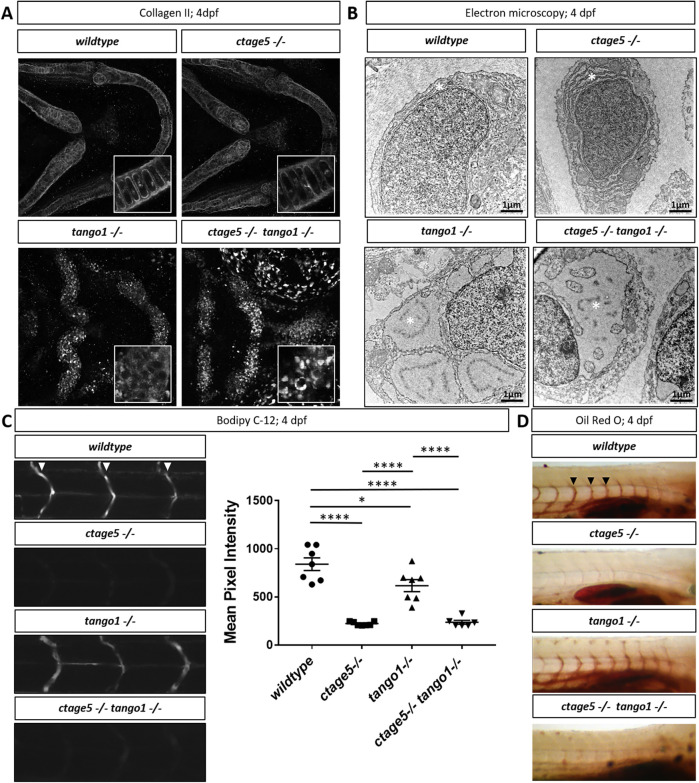
TANGO1 has been well established as a cargo receptor for collagen trafficking in various cell types in vitro and in vivo (Saito et al., 2009; Wilson et al., 2011; Maeda et al., 2016; Maiers et al., 2016; Gorur et al., 2017; Rios-Barrera et al., 2017). CTAGE5 is necessary for collagen VII trafficking in an epidermal carcinoma human cell line (Saito et al., 2011), but its role in secreting other collagens and in other cell lines or in vivo is still largely unknown. Lipoprotein trafficking is also altered with loss of CTAGE5 or TANGO1 (Santos et al., 2016; Wang et al., 2016; Zhang et al., 2018). To begin investigating large-molecule trafficking in zebrafish, we performed wholemount staining for Collagen II. The branchial arch cartilage of wild-type and ctage5 mutant animals showed extracellular Collagen II surrounding chondrocytes. In contrast, in tango1 and double mutants, Collagen II was found in intracellular puncta within chondrocytes (Figure 3A). With decreased trafficking of large molecules, we hypothesized that protein buildup occurred within the ER, which would be consistent with previous reports of distended ER after loss of Tango1 (Wilson et al., 2011; Petley-Ragan et al., 2016; Rios-Barrerra et al., 2017; Maiers et al., 2016). Indeed, we observed distended ER in tango1 and double mutant chondrocytes, while the ER in ctage5 mutants and wild-type chondrocytes was normal (Figure 3B). We next investigated lipoprotein trafficking using a lipid analog, BODIPY-c12, that has previously been shown to require β-lipoproteins for trafficking from the yolk into the zebrafish vasculature (Miyares et al., 2014). Efficient β-lipoprotein trafficking was observed in wild-type larvae by monitoring BODIPY-c12 vascular fluorescence following injection into the yolk. However, BODIPY-c12 trafficking was decreased in tango1 mutants, and nearly undetectable in ctage5 or double mutant embryos (Figure 3C). Oil Red O staining confirmed this significant loss of serum lipids in ctage5 single and ctage5;tango1 double mutant embryos (Figure 3D). Together these results suggest that lipoprotein trafficking is significantly affected after deletion of Ctage5, and to a lesser extent with loss of Tango1, consistent with results reported from the Malhotra Lab (Santos et al. 2016). Conversely, in cell types that secrete large amounts of collagen, such as chondrocytes, deletion of tango1, but not ctage5 severely affected protein export and ER maintenance.
FIGURE 3:
Trafficking differences in ctage5 vs. tango1 mutant embryos. (A) Representative 40× images showing collagen 2 wholemount immunohistochemistry in 4dpf larvae chondrocytes (n = 3–4 per group). Inserts are 40× images with 3× zoom and cropped to show higher-resolution chondrocyte collagen 2 staining. (B) Representative electron microscopy images of endoplasmic reticulum (*) in 4dpf larvae chondrocytes (n = 3 per group). (C) Representative images and quantification of BODIPY C-12 fluorescence in the intersegmental vessels (arrowheads; one-way ANOVA, F = 39.36, p < 0.0001). (D) Representative images of Oil Red O staining for lipids in 4dpf zebrafish larvae intersegmental vessels (arrowheads). * = p < 0.05, **** = p < 0.0001.
Cell stress is differentially activated in tissues after ctage5 or tango1 deletion
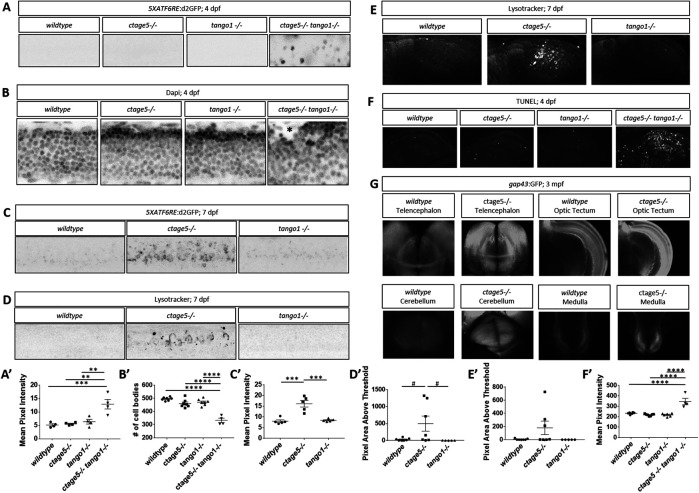
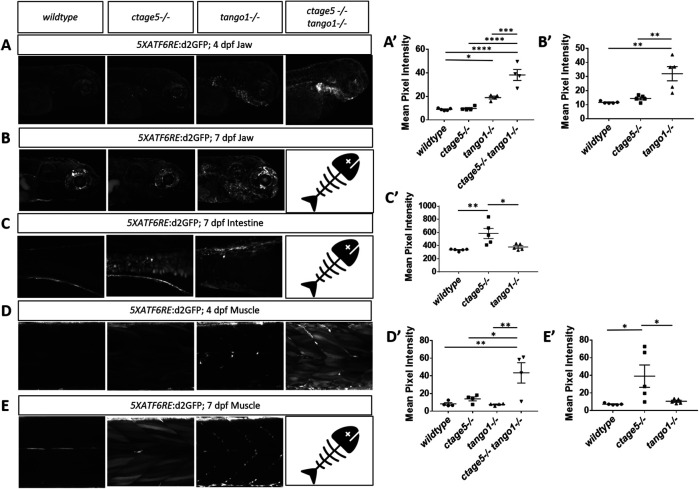
When cellular trafficking is affected and cargo builds up at the ER, the evolutionarily conserved unfolded protein response (UPR) can be activated (Walter and Ron, 2011). Numerous reports suggest ER stress and autophagy can be altered after deletion of CTAGE5 or TANGO1 (Wilson et al., 2011; Maiers et al., 2016; Petley-Ragan et al., 2016; Santos et al., 2016; Fan et al., 2017; Ge et al., 2017; Rios-Barrerra et al., 2017), but a direct comparison of either factor on the ATF6 ER stress or autophagy has not been carried out. We recently described a transgenic zebrafish line to measure the ATF6 activation, which mediates the major transcriptional response to ER stress (Clark et al., 2020). Using these transgenic fish, we observed that ctage5-/-; tango1-/- larvae have significantly increased ATF6 reporter fluorescence in the spinal cord at 4dpf (Figure 4, A and A’). As assessed by nuclei packing and cell body morphology, spinal cord cells in the double mutants are disorganized and in some instances mislocalized from the normal lamination pattern (Figure 4, B and B’, asterisk). Although spinal cord cells of ctage5 single mutants were unaffected at 4dpf, by 7dpf these larvae had significantly increased ATF6 reporter activation (Figure 4, C and C’). Furthermore, 7dpf ctage5 mutant larvae had increased lysosomal volume in the spinal cord and brain compared with wild-type or tango1 mutants (Figure 4, D, D’, E, and E’). In addition, within the brain, ctage5;tango1 double mutant embryos showed significant increase in cell death markers, while single mutations were similar to wild-type brains (Figure 4, F and F’). As adults, ctage5 mutant zebrafish show elevated gap43:GFP expression (Figure 4G). Expression of gap43 is known to be induced with neuronal stress (Skene, 1989; Benowitz and Routtenberg, 1997; Bormann et al., 1998; Kaneda et al., 2008; Udvadia 2008; Diekmann et al., 2015). Therefore, ctage5 mutant larvae undergo cellular stress throughout the nervous system, as evidenced by activated ER stress, altered cell morphology, and elevated autophagy and apoptosis, which continue into adulthood. Our observations are consistent with the axonal trafficking defects described in mice with conditional Ctage5 deletion from the nervous system (Zhang et al., 2018). We also noticed ER stress in other tissues including the jaw in tango1 mutant and double mutant larvae, but not in ctage5 mutant embryos, consistent with craniofacial defects observed in Figure 2 (Figure 5, A, A’, B, and B’). Conversely, ER stress was observed in the intestine of ctage5 mutant larvae but not tango1 mutants at 7dpf (Figure 5, C and C’). Within trunk skeletal muscle, both ctage5 and tango1 mutant embryos showed ATF6-ER stress (Figure 5, D, D’, E, and E’). Interestingly, however, ctage5 mutants show significantly elevated ATF6 activation in the muscle fibers, whereas in tango1 mutants the ER stress is activated along cells lining the chevron-shaped muscle segments. The role for Ctage5 or Tango1 in muscle tissue has not been investigated previously, but these data suggest that Ctage5/Tango1 family of proteins also have divergent functions in that tissue type.
FIGURE 4:
Stress pathway activation in the nervous system is specific for ctage5 -/- and amplified by tango mutation. (A, A’) Representative images, A, and quantification, A’, of 5XATF6RE:d2GFP activation in 4dpf spinal cords (one-way ANOVA, F = 12.42, p = 0.0005). (B, B’) Representative images of DAPI staining, B, and quantification of brightfield images, B’, in 4dpf spinal cords to show cellular organization (asterisk shows cells mislocalized from the normal lamination pattern; one-way ANOVA, F = 37.51, p < 0.0001). (C, C’) Representative images, C, and quantification, C’, of 5XATF6RE:d2GFP activation in 7dpf spinal cords (one-way ANOVA, F = 22.22, p < 0.0001). (D, D’) Representative images, D, and quantification, D’, of the lysotracker staining area above threshold for acidic compartments in 7dpf spinal cords (one-way ANOVA, F = 3.881, p = 0.0423). (E, E’) Representative images, E, and quantification, E’, of the lysotracker staining area above threshold of lysotracker for acidic compartments in 7dpf brains (one-way ANOVA, F = 2.479, p = 0.1154). (F, F’) Representative images, F, and quantification, F’, of TUNEL staining for cell death in 4dpf brains (one-way ANOVA, F = 24.33, p < 0.0001). (G) Representative images of gap43:GFP in 3–mo old PACT cleared ctage5 wildtype (WT) or homozygous mutant brains. # = p < 0.08, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
FIGURE 5:
ER stress is prevalent in tissues outside of the nervous system. Representative images and quantification of 5XATF6RE:d2GFP ER stress in the jaw at 4dpf (A, A’; one-way ANOVA, F = 34.44, p < 0.0001) and 7dpf (B, B’; one-way ANOVA, F = 13.86, p < 0.0008), intestine at 7dpf (C, C’; one-way ANOVA, F = 8.358, p = 0.0053), and muscle at 4dpf (D, D’; one-way ANOVA, F = 8.205, p = 0.0031) and 7dpf (E, E’: F = 5.624, p = 0.0189). * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.  = indicates ctage5-/- tango1-/- mutants do not survive to 7dpf.
= indicates ctage5-/- tango1-/- mutants do not survive to 7dpf.
DISCUSSION
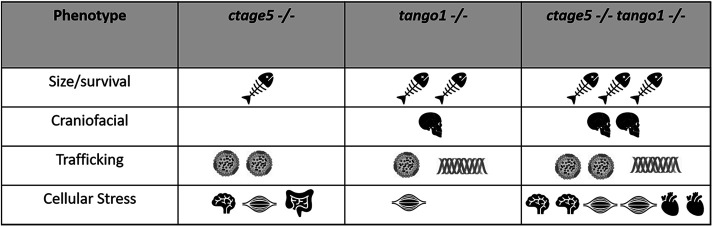
CTAGE5 and TANGO1 are both known to act in the COPII trafficking pathway (Saito et al., 2009, 2011), but until now the effects of deleting these proteins in the same model system have not been investigated. Our results suggest that zebrafish Ctage5 and Tango1 have complementary, but also divergent roles across multiple tissues (Figure 6). For example, we found that both factors affect fish growth when deleted. tango1 mutant fish are shorter at larval stages, whereas ctage5 mutant larvae are morphologically normal initially, but those that survive are much smaller than their wild-type siblings. Consistent with our results, global knockout of Tango1 inhibits growth in mouse embryos (Wilson et al., 2011). Interestingly, conditional knockout of Ctage5 in the mouse brain or liver also results in decreased animal size (Wang et al., 2016; Zhang et al., 2018), indicating that Ctage5 function in multiple organ systems is important for embryonic growth and development.
FIGURE 6:
Summary of phenotype magnitude in mutant fish. Table summarizing phenotype severity. Any symbol represents an elevated severity compared with wild-type. More symbols represent increased severity.  Survival/size defects,
Survival/size defects, 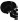 craniofacial defects,
craniofacial defects,  lipoprotein,
lipoprotein,  collagen,
collagen, 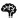 brain,
brain,  muscle,
muscle, 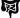 intestine,
intestine,  heart.
heart.
More striking differences in tango1 versus ctage5 mutant phenotypes were characterized for collagen secretion. Collagen II export was blocked in tango1 mutant larvae, most notably in the chondrocytes, resulting in distended ER and craniofacial defects. ctage5 mutants, however, have normal collagen II trafficking in chondrocytes and craniofacial structures appeared normal. These results are in line with findings from Wilson et al. (2011), where trafficking of multiple collagens were affected with loss of TANGO1 resulting in severe craniofacial defects. In our study, we found similar results by investigating Collagen II trafficking. Prior to our study, the role of CTAGE5 in collagen trafficking in vivo had not been investigated. In vitro studies, however, showed that collagen VII trafficking is disrupted by deletion of CTAGE5 (Saito et al., 2011). Our results suggest that collagen II trafficking is not affected with ctage5 deletion alone, indicating that Tango1 has a more significant role in Collagen II trafficking and craniofacial development.
In contrast to collagen trafficking, lipoprotein trafficking was predominantly affected by mutation to ctage5, where lipoprotein trafficking from the yolk into the zebrafish embryo was completely disrupted. Previous studies have shown that lipoprotein trafficking and lipid profiles are altered with loss of CTAGE5 or TALI (Wang et al., 2016; Santos et al., 2016, Zhang et al., 2018). Our results are consistent with these studies and extend these findings through a direct comparison of Ctage5 and Tango1 for lipid trafficking in vivo. These results suggest that Tango1 plays a primary role in collagen trafficking, whereas Ctage5 is more important for lipoprotein trafficking.
While our studies addressed the shared and unique functions between Ctage5 and Tango1 with respect to trafficking and cellular homeostasis, the experiments in zebrafish also demonstrated that Tango1 plays an important role in maintaining ER-Golgi morphology, consistent with observations by others (Bard et al., 2006; Liu et al., 2017; Rios-Barrerra et al., 2017; Reynolds et al., 2019). CTAGE5 also remodels ER-exit sites and plays a role in the ER and ER Golgi intermediate compartment (ERGIC), creating an autophagy supply pathway (Ge et al., 2017). In this regard, our observation of increased lysosomal volume in ctage5 mutants is consistent with compensatory up-regulation of lysosome-mediated protein degradation. As lysosomal volume was unchanged in tango1 mutants, these data suggest a specific role of Ctage5 in autophagosome formation. Overall, our data confirm and extend the important, but surprisingly divergent and cell type–specific roles of Ctage5 and Tango1 in large–cargo trafficking and intracellular membrane dynamics.
In our experiments, there were some limitations on separating complementary and divergent functions from tissue and cellular expression differences. In mouse and human studies, Tango1 and Ctage5 are expressed ubiquitously (Maeda et al., 2016; Santos et al., 2016). Bulk tissue RNAseq (Askary et al., 2017; White et al., 2017; Burkhard and Bakkers, 2018; San et al., 2018) and the scRNA seq EMBL-EBL atlas also suggest ubiquitous expression, but that different cell types within a tissue could have different expression levels of ctage5 and tango1. The cell-type differences in expression levels could be coupled to cellular trafficking demand, potentially explaining some of the differences in phenotypes with loss of either ctage5 or tango1. Zebrafish also receive maternal (wild-type) proteins from the yolk, which can complicate loss-of-function phenotypes in embryos derived from heterozygous incrosses. However, yolk trafficking defects observed in the mutants indicate maternal protein rescue is not a significant confound. Finally, it is possible that Tango1s protein is generated in the tango1 mutant used for analysis (Supplemental Figure S1). In future studies, it will be interesting to address potential functional redundancy and differences in Tango1l versus Tango1s.
With the described results and limitations in mind, there are a couple of hypotheses for complementary and divergent functions at the ER: 1) Expression-based explanation: Some cell types express more ctage5 or tango1, and so different proportions of protein are localized at the ER. Loss of the predominant protein would have the largest effect on the cell type, while loss of the minor abundant protein may have a smaller effect. 2) Function-based explanation: 2A) ER subdomains: Even if ctage5 and tango1 are expressed at similar levels, one protein could predominate at ERESs or subdomains within an ERES (Maeda et al., 2019). ER domains that are more concentrated with either Ctage5 or Tango1 could be related to the primary molecule being secreted from that domain. Deletion of one factor could cause a specific effect through elimination from the subdomain, but residual activity could persist until the other factor is deleted causing a complementary effect. 2B) ERES interactions: Ctage5 and Tango1 could always be located in the same ERES but interact in different homo- and heterodimers, resulting in divergent functions. Therefore, deletion of one component could eliminate a subset of interactions and lead to specific trafficking defects. Deletion of an additional factor would eliminate more complexes at the ERES causing a complementary effect. Aside from interactions within the family, CTAGE5 and TANGO1 have unique interactions with many other protein families (Stark et al., 2006), further supporting specific cellular functions. Investigating expression coupled with specific interactions and ERES subdomain localization in the cell types that were described in this study will be an interesting to pursue with future experimentation.
Several observations from our studies have relevance to human disease. The nervous system was severely affected by ctage5 deletion, consistent with descriptions in mice (Zhang et al., 2018). We found neuronal stress was not observed with deletion of tango1 alone, but ctage5 mutant–dependent stress was exacerbated with tango1 deletion. Interestingly, in humans, the CTAGE5 variant P521A was identified as a risk factor for Fahr’s disease, a rare neurologic disorder (Oliveira et al., 2007; Lemos et al., 2011). Furthermore, by investigating organ systems not previously characterized, we discovered that tango1 and ctage5 mutant cells in the intestine, muscle, and heart show elevated ER stress and morphological abnormalities. Cardiac defects in tango1 mutants are particularly interesting as the European Biomedical Institute GWAS catalogue reports that multiple heart diseases, including coronary artery disease, coronary heart disease, and myocardial infarction, are associated with TANGO1 intron variants (Buniello et al., 2019). Very recently, homozygous hypomorphic coding mutations in human TANGO1 were shown to result in collagen secretion defects leading to short stature and skeletal and neuronal defects, as well as insulin-dependent diabetes (Lekszas et al., 2020). Establishing the role of Ctage5 and Tango1 in multiple cellular processes and specific cell types in zebrafish suggests that this model system will be valuable to assess other candidate disease variants in human CTAGE5 and TANGO1.
In conclusion, our study uses the zebrafish model system to define the in vivo consequences when the cellular functions of cTAGE5 and TANGO1 are disrupted. Through this analysis, complementary and divergent roles for these proteins have been discovered, including identification of novel physiological processes and tissue dependences on these factors.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Mutant generation
Clustered regularly interspaced short palindromic repeats (CRISPR) guides were designed using ZiFiT Targeter Version 4.2 (http://zifit.partners.org/ZiFiT, in the public domain) against regions of zebrafish ctage5 exon14, 5′-GGCCGCAAGGGCAGCTGATC-3′, and exon18 5′-GGCCTCCTATATGGCAAACC-3′ (ensemble transcript mia2-205). CRISPR guides targeting tango1 were designed against exon1, 5′-TGGCTGCACCGAATAATCCC-3′, and exon7, 5′-ATCTGAAGTCGATGAGTCGG-3′ (ensemble transcript mia3-202). CRISPR gRNA templates were generated by cloning annealed oligonucleotides with appropriate overhangs into BsaI-digested pDR274 plasmid. CRISPR gRNAs were synthesized using a MEGAshortscript T7 Transcription Kit and purified using a mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA). Zebrafish codon-optimized cas9 was synthesized using a mMESSAGE mMACHINE Kit (Ambion) and polyadenylated using a Poly(A) Tailing Kit (Ambion). CRISPR gRNAs and cas9 mRNA were coinjected into one- to four-cell zebrafish embryos from wild-type ZDR fish maintained internally in the Link lab at 10 and 50 ng/μl, respectively, and surviving embryos were raised to adulthood before outcrossing to identify the founder fish carrying germline edits in ctage5 or tango1. Offspring from these fish were raised to adulthood and then fin-clipped for genotyping (see below for details). The resulting 6285-bp deletion ctage5 mutant and 14,043-bp deletion tango1 mutant described here were identified via sequencing (Retrogen, San Diego, California, USA).
Mutant and transgenic fish used for experiments:
ctage5 mutant: mia2mw91 (this study)
tango1l mutant: mia3mw92 (this study)
Tg(Tru.Gap43:EGFP) (Udvadia 2008)
Tg(Hsa.ATF6RE-fasab:d2eGFP)mw85Tg (Clark et al., 2020)
Genotyping
Genomic DNA was extracted from zebrafish tissue using a Puregene Core Kit (Qiagen, Hilden, Germany). The genomic region containing either the ctage5 or tango1 mutation was amplified by PCR. The PCR protocol utilized primers flanking the expected deletion to identify the mutant allele and one primer flanking the expected deletion and one inside the deleted region to identify the wildtype allele.
To identify the ctage5 wildtype allele, a forward primer for exon 13, 5′-AAGGCTCATGATAACTGG-3′, and a reverse primer for exon 14, 5′-GAGCGTTTTCTCTCTTGA-3′, creating a 171-bp amplicon. For the ctage5 mutant allele, the exon 13 forward primer was paired with a reverse primer for exon 18, 5′-AGGAAGTGGAGGTCTAGG-3′, creating a 204-bp amplicon. To identify the tango1 wildtype allele, a forward primer for exon 1, 5′-TGTTGGTGCTGCTATCTC-3′, and a reverse primer for exon 1, 5′-CTGCTGCATTCTTCATCCG-3′ were used to generate a 262-bp amplicon. For the tango1 mutant allele, the exon1 forward primer was paired with a reverse primer for exon 7, 5′-GTAGTACTCACAATCTCTCC-3′, creating a 237-bp amplicon.
Fish maintenance
Zebrafish (Danio rerio) were maintained at 28.5°C in an Aquatic Habitats recirculating filtered water system (Aquatic Habitats, Apopka, FL) in reverse-osmosis purified water supplemented with Instant Ocean salts (60 mg/l) on a 14-h light:10-h dark lighting cycle and fed a standard diet (Westerfield, 1995). All animal husbandry and experiments were approved and conducted in accordance with the guidelines set forth by the Institutional Animal Care and Use Committee of the Medical College of Wisconsin.
Survival analysis and size measurements
Zebrafish embryos were added to the Aquatic Habitats system at 5dpf at either high density (30 embryos per tank) or low density (10 embryos per tank). At 2 months, zebrafish were anesthetized in Tricaine and blotted with paper towels, mass length was calculated, and fin clips were collected. gDNA was extracted from the fin clips and genotyped.
Plastic sectioning and electron microscopy
4dpf embryos were fixed in cacodylate fix (0.1M cacodylate, 2% PFA, 2% glutaraldehyde [pH 7.4]) at 4°C overnight, postfixed in 1% osmium buffer solution, dehydrated through a series of methanol and acetonitrile washes, and infiltrated with Embed 812 resin (14120; Electron Microscopy Sciences, Hatfield, PA). Semithin transverse sections (1 µm) were cut with glass knives on a Leica RM2255 microtome, heat-fixed to glass slides, stained with 1% toluidine blue in 1% borax buffer, and imaged on a Nikon Eclipse E800 light microscope with an attached Sony DSC HX1 camera to capture craniofacial images. Plastic blocks were then submitted to the Electron Microscopy Facility at the Medical College of Wisconsin for electron microscopy. Ultrathin sections (70–80 nm) were collected using a DiATOME Ultra 45° diamond knife (MT7376; DiATOME), collected on copper hexagonal mesh coated grids (G200H-Cu; Electron Microscopy Sciences), and stained with uranyl acetate and lead citrate for contrast. Images were captured using a Hitachi H600 TEM microscope (Hitachi, Tokyo, Japan).
TUNEL
4dpf zebrafish embryos were fixed with 4% PFA overnight at 4°C. Fixed embryos were washed three times in phosphate-buffered saline (PBS) for 10 min, rinsed with dH20, and incubated with dH20 for 30 min at room temperature followed by acetone at –20°C for 30 min and 30 min dH20 to permeabilize embryos. Further, embryos were washed three times with PBST (PBS + 1% TritonX-100) and then incubated in 1 mg/ml Collagenase II in PBST for 90 min at room temperature. Embryos were washed three times for 15 min in PBST and TUNEL reaction mixture (Roche), or the label solution alone was added for 2 h at 37°C as a control. Embryos were washed 3 × 15 min in PBS and then imaged using confocal microscopy. Mean pixel intensity was measured using ImageJ (Rasband, W.S. ImageJ, U.S. National Institutes of Health, MD), and quantified using GraphPad Prism software (GraphPad, La Jolla, CA).
Alcian blue
Alcian blue cartilage staining was performed similarly to Hendee et al. (2018). 4dpf zebrafish embryos were fixed in 4% PFA overnight at 4°C. Fixed embryos were washed two times with 1× PBS for 5 min and bleached in 1 ml 10% hydrogen peroxide (H2O2) in ddH2O supplemented with 50 µl of 2M KOH for 1 h at room temperature with rotation. Embryos were stained with 0.1% Alcian blue solution composed of Alcian blue 8GX (Sigma-Aldrich, St. Louis, MO) dissolved in acidic ethanol. Stained embryos were briefly rinsed with acidic ethanol and then washed 4 × 60 min with acidic ethanol. Following washes, embryos were digested in 10 µg/mL proteinase K diluted in PBST (PBS + 0.1% Tween20) for 1 h at room temperature, refixed in 4% PFA, progressively dehydrated with ethanol, and stored in 80% glycerol. Wholemount images were obtained on a Leica MZFLIII dissecting scope with an attached Nikon E995 camera.
Lysotracker red
7dpf zebrafish were treated with 10 µM LysoTracker (LysoTracker Red DND-99; L7528) in 1-phenyl 2-thiourea (PTU) or 1% DMSO in PTU for control embryos for 1 h at 28.5°C. Embryos were washed 3 × 15 min with PTU and then fixed with 4% PFA and imaged using confocal microscopy. Using ImageJ, a threshold was set to identify only high lysotracker staining, and the area was measured. Data were processed using Microsoft Excel (Microsoft, Redmond, WA) and graphed using GraphPad Prism (GraphPad, La Jolla, CA).
Oil Red O
4dpf larvae were fixed with 4% PFA overnight at 4°C. Larvae were washed three times with PBS, rinsed with 60% isopropanol, and then incubated in 60% isopropanol for 30 min on a rotator at room temperature. Next, larvae were dyed in fresh filtered 0.3% Oil Red O in 60% isopropanol for 3 h and then washed multiple times with 60% isopropanol and dH20 until the nonspecific staining was removed. Washed larvae were mounted in agarose and imaged on a Leica MZFLIII dissecting scope with an attached Nikon E995 camera.
Immunofluorescence
4dpf larvae were fixed overnight with 4% PFA at 4°C. For notochord staining, larvae were left intact, but for chondrocyte staining, trunks were removed and used for DAPI staining and only the head was stained. Antigen retrieval was used for chondrocyte staining by incubating larvae in antigen retrieval buffer (150 mM Tris-HCL, pH 9), followed by heating to 70°C in antigen retrieval buffer for 15 min, and washed two times for 10 min in PBST (0.1–1% Triton in PBS) at room temperature. All following steps were completed on a rotator when possible. Larvae were permeabilized by washing once with dH20 followed by dH20 for 30 min at room temperature, acetone for –20°C for 30 min, dH20 for 30 min, and 10 µg/ml proteinase K in PBST at room temperature. Then 4% PFA was added for 20 min at room temperature to postfix and washed three times for 15 min in PBST. Larvae were incubated in PBDT buffer (1% DMSO, 1% BSA, 0.5–1% Triton X-100, 1× PBS) with 5% goat serum (NGS) for 60 min at room temperature. Collagen II (II-II6B3; 1:100; Developmental Studies Hybridoma Bank) was stained overnight in the chondrocytes in PBDT/5% NGS at 4°C. Larvae were washed with PBST six times for 15 min at room temperature and then incubated with secondary antibody (1:400) in 2% NGS/PBDT for 4 h at room temperature. Larvae were washed six times with PBST for 15–20 min, and if necessary, were stained with DAPI (1:1000 in PBST) for 30 min at room temperature and washed three times in PBS. Larvae were then mounted and imaged using confocal microscopy.
Adult lens dissection
Three-month-old zebrafish were fixed overnight with 2% PFA at 4°C. Zebrafish lenses were dissected from the eye globe and imaged using a Leica MZFLIII dissecting scope with an attached Nikon E995 camera.
Labeled fatty acid injection into yolk
To analyze lipid transport from the zebrafish embryo yolk into the body, fluorescently tagged BODIPY (BODIPY FL C12; Invitrogen D3822) stock was dried completely in a speed vacuum and resuspended in canola oil to a final concentration of 1 mg/ml. The BODIPY oil mixture (4.6 nl) was injected into the zebrafish yolk at 4dpf. Four hours after injection, zebrafish intersegmental vessels in the trunk were imaged using confocal microscopy. For measuring the mean pixel intensity of BODIPY- C12 in the intersegmental vessels, three vessels were averaged as a single data point using Microsoft Excel (Microsoft, Redmond, WA) and graphed using GraphPad Prism (GraphPad, La Jolla, CA).
RNA extraction and real-time PCR
Analysis of ctage5 and tango1 transcripts was performed on whole embryos. mRNA was extracted from dechorionated embryos at 7dpf using TRizol–chloroform treatment. The isolated aqueous phase from the resulting TRizol–chloroform gradient is transferred to a new tube and incubated with isopropanol at room temperature for 10 min. Centrifugation at 4°C for 10 min results in formation of an RNA pellet, which is then washed with 75% ethanol in diethyl procarbonate (DEPC)-treated water. Following this wash, the pellet is allowed to dry and is resuspended in DEPC-treated water with a 10-min incubation at 60°C. Resuspended RNA is then subjected to a DNAse treatment and concentration is quantified.
cDNA was generated using the Superscript III First-Strand Synthesis System for RT-PCR Kit (Invitrogen) per manufacturer’s instructions, and all qRT-PCR was performed on a CFX Connect Real-Time System (Bio-Rad) using PrimeTime Gene Expression Master Mix (IDT). qRT-PCR was performed on five biological replicates, and all biological replicates were run in triplicate for each transcript. The zebrafish housekeeping gene ef1a was used for normalization. Statistical significance of differences in transcript abundance was calculated using Welch’s t test. qPCR sequences are as follows:
ctage5 exon 7–9:
Primer 1: CTCATCTTGGCCGCTTCTAT
Primer 2: CATCGACGGCAGCACTAATA
ctage5 exon 19–20:
Primer 1: GGCGGAGGCATTGACATTA
Primer 2: AGAGAAGGCTCTGGAGATATGA
tango1 exon 28–29:
Primer 1: AGAGGTCCAGGCGGAAA
Primer 2: CACACATCGGCCCGTTT
Imaging and data analysis
Fluorescence was detected using Nikon Eclipse E600FN or C2 Nikon Eclipse 80i confocal systems. Mean pixel intensity was measured using ImageJ (Rasband, W.S. ImageJ, U.S. National Institutes of Health, MD). To measure spinal cord cell number, spot detection software was used in Imaris (Bitplane, Zurich, Switzerland). For survival analysis, a χ2 analysis was performed with the expected numbers calculated from the total number of fish collected at the 2-mo timepoint when fish were genotyped and counted. Data were processed using Microsoft Excel (Microsoft, Redmond, WA) and graphed using GraphPad Prism (GraphPad, La Jolla, CA). An unpaired two-tailed t test was used to analyze graphs with two groups. For three or more groups, a one-way ANOVA was conducted with Tukey’s post hoc analysis for pairwise comparisons.
Adult brain clearing
PACT clearing was performed based on Cronan et al. (2015) with slight modifications. Briefly, zebrafish were fixed in 2% paraformaldehyde (PFA) for 1 day at 4°C. The fixed whole adult fish brain was dissected and incubated at 4°C for 1 day in ice-cold, freshly made hydrogel monomer solution of A4P0 (4% acrylamide in PBS) supplemented with 0.25% VA-044. A4P0-infused samples were incubated for 3 h at 37°C to initiate tissue-hydrogel hybridization. Hydrogel monomer solution was removed and washed three times with PBS. Whole brains were then incubated in 8% SDS in 200 mM boric acid, pH 8.5, at 37°C under shaking for 5 h or until the brain periphery was clear. This solution was replaced with PBST and samples were left to shake overnight at room temperature, allowing the center of the brain to clear overnight. Samples were then washed throughout the day with four changes of PBS, 0.1% Triton X-100 at room temperature, and then incubated overnight in RIMS imaging media (Yang et al., 2014) at room temperature on a rotator. Samples were stored in RIMS at room temperature until imaged using confocal microscopy.
Supplementary Material
Acknowledgments
The authors thank Michael Cliff, William Hudzinski, and Edi Kuhn for zebrafish husbandry, Jon Bostrom for help with CRISPR design, and Elena Semina and her laboratory for assistance with Alcian blue staining. We are also grateful to Clive Wells for help with electron microscopy experiments. This work was supported by the National Institutes of Health/National Aging Institute (R21AG066038 to B.A.L.), the Clinical and Translational Science Institute, Medical College of Wisconsin (TL1TR001437 Award to E.M.C.), and the Foundation Fighting Blindness (PPA-0617-0718).
Abbreviations used:
- APOB
apolipoprotein B
- ATF4
activating transcription factor 4
- ATF6
activating transcription factor 6
- BODIPY
boron-dipyrromethene
- BSA
bovine serum albumin
- CAS9
CRISPR associated protein 9
- COPII
coat protein complex II
- CRISPR
clustered regularly interspaced short palindromic repeats
- CTAGE5
cutaneous T-cell lymphoma-associated antigen 5
- D2GFP
destabilized green fluorescent protein
- DAPI
4’,6-diamidino-2-phenylindole
- DEPC
diethyl pyrocarbonate
- dH 2O
distilled H 2O
- DMSO
dimethyl sulfoxide
- DNA
deoxyribonucleic acid
- DPF
days post-fertilization
- EMBL-EBI
European bioinformatics institute
- ER
endoplasmic reticulum
- ERES
endoplasmic reticulum exit sites
- ERGIC
endoplasmic reticulum Golgi intermediate compartment
- GAP43
growth associated protein 43
- gDNA
genomic deoxyribonucleic acid
- gRNA
guide ribonucleic acid
- GWAS
genome-wide association study
- MIA
melanoma inhibitory activity
- mRNA
messenger ribonucleic acid
- NGS
normal goat serum
- PACT
passive clarity technique
- PBS
phosphate buffered saline
- PCR
polymerase chain reaction
- PERK
PKR-like endoplasmic reticulum kinase
- PFA
paraformaldehyde
- PTU
1-phenyl 2-thiourea
- RNAseq
ribonucleic acid sequencing
- SAR1
secretion associated RAS related GTPase 1
- SAR1
secretion-associated RAS-related 1
- scRNAseq
single cell ribonucleic acid sequencing
- TALI
TANGO-like
- TANGO1l
transport and Golgi organization protein 1 long
- TANGO1s
TANGO1 short
- TUNEL
terminal deoxynucleotidyl transferase biotin-dUTP nick end labeling
- UPR
unfolded protein response
- VLDL
very-low density lipoprotein
- XBP1
x-box binding protein 1
- ZDR
zebrafish Danio rerio.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-11-0745) on January 13, 2021.
REFERENCES
- Askary A, Xu P, Barske L, Bay M, Bump P, Balczerski B, Bonaguidi MA, Crump JG (2017). Genome-wide analysis of facial skeletal regionalization in zebrafish. Development 144, 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard F, Casano L, Mallabiabarrena A, Wallace E, Saito K, Kitayama H, Guizzunti G, Hu Y, Wendler F, DasGupta R, et al. (2006). Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature 439, 604–607. [DOI] [PubMed] [Google Scholar]
- Benowitz LI, Routtenberg A (1997). GAP-43: an intrinsic determinant of neuronal development and plasticity. Trends Neurosci 20, 84–91. [DOI] [PubMed] [Google Scholar]
- Bonifacio JS, Glick BS (2004). The mechanisms of vesicle budding and fusion. Cell 116, 153–166. [DOI] [PubMed] [Google Scholar]
- Bormann P, Zumsteg VM, Roth LWA, Reinhard E (1998). Target contact regulates GAP-43 and α-tubulin mRNA levels in regenerating retinal ganglion cells. J Neurosci Res 52, 405–419. [DOI] [PubMed] [Google Scholar]
- Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, et al. (2019). The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47, D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard SB, Bakkers J (2018). Spatially resolved RNA-sequencing of the embryonic heart identifies a role for Wnt/β-catenin signaling in autonomic control of heart rate. eLife 7, e31515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark EM, Nonarath HJT, Bostrom JR, Link BA. (2020) Establishment and validation of an endoplasmic reticulum stress reporter to monitor zebrafish ATF6 activity in development and disease. Dis Models Mech 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan MR, Rosenberg AF, Oehlers SH, Saelens JW, Sisk DM, Smith KLJ, Lee S, Tobin DM (2015). CLARITY and PACT-based imaging of adult zebrafish and mouse for whole-animal analysis of infections. Dis Model Mech 8, 1643–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann H, Kalbhen P, Fischer D (2015). Characterization of optic nerve regeneration using transgenic zebrafish. Front Cell Neurosci 9, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Wang Y, Liu L, Zhang H, Zhang F, Shi L, Yu M, Gao F, Xu Z (2017). cTAGE5 deletion in pancreatic β cells impairs proinsulin trafficking and insulin biogenesis in mice. J Cell Biol 216, 4153–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Zhang M, Kenny SJ, Liu D, Maeda M, Saito K, Mathur A, Xu K, Schekman R (2017). Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep 18, 1586–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorur A, Yuan L, Kenny SJ, Baba S, Xu K, Schekman R (2017). COPII-coated membranes function as transport carriers of intracellular procollagen I. J Cell Biol 216, 1745–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendee KE, Sorokina EA, Muheisen SS, Reis LM, Tyler RC, Markovic V, Cuturilo G, Link BA, Semina EV (2018). PITX2 deficiency and associated human disease: insights from the zebrafish model. Hum Mol Genet 27, 1675–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir MF, Kim H-R, Chae H-J (2018). Endoplasmic Reticulum Stress and Autophagy. Endoplasmic Reticulum, DOI:10.5772/INTECHOPEN.81381. [Google Scholar]
- Kaneda M, Nagashima M, Nunome T, Muramatsu T, Yamada Y, Kubo M, Muramoto K, Matsukawa T, Koriyama Y, Sugitani K, et al. (2008). Changes of phospho-growth-associated protein 43 (phospho-GAP43) in the zebrafish retina after optic nerve injury: A long-term observation. Neurosci Res 61, 281–288. [DOI] [PubMed] [Google Scholar]
- Kim S-D, Pahuja KB, Ravazzola M, Yoon J, Boyadjiev SA, Hammamoto S, Schekman R, Orci L, Kim J (2012). SEC23-SEC31 the interface plays a critical role for export of procollagen from the endoplasmic reticulum. J Biol Chem 287, 10134–10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Okamoto M, Nakano A (2014). Contact of cis- Golgi with ER exit sites executes cargo capture and delivery from the ER. Nat Commun 5, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang MR, Lapierre LA, Frotscher M, Goldenring JR, Knapik EW (2006). Secretory COPII coat component Sec23a is essential for craniofacial chondrocyte maturation. Nat Genet 38, 1198–1203. [DOI] [PubMed] [Google Scholar]
- Lee MCS, Miller EA, Goldberg J, Orci L, Schekman R. (2004). Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol 20, 87–123. [DOI] [PubMed] [Google Scholar]
- Lekszas C, Foresti O, Raote I, Liedtke D, König E-M, Nanda I, Vona B, Coster PD, Cauwels R, Malhotra V, et al. (2020). Biallelic TANGO1 mutations cause a novel syndromal disease due to hampered cellular collagen secretion. Elife 9, e51319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos RR, Oliveira DF, Zatz M, Oliveira JRM (2011). Population and computational analysis of the MGEA6 P521A variation as a risk factor for familial idiopathic basal ganglia calcification (Fahr’s Disease). J Mol Neurosci 43, 333–336. [DOI] [PubMed] [Google Scholar]
- Liu M, Feng Z, Ke H, Liu Y, Sun T, Dai J, Cui W, Pastor-Pareja JC (2017). Tango1 spatially organizes ER exit sites to control ER export. J Cell Biol 216, 1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda M, Katada T, Saito K (2017). TANGO1 recruits Sec16 to coordinately organize ER exit sites for efficient secretion. J Cell Biol 216, 1731–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda M, Kurokawa K, Katada T, Nakano A, Saito K (2019). COPII proteins exhibit distinct subdomains within each ER exit site for executing their functions. Sci Re 9, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda M, Saito K, Katada T (2016). Distinct isoform-specific complexes of TANGO1 cooperatively facilitate collagen secretion from the endoplasmic reticulum. Mol Biol Cell 27, 2688–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiers JL, Kostallari E, Mushref M, deAssuncao TM, Li H, Jalan-Sakrikar N, Huebert RC, Cao S, Malhi H, Shah VH (2016). The unfolded protein response mediates fibrogenesis and collagen I secretion through regulating TANGO1 in mice. Hepatology 65, 983–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra V, Erlmann P (2015). The pathway of collagen secretion. Annu Rev Cell Dev Biol 31, 109–124. [DOI] [PubMed] [Google Scholar]
- McCaughey J, Stephens DJ (2019). ER-to-Golgi transport: a sizeable problem. Trends Cell Biol 29, 940–953. [DOI] [PubMed] [Google Scholar]
- Melville DB, Montero-Balaguer M, Levic DS, Bradley K, Smith JR, Hatzopoulos AK, Knapik EW (2011). The feelgood mutation in zebrafish dysregulates COPII-dependent secretion of select extracellular matrix proteins in skeletal morphogenesis. Dis Models Mech 4, 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EA, Schekman R (2013). COPII—a flexible vesicle formation system. Curr Opin Cell Biol 25, 420–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyares RL, de Rezende VB, Farber SA (2014). Zebrafish yolk lipid processing: a tractable tool for the study of vertebrate lipid transport and metabolism. Dis Models Mech 7, 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira JRM, Sobrido MJ, Spiteri E, Hopfer S, Meroni G, Petek E, Baquero M, Geschwind DH (2007). Analysis of candidate genes at the IBGC1 locus associated with idiopathic basal ganglia calcification (“Fahr” Disease’). J Mol Neurosci 33, 151–154. [DOI] [PubMed] [Google Scholar]
- Omari S, Makareeva E, Roberts-Pilgrim A, Mirigian L, Jarnik M, Ott C, Lippincott-Schwartz J, Leikin S (2018). Noncanonical autophagy at ER exit sites regulates procollagen turnover. Proc Natl Acad Sci USA 115, E10099–E10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petley-Ragan LM, Ardiel EL, Rankin CH, Auld VJ (2016). Accumulation of laminin monomers in Drosophila glia leads to glial endoplasmic reticulum stress and disrupted larval locomotion. J Neurosci 36, 1151–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman JL, Bonnet DJ, Curtiss LK, Gekakis N (2011). Reduced cholesterol and triglycerides in mice with a mutation in Mia2, a liver protein that localizes to ER exit sites. J Lipid Res 52, 1775–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raote I, Ernst AM, Campelo F, Rothman JE, Pincet F, Malhotra V (2020). TANGO1 membrane helices create a lipid diffusion barrier at curved membranes. eLife 9, e57822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raote I, Malhotra V (2019). Protein transport by vesicles and tunnels. J Cell Biol 218, 737–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raote I, Ortega Bellido M, Pirozzi M, Zhang C, Melville D, Parashuraman S, Zimmermann T, Malhotra V (2017). TANGO1 assembles into rings around COPII coats at ER exit sites. J Cell Biol 216, 901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds HM, Zhang L, Tran DT, Hagen KGT (2019). Tango1 coordinates the formation of ER/Golgi docking sites to mediate secretory granule formation. J. Biol. Chem. 295, 19498–19510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos-Barrera LD, Sigurbjörnsdóttir S, Baer M, Leptin M (2017). Dual function for Tango1 in secretion of bulky cargo and in ER-Golgi morphology. Proc Natl Acad Sci U S A 114, E10389–E10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Chen M, Bard F, Chen S, Zhou H, Woodley D, Polischuk R, Schekman R, Malhotra V (2009). TANGO1 facilitates cargo loading at endoplasmic reticulum exit sites. Cell 136, 891–902. [DOI] [PubMed] [Google Scholar]
- Saito K, Yamashiro K, Ichikawa Y, Erlmann P, Kontani K, Malhotra V, Katada T (2011). cTAGE5 mediates collagen secretion through interaction with TANGO1 at endoplasmic reticulum exit sites. Mol Biol Cell 22, 2301–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Yamashiro K, Shimazu N, Tanabe T, Kontani K, Katada T (2014). Concentration of Sec12 at ER exit sites via interaction with cTAGE5 is required for collagen export. J Cell Biol 206, 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San B, Aben M, Elurbe DM, Voeltzke K, Den Broeder MJ, Rougeot J, Legler J, Kamminga LM (2018). Genetic and Epigenetic Regulation of Zebrafish Intestinal Development. Epigenomes 2, 19. [Google Scholar]
- Santos AJM, Nogueira C, Ortega-Bellido M, Malhotra V (2016). TANGO1 and MIA2/cTAGE5 (TALI) cooperate to export bulky pre-chylomicrons/VLDLs from the endoplasmic reticulum. J Cell Biol 213, 343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Nakano A (2005). Dissection of COPII subunit-cargo assembly and disassembly kinetics during Sar1p-GTP hydrolysis. Nat Struct Mol Biol 12, 167–174. [DOI] [PubMed] [Google Scholar]
- Skene JHP (1989). Axonal growth-associated proteins. Annu Rev Neurosci 12, 127–156. [DOI] [PubMed] [Google Scholar]
- Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M (2006). BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34, D535–D539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townley AK, Feng Y, Schmidt K, Carter DA, Porter R, Verkade P, Stephens DJ (2008). Efficient coupling of Sec23-Sec24 to Sec13-Sec31 drives COPII-dependent collagen secretion and is essential for normal craniofacial development. J Cell Sci 121, 3025–3034. [DOI] [PubMed] [Google Scholar]
- Udvadia AJ (2008). 3.6kb genomic sequence from Takifugu capable of promoting axon growth-associated expression in developing and regenerating zebrafish neurons. Gene Expression Patterns 8, 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D (2011). The unfolded protein response: From stress pathway to homeostatic regulation. Science 334, 1081–1086. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu L, Zhang H, Fan J, Zhang F, Yu M, Shi L, Yang L, Lam SM, Wang H, et al. (2016). Mea6 controls VLDL transport through the coordinated regulation of COPII assembly. Cell Res 26, 787–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P, Townley AK, Koka P, Palmer KJ, Stephens DJ (2006). Sec16 defines endoplasmic reticulum exit sites and is required for secretory cargo export in mammalian cells. Traffic 7, 1678–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M (1995). The zebrafish Book. Eugene, Oregon: University of Oregon Press. [Google Scholar]
- White RJ, Collins JE, Sealy IM, Wali N, Dooley CM, Digby Z, Stemple DL, Murphy DN, Billis K, Hourlier T, et al. (2017). A high-resolution mRNA expression time course of embryonic development in zebrafish. eLife 6, e30860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DG, Phamluong K, Li L, Sun M, Cao TC, Liu PS, Modrusan Z, Sandoval WN, Rangell L, Carano RAD, et al. (2011). Global defects in collagen secretion in a Mia3/TANGO1 knockout mouse. J Cell Biol 193, 935–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen C-K, Lubeck E, Shah S, Cai L, Gradinaru V (2014). Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang Y, Wang T, Yao L, Lam SM, Huang X, Fan J, Wang Q, Liu L, Jiang Y, et al. (2018). cTAGE5/MEA6 plays a critical role in neuronal cellular components trafficking and brain development. Proc Natl Acad Sci USA 115, E9449–E9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.