Abstract
Nuclear RNA exosome is the main 3′→5′ RNA degradation and processing complex in eukaryotic cells and its dysregulation therefore impacts gene expression and viability. In this work we show that RNA exosome activity is necessary for maintaining cell wall stability in yeast Saccharomyces cerevisiae. While the essential RNA exosome catalytic subunit Dis3 provides exoribonuclease catalytic activity, the second catalytic subunit Rrp6 has a noncatalytic role in this process. RNA exosome cofactors Rrp47 and Air1/2 are also involved. RNA exosome mutants undergo osmoremedial cell lysis at high temperature or at physiological temperature upon treatment with cell wall stressors. Finally, we show that a defect in protein glycosylation is a major reason for cell wall instability of RNA exosome mutants. Genes encoding enzymes that act in the early steps of the protein glycosylation pathway are down-regulated at high temperature in cells lacking Rrp6 protein or Dis3 exoribonuclease activity and overexpression of the essential enzyme Psa1, that catalyzes synthesis of the mannosylation precursor, suppresses temperature sensitivity and aberrant morphology of these cells. Furthermore, this defect is connected to a temperature-dependent increase in accumulation of noncoding RNAs transcribed from loci of relevant glycosylation-related genes.
INTRODUCTION
In eukaryotic cells, 3′→5′ RNA degradation and processing is accomplished through activity of the RNA exosome complex (Chlebowski et al., 2013; Zinder and Lima, 2017; Lingaraju et al., 2020). It plays a major part in RNA metabolism in the nucleus and cytoplasm because it targets almost all RNA classes: its roles include RNA surveillance; mRNA turnover; processing and maturation of rRNAs, snRNAs, and snoRNAs; and degradation of noncoding transcripts (Allmang et al., 1999a; Hilleren et al., 2001; Wyers et al., 2005). It is therefore not surprising that dysregulation of RNA exosome activity broadly impacts gene expression (Van Dijk et al., 2007; Lardenois et al., 2011; Gudipati et al., 2012; Schneider et al., 2012; Bresson et al., 2017; Davidson et al., 2019) and is also implicated in various human malignancies and disorders (Fasken et al., 2020). Rare diseases caused by mutations in genes that encode human exosome subunits (EXOSC proteins) have been termed exosomopathies. They usually encompass single amino acid substitutions rather than more substantial mutations, as RNA exosome activity is essential for viability (de Amorim et al., 2020).
The central part of the highly conserved RNA exosome complex is the exosome core (Exo9). It encompasses nine subunits that form a doughnut-shaped channel that has a structural and regulatory role (Wasmuth and Lima, 2012; Wasmuth et al., 2014). Catalytic activity is provided by two additional subunits: Rrp6, which has exonuclease activity, and Dis3/Rrp44, which has exonuclease and endonuclease activities (Briggs et al., 1998; Dziembowski et al., 2007; Lebreton et al., 2008). In yeast, Dis3 is found in both the nuclear and the cytoplasmic isoforms of the exosome complex, whereas Rrp6 is only found in the nuclear isoform, where it additionally associates with its stabilization partner Rrp47 to form the 12-subunit complex Exo12Dis3/Rrp6/Rrp47 (Feigenbutz et al., 2013; Makino et al., 2015). Activity of the nuclear RNA exosome is also stimulated by its cofactors Mpp6 and the TRAMP complex, which function to guide substrate specificity and aid RNA degradation (Schilders et al., 2005; Stuparevic et al., 2013; Wasmuth et al., 2017). The three-subunit TRAMP complex provides RNA-binding (Air1 or Air2), RNA-helicase (Mtr4), and poly(A)-polymerase (Trf4 or Trf5) activities, which play a major role in noncoding RNA degradation (LaCava et al., 2005; Wyers et al., 2005).
All core exosome subunits, as well as catalytic subunit Dis3, are essential in yeast (Mitchell et al., 1997). In contrast, deletion of the gene encoding the catalytic subunit Rrp6 is viable; however, it results in slow growth at physiological temperature and temperature sensitivity (Allmang et al., 1999b; Phillips and Butler, 2003). Interestingly, these two phenotypes are not both caused by the lack of Rrp6 catalytic activity, as Rrp6 catalytic mutants also grow slowly at physiological temperature, but are not temperature sensitive (Phillips and Butler, 2003). Because of that, it has long been clear that Rrp6 has a noncatalytic role in maintaining cell viability upon heat stress, but the molecular nature of this predicament has not been explained. Recent work connected RNA degradation to the cell wall integrity (CWI) pathway, which regulates gene expression to ensure cellular integrity upon stress, through a MAPK signaling cascade (Catala et al., 2012; Wang et al., 2020). Involvement of Rrp6 in this process was inferred from the additive cell wall instability phenotype of rrp6Δ mpk1Δ mutant cells, in which CWI signal transduction is inhibited (Wang et al., 2020). Specifically, a role was proposed for a solitary “moonlighting” function of Rrp6, independent of other exosome subunits and its interactors Rrp47 and Isw1, in maintaining CWI at high temperature (Wang et al., 2020).
In this work, we show that the RNA exosome complex is a major regulator of yeast cell wall stability. Exoribonuclease catalytic activity of the Dis3 subunit is essential for maintaining cellular integrity upon heat stress or treatment with cell wall stressors, together with the second catalytic subunit Rrp6 that has a noncatalytic role in this process. The RNA exosome cofactors Rrp47 and Air1/2 also contribute in a significant way. Cells lacking these proteins or Dis3 exoribonuclease activity are not viable at high temperature because of compromised cell wall stability. Importantly, cell bursting and aberrant cell morphology of RNA exosome mutants are suppressed by osmotic support, as well as by overexpression of the Psa1 enzyme, which enables increased production of GDP-mannose that is incorporated into mannoproteins, indicating that protein glycosylation is a major reason for cell wall instability of RNA exosome mutants. Expression of protein glycosylation-related genes PSA1, DPM1, and ALG7 is dysregulated in these mutants at high temperature, presumably through mechanisms that involve accumulation of specific noncoding RNAs transcribed from their gene loci.
RESULTS
RNA exosome mutants undergo osmoremedial cell lysis at high temperature
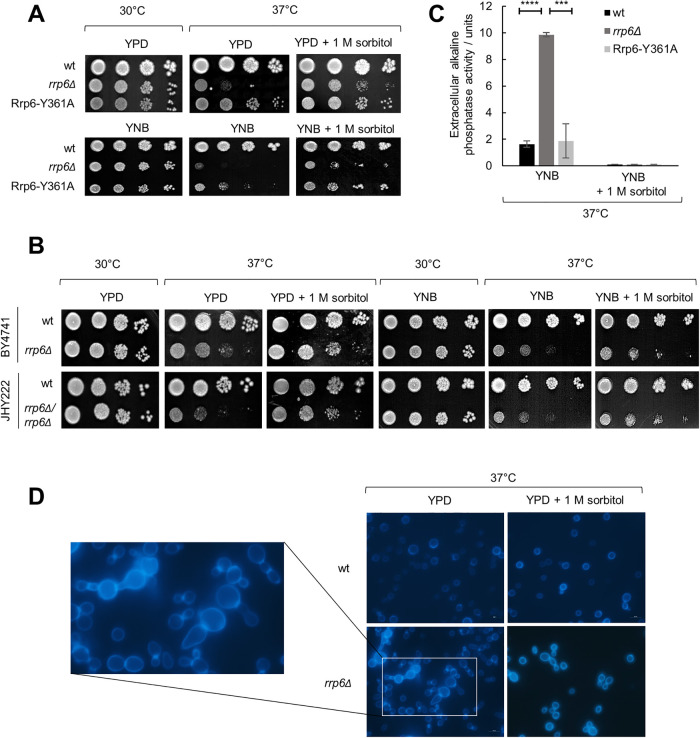
All subunits of yeast RNA exosome complex are essential for viability except for the nuclear-specific catalytic subunit Rrp6, whose inactivation is lethal only above 37°C (Allmang et al., 1999b; Phillips and Butler, 2003). The reason for temperature sensitivity of the rrp6Δ mutant remained unknown, especially because Rrp6 catalytic mutants are not temperature sensitive (Phillips and Butler, 2003), and lack of Rrp6 was not linked to significant RNA processing defects at high temperature (Allmang et al., 2000). To test whether rrp6Δ cells are inviable at 37°C due to compromised cellular integrity, we supplemented the growth medium with 1 M sorbitol, which acts as osmotic support. We performed all experiments in the W303-derived BMA41 genetic background in which Rrp6-related phenotypes are most pronounced (Klauer and Van Hoof, 2013; Wasmuth and Lima, 2017). Interestingly, osmotic stabilization of the growth medium completely suppressed its temperature-sensitive phenotype and enabled wild-type level of growth after 3 d at 37°C on both YPD and synthetic YNB mediums (Figure 1A). This was due to osmotic stabilization and not sorbitol itself, because the addition of 1 M sucrose, NaCl, or KCl led to a similar level of suppression (Supplemental Figure S1). Also, this effect was not specific to BMA41 genetic background, as growth at 37°C could also be restored with the less temperature-sensitive rrp6Δ haploid BY4741 and diploid JHY222 genetic backgrounds (Figure 1B).
FIGURE 1:
Cells lacking Rrp6 display phenotypes indicative of cell wall instability. Strains are BMA41 wild type (wt) and isogenic mutants, unless noted otherwise. Tenfold serial dilutions of cells were spotted on plates and were photographed after 3 d at indicated temperature. (A) Osmotically supporting medium with 1 M sorbitol rescues growth of rrp6Δ cells at high temperature. (B) Osmotic support rescues growth of rrp6Δ mutants of other genetic backgrounds (haploid BY4741 and diploid JHY222) at high temperature. (C) rrp6Δ cells burst at high temperature, unless osmotic support is provided. Strains were grown for 3 d at 37°C and activity of alkaline phosphatase was measured in growth medium. Measurements were performed in duplicate, and reported values represent the means and standard deviations of three independent experiments (n = 3). Indicated differences show the significant differences using an unpaired Student’s t test. Three (***) and four (****) asterisks denote a p-value lower than or equal to 0.001 and 0.0001, respectively. (D) Aberrant cellular morphology and cell separation defect of rrp6Δ cells at high temperature, visualized by fluorescent microscopy after Calcofluor White staining.
Osmotic instability results in cell lysis, so we grew cells in liquid medium for 3 d at 37°C and measured the activity of alkaline phosphatase released into the medium. Alkaline phosphatase is an intracellular enzyme so its release into the medium implies membrane and cell wall lysis. Cells lacking Rrp6 released almost a fivefold higher amount of alkaline phosphatase than wild-type or Rrp6-Y361A catalytic mutant cells, and cell lysis in all strains was completely suppressed by the addition of 1 M sorbitol (Figure 1C) or upon growth at 30°C regardless of sorbitol addition (unpublished data). As higher activity measured with rrp6Δ cells could also be due to a change in expression of alkaline phosphatase, we measured intracellular alkaline phosphatase activity with the same cells and found practically no differences between the strains (Supplemental Figure S2), confirming that the extracellular activity observed for the rrp6Δ strain is indicative of cell lysis. We also examined the cells by fluorescent microscopy after Calcofluor White (CFW) staining. CFW stains chitin, which in yeast is localized primarily in bud necks and bud scars, as it forms the primary septum (Klis et al., 2002). It was revealed that morphology of rrp6Δ cells without osmotic support at 37°C was also consistent with weakened cellular integrity, as the cells were enlarged, unevenly shaped, and grew in clumps (Figure 1D). Based on the intensive staining of cell septa by CFW, it was clearly visible that two or more rrp6Δ cells stuck together at their bud necks, meaning the clumps result from a defect in cell separation after division (Figure 1D).
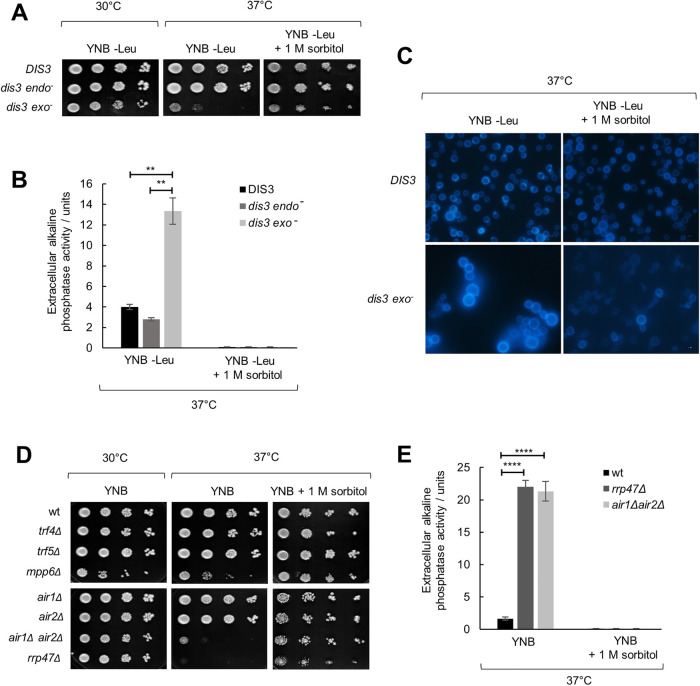
Deletion of the DIS3 gene encoding the second exosome catalytic subunit is lethal, but it is possible to generate mutants deficient in Dis3 exo- or endoribonuclease activity (Dziembowski et al., 2007; Lebreton et al., 2008). Exo− (dis3-D551N) mutant displays temperature sensitivity (Dra̧zkowska et al., 2013; Milbury et al., 2019), so we wondered whether the cause is similar as for rrp6Δ. Indeed, the addition of 1 M sorbitol restored growth and morphology of this mutant at 37°C and suppressed its cell lysis, as measured by the release of alkaline phosphatase (Figure 2, A–C). Furthermore, we tested viability at 37°C of mutants in monomeric cofactors Mpp6 and Rrp47, as well as viable mutants in subunits of the TRAMP complex, which function as coactivators of the nuclear exosome, and found that the temperature sensitivity and the temperature-induced cell lysis of air1Δair2Δ and rrp47Δ mutants are also suppressed by osmotic stabilization (Figure 2, D and E). Taken together, temperature-sensitive mutants of RNA exosome catalytic subunits rrp6Δ and dis3 exo−, as well as mutants in exosome cofactors Rrp47 and Air1/2, undergo osmoremedial cell lysis at 37°C, which is a phenotype indicative of a weakened cell wall.
FIGURE 2:
Inactivation of Dis3 exonuclease activity and certain exosome cofactors leads to cell wall instability. Strains are W303-derived with genomic copy of DIS3 gene deleted but bearing a centromeric plasmid that carries the wild-type copy of the DIS3 gene (DIS3) or its alleles with abolished endonuclease (dis3 endo−, D171N) or exonuclease (dis3 exo−, D551N) activity. Tenfold serial dilutions of cells were spotted on plates and were photographed after 3 d at indicated temperature. (A) Osmotically supporting medium with 1 M sorbitol rescues growth of dis3 exo− at high temperature. (B) dis3 exo− cells burst at high temperature, unless osmotic support is provided. Strains were grown for 3 d at 37°C and activity of alkaline phosphatase was measured in growth medium. Measurements were performed in duplicate, and reported values represent the means and standard deviations of three independent experiments (n = 3). Indicated differences show the significant differences using an unpaired Student’s t test. Two (**) asterisks denote a p-value lower than or equal to 0.01. (C) Aberrant cellular morphology and cell separation defect of dis3 exo− cells at high temperature, visualized by fluorescent microscopy after Calcofluor White staining. (D) Strains are BMA41 wild-type (wt) and isogenic mutants. Mutants in exosome cofactors rrp47Δ and air1Δair2Δ also show osmoremedial temperature sensitivity. (E) Strains were grown for 3 d at 37°C and activity of alkaline phosphatase was measured in growth medium. Measurements were performed in duplicate, and reported values represent the means and standard deviations of three independent experiments (n = 3). Indicated differences show the significant differences using an unpaired Student’s t test. Four (****) asterisks denote a p-value lower than or equal to 0.0001.
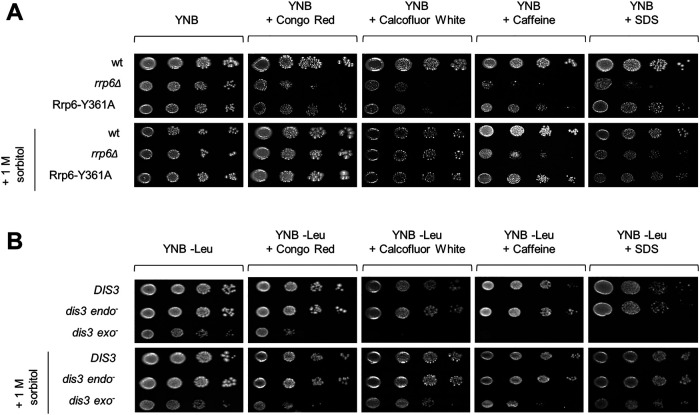
RNA exosome mutants are hypersensitive to cell wall stressors
To investigate whether it is possible to detect cell wall–related phenotypes in RNA exosome mutants at the physiological temperature of 30°C, we examined their growth on media containing known cell wall stressors Congo Red (CR), CFW, caffeine, and SDS. CR and CFW interfere with glucan and chitin assembly, respectively (Roncero and Duran, 1985; Kopecká and Gabriel, 1992); caffeine primarily affects TOR signaling (Kuranda et al., 2006); and SDS is a general cell wall and membrane destabilizer (Popolo et al., 2001). rrp6Δ and dis3 exo− mutants were hypersensitive to all of these compounds, thereby demonstrating that their cell walls are weaker than those of the corresponding wild-type cells even at the permissive temperature of 30°C when faced with cell wall stressors (Figure 3). Furthermore, their growth was significantly restored by the addition of 1 M sorbitol, which strengthens the argument that the effect is related to cell wall stability (Figure 3).
FIGURE 3:
Cells lacking Rrp6 protein or Dis3 exoribonuclease activity are hypersensitive to cell wall stressors. Strains are described in Figures 1 and 2. Tenfold serial dilutions of cells were spotted on plates and were photographed after 3 d at 30°C. Concentrations of compounds used: Congo Red 10 μg/ml, Calcofluor White 20 μg/ml, caffeine 6 mM, SDS 0.0075%.
Regarding mutants in RNA exosome cofactors, for air1Δair2Δ and rrp47Δ that are temperature sensitive we found that they are also hypersensitive to all tested cell wall stressors, while mpp6Δ showed specific sensitivity to caffeine (Supplemental Figure S3). The fact that single mutants in the TRAMP RNA-binding subunits air1Δ and air2Δ did not show cell wall–related phenotypes, but their combined inactivation in the double air1Δair2Δ mutant did, indicates their functional redundancy. Inactivation of either Trf4 or Trf5 TRAMP poly(A)-polymerase also did not lead to any cell wall–related phenotypes, while in this case it was not possible to explore whether it is due to functional redundancy because the double mutant is not viable.
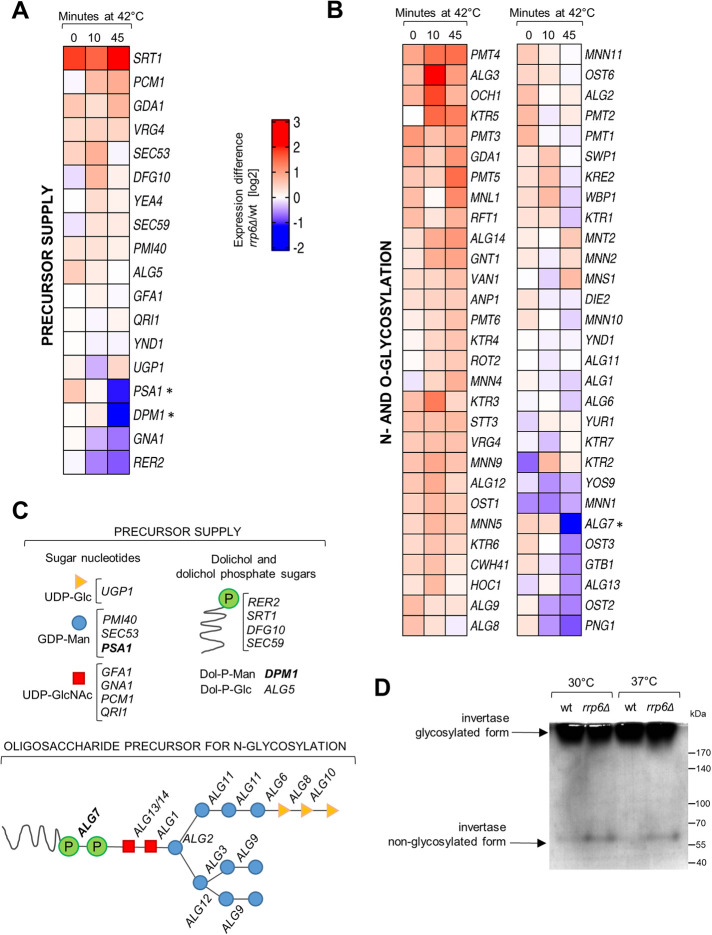
Genes involved in protein glycosylation are dysregulated in RNA exosome mutants at high temperature and aiding this process suppresses their temperature sensitivity
The Rrp6-containing RNA exosome is located in the nucleus of the yeast cells, which precludes any direct link to the cell periphery. Instead, given the ubiquitous role of the RNA exosome in gene expression, its role in maintaining cell wall stability upon stress should be visible at the level of mRNAs encoding proteins that are important for cell wall biosynthesis and remodeling. To this aim, we made use of the recently published genome-wide RNA-sequencing analysis that included the datasets of rrp6Δ mutant before and after a 45-min heat shock at 42°C (Wang et al. 2020). We inspected gene expression profiles of ∼180 genes involved in cell wall biogenesis (Orlean, 2012) and visualized them as rrp6Δ/wt mRNA ratios on a log2 scale (Figure 4, A and B, and Supplemental Figure S4). Heat shock–dependent down-regulation in rrp6Δ cells as compared with wild-type cells could be seen for a number of genes, such as GPI12, encoding an essential protein involved in GPI anchor assembly, and YPS3, encoding an aspartic protease (Supplemental Figure S4). However, cell wall–related gene subcategories that encompassed genes with most prominent transcript down-regulation in rrp6Δ cells compared with wild-type cells at high temperature were the precursor supply gene category, which includes enzymes involved in the synthesis of sugar nucleotides and dolichol phosphate sugars that are precursors for cell wall components, and the N- and O-glycosylation category (Figure 4, A and B). In the precursor supply gene category, we noticed a strong heat shock–dependent down-regulation of PSA1 and DPM1 genes in rrp6Δ cells as compared to wild-type cells (Figure 4A). These genes are involved in the synthesis of GDP-mannose and its binding to the dolichol carrier, respectively (Figure 4C). Mannose is exclusively bound to cell wall proteins through N- or O-linked glycosylation in the endoplasmic reticulum and Golgi (Klis et al., 2002). Inspection of the N- and O-glycosylation category revealed that ALG7, which catalyzes the initial step in synthesis of the oligosaccharide precursor for N-glycosylation (Figure 4C), also showed heat shock–dependent down-regulation in rrp6Δ cells compared to wild-type cells (Figure 4B). Even though a large number of genes in this category seemed to be up-regulated in rrp6Δ relative to wild-type cells, we hypothesized that protein glycosylation in this mutant should nevertheless be affected, because precursor synthesis and the very early steps in the glycosylation pathway are severely impaired. To experimentally verify whether protein glycosylation is affected in RNA exosome mutants, we analyzed the degree of glycosylation of periplasmic invertase, normally a heavily N-glycosylated protein, by following its electrophoretic mobility with subsequent in-gel activity staining. Periplasmic invertase is easily inducible and is secreted even upon glycosylation defects so it provides a simple readout of the glycosylation status of the cell (Esmon et al., 1987; Belcarz et al., 2002). Positively, we noticed the appearance of a nonglycosylated form of invertase in periplasmic extracts of rrp6Δ cells after staining the gel for invertase activity (Figure 4D). This form was also present in periplasmic extracts of other RNA exosome mutants whose cell wall is destabilized: rrp47Δ, air1Δair2Δ, and dis3 exo−, and was mostly absent from periplasmic extracts of wild-type and dis3 endo− cells (Supplemental Figure S5). Because protein mannosylation is essential for cell viability and its impairment leads to cell wall defects (Janik et al., 2012), this analysis opened the possibility that a general defect in protein glycosylation may be the cause of cell wall instability and therefore temperature sensitivity of RNA exosome mutant cells.
FIGURE 4:
Protein glycosylation is dysregulated in cells lacking Rrp6. (A) RNA-seq heat map showing the expression difference of mRNAs encoding genes important for precursor synthesis of cell wall components, visualized as rrp6Δ/wt mRNA ratio on a log2 scale. Data is from Wang et al. (2020). (B) Same as for A, but for the N- and O-glycosylation gene category. (C) Scheme of the genes involved in the synthesis of sugar nucleotides, dolichol and dolichol phosphate sugars, that act as cell wall precursors (above), and in the synthesis of the oligosaccharide precursor for N-glycosylation (below). Genes that are down-regulated in rrp6Δ cells upon 45 min at 42°C are marked in bold. (D) Activity staining of invertase from periplasmic extracts. Extracts of rrp6Δ cells contain an additional nonglycosylated form of periplasmic invertase, revealing that protein glycosylation is affected in this mutant.
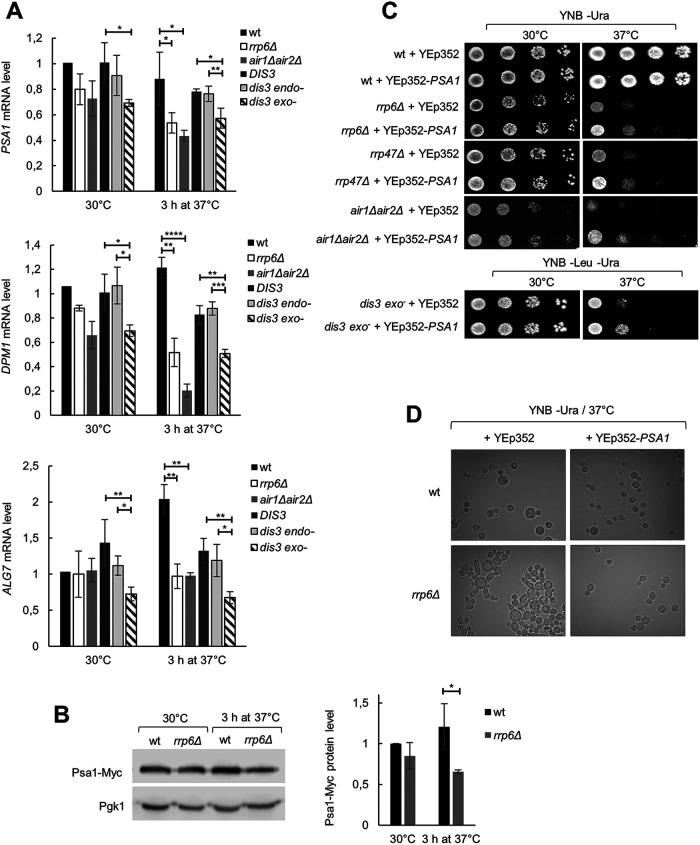
Quantification of PSA1, DPM1, and ALG7 mRNAs by reverse-transcription quantitative PCR (RT-qPCR) showed that their levels are lower in rrp6Δ, air1Δair2Δ, and dis3 exo− cells than in corresponding wild-type and dis3 endo− cells at high temperature (3 h at 37°C; Figure 5A), in line with their down-regulation observed with rrp6Δ cells as compared with wild-type cells upon 45 min of heat shock at 42°C (Figure 4, A and B). For some of these genes, down-regulation in certain RNA exosome mutant cells in comparison to wild-type cells could be observed already at 30°C (Figure 5A), which could explain why glycosylation defects can be detected already at this temperature (Figure 4D and Supplemental Figure S5), even though the effect is not strong enough to cause a detectable phenotype. Out of these three genes, PSA1 acts most upstream in the protein glycosylation pathway, as it encodes the enzyme GDP-mannose pyrophosphorylase, which synthesizes the activated form of mannose that is incorporated into N- and O-linked glycoproteins (Hashimoto et al., 1997). Psa1 is essential, but partial loss of function of this enzyme or its down-regulation result in phenotypes such as sensitivity to hyposmolarity, cell leakage, and cell separation defects (Zhang et al., 1999; Tomlin et al., 2000; Warit et al., 2000), that are reminiscent of those noticed with the rrp6Δ mutant at 37°C. To explore whether the decrease in the PSA1 mRNA level is reflected by a decrease in the Psa1 protein level in rrp6Δ mutant, we C-terminally tagged Psa1 with a Myc tag at its genomic locus and quantified the protein by Western blotting. The Psa1 protein level corresponded well with the mRNA level and confirmed a decrease in the Psa1 level in rrp6Δ cells after incubation at 37°C (Figure 5B). Therefore, a low expression level of the essential Psa1 enzyme, or a more general and additive defect in protein glycosylation, could be the reason for the lethality of rrp6Δ and other RNA exosome mutant cells at 37°C. To test this hypothesis, we overexpressed Psa1 in these cells from a 2μ plasmid under regulation of its own promoter. Interestingly, overexpression of Psa1 restored the viability of rrp6Δ, rrp47Δ, and air1Δair2Δ mutants at 37°C to a similar degree as osmotic stabilization (Figure 5C), confirming that protein glycosylation was limiting for growth of these cells at 37°C. Overexpression of PSA1 also partially suppressed the temperature sensitivity of dis3 exo− cells (Figure 5C), while in this case the incomplete suppression could be due to the necessity to stably replicate two plasmids (the centromeric plasmid carrying the DIS3-D551N allele and the 2μ plasmid carrying the PSA1 gene) in order to survive. Additionally, overexpression of Psa1 completely suppressed aberrancies in the cell morphology of rrp6Δ cells, such as the enlargement of cells and the defect in cell separation (Figure 5D). Taken together, these results demonstrate a role for the RNA exosome in enabling proper protein mannosylation that is needed to preserve cell viability upon temperature-induced stress.
FIGURE 5:
Overexpression of PSA1 rescues temperature sensitivity of RNA exosome mutants. The strains are described in Figures 1 and 2. (A) Levels of PSA1, DPM1, and ALG7 mRNAs are lower in rrp6Δ, air1Δair2Δ, and dis3 exo− cells than in the corresponding wild-type cells at high temperature. RT-qPCR values are normalized to PMA1 mRNA and expressed relative to transcript abundance in wild-type cells at 30°C, which is set as 1. Reported values represent the means and standard deviations of three independent experiments (n = 3). Indicated differences show the significant differences using an unpaired Student’s t test. One (*), two (**), three (***), and four (****) asterisks denote a p-value lower than or equal to 0.05, 0.01, 0.001, and 0.0001, respectively. (B) Psa1 protein level is lower in rrp6Δ than in wild-type cells at high temperature. Myc-tagged Psa1 was quantified by Western blotting. Values are normalized to Pgk1 and expressed relative to protein abundance in wild-type cells at 30°C, which is set as 1. Reported values represent the means and standard deviations of three independent experiments (n = 3). Indicated differences show the significant differences using an unpaired Student’s t test. One (*) asterisk denotes a p-value lower than or equal to 0.05. (C) Overexpression of Psa1 from a multicopy plasmid (YEp352-PSA1) fully rescues temperature sensitivity of rrp6Δ, rrp47Δ, and air1Δair2Δ cells and partially of dis3 exo− cells. Tenfold serial dilutions of cells were spotted on plates and were photographed after 3 d at indicated temperature. Control cells were transformed with the empty vector (YEp352). (D) Overexpression of Psa1 from a multicopy plasmid (YEp352-PSA1) rescues aberrant phenotype of rrp6Δ cells at high temperature.
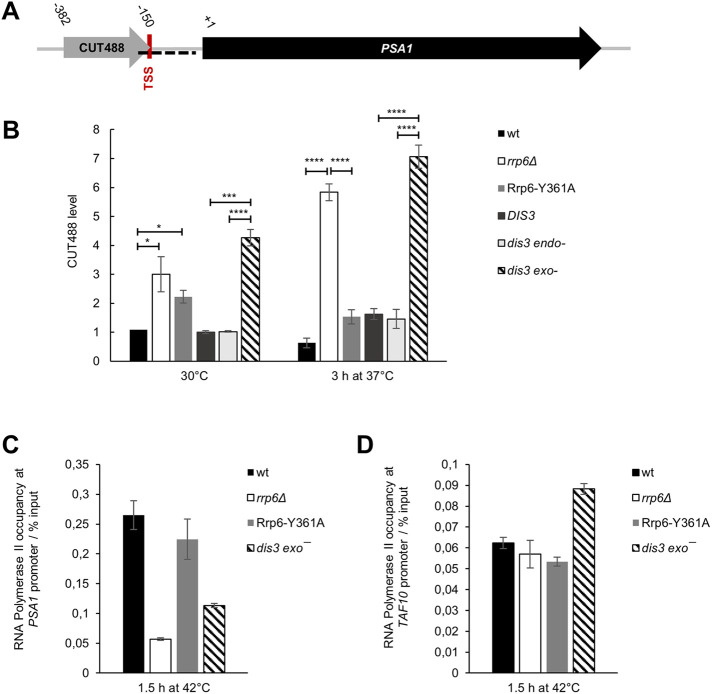
The Rrp6-containing RNA exosome is responsible for the degradation of a class of noncoding RNA transcripts termed CUTs (cryptic unstable transcripts), and inactivation of Rrp6 therefore results in increased CUTs accumulation (Xu et al., 2009). Intriguingly, CUT488 is transcribed in the sense direction through the PSA1 gene promoter and the 3′ end of this transcript overlaps with the PSA1 transcription start site (Figure 6A). Quantification of CUT488 by RT-qPCR showed its stabilization in rrp6Δ and dis3 exo− cells compared with wild-type cells and revealed an additional increase in its level at 37°C (Figure 6B). Because promoter sense transcripts have previously been shown to have gene regulatory roles in yeast (Hainer et al., 2011; Van Werven et al., 2012; Yu et al., 2016), this represents a possible mechanism for PSA1 down-regulation in rrp6Δ and dis3 exo− cells. We also found that recruitment of RNA polymerase II to the PSA1 gene promoter was not significantly changed in these mutant cells compared with wild-type cells at physiological temperature (Supplemental Figure S6). However, at high temperature the occupancy of RNA polymerase II at the PSA1 gene promoter was drastically decreased in rrp6Δ and dis3 exo− mutants as compared with wild-type cells or the Rrp6-Y361A catalytic mutant cells (Figure 6C). This was not due to a general effect on gene transcription in these mutant cells at high temperature, as this effect was not present when probing for RNA polymerase II occupancy at the promoter of the TAF10 gene (Figure 6D), which is constitutively expressed and does not show any noncoding transcription at its locus. This is conceivably in line with a regulatory mechanism in which accumulation of the normally unstable noncoding RNAs in rrp6Δ and dis3 exo− cells out-titrates the NNS termination system, thereby promoting read-through of CUT488 into the PSA1 promoter region, which was recently shown to be a transcriptome-wide phenomenon (Moreau et al., 2019; Villa et al., 2020). Read-through of CUT488 could limit transcription factor and/or RNA polymerase II recruitment to the PSA1 promoter region and negatively influence transcription of the PSA1 gene. We also found that the gene loci of the two other down-regulated glycosylation-related genes, DPM1 and ALG7, show transcription of noncoding antisense transcripts at their genomic loci, which are stabilized in rrp6Δ cells at high temperature (Supplemental Figure S7). The antisense transcript at the DPM1 locus was previously mapped as CUT923, while the antisense transcript at the ALG7 locus was not mapped but can be seen upon inspection of whole-transcriptome tiling array datasets (Xu et al., 2009). Taken together, a possible mechanism for dysregulation of glycosylation-related genes in RNA exosome mutants involves a temperature-dependent increase in accumulation of noncoding transcripts transcribed from their genomic loci.
FIGURE 6:
Noncoding transcript CUT488 accumulates in cells lacking Rrp6 or Dis3 exoribonuclease activity at high temperature. Strains are described in Figures 1 and 2. (A) Scheme of PSA1 locus, showing sense transcription of a noncoding transcript CUT488 at its promoter region. Transcription start site (TSS) of PSA1 is located at position −149 relative to the start of the ORF. The region used for ChIP is marked as a dashed black line. (B) Level of CUT488 RNA is higher in rrp6Δ and dis3 exo− cells than in corresponding wild-type and Rrp6-Y361A or dis3 endo− cells, and that difference is even greater at high temperature. RT-qPCR values are normalized to PMA1 mRNA and expressed relative to transcript abundance in wild-type cells at 30°C, which is set as 1. Reported values represent the means and standard deviations of three independent experiments (n = 3). Indicated differences show the significant differences using an unpaired Student’s t test. One (*), three (***), and four (****) asterisks denote a p-value lower than or equal to 0.05, 0.001, and 0.0001, respectively. (C) Recruitment of RNA polymerase II to PSA1 gene promoter is decreased in rrp6Δ and dis3 exo− cells compared with wild-type and Rrp6-Y361A cells at high temperature. Quantification was performed by ChIP of RNA polymerase II using specific antibodies 8WG16. Immunoprecipitated samples (output) were normalized to input following quantification by qPCR. Reported values represent the means and range of two independent experiments (n = 2). (D) The decrease of RNA polymerase II occupancy over the PSA1 promoter observed for rrp6Δ and dis3 exo− cells was not due to a general effect on transcription in these cells because this difference was not present for TAF10 gene promoter. Quantification was performed as in C.
DISCUSSION
In this work, we demonstrate that the activity of RNA exosome is necessary for maintaining cell wall stability in yeast Saccharomyces cerevisiae. RNA exosome mutants undergo osmoremedial cell lysis and show numerous cell wall–related phenotypes that are exacerbated at high temperature. Importantly, this explains that aberrancies in cell wall structure are the reason for temperature sensitivity of these mutants. The essential RNA exosome catalytic subunit Dis3 provides exoribonuclease catalytic activity, while the second catalytic subunit Rrp6 has a noncatalytic role in this process. Besides RNA exosome catalytic subunits, exosome cofactors Rrp47 and Air1/2 are also involved. We show a role for these proteins in maintaining cellular integrity upon heat stress, but also upon treatment with cell wall stressors at physiological temperature, clearly showing that their role is not specific to temperature but to conditions of cell wall stress. Importantly, we provide mechanistic insight into cell wall instability of RNA exosome mutants, as we highlight differential expression of protein glycosylation genes as the factor that disrupts their CWI. Specifically, down-regulation of genes encoding proteins that act in the early steps of the protein mannosylation pathway (PSA1, DPM1, and ALG7) in RNA exosome mutant cells compared with wild-type cells leads to aberrant morphology and temperature sensitivity of these mutants. In addition, artificially aiding protein glycosylation through overexpression of Psa1 suppresses their temperature-sensitive phenotypes, which were previously shown to be due to cell wall instability.
Our results partially contrast with a study that was published during the preparation of this article, which highlighted the role of RNA exosome catalytic subunit Rrp6 in promoting cell survival during heat stress, but argued against involvement of other RNA exosome subunits and cofactors (Wang et al., 2020). They proposed that Rrp6 alone has a highly specialized “moonlighting” function in this process, that is independent of all of its currently known interactors, including its stabilization partner Rrp47 (Wang et al., 2020). Our results clearly show the importance of the essential RNA exosome catalytic subunit Dis3 in this process, as the catalytically inactive dis3 exo− (dis3-D551N) mutant displays practically identical cell wall aberrancies as rrp6Δ mutant (Figures 1–3), which is also the case for mutants in exosome cofactors Rrp47 and Air1/2 (discussed below). This challenges the idea of a highly specialized Rrp6 function in maintaining CWI and is important to delineate, especially as exosome-independent roles of Rrp6 are a highly debated topic in the field (Callahan and Butler, 2008). Furthermore, the potential role of Rrp6 in the CWI pathway was inferred from the additive cell wall instability phenotype of the double mutant rrp6Δ mpk1Δ, in which a major CWI signaling component was inactivated (Wang et al., 2020). Additivity is suggestive of parallel and redundant functions, and this interpretation was previously applied to the equally severe phenotype of the rnt1Δ mpk1Δ mutant, which harbors deletion of the dsRNA-specific ribonuclease Rnt1 (Catala et al., 2012). It is, however, clear that Rrp6 has a noncatalytic role in maintaining cellular integrity upon heat stress, as previously implied by the fact that all tested Rrp6 catalytic mutants grow normally at high temperature (Phillips and Butler, 2003). The most straightforward explanation, which fits well with our results, could lie in the well-documented role of Rrp6 in allosterically stimulating the activity of RNA exosome through its C-terminal domain, a process which is independent of Rrp6 catalytic activity (Makino et al., 2015; Wasmuth and Lima, 2017). In line with this, the deletion of only the Rrp6 EAR (exosome-interacting region) domain leads to temperature sensitivity, which pinpoints it as the region of Rrp6 that is necessary for stress resistance (Wasmuth and Lima, 2017).
Besides mutants in the RNA exosome catalytic subunits, we show that for mutants in RNA exosome cofactors temperature sensitivity is also associated with cell wall instability (Figure 2, D and E, and Supplemental Figure S3). Inactivation of the RRP47 gene, encoding the obligate stabilization partner of Rrp6, results in osmoremedial temperature sensitivity and hypersensitivity to cell wall stressors. Because Rrp47 is critical for Rrp6 protein stability (Feigenbutz et al., 2013; Stuparevic et al., 2013), this result confirms the necessity of Rrp6 protein presence for maintaining cellular integrity upon heat stress. Also, simultaneous inactivation of two homologous genes that encode the TRAMP complex subunits Air1 and Air2 results in cell wall–related phenotypes, in contrast to their individual inactivation. Because Air1 and Air2 function as RNA-binding subunits in different isoforms of the TRAMP complex, this indicates that these isoforms have fully redundant roles in ensuring cellular integrity, which is interesting considering that these isoforms were previously shown to have some nonoverlapping roles based on differential substrate specificity (Schmidt et al., 2012; Stuparevic et al., 2013), somehow similar to what has been recently shown for Trf4 and Trf5 (Delan-Forino et al., 2020). Finally, cell wall instability is an elegant explanation for the observation that rrp6Δ phenotype, that is, its temperature sensitivity, is most pronounced in W303 and its derived genetic backgrounds, as wild type of this strain was shown to have an already more destabilized cell wall compared with wild types of other backgrounds (Trachtulcová et al., 2003; Schroeder and Ikui, 2019).
Yeast cell wall is the outermost part of the cell, which determines its shape and provides physical and osmotic protection. It is a polysaccharide network built out of glucan and chitin to which cell wall proteins are bound (Klis et al., 2002). Cell wall proteins function as structural components of the cell wall or enzymes that modify cell wall composition and are often heavily mannosylated through N- or O-linked glycosylation. This modification is vital for yeast, as well as for humans, because it ensures proper protein activity, stability, and localization (Lehle et al., 2006). The essential cytoplasmic enzyme GDP-mannose pyrophosphorylase Psa1 catalyzes the production of GDP-mannose, which is the activated form of mannose that gets incorporated into glycoproteins (Hashimoto et al., 1997). Its partial loss of function or down-regulation leads to multiple strong cell wall–related phenotypes such as sorbitol-dependence, cell rupture, and cell separation defects (Zhang et al., 1999; Tomlin et al., 2000; Warit et al., 2000). Importantly, we found that transcription of the PSA1 gene is down-regulated in RNA exosome mutant cells at high temperature, leading to a lower Psa1 protein level and potentially resulting in psa1 phenotypes (Figures 4A and 5, A and B). This is strongly supported by rescuing the temperature-sensitive growth and aberrant morphology of these mutants through overexpression of Psa1 (Figure 5, C and D). Because of the previously mentioned strong effects of PSA1 down-regulation, it is plausible that about a 50% down-regulation of the Psa1 protein level observed in rrp6Δ compared with wild-type cells at high temperature can push the protein’s enzymatic activity below a physiologically critical level. However, we cannot exclude the possibility that dysregulating expression of genes encoding other proteins involved in the early steps of the glycosylation pathway, such as DPM1 and ALG7 (Figures 4, A and B, and 5A) plays a significant contribution in cell wall phenotypes of RNA exosome mutants and that Psa1 overexpression rescues them by generating more precursor supply (Janik et al., 2003). We also hypothesize that a possible reason for transcriptional down-regulation of the PSA1 gene in RNA exosome mutant cells compared with wild-type cells is the temperature-induced increased accumulation of CUT488, a noncoding transcript transcribed through the PSA1 gene promoter, which is accompanied by a decrease in RNA polymerase II recruitment at this promoter (Figure 6). At the PHO84 gene, which is regarded as a model gene for transcriptional regulation through noncoding RNA transcription, loss of Rrp6 was shown to lead to higher production of the antisense transcript due to the decreased recruitment of the NNS complex that normally terminates its transcription (Castelnuovo et al., 2013). Recent transcriptome studies from ours and the D. Libri laboratory showed that out-titration of the NNS complex, accomplished either by perturbation of mRNP biogenesis or inactivation of the RNA exosome, leads to termination defects at ncRNA-producing targets (Moreau et al., 2019; Villa et al., 2020). In line with this mechanism, out-titration of the NNS complex in rrp6Δ and dis3 exo− cells, because of the more prominent accumulation of ncRNAs in these mutant cells at high temperature, could lead to transcriptional read-through of CUT488 through the PSA1 gene promoter and negatively influence PSA1 gene transcription. Notable examples of loci regulated by noncoding promoter transcription in yeast include SER3, HO, and IME1 genes, which are all negatively regulated by transcription of a sense transcript at their promoter regions (Winston et al., 2005; Hainer et al., 2011; Van Werven et al., 2012; Yu et al., 2016). While these are nonessential genes expressed specifically under a certain physiological or life/cell cycle condition, PSA1 is essential and constitutively expressed. However, it is strongly cell cycle regulated, peaking at the START phase of the cell cycle (Benton et al., 1996), so the possibility of a cell cycle–based regulation of its transcription through noncoding RNA transcription could be an exciting subject for future investigation. Of course, such broad dysregulation of cell wall stability in rrp6Δ cells is most probably due to effects on expression of multiple cell wall–related genes and could involve regulatory roles of noncoding transcription (Novačić et al., 2020), as well as the CWI pathway (Wang et al., 2020). In line with this, we also noticed increased accumulation of noncoding antisense transcripts that are transcribed at DPM1 and ALG7 gene loci in rrp6Δ cells at high temperature (Supplemental Figure S7).
Another interesting point is that the increase in accumulation of CUT488 at high temperature is independent of Rrp6 catalytic activity but is dependent on the presence of Rrp6 protein and the exoribonuclease activity of Dis3 (Figure 6B). This implies that this CUT is degraded primarily by Dis3 at high temperature and that Rrp6 provides a noncatalytic function in this process, probably that of an equivalent of an RNA exosome cofactor (as discussed in the second paragraph of the Discussion). Allosteric stimulation of Dis3 activity by Rrp6 probably happens by direct RNA binding, as well as the widening of the RNA exosome channel through which RNAs need to be threaded to reach the active site of Dis3 (Kilchert et al., 2016). Transcripts termed as CUTs were originally identified as ones that accumulate in rrp6Δ deletion mutant (Xu et al., 2009) and comparison of the transcriptome of this mutant with that of Dis3 catalytic mutants revealed some unique and some specific roles of the two catalytic subunits (Gudipati et al., 2012); however, the transcriptome of the Rrp6 catalytic mutant was studied only with Schizosaccharomyces pombe cells (Mukherjee et al., 2016). The study with S. pombe revealed that some RNA targets of Rrp6 depended mainly on its structural role, such as RNAs of early meiotic and iron metabolism genes (Mukherjee et al., 2016). Noncatalytic roles of Rrp6 have not yet been explored transcriptome-wide in S. cerevisiae but could be an interesting subject to study, especially with relation to unique cellular states, such as meiosis or conditions of heat shock.
The cell wall structure is absent from mammalian cells; however, protein glycosylation is conserved and essential for viability from yeast to human. The importance of protein glycosylation is underscored by the congenital disorders of glycosylation syndrome, which encompasses multisystemic diseases in children that result from defects in various steps along glycan modification pathways (Chang et al., 2018). While final sugar composition and branching differs between yeast and human, the earliest steps in the glycosylation pathway, precursor synthesis and initial N-glycosylation reactions, are highly conserved (Lehle et al., 2006). Our work in yeast clearly shows that one of the molecular consequences of RNA exosome inactivation is impairment of protein glycosylation at these early steps. Given the high conservation of both the RNA exosome complex and the glycosylation pathway, as well as the association of both with human diseases, this study opens the possibility for future investigation with human cells.
Summary
RNA exosome activity, accomplished through Dis3 exonuclease activity and a noncatalytic function of Rrp6, is necessary for maintaining cell wall stability in yeast Saccharomyces cerevisiae.
A defect in protein glycosylation is a major reason for cell wall instability of RNA exosome mutants.
Genes encoding proteins involved in the early steps of protein glycosylation are dysregulated in RNA exosome mutants through mechanisms that involve increased accumulation of noncoding RNAs at high temperature.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Strains, media, plasmids, and strain construction
Yeast strains and primers used in this study are listed in Supplemental Tables S1 and S2, respectively. Experiments were performed with the BMA41 (W303-derived) strain background, unless noted otherwise. Yeast strains were grown in YPD (containing per liter 20 g peptone, 10 g yeast extract, 20 g glucose, 0.1 g adenine) or YNB medium (containing per liter 6.7 g yeast nitrogen base without amino acids, 2 g drop-out mix as in Musladin et al., 2014, 20 g glucose) supplemented with the required amino acids and uracil (80 mg/l each). Plasmid YEp352-PSA1 is a high copy vector that carries the PSA1/MPG1 gene (Janik et al., 2003).
Psa1 was tagged at its genomic locus with a C-terminal 9xMyc tag. The tagging cassette was PCR amplified from plasmid pYM20 (Janke et al., 2004) using the primer pair PSA1Ctag_fwd/ PSA1Ctag_rev and transformed into BMA41 wild-type and rrp6Δ strain by a standard lithium acetate procedure. Transformants were selected on Hygromycin B (0.3 mg/ml, Roche) plates and the presence of the tag was confirmed by Western blotting. The RRP6 gene was deleted in BY4741 strain using a disruption cassette generated by PCR with primers RRP6-Kan1 and RRP6-Kan2 (Mosrin-Huaman et al., 2009).
Phenotypic assays
Sensitivities to CR, CFW, caffeine, and SDS were tested by spotting assays. Exponential phase cultures were adjusted to an OD600 of 1 and four 10-fold serial dilutions of that sample were spotted onto plates supplemented with indicated amounts of each compound. Plates were incubated at 30°C or 37°C for 3 d and photographed using a UVIDOC HD6 camera (Uvitec, Cambridge).
Alkaline phosphatase activity assays
Activity of alkaline phosphatase released into the medium was measured as in Molina et al. (1998) with slight modifications. Supernatant (500 μl) from liquid culture was mixed with equal volume of 20 mM p-nitrophenylphosphate in Tris-HCl buffer, pH 8.8 and assayed for alkaline phosphatase activity. The reaction was performed at 30°C, stopped by the addition of 500 μl of 1 M NaOH, and absorbance of liberated p-nitrophenol was measured at 420 nm using a Helios Gamma spectrophotometer (Thermo Fisher). Enzyme activity was normalized to OD600 of the culture and the assay time in minutes and was expressed in arbitrary units: A420*10,000/[OD600*(t/min)*(Vsample/Vtotal)]. Intracellular activity of alkaline phosphatase was measured exactly as described in Münsterkötter et al. (2000).
Fluorescence microscopy
Cells were stained with CFW stain (Sigma) and observed with an Olympus BX51 fluorescence microscope. The fluorescence from CFW was filtered with a DAPI filter.
RNA-seq data processing and computational analysis
Raw data from Wang et al. (2020) were downloaded via GEO (accession number GSE140504). Alignment and reads abundance estimation were conducted as described in the original publication. In short, Hisat2 was used to align reads against S. cerevisiae reference genome (taken from SGD, release R64-1-1); read abundance for mRNAs was estimated with HTseq-count (with the option –s reverse). Differential analysis between wt and rrp6Δ strain was conducted under the R environment using the DESeq2 package. Resulting log2FC were used to construct heatmaps using the ggplot2 and complexHeatmap packages.
Analysis of the degree of glycosylation of periplasmic invertase
The secretory invertase was analyzed as described in Hashimoto et al. (1997). Briefly, invertase expression was induced by incubating midlogarithmic phase cells in medium that contains sucrose instead of glucose for 2 h at 30°C or 37°C. Cells were treated with zymolyase and the periplasmic fraction containing invertase was separated from spheroplasts by centrifugation. The periplasmic fraction was subjected to 7.5% SDS–PAGE, gel was bathed in 0.1 M sodium acetate, pH 5.1, containing 0.1 M sucrose at 37°C for 1 h to carry out the enzymatic reaction of invertase, and then washed with water, placed in 0.1% 2,3,5- triphenyltetrazolium chloride, 0.5 M NaOH, and boiled to detect red bands. After staining, gel was washed with 7.5% acetic acid.
RNA isolation and RT-qPCR analysis
Total RNA was extracted by the hot phenol method (Schmitt et al., 1990) and column purified with DNase treatment using a NucleoSpin RNA kit (Macherey Nagel) according to manufacturer instructions. RNA was quantified with a Nanodrop spectrophotometer and 1 μg was used in a strand-specific reverse-transcription reaction with a ProtoScript First Strand cDNA Synthesis Kit (New England Biolabs) with 0.1 μM gene-specific oligonucleotides and supplemented with actinomycin D (Sigma) to final concentration 5 μg/ml to ensure strand specificity. Twofold diluted cDNA (1 μl) was then amplified in Roche LightCycler 480 with the Maxima SYBR Green qPCR Master Mix detection kit from Thermo Scientific as recommended by the supplier. The qPCR datasets were analyzed using the ΔΔCt method, and the results were normalized to PMA1 mRNA RT-qPCR amplification, which was used as internal control. The level of a certain transcript for each sample was expressed relative to its abundance in wild-type cells at 30°C, which was set as 1. Amplifications were done in duplicate for each sample, and three independent RNA extractions were analyzed.
Western blot analysis
Total proteins were obtained as described in Kushnirov (2000), resolved on SDS 10% polyacrylamide electrophoresis gels, and analyzed by Western blotting according to standard procedures. Myc-tagged Psa1 was probed with anti-c-Myc (9E10; Santa Cruz Biotechnology) at 1:1000 dilution and Pgk1 with anti-PGK1 (22C5D8; Abcam) at 1:5000 dilution. In both cases, mouse IgG kappa-binding protein HRP (Santa Cruz Biotechnology) at 1:50,000 dilution was used to detect the primary antibody. Blots were developed using Biorad Clarity Western ECL substrates and visualized with a C-DiGit Blot scanner (LI-COR Biosciences). Band intensity was quantified with GelAnalyzer 19.1 software and the results were normalized to Pgk1. The level of protein was expressed relative to its abundance in wild-type cells at 30°C, which was set as 1. Three independent protein extractions were analyzed for each sample.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed similarly as described in Stuparevic et al. (2013). Forty milliliters of cells were fixed with 1% formaldehyde for 20 min. After glycine addition to stop the reaction, the cells were washed and lysed with glass beads to isolate chromatin. The cross-linked chromatin was sheared by sonication with a Vibra-Cell sonicator to reduce average fragment size to approximately 500 base pairs. Chromatin fractions of 400 μl were taken for each immunoprecipitation reaction and incubated with 4 μl of anti-RNA polymerase II antibodies (8WG16, sc-56767; Santa Cruz Biotechnology) at 4°C overnight. After incubation, 40 μl of protein G PLUS-agarose beads (sc-2002; Santa Cruz Biotechnology) were added and incubated for 2 h at 4°C. The beads were then washed extensively, and the chromatin was eluted. Eluted supernatants (output) and the input controls were hydrolyzed with Pronase (0.8 mg/ml final concentration; Sigma) for 2 h at 42°C, followed by 7 h incubation at 65°C to reverse cross-linked DNA complexes. DNA was extracted using the Macherey Nagel Nucleospin Gel & PCR Cleanup Kit. The immunoprecipitated DNAs (output) were quantified by qPCR in Roche LightCycler 480 with the Maxima SYBR Green qPCR Master Mix detection kit from Thermo Scientific as recommended by the supplier. Amplifications were done in triplicate for each sample. Immunoprecipitated samples (output) were normalized to input.
Supplementary Material
Acknowledgments
This work was supported by Croatian Science Foundation Grant no. UIP-2017-05-4411 to I.S., funding from la Ligue Contre le Cancer (comités du Loiret et de l’Indre) to A.R.R., and funding from the Croatian Academy of Sciences and Arts to A.N. V.B. is a recipient of a PhD fellowship from the Région Centre-Val de Loire and A.N.’s research stay at the CBM, CNRS in France was funded by a Scholarship of the French Government. We are grateful to Michael Primig and Vladimir Mrša for helpful discussions. We are also grateful to Sandi Orlić for help with fluorescence microscopy and to Grazyna Palamarczyk for the gift of YEp352- MPG1 plasmid.
Abbreviations used:
- CFW
Calcofluor White
- ChIP
chromatin immunoprecipitation
- CR
Congo Red
- CUTs
cryptic unstable transcripts
- CWI
cell wall integrity
- EAR
exosome interacting region
- ncRNAs
noncoding RNAs
- RT-qPCR
reverse-transcription quantitative PCR
- TSS
transcription start site.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-08-0544-T) on January 13, 2021.
REFERENCES
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D (1999a). Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 18, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Mitchell P, Petfalski E, Tollervey D (2000). Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res 28, 1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P (1999b). The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes Dev 13, 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim J De, Slavotinek A, Fasken MB, Corbett AH, Morton DJ, Corbett AH (2020). Modeling pathogenic variants in the RNA exosome. RNA Dis 7, 1–9. [PMC free article] [PubMed] [Google Scholar]
- Belcarz A, Ginalska G, Lobarzewski J, Penel C (2002). The novel non-glycosylated invertase from Candida utilis (the properties and the conditions of production and purification). Biochim Biophys Acta - Protein Struct Mol Enzymol 1594, 40–53. [DOI] [PubMed] [Google Scholar]
- Benton BK, Plump SD, Roos J, Lennarz WJ, Cross FR (1996). Over-expression of S. cerevisiae G1 cyclins restores the viability of alg1 N-glycosylation mutants. Curr Genet 29, 106–113. [PubMed] [Google Scholar]
- Bresson S, Tuck A, Staneva D, Tollervey D (2017). Nuclear RNA decay pathways aid rapid remodeling of gene expression in yeast. Mol Cell 65, 787–800.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs MW, Burkard KTD, Butler JS (1998). Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5. 8 S rRNA 3. J Biol Chem 273, 13255–13263. [DOI] [PubMed] [Google Scholar]
- Callahan KP, Butler JS (2008). Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res 36, 6645–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelnuovo M, Rahman S, Guffanti E, Infantino V, Stutz F, Zenklusen D (2013). Bimodal expression of PHO84 is modulated by early termination of antisense transcription. Nat Struct Mol Biol 20, 851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala M, Aksouh L, Elela SA (2012). RNA-dependent regulation of the cell wall stress response. Nucleic Acids Res 40, 7507–7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang IJ, He M, Lam CT (2018). Congenital disorders of glycosylation. Ann Transl Med 6, 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski A, Lubas M, Jensen TH, Dziembowski A (2013). RNA decay machines: The exosome. Biochim Biophys Acta - Gene Regul Mech 1829, 552–560. [DOI] [PubMed] [Google Scholar]
- Davidson L, Francis L, Cordiner RA, Eaton JD, Estell C, Macias S, Cáceres JF, West S (2019). Rapid depletion of DIS3, EXOSC10, or XRN2 reveals the immediate impact of exoribonucleolysis on nuclear RNA metabolism and transcriptional control. Cell Rep 26, 2779–2791.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delan-Forino C, Spanos C, Rappsilber J, Tollervey D (2020). Substrate specificity of the TRAMP nuclear surveillance complexes. Nat Commun 11, 3122–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dra˛zkowska K, Tomecki R, Stodus´ K, Kowalska K, Czarnocki-Cieciura M, Dziembowski A (2013). The RNA exosome complex central channel controls both exonuclease and endonuclease Dis3 activities in vivo and in vitro. Nucleic Acids Res 41, 3845–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Séraphin B (2007). A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol 14, 15–22. [DOI] [PubMed] [Google Scholar]
- Esmon PC, Esmon BE, Schauer IE, Taylor A, Schekman R (1987). Structure, assembly, secretion of octameric invertase. J Biol Chem 262, 4387–4394. [PubMed] [Google Scholar]
- Fasken MB, Morton DJ, Kuiper EG, Jones SK, Leung SW, Corbett AH (2020). The RNA exosome and human disease. Methods Mol Biol 2062, 3–33. [DOI] [PubMed] [Google Scholar]
- Feigenbutz M, Garland W, Turner M, Mitchell P (2013). The exosome cofactor Rrp47 is critical for the stability and normal expression of its associated exoribonuclease Rrp6 in Saccharomyces cerevisiae. PLoS One 8, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati RK, Xu Z, Lebreton A, Séraphin B, Steinmetz LM, Jacquier A, Libri D (2012). Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol Cell 48, 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer SJ, Pruneski JA, Mitchell RD, Monteverde RM, Martens JA (2011). Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev 25, 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Sakakibara A, Yamasaki M, Yoda K (1997). Saccharomyces cerevisiae VIG9 encodes GDP-mannose pyrophosphorylase, which is essential for protein glycosylation. J Biol Chem 272, 16308–16314. [DOI] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH (2001). Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413, 538–542. [DOI] [PubMed] [Google Scholar]
- Janik A, Juchimiuk M, Kruszewska J, Orowski J, Pasikowska M, Palamarczyk G (2012). Impact of yeast glycosylation pathway on cell integrity and morphology. Glycosylation 11, 259–272. [Google Scholar]
- Janik A, Sosnowska M, Kruszewska J, Krotkiewski H, Lehle L, Palamarczyk G (2003). Overexpression of GDP-mannose pyrophosphorylase in Saccharomyces cerevisiae corrects defects in dolichol-linked saccharide formation and protein glycosylation. Biochim Biophys Acta - Gen Subj 1621, 22–30. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. (2004). A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962. [DOI] [PubMed] [Google Scholar]
- Kilchert C, Wittmann S, Vasiljeva L (2016). The regulation and functions of the nuclear RNA exosome complex. Nat Rev Mol Cell Biol 17, 227–239. [DOI] [PubMed] [Google Scholar]
- Klauer AA, Van Hoof A (2013). Genetic interactions suggest multiple distinct roles of the arch and core helicase domains of Mtr4 in Rrp6 and exosome function. Nucleic Acids Res 41, 533–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klis FM, Mol P, Hellingwerf K, Brul S (2002). Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol Rev 26, 239–256. [DOI] [PubMed] [Google Scholar]
- Kopecká M, Gabriel M (1992). The influence of Congo red on the cell wall and (1 → 3)-β-d-glucan microfibril biogenesis in Saccharomyces cerevisiae. Arch Microbiol 158, 115–126. [DOI] [PubMed] [Google Scholar]
- Kuranda K, Leberre V, Sokol S, Palamarczyk G, François J (2006). Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol Microbiol 61, 1147–1166. [DOI] [PubMed] [Google Scholar]
- Kushnirov VV (2000). Rapid and reliable protein extraction from yeast. Yeast 16, 857–860. [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D (2005). RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121, 713–724. [DOI] [PubMed] [Google Scholar]
- Lardenois A, Liu Y, Walther T, Chalmel F, Evrard B, Granovskaia M, Chu A, Davis RW, Steinmetz LM, Primig M (2011). Execution of the meiotic noncoding RNA expression program and the onset of gametogenesis in yeast require the conserved exosome subunit Rrp6. Proc Natl Acad Sci 108, 1058–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton A, Tomecki R, Dziembowski A, Séraphin B (2008). Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456, 993–996. [DOI] [PubMed] [Google Scholar]
- Lehle L, Strahl S, Tanner W (2006). Protein glycosylation, conserved from yeast to man: A model organism helps elucidate congenital human diseases. Angew Chemie - Int Ed 45, 6802–6818. [DOI] [PubMed] [Google Scholar]
- Lingaraju M, Schuller JM, Falk S, Gerlach P, Bonneau F, Basquin J, Benda C, Conti E (2020). To process or to decay: a mechanistic view of the nuclear RNA exosome. Cold Spring Harb Symp Quant Biol LXXXIV, 040295. [DOI] [PubMed] [Google Scholar]
- Makino DL, Schuch B, Stegmann E, Baumgärtner M, Basquin C, Conti E (2015). RNA degradation paths in a 12-subunit nuclear exosome complex. Nature 524, 54–58. [DOI] [PubMed] [Google Scholar]
- Milbury KL, Paul B, Lari A, Fowler C, Montpetit B, Stirling PC (2019). Exonuclease domain mutants of yeast DIS3 display genome instability. Nucleus 10, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D (1997). The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91, 457–466. [DOI] [PubMed] [Google Scholar]
- Molina M, Martín H, Sánchez M, Nombela C (1998). MAP kinase-mediated signal transduction pathways. Methods Microbiol 26, 375–393. [Google Scholar]
- Moreau K, Le Dantec A, Mosrin-Huaman C, Bigot Y, Piégu B, Rahmouni AR (2019). Perturbation of mRNP biogenesis reveals a dynamic landscape of the Rrp6-dependent surveillance machinery trafficking along the yeast genome. RNA Biol 16, 879–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosrin-Huaman C, Honorine R, Rahmouni AR (2009). Expression of bacterial Rho factor in yeast identifies new factors involved in the functional interplay between transcription and mRNP biogenesis. Mol Cell Biol 29, 4033–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K, Gardin J, Futcher B, Leatherwood J (2016). Relative contributions of the structural and catalytic roles of Rrp6 in exosomal degradation of individual mRNAs. RNA 22, 1311–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münsterkötter M, Barbaric S, Hörz W (2000). Transcriptional regulation of the yeast PHO8 promoter in comparison to the coregulated PHO5 promoter. J Biol Chem 275, 22678–22685. [DOI] [PubMed] [Google Scholar]
- Musladin S, Krietenstein N, Korber P, Barbaric S (2014). The RSC chromatin remodeling complex has a crucial role in the complete remodeler set for yeast PHO5 promoter opening. Nucleic Acids Res 42, 4270–4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novačić A, Vučenović I, Primig M, Stuparević I (2020). Non-coding RNAs as cell wall regulators in Saccharomyces cerevisiae. Crit Rev Microbiol 0, 1–11. [DOI] [PubMed] [Google Scholar]
- Orlean P (2012). Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 192, 775–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S, Butler JS (2003). Contribution of domain structure to the RNA 3′ end processing and degradation functions of the nuclear exosome subunit Rrp6p. RNA 9, 1098–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popolo L, Gualtieri T, Ragni E (2001). The yeast cell-wall salvage pathway. Med Mycol 39, 111–121. [PubMed] [Google Scholar]
- Roncero C, Duran A (1985). Effect of Calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J Bacteriol 163, 1180–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilders G, Raijmakers R, Raats JMH, Pruijn GJM (2005). MPP6 is an exosome-associated RNA-binding protein involved in 5.8S rRNA maturation. Nucleic Acids Res 33, 6795–6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K, Xu Z, Mathews DH, Butler JS (2012). Air proteins control differential TRAMP substrate specificity for nuclear RNA surveillance. RNA 18, 1934–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL (1990). A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18, 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Kudla G, Wlotzka W, Tuck A, Tollervey D (2012). Transcriptome-wide analysis of exosome targets. Mol Cell 48, 422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder L, Ikui AE (2019). Tryptophan confers resistance to SDS-associated cell membrane stress in Saccharomyces cerevisiae. PLoS One 14, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuparevic I, Mosrin-Huaman C, Hervouet-Coste N, Remenaric M, Rahmouni AR (2013). Cotranscriptional recruitment of RNA exosome cofactors Rrp47p and Mpp6p and two distinct Trf-Air-Mtr4 polyadenylation (TRAMP) complexes assists the exonuclease Rrp6p in the targeting and degradation of an aberrant messenger ribonucleoprotein particle (mRNP) in yeast. J Biol Chem 288, 31816–31829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlin GC, Hamilton GE, Gardner DCJ, Walmsley RM, Stateva LI, Oliver SG (2000). Suppression of sorbitol dependence in a strain bearing a mutation in the SRB1/PSA1/VIG9 gene encoding GDP-mannose pyrophosphorylase by PDE2 overexpression suggests a role for the Ras/cAMP signal-transduction pathway in the control of yeast cell-wall biogenesis. Microbiology 146, 2133–2146. [DOI] [PubMed] [Google Scholar]
- Trachtulcová P, Frýdlová I, Janatová I, Dorosh A, Hašek J (2003). The W303 genetic background affects the isw2Δ mutant phenotype in Saccharomyces cerevisiae. Folia Microbiol (Praha) 48, 745–753. [DOI] [PubMed] [Google Scholar]
- Villa T, Barucco M, Martin-Niclos MJ, Jacquier A, Libri D (2020). Degradation of non-coding RNAs promotes recycling of termination factors at sites of transcription. Cell Rep 32, 107942– 107957. [DOI] [PubMed] [Google Scholar]
- Wang C, Liu Y, DeMario SM, Mandric I, Gonzalez-Figueroa C, Chanfreau GF (2020). Rrp6 Moonlights in an RNA exosome-independent manner to promote cell survival and gene expression during stress. Cell Rep 31, 107754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warit S, Zhang N, Short A, Walmsley RM, Oliver SG, Stateva LI (2000). Glycosylation deficiency phenotypes resulting from depletion of GDP-mannose pyrophosphorylase in two yeast species. Mol Microbiol 36, 1156–1166. [DOI] [PubMed] [Google Scholar]
- Wasmuth EV, Januszyk K, Lima CD (2014). Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature 511, 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth EV, Lima CD (2012). Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol Cell 48, 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth EV, Lima CD (2017). The Rrp6 C-terminal domain binds RNA and activates the nuclear RNA exosome. Nucleic Acids Res 45, 846–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth EV, Zinder JC, Zattas D, Das M, Lima CD (2017). Structure and reconstitution of yeast Mpp6-nuclear exosome complexes reveals that mpp6 stimulates RNA decay and recruits the Mtr4 helicase. Elife 6, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk EL, Schilders G, Pruijn GJM (2007). Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. RNA 13, 1027–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, Van Oudenaarden A, Primig M, Amon A (2012). Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell 150, 1170–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Martens JA, Wu P-YJ (2005). Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev 19, 2695–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, et al. (2005). Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121, 725–737. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM (2009). Bidirectional promoters generate pervasive transcription in yeast. Nature 457, 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yarrington RM, Chuong EB, Elde NC, Stillman DJ (2016). Disruption of promoter memory by synthesis of a long noncoding RNA. Proc Natl Acad Sci USA 113, 9575–9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Gardner DCJ, Oliver SG, Stateva LI (1999). Down-regulation of the expression of PKC1 and SRB1/PSA1/VIG9, two genes involved in cell wall integrity in Saccharomyces cerevisiae, causes flocculation. Microbiology 145, 309–316. [DOI] [PubMed] [Google Scholar]
- Zinder JC, Lima CD (2017). Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev 31, 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.